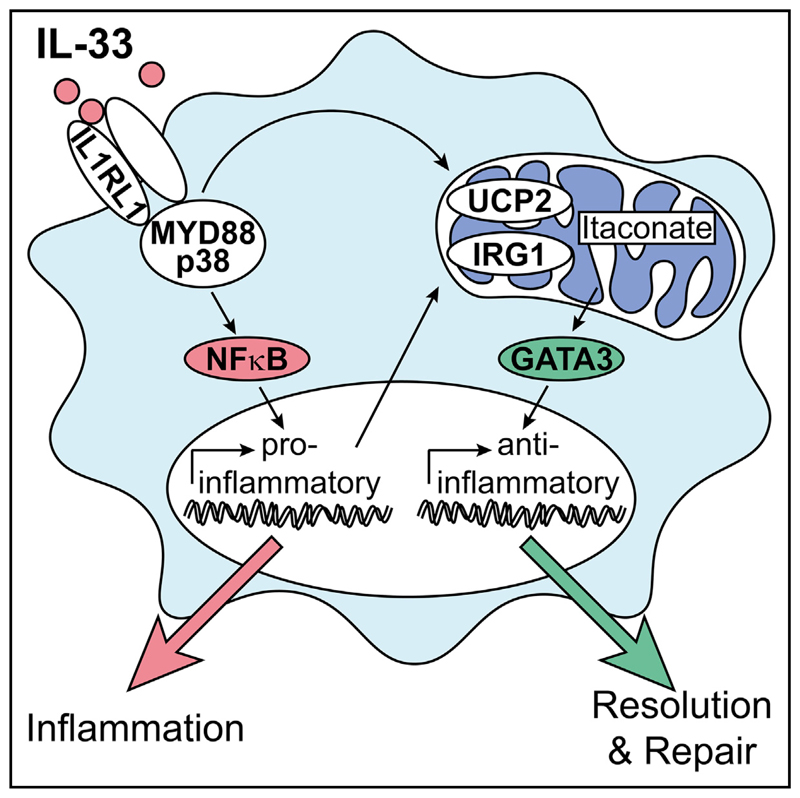
Summary
Alternatively activated macrophages (AAMs) contribute to the resolution of inflammation and tissue repair. However, molecular pathways that govern their differentiation have remained incompletely understood. Here, we show that uncoupling protein-2-mediated mitochondrial reprogramming and the transcription factor GATA3 specifically controlled the differentiation of pro-resolving AAMs in response to the alarmin IL-33. In macrophages, IL-33 sequentially triggered early expression of pro-inflammatory genes and subsequent differentiation into AAMs. Global analysis of underlying signaling events revealed that IL-33 induced a rapid metabolic rewiring of macrophages that involved uncoupling of the respiratory chain and increased production of the metabolite itaconate, which subsequently triggered a GATA3-mediated AAM polarization. Conditional deletion of GATA3 in mononuclear phagocytes accordingly abrogated IL-33-induced differentiation of AAMs and tissue repair upon muscle injury. Our data thus identify an IL-4-independent and GATA3-dependent pathway in mononuclear phagocytes that results from mitochondrial rewiring and controls macrophage plasticity and the resolution of inflammation.
Graphical abstract.
Introduction
Macrophages constitute a heterogeneous population of mononuclear phagocytes that fulfill important roles during tissue homeostasis, inflammation, and host defense (Ginhoux et al., 2016). These cells can directly originate from infiltrating blood monocytes or self-maintain their numbers within the respective tissue in a monocyte-independent manner (Ginhoux et al., 2010; Schulz et al., 2012; Yona et al., 2013). During steady state, the specific local micro-environmental niche seems to exert a dominant influence on the macrophage’s phenotype, resulting in tissue-specific transcriptional and epigenetic signatures (Okabe and Medzhitov, 2014). Under states of increased stress and inflammation, however, newly recruited macrophages show a considerable degree of context-dependent plasticity by which they rapidly respond to challenges within their environment and enter common and conserved polarized activation states that are observed across different tissues and organs (Murray, 2017).
Pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) as well as cytokines such as interferon-γ promote the differentiation of classically activated macrophages, which express high amounts of inducible nitric oxide synthase and/or pro-inflammatory cytokines such as TNF or interleukin (IL)-1β. These cells dominate during early stages of inflammation and have been shown to essentially contribute to the host defense against different extra- and intracellular pathogens (Murray, 2017). Increased differentiation and persistence of such pro-inflammatory macrophages, on the other hand, might contribute to the pathogenesis of inflammatory diseases and inflammation-associated tissue damage (Murray and Wynn, 2011). Other cytokines such as IL-4 have been shown to induce the differentiation of an alternatively activated macrophage (AAM) phenotype that displays expression of a characteristic set of proteins including arginase-1 (ARG1), resistin-like molecule (RELM)-alpha or chitinase 3-like 1 (Gordon, 2003). AAMs have been primarily implicated in the protection against helminths and other parasites. However, macrophages displaying AAM-related signatures dominate during later stages of various types of infectious and sterile inflammation, where they seem to facilitate the clearance of dying cells, foster the resolution of inflammation, and initiate tissue repair (Allen and Sutherland, 2014; Arnold et al., 2007; Coakley and Harris, 2020; Murray, 2017). The exact nature of signals and mechanisms that control differentiation and polarization of such pro-resolving and reparative macrophages are incompletely defined. In particular, the in vivo relevance of the originally described IL-4-mediated pathway during the resolution of injury-induced inflammation remains largely unclear.
Potential signals that affect macrophage polarization and induce an IL-4-independent differentiation of AAMs in response to tissue injury include the IL-1 family member IL-33, a cytokine that is released from cells undergoing necrosis. IL-33 has been originally described as an “alarmin” that triggers a MyD88- and NF-κB-dependent acute inflammatory response via its surface receptor IL1RL1 (also known as ST2) (Liew et al., 2016). However, increasing evidence suggests a more complex and dual role of IL-33, whereby this cytokine simultaneously induces the expression of various pro-inflammatory genes as well as differentiation of macrophages with an AAM-related signature (Kurowska-Stolarska et al., 2009). Although the underlying molecular mechanisms involved remain incompletely understood, IL-33 might thereby contribute both to the onset and to the resolution of inflammation in response to cell death and injury.
In the present study, we aimed to disentangle the molecular basis of the pro- and anti-inflammatory properties of IL-33 in macrophages and thereby sought to identify molecular mechanisms and signaling hubs controlling macrophage plasticity in the context of tissue damage and the resolution of sterile inflammation. Detailed molecular profiling revealed an IL-33-induced metabolic reprogramming of macrophages that resulted in uncoupling of the mitochondrial respiratory chain and increased production of the mitochondria-derived metabolite itaconate, which resulted in a consecutive expression of the transcription factor GATA3. GATA3, in turn, orchestrated an IL-4-independent differentiation of AAMs and promoted the resolution of inflammation and initiation of repair in response to tissue injury.
Results
IL-33 induces the delayed differentiation of a pro-resolving AAM phenotype
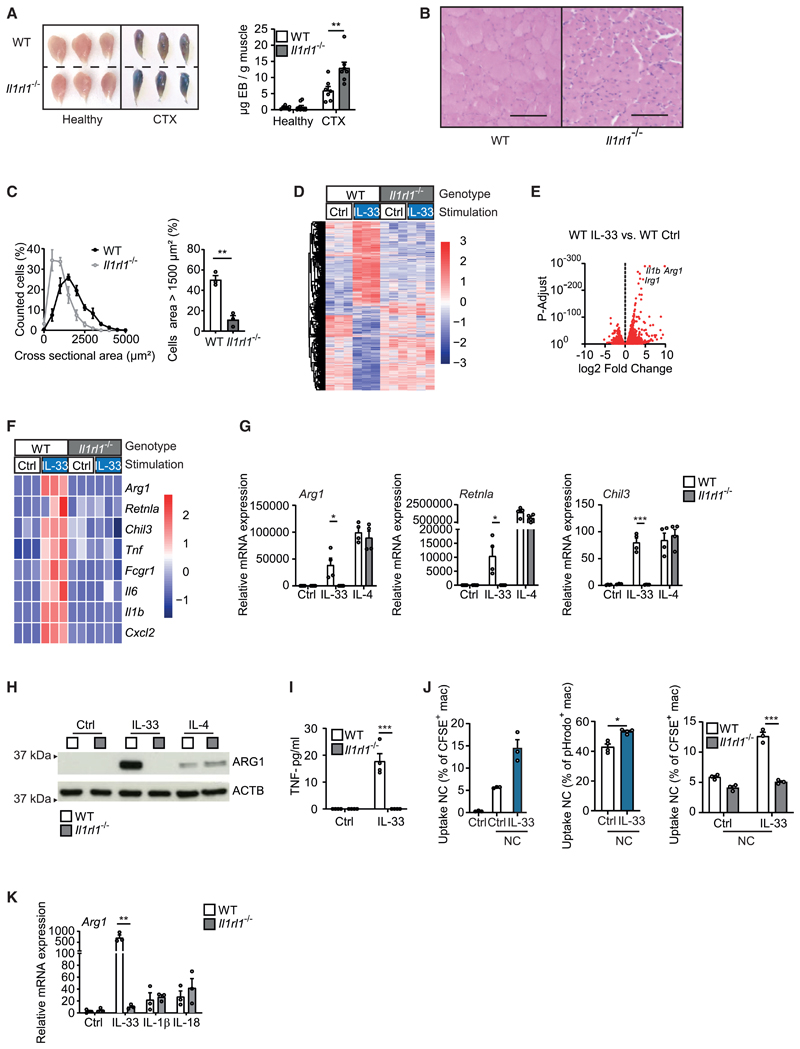
Injection of cardiotoxin (CTX) into the anterior tibial muscle of mice results in local muscle cell necrosis and tissue damage-induced inflammation that is characterized by the infiltration of large numbers of monocytes as well as the subsequent appearance of monocyte-derived macrophages (Figures S1A and S1B). Clearance of cellular debris, resolution of inflammation, and muscle fiber regeneration all heavily depend on the subsequent differentiation of pro-resolving AAMs (Arnold et al., 2007; Chazaud, 2020). To assess the role of IL-33 during these AAM-mediated events, we confirmed IL-33 expression in healthy muscle tissue, where this cytokine showed a nuclear localization pattern, which involved, among others, vessel-forming podoplanin+ lymphatic endothelial cells (Figures S1C and S1D; Video S1). As previously described, the number of IL-33-expressing cells increased in response to injury (Kuswanto et al., 2016), where the expression pattern became increasingly dispersed (Figure S1C). Next, we analyzed the phenotype of wild-type (WT) and IL-33 receptor-deficient Il1rl1−/− mice during the resolution phase of CTX-induced muscle injury, where we determined the extent of Evans blue (EB) staining, a dye that accumulates in damaged and necrotic muscle fibers, and analyzed the number of leukocytes in injured muscle. Notably, Il1rl1−/− mice displayed both delayed clearance of EB staining and a persistently elevated number of CD45+CD11B+LY6G− mononuclear phagocytes in the injured muscle, pointing toward an impaired clearance of damaged muscle cells and a defective resolution of inflammation (Figures 1A and S1E). Analysis of the morphology and cross-sectional area of muscle fibers at day 14 after CTX injury confirmed that muscle fiber regeneration in response to injury was impaired in Il1rl1−/− mice, which showed both a decreased cross-sectional area as well as centralized nuclei indicative of altered repair processes (Folker and Baylies, 2013) (Figures 1B and 1C). These data supported a central role of the IL-33-IL1RL1 signaling axis during the resolution of injury-induced inflammation and the consecutive process of tissue repair. They additionally suggested impaired functional properties of IL1RL1-deficient mononuclear phagocytes.
Figure 1. IL-33 promotes the resolution of injury-induced inflammation and the differentiation of AAMs.
(A) Determination of Evans blue accumulation in healthy and CTX-injected muscles of WT (n = 7) and Il1rl1−/− (n = 7) mice on day 7 after injury.
(B and C) Example H&E staining (B) and quantification and comparison of cross-sectional areas from muscle sections (C) of CTX-injected WT (n = 3) and Il1rl1−/− (n = 3) muscles on day 14 after injury. Scale bar indicates 100 μm.
(D–F) Bulk mRNA sequencing data of WT and Il1rl1−/− BMDMs that were treated with vehicle (Ctrl) or with IL-33 (10 ng/mL for 36 h). Data are presented as a heatmap illustrating differential gene expression (D), a volcano plot in which each dot represents a gene with a adjusted p < 0.05 (E), and a heatmap of genes encoding for alternative and classic macrophage activation markers (Z scores) (F).
(G–I) Quantitative real-time PCR (G), western blot (H), and ELISA (I) showing mRNA and protein expression of indicated markers of alternative activation (Arg1, Retnla, Chil3, ARG1) and classic activation (TNF) in BMDMs of WT and Il1rl1−/− BMDMs upon stimulation with vehicle (Ctrl) or IL-33 (10 ng/mL for 5 days) and IL-4 (20 ng/mL for 24 h). Data are representative of three individual experiments.
(J) Uptake of necrotic C2C12 cells (NC) after 1 h (CFSE-labeled) or 2 h (pHrodo-labeled) of phagocytosis. Data are representative of three individual experiments.
(K) Arg1 mRNA expression in response to IL-33 (10 ng/mL), IL-1β (10 ng/mL), or IL-18 (10 ng/mL) after 5 days of stimulation.
Data are presented as mean + SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. See also Figures S1 and S2 and Video S1.
We consequently sought to better characterize molecular pathways contributing to the pro-resolving effects of IL-33 in mononuclear phagocytes and performed bulk RNA sequencing of IL-33-stimulated WT and Il1rl1−/− bone marrow-derived macrophages (BMDMs). This analysis confirmed that the IL-33-IL1RL1 axis specifically induced the concomitant expression of pro- and anti-inflammatory genes (Figures 1D–1F). These included genes encoding for pro-inflammatory cytokines and chemokines such as Il1b, Il6, Tnf, and Cxcl2 but also genes encoding for enzymes and proteins previously implicated in AAM differentiation and the resolution of inflammation such as Arg1 and Retnla (encoding for ARG1 and RELM-alpha) (Nair et al., 2009; Yurdagul et al., 2020). Additional experiments confirmed the simultaneous and IL1RL1-dependent induction of these pro-inflammatory cytokines and of markers of AAM differentiation in response to IL-33 via quantitative real-time PCR as well as immunoblot blot analysis and ELISA at the mRNA and protein levels, respectively (Figures 1G–1I, S2A, and S2B). In accordance with a switch toward a pro-resolving macrophage phenotype, IL-33-activated macrophages also displayed an increased capacity to ingest and clear necrotic cells (Figures 1J and S2C).
Other pro-inflammatory alarmins and IL-1 family members such as IL-1β and IL-18 that equally use MyD88-dependent signaling, in turn, showed no impact on AAM markers (Figure 1K), while IL-4 similarly induced markers of AAM differentiation (Figures 1G and 1H) but failed to induce pro-inflammatory cytokines (data not shown). Notably, expression of distinct IL-33-induced genes followed different kinetics (Figure S2D). IL-33-induced pro-inflammatory genes showed an early expression peak, which resembled the response observed after stimulation with LPS and/or interferon-γ and was in accordance with an activation of common signaling pathways such as NF-κB. Proresolving genes characteristic of IL-33-induced AAMs, in contrast, displayed a delayed peak of expression and thus sharply differed from the transcriptional kinetics observed upon IL-4 stimulation (Figure S2D). These data indicate the consecutive activation of IL-33-induced pro- and anti-inflammatory signaling and transcriptional modules. Moreover, they suggest the presence of specific IL-33-induced and intrinsically imprinted molecular switches that trigger the delayed transition from a pro-inflammatory to a pro-resolving AAM-like macrophage phenotype upon encounter of tissue damage-derived signals.
IL-33 and IL-4 induce distinct AAM phenotypes
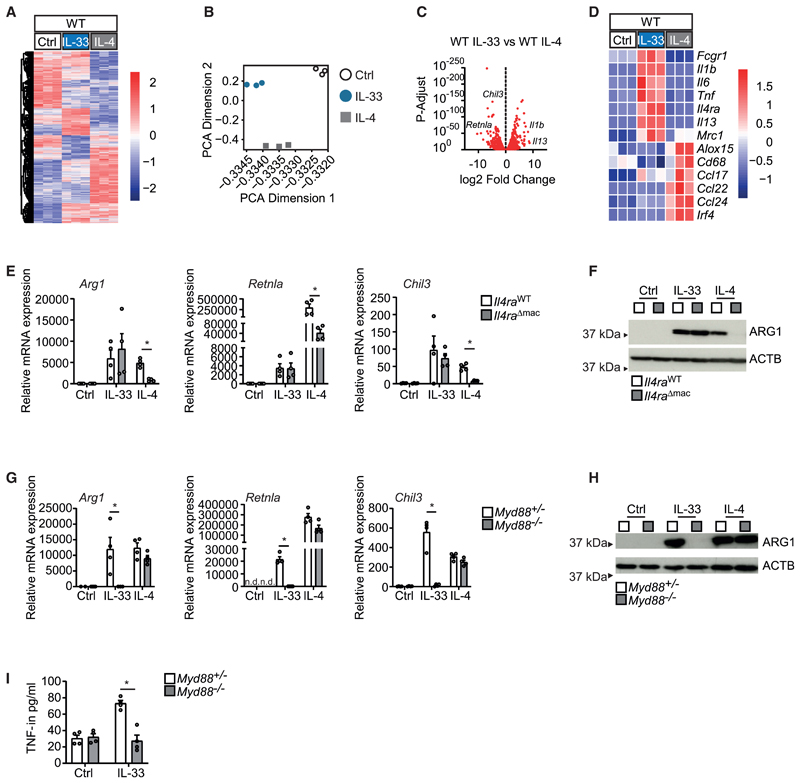
To identify such IL-33-specific molecular pathways that would promote an IL-4-independent pro-resolving AAM phenotype in response to injury, we performed a parallel comparative analysis of the global mRNA expression in IL-33 and IL-4-induced AAMs by bulk RNA sequencing. This approach showed that IL-33 and IL-4 induced partially overlapping but mostly distinct gene expression patterns in these cells (Figures 2A–2D). The direct comparison of the IL-33- and IL-4-induced gene expression profiles in BMDMs demonstrated that both cytokines triggered the expression of genes encoding for typical AAM markers such as Arg1 and Retnla. Among genes specifically induced by IL-33, in turn, we identified mostly mRNAs encoding for pro-inflammatory cytokines such as IL-1β, IL-6, and TNF but also certain previously described AAM marker genes including those encoding for the mannose receptor CD206 (encoded by Mrc1) and IL-13 (Figures 2C and 2D). Deletion of the gene encoding for the IL-4 receptor alpha (Il4ra) accordingly blocked IL-4-induced AAM differentiation but did not impair the IL-33-induced expression of Arg1, Retnla, and Chil3 (Figures 2E and 2F). Deletion of the gene encoding for the signaling adaptor MyD88, in turn, specifically inhibited both the differentiation of AAM and expression of pro-inflammatory cytokines in response to IL-33, whereas IL-4-induced gene expression was not affected (Figures 2G–2I). These results confirm that IL-33 and IL-4 use different receptors and proximal signaling adaptors to imprint an AAM phenotype.
Figure 2. IL-33 triggers the IL-4-independent differentiation of AAMs.
(A–D) Bulk mRNA sequencing data comparing BMDMs stimulated with vehicle (Ctrl), IL-33 (10 ng/mL for 36 h), and IL-4 (20 ng/mL for 36 h). Data are presented as a heatmap illustrating differential gene expression (A), PCA plot (B), volcano plot (C), and heatmap of genes for alternative and classic macrophage activation derived from bulk sequencing data in the different conditions (D).
(E–I) Quantitative real-time PCR (E and G), immunoblot (F and H), and ELISA (I) showing mRNA and protein expression of indicated markers of alternative activation (Arg1, Retnla, Chil3, ARG1) and classic activation (TNF) in BMDMs of Il4raWT and Il4raΔmac (E and F) or Myd88+/− and Myd88−/− (G–I) upon stimulation with vehicle (Ctrl) or IL-33 (10 ng/mL for 5 days) or IL-4 (20 ng/mL for 24 h). Data are representative of two individual experiments. Not detected (n.d.).
Data are presented as mean + SEM. *p < 0.05, **p < 0.01, and ***p < 0.001.
IL-33-induced AAM differentiation depends on mitochondrial uncoupling
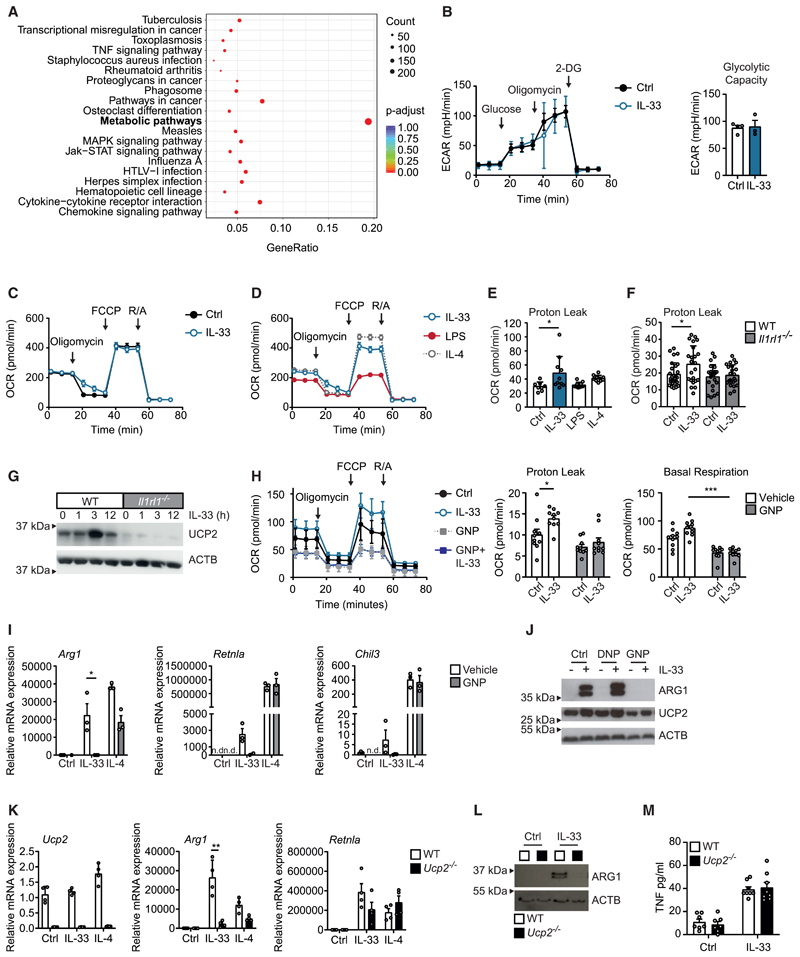
To better narrow down molecular pathways that mediated the IL-4-independent differentiation of pro-resolving AAMs in response to IL-33 and tissue injury, we next performed a Gene Ontology enrichment analysis on the basis of our bulk RNA sequencing data. This analysis unexpectedly suggested that the primary consequence of IL-33 stimulation was a metabolic reprogramming of macrophages (Figure 3A; Figure S3A). As cellular metabolism and polarization of macrophages are mutually linked (Mills et al., 2017), we decided to dissect these IL-33-induced metabolic changes in macrophages in more detail. Extracellular flux analysis showed that in contrast to IL-4 or lipopolysaccharide (LPS), IL-33 did not directly affect the extracellular acidification rate and changed neither the basal nor the maximal respiratory capacity in macrophages after a stimulation of 6 h (Figures 3B–3D). However, IL-33-treated macrophages rapidly developed a proton leak after IL-33 stimulation, which indicated an IL-33-induced and IL1RL1-dependent uncoupling of the mitochondrial respiratory chain in macrophages (Figures 3C–3F). Uncoupling was not associated with transcriptional changes in the expression of genes encoding for uncoupling proteins (UCPs) such as UCP1 or UCP2. However, we observed accumulation of UCP2 protein in the mitochondria of IL-33-treated macrophages, an effect that was absent in Il1rl1−/− cells and abrogated upon inhibition of p38 MAPK signaling by SB 203580, suggestive of an IL-33-induced and p38 MAPK-mediated translocation of preformed UCP2 into the mitochondrial compartment (Figures 3G, S3B, and S3C). Mitochondrial uncoupling has been previously shown to limit excessive production of mitochondrial reactive oxygen species (ROS) that derive from reverse electron transport (Sluse et al., 2006). Such ROS are otherwise characteristic for inflammatory macrophages, where they cause mitochondrial damage, impaired mitochondrial function, and/or a block in the tricarboxylic acid (TCA) cycle as observed, for example, in LPS-activated macrophages (Mills and O’Neill, 2016). Measurement of ROS production in response to prolonged IL-33 treatment (24 h) indeed showed an exacerbated ROS production upon pharmacological blockade of mitochondrial uncoupling by the compound genipin (GNP) (Figure S3D). Apart from reducing the IL-33-induced proton leak and in accordance with cumulative ROS-induced mitochondrial damage, we observed that GNP treatment of IL-33-stimulated macrophages resulted in increasingly impaired mitochondrial function and a decrease in the oxygen consumption rate after prolonged stimulation for 24 h (Figure 3H). In summary, these data thus indicated that IL-33-activated macrophages displayed a metabolic rewiring that was distinct from IL-4- or LPS-treated macrophages and characterized by UCP2-mediated uncoupling of the respiratory chain, which blocked the generation of ROS and allowed sustained mitochondrial respiration as well as an intact TCA cycle in otherwise pro-inflammatory macrophages. To address a potential relevance of the UCP2-mediated mitochondrial uncoupling and the sustained mitochondrial respiration for the IL-33-induced polarization of macrophages, we subsequently studied the consequence of a GNP-mediated pharmacological inhibition of mitochondrial uncoupling and of a genetic deletion of Ucp2 on macrophage polarization. Notably, both addition of GNP and genetic absence of UCP2 in Ucp2−/− macrophages interfered with AAM differentiation in response to IL-33, whereas AAM differentiation in response to IL-4 was largely unaffected (Figures 3I–3L). IL-33 stimulation of Ucp2−/− macrophages additionally resulted in upregulation of Ucp3 and Ucp4, suggesting a compensatory proton leak via alternative UCPs in these IL-33-stimulated macrophages (Figure S3E). Forced uncoupling induced by 2,4-dinitrophenol (DNP) further increased IL-33-mediated ARG1 expression but did not promote AAM differentiation in the absence of IL-33 (Figure 3J). The IL-33-induced expression of pro-inflammatory cytokines such as TNF, in turn, did not depend on UCP2 expression (Figures 3M and S3F).
Figure 3. IL-33-induced mitochondrial rewiring promotes AAM differentiation.
(A) KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis of bulk mRNA sequencing data comparing IL-33 (10 ng/mL for 36 h) and IL 4 (20 ng/mL for 36 h) treated BMDMs.
(B) Extracellular acidification rate (ECAR) of glycolysis stress test (extracellular flux assay) of BMDMs either vehicle treated (Ctrl) or treated with IL-33 (10 ng/mL for 6 h).
(C) Oxygen consumption rate (OCR) of Mito Stress Test (extracellular flux assay) of BMDMs either vehicle treated (Ctrl) or treated with IL-33 (10 ng/mL for 6 h). Data are representative of three individual experiments.
(D) OCR of BMDMs treated with IL-33 (10 ng/mL for 6 h), IL-4 (20 ng/mL for 6 h), or LPS (10 ng/mL for 6 h).
(E) Proton leak calculated from Mito Stress Test (extracellular flux assay) of BMDMs either vehicle treated (Ctrl) or treated with IL-33 (10 ng/mL for 6 h), IL-4 (20 ng/ mL for 6 h), or LPS (10 ng/mL for 6 h).
(F) Proton leak calculated from Mito Stress Test (extracellular flux assay) of WT or Il1rl1−/− BMDMs either vehicle treated (Ctrl) or treated with IL-33 (10 ng/mL for 6 h).
(G) Western blot of UCP2 protein from mitochondrial fractions of WT or Il1rl1−/− BMDMs treated with IL 33 (10 ng/mL) for the indicated time points. Data are representative of three individual experiments.
(H) Oxygen consumption rate (OCR) of Mito Stress Test (extracellular flux assay) of BMDMs vehicle treated (Ctrl) or treated with IL-33 (10 ng/mL for 24 h) and/or GNP (100 μM for 24 h). Proton leak and basal respiratory capacity calculated from Mito Stress Test.
(I and J) Quantitative real-time PCR (I) and western blot (J) showing mRNA and protein expression of indicated markers of alternative activation (Arg1, Retnla, Chil3, ARG1) in BMDMs treated with IL-33 (10 ng/mL for 5 days), IL-4 (20 ng/mL for 5 days), genipin (GNP; 100 μM for 5 days), and/or DNP (50 μM for 5 days). Data are representative of three individual experiments.
(K–M) Quantitative real-time PCR (K), western blot (L), and ELISA (M) showing mRNA and protein expression of indicated markers of alternative activation (Arg1, Retnla, Chil3, ARG1), Ucp2, and TNF in WT and Ucp2−/− BMDMs upon stimulation with vehicle (Ctrl) or IL-33 (10 ng/mL for 5 days). Data are representative of three individual experiments.
Data are presented as mean + SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. See also Figure S3.
IL-33-mediated mitochondrial uncoupling results in a shift of metabolites and a subsequent expression of the transcription factor GATA3
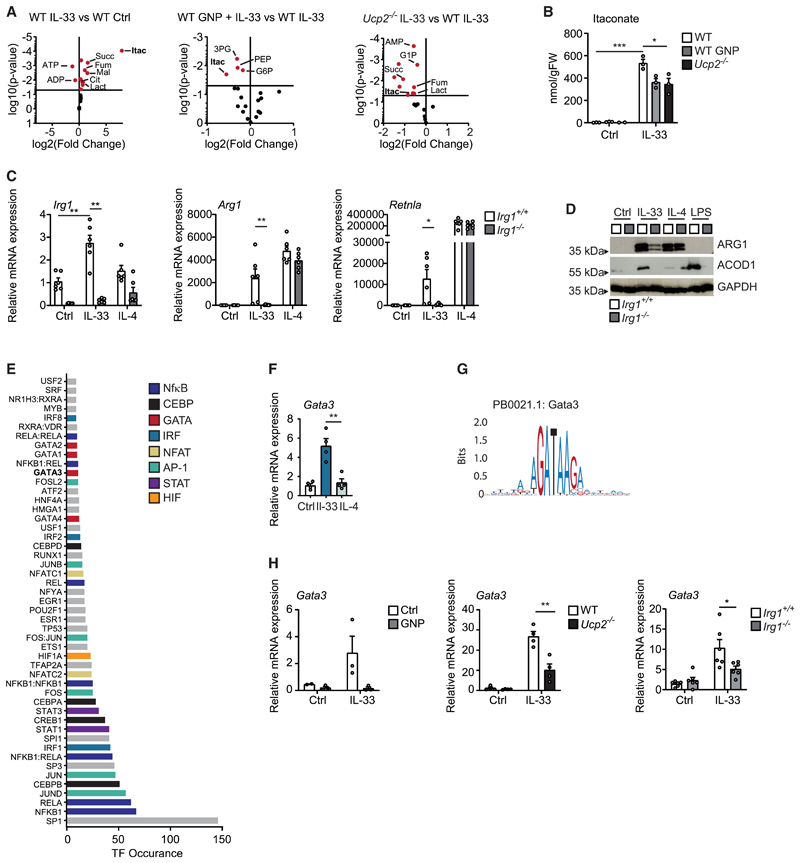
As these data suggested that UCP2-mediated uncoupling of the respiratory chain was critical for mitochondrial functionality, macrophage plasticity, and AAM differentiation in response to IL-33, we subsequently sought to further narrow down underlying mechanisms and performed mass spectrometry (MS)-based metabolomic fingerprinting of IL-33-stimulated WT, GNP-treated, and Ucp2−/− macrophages. In accordance with a central metabolic role of mitochondrial uncoupling during mitochondrial fitness and function, this approach confirmed significant IL-33-induced and UCP2-dependent shifts in intracellular metabolites. Such UCP2-dependent changes included an IL-33-induced increase in itaconate and other TCA metabolites such as succinate and fumarate, which, in turn, decreased upon block or deletion of UCP2 (Figures 4A and 4B).
Figure 4. IL-33-mediated mitochondrial uncoupling results in the induction of the transcription factor GATA3.
(A) Volcano plots illustrating the results of targeted metabolomics profiling comparing (left) IL-33 (10 ng/mL) treated and untreated (Ctrl) BMDMs 24 h after stimulation, (middle) IL-33 (10 ng/mL) treated BMDMs in the presence of GNP (100 μM) or vehicle 24 h after stimulation, and (right) IL-33 (10 ng/mL) treated Ucp2−/− BMDMs and WT BMDMs 24 h after stimulation.
(B) Itaconate concentration in nmol/g fresh weight (FW) in untreated (Ctrl) IL-33 (10 ng/mL) and/or GNP (100 μM) treated BMDMs of WT and/or BMDMs of Ucp2−/− mice.
(C and D) Quantitative real-time PCR (C) and western blot (D) showing mRNA and protein expression of indicated markers of alternative activation (Arg1, Retnla, Chil3, ARG1) and ACOD1 in BMDMs treated with vehicle (Ctrl), IL-33 (10 ng/mL for 5 days), or IL-4 (20 ng/mL for 24 h) in BMDMs of Irg1+/+ and/or Irg1−/− mice. Data are representative of two individual experiments.
(E) Transcription factor (TF) prediction for genes differentially expressed between IL-33- and IL-4-treated macrophages and significantly upregulated by IL-33. Bars are color coded according to indicated TF families.
(F) Relative Gata3 mRNA expression in BMDMs treated with vehicle (Ctrl) or IL-33 (10 ng/mL for 5 days) or IL-4 (20 ng/mL for 24 h). Data are representative of three individual experiments.
(G) JASPAR binding motif of GATA3.
(H) Relative Gata3 mRNA expression in BMDMs from WT, Ucp2−/−, or Irg1−/− mice treated with vehicle or IL-33 (10 ng/mL for 5 days) in the presence and absence of GNP (100 μM for 5 days) as indicated. Data are representative of two individual experiments.
Data are presented as mean + SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. See also Figure S4.
Of particular interest was the observed IL-33- and UCP2-dependent increase in the mitochondria-derived metabolite itaconate. Itaconate derives from the TCA cycle intermediate cis-aconitate that is converted by the enzyme aconitate decarboxylase 1 (ACOD1), which is encoded by the immune responsive gene 1 (Irg1). Itaconate is likewise able to inhibit mitochondrial ROS production by limiting the activity of succinate dehydrogenase (SDH) and can transfer into the cytoplasm, where it exerts multiple anti-inflammatory actions that depend on the activation of transcription factors such as ATF3 and Nrf2 (Murphy and O’Neill, 2018). Our data confirmed that IL-33, but not IL-4, strongly induced Irg1 mRNA expression, which reached an early peak after 3 h (Figure S4). Our experiments revealed that the sustained itaconate production that depended on IL-33 stimulation and UCP2-mediated uncoupling was crucially involved in the IL-33-induced AAM differentiation as Irg1−/− macrophages failed to respond to IL-33 in terms of an expression of AAM-related markers such as Arg1 and Retnla. IL-4-induced AAM differentiation, in contrast, was un-affected in Irg1−/− BMDMs (Figures 4C and 4D).
We consequently sought to identify potential transcriptional mediators of such an IL-33-induced mitochondrial rewiring of macrophages and tried to predict transcriptional modules involved in the regulation of IL-33-dependent and IL-4-independent genes we had previously determined in macrophages by bulk RNA sequencing. As expected, this analysis suggested that IL-33-induced gene expression would specifically involve several transcription factors previously described to mediate MyD88-dependent pro-inflammatory signaling such as members of the NF-κB, AP-1, IRF, and NFAT families of transcription factors (Figure 4E). However, this approach also predicted the additional involvement of members of the GATA family of transcription factors as mediators of the IL-33-induced transcriptional changes in macrophages (Figure 4E). Notably, GATA3 expression in T helper-2 (Th2) cells is regulated by Nrf2 as one of several itaconate-responsive transcription factors described so far (Rockwell et al., 2012). This finding thus provided a potential link between the observed IL-33-induced and UCP2- and ACOD1-mediated mitochondrial reprogramming and GATA3 expression. Our subsequent analyses indeed showed that IL-33 specifically induced expression of Gata3 in BMDMs, whereas IL-4 did not promote expression of this transcription factor (Figures 4F and 4G). We additionally observed that IL-33-induced expression of GATA3 was dependent on both UCP2-mediated mitochondrial uncoupling and ACOD1-mediated itaconate production, as addition of GNP as well as deletion of Ucp2 or Irg1 inhibited IL-33-mediated GATA3 expression in macrophages (Figure 4H). These data accordingly suggest a central role of GATA3 as metabolic signaling hub in macrophages that responds to an IL-33-induced mitochondrial reprogramming and translates it into global changes in gene expression.
GATA3 controls the differentiation of AAMs in response to IL-33
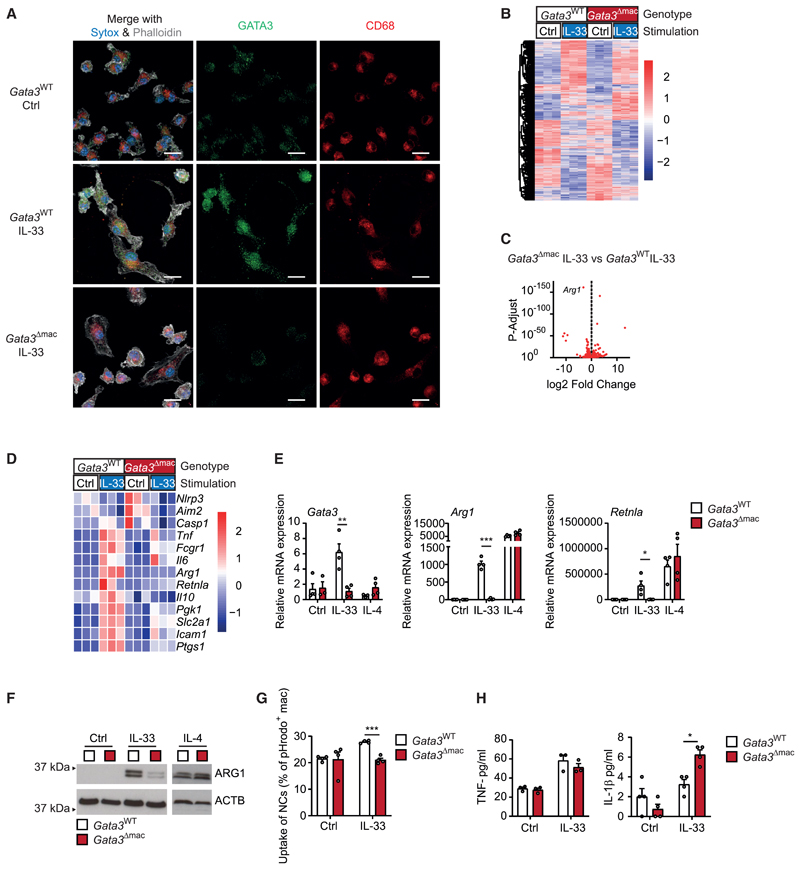
We subsequently crossed mice carrying floxed alleles of Gata3 with mice expressing a Cx3cr1-driven Cre recombinase to receive animals with a monocyte- and macrophage-specific deletion of this transcription factor (Gata3Δmac mice). Immunofluorescence microscopy confirmed that IL-33 induced a nuclear accumulation of GATA3 protein in BMDMs derived from WT mice (Gata3WT mice), whereas nuclear GATA3 was absent in IL-33-stimulated BMDMs of littermate Gata3Δmac mice (Figure 5A). Bulk RNA sequencing of these macrophages showed that absence of GATA3 indeed abrogated IL-33-mediated induction of a subset of IL-33-responsive genes in macrophages. Such GATA3-dependent genes included those encoding for AAM markers such as ARG1 and RELM-alpha (Figures 5B–5D; Table S1). Quantitative real-time PCR and western blot analysis confirmed the GATA3-dependent control of AAM differentiation in response to IL-33 on the mRNA and protein level (Figures 5E and 5F) and additionally showed that IL-4-induced AAM differentiation was intact and thus independent of GATA3. Notably, IL-33 also failed to increase the capacity of GATA3-deficient macrophages to ingest and clear necrotic cells demonstrating that this transcription factor coordinated various pro-resolving properties of macrophages in response to IL-33 (Figure 5G). The defective IL-33-induced AAM differentiation in GATA3-deficient BMDMs, however, did not alter expression of IL-33-induced Irg1 mRNA and of pro-inflammatory cytokines such as TNF, IL-6, or IL-1β (Figures 5H and S5). Production of IL-1β was even increased (Figure 5H), a phenomenon potentially related to an increased expression of different inflammasome subunits in GATA3-deficient macrophages (Figure 5D).
Figure 5. GATA3 controls the IL-33-induced polarization of AAMs.
(A) Immunofluorescence microscopy of GATA3 protein in BMDMs upon stimulation with vehicle (Ctrl) or IL-33 (10 ng/mL for 5 days); GATA3 depicted in green, CD68 in red, Phalloidin in white, and Sytox blue in blue. Data are representative of two individual experiments. Scale bar indicates 10 μm.
(B–D) Bulk mRNA sequencing data of BMDMs isolated from Gata3WT or Gata3Δmac mice upon stimulation with vehicle (Ctrl) or IL-33 (10 ng/mL for 36 h). Data are presented as heatmap (B) and volcano plot (C) of differential gene expression as well as heatmap of gene expression of alternative and classic macrophage activation markers and of genes involved in inflammasome activation (D).
(E–H) Quantitative real-time PCR-based measurement of mRNA expression of indicated genes (E), (F) immunoblot-based determination of ARG1 expression, (G) uptake of pHrodo-stained necrotic cells after 2 h of phagocytosis, and (H) ELISA-based measurement of TNF and IL-1β secretion in BMDMs isolated from Gata3WT or Gata3Δmac mice upon stimulation with vehicle (Ctrl), IL-33 (10 ng/mL for 5 days), or IL-4 (20 ng/mL for 24 h). Data (E–G) and TNF ELISA (H) are representative of three individual experiments.
Data are presented as mean + SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. See also Figure S5 and Table S1.
Tissue damage results in the differentiation of pro-inflammatory and pro-resolving mononuclear phagocytes
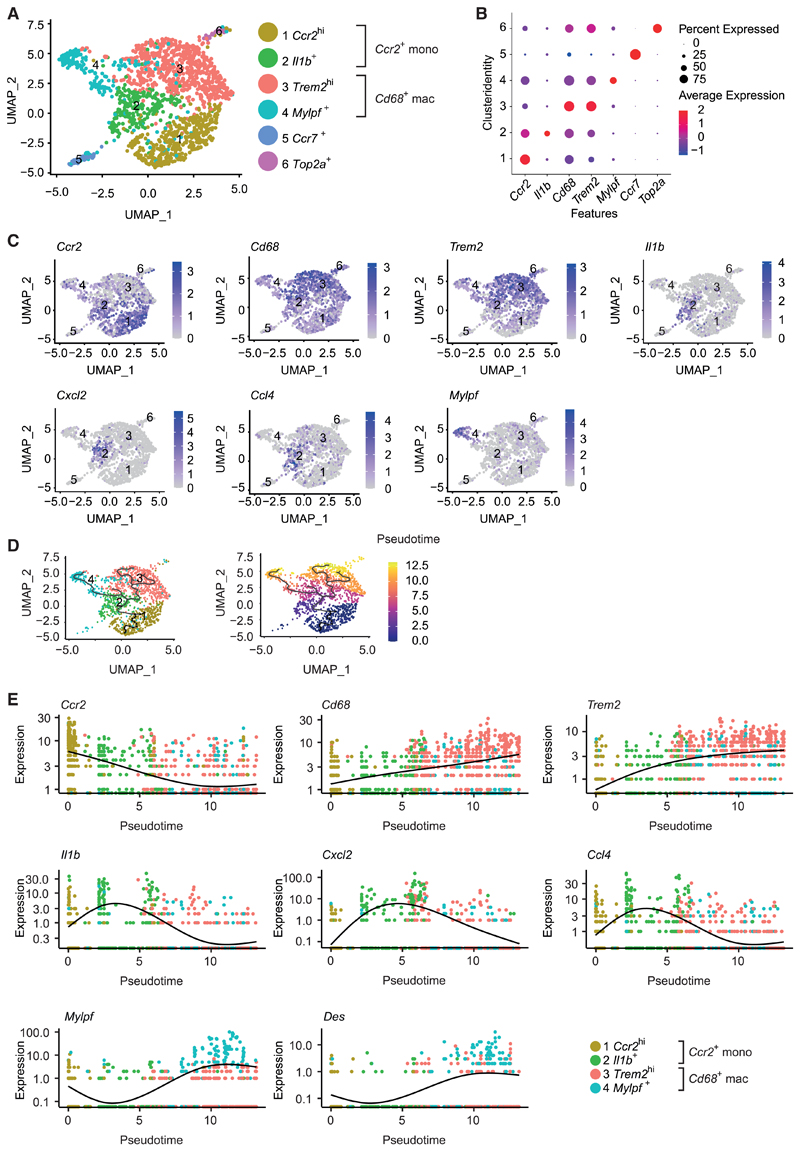
Consequently, we sought to determine the role of GATA3 in the function and plasticity of mononuclear phagocytes during their response to tissue damage in vivo. We therefore revisited the model of CTX-induced muscle injury in Gata3Δmac mice harboring a conditional deletion of GATA3 in mononuclear phagocytes and in their Gata3WT littermates, respectively. Single-cell RNA sequencing (scRNA-seq) of total CD45+CD11B+LY6G− mononuclear phagocytes sorted from the CTX-injected muscles allowed us to obtain an in-depth look and unbiased comparison of the distribution and activation profile of monocytes and macrophages infiltrating the injured tissue (Figure S6A). This analysis revealed the presence of four major and two smaller cellular clusters of monocytes and macrophages that appeared in the damaged muscle (Figures 6A–6C and S6B). These corresponded to two large clusters of Ccr2-expressing infiltrating monocytes (clusters 1 and 2) and two large clusters of Cd68-expressing macrophages (clusters 3 and 4). We additionally identified two smaller clusters, including a population of Ccr7+ mononuclear phagocytes (cluster 5) resembling dendritic cells and a cluster of Top2a+ proliferating cells (cluster 6) (Figures 6A–6C, S6C, and S6D). Ccr2-expressing monocytes could be further divided into a Ccr2hi cluster (cluster 1) that likely represented recently infiltrated blood monocytes as well as Il1b+ monocytes (cluster 2) that showed a highly inflammatory phenotype, including strong expression of genes encoding for various pro-inflammatory cytokines such as Il1b, Cxcl2 (encoding for MIP1alpha), and Ccl4 (encoding for MIP1beta) (Figures 6A–6C and S6D). Among Cd68-expressing macrophages, in turn, we could distinguish a large population of Trem2hi macrophages (cluster 3) and a cluster of Mylpf+ macrophages (cluster 4). Mylpf+ macrophages seemingly expressed both macrophage and skeletal muscle cell markers and thus presumably represented proresolving macrophages that were actively phagocytosing and clearing injured muscle cells, thereby ingesting muscle-specific mRNA (Figures 6B, 6C, S6B, and S6D), a phenomenon that has been described in scRNA-seq datasets before (Lantz et al., 2020). A pseudotime trajectory analysis generated with these scRNA-seq datasets suggested that freshly recruited Ccr2hi monocytes initially transformed into inflammatory Il1b+ monocytes before transitioning into Trem2hi and Mylpf+ macrophages (Figures 6D and 6E). This gradual transition from monocytes to pro-resolving macrophages was paralleled by downregulation of Ccr2 as well as upregulation of Cd68 and Trem2 (Figure 6E). Expression of pro-inflammatory genes such as Il1b, Ccl4, or Cxcl2 peaked at the intermediary stage of pro-inflammatory Il1b+ monocytes and decreased again in Trem2hi and Mylpf+ macrophages (Figure 6E). mRNAs such as Mylpf and Des encoding for muscle cell markers accordingly appeared primarily in differentiated pro-resolving macrophages (Figure 6E).
Figure 6. Muscle injury results in the differentiation of pro-resolving macrophages.
(A–E) Data derived from single-cell mRNA sequencing (scRNA-seq) from Zombie Aqua−CD45+CD11B+LY6G− mononuclear phagocytes from the injured muscle on day 4 after CTX injection of Gata3WT (n = 1) and Gata3Δmac (n = 1) mice. (A) Uniform manifold approximation and projection (UMAP) visualization of scRNA-seq cells of Gata3WT and Gata3Δmac origin that were clustered together. Each point represents an individual cell that is colored by cluster identity. (B) Dot-plot visualization of cluster markers Ccr2, Il1b, Cd68, Trem2, Mylpf, Ccr7, and Top2a. (C) UMAP visualization of cluster markers Ccr2, Cd68, Trem2, Il1b, Cxcl2, Ccl4, and Mylpf. (D) Pseudotime trajectory analysis determining the developmental relationship of identified cellular clusters (left) or as function of pseudotime (right) on the basis of scRNA-seq data. (E) Gene expression changes of selected marker genes as a function of pseudotime reflecting the cellular differentiation of Ccr2hi monocytes into Cd68+ macrophages.
See also Figure S6.
GATA3 controls the differentiation of pro-resolving macrophages
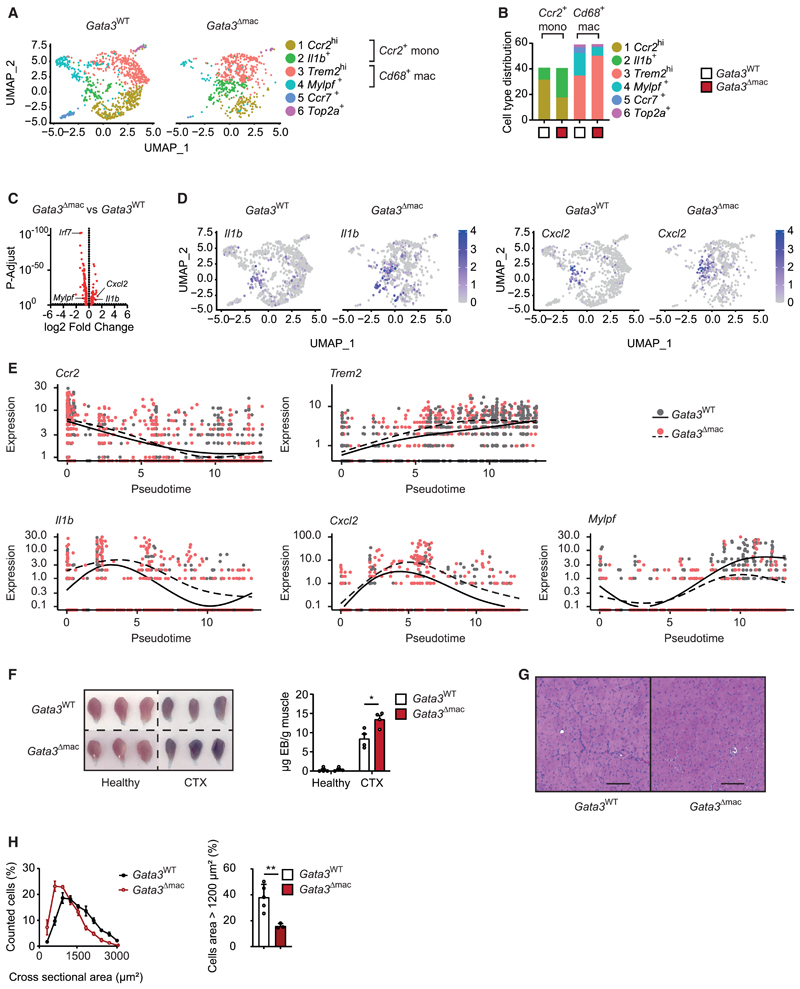
Loss of GATA3 in mononuclear phagocytes did not change the overall numbers of infiltrating monocytes or macrophages but resulted in an altered distribution of the different identified clusters of mononuclear phagocytes within the damaged tissue. Gata3Δmac mice displayed drastically increased numbers of pro-inflammatory Il1b+ monocytes upon CTX-induced muscle injury, while proresolving Mylpf+ macrophages were almost absent (Figures 7A, 7B, and S7A–S7C). In conjunction with our previous pseudotime trajectory analysis, these findings suggested an impaired trans-differentiation from pro-inflammatory Il1b+ monocytes to proresolving Mylpf+ macrophages as well as an impaired phagocytosis of necrotic muscle cells in the absence of GATA3. This partial differentiation arrest of mononuclear phagocytes at a pro-inflammatory activation state in Gata3Δmac mice accordingly correlated with increased overall expression of genes encoding for pro-inflammatory cytokines such as Cxcl2 and Il1b in differentiating GATA3-deficient mononuclear phagocytes that entered the injured muscle tissue (Figures 7C, 7D, and S7A; Tables S2 and S3). Among others, absence of GATA3 additionally resulted in reduced expression of Irf7 (encoding for the interferon regulatory factor 7 [IRF-7]) as well as of Ly6e (encoding for Sca-1). A comparative pseudotime analysis confirmed that infiltrating mononuclear phagocytes in Gata3Δmac animals displayed an exacerbated and prolonged inflammatory response during their differentiation into macrophages (Figure 7E). Accumulation of skeletal muscle mRNAs (e.g., Mylpf) in differentiated macrophages as readout for their pro-resolving and phagocytic activity, in turn, was reduced in Gata3Δmac mice, confirming the decreased efferocytosis observed in the absence of GATA3 in vitro (Figures 7E and S7B). In accordance with a GATA3-dependent differentiation of proresolving and reparative macrophages in response to tissue injury, loss of GATA3 in mononuclear phagocytes also resulted in persistent EB staining of injured muscle as well as altered central positioning of nuclei and decreased size of muscle fibers upon resolution of CTX-induced inflammation (Figures 7F–7H). These data thus demonstrated the functional relevance of GATA3 as transcriptional hub in macrophages that controlled their pro-resolving and reparative properties in response to tissue damage.
Figure 7. GATA3 controls the differentiation of pro-resolving macrophages upon muscle injury.
(A–E) Data derived from single-cell mRNA sequencing (scRNA-seq) from Zombie Aqua−CD45+CD11B+LY6G− mononuclear phagocytes from the injured muscle on day 4 after CTX injection of Gata3WT (n = 1) and Gata3Δmac (n = 1) mice. (A) Split UMAP visualization of identified clusters and (B) relative cell type distribution per cluster in the respective genotypes. (C) Volcano plot of differentially expressed genes and (D) split UMAP visualization of Il1b and Cxcl2 expression in mononuclear phagocytes from Gata3WT and Gata3Δmac mononuclear phagocytes. (E) Gene expression changes of selected marker genes as a function of pseudotime during the cellular differentiation of Ccr2hi monocytes into Cd68+ macrophages in Gata3WT (solid line) and Gata3Δmac (dashed line) mice.
(F) Macroscopic picture and quantification of Evans blue accumulation in healthy and CTX-injected muscles of Gata3WT and Gata3Δmac mice on day 7 after injury. (G and H) Histology (G) and (H) quantification and comparison of cross-sectional areas (CSAs) from muscle sections of CTX-injected Gata3WT and Gata3Δmac mice on day 14 after injury. Scale bar indicates 100 μm.
Data are presented as mean + SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. See also Figure S7 and Tables S2 and S3.
Finally, we sought to understand the relative contribution of this IL-4-independent, GATA3-mediated pathway and of IL-4 signaling during the macrophages’ response to tissue damage. Therefore, we additionally analyzed mice that expressed a Cx3cr1-driven Cre recombinase and carried floxed alleles of Il4ra, which accordingly resulted in a deletion of Il4ra in mononuclear phagocytes of these animals (Il4raΔmac). Notably, however, we did not detect significant differences in EB staining or in the size of regenerating muscle fibers when comparing Il4raΔmac mice and their Il4raWT littermates (Figures S7D and S7E), suggestive of an inferior role of macrophage-intrinsic IL-4 signaling during the injury-induced pro-resolving and reparative response of macrophages.
Discussion
Macrophages displaying an AAM signature accumulate during the resolution of tissue injury-induced inflammation (Arnold et al., 2007; Chazaud, 2020). However, it has remained incompletely understood which molecular pathways coordinate their differentiation and function in vivo. The IL-4 receptor alpha has been implicated in the differentiation of reparative macrophages during skin repair (Knipper et al., 2015), but previously published data and our present study show that conditional deletion of the IL-4 receptor alpha in myeloid cells and mononuclear phagocytes, respectively, did not affect tissue regeneration in response to muscle injury (Heredia et al., 2013), suggesting that IL-4-independent pathways might promote AAM differentiation and macrophage-mediated repair processes in response to cell death and trauma. Although crosstalk between IL-33 and IL-4 signaling has been reported (Jackson-Jones et al., 2016), our present data identify an IL-4-independent, IL-33-induced signaling axis in macrophages that involves an UCP2-mediated uncoupling of the mitochondrial respiratory chain and a metabolic reprogramming of macrophages that culminated in an increased production of itaconate and the GATA3-dependent differentiation of AAMs. GATA3 thereby acted as a transcriptional hub that integrated immune-metabolic changes and coordinated the IL-4-independent differentiation of pro-resolving AAMs in response to injury. These findings also show that IL-33, although initially serving as a pro-inflammatory “alarmin,” directly imprinted a mitochondria- and GATA3-mediated pro-resolving program that promoted the delayed differentiation of AAMs, which mediated the resolution of inflammation and macrophage-mediated tissue repair. Our present findings are in accordance with previous studies that have highlighted the importance of metabolic rewiring in mononuclear phagocytes responding to sterile muscle injury and which observed both an increase in oxidative phosphorylation and changes in lipid mediators in macrophages during muscle regeneration (Giannakis et al., 2019; Varga et al., 2016).
Although MyD88 and NF-κB are known to transduce the early IL-33-mediated pro-inflammatory response (Liew et al., 2016), the molecular mechanisms controlling IL-33-mediated proresolving responses associated with an AAM signature have remained less well understood. The observed role of an initial IL-33-induced metabolic reprograming that subsequently promotes a delayed GATA3-dependent differentiation of AAMs is in line with the emerging concept of a mutual influence of cellular metabolism and the polarization of immune cells. In particular, mitochondrial dynamics and activity have been closely linked to the polarization and activation state of macrophages (Mills et al., 2017). The observation that IL-33 acts as an endogenous inducer of the enzyme ACOD1 and exerts strong metabolic effects in macrophages that substantially differ from both IL-4- or LPS-induced metabolic changes additionally stresses the role of this cytokine during onset and resolution of inflammation. Our present data suggest that UCP2-dependent uncoupling of the respiratory chain in response to IL-33 reduces ROS production and preserves mitochondrial respiratory function. IL-33-activated macrophages thus display not only high expression of ACOD1 but also (and in contrast to classically activated macrophages) integrity of their TCA cycle, which, among others, seems to be required to fuel persistent production of itaconate and the subsequent GATA3-dependent differentiation of AAMs. Itaconate has recently emerged as critical anti-inflammatory metabolite that acts as intracellular messenger and links mitochondrial metabolism and immune cell function, thereby exerting important tissue protective roles (Mills and O’Neill, 2016), although the factors that control itaconate production during sterile inflammation have remained largely unclear. Within mitochondria, itaconate blocks SDH and thereby functionally synergizes with UCP2, which reduces SDH-mediated reverse electron transport, in ameliorating mitochondrial ROS production. Within the cytoplasm, itaconate additionally activates the transcription factors Nrf2 and ATF3 that exert potent anti-inflammatory effects and likewise promote an anti-oxidative response program within the cell. Notably, Nrf2 also acts as direct transcriptional inducer of GATA3 expression (Rockwell et al., 2012), suggesting a potential role of this and related transcription factors during the induction of GATA3 expression in response to IL-33.
In this study we identify GATA3 as an IL-33-responsive transcriptional hub that links cellular metabolism and differentiation of macrophages. GATA3 has been described as a central transcription factor during the differentiation of Th2 cells and innate lymphocyte type 2 cells (Wan, 2014). An important feature of GATA3 is its ability to auto-amplify its expression (Wan, 2014), which explains the gradual increase in GATA3 expression and the continuous shift toward a pro-resolving phenotype that is observed in IL-33-activated macrophages and during tissue injury over several days. Although its role in macrophage function is poorly studied, a previous study identified GATA3-positive macrophages in hypertrophic and regenerating myocardium and suggested that such GATA3-positive macrophages might be involved in pathologic IL-4-mediated effects, thereby adversely affecting cardiac remodeling (Yang et al., 2018). Our present study, however, clearly excludes a role of GATA3 in IL-4-mediated effects in macrophages and additionally shows that IL-33-induced GATA3 expression primarily promotes pro-resolving properties of macrophages in response to tissue injury, which involves an increased ability of macrophages to ingest and clear dying cells, an important prerequisite for the resolution of inflammation and initiation of tissue repair. Irrespective of IL-33 signaling and GATA3 expression, UCP2-mediated mitochondrial uncoupling itself has been previously shown to enable a continuous phagocytosis of apoptotic cells by macrophages (Park et al., 2011). Together with our present findings on a critical role of GATA3 in promoting the clearance of necrotic cells by macrophages, these observations additionally stress the close interconnection of metabolism, polarization, and functional properties in macrophages during the resolution of injury-induced inflammation. Although functional data are lacking so far, very recent data suggest that the related transcription factor GATA2 might regulate the IL-33-induced control of iron recycling in macrophages (Lu et al., 2020).
Our current scRNA-seq data show a considerable degree of plasticity of mononuclear phagocytes in vivo and largely support a scenario in which monocytes recruited to sites of injury undergo an initial inflammatory activation boost that is paralleled by expression of cytokines such as IL-1β before they differentiate into pro-resolving and anti-inflammatory macrophages. By accumulating muscle-specific mRNAs, Mylpf+ macrophages show indirect evidence for the phagocytosis of dead muscle cells, a phenomenon that has allowed assessing increased efferocytosis in scRNA datasets before (Lantz et al., 2020). The GATA3-dependent regulation of the IRF-7 is in accordance with its role in the IL1RL1 signaling cascade (He et al., 2019). Notably, IRF-7 has been additionally implicated in monocyte to macrophage differentiation and a shift to an AAM-related phenotype (Cohen et al., 2014; Lu and Pitha, 2001) that dampens inflammation (Salem et al., 2011). The molecular signature of the identified Trem2hi and Mylpf+ macrophage clusters, which dominated during muscle injury and repair, largely matches the signature of related Trem2-expressing macrophages reported during diverse inflammation-and damage-associated pathologies in other organs. scRNA-seq profiling has identified closely related macrophage populations in Alzheimer’s disease, arthritis, atherosclerosis, metabolic disease, and within the tumor microenvironment, where they were shown to exert protective and anti-inflammatory roles or interfere with anti-tumor immunity (Cochain et al., 2018; Culemann et al., 2019; Jaitin et al., 2019; Keren-Shaul et al., 2017; Molgora et al., 2020). This indicates common pathways that control macrophage polarization and function under diverse conditions of cellular stress, inflammation, and tissue damage. The full spectrum of signals and transcription factors involved in the polarization and function of such macrophages in these different settings is still unclear. Our present data, however, suggest an important role of an immune-metabolic IL-33-GATA3 axis during the transition from pro-inflammatory monocytes to such pro-resolving macrophages as well as during the control of the clearance of cellular debris and tissue regeneration upon tissue injury. These data additionally shed light on the double-edged role of macrophages during injury and inflammation and identify a potential molecular target that impedes inflammation-associated tissue damage and simultaneously fosters resolution of inflammation and repair processes. It remains to be determined whether GATA3 also contributes to the plasticity and function of mononuclear phagocytes in other settings, such as during parasite defense, metabolic disorders, cancer, or Alzheimer’s disease.
Limitations of study
A clear limitation of our present study is the use of an in vitro modeling system, which allowed us to dissect IL-33-induced metabolic rewiring in macrophages. Future studies are therefore needed to better address the in vivo significance of these findings and to determine in particular the role of mitochondrial uncoupling and ACOD1 during the resolution of injury-induced inflammation.
Star★Methods
Detailed methods are provided in the online version of this paper and include the following:
-
-
○
Lead contact
-
○
Materials availability
-
○
Data and code availability
-
○
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○
Primary cell culture
-
○
Sterile muscle injury
-
○
-
-
○
qPCR
-
○
WB
-
○
Phagocytosis assay
-
○
ELISA
-
○
IF
-
○
BulkSeq
-
○
Transcription factor enrichment analysis
-
○
Extracellular flux assay
-
○
Metabolomics
-
○
Flow cytometry
-
○
Single cell sequencing
-
○
Star⋆Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies and fluorescence dyes | ||
| Monoclonal Anti-β-Actin antibody | Sigma-Aldrich | Cat# A5441; RRID:AB_476744 |
| Arginase I (V-20) | Santa Cruz | Cat#sc-18354; RRID:AB_2227469 |
| Donkey Anti-Goat IgG H&L (Alexa Fluor 647) | Abcam | Cat#ab150131 |
| Alexa Fluor 594 anti-mouse CD68 | BioLegend | Cat#137020; RRID:AB_2563305 |
| Gata3 (H-48) | Santa Cruz | Cat#sc-9009; RRID:AB_640893 |
| Alexa Fluor 488 Phalloidin | Invitrogen | Cat#A12379 |
| PE/Cy7 anti-mouse CD11b | BioLegend | Cat#101215; RRID:AB_312798 |
| Alexa Fluor 488 anti-mouse Ly6C | BioLegend | Cat#128021; RRID:AB_10640820 |
| Alexa Fluor 700 anti-mouse CD45.2 | BioLegend | Cat#109822; RRID:AB_493731 |
| Brilliant Violet 421 anti-mouse Ly6G | BioLegend | Cat#127627; RRID:AB_10897944 |
| TruStain FcX (anti-mouse CD16/32) Antibody | BioLegend | Cat#101319; RRID:AB_1574973 |
| Dapi | Roche | Cat#10236276001 |
| Sytox Blue | ThermoFisher | Cat#S34857 |
| Zombie Aqua Fixable Viability Kit | BioLegend | Cat#423101 |
| Mouse IL-33 Antibody (goat) | R&D Systems | Cat#AF3626-SP; RRID:AB_884269 |
| Donkey anti goat AF647 | Invitrogen | Cat#A-21447 |
| Donkey anti goat AF555 | Invitrogen | Cat#A-21432 |
| POPO-1 Iodide | ThermoFisher | Cat#P3580 |
| Alexa Fluor® 488 anti-mouse Ly-6A/E (Sca-1) Antibody | BioLegend | Cat#108115; RRID:AB_493270 |
| Alexa Fluor® 594 anti-mouse Podoplanin | BioLegend | Cat#156205; RRID:AB_2800698 |
| Wheat Germ Agglutinin (AF647) | ThermoFisher | Cat#W32466 |
| IRG1 Antibody (detects ACOD1) | Cell Signaling Technology | Cat#17805 |
| UCP2 (D1O5V) Rabbit mAb | Cell Signaling Technology | Cat#89326 S; RRID:AB_2721818 |
| Recombinant Anti-TOMM20 antibody | Abcam | Cat# ab186735; RRID:AB_2889972 |
| Rabbit Anti-GAPDH Monoclonal Antibody | Cell Signaling Technology | Cat#2118S; RRID:AB_561053 |
| APC anti-mouse IL-33Rα (ST2) Antibody (detects IL1RL1) | BioLegend | Cat#146605; RRID:AB_2728174 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse IL-33 | BioLegend | Cat#580502 |
| rmIL-33 | R&D Systems | Cat#3626-ML |
| Recombinant Mouse IL-4 | Biolegend | Cat#574302 |
| Cardiotoxin Naja pallida | Latoxan | Cat#L8102-1MG |
| LPS | Sigma-Aldrich | Cat#L-2630 |
| IL1b | R&D Systems | Cat#401-ML-005 |
| IL18 | MBL | Cat#B002-5 |
| Genipin (GNP) | Wako | Cat#078-03021 |
| 2,4-Dinitrophenol (DNP) | Sigma-Aldrich | Cat# D198501-5G |
| SB203580 | biotechne (tocris) | Cat#1202 |
| Critical commercial assays | ||
| Mouse IL1β/IL-1F2 DuoSet ELISA | R&D Systems | Cat#DY401-05 |
| Mouse TNF-α DuoSet ELISA | R&D Systems | Cat#DY410-05 |
| RNeasy Mini Kit (50) | QUIAGEN | Cat#74104 |
| Deposited data | ||
| Bulk Sequencing Data Set | This paper | GEO: GSE151655 |
| Single Cell Sequencing Data Set | This paper | GEO: GSE151655 |
| Experimental models: Organisms/strains | ||
| WT | Charles River | C57BL/6J |
| Il1rl1 –/– | Il1rl1tm1Aki | MGI:3623124 (backcrossed to C57BL/6) |
| MydSS –/– | Myd88tm1Aki | MGI:2385681 (backcrossed to C57BL/6) |
| Il4ra Δmac | Il4ratm2Fbb X STOCK Tg(Cx3cr1-cre) MW126Gsat | MGI:3689995 X RRID:MMRRC_036395-UCD |
| Ucp2 –/– | B6;129S-Ucp2tm2.1Lowl/J X B6.FVB-Tg (Ella-cre)C5379Lmgd/J | MGI:4887236 X MGI:2137691 |
| Irg1 –/– | Irg1tm1a(KOMP)Wtsi | MGI:4362417 |
| Gata3 Δmac | Gata3tm1Iho X STOCK Tg(Cx3cr1-cre) MW126Gsat | MGI:3719568 X RRID:MMRRC_036395-UCD |
| Oligonucleotides | ||
| See Table S4 for primer sequences. | N/A | |
| Software and algorithms | ||
| DESeq2 v.1.22.2 & v.1.24.0 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ggplot2 v.3.3.0 | Wickham, 2016 | https://cran.r-project.org/web/packages/ggplot2/index.html |
| pheatmap v.1.0.12 | Kolde, 2019 | https://cran.r-project.org/web/packages/pheatmap/index.html |
| Seurat v.3.0.2 | Stuart et al., 2019 | https://cran.r-project.org/web/packages/Seurat/index.html |
| monocle3 v.0.1.2 | Trapnell et al., 2014 | https://github.com/cole-trapnell-lab/monocle3 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Gerhard Krönke (gerhard.kroenke@uk-erlangen.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The single-cell RNA-seq data as well as the bulk RNA-seq data generated during this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model And Subject Details
Primary cell culture
Bone marrow was flushed from femurs and tibias of 8- to 12-week old mice (Unique identifiers are listed in the Key Resources Table). Following erylysis cells were incubated at 37°C in BMDM-medium consisting of DMEM (GIBCO), 10% FCS (Biochrom), 1% Penicillin/Streptomycin (GIBCO) and 10% M-CSF-containing L929-supernatants. L929 cells were kindly provided by Prof. Dr. R. Lang (Erlangen). The following day, non-adherent cells were collected and differentiated for 5 days in fresh BMDM-medium. In the experiments, BMDM were stimulated with 10 ng/ml IL-33, 10 ng/ml LPS, 10 ng/ml or 20 ng/ml IL-4, 10 ng/ml IL-1β, 10 ng/ml IL-18, 100 μM Genipin, 50μM DNP, 10 ng/ml INFγ,1 μM or 10 μM SB 203580 for the indicated time periods. The details of the used stimulants are listed in the Key Resources Table.
Sterile muscle injury
The animal experiments were performed in accordance with German guidelines and laws and were approved by the local animal ethics committees of the Regierung von Mittelfranken. Gata3fl/fl mice were kindly provided by B. Weigmann and crossed to Cx3cr1CRE mice obtained from the Mutant Mouse Regional Resource Center (MMRC), a National Institutes of Health (NIH)-funded strain repository, and were donated to the MMRRC by the National Institute of Neurological Disorders and Stroke (NINDS)-funded GENSAT BAC transgenic project. In experiments with Cx3cr1CRE Gata3fl/fl (Gata3Δmac) mice Cx3cr1WT Gata3fl/fl (Gata3WT) littermates were used as WT controls. All transgenic mice were bred and housed at the animal facilities of the Friedrich-Alexander University of Erlangen-Nürnberg under specific-pathogen-free conditions.
Muscle injury in 8- to 12-week old mice of both sexes was induced by the injection of 50 μL of 10 μM solution of cardiotoxin (CTX) from Naja pallida (Latoxan) into the mid-belly of the tibialis anterior muscle. To assess muscle damage on day 7, mice were given an i.v. injection of Evans blue dye (25 mg/kg) 35 min prior to sacrifice. To quantify Evans Blue enrichment muscles were incubated over night at 60°C in 5 μl Formamid (Carl Roth) per mg muscle tissue. Adsorption was measured at a wavelength of 620 nm. Cross sectional area was determined on day 14 after CTX injection. Muscles were isolated, fixed in 4% buffered formalin (pH 7.4) and embedded in paraffin before sectioning. 2 μm sections were stained for H&E using standard procedures. Pictures were taken with a Zeiss Axio Lab.A1 Laboratory Microscope using cellSens Entry Software (OLYMPUS) at 10x optical zoom (Zeiss Nachroplan 10x/0,25 Ph1 ∞/- objective) at the center of each muscle cross section. The cross sectional area of muscle fibers was measured using ImageJ Software. To avoid bias in measurements the experimentator manually identifying the circumferences of the muscle fibers was not aware of the underlying genotypes. Depending on the state of the muscle regeneration between approximately 400 and 850 individual muscle fibers were measured per animal.
Method Details
qPCR
Total RNA was isolated according to manufacturer’s protocol using peqGOLD TriFast (Peqlab). 500 ng of isolated RNA was reverse transcribed into cDNA. mRNA was quantified using a QuantStudio Real Time Systems LightCycler instrument (ThermoFisher) and SYBR Green I kit (Roche). mRNA amounts are presented as ddCT-Values normalized to β-actin expression as well as the untreated WT control. Primers used are listed in the Key Resources Table.
WB
After lysis of the cells in Lämmli-Buffer (BioRad) containing β-mercaptoethanol, the protein extracts were separated on 10% SDS–polyacrylamide gel or 4%–20% precast gels (BioRad) transferred onto a polyvinylidene difluoride membrane (BioRad) and immunoblotted with the antibodies listed in the Key Ressources Table.
Phagocytosis assay
Necrotic cells (NC) were generated from either CFSE- (BioLegend) or pHrodo-stained (Life Technologies) C2C12 cells by two cycles of freeze-thawing for 15 min at −80°C and 5 min at 37°C. 12 h before measurement BMDM-medium was replaced with fresh BMDM-medium not containing any stimulants. The day of the assay BMDMs were confronted with NCs in a ratio of 1:3 for 1 h if stained with CFSE or 2 h if stained with pHrodo. Cells were washed 2 times with PBS and detached using Cellstripper (Corning) for 10 min at 37°C. Additional staining and flow cytometry measurements were performed as described below.
ELISA
ELISA based measurements of undiluted supernatants were performed according to the manufacturer’s protocol using the DuoSet ELISA development System (R&D) for mouse TNF-α (DY410) and mouse IL-1β (DY401). Optical density was determined at 450 nm with a wavelength correction at 540 nm.
IF
From cell culture
Cells were grown on glass coverslips and fixed for 24 h with 4% PFA (pH 7,4). After washing with PBS, cells were permeabilized using 0.2% Triton X-100 (Merck Millipore) in PBS for 20 min at room temperature and were blocked with 0.05% Tween 20 (Sigma Aldrich), 1% BSA (Sigma Aldrich) and 10% rat serum in PBS for 1 h at room temperature. Staining with primary antibodies was performed in blocking buffer at 4°C over night. Secondary staining was executed at room temperature for 4 h. The antibodies used in the experiments are listed in the Key Resources Table.
To produce high-magnification imaging of BMDM, a Leica TCS SP 5 II confocal microscope (Software Las AF, Version 2.7.3.9723, Leica) with acousto-optic tunable filter and acousto-optical beam splitter, and hybrid detector (HyD) on a DMI6000 CS frame was used with 1x digital zoom. Imaging of coverslip-embedded samples was executed using an HCX PL APO 100x/1.44 Oil CORR CS objective. Alexa Fluor 595 was excited using a diode-pumped solid-state laser at 561 nm and was detected with a HyD at 600–650 nm. Alexa Fluor 488 staining was visualized using an argon laser at 488 nm for excitation and a HyD detector at 500–550 nm. Detection of SYTOX Blue involved a 458-nm argon laser and a HyD at 470–520 nm. Alexa Fluor 647 was excited with a 633-nm helium-neon laser an Immunity 54, 2531–2546.e1–e5, November 9, 2021 e3 detected by HyD at 650–700 nm. The generated images were deconvoluted with Huygens Professional (Version 17.10, Scientific Volume Imaging B.V.) and visualized with Imaris software (Version 9.3, Bitplane).
From tissue
Tibialis anterior muscles were harvested from WT mice (male or female, 8-10 weeks of age), manually cut into 1 mm chunks and fixated in 1% PFA overnight at 4°C. Samples were thoroughly washed with PBS and either stored in PBS/NaN3 or directly processed for immuno-imaging as previously described (Uderhardt et al., 2019). In brief, samples were blocked in PBS + 0.1% Triton-X + 5% normal rat serum + 1% bovine serum albumin at room temperature for at least 3 hours. Muscle tissues were incubated with primary antibodies in PBS + 0.1% Triton-X + 1% normal rat serum for 2 nights at room temperature on an overhead mixer with the following antibodies or reagents: POPO-1 (ThermoFisher), anti-Podoplanin Alexa594 (BioLegend), goat anti-IL33 (R&D) + donkey anti-goat Alexa647 or donkey anti-goat Alexa555 (Invitrogen), wheat germ agglutinin Alexa647 (ThermoFisher). Samples were then thoroughly washed with PBS + 0.1% Triton-X and optically cleared overnight at room temperature on an overhead mixer following the Ce3D protocol (Li et al., 2019). Samples were individually mounted in Ce3D clearing buffer and imaged using a Zeiss LSM 880 laser scanning microscope equipped with a 20x / NA 1.0 Plan-Apochromat water immersion objective (Zeiss) and an Coherent Chameleon Ultra II femtosecond Ti:sapphire laser. Autofluorescence of muscle fibers was recorded using the green channel (450-520 nm). 200-250 μm of tissue volumes were recorded as tiled stacks (zoom of 1.5x; 16bit; 512x512; z stack stepsize 1.5 um), stitched using ZEN Blue 3.1 (Zeiss) and visualized using IMARIS software (Version 9.3, Bitplane).
BulkSeq
Total RNA from BMDMs was isolated using the RNeasy Mini Kit (QIAGEN). The sequencing steps were performed at Novogene Co., Ltd. A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB) following manufacturer’s instructions and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s recommendations. After cluster generation, the library preparations were sequenced on an Illumina PE150 platform (HWI-ST1276) and paired-end reads were generated. Reference genome and gene model annotation files were downloaded from genome website browser (NCBI/UCSC/Ensembl) directly. Paired-end clean reads were mapped to the mouse reference genome (GRCm38/mm10) using HISAT2 (v.2.0.5) software. HTSeq (v.0.6.1) was used to count the read numbers mapped of each gene. Subsequently, RPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis between two conditions with three biological replicates per condition was performed using DESeq2 (v.1.22.2) (Love et al., 2014). The resulting p values were adjusted using Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR). Genes with an adjusted p value < 0.05 found by DESeq2 were assigned as differentially expressed. To visualize data, the R packages ggplot2 (v.3.3.0) (Wickham, 2016), DESeq2 (v.1.24.0) and pheatmap (v.1.0.12) were used.
The DEGs were analyzed for functional enrichment using g:Profiler web tool (Raudvere et al., 2019) and selected significantly enriched processes (adjusted p value < 0.05) were plotted using ggplot2 package version 3.3.23 in R (version 4.0.2).
Transcription factor enrichment analysis
Transcriptional interactions were investigated with the TRANSFAC (Matys et al., 2006) database (release 2015/1), which curates and annotates experimentally validated interactions from literature. All transcription factor-gene pairs from mouse or human were considered eligible. For a query list of target genes, transcription factors were collected, and their frequency of appearance (termed occurrence) was tabulated and ranked.
Extracellular flux assay
Real-time bioenergetic profiles of OCPs were observed by measuring oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) using the XFe96 extracellular flux analyzer (Seahorse Bioscience, Agilent Technologies, North Billerica, MA). BMDMs were plated at a density of 60,000 cells per well and left untreated or treated with IL-33 (10 ng/ml, 6 h or 24 h), IL-4 (10 ng/ml, 6 h), LPS (10ng/ml, 6 h) or Genipin (100 μM, 24 h). Assays were performed following the manufacturers protocol. In brief, a Cell Mito Stress Test was performed by sequential titration to 1 μM oligomycin, 1.5 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) and 0.5 μM rotenone/antimycin A (R/A). A Cell Glyco Stress Test was performed by sequential titration to 10 mM glucose, 1 μM oligomycin and 50 mM 2dG. Proton leak was calculated by subtracting the minimum rate measurement after R/A injection from the minimum rate measurement after Oligomycin injection.
Metabolomics
Phosphorylated intermediates and carboxylates were extracted with perchloric acid from 50 mg samples of shock-frozen BMDMs for four biological replicates as described previously (Hofmann et al., 2011) applying ionchromatography with an ICS3000 HPLC-system (Dionex) and ESI/MS/MS detection using a QTrap3200 Triple-Quadrupole mass spectrometer with turbo V ion source (Applied Bio-systems) operated in multiple reaction monitoring mode.
Flow cytometry
For isolation of macrophages, muscles were dissected on day 4 after CTX injection and dissociated by incubation for 45 min at 37°C in a digestion solution consisting of RPMI medium (GIBCO), 10% heat-inactivated fetal calf serum (FCS), collagenase (2 mg/ml) from Clostridium histolyticum (Sigma-Aldrich, C5138-1G) and 0.03 mg/ml DNase (Sigma-Aldrich, 9003-98-9). After washing with PBS containing 2% heat-inactivated FCS and 2 mM EDTA, cells were blocked with 10% rat serum, 2% heat-inactivated FCS and 1% Fc Block (BioLegend) in PBS for 10 min at room temperature and stained with fluorophore-conjugated antibodies for 20 min at 4°C. Antibodies and dyes used, are listed in the Key Resources Table. After washing, cells were resuspended in 2% heat-inactivated FCS in PBS. Flow cytometry was performed with a CytoFLEX S, Beckman Coulter. Sorting of cells was performed with a MoFlo XDP, Beckman Coulter and the Summit Software System. Data was analyzed with CytExpert (Beckman Coulter, v2.4) or FlowJo (v.10.6.1).
Single cell sequencing
Sorted cells were subjected to 10x Chromium Single Cell 3′ Solution v3 library preparation according to the manufacturer’s instructions. Library sequencing was performed on an Illumina HiSeq 2500 sequencer to a depth of 200 million reads each. In all samples, the mean number of reads per cell was greater than 45000. Reads were converted to FASTQ format using mkfastq from Cell Ranger 3.0.1 (10x Genomics). Reads were then aligned to the mouse reference genome (mm10 3.0.0, Ensembl annotation release 93). Alignment was performed using the count command from Cell Ranger 3.0.1 (10x Genomics). Primary analysis, quality control filtering (gene count per cell, unique molecular identifier count per cell, percentage of mitochondrial as well as ribosomal transcripts), clustering, identification of cluster markers and visualization of gene expression were performed using the Seurat (v.3.0.2) package for R. Pseudotime was analyzed using monocle3 (v.0.1.2) (Stuart et al., 2019; Trapnell et al., 2014).
Quantification And Statistical Analysis
For calculations of statistical significance, GraphPad Prism 8.3 was used. Data are presented as mean + s.e.m. and were analyzed using the two-sided Student’s t test unless stated otherwise. P values less than 0.05 were considered significant. All in vitro results are representative up to three 3 individual experiments.
Supplementary Material
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.09.010.
Highlights.
IL-33 induces rapid mitochondrial rewiring in macrophages
Uncoupling of the respiratory chain promotes IL-33-induced AAM differentiation
GATA3 links macrophage metabolism and differentiation in response to IL-33
GATA3 orchestrates the differentiation of pro-resolving macrophages in vivo
In brief.
Alternatively activated macrophages (AAMs) promote the resolution of inflammation and tissue repair. Faas et al. show that the alarmin IL-33 promotes a mitochondrial rewiring of macrophages and the consecutive activation of the transcription factor GATA3, which orchestrates the IL-4-independent differentiation of AAMs and the resolution of inflammation upon tissue injury.
Acknowledgments
We thank A. Klej, R. Weinkam, and R. Palmisano as well as the Optical Imaging Center Erlangen (OICE) for excellent technical assistance. M. Mroz and U. Appelt provided help during cell sorting. We want to additionally thank Yolanda Pires-Afonso from the Luxembourg Institute of Health (LIH) for help during isolation of macrophages from Irg1−/− mice. This work was supported by Deutsche Forschungsgemeinschaft (DFG; CRC1181-A03/A01/A02/Z2 to G.K., G.S., and D.V.; GK1660 to G.K.; and FG 2886 “PANDORA” – B01/A03 to G.K. and G.S.), the Emerging Field Initiative (EFI) of Friedrich-Alexander University Erlangen-Nürnberg (FAU) (EFI_Verbund_Med_05_MIRACLE to G.K.), Bundesministerium für Bildung und Forschung (BMBF) (MASCARA to G.K. and G.S., MelAutim (FKZ:01ZX1905A) to G.K. and J.V.-G.), and the European Union (Horizon 2020 ERC-2014-StG 640087 – SOS and Horizon 2020 ERC-2020-CoG 101001866 to G.K. as well as Horizon 2020 ERC-2018-SyG nano-SCOPE and RTCure to G.S.). Volume imaging was performed on a Zeiss LSM 880 system, funded by DFG (German Research Foundation), project 261193037.
Footnotes
Author Contributions
M. Faas., N.I., J.A., S.C., and S.U. designed the study, performed experiments, interpreted results, and wrote the manuscript. A.G., F.S., J.H., C. Scholtysek, C. Stoll, and M.B. performed experiments, collected data, and interpreted results. B.W., S.W., D.V., R.L., and A.M. provided expertise, essential material, and input and wrote the manuscript. P.K., M.E., A.E., M. Fuchs., M.K., and J.V.-G. performed bioinformatics analysis and interpreted the data. D.M. and G.S. wrote the manuscript and provided valuable input. G.K. designed the study and experiments and wrote the manuscript. All authors read and commented on the manuscript.
Declaration of Interests
The authors declare no competing interests
References
- Allen JE, Sutherland TE. Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin Immunol. 2014;26:329–340. doi: 10.1016/j.smim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud B. Inflammation and skeletal muscle regeneration: leave it to the macrophages! Trends Immunol. 2020;41:481–492. doi: 10.1016/j.it.2020.04.006. [DOI] [PubMed] [Google Scholar]
- Coakley G, Harris NL. Interactions between macrophages and helminths. Parasite Immunol. 2020;42:e12717. doi: 10.1111/pim.12717. [DOI] [PubMed] [Google Scholar]
- Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–1674. doi: 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- Cohen M, Matcovitch O, David E, Barnett-Itzhaki Z, Keren-Shaul H, Blecher-Gonen R, Jaitin DA, Sica A, Amit I, Schwartz M. Chronic exposure to TGFβ1 regulates myeloid cell inflammatory response in an IRF7-dependent manner. EMBO J. 2014;33:2906–2921. doi: 10.15252/embj.201489293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culemann S, Grüneboom A, Nicolás-Ávila JÁ, Weidner D, Lämmle KF, Rothe T, Quintana JA, Kirchner P, Krljanac B, Eberhardt M, et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 2019;572:670–675. doi: 10.1038/s41586-019-1471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Baylies MK. Nuclear positioning in muscle development and disease. Front Physiol. 2013;4:363. doi: 10.3389/fphys.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis N, Sansbury BE, Patsalos A, Hays TT, Riley CO, Han X, Spite M, Nagy L. Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration. Nat Immunol. 2019;20:626–636. doi: 10.1038/s41590-019-0356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17:34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- He J, Yang Q, Xiao Q, Lei A, Li X, Zhou P, Liu T, Zhang L, Shi K, Yang Q, et al. IRF-7 is a critical regulator of type 2 innate lymphoid cells in allergic airway inflammation. Cell Rep. 2019;29:2718–2730.:e6. doi: 10.1016/j.celrep.2019.10.077. [DOI] [PubMed] [Google Scholar]
- Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, Chawla A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, Börnke F, Schmiedl A, Kleine T, Sonnewald U. Detecting functional groups of Arabidopsis mutants by metabolic profiling and evaluation of pleiotropic responses. Front Plant Sci. 2011;2:82. doi: 10.3389/fpls.2011.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Jones LH, Rückerl D, Svedberg F, Duncan S, Maizels RM, Sutherland TE, Jenkins SJ, McSorley HJ, Bénézech C, MacDonald AS, Allen JE. IL-33 delivery induces serous cavity macrophage proliferation independent of interleukin-4 receptor alpha. Eur J Immunol. 2016;46:2311–2321. doi: 10.1002/eji.201646442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178:686–698.:e14. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.:e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Knipper JA, Willenborg S, Brinckmann J, Bloch W, Maaß T, Wagener R, Krieg T, Sutherland T, Munitz A, Rothenberg ME, et al. Interleukin-4 receptor α signaling in myeloid cells controls collagen fibril assembly in skin repair. Immunity. 2015;43:803–816. doi: 10.1016/j.immuni.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps R package version 1.0.12. 2019. https://CRAN.R-project.org/package=pheatmap .
- Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, Pitman N, Mirchandani A, Rana B, van Rooijen N, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- Kuswanto W, Burzyn D, Panduro M, Wang KK, Jang YC, Wagers AJ, Benoist C, Mathis D. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44:355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz C, Radmanesh B, Liu E, Thorp EB, Lin J. Single-cell RNA sequencing uncovers heterogenous transcriptional signatures in macrophages during efferocytosis. Sci Rep. 2020;10:14333. doi: 10.1038/s41598-020-70353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Germain RN, Gerner MY. High-dimensional cell-level analysis of tissues with Ce3D multiplex volume imaging. Nat Protoc. 2019;14:1708–1733. doi: 10.1038/s41596-019-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew FY, Girard JP, Turnquist HR. Interleukin-33 in health and disease. Nat Rev Immunol. 2016;16:676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Pitha PM. Monocyte differentiation to macrophage requires interferon regulatory factor 7. J Biol Chem. 2001;276:45491–45496. doi: 10.1074/jbc.C100421200. [DOI] [PubMed] [Google Scholar]
- Lu Y, Basatemur G, Scott IC, Chiarugi D, Clement M, Harrison J, Jugdaohsingh R, Yu X, Newland SA, Jolin HE, et al. Interleukin-33 signaling controls the development of iron-recycling macrophages. Immunity. 2020;52:782–793.:e5. doi: 10.1016/j.immuni.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the power-houses of immunity. Nat Immunol. 2017;18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- Mills EL, O’Neill LA. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur J Immunol. 2016;46:13–21. doi: 10.1002/eji.201445427. [DOI] [PubMed] [Google Scholar]
- Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, Brioschi S, Bugatti M, Omodei AS, Ricci B, et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell. 2020;182:886–900.:e17. doi: 10.1016/j.cell.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP, O’Neill LAJ. Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell. 2018;174:780–784. doi: 10.1016/j.cell.2018.07.030. [DOI] [PubMed] [Google Scholar]
- Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair MG, Du Y, Perrigoue JG, Zaph C, Taylor JJ, Goldschmidt M, Swain GP, Yancopoulos GD, Valenzuela DM, Murphy A, et al. Alternatively activated macrophage-derived RELM-alpha is a negative regulator of type 2 inflammation in the lung. J Exp Med. 2009;206:937–952. doi: 10.1084/jem.20082048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Han CZ, Elliott MR, Kinchen JM, Trampont PC, Das S, Collins S, Lysiak JJ, Hoehn KL, Ravichandran KS. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature. 2011;477:220–224. doi: 10.1038/nature10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell CE, Zhang M, Fields PE, Klaassen CD. Th2 skewing by activation of Nrf2 in CD4(+) T cells. J Immunol. 2012;188:1630–1637. doi: 10.4049/jimmunol.1101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M, Mony JT, Løbner M, Khorooshi R, Owens T. Interferon regulatory factor-7 modulates experimental autoimmune encephalomyelitis in mice. J Neuroinflammation. 2011;8:181. doi: 10.1186/1742-2094-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Sluse FE, Jarmuszkiewicz W, Navet R, Douette P, Mathy G, Sluse-Goffart CM. Mitochondrial UCPs: new insights into regulation and impact. Biochim Biophys Acta. 2006;1757:480–485. doi: 10.1016/j.bbabio.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uderhardt S, Martins AJ, Tsang JS, Lämmermann T, Germain RN. Resident macrophages cloak tissue microlesions to prevent neutrophil-driven inflammatory damage. Cell. 2019;177:541–555.:e17. doi: 10.1016/j.cell.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga T, Mounier R, Horvath A, Cuvellier S, Dumont F, Poliska S, Ardjoune H, Juban G, Nagy L, Chazaud B. Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. J Immunol. 2016;196:4771–4782. doi: 10.4049/jimmunol.1502490. [DOI] [PubMed] [Google Scholar]
- Wan YY. GATA3: a master of many trades in immune regulation. Trends Immunol. 2014;35:233–242. doi: 10.1016/j.it.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- Yang M, Song L, Wang L, Yukht A, Ruther H, Li F, Qin M, Ghiasi H, Sharifi BG, Shah PK. Deficiency of GATA3-positive macrophages improves cardiac function following myocardial infarction or pressure overload hypertrophy. J Am Coll Cardiol. 2018;72:885–904. doi: 10.1016/j.jacc.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurdagul A, Jr, Subramanian M, Wang X, Crown SB, Ilkayeva OR, Darville L, Kolluru GK, Rymond CC, Gerlach BD, Zheng Z, et al. Macrophage metabolism of apoptotic cell-derived arginine promotes continual efferocytosis and resolution of injury. Cell Metab. 2020;31:518–533.:e10. doi: 10.1016/j.cmet.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell RNA-seq data as well as the bulk RNA-seq data generated during this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.