Abstract
Methylation of specific nucleotides is integral for ribosomal biogenesis and also serves as a common mechanism to confer antibiotic resistance by pathogenic bacteria. Here, by determining the high-resolution structure of the 30S−KsgA complex by cryo-electron microscopy, a state was captured, where KsgA juxtaposes between helices h44 and h45 of the 30S ribosome, separating them, thereby enabling remodeling of the surrounded rRNA and allowing the cognate site to enter the methylation pocket. With the structure as a guide, several mutant versions of the ribosomes, where interacting bases in the catalytic helix h45 and surrounding helices h44, h24, and h27, were mutated and evaluated for their methylation efficiency revealing factors that direct the enzyme to its cognate site with high fidelity. The biochemical studies show that the three-dimensional environment of the ribosome enables the interaction of select loop regions in KsgA with the ribosome helices paramount to maintain selectivity.
Introduction
Ribosomes, the central protein synthesis machinery, serve as a prime target for several antibiotics that bind to various positions within them, thereby stalling one of the many steps in translation.1–3 Naturally, pathogens have developed several resistance mechanisms to evade the inhibitory action of these drugs, leading to the evolution of drug resistance.4–7 One prevalent mechanism for antibiotic resistance is by the covalent modification of the ribosomal bases at specific positions that prevents the binding of these drugs. Acetylation and methylation of conserved bases within the ribosome by specific enzymes in pathogens are the most common mode for developing resistance.8–10
Strikingly, the pathogenic methylases such as erythromycin resistance methyltransferase (Erm) that induces resistance to erythromycin exclusively methylate at position A2058 [Escherichia coli (E. coli) numbering] in the 50S subunit and aminoglycoside resistance methyltransferase (NpmA and ArmA) that modifies A1408 and G1405, respectively, in the 30S subunit exhibit evolutionary similarity with the ribosomal methylases (rMTases) that are essential for biogenesis.11–15 Ribosomal MTases have evolved high specificity toward their substrate likely because they participate in the important process of ribosomal biogenesis. Perhaps, this specificity could be one of the primary reasons why methylation has also been chosen as the prevalent resistance modification by pathogens.15,16 For instance, KsgA, which is involved in ribosome biogenesis, dimethylates A1518 and A1519 in the 16S rRNA, near the decoding center within the 30S ribosomal subunit, exhibiting both structural and sequence similarity to the pathogenic Erm MTase. Both these enzymes harbor a conserved S-adenosine methionine (SAM) binding Rossmann fold and carry out N6 methylation of adenine (N6A) but with different substrate specificities.17,18 For example, Erm MTase is able to methylate short RNA stretches but unable to act on the assembled 50S subunit.19,20 In contrast, KsgA does not methylate short RNAs but acts only on the 30S subunit.21 It has been recently shown that a chimeric version of KsgA, where certain structural elements from Erm were introduced into the KsgA scaffold, altered the reaction profile of KsgA, allowing it to act on a short Erm substrate.22 Thus, the ability of rMTases to recognize their cognate substrates is encoded within the enzyme and how they achieve this is an interesting question that remains to be addressed.
In order to further understand the mechanism of substrate specificity and targeting and the molecular details of N6 adenosine methylation, KsgA was chosen as a target as the absence of KsgA-mediated methylation at position 1518 and 1519 in 16S rRNA entails aminoglycoside kasugamycin resistance in E. coli and other bacteria due to conformational rearrangement in the binding site of the antibiotic.23 Furthermore, KsgA deletion strains exhibit decoding errors during elongation and initiation from non-AUG codon.23,24 In addition, the ksgA knockout strains manifest cold-sensitive phenotype and other 16S rRNA-processing defects.25
A cryo-electron microscopy (EM) structure of 30S subunit in complex with E. coli KsgA was reported to 13 Å, and the high-resolution structure of the same complex was reported very recently.26,27 This structure captured the complex in a state where h45 is anchored into KsgA with one of the target bases A1519, and the terminal tetraloop base, G1516, being flipped-out interacting with the enzyme.27 In this structure, the adjacent ribosomal helix that undergoes substantial motion to allow KsgA to enter, h44, was disordered. Therefore, details as to how KsgA inserts itself onto the 30S platform remained elusive. Here, we have determined the cryo-EM structure of the Thermus thermophilus (Tt) immature 30S ribosomal subunit in complex with KsgA to an overall resolution of ~3.2 Å. Three-dimensional (3D) classification of the data allowed us to separate multiple conformers of the complex. Among these, one state is similar to the recently reported structure by Stephan et al., while another state with ordered h44 was also captured. The trapped second state enabled us to visualize the distortions and interactions in the ribosomal rRNA that enables helix h44 to position the catalytic helix h45 into the active site pocket of KsgA. Furthermore, to delve deeper into the mechanism of substrate selection as well as to discern the targeting determinants, using the structure as a guide, we designed multiple mutant versions of the 30S ribosomal rRNA and investigated their methylation potential. Methylation activity on these site-directed mutants revealed the importance of the interaction of select loop elements on KsgA with specific rRNA helices that are crucial for regulating enzymatic activity as well as targeting. Overall, the structural and biochemical data allow us to determine the general strategies adopted by rRNA N6 adenosine MTases (N6A-MTases) for accurate methylation and opens avenues for targeted inhibitor design.
Results
Cryo-EM Maps of KsgA and the Small Ribosomal Subunit
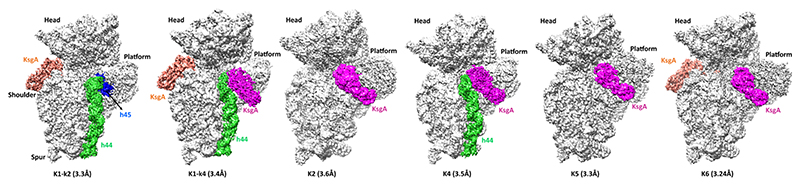
The 30S subunits were isolated from ΔksgA T. thermophilus (Tt) HB8 strain, and a complex with Bacillus subtilis (Bs) KsgA was prepared at low-magnesium concentration (details are given in Material and Methods) (Figure S1A). Using tritium-labeled SAM as a cofactor, scintillation assay was used to confirm that BsKsgA is also able to methylate the Tt 30S with equal efficiency as the E. coli ribosome (Figure S1B). The map of the KsgA−30S complex from the full data set showed heterogeneity (Figures 1 and S2), and subsequent 3D classification yielded five main classes (K1, K2, K4, K5, and K6) with KsgA bound to the ribosomes and the overall resolutions of the cryo-EM maps range between 3.17 and 3.6 Å (Figures S2− S4 and Table S1). Furthermore, based on the difference in the density of KsgA near h45 region, class K1 was subclassified into two classes K1-k2 and K1-k4. It was observed that classes K2, K5, and K6 have h44 in a disordered state with KsgA bound to the platform region of the 30S ribosome (Figures S2 and S5). Superposition with the recently published E. coli 30S−KsgA structure (PDB ID: 7O5H) reveals that the structure of the complex is analogous to that observed by Stephan et al. However, in classes K1-k4 and K4, we were able to observe clear density for the remodeled h44 helix along with the KsgA bound at the cognate site. In class K1-k4 and class K6, an additional KsgA monomer is bound to a secondary site near helix h16. The primer extension assays reveal that this KsgA is unable to modify the ribosome at the secondary site (Figure S6F,G). Biochemical and scintillation assay showed that KsgA is unable to methylate any other site apart from the A1518 and A1519 position (Figure S6E). Moreover, structural analysis reveals that KsgA is not positioned favorably for competent methylation (details are given in the Supporting Information) (Figure S6). We hence conclude that this second site is an artifact and binding at this site is due to the usage of 20-fold excess of KsgA during the preparation of the 30S−KsgA complex and is not a functionally relevant site (Figure S6). Class K4 and sub class K1-k4 with ordered h44 were chosen for all further analysis. Global analysis of the structure shows that KsgA is bound on the platform with its head docked over the negatively charged platform region formed by the interaction of h27, h24, h44, and h45 loops (Figure 2). For the structures reported in this study, density for the 30S head region is more pronounced in all the classes (Figures S3 and S4), and the comparison of the various populations shows that the head region exhibits heterogeneity and its swiveling movements are attributable to h28 (neck) flexibility (Figure S9).
Figure 1. Cryo-EM structure of the KsgA−30S complex.
The maps of cryo-EM classes reported in this study (sharpened map from the Relion postprocess procedure). Class K1-k2 has KsgA density near helix 16, and very weak density at the platform region indicates a potential initial scanning of protein on the 30S platform. In class K1-k4, KsgA density is present at the platform as well as near h16 of the 30S ribosome. Additionally, remodeled helix 44 is unique for this state, which enabled us to understand the mode of KsgA binding on the 30S platform region. Classes K2 and K5 show KsgA density at cognate position, similar to that reported by Stephan et al. Class K4 is similar to K1-k4 class, but KsgA is observed at only cognate position. Class K6 has an additional KsgA near h16 but otherwise similar to K2 and K5 classes. These classes also show swiveling motion in the 30S ribosomal subunit head.
Figure 2. Cryo-EM structure of the KsgA−30S complex.
(A) Cryo-EM reconstruction corresponding to K1-k4 class; KsgA (magenta) binds to the platform and is surrounded by rRNA: h45 (blue), h27 (orange), h24 (yellow), and the upper region (green) of h44. (B) Electrostatic surface potential map of KsgA colored by surface charge [calculated with adaptive Poisson−Boltzmann solver; the scale ranges from −5 kT/e (red) to 5 kT/e (blue)] in complex with h45 (dark blue), h44, h24, and h27. (C) Cryo-EM densities of h44 and h45 showing the arrangement of helices in the 30S−KsgA complex. (D) The panel shows the displacement of h44 compared to the unbound state (gray) vs bound (green) to accommodate the KsgA β6/7 linker and loop1 onto the 30S platform. KsgA is shown in magenta with highlighted loop1, and C-domain is shown in firebrick red. (E) In order to present the target residue (red) to KsgA, the displacement of h44 and h45 is depicted. The unbound 30S h44 and h45 helices are shown in gray (PDB ID: 3OTO), whereas the bound forms of h44 and h45 are shown in green and blue, respectively (numbering shown in bracket is for Tt). To clearly visualize the ribosomal structural rearrangement, the model of KsgA is not shown.
Role of h44 in the Recognition of KsgA
Helix 44 is one of the important RNA helices involved in decoding and harbors two bases at 1492 and 1493 (E. coli numbering) that monitors the correct interaction of codon−anticodon helix by stabilizing the first two Watson−Crick base pairs.28,29 In the h44 ordered structure (classes K1-k4 and K4), substantial conformational changes were observed upon the binding of KsgA with the 30S, which opens up the canonical engaged conformation of h44 and h45 to a conformation where they are separated by more than 20 Å with respect to the unmethylated immature ribosomal structure (Figure 2C). The flexibility of h44 in the immature 30S subunit is possibly one of the reasons for the inability to trap this state in the recently reported structure by Stephan et al.27 Similar flexibility in the h44−45 linker is also observed in the low-resolution cryo-EM structure of the eukaryotic pre-40S-assembly factor complex where the human homolog of KsgA, Dim1, was found bound as one of the proteins.30,31
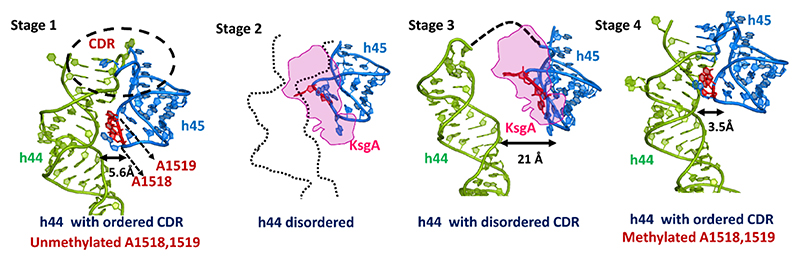
The structures of the 30S−KsgA complex (K1-k4 and K4) reveal that distortions in both h44 and h45 are introduced by the steric interaction of the N-terminal β6/7 linker of KsgA with the h44−45 linker, in order to present the target residue to the active pocket of the enzyme (Figure 2D,E). In this conformation, h44 has bent away from the 30S subunit platform, and as a result, the decoding site, importantly A1492/A1493 and C1054, moved away, thereby precluding the interaction with mRNA/tRNA duplex. Comparison of the unmethylated and methylated 30S subunits reveals that dimethylation at h45 by KsgA results in the closure of the gap between h45 and h44 that finally orders h44, maturing the 30S ribosomal subunit (Figure 3). Thus, before methylation, h44 is largely flexible and allows the entry of KsgA (Figure 3, stage 2). KsgA inserts itself between h44 and h45, separating these helices, and gets anchored onto the immature 30S through its C-terminal domain, which has several positively charged residues (Figures 2B, 3, stage 3, and S7). This enables the interaction of KsgA and positions the correct substrate, h45, into the catalytic center via flipping of G1516 into the exclusive pocket prepping the catalytic pocket for methylation by allowing the flip of A1518/A1519 (Figure 4A−C). In the final methylated form, both helices h44 and h45 come closer and are stabilized by both hydrogen bonding and van der Waals interactions, thereby sealing the interface that results in a structured h44 such that the biogenesis is complete and the ribosome is primed for further assembly (Figure 3, stage 4).
Figure 3.
Steps involved in KsgA methylation during biogenesis to achieve mature 30S. Stage 1 is the premethylated stage where h44 (green) and h45 are loosely packed. At stage 2, h44 adopts a flexible conformation (shown as dotted line) allowing the entry of KsgA (magenta) (similar to the state observed by Stephan et al.). This later rearranges in stage 3 to a conformation where h45 (marine blue) presents A1518 and A1519 (red) to the KsgA catalytic pocket for methylation. In stage 4, methylation leads to the dissociation of KsgA and orders the h44 and h45 conformation so as to achieve an active central decoding center. PDB ID: 3OTO (unmethylated A1518/19, stage 1) and 4B3R (methylated A1518/19, stage 4). Stage 2 (K5) and stage 3 (K1-k4) are experimentally observed as cryo-EM maps in this work.
Figure 4. Effect of mutagenesis of select bases in h45 and h44 of 30S present near the KsgA interaction region.
(A) Schematic diagram depicting the interaction of the GGAA tetraloop of h45 and KsgA from class K1-k4. (B) The active site of KsgA with key residues shown in stick representation pockets with flipped A1519 (red) (numbering shown in bracket is for Tt) into the catalytic pocket; carbon atoms of the interacting residue are shown in magenta and salmon and those of SAM are shown in yellow, while oxygen and nitrogen atoms are colored in red and blue, respectively. SAM is modeled by using PDB ID: 6IFT, not present in the 30S−KsgA complex structure. (C) Interaction of G1516 (blue) (numbering shown in bracket is for Tt) with KsgA via a pocket formed at the interface of its N- and C-terminal domain. Loop12, C-terminal domain residues are shown in salmon and N-terminal residues are shown in magenta. (D) Design of h45 and h44 mutants; KsgA target residues are highlighted in red, and residues encircled are subjected to mutagenesis. (E) In vitro KsgA methylation assay with constructs harboring mutation in the h45 tetraloop. (F) In vitro KsgA methylation assay with altered target site constructs of h44. Data for three experimental replicates are shown with the mean and standard deviation. Histogram colors are as per the scheme chosen for each 30S helix in Figure 1. Student’s t-tests were used to calculate P-values (**P < 0.01, ***P < 0.001) for the comparisons of 30S_WT (wild type) with the 30S mutant variant. Results show that methylation efficiency is significantly affected on perturbation of these important regions.
Mechanism of Substrate Fidelity of KsgA
Analyzing cryo-EM maps and models generated in this work as well as the previously reported structure by Stephan et al.27 reveals that for the productive methylation of the two adenines, KsgA positions itself by establishing extensive interactions with the 16S rRNA formed via the apposition of helices h27, h24, h45, and h44 (Figure 2B). Although these four helices are distantly placed in the secondary structure but upon folding and assembly in the ribosomes, these helices come close together to form a complex interface recognized by KsgA (Figure 2A,B). These rRNA helices surround KsgA, and this extensive binding mode clearly supports the rationale for the requirement of fully assembled immature 30S rather than a short fragment of RNA or 16SrRNA as a substrate. The conserved h45 “GGAA” tetraloop predominantly mediates the interaction of the ribosome with the N-terminal domain of KsgA by bending and inducing a 12 Å shift from its unbound form (Figure 2E), such that it positions A1519 into the active site cavity. The complexation results in the flipping out of A1519 (Figures 4A,B and S7), a conformation commonly observed in several MTases and necessary to facilitate catalysis.32–36
Comparison of the 30S apo structure with the 30S−KsgA complex (classes K1-k4 and K4) further shows that the entry of KsgA pushes h44 inward and its upper portion swings by ~9.4 Å to accommodate the linker that connects β6−β7 as well as loop1 and loop12 of KsgA (Figure 2D,E). This rearrangement is in accordance with previous reports from chemical probing studies, where this region of h44 is protected in the presence of KsgA.37
Thus, in order to present the conserved adenine to the MTase active site, displacement of both h44 and h45 from the canonical conformation occurs. Furthermore, to anchor itself to the ribosome, KsgA undergoes a substantial change in the SAM binding loop1 [root-mean-square deviation (rmsd) 5.0 Å, between 30 N-terminal amino acids], the interface loop12 (rmsd 1.3 Å, between 13 amino acids), as well as C-terminal domain (rmsd 2.48 Å, between 75 C-terminal amino acids). Several positively charged residues contribute to the interaction interface via contact with the stem loop of h45 (1513−1515) (Figure S10A). In addition, G1516 inserts into a shallow cleft that lies at the interface of the head and loop12 in KsgA (Figures 4A,C and S7). Comparison with the published structure by Stephan et al. reasserts that the G1516 region is indeed important for the positioning of the target RNA.27
To further understand the importance of base conservation in the stabilization of the tetraloop and its sensitivity toward perturbation to catalysis, we constructed several variant versions of the E. coli ribosome. The sequence alignment of KsgA-binding regions of E. coli 30S and Tt 30S (Figure S11) shows high conservation in the functionally relevant regions between the two species, and thus, the cryo-EM structures reported here can be used as a guide for functional analysis. As a positive control, we mutated A1518 and A1519 to uracil, and as expected, methylation activity was obliterated, and no incorporation of tritium-labeled SAM was observed (Figure 4E). Mutating the other important interacting tetraloop base G1516 to an alternate purine base, adenosine, resulted in 50% loss in activity (Figure 4D,E), iterating the importance of the correct base stabilization in the loop12-C-terminal cleft of KsgA. We also find that when the C-terminal domain of KsgA is deleted (ΔC-KsgA), the activity is reduced close to 50% of the native enzyme, which is similar to the G1516A mutant (Figure 5D). This is because in ΔC-KsgA, the cleft is not fully formed resulting in the incomplete stabilization of G1516.
Figure 5. Probing tertiary interactions of KsgA with h24 and h27.
(A) The highly basic loop14 of KsgA-targeting domain recognizes the juxtaposition of helix 27 and 24 through multiple interactions with the rRNA backbone (numbering shown in bracket is for Tt). (B) Loop1 interaction with the helix 24 (790-loop) phosphate backbone (numbering shown in bracket is for Tt). The density for the residues 773−779 is poor in all classes with KsgA bound and not modeled. In this figure, using the K1-k2 model, this region has been extrapolated to show potential interaction built on the K1-k4 map. (C) Design of h27 (GCAA, green, a conserved 900-loop) and h24 mutants. The residues that were mutated/inserted/deleted are highlighted in maroon and black for h27 and h24, respectively. (D) In vitro methylation assay using 3H-SAM with native and ΔC-KsgA. Histogram colors are as per the scheme chosen for each 30S helix in Figure 1. Data for three experimental replicates are shown with the mean and standard deviation. Student’s t-tests were used to calculate P-values (**P < 0.01, ***P < 0.001) for the comparisons of 30S_WT (wild type) with the 30S mutant variant.
The terminal tetraloop bases in RNA tetraloop-recognizing enzymes have been shown to be important in establishing the binding surface with the protein, and any mutation in these bases results in significant decrease in the binding affinity.38 The base G1516 is ~15 Å away from the main catalytic pocket and yet has a marked effect on the activity, signifying that a long-distance recognition element is crucial for the activity of the enzyme. When an adjacent base in the tetraloop, G1515, is mutated, either to adenine or cytosine, these ribosomal mutants exhibit 60 and 80% reduced activity, respectively (Figure 4D,E), strengthening the importance that the stem region adjacent to the catalytic tetraloop plays an important role not only in interacting with KsgA but also for the stabilization of the tetraloop conformation itself. The significant loss of activity observed for the G1515C mutant possibly results due to the disruption of the Watson−Crick base-pairing interactions at this site and results in the widening of the tetra loop that subsequently results in the disruption of the overall interactions of h45 with KsgA (Figure 4D,E). Note that in the Tt rRNA, the equivalent residue of G1515 is C1493 and it base pairs with G1498.
Mutation of the h44 Region and Shuffling of the Target Site
Helix h44 of 16S rRNA contacts KsgA via a distorted bulge (1485−1486), an interaction responsible for h44 to pivot toward KsgA. Thus, we asked if the ability of methylation is affected with the removal of the bulge. The bulge was replaced with a perfect Watson−Crick match at this position, and it was observed that the activity was reduced by 40% (Figure 4D,F), showing that the loss of anchoring at this position does affect methylation efficiency. Previous chemical probing experiments have shown that this area is protected in the presence of KsgA,37 which is not surprising as the structure shows that the region 1482−1497 of h44 indeed comes close to KsgA (Figure S10C) forming an interaction interface. We conclude that this region is important in providing the flexibility to h44 during methylation and also for its interaction with KsgA. The above observations raise the question if the tetraloop architecture is sufficient or the surrounding h44−h45 conformation is also essential for methylation. To address this, the tetraloop and stem region were stappled at other positions within the ribosome and methylation efficiency was gauged. Chimeric ribosomes where a portion of h44 was replaced by h45 such that the ribosome had two catalytic tetraloops were created, and they were found to be completely inactive (Figure 4D,F). In the absence of properly positioned h44 and h45, the ribosome is unable to support the proper binding of KsgA at either of the two catalytic centers. The 3D RNA composer predicts that the replacement of the h44 region by h45 completely melts the overall structure of h44, which then changes the orientation of h45 away from the canonical conformation resulting in a different 3D architecture (Figure S10D) that is not recognized by KsgA. The fact that this Shuffling (h44-LM) has resulted in the reorganization of the h44−h45 interface is supported by the nonviability of Δ7prrn strain as opposed to other minor variant ribosomes (Table S2). Another chimera was created where h45- and h44-interacting regions were swapped. However, this shuffled ribosome also showed no activity, suggesting that the order and appropriate placement of peripheral regions are paramount for the efficient functioning of KsgA. In both cases, production of ribosome was confirmed by the sucrose gradient in the MS2-tagged system, which harbors both plasmid-born tagged and intrinsic untagged ribosomes (see Material and Methods). In summary, the Shuffling studies strengthen the idea that KsgA recognizes the mini-tetraloop structure in a complex environment with correctly placed h44 and h45 and other surrounding helices to enable the catalytic conformation, explaining why it does not methylate mini-RNA templates.
Interaction of KsgA with Peripheral Helices h27 and h24
As mentioned earlier, KsgA interacts with other important helices such as the h27 switch and h24 that are present near the decoding site. These helices along with h44 rearrange during biogenesis to create a conducive environment for the binding of the initiation factors as well as 50S such that the catalytic machinery can be assembled for protein synthesis.39,40 Three loop structures of KsgA, loop1 (N-terminal loop), loop12 (peripheral loop at the opposite end of the N-terminal domain), and loop between α10 and α12 (loop 14) that lies in the C-terminal domain primarily interact with the peripheral rRNA. While loop12 holds both h45 and h27 on the front interface (Figure S10B), loop14 anchors h24 and h27 tethering them via a string of positively charged interactions (Figure 5A). Here, bases C899−A901 in h27 as well as bases G769, G770, and A802 in h24 juxtapose via a bridging interaction of loop14 residues R236−K237 (Figure 5A). Moreover, R271 from loop15 can form potential hydrogen bonds with both the helices further strengthening the interface (Figure 5A). The highly extended conserved basic surface of loop14 seems to impart specificity of recognition to KsgA for this site. While one end of helix h24 is tethered by loop14, it extends further and runs across the entire backside of KsgA making multiple contacts (Figure 2B). More importantly, h24 makes important hydrogen-bonding contacts with loop1. Loop1 acts as a lid; its opening and closing serves as an entry path for SAM. Here, bases A790 and G791 on h24 can form a potential hydrogen bond with residues K24 as well as the backbone of L26 and G27 on KsgA (Figure 5B). It has been reported earlier that the mutation of K24 results in a substantial loss of activity,22 and therefore, h24 plays an indirect role in controlling reactivity via interaction with loop1.
To access the importance of some of these peripheral interactions, multiple MS2-tagged variants were constructed where specific bases were either replaced or deleted. The aim was to disrupt the interactions that hold helices h27 and h24 to KsgA. A point mutation involving A901G in h27 was performed as it contacts both loop12 and loop14. We observed 70% activity (Figure 5C,D), hinting that a purine-to-purine exchange does not alter the conformation much as most of the contacts are via the phosphate backbone interactions. On the other hand, A900G/A901C mutation has only 40% activity as changes in these bases disturb the overall conformation of this region (Figure 5C,D). Shortening of helix h27 by creating a deletion at position 898−901 also results in 50% loss of activity (Figure 5C,D). We believe that the replacement of the bases in this region results in nonconducive interactions with the head, reiterating the importance of a C-terminal anchor point. In fact, a mutation that has an increased length via the insertion of a GC pair at the position between 901 and 902 results in the complete loss of activity for the full-length KsgA (Figure 5C,D). Lengthening of this region results in a direct steric clash with the C-terminal domain of KsgA and perhaps does not allow KsgA to position itself at its cognate position. The activity of the longer h27 is restored to 50% of the native when ΔC-KsgA was used for methylation reasserting that steric clash with the head domain was the reason for the loss of activity.
Mutations on h24 that run along the entire backside of KsgA were also engineered. Several mutations in order to shorten or lengthen h24 to disrupt individual contacts at the N-terminal region were made. Here, the most significant effect was on lengthening the helix h24 by two base pairs, where the methylation efficiency was reduced by 60% both with native and ΔC-KsgA (Figure 5C,D).
This is because the longer h24 again causes a steric clash with loop1 displacing it from its original position and thus likely to affect the entry and exit of SAM. A similar loss in the activity of about 60% was reported when G791 which contacts loop1 via K24 was disrupted.41 To investigate the importance of the flexibility of h24, the hinge region at position GGA775-777, observed below the 790 loop of h24, which probably imparts motion, was deleted and a shortened h24 G785Δ/C797Δ construct was created. It was observed that this shortened and rigid version of h24 again shows 30 and 60% of activity (Figure 5C,D), respectively. Since h24 anchors at both ends of KsgA, shortening of h24 induces strain in the helix and this shorter version is neither able to reach loop1 and help in its placement effectively nor is it able to properly engulf KsgA in this strained form. Corroborating this data, it was observed that in the ΔC-KsgA version where the C-terminal anchor point is deleted, methylation activity no longer depends on the perturbation of h24 length and is always reduced to 50% of the native. These data support the fact that the role of h24 and h27 helps in the proper positioning of the head of KsgA and enhances its methylation potential by serving as a peripheral anchor that prevents dislodging of KsgA from the ribosome.
Discussion
Insights into the Mechanism of Methylation
Riboso-mal N6A-MTases whether from pathogenic organisms or those involved in biogenesis display high fidelity and only methylate the cognate bases.14,42 Thus, it appears that they have been programmed to accurately recognize the correct sequence and this information is embedded in their structure. Comparison of the Rossmann fold harboring N6A-MTases reveals that the substrate enters perpendicular to the body of the enzyme and inserts itself into the catalytic pocket by inducing a base flip (Figure S12). The flipping of the methylating base is a common feature observed in several other MTases including N6A-DNA MTases and seems to be a prerequisite for the proper binding of the substrate.43–45 In all these cases, the flipped base is stabilized in the catalytic site via the π−π interaction of the adenine moiety with tyrosine or a phenylalanine residue (Figure S13D). One of the questions that remains a matter of debate is how do these enzymes enable dimethylation? In each round, a fresh SAM has to enter to execute the reaction; therefore, the entry of SAM and exit of SAH require a substantial motion of the protein or the RNA architecture. Furthermore, in the case of KsgA, successive bases are methylated, thus a shift in the base that is to be methylated has to also occur.
Analysis of the conformation of the tetraloop in the unmethylated versus protein bound and the postmethylated versions points to the hypothesis that it is the malleability of the tetraloop structure and the overall backbone conformations that determine the manner in which the bases are presented to the active site. In the presence of the protein, the mix of hydrophobic and electrostatic interactions alters the conformation of the tetraloop by a rotation of approximately 90° (Figure 6A), inducing significant distortions in the backbone such that it fits into the active site pocket. In this new conformation, both bases G1516 and A1519 flip out and occupy tailored protein pockets. Surprisingly, the h45 tetraloop adopts a similar conformation in the human processome46 which indicates that this tetraloop region can exist in multiple conformations. Other studies have also shown that tetraloop structures are malleable for instance, the ribosomal sarcin tetraloop, also significantly changes conformation in the presence of the protein.38 Thus, this malleability allows the 3D structure of the tetraloop to vacillate between the various partially stacked states as is seen in KsgA and TFB1M complexes. Based on the cryo-EM structure, Stephan et al. proposed a mechanism of successive base flipping for A519 and A1518 at the target site. They proposed that after A1519 gets dimethylated, A1518 also flips out for methylation. However, considering the steric requirements, we propose that tandem methylation can also be facilitated via local flipping motions concentrated around the bases which undergo methylation. Therefore, in order to facilitate consecutive methylation after A1519 gets dimethylated, we propose it flips inward, which induces A1518 to flip out and get methylated. This hypothesis is supported by the NMR structure of the tetraloop, where the analogous base position to A1519 is methylated and the state we propose is captured experimentally.
Figure 6. Tetraloop conformational flexibility.
(A) An overlay of the h45 tetraloop bound to KsgA (marine blue) and the unbound state (green) (PDB ID: 3OTO) of the Tt ribosome showing the conformational differences. (B) Conformational difference of the h45 tetraloop in the bound state with KsgA (marine blue) and TFB1M (yellow) (PDB ID: 6AAX). (C) Overlay of the Tt h45 tetraloop (marine blue) in the bound state with BsKsgA and the Ec h45 tetraloop (salmon) with EcKsgA (PDB ID: 7O5H) (numbering shown in bracket is for Tt). (D) Proposed mechanism of dimethylation. Tetraloop conformational change induces A1519 flips into the active pocket. Dimethylation of A1519 results in the further local flipping of the adjacent A1518 base. Finally, dimethylation of A1518 dissociates the enzyme from 30S and h45 resumes its native conformation as observed in the mature 30S ribosomal subunit.
In this structure, Liu et al. show that the methylated h45 tetraloop segment after the methylation of A14 (A1519) is flipped in, whereas (A13) A1518 is flipped out.47 After dimethylation is complete, we further suggest that the methylation of A1518 triggers the release of the RNA from the protein, purporting a reorganization of the tetraloop resulting in a conformation where both the methylated bases are flipped out such that they engage closely with h44 (Figure 6B).
Stephan et al. using their recent structure of the E. coli KsgA− 30S complex as a guide also hypothesized that within the tetraloop, G1516 acts as an anchor. Our structure also corroborates this observation, and the insertion of G1516 in the positively charged pocket formed within KsgA can be visualized (Figures 4B, S7, and S8). Mutagenesis of this region asserts that the flipping-in of G1516 into KsgA is important for methylation and this base likely serves as a selectivity checks to allow for the correct RNA to bind the protein. It was also observed that KsgA is tightly anchored near the methylation site via the extensive interaction of its C-terminal region with the surrounding RNA helices h24 and h27, and mutagenesis of this region also affected the methylation potential of KsgA. Thus, C-terminal along with G1516 insertion within KsgA likely serves as a pivot that allows the h45 tetraloop stretch to slightly reconfigure after each methylation, thereby enabling successive methylations without the enzyme leaving the vicinity of the rRNA. The importance of C-terminal in imparting methylation efficiency is reasserted as KsgA shows only 50% activity in the absence of its head domain. Moreover, it has been reported that the headless Erm37 from Mycobacterium tuberculosis shows a mixed methylation profile, and in addition, it is error prone, likely due to insufficient anchoring.48 Sequence analysis confirms that the C-terminal of ribosomal-methylating N6A-MTases is not conserved across these enzymes. Since it appears to be important for anchoring, we propose that anchoring via this region could be a common mechanism across N6A-MTases to facilitate site-specific methylation.
Selectivity of the Cognate Substrate via Divergent Loop Elements
Comparison of various ribosomal N6A-MTases shows that both loop1 and loop12 vary in both sequence and size across these enzymes. An earlier study demonstrated that KsgA can be altered to methylate Erm substrates by swapping loop1 and loop12. In an in vitro setting, chimera’s which consisted of loop1 and loop12 of Erm onto an otherwise KsgA scaffold made it capable of methylating Erm mini-RNA substrates.22 Furthermore, saturation mutagenesis of loop12 also confirmed that positively charged residues in loop12 of Erm are important for substrate recognition.49 Thus, it appears that these important loop elements are crucial in tunning specificity. The 30S−KsgA cryo-EM structure shows that loop 12 and another loop14 that resides in the C-terminal head domain serving as anchor points via interaction with the peripheral RNA segments provide an additional layer of stringency to the recognition process. To explain how these loops are tailored to maintain specificity, Erm was modeled in place of KsgA in the 30S complex. As shown in Supporting Information, Figure S13B,C, due to short loops 12 and 14, in Erm, its binding in the ribosomal 30S region, where KsgA docks, is not favorable, and as a result, it is unable to effectively interact and stabilize h45 into its catalytic center. Moreover, the structure of the 30S−KsgA complex also reveals that loop12 is the region which along with the C-terminal forms the exclusive pocket where G1516 flips in. A similar flip is also observed in the TFB1M RNA protein complex where the anchoring base G934 is also approximately 15 Å from the methylating base A93747 (Figure S12B). This peripheral base flipping seems to be a common targeting mechanism in other RNA N6A-MTases, for instance, in the mRNA-recognizing protein METTL16, multiple base flips are observed to seal the interaction interface (Figure S12B). It is also observed that in METTL16 where mRNA is the primary substrate, loop12 extends into the RNA and holds it at multiple positions highlighting the evolutionary diversity of this loop segment50 (Figure S12A).
The other region that plays an important role is loop1. In KsgA, this loop interacts with h24 and the interaction of h24 via K24 and G791 has been shown to be a very important anchor point to maintain catalytic activity. Loop1 is found to be variable across N6A-MTases; for instance, loop1 is much shorter in ErmC′ than in KsgA (Figure 7C). It appears that N6A-MTase accesses particular rRNA stretch via tailor-made loops that harbor specificity determinants and these loops act as tweezers, holding the rRNA so that a catalytic-competent conformation is enabled.
Figure 7. Targeting determinant of methyltransferase.
(A) Comparison of longer loop12 and loop14 of KsgA (firebrick red) with shorter respective loops of ErmC′ (green, PDB ID: 1QAM), interacting with the h45 stem region and (B) other 30S platform helices h27 and h24. (C) Similarly, longer loop1 of KsgA (firebrick red) than ErmC′ (green) interacts with h24. Respective sequence differences of KsgA and ErmC′ loops are highlighted in the inset.
In conclusion, the cryo-EM structures of KsgA in complex with 30S ribosome along with the mutagenesis studies performed here shed light on both the mechanism of targeting and recognition adopted by N6 Rossmann fold rMTases to achieve specificity. The study asserts that the binding of KsgA onto the 30S platform is stabilized by interaction with several rRNA helices, which play an important role in tuning its methylation potential. It was established that specificity elements in KsgA such as tailored loops and the C-terminal head domain together act as a robust anchor and allow the enzyme to properly latch onto the ribosome. Furthermore, the two successive states, where the 30S−KsgA complex harbors h44 in either a disordered or in an ordered configuration, aid in capturing events that lead to the final step of biogenesis. These findings enhance our overall understanding as to how N6A-rMTases select their target substrates, a knowledge that can be further exploited for the design of selective inhibitors, tailored to inhibit a particular N6A-rMTase.
Supplementary Material
Acknowledgments
The authors acknowledge the National CryoEM Facility for EM data collection and the computing facility for data processing at the Bangalore Life Science Cluster and Scintillation facility at IIT Bombay. We thank G. Culver (University of Rochester) and R. Green (Johns Hopkins University) for sharing strains and plasmid. J.S. thanks University Grants Commission (UGC) and IIT Bombay for PhD fellowship.
Funding
R.A. acknowledges DBT/Wellcome Trust-India Alliance Fellowship [grant number IA/S/19/1/504293] and STARS [grant number MoE/STARS-1/APR2019/BS/523/FS]. K.R.V. acknowledges SERB, India, for the Ramanujan Fellowship (RJN-094/2017), Department of Atomic Energy, Government of India, under project identification no. RTI4006 and the DBT B-Life grant DBT/PR12422/MED/31/287/2014, which supports the National cryoEM Facility.
Footnotes
Author Contributions
J.S. performed all the experiments including sample preparation, biochemical assay, and data analysis under the supervision of R.A. J.S., R.R., and K.R.V. performed cryo-EM work. All authors have contributed equally in the manuscript preparation. J.S. and R.R. are considered the first author.
Notes
The authors declare no competing financial interest.
Contributor Information
Juhi Singh, Department of Chemistry, Indian Institute of Technology Bombay, Mumbai 400076, India.
Rahul Raina, National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bengaluru 560065, India.
References
- (1).Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- (2).Arenz S, Wilson DN. Blast from the Past: Reassessing Forgotten Translation Inhibitors, Antibiotic Selectivity, and Resistance Mechanisms to Aid Drug Development. Mol Cell. 2016;61:3–14. doi: 10.1016/j.molcel.2015.10.019. [DOI] [PubMed] [Google Scholar]
- (3).Kavčič B, Tkačik G, Bollenbach T. Mechanisms of drug interactions between translation-inhibiting antibiotics. Nat Commun. 2020;11:4013. doi: 10.1038/s41467-020-17734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- (5).Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- (6).Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- (7).Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Schaenzer AJ, Wright GD. Antibiotic Resistance by Enzymatic Modification of Antibiotic Targets. Trends Mol Med. 2020;26:768–782. doi: 10.1016/j.molmed.2020.05.001. [DOI] [PubMed] [Google Scholar]
- (9).Ahmed S, Sony SA, Chowdhury MB, Ullah MM, Paul S, Hossain T. Retention of antibiotic activity against resistant bacteria harbouring aminoglycoside-N-acetyltransferase enzyme by adjuvants: a combination of in-silico and in-vitro study. Sci Rep. 2020;10:19381. doi: 10.1038/s41598-020-76355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Luthra S, Rominski A, Sander P. The Role of Antibiotic-Target-Modifying and Antibiotic-Modifying Enzymes in Mycobacterium abscessus Drug Resistance. Front Microbiol. 2018;9:2179. doi: 10.3389/fmicb.2018.02179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Svetlov MS, Syroegin EA, Aleksandrova EV, Atkinson GC, Gregory ST, Mankin AS, Polikanov YS. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat Chem Biol. 2021;17:412–420. doi: 10.1038/s41589-020-00715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Husain N, Obranić S, Koscinski L, Seetharaman J, Babić F, Bujnicki JM, Maravić-Vlahoviček G, Sivaraman J. Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit. Nucleic Acids Res. 2011;39:1903–1918. doi: 10.1093/nar/gkq1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Husain N, Tkaczuk KL, Shenoy RT, Kaminska KH, Čubrilo S, Maravić-Vlahoviček G, Bujnicki JM, Sivaraman J. Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m 7 G methyltransferases. Nucleic Acids Res. 2010;38:4120–4132. doi: 10.1093/nar/gkq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sergiev PV, Aleksashin NA, Chugunova AA, Polikanov YS, Dontsova OA. Structural and evolutionary insights into ribosomal RNA methylation. Nat Chem Biol. 2018;14:226–235. doi: 10.1038/nchembio.2569. [DOI] [PubMed] [Google Scholar]
- (15).Davis JH, Williamson JR. Structure and dynamics of bacterial ribosome biogenesis. Philos Trans R Soc London, Ser B. 2017;372:20160181. doi: 10.1098/rstb.2016.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Park AK, Kim H, Jin HJ. Phylogenetic analysis of rRNA methyltransferases, Erm and KsgA, as related to antibiotic resistance. FEMS Microbiol Lett. 2010;309:151–162. doi: 10.1111/j.1574-6968.2010.02031.x. [DOI] [PubMed] [Google Scholar]
- (17).O’Farrell HC, Scarsdale JN, Rife JP. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J Mol Biol. 2004;339:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- (18).Bussiere DE, Muchmore SW, Dealwis CG, Schluckebier G, Nienaber VL, Edalji RP, Walter KA, Ladror US, Holzman TF, Abad-Zapatero C. Crystal Structure of ErmC’, an rRNA Methyltransferase Which Mediates Antibiotic Resistance in Bacteria. Biochemistry. 1998;37:7103–7112. doi: 10.1021/bi973113c. [DOI] [PubMed] [Google Scholar]
- (19).Champney WS, Chittum HS, Tober CL. A 50S ribosomal subunit precursor particle is a substrate for the ErmC methyltransferase in Staphylococcus aureus cells. Curr Microbiol. 2003;46:453–460. doi: 10.1007/s00284-002-3901-8. [DOI] [PubMed] [Google Scholar]
- (20).Hansen LH, Lobedanz S, Douthwaite S, Arar K, Wengel J, Kirpekar F, Vester B. Minimal substrate features for Erm methyltransferases defined by using a combinatorial oligonucleotide library. Chembiochem. 2011;12:610–614. doi: 10.1002/cbic.201000606. [DOI] [PubMed] [Google Scholar]
- (21).Desai PM, Rife JP. The adenosine dimethyltransferase KsgA recognizes a specific conformational state of the 30S ribosomal subunit. Arch Biochem Biophys. 2006;449:57–63. doi: 10.1016/j.abb.2006.02.028. [DOI] [PubMed] [Google Scholar]
- (22).Bhujbalrao R, Anand R. Deciphering Determinants in Ribosomal Methyltransferases That Confer Antimicrobial Resistance. J Am Chem Soc. 2019;141:1425–1429. doi: 10.1021/jacs.8b10277. [DOI] [PubMed] [Google Scholar]
- (23).Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzu M, et al. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol. 2006;13:871–878. doi: 10.1038/nsmb1145. [DOI] [PubMed] [Google Scholar]
- (24).Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JHD, Vila-Sanjurjo A. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol. 2006;13:879–886. doi: 10.1038/nsmb1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Connolly K, Rife JP, Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Boehringer D, O’Farrell HC, Rife JP, Ban N. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem. 2012;287:10453–10459. doi: 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Stephan NC, Ries AB, Boehringer D, Ban N. Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA. Nucleic Acids Res. 2021;49:6389–6398. doi: 10.1093/nar/gkab430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- (29).Svidritskiy E, Korostelev AA. Ribosome Structure Reveals Preservation of Active Sites in the Presence of a P-Site Wobble Mismatch. Structure. 2015;23:2155–2161. doi: 10.1016/j.str.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Johnson MC, Ghalei H, Doxtader KA, Karbstein K, Stroupe ME. Structural Heterogeneity in Pre-40S Ribosomes. Structure. 2017;25:329–340. doi: 10.1016/j.str.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Scaiola A, Peña C, Weisser M, Böhringer D, Leibundgut M, Klingauf-Nerurkar P, Gerhardy S, Panse VG, Ban N. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 2018;37:e98499. doi: 10.15252/embj.201798499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Liu R-J, Long T, Li J, Li H, Wang E-D. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017;45:6684–6697. doi: 10.1093/nar/gkx473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lin C-C, Chen Y-P, Yang W-Z, Shen JCK, Yuan HS. Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B. Nucleic Acids Res. 2020;48:3949–3961. doi: 10.1093/nar/gkaa111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- (35).Jiang Y, Li F, Wu J, Shi Y, Gong Q. Structural insights into substrate selectivity of ribosomal RNA methyltransferase RlmCD. PLoS One. 2017;12:e0185226. doi: 10.1371/journal.pone.0185226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ma B, Ma J, Liu D, Guo L, Chen H, Ding J, Liu W, Zhang H. Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori. Oncotarget. 2016;7:40965–40977. doi: 10.18632/oncotarget.9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Xu Z, O’Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- (38).Thapar R, Denmon AP, Nikonowicz EP. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip Rev: RNA. 2014;5:49–67. doi: 10.1002/wrna.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- (40).Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Desai PM, Culver GM, Rife JP. Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly. Biochemistry. 2011;50:854–863. doi: 10.1021/bi101005r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Pletnev P, Guseva E, Zanina A, Evfratov S, Dzama M, Treshin V, Pogorel’skaya A, Osterman I, Golovina A, et al. Comprehensive Functional Analysis of Escherichia coli Ribosomal RNA Methyltransferases. Front Genet. 2020;11:97. doi: 10.3389/fgene.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wilson KA, Kellie JL, Wetmore SD. DNA-protein π-interactions in nature: abundance, structure, composition and strength of contacts between aromatic amino acids and DNA nucleobases or deoxyribose sugar. Nucleic Acids Res. 2014;42:6726–6741. doi: 10.1093/nar/gku269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J Mol Biol. 2006;358:559–570. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat Struct Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- (46).Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S. Nucleolar maturation of the human small subunit processome. Science. 2021;373:eabj5338. doi: 10.1126/science.abj5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Liu X, Shen S, Wu P, Li F, Liu X, Wang C, Gong Q, Wu J, Yao X, Zhang H, et al. Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function. Nucleic Acids Res. 2019;47:7648–7665. doi: 10.1093/nar/gkz505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Madsen CT, Jakobsen L, Buriánková K, Doucet-Populaire F, Pernodet J-L, Douthwaite S. Methyltransferase Erm(37) slips on rRNA to confer atypical resistance in Mycobacterium tuberculosis. J Biol Chem. 2005;280:38942–38947. doi: 10.1074/jbc.M505727200. [DOI] [PubMed] [Google Scholar]
- (49).Rowe SJ, Mecaskey RJ, Nasef M, Talton RC, Sharkey RE, Halliday JC, Dunkle JA. Shared requirements for key residues in the antibiotic resistance enzymes ErmC and ErmE suggest a common mode of RNA recognition. J Biol Chem. 2020;295:17476–17485. doi: 10.1074/jbc.RA120.014280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y. Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol Cell. 2018;71:1001–1011.:e4. doi: 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.