Abstract
Blockade of epidermal growth factor receptor (EGFR) causes tumor regression in some patients with metastatic colorectal cancer (mCRC). However, residual disease reservoirs typically remain even after maximal response to therapy, leading to relapse. Using patient-derived xenografts (PDXs), we observed that mCRC cells surviving EGFR inhibition exhibited gene expression patterns similar to those of a quiescent subpopulation of normal intestinal secretory precursors with Paneth-cell characteristics. Compared with untreated tumors, these pseudodifferentiated tumor remnants had reduced expression of genes encoding EGFR-activating ligands, enhanced activity of human epidermal growth factor 2 (HER2) and human epidermal growth factor receptor 3 (HER3), and persistent signaling along the phosphatidylinositol-3-kinase (PI3K) pathway. Clinically, properties of residual disease cells from the PDX models were detected in lingering tumors of responsive patients and in tumors of individuals who had experienced early recurrence. Mechanistically, residual tumor reprogramming after EGFR neutralization was mediated by inactivation of Yes-associated protein (YAP), a master regulator of intestinal epithelium recovery from injury. In preclinical trials, pan-HER antibodies minimized residual disease, blunted PI3K signaling, and induced long-term tumor control after treatment discontinuation. We found that tolerance to EGFR inhibition is characterized by inactivation of an intrinsic lineage program that drives both regenerative signaling during intestinal repair and EGFR-dependent tumorigenesis. Thus, our results shed light on CRC lineage plasticity as an adaptive escape mechanism from EGFR-targeted therapy and suggest opportunities to preemptively target residual disease.
Introduction
Although many advanced-stage solid cancers regress when treated with therapies that target drivers of tumor growth, responses are usually incomplete, owing to the presence of residual cells that withstand initial treatment (1). Clinical studies have documented a correlation between depth of response to anticancer therapies and progression-free survival (PFS) (2–4), supporting the notion that the pool of cancer cells that persist with therapy can be responsible for therapy-resistant tumor progression.
The mechanisms that sustain residual tumor burden remain unclear. In cell line-based experiments, exposure to potentially lethal growth-inhibitory cues (such as anti-cancer agents) prompts the emergence of a drug-tolerant state, in which drug-resistant subpopulations arise de novo from a drug-sensitive population (5,6). These ‘drug-tolerant persisters’ (5) occasionally divide and fail to undergo cell death under drug pressure. Moreover, they resume vigorous proliferation when therapy is withdrawn and regain drug sensitivity after a washout period, indicating a reversible phenotype that stochastically appears and disappears in a bulk of heterogeneous cells. The pool of drug-tolerant cancer cells that survive initial therapy can also evolve subclones with more stable, genetically determined mechanisms of acquired resistance. For example, after prolonged drug exposure, non-small cell lung cancer (NSCLC) cells with mutant epidermal growth factor receptor (EGFR) and that tolerate EGFR blockade give rise to newly emerging subpopulations with a gatekeeper resistance mutation (6,7).
More clinically relevant attempts to understand how these principles from the in vitro studies apply to residual disease in human tumors are limited (8–10). Hurdles to using tumor material from patients include the difficulty in collecting and analyzing residual cells at maximal tumor response, the paucity of clinical specimens from treated patients, and a dearth of experimental models that accurately recapitulate fundamental aspects of residual disease and recurrence (1). We addressed this knowledge gap with a focus on metastatic colorectal cancer (mCRC), a tumor setting for which the clinical challenge of residual disease is particularly important. Monoclonal antibodies that inhibit EGFR (cetuximab and panitumumab) achieve clinically meaningful rates of response in chemorefractory patients with inoperable mCRC tumors (11–15). Several laboratories, including ours, have identified genetic alterations that are associated with and causally responsible for innate resistance to EGFR blockade in mCRC (16–20). Although omitting anti-EGFR therapy in mCRC patients with these resistance-associated genotypes has refined the identification of sensitive cases, complete regressions remain exceptions. Consequently, even responsive patients have a suboptimal outlook, with only 2-month improvement in PFS and 6-month improvement in overall survival compared with chemotherapy (15). Thus, understanding the molecular bases of drug tolerance in EGFR-inhibited mCRC is key to limiting residual disease and delaying the development of resistance.
Here, we used mCRC patient-derived xenografts (PDXs), analyzed at maximal response to cetuximab, to disentangle the phenotypic and functional ramifications of EGFR inhibition in drug-tolerant tumor cells. We found that EGFR blockade in CRC (i) disrupts a feed-forward homeostatic circuit that maintains tumor reliance on EGFR signals by co-opting pro-regenerative programs active in the normal intestine after injury and (ii) unleashes pseudodifferentiation towards an EGFR-independent, Paneth cell-like phenotype with alternate therapeutic vulnerabilities. This study illustrates the importance of lineage-based adaptive reprogramming as a means for cancer cells to evade pathway dependency and eradication by therapy.
Results
Residual colorectal tumors after EGFR blockade are made of slowly cycling cells that resemble EGFR-inhibited normal intestinal cells
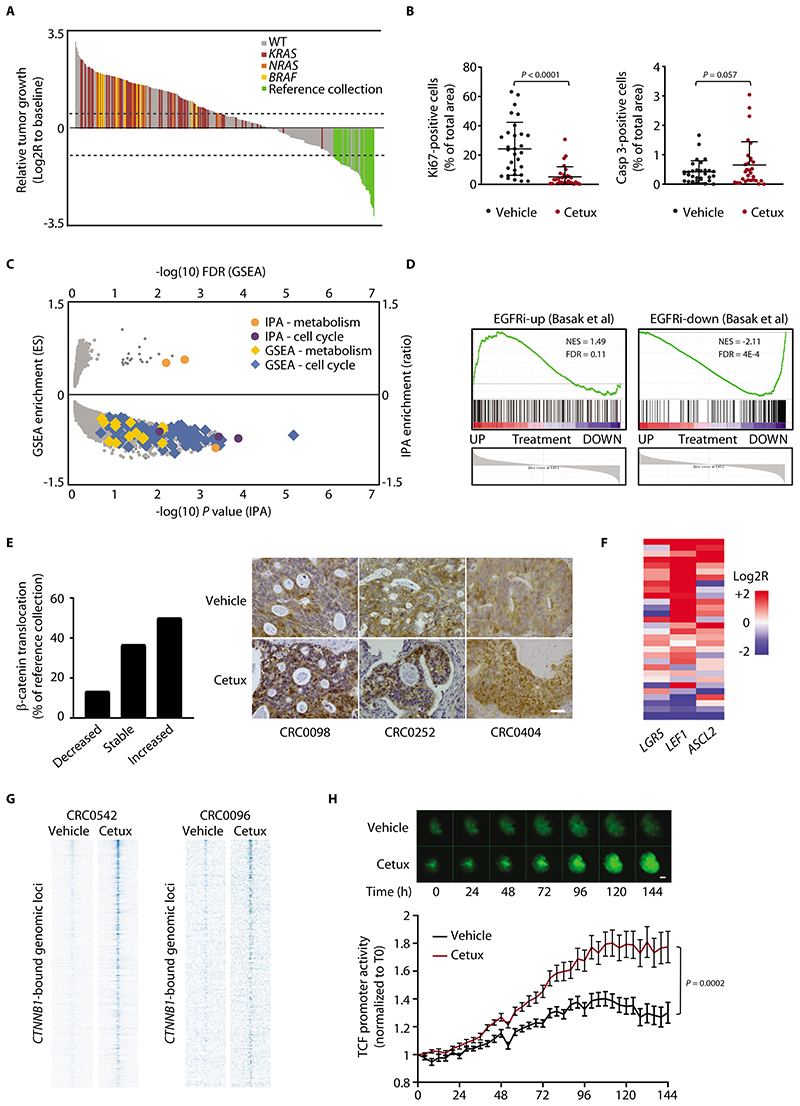
For population-level assessment of residual disease after cetuximab therapy, we used a set of 279 mCRC PDXs, derived from as many patients, part of which had been used in previous independent studies (16,20,21). In 40 cases (14.3%), tumor regression (designated as at least 50% mean tumor volume reduction compared with pre-treatment volumes) suitable for analysis of residual disease occurred (Fig. 1A). Consistent with observations in patients (15), all these responsive cases were wildtype for clinically validated resistance mutations (KRAS, NRAS, and BRAF) (Fig. 1A) and for additional alterations that are known to confer resistance or attenuate response (16–21) but have not yet entered routine determination.
Figure 1. Residual tumors after EGFR blockade are made of slowly cycling cells with high Wnt signaling.
(A) Waterfall plot of response after 3 weeks of treatment with cetuximab (20 mg/kg, intraperitoneal injection twice a week), compared with tumor volume at baseline, in a population of 279 PDX models (n = 6 or 12 mice for each bar, depending on whether initial engraftment was successful in one or two mice). Dotted lines indicate the cut-off values for arbitrarily defined categories of therapy response (16): regression (below the lower line, -50%), progressive disease (above the upper line, +35%), and stabilization (between the lines). WT represents cases with no mutations in KRAS, NRAS, or BRAF. Reference collection represents responder cases selected for characterization of residual disease (all wild-type for KRAS, NRAS, and BRAF). (B) Morphometric quantification of proliferation (left panel) and apoptosis (right panel) in PDXs from the reference collection after treatment with vehicle (until tumors reached an average volume of 1500 mm3) or cetuximab (for 6 weeks). Each dot represents the average of 10 optical fields (Ki67, 40X) or 5 optical fields (caspase 3, 20X) in a section from randomly chosen tumors from vehicle-treated and cetuximab-treated mice bearing a PDX from the same original patient (n = 30). The plots show means ± SD. Statistical analysis by two-tailed paired Student’s t-test. Casp, caspase. Cetux, cetuximab. (C) Scatter plot showing GSEA and IPA results of differential gene expression profiles obtained by comparing PDXs treated with cetuximab with their matched vehicle-treated controls (GSE108277). Enrichments are plotted on the y axes, and their significance is plotted on the x axis. Gene sets related to cell cycle and metabolism are shown by the indicated colors. ES, enrichment score. (D) GSEA plots showing modulation of the EGFR inhibition signature in PDXs treated with cetuximab (GSE108277). EGFR inhibition signature was defined from LGR5+ normal intestinal cells (24). EGFRi-up, genes upregulated by EGFR inhibition; EGFRi-down, genes downregulated by EGFR inhibition; NES, normalized enrichment score; FDR, false discovery rate. (E) (Left) Qualitative morphometric assessment of the extent of nuclear β-catenin translocation in PDXs from the reference collection after treatment with cetuximab for 6 weeks, compared with vehicle-treated counterparts. (Right) Representative images in PDXs from the reference collection after treatment with vehicle or cetuximab. Scale bar, 50 μm. (F) Heatmap showing expression changes for the indicated Wnt target genes in PDXs of the reference collection after treatment with cetuximab for 6 weeks. Average gene expression, Log2R relative to vehicle-treated tumors: LGR5 0.46, P = 0.029; LEF1 1.98, P < 0.0001; ASCL2 0.61, P = 0.011 by two-tailed Wilcoxon test. Benjamini-Hochberg FDR < 0.1 for all genes. (G) Heat maps of ChIP-seq signal in the indicated tumors and experimental conditions at CTNNB1 (β-catenin)-enriched genomic loci +/-4kb to the peak center. (H) Longitudinal time-lapse monitoring of β-catenin transcriptional activity in CRC0078 colospheres transduced with a TOP-GFP lentiviral construct containing a TCF/LEF-responsive promoter. PuraMatrix-embedded colospheres were treated with vehicle or cetuximab (20 μg/ml) in EGF-deprived medium (EGF = 0.4 ng/ml) for 6 days. Upper panel, representative images (scale bar, 50 μm); lower panel, morphometric quantification of GFP fluorescence. Values are the means ± SEM from one experiment (n = 18 for vehicle-treated colospheres and 20 for cetuximab-treated colospheres). Statistical analysis by two-way ANOVA.
Among the 40 responsive PDXs, we conducted systematic studies for specific biomarkers on 30 cases (hereafter referred to as the ‘reference collection’) (Fig. 1A). We compared cells from untreated tumors in the reference collection with residual disease cells from matched tumors from animals treated for 6 weeks. The residual disease cells from the treated tumors exhibited widespread reduction (but not complete abrogation) of cell proliferation and a slight increase in apoptosis (Fig. 1B). Longitudinal evaluation for 12 weeks in 2 cases (CRC0078 and CRC0096) revealed a drastic drop in cell proliferation immediately after therapy initiation, with later resumption of low but continuous mitotic activity beginning 1 week (CRC0078) or 3 weeks (CRC0096) after treatment onset (fig. S1A). The reduction in cell proliferation after prolonged treatment with cetuximab was confirmed by reduction of 5-ethynyl-2’-deoxyuridine (EdU) incorporation in both PDXs and PDX-derived organoids (cancer cell assemblies embedded in an extracellular matrix gel) (fig. S1B). The number of apoptotic cells fluctuated only marginally over time, without an apparent trend (fig. S1A). Finding tumor shrinkage that was accompanied by reduced cancer cell proliferation without pronounced apoptosis suggested that cetuximab produced a new equilibrium on the spontaneous growth and death dynamics of the tumormass, shifting the net balance between cell production and cell loss to one that favored basal tumor attrition without increasing the number of apoptotic cells (22).
We also monitored the response to a long-term stop-and-go schedule of cetuximab in 1 case. Antibody administration in mice with exponentially growing tumors resulted in a rapid (4 weeks) and massive (approximately 80%) plateau of tumor regression but failed to achieve tumor eradication (fig. S1C). Therapy was maintained for additional 17 weeks and then suspended. Upon treatment withdrawal, and following a latency of 5 weeks, tumors started regrowing with kinetics and phenotypes similar to treatment-naïve counterparts and were sensitive to antibody rechallenge (fig. S1C).
We explored whether the residual cells that persist at maximal response originate from genetic selection or stochastic plasticity. Genetic selection of defined pre-existing subclones would produce a population enriched in high-frequency somatic mutations (fig. S2A). Conversely, a lack of substantial modifications in the frequency distribution of sequence variants would be compatible with functional plasticity (fig. S2B). Whole-exome sequencing (WES) in 2 models of the reference collection did not reveal an enrichment for somatic mutations either in cetuximab-treated samples or in tumors that had relapsed following drug withdrawal, when compared to untreated tumors (fig. S2, C and D). Because our survey was restricted to protein-coding genes, we cannot exclude therapy-induced changes in the mutational profile of DNA regulatory regions. However, our results suggested that cetuximab pressure causes residual cancer cells to undergo plastic phenotypic changes in the absence of considerable genetic selection.
We also investigated whether rare subclones with known cetuximab-resistance mutations emerged during prolonged antibody treatment in 5 PDX models. Sequenom genotyping for hotspot mutations in 9 genes and Nanostring evaluation of copy number alterations in 11 genes documented a wild-type euploid or nearly euploid status for established resistance-associated genotypes in residual tumors after therapy (data file S1). A hitherto unrecognized Y1021S mutation in the PIK3CA gene, with no demonstrated connection to cetuximab resistance, was present at a 10% allele frequency in 1 of 2 post-cetuximab samples obtained from 1 model. Collectively, these results indicate that residual disease in PDXs has the identifying characteristics of drug tolerance, as observed in cultured cell lines (5): smoldering growth and minimal apoptosis under drug pressure; a metastable state of reversible sensitivity to therapy; and no acquisition or selection of resistance-conferring genetic alterations.
To better understand the molecular features of residual disease, we performed microarray-based gene expression analysis in 18 cetuximab-sensitive PDX models (including 12 models from the reference collection) and extracted paired information on vehicle-treated versus cetuximab-treated tumors (GSE108277). Combined gene set enrichment analysis (GSEA) and Ingenuity Pathway Analysis (IPA) confirmed the observed reduction of cell proliferation in treated tumors, with no evident signs of apoptosis, and revealed a metabolic switch from glycolysis to lipid metabolism and oxidative phosphorylation (Fig. 1C and data files S2 and S3). This conforms to findings in pancreatic cancer, inwhich reliance on mitochondrial respiration and impaired glycolytic function have been described as hallmarks of a subpopulation of quiescent tumor cells that survive extinction of KRAS oncogenic signals and are eventually responsible for tumor relapse (23).
Normal murine intestinal stem cells undergo a state of reversible quiescence after EGFR inhibition by the small-molecule gefitinib (24), similar to the growth disadvantage observed in cetuximab-treated tumors. Accordingly, the transcriptional rewiring triggered by EGFR blockade in tumors echoed that of EGFR-inhibited normal stem cells (24), with a substantial overlap between the set of genes modulated by cetuximab in human tumors and those modulated by gefitinib in normal murine intestinal stem cells (Fig. 1D). Similarities also occurred at the functional level. EGFR-inhibited quiescent stem cells of the normal mouse intestine display enhanced Wnt/β-catenin signaling compared to rapidly cycling cells with active EGFR signaling (24). Likewise, nuclear localization of β-catenin (Fig. 1E) and expression of the canonical Wnt target genes LGR5, LEF1, and ASCL2 (Fig. 1F), all indicative of Wnt activation, were generally higher in residual tumors than in vehicle-treated controls of the reference collection. Stronger Wnt signaling in drug-tolerant cells was confirmed by ChIP-seq analysis in 2 representative models, in which prolonged cetuximab treatment potentiated β-catenin binding to DNA targets (Fig. 1G and fig. S3). Similarly, TCF/LEF reporter assays in PDX-derived colospheres (cancer cell spheroids cultured in suspension) revealed increased β-catenin-dependent transcriptional activity over time after EGFR inhibition (Fig. 1H). Time-course experiments in the 2 PDX models treated with cetuximab for 12 weeks revealed an increase in the number of cells with higher abundance and stronger nuclear staining of β-catenin starting 1 week (CRC0078) or 2 weeks (CRC0096) after treatment initiation and persisting until therapy was terminated (fig. S4).
Cancer cells that withstand EGFR inhibition are similar to secretory precursors expressing markers of the Paneth-cell lineage
The gene expression profile induced by EGFR blockade in normal intestinal stem cells shares traits with the profile of slowly cycling, DNA-label-retaining cells (LRCs) (24), a reserve population that accounts for tissue regeneration after damage (25). In agreement with the observation that cetuximab-tolerant cancer cells resembled rarely dividing cells in the normal tissue, we observed upregulation of 3 typical LRC markers (BMI1, HOPX, LRIG1) (26) in treated tumors of the reference collection (Fig. 2A). However, TERT, another LRC marker (26), was downregulated in EGFR-inhibited tumors (Fig. 2A), consistent with reports that EGFR blockade decreases TERT expression (27).
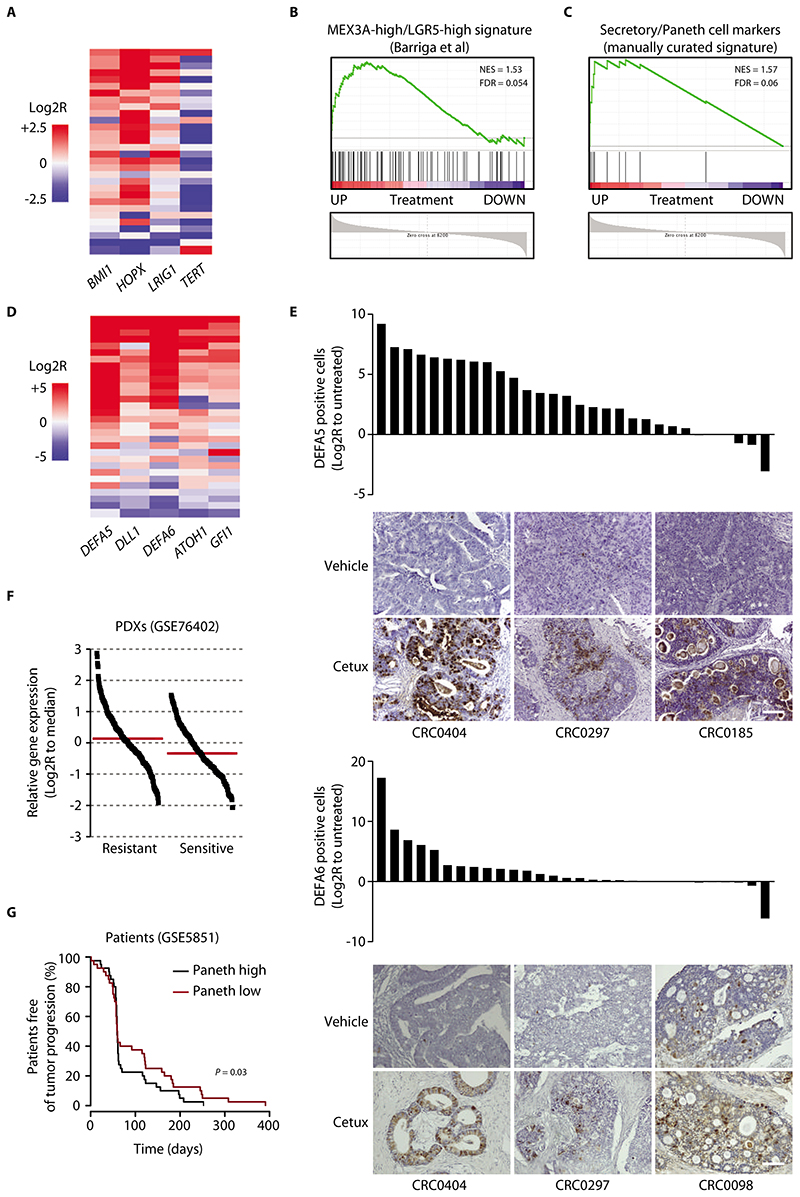
Figure 2. Cancer cells that withstand EGFR inhibition express markers of the Paneth cell lineage.
(A) Heatmap showing expression changes for the indicated LRC markers in PDXs of the reference collection after treatment with cetuximab (20 mg/kg, intraperitoneal injection twice a week for 6 weeks). Average gene expression, Log2R relative to vehicle-treated tumors: BMI1 0.57, P = 0.0002; HOPX 1.41, P < 0.0001; LRIG1 0.42, P = 0.006; TERT -1.41, P = 0.0071 by two-tailed Wilcoxon test. Benjamini-Hochberg FDR < 0.1 for all genes. (B) GSEA plot showing positive modulation of the MEX3A-high/LGR5-high signature (29) in PDXs treated with cetuximab (GSE108277). NES, normalized enrichment score; FDR, false discovery rate. (C) GSEA plot showing upregulation of a manually curated subset of secretory/Paneth cell genes in PDXs treated with cetuximab (GSE108277). (D) Heatmap showing expression changes for the indicated secretory/Paneth-cell markers in PDXs of the reference collection after treatment with cetuximab. Average gene expression, Log2R relative to vehicle-treated tumors: DEFA5 4.03, P < 0.0001; DLL1 1.12, P = 0.002; ATOH1 1.26, P < 0.0001; DEFA6 2.95, P = 0.0001; GFI1 1.04, P = 0.0006 by two-tailed Wilcoxon test. Benjamini-Hochberg FDR < 0.1 for all genes. (E) Morphometric quantification of DEFA5 and DEFA6 protein abundance changes in PDXs of the reference collection after treatment with cetuximab. Each bar represents the change of average protein abundance in 5 optical fields (20X) in a section from randomly chosen tumors from cetuximab-treated mice compared with a tumor from matched vehicle-treated mice (n = 5 for each bar). Representative images are also shown. P = 0.028 for DEFA5 and 0.0131 for DEFA6 by two-tailed paired Student’s t-test. Scale bar, 100 μm. Cetux, cetuximab. (F) Expression of secretory/Paneth cell markers in a dataset of mCRC PDXs annotated for response to cetuximab (GSE76402). Each dot represents the average expression of the secretory/Paneth cell signature metagene (ATOH1, GFI1, SOX9, XBP1, DEFA5, DEFA6, LYZ, SPINK4, DLL1, DLL4) in individual tumors. Log2R relative to the dataset median (red line). P < 0.0001 by two-tailed unpaired Student’s t-test. (G) PFS of patients with CRC metastases treated with cetuximab monotherapy (34), divided into two groups using the median signal intensity of the secretory/Paneth cell signature metagene as a cutoff (n = 40 patients with high signature and 40 patients with low signature). Statistical analysis by Cox regression.
LRCs are secretory precursors that are committed to mature into Paneth cells; thus, LRCs express some genes of the Paneth-cell lineage (28). Accordingly, analysis of our global transcriptomic data revealed that residual PDXs were enriched for a signature previously identified in mouse MEX3A-high/LGR5-high cells (Fig. 2B), which are a subpopulation of slowly dividing intestinal stem cells with increased Wnt signaling and expression of Paneth cell genes (29) (Fig. 2B). The gene expression-profile of residual tumors was also enriched for a signature of the Paneth cells of the mouse small intestine (30) but was different from that of Reg4+ deep crypt secretory cells, which serve as Paneth cell equivalents in the murine colon (31) (fig. S5). Therefore, we derived a manually curated gene subset that includes established markers of classical Paneth-cell commitment and differentiation— in particular, transcription factors that specify the secretory fate of intestinal progenitors (ATOH1, GFI1, SOX9, XBP1) and markers of Paneth-cell terminal differentiation (DEFA5, DEFA6, LYZ, SPINK4, DLL1, DLL4) (referred to as the ‘secretory/Paneth-cell signature’) (32). By GSEA, we found that this signature was upregulated in cetuximab-treated PDXs (Fig. 2C). For 5 genes with strong induction by cetuximab (‘core signature’) (data file S4), results were validated by RT-qPCR in the reference collection (Fig. 2D). Finally, the top-ranked markers defensin-5 (DEFA5) and defensin-6 (DEFA6) in residual tumors were substantiated as more abundant at the protein level, with a 50-fold increase for DEFA5 and an 8-fold increase for DEFA6 in the average percentage of protein positivity relative to that in untreated tumors (Fig. 2E). DEFA5 positivity in treated tumors was associated with β-catenin positivity (fig. S6), compatible with the gene expression data showing the co-occurrence of high Wnt signaling and Paneth cell-like pseudodifferentiation in residual tumors.
To analyze the temporal dynamics of Paneth cell-like phenotypic rewiring, we used 2 different observation time frames and treatment schedules. First, we analyzed the expression of the secretory/Paneth cell core signature genes over time in the 2 models in which cetuximab was administered for 12 weeks. Like the increase in β-catenin abundance (fig. S4), many of the core signature genes were upregulated soon after the first dose of cetuximab and continued to be more expressed than in control tumors during the whole period of antibody administration (fig. S7A). At the protein level, the percentage of DEFA6 positivity increased by 3 fold after 2 weeks of treatment and remained higher at subsequent time points (fig. S7B). Second, we assessed the expression of the core signature genes at an initial time point (after 3 days of treatment) in the whole reference collection and after cetuximab suspension (when regrown tumors had reached volumes of around 750 mm3, typically several weeks after therapy interruption) in 5 representative cases. Overall, the core signature gene transcripts were upregulated as early as 3 days after the first antibody dose (fig. S7C) – although for some transcripts the extent of gene modulation was lower than that detected in long-treated tumors – and most of them reverted after antibody withdrawal (fig. S7D). Hence, we found that cetuximab induces reversible changes that are rapidly acquired after therapy onset, persist in residual disease under continued drug pressure, and dissipate after therapy discontinuation.
Principal component analysis of global RNA-seq data from untreated, cetuximab-treated, and regrown samples in 1 PDX model confirmed that tumors that had relapsed after therapy discontinuation showed a transcriptomic profile indistinguishable from that of their untreated counterparts but radically different from that of on-treatment samples (fig. S7E). This result supports the hypothesis that thephenotypic rewiring that occurs in residual tumors is driven by cellular functional plasticity rather than selection of pre-existing resistant cells.
Analysis of the baseline (pre-treatment) gene expression profiles of our set of therapeutically annotated PDXs (n = 241; GSE76402) (33) revealed higher expression of the secretory/Paneth-cell signature genes in tumors with intrinsic resistance to cetuximab than in sensitive cases (Fig. 2F). Using a publicly available gene expression dataset comprising CRC metastases from 80 individuals treated with cetuximab monotherapy (GSE5851) (34), we found that patients bearing tumors with high expression of the signature genes had shorter PFS than patients bearing tumors with low signature expression (Fig. 2G). Overall, data in PDXs and in patients suggested that secretory/Paneth-cell like traits may be general hallmarks of tolerance to EGFR blockade not only in cancer cell persisters after adaptation to antibody treatment but also in tumors that are intrinsically poorly sensitive at treatment outset.
Acquisition of the Paneth cell-like phenotype in residual cells is regulated by inactivation of YAP
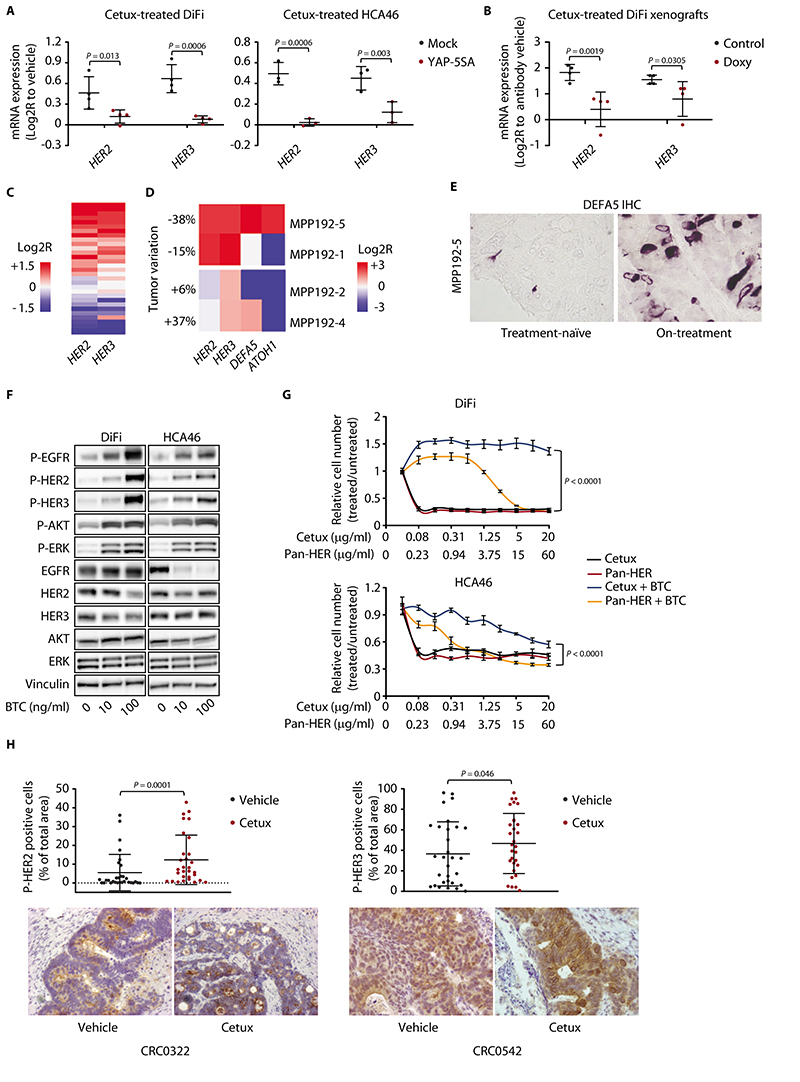
In mice, YAP, a downstream transcriptional effector of the Hippo pathway, promotes intestinal regeneration after injury by enhancing EGFR pathway activity and transiently suppressing Wnt signaling and Paneth cell differentiation of stem cells (35). From the phenotype of high Wnt/β-catenin signaling and Paneth cell-like properties displayed by drug-tolerant tumors, we hypothesized that residual disease could be sustained by reduced activation of YAP in EGFR-inhibited cancer cells. We thus compared the gene expression profiles of residual mCRC PDXs after cetuximab treatment with those of mice with YAP conditional deletion in the gut (35). The genes with the greatest extent of transcriptional regulation in YAP-knockout mice were highly correlated with the genes most strongly modulated by cetuximab in PDXs (Fig. 3A). Furthermore, a signature of YAP-inducible target genes (36) was downmodulated in cetuximab-treated tumors (Fig. 3B). In accordance with a phenotype of signal deactivation, the abundance or nuclear accumulation of YAP, or both, was reduced in most residual tumors of the reference collection (Fig. 3C). Time-course analysis of 2 PDXs from the reference collection indicated that the reduction in YAP protein, along with transcriptional downregulation of YAP target genes, was detectable within one week after cetuximab administration and was maintained for several weeks until the end of the experiment (fig. S8, A and B). After therapy suspension, YAP immunoreactivity increased to amounts similar to those in samples prior to treatment (fig. S8C).
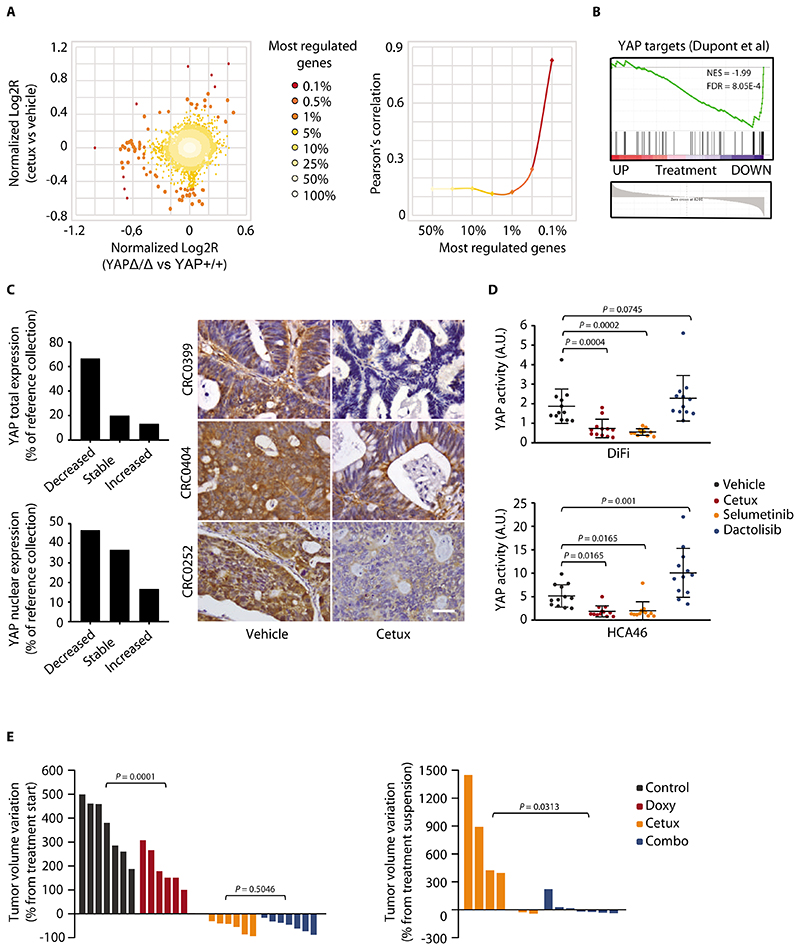
Figure 3. Cetuximab inactivates YAP.
(A) Correlation between gene expression changes induced by cetuximab (20 mg/kg, intraperitoneal injection twice a week for 6 weeks) in CRC PDXs and those induced by YAP knockout (YAPΔ/Δ) in the normal mouse intestine (35). Scatter plot of gene expression changes (left) and Pearson’s correlationof differentially regulated genes (right) are shown. Cetux, cetuximab. (B) GSEA plot showing the regulation of YAP-inducible target genes (36) in PDXs from mice treated with cetuximab. NES, normalized enrichment score; FDR, false discovery rate. (C) Graphs show qualitative morphometric assessment of YAP total and nuclear abundance in PDXs from the reference collection after treatment with cetuximab, compared with vehicle-treated counterparts. Representative images of YAP staining in PDXs from the reference collection after treatment with vehicle (until tumors reached an average volume of 1500 mm3) or cetuximab (for 6 weeks) are shown. Scale bar, 50 μm. (D) Measurement of YAP transcriptional activity in DiFi and HCA46 cell lines after treatment for 48h with vehicle (DMSO), cetuximab (20 μg/ml), selumetinib (1 μM), or dactolisib (250 nM). Luciferase activity was normalized against reporter plasmid concentrations as determined by DNA qPCR and expressed as arbitrary units (A.U.). Three independent experiments were performed in biological quadruplicates (n = 12). The plots show means ± SD. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. (E) Effect of doxycycline-regulated YAP activation, with or without concomitant EGFR inhibition, in established DiFi xenografts. (Left) Tumor volume changes after 3 weeks of treatment with cetuximab (20 mg/kg, intraperitoneal injection twice a week), doxycycline (50 mg/kg, daily oral gavage), or both. (Right) Tumor volume changes after 4 weeks from treatment suspension. n = 7 animals per each treatment arm. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction (left) or two-tailed Wilcoxon test (right). Control, doxycycline vehicle + cetuximab vehicle; Doxy, doxycycline; Combo, doxycycline + cetuximab.
We confirmed the inactivation of YAP by EGFR inhibition in cetuximab-sensitive CRC cell lines (DiFi and HCA46). EGFR blockade diminished YAP activity, as indicated by reduced YAP-dependent transcriptional activity (Fig. 3D). Cetuximab treatment also appeared to increase the phosphorylation of two inhibitory serine residues involved in YAP cytoplasmic retention and degradation (fig. S9A) andreduced the expression of established YAP target genes (fig. S9B). YAP negatively regulates Wnt signaling (35), and we found that Wnt target genes were upregulated after RNAi-mediated YAP knockdown (fig. S10) and were downregulated after transduction of a constitutively active form of YAP (YAP-5SA) or wild-type YAP (fig. S11).
Among the upstream signals that activate YAP, the mitogen-activated protein kinase (MAPK) cascade and the PI3K/AKT axis play prominent roles (37–39). Because these pathways are typically triggered by EGFR (40), we investigated the contribution of either pathway to cetuximab-induced YAP inhibition. MAPK kinase (MEK) blockade by selumetinib or trametinib was as effective as EGFR inhibition in abating YAP reporter activity (Fig. 3D and fig. S12). However, this effect was not recapitulated by PI3K pathway inhibition by dactolisib (targeting all PI3K isoforms and mTOR), buparlisib (targeting all PI3K isoforms), or alpelisib (targeting PI3Kα) (Fig. 3D and fig. S12). Similarly, the modulation of established YAP target genes was similar after EGFR (cetuximab) or MEK (selumetinib) inhibition, whereas it was less profound or more heterogeneous, or both, after PI3K pathway inhibition with dactolisib (fig. S9B). Collectively, these results indicated that cetuximab mainly affects the MAPK pathway to mediate YAP inactivation.
To explore whether cetuximab-dependent YAP inhibition has a role in cancer cell persistence, we simulated a therapeutic setting in established cell-line xenografts. We analyzed tumor growth 3 weeks after doxycycline-induced expression of constitutively active YAP-5SA (fig. S13, A and B) with or without administration of cetuximab. After 3 weeks, DiFi tumors with induced expression of active YAP were smaller than the tumors from non-induced (YAP-5SA-negative) controls (Fig. 3E, left). Treatment with cetuximab alone or cetuximab and doxycycline caused tumor regression (Fig. 3E, left). Moreover, 4 weeks after treatment withdrawal, tumors from mice treated with cetuximab only showed a rebound in growth; whereas those from mice treated with cetuximab and induced with doxycycline had impaired growth resumption (Fig. 3E, right). Growth retardation in xenografts expressing YAP-5SA suggested that YAP hyperactivation decreases the fitness of EGFR-dependent cells. In drug-tolerant cells surviving prolonged treatment with cetuximab, this reduction in fitness translates into weakened restoration of tumor growth following therapy discontinuation. Thus, we hypothesized that YAP inhibition enhances the viability of EGFR-inhibited cells, resulting in cancer cell persistence and residual disease.
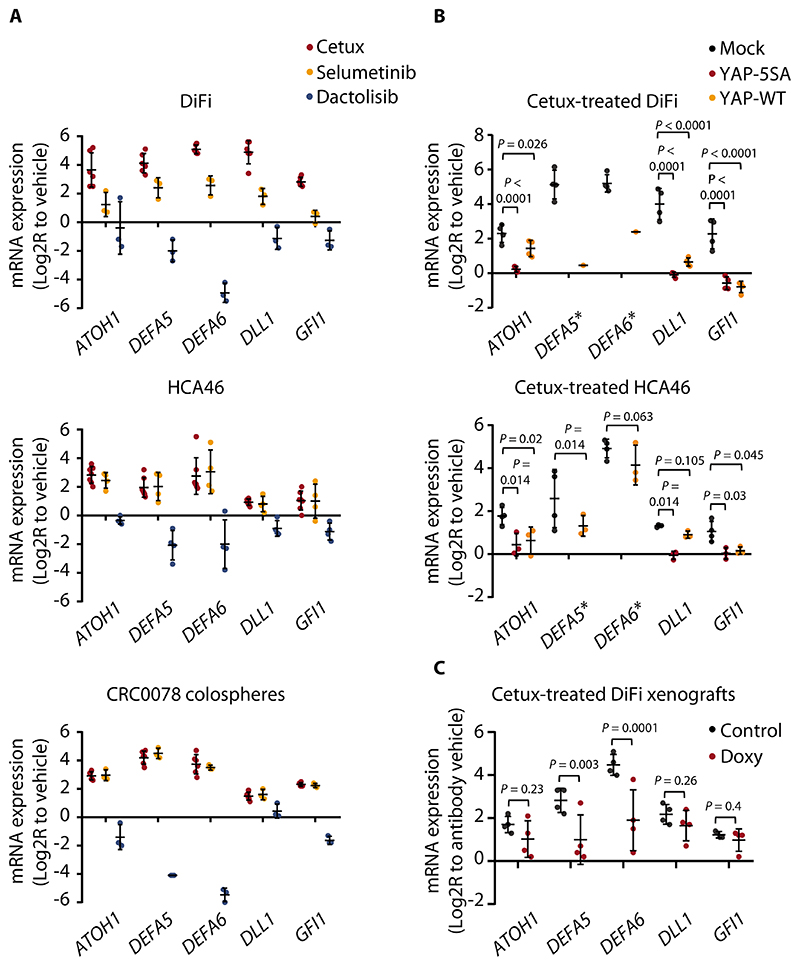
To causally link EGFR blockade, YAP inhibition, and acquisition of the Paneth cell-like phenotype, we modulated EGFR pathway and YAP activity in cell lines and patient-derived models. We confirmed increased expression of the core signature secretory/Paneth cell genes after EGFR inhibition in cell lines and colospheres, as observed in PDXs (fig. S13C). Consistent with the effect of the pathway inhibitors in the YAP reporter assay (Fig. 3D), upregulation of the secretory/Paneth cell genes was induced by EGFR inhibition or MEK inhibition, but not by agents targeting the PI3K pathway, in cultured cells, colospheres, organoids, and xenografts (Fig. 4A and fig. S13, D and E). This indicates thatcetuximab-regulated YAP inactivation and induction of the Paneth cell-like phenotype are both primarily mediated by inhibition of the MAPK cascade.
Figure 4. Acquisition of the Paneth cell-like phenotype in residual cells involves YAP inactivation.
(A) RT-qPCR analysis of the indicated secretory/Paneth-cell markers in DiFi and HCA46 cells and in CRC0078 colospheres treated with cetuximab (20 μg/ml), selumetinib (1 μM), or dactolisib (250 nM) for 72h. Independent experiments (n = 3 for DiFi and CRC0078; n = 4 for HCA46) were performed in technical triplicates. In some experiments, controls (vehicle) with untreated cells were repeated. The plots show means ± SD. The effects of cetuximab and selumetinib were positively correlated (DiFi, r = 0.857; HCA46, r = 0.963; CRC0078, r = 0.984, Pearson’s correlation coefficient), whereas dactolisib-induced transcriptional changes tended to negatively correlate with those triggered by cetuximab (DiFi, r = -0.592; HCA46, r = -0.104; CRC0078, r = -0.905). Cetux, cetuximab. (B) RT-qPCR analysis of the indicated secretory/Paneth-cell genes in DiFi and HCA46 cell lines transduced with a pLVX-control vector (mock), the constitutively active YAP variant (YAP-5SA), or wild-type YAP (YAP-WT) and treated with cetuximab (20 μg/ml) for 72h. Independent experiments (n = 3 for HCA46 and n = 4 for DiFi) were performed in technical triplicates. In some experiments, controls (vehicle) with untreated cells were repeated. The plots show means ± SD. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. Stars next to DEFA5 and DEFA6 indicate loss ofbasal expression in the YAP-5SA experimental group, which prevented statistical comparison against mock controls for these genes. In the DiFi YAP-WT group, DEFA5 and DEFA6 expression was above detection in one replicate. (C) RT-qPCR analysis of the indicated secretory/Paneth-cell genes in established DiFi xenografts transduced with doxycycline-inducible YAP-5SA and treated with vehicle (control), cetuximab (20 mg/kg, intraperitoneal injection, 1 administration), doxycycline (Doxy, 50 mg/kg, daily oral gavage), or the combination of cetuximab and doxycycline for 1 week. Results from animals treated with cetuximab were normalized against those from animals treated with vehicle. Results from animals treated with cetuximab and doxycycline were normalized against those from animals treated with doxycycline. Four samples for each condition were analyzed in technical triplicates. The plots show means ± SD. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction.
We hypothesized that YAP suppresses Paneth cell pseudodifferentiation in CRC cells. Indeed, secretory/Paneth cell genes were upregulated after RNAi-mediated silencing of YAP in CRC cell lines (fig. S14A). Either overexpression of constitutively active YAP-5SA or wild-type YAP caused a reduction in the basal expression of some secretory/Paneth cell genes (fig. S14B). Enforced YAP-5SA expression prevented or impaired cetuximab-induced upregulation of secretory/Paneth cell markers (Fig. 4B). Indeed, YAP-5SA suppressed DEFA5 and DEFA6 so profoundly that even in the presence of cetuximab, their expression was undetectable. Overexpression of wild-type YAP, which is regulated by upstream signals, was not as effective as YAP-5SA in preventing cetuximab-induced acquisition of the Paneth cell-like phenotype (Fig. 4B). Similar to in vitro data, established DiFi xenografts had increased expression of secretory/Paneth cell genes after cetuximab treatment and decreased expression of the same genes after doxycycline-regulated expression of YAP-5SA (Fig. 4C and fig. S14C). Moreover, conditional YAP-5SA induction in xenografts attenuated cetuximab-dependent upregulation of the secretory/Paneth cell markers, again with a particularly pronounced effect for DEFA5 and DEFA6 (Fig. 4C). Together, these findings supported a causal role for YAP inhibition in mediating the lineage rewiring triggered by EGFR blockade.
The pro-regenerative program driven by YAP in the normal intestine entails not only suppression of Wnt signaling and Paneth cell differentiation but also transcriptional induction of EGFR ligands. These induced ligands are amphiregulin (encoded by AREG), epiregulin (EREG), and heparin-binding EGF (HBEGF) (35). Another ligand of the EGFR family is betacellulin (encoded by BTC), which functions as a HER3 ligand and activates the HER2/HER3 heterodimer (41,42). We found that, among the EGFR-inhibited residual PDXs of the reference collection, some tumors had reduced expression of AREG, HBEGF, and EGF, most had reduced expression of EREG, and most had increased expression of BTC (fig. S15). High amounts of AREG and EREG correlate with response to EGFR antibodies in mCRC patients (34). Therefore, our results indicated that the drug-tolerant state in CRC is characterized by reduced expression of genes encoding the ligands that sustain EGFR dependency and increased expression of the gene encoding a growth factor that can activate alternate survival pathways.
Combined EGFR/PI3K inhibition reduces residual disease in vitro but has little impact on tumor control in vivo
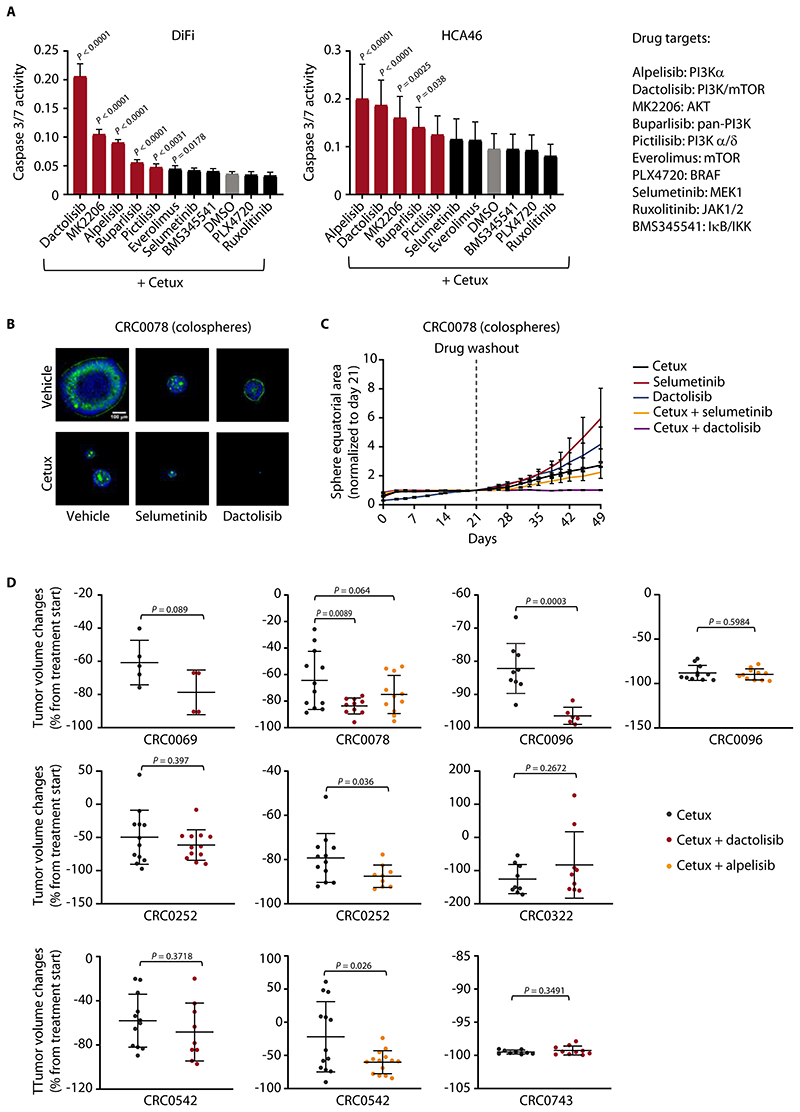
Analysis of residual tumors after EGFR inhibition revealed widespread reduction of cell proliferation without signs of apoptosis (Fig. 1B). Accordingly, cetuximab did not trigger considerable apoptosis (as assessed by caspase-3/7 activation) in DiFi and HCA46 cells (fig. S16A). We sought to identify therapeutic vulnerabilities in residual disease by testing for synergistic enhancement of apoptosis bycombination treatments with inhibitors of EGFR downstream effectors. EGFR mainly signals through the PI3K-AKT, MAPK, JAK-STAT, and NF-κB pathways (40). We selected inhibitors of each of these pathways and determined biologically relevant concentrations by mining a publicly available, large-scale data set reporting the sensitivity of several hundred cancer cell lines to hundreds of drugs (43). We used each drug at least twice the concentration of the minimum IC50 extracted from the database. The selected drug concentrations are active in various cell lines as reported in independent studies (44–48).
When used in monotherapy, specific inhibitors against each of these pathways did not induce apoptosis (fig. S16A). When combined with cetuximab, the drugs with the strongest pro-apoptotic activity were those selectively intercepting the PI3K-AKT signaling axis (as evidenced by abrogation of AKT phosphorylation after treatment with representative PI3K inhibitors) (Fig. 5A and fig. S16B). Similar to that observed in cell lines, co-inhibition of EGFR and PI3K in colospheres had a greater synergistic effect on reducing cell viability than combined blockade of EGFR and MEK (fig. S16C). Consistently, although concomitant neutralization of EGFR and MEK led to smaller colospheres than cetuximab alone, only co-inhibition of EGFR and PI3K almost completely obliterated the colonies (Fig. 5B). We also evaluated the colospheres for their latency before resuming proliferation after treatment cessation. Cells that survived prolonged treatment with cetuximab alone or cetuximab and the MEK inhibitor selumetinib started growing after drug washout; conversely, concomitant blockade of EGFR and PI3K prevented regrowth after treatment withdrawal (Fig. 5C).
Figure 5. Combined EGFR/PI3K inhibition unleashes apoptosis and reduces residual disease in vitro but has suboptimal activity in vivo.
(A) Luciferase-based evaluation of apoptosis (assessed by caspase 3/7 activity) in DiFi and HCA46 cell lines treated for 24h with the indicated drugs at the following concentrations: cetuximab, 20 μg/ml; BMS345541, ruxolitinib, selumetinib, PLX4720, alpelisib, 1 μM; dactolisib, 250 nM; everolimus, 50 nM; pictilisib, 100 nM; MK2206, 0.5 μM; buparlisib, 0.5 μM. The effect of cetuximab alone is indicated by the grey bar. The effect of combined treatment with cetuximab and PI3K/AKT inhibitors is indicated by red bars. Results represent the means ± SD of 2 independent experiments performed in biological quintuplicates (n = 10). Statistical analysis (combinatorial treatment versus cetuximab alone) by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. Cetux, cetuximab. (B) Representative confocal images of PuraMatrix-embedded CRC0078 colospheres treated for 2 weeks with vehicle (DMSO), cetuximab (10 μg/ml), selumetinib (300 nM), or dactolisib (100 nM), alone or in combination, prior to fixation and staining for F-actin (green) and DNA (blue). (C) Growth curves of PuraMatrix-embedded CRC0078 colospheres treated with cetuximab (30 μg/ml), selumetinib (1 μM), or dactolisib (300 nM), alone or in combination, for 21 days and then kept without drugs for additional 28 days. Time-lapse imaging was carried out by bright field microscopy, and individual equatorial areas were measured with Image J software. Average time-dependent area variation is represented as fold change relative to area value at day 21. Results are the means ± SEM of at least 15 colospheres for each experimental condition. Statistical analysis by two-way ANOVA (calculated after drug withdrawal) followed by Benjamini, Krieger and Yekutieli FDR correction: cetuximab + selumetinib versus cetuximab alone, P = 0.0138; cetuximab + dactolisib versus cetuximab alone, P < 0.0001. (D) Tumor volume changes in PDXs from mice treated with the indicated modalities for 4 weeks. Cetuximab, 20 mg/kg (intraperitoneal injection twice a week); dactolisib, 35 mg/kg (daily oral gavage); alpelisib, 25 mg/kg (daily oral gavage). Dots represent volume changes of PDXs from individual mice, and plotsshow the means ± SD for each treatment arm. n = 4 to 14 animals per each treatment arm. For CRC0069, CRC0096, CRC0252, CRC0322, CRC0542, and CRC0743, statistical analysis was performed by two-tailed unpaired Welch’s t-test. For CRC0078, statistical analysis was performed by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. Tumor variation outliers were excluded using the ROUT method (GraphPad Prism). Pre- and end-of-treatment tumor volumes in individual mice are reported in fig. S20.
Because apoptosis was unleashed only when EGFR and PI3K were concomitantly inhibited, we hypothesized that persistent activation of the PI3K pathway attenuates the antitumor effects of cetuximab. Indeed, cetuximab treatment inhibited extracellular signal-regulated kinase (ERK) phosphorylation in cell lines and colospheres, whereas AKT phosphorylation was less affected (fig. S17). Similarly, in cetuximab-treated PDXs of the reference collection, the phosphorylation of ERK was more markedly reduced than that of ribosomal protein S6 (S6), a downstream effector of PI3K/AKT (fig. S18A). Both ERK and S6 phosphorylation were regained in tumors that had resumed growth after cetuximab discontinuation (fig. S18B). A dichotomy between MAPK and PI3K signaling was also observed when delineating the pathways that transduce cetuximab-dependent impairment of YAP activity and upregulation of secretory/Paneth cell genes: MAPK inhibition, instead of PI3K inhibition, better recapitulated the effects of cetuximab (Fig. 3D, Fig. 4A, fig. S9B, fig. S12, fig. S13, D and E). Collectively, these results indicated that the outcomes of EGFR inhibition in CRC largely result from inactivation of the MAPK pathway and are countered by persistent PI3K signaling in residual cells.
Although concomitant inactivation of the EGFR and PI3K pathways increased cancer cell apoptosis in vitro, the burden of residual disease was only partially lessened in mice treated with both EGFR and PI3K inhibitors with respect to the residual disease in mice treated with cetuximab alone. We used the combination of cetuximab and dactolisib or the combination of cetuximab and alpelisib,as an alternative PI3K inhibitor, and compared these combinations to cetuximab monotherapy in PDX models. Both compounds were administered at pharmacodynamically active concentrations, as indicated by the specific reduction of phospho-S6 in tumors treated with either inhibitor (fig. S19A) and the decrease of both phospho-S6 and phospho-ERK in tumors treated also with cetuximab (fig. S19B). When used as monotherapies in 2 PDX models, neither dactolisib nor alpelisib induced tumor shrinkage (fig. S19C). We tested cetuximab and dactolisib in 7 PDX models, all with wild-type forms of the PIK3CA and PTEN genes, and cetuximab and alpelisib in 4 of the PDX models. The combination of dactolisib and cetuximab was more effective than cetuximab alone in regressing palpable masses in the CRC0078 and CRC0096 models, and the combination of alpelisib and cetuximab was superior to cetuximab monotherapy in the CRC0252 and CRC0542 models (Fig. 5D and fig. S20). Overall, the response to cetuximab and either dactolisib or alpelisib was better than response to cetuximab alone in 4/7 models (57%). Thus, susceptibility to dactolisib or alpelisib was heterogeneous in the models treated with both compounds, likely due to the different targets of the two inhibitors (all PI3K isoforms and mTOR in the case of dactolisib versus only PI3Kα for alpelisib) and differences in the ensuing signaling feedback loops (49).
Inspection of cancer cell density in PDXs with reduction in tumor volume, a macroscopic response, revealed decreased representation of epithelial neoplastic islets in 3/5 (60%) tumors, which suggests some therapeutic efficacy (fig. S21A). Dual inhibition of EGFR and PI3K induced a slight increase in the number of apoptotic cells compared with individual EGFR blockade in some PDX models in which the combination therapy was more effective than cetuximab alone (Fig. 5D and fig. S21B). However, tumor shrinkage was not invariably accompanied by apoptosis; some tumors with stronger induction of apoptosis after combination therapy did not respond better in terms of tumor regression, and others with a more pronounced size reduction were not more apoptotic (Fig. 5D and S20B). Hence, in models sensitive to the combination therapy, we propose that the strong antiproliferative effect of dual EGFR-PI3K inhibition shifted the steady-state equilibrium between new cell production and cell loss toward tumor breakdown, without induction of overt apoptosis.
The limited extent of residual disease reduction documented above was not sufficient to increase animals’ survival; indeed, PI3K blockade combined with cetuximab did not affect time to relapse after treatment discontinuation in the 4 PDX models examined (fig. S21C). In summary, combined EGFR and PI3K blockade was more effective than cetuximab alone in impairing cell viability in vitro; however, this therapy provided suboptimal and model-specific benefits in reducing residual disease at the end of treatment and did not have long-term effects on tumor control after therapy suspension.
Inhibition of the EGFR-YAP axis leads to increased HER2/HER3 signaling in PDX models and in patients
To identify more effective therapies against residual disease in vivo we searched for compensatory cues triggered by inhibition of the EGFR-YAP axis and expected to sustain persistent PI3K signaling. We focused on HER2 and HER3, because tumor models treated with EGFR or PI3K inhibitors exhibit feedback activation of such receptors and HER2/HER3 signaling preferentially stimulates the PI3K pathway (50–53). Additionally, the regulatory regions of the HER2 and HER3 genes have binding site peaks for TEAD4 (a canonical transcriptional partner of YAP), as shown by analysis of the ENCODE ChIP-seq datasets (54).
We evaluated HER2 and HER3 expression in DiFi and HCA46 cells transduced with either a control vector or various YAP constructs and the effect of cetuximab in these different conditions. EGFR blockade resulted in increased expression of HER2 and HER3 in control cells (Fig. 6A). Overexpression of constitutively active YAP-5SA reduced the basal amounts of HER2 or HER3, or both (fig. S22A), and blunted their upregulation by cetuximab (Fig. 6A). Likewise, doxycycline-regulated activation of YAP in established DiFi xenografts reduced the extent of HER2 and HER3 induction triggered by cetuximab (Fig. 6B). Introduction of wild-type YAP reduced HER3 basal expression (fig. S22B). However, wild-type YAP can be inhibited by upstream signals, and its exogenous overexpression was not sufficient to prevent HER2 and HER3 upregulation by cetuximab (fig. S22C). In loss-of-function experiments, we found that only the more effective shRNA construct against YAP (fig. S10) caused HER3 upregulation while HER2 was downregulated (fig. S22D). Thus, hyperactive YAP prevented the induction of HER2 and HER3 expression triggered by EGFR inhibition, but YAP silencing was insufficient to increase both HER2 and HER3 expression, suggesting that YAP is not the only regulator affecting EGFR-dependent modulation of HER2 and HER3.
Figure 6. EGFR blockade leads to increased HER2 and HER3 signaling in PDXs and in patients.
(A) RT-qPCR analysis of HER2 and HER3 expression in DiFi and HCA46 cell lines transduced with either a pLVX-control vector (mock) or the constitutively active YAP variant (YAP-5SA) and treated with cetuximab (20 μg/ml) for 48h. Independent experiments (n = 4 for DiFi and n = 3 for HCA46) were performed in technical triplicates. The plots show means ± SD. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. Cetux, cetuximab. (B) RT-qPCR analysis of HER2 and HER3 expression in established DiFi xenografts transduced with doxycycline-inducible YAP-5SA and treated with vehicle (control), cetuximab (20 mg/kg, intraperitoneal injection, 1 administration), doxycycline (Doxy, 50 mg/kg, daily oral gavage), or the combination of cetuximab and doxycycline for 1 week. Results from animals treated with cetuximab were normalized against those from animals treated with vehicle. Results from animals treated with cetuximab and doxycycline were normalized against those from animals treated with doxycycline. Four samples for each condition were analyzed in technical triplicates. The plots show means ± SD. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. (C) Heatmap showing expression changes for HER2 and HER3 in PDXs of the reference collection, after treatment with cetuximab for 6 weeks (20 mg/kg, intraperitoneal injection twice a week). Average gene expression, Log2R relative to vehicle-treated tumors: HER2 0.29, P = 0.029; HER3 0.31, P = 0.014 by two-tailed Wilcoxon test. Benjamini-Hochberg FDR < 0.1 for both genes. (D) Heatmap showing expression changes for HER2, HER3, DEFA5, and ATOH1 in tumors from patients after 4 weeks of treatment with Sym004. Changes in the sum of diameters of target lesions, as per RECIST criteria, are indicated as tumor variations. (E) Abundance of DEFA5 in the treatment-naïve sample and on-treatment biopsy of patient MPP192-5. Scale bar, 20 μm. IHC, immunohistochemistry. (F) Western blot analysis of the indicated proteins and phosphoproteins in DiFi and HCA46 cells treated for 10 min with BTC at the indicated concentrations. Total proteins were used for normalization; vinculin was used as a loading control. Western blots for total proteins were run with the same lysates as those used for anti-phosphoprotein detection but on different gels. The images shown are representative of 2 experiments on independent biological replicates. P-EGFR, phospho-EGFR; P-HER2, phospho-HER2; P-HER3, phospho-HER3; P-ERK, phospho-ERK; P-AKT, phospho-AKT. (G) Quantification of cell number (assessed by ATP content) in DiFi and HCA46 cell lines treated for 72h with the indicated antibodies at the indicated concentrationsin the absence or presence of 10 ng/ml BTC. Results represent the means ± SD of 2 independent experiments performed in biological triplicates (n = 6). Statistical analysis by two-way ANOVA. (H) Morphometric quantification of phospho-HER2 and phospho-HER3 immunoreactivity in PDXs from the reference collection after treatment with vehicle (until tumors reached an average volume of 1500 mm3) or cetuximab (for 6 weeks). Each dot represents the average of 10 optical fields (40X) in a section from randomly chosen tumors from vehicle-treated and cetuximab-treated mice bearing a PDX from the same original patient (n = 30). The plots show means ± SD. Statistical analysis by ratio paired t-test. Scale bar, 50 μm.
We analyzed HER2 and HER3 expression in treatment-naïve and cetuximab-treated PDXs of the reference collection. We detected moderate-to-strong overexpression of HER2 and HER3 in many of EGFR-inhibited models (Fig. 6C). Similar to what we observed for Paneth cell genes (fig. S7A), HER2 and HER3 upregulation was evident as early as one week after the first cetuximab administration and was maintained for 12 weeks of antibody treatment (fig. S23). DEFA5, HER2 and HER3 are upregulated by cetuximab and under the transcriptional control of YAP. Therefore, we evaluated the abundance of DEFA5 and the activation of HER2 and HER3 (phospho-HER2 and phospho-HER3) in 2 PDX models. Inspection of tissue sections revealed the presence of cytoplasmic DEFA5 in some, but not all, cells positive for phospho-HER2 or phospho-HER3 (fig. S24), suggesting that HER2/HER3 activation after cetuximab treatment is regulated by YAP-dependent but also YAP-independent pathways. For example, HER2/HER3 could be activated in “bystander” DEFA5-negative cells due to the observed surge in BTC expression after cetuximab treatment (fig. S15).
The transcriptional rewiring that occurred in EGFR-inhibited cell lines and PDXs was also observed in patient tumors. The present standard of care for patients with KRAS/NRAS-wild-type mCRC is combinatorial treatment with cetuximab or panitumumab and chemotherapy (14,15). Although this regimen is typically administered in the chemorefractory setting, residual reactions to cytotoxic agents may influence the response to EGFR inhibition. To overcome this hurdle, we evaluated samples from patients with KRAS/NRAS/BRAF wild-type tumors who had undergone single-agent therapy with Sym004, a mixture of two recombinant antibodies against EGFR that induces tumor shrinkage in mCRC patients with acquired resistance to cetuximab or panitumumab (55). Patients had been monitored in the frame of a controlled clinical trial with planned on-treatment biopsies immediately before first clinical restaging, with paired tumor tissues including the treatment-naïve surgical specimen after primary tumor resection and the on-treatment biopsy of a metastasis after 4 weeks of therapy (data file S5). We detected strong transcriptional induction of HER2 and HER3 in the on-treatment samples from 2 individuals who experienced a RECIST partial response (-38%; patient MPP192-5) or a minor tumor shrinkage (-15%; patient MPP192-1) at the time of metastatic biopsy (Fig. 6D). In patient MPP192-5, who responded better to Sym004, we also observed transcriptional upregulation of the secretory/Paneth-cell markers DEFA5 and ATOH1 (Fig. 6D) and confirmed an increase in DEFA5 abundance at the protein level (Fig. 6E). Such modulations appeared specific to sensitive cases, because the transcripts were not upregulated in samples from 2 patients in whom Sym004 was poorly active (+6.4%; patient MPP192-2) or ineffective (+37.8%; patient MPP192-4) (Fig. 6D).
Consistent with EGFR inhibition in sensitive tumors triggering an adaptive shift from high EGFR pathway activity to high HER2 and HER3 signaling, we found that residual PDXs not only showed HER2 and HER3 upregulation but also higher expression of the gene encoding the promiscuous HER ligand BTC (fig. S15). Indeed, CRC cells exposed to BTC showed activation of EGFR, HER2, and HER3, as well as increased phosphorylation of downstream transducers (Fig. 6F). BTC-stimulated cells were also less responsive (or, for DiFi, totally refractory) to cetuximab (Fig. 6G), suggesting that BTC counteracts EGFR blockade through compensatory HER2/HER3 activation. To test this hypothesis, we incubated cells with BTC and treated them with Pan-HER, a mixture of 6 antibodies against the 3 HER receptors (56). For stoichiometric targeting of EGFR, the Pan-HER concentration was adjusted to achieve equimolar concentrations of cetuximab and the anti-EGFR constituent of Pan-HER. Different from cetuximab, Pan-HER effectively reduced the viability of BTC-treated cells (Fig. 6G), consistent with HER2, HER3, and BTC limiting responsiveness to individual EGFR blockade and imparting a new signal dependency to EGFR-inhibited cells. Selective inhibition of HER2 or HER3 with trastuzumab or the isolated anti-HER components of Pan-HER did not affect cell viability either in the presence or in the absence of BTC, indicating that targeting of all HER family members is required to thwart BTC-dependent survival signals (fig. S25). In agreement with increased HER2 and HER3pathway activity in cultured cells, residual tumors in PDXs of the reference collection displayed increased phosphorylation of HER2 and HER3 (Fig. 6H).
Targeting HER2, HER3, and EGFR provides long-term therapeutic benefits in PDX trials
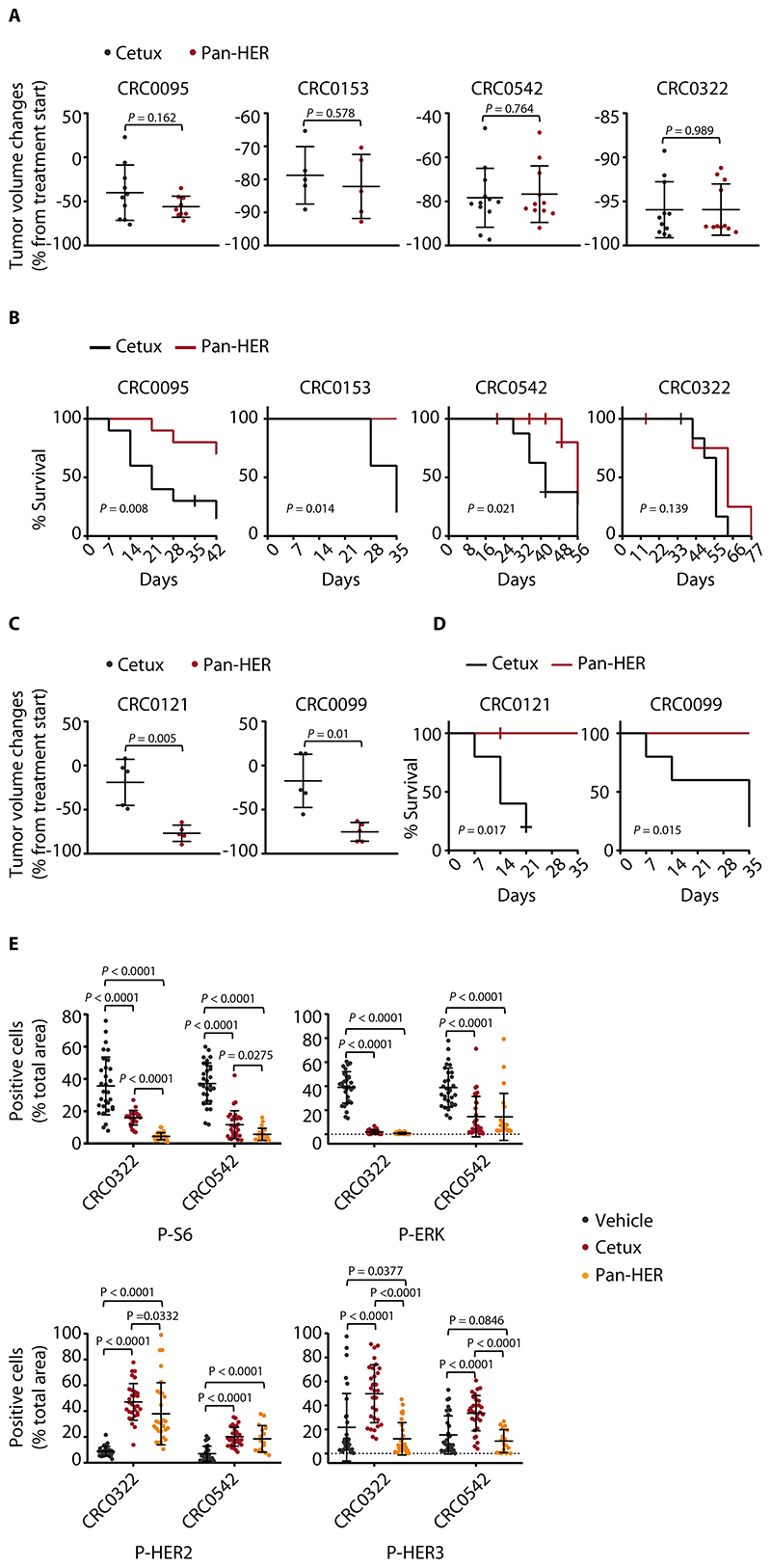
To explore whether compensatory HER2/HER3 signaling contributes to residual disease in vivo, we treated 6 PDX models with cetuximab or Pan-HER. In the 4 PDXs in which cetuximab induced clear regressions, the extent of tumor shrinkage obtained with Pan-HER was similar to that achieved by cetuximab (Fig. 7A). However, resumption of tumor growth after antibody withdrawal was delayed in 3/4 (75%) models treated with Pan-HER compared with those treated with cetuximab (Fig. 7B). In 2 models in which cetuximab left a higher burden of residual disease, Pan-HER was much more effective in reducing tumor volumes (Fig. 7C) and in retarding tumor relapse after treatment suspension (Fig. 7D). Of the 6 PDX cases tested, mouse cohorts implanted with 5 of the models (83%) experienced a significant survival benefit after therapy cessation.
Figure 7. Targeting HER2/HER3 together with EGFR provides long-term therapeutic benefits in PDX trials.
(A) Tumor volume changes in PDXs treated with the indicated modalities for 4 weeks. Cetuximab, 20 mg/kg (intraperitoneal injection twice a week); Pan-HER, 60 mg/kg (intraperitoneal injection twice a week). Dots represent volume changes of individual mice, and plots show the means ± SD for each treatment arm. n = 5 to 12 animals per each treatment arm. Statistical analysis by two-tailed unpaired Welch’s t-test. Tumor variation outliers were excluded using the ROUT method (GraphPad Prism). Cetux, cetuximab. (B) Kaplan-Meier survival curves in PDXs after discontinuation of the indicated treatments. n = 5 to 10 animals per each treatment arm. Statistical analysis by Log-rank (Mantel-Cox) test. (C) Tumor volume changes in PDXs treated with the indicated modalities for 4 weeks. n = 5 animals per each treatment arm. Statistical analysis as in A. (D) Kaplan-Meier survival curves in PDXs after discontinuation of the indicated treatments. n = 5 animals per each treatment arm. Statistical analysis as in B. (E) Morphometric quantification of the indicated phosphoproteins in PDXs CRC0322 and CRC0542 after treatment with vehicle (until tumors reached an average volume of 1500 mm3), cetuximab (5 weeks), or Pan-HER (5 weeks). At the end of treatment, 3 tumors from 3 different mice were explanted and subjected to immunohistochemical analysis. Each dot represents the value measured in one optical field (40x), with 2 to 10 optical fields per tumor depending on the extent of section area (n = 16 to 30). The plots show means ± SD. Statistical analysis by one-way ANOVA followed by Benjamini, Krieger and Yekutieli FDR correction. P-S6, phospho-S6; P-ERK, phospho-ERK; P-HER2, phospho-HER2; P-HER3, phospho-HER3. Representative images for phospho-S6 and phospho-ERK immunoreactivity in CRC0542 are shown in fig. S26.
The prominent activity of Pan-HER was also evident at the signaling level. Consistent with HER2 and HER3 cooperating to preferentially stimulate the PI3K-AKT pathway, Pan-HER induced stronger inhibition of PI3K-dependent signals than did cetuximab – as assessed by S6 phosphorylation – whereas the effects on the MAPK cascade were similar (Fig. 7E and fig. S26). Moreover, Pan-HER reduced the feedback activation of HER2 and HER3 triggered by individual EGFR blockade (Fig. 7E). In summary, Pan-HER minimized residual disease and reduced EGFR downstream signaling in EGFR-dependent mCRC PDXs. This effect translated into tumor control after therapy discontinuation, with longer time to progression than that obtained with cetuximab.
Discussion
Following EGFR blockade, actively dividing stem cells of the normal mouse intestine morph into quiescent cells that maintain robust Wnt signaling and share attributes with DNA-label-retaining precursors expressing some secretory markers of the Paneth-cell lineage (24,28,29). Using paired samples of untreated and cetuximab-treated PDXs, we showed that EGFR inhibition in mCRC tumors results in the emergence of residual cancer cell subpopulations that display hyperactive Wnt signaling and exhibit traits of secretory commitment and Paneth cell-like differentiation. Paneth cells are absent from colons of humans and rodents. Hence, a functional reprogramming like that driving quiescence in the normal small intestine of the mouse is activated during acquisition of drug tolerance by colon cancer cells in humans, suggesting a higher-order lineage plasticity that goes beyond species, anatomical location, and tissue functional state.
We provide evidence that the Paneth cell-like phenotypic rewiring induced by EGFR inhibition in colorectal tumors is an effect of YAP inactivation. During intestinal regeneration after injury, YAP reprograms intestinal stem cells by transiently suppressing Paneth cell differentiation, while bolstering EGFR-dependent proliferation mediated by increased production of EGFR ligands (35). In a reciprocal scenario, we found that residual tumors after EGFR inhibition had reduced abundance and nuclear localization of YAP, increased expression of Paneth-cell markers, and downregulation of genes encoding EGFR-activating ligands. Our data indicated that these features are likely causally linked, because exogenous introduction of a hyperactive form of YAP prevented the induction of the Paneth cell-like phenotype by cetuximab in CRC cell lines. We also showed that EGFR activation is not only a consequence of YAP function but is also an upstream regulatory event. This module represents a bistable, feed-forward autoregulatory loop that potentiates EGFR activation when the kinase is not constrained, but triggers EGFR neutralization and fosters evasion from EGFR dependence as soon as the receptor activity is restricted, for example by low ligand availability or by pharmacological inhibition.
Many cetuximab-treated tumors also featured HER2 and HER3 overexpression, which, similar to manifestation of the Paneth cell-like phenotype, can be ascribed to cetuximab-induced inhibition of YAP. YAP inactivation appears therefore to orchestrate both a lineage shift toward the secretory/Paneth cell-like fate and a signaling shift from high EGFR activity to high HER2/HER3 activity. Our results indicated that EGFR pathway rewiring sustains residual disease, because targeting of EGFR, HER2, and HER3 induced more pronounced tumor regressions and delayed tumor relapse after treatment cessation in several PDX models than targeting EGFR alone. We propose that the materialization of Paneth-cell-like characteristics represents the accompanying phenotype of the change in signal dependency. We detected HER2 and HER3 phosphorylation, indicative of activation, also in non-Paneth-like (DEFA5-negative) residual cells, suggesting that EGFR family adaptive reprogramming after EGFR inhibition may occur through additional mechanisms, for example through BTC upregulation as we observed here or YAP-independent, MYC-dependent modulation of HER3 expression (57). HER2/HER3 activation, besides driving tolerance in cetuximab-sensitive tumors as a consequence of compensatory gene overexpression, may also sustain intrinsic antibody resistance on a genetic basis, as shown for mCRC subsets with ERBB2 gene amplification or mutations (16,19,20). Finally, residual disease may be further fostered by implementation of a cancer stem cell program, which is consistent with our results that both EGFR inhibition and YAP inactivation increase expression of Wnt target genes known to control stem cell maintenance in the normal intestine and colorectal cancer (25).
When translated to the clinic, our results advocate the use of agents targeting functional dependencies in residual disease as either first-line therapies or as sequential therapies immediately after maximal response. Such treatments may complement other pre-emptive strategies aimed to neutralize genetic mechanisms of resistance before clinical manifestation of disease progression, forexample by combining EGFR antibodies and MEK inhibitors to limit the outgrowth of RAS-mutant subclones (58). Approaches of this kind would be a paradigm shift in clinical decision making by repositioning rational therapeutic intervention from the refractory state, when multiple genetic mechanisms of resistance reduce the chances of clinical benefit, to the phase of treatment when massive regression occurs.
Our findings have certain limitations. For example, we have not tested whether Paneth cell-like cells act as tumor-(re)initiating cells following cetuximab cessation, mainly because PDXs are hardly amenable to lineage-tracing approaches. Moreover, we acknowledge that the clinical validation of our results is restricted to a small number of patients. The procurement of paired pre- and on-treatment samples to investigate early adaptive response to therapy remains a hurdle in mCRC, not only for ethical and logistical reasons but also due to the difficulty of selecting patients with a treatment history and clinical monitoring that enable unequivocal interpretation of the results. For accurate clinical investigation of residual disease, we decided to evaluate samples from mCRC patients who had received a ‘clean’ treatment (EGFR antibody monotherapy without concomitant chemotherapeutic agents), even though this conservative choice limited the analysis to 4 patients. Albeit circumscribed to a small cohort of individuals, the results confirmed HER2 and HER3 upregulation in the residual tumors of sensitive patients and overexpression of secretory/Paneth-cell markers in 1 good responder. The clinical relevance of our findings is also backed by evidence that patients experiencing earlier relapse during cetuximab therapy had tumors with higher amounts of secretory/Paneth cell markers than individuals with longer PFS, providing clinical support to the notion that this cell fate reprogramming typifies lingering cells that tend to endure EGFR inhibition.
Transdifferentiation has been associated with drug resistance in NSCLC, prostate cancer, and melanoma (9,59,60). By extending the generality of this mechanism to other tumor types, our results illustrate how perturbations of conserved developmental pathways that control cellular plasticity may represent key factors shaping drug response in cancer. Such knowledge can motivate the design of therapeutic strategies to target the reversible phase of drug tolerance preceding the onset of mutational resistance.
Materials and Methods
Study design
This study was designed to identify mechanisms that sustain drug tolerance in residual CRC tumors at maximal response to the EGFR antibody cetuximab and find therapies that, when combined with EGFR blockade, may minimize residual disease and delay tumor regrowth after treatment discontinuation. In the first part of the study, cohorts of nonobese diabetic-severe combined immunodeficient (NOD-SCID) mice bearing tumors expanded from 279 independent PDXs (12 or 24 mice for each original sample) were treated with cetuximab monotherapy to identify cases with objective but incomplete response. In the second part, molecular profiles (gene expression data obtained from oligonucleotide microarrays or RT-qPCR) were analyzed to extract the identifying features of residual disease and generate hypotheses about potential regulatory mechanisms. In the third part, mechanistic studies based on the working hypotheses that emerged from the second part were conducted using gain-of-function and loss-of-function approaches in cell lines. Finally, candidate targets likely to sustain residual disease were inhibited with specific therapies, alone or in combination with cetuximab, in PDXs. Tumor-bearing mice were randomized to treatment cohorts. Cohort sample sizes were calculated to have 80% power to detect a twofold change in tumor size between groups, using a two-group t test with a two-sided type I error rate of 0.05. End points for animal experiments were selected in accordance with institutional-approved criteria and guidelines from the Italian National Institute of Health. Information on immunohistochemistry and morphometric quantifications, molecular and bioinformatics analyses, and cell-based experimentation can be found in the Supplementary Materials. Details regarding sample number and replication in assays, as well as statistical analysis, are given in figure legends and in the Methods below. All values for quantification of immunohistochemistry images and tumor growth curves in animal experiments were recorded blindly.
Specimen collection and annotation
Tumor and matched normal samples were obtained from patients treated by liver metastasectomy at the Candiolo Cancer Institute (Candiolo, Torino, Italy), Mauriziano Umberto I (Torino), and San Giovanni Battista (Torino). All patients provided informed consent. Samples were procured and the study was conducted under the approval of the Review Boards of the Institutions.
PDX models and in vivo treatments
Tumor implantation and expansion were performed in 6-week-old male and female NOD/SCID mice as previously described (16). Once tumors reached an average volume of ~400 mm3, mice were randomized into treatment arms, with at least n = 5 per group, and were treated with the modalities indicated in the figures. Dactolisib (Carbosynth) was dissolved in 1 volume of N-methyl-2-pyrrolidone(Sigma Aldrich) and 9 volumes of PEG300 (Fluka). Alpelisib (Carbosynth) was dissolved in 0.5% carboxymethylcellulose (Fluka). Doxycycline was dissolved in water. Tumor size was evaluated once-weekly by caliper measurements, and the approximate volume of the mass was calculated using the formula 4/3π · (d/2)2 · D/2, where d and D are the minor tumor axis and the major tumor axis, respectively. Results were considered interpretable when a minimum of 4 mice per treatment group reached the pre-specified endpoints (at least 3 weeks on therapy or development of tumors with average volumes larger than 1500 mm3 within each treatment group in trials aimed to assess drug efficacy; at least 3 weeks after treatment cessation or development of individual tumors with volumes larger than 750 mm3 in survival experiments aimed to assess tumor control by therapy). Operators were blinded during measurements. In vivo procedures and related biobanking data were managed using the Laboratory Assistant Suite (61). Animal procedures were approved by the Italian Ministry of Health (authorization 806/2016-PR).
Statistics
Error bars indicate standard deviations (SD) unless otherwise indicated. The number of biological (nontechnical) replicates for each experiment is reported in the figure legends. For experiments with two groups, statistical analysis was performed using two-tailed Student’s t-test, Welch’s t-test or Wilcoxon matched pairs signed-rank test unless otherwise noted. For experiments with more than two groups, one-way ANOVA was used. Two-way ANOVA was applied for experiments in which the determinations were considered interdependent. In case of multiple testing, we adopted the Benjamini, Krieger and Yekutieli false discovery rate (FDR) two-stage step-up method or the Benjamini-Hochberg FDR test. Correlations were calculated by Pearson’s coefficients. Statistical analyses in the survival experiments were performed by Log-rank (Mantel-Cox) test. The level of statistical significance was set at P < 0.05. For multiple comparisons, the results were considered significant when the Benjamini, Krieger and Yekutieli method was < 0.05 and when the Benjamini–Hochberg FDR was < 0.1. Graphs were generated and statistical analyses performed using the GraphPad Prism (v8.0) statistical package. Original data are provided in data file S6.
Supplementary Material
Single sentence summary.
Drug tolerance in EGFR-inhibited mCRC involves secretory pseudodifferentiation and rewiring of oncogenic dependencies.
Acknowledgments
We acknowledge Merck KGaA (Darmstadt, Germany) for a generous gift of cetuximab. We thank L. Ailles for providing the TOP-GFP reporter construct; J. Baselga for DiFi cells; L. Azzolin, E. Berrino, I. Catalano, M. Cordenonsi, F. Cottino, S.M. Leto, S. Marsoni, F. Orzan, and G. Siravegna for discussion and sharing reagents; A. Ferrero, C. Marchiò, A. Mellano, A. Muratore, M. Papotti, G. Paraluppi, S. Perotti, and A. Sapino for sample acquisition; M. Buscarino and B. Martinoglio for support with real-time PCR and Sanger sequencing; A. Giove, F. Maina, R. Machiorlatti and C. Zerbone for animal husbandry; R. Albano, S. Giove, L. Fontani, and L. Palmas for technical assistance; and D. Gramaglia and G. Falca for secretarial assistance. Editorial services were provided by Nancy R. Gough (BioSerendipity, LLC, Elkridge, MD).
Funding
Supported by AIRC, Associazione Italiana per la Ricerca sul Cancro, Investigator Grants 20697 (to A.B.), 12944 (to E.M.), 20290 (to D.P.), and 22802 (to L.T.). AIRC 5x1000 grant 21091 (to A.B., E.M. and L.T). AIRC/CRUK/FC AECC Accelerator Award 22795 (to L.T.). My First AIRC Grant 19047 (to C.I.). European Research Council Consolidator Grant 724748 – BEAT (to A.B.). H2020 grant agreement no. 754923 COLOSSUS (to L.T.). H2020 INFRAIA grant agreement no. 731105 EDIReX (to A.B. and E.M.). Transcan, TACTIC (to L.T.). Fondazione Piemontese per la Ricerca sul Cancro-ONLUS, 5x1000 Ministero della Salute 2015 (to E.M. and L.T.), 2014 and 2016 (to L.T.). A.B., E.M. and L.T. are members of the EurOPDX Consortium. A.P. is the recipient of a fellowship from Fondazione Umberto Veronesi.
Footnotes
Author contributions: B.L. was responsible for study design, biological experimentation in vitro, data interpretation, and manuscript writing. F.S. performed immunohistochemistry and analyzed and interpreted morphological data. M.P. assisted with reporter assays and RT-qPCR experiments. F.G. conducted mutational and gene expression analyses. E.R.Z. coordinated the biobank of viable CRC specimens and molecular derivatives. V.V. performed experiments with CRC organoids. G.M. designed and conducted PDX experimentation. P.A.M. designed, performed, and interpreted experiments with colospheres under the supervision of A.P. and L.P. D.M. performed ChIP-seq experiments under the supervision of D.P. P.L. and C.B. provided colospheres. M.K., M.W.P. and I.H. developed, characterized, and provided the Pan-HER antibody. E.M and C.I. analyzed gene expression data. D.N., I.S., T.H., and A.V. performed and interpreted WES and RNA-seq analyses under the supervision of A.S and T.A.G. E.E., G.A., P.N. and R.D. selected, annotated and provided clinical samples. E.G. analyzed gene expression data and supervised statistical analyses. A.B. and L.T. conceptualized the study, coordinated, and supervised overall research, and wrote the manuscript. All authors have critically read, edited, and approved the final version of the manuscript.
Competing interests: L.T. receives research grants from Symphogen,which has developed the Pan-HER antibody mixture used in this study. M.W.P. is, and M.K. and I.D.H. have been, employed at Symphogen and have ownership interest (including patents) in the same. L.T. also receives research grants from Servier, Pfizer, Menarini, and Merus, and he is in the speakers’ bureau of Eli Lilly, AstraZeneca, and Merck KGaA.
Data and materials availability
Microarray data have been deposited in GEO with accession code GSE108277. RNA-seq data have been deposited in the Sequence Read Archive (SRA) with accession code EGAS00001003765. WES data have been deposited in SRA with accession code EGAS00001003764. PDX models and derivatives thereof are available for solely academic purposes from L.T. or A.B. under a material transfer agreement with the University of Torino. All data associated with this study are present in the paper or the Supplementary Materials.
References
- 1.Bivona TG, Doebele RC. A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nat Med. 2016;22:472–478. doi: 10.1038/nm.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenthal GM, Karuri SW, Zhang H, Zhang L, Khozin S, Kazandjian D, Tang S, Sridhara R, Keegan P, Pazdur R. Overall response rate, progression-free survival, and overall survival with targeted and standard therapies in advanced non-small-cell lung cancer: US Food and Drug Administration trial-level and patient-level analyses. J Clin Oncol. 2015;33:1008–1014. doi: 10.1200/JCO.2014.59.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piessevaux H, Buyse M, De Roock W, Prenen H, Schlichting M, Van Cutsem E, Tejpar S. Radiological tumor size decrease at week 6 is a potent predictor of outcome in chemorefractory metastatic colorectal cancer treated with cetuximab (BOND trial) Ann Oncol. 2009;20:1375–1382. doi: 10.1093/annonc/mdp011. [DOI] [PubMed] [Google Scholar]
- 4.Piessevaux H, Buyse M, Schlichting M, Van Cutsem E, Bokemeyer C, Heeger S, Tejpar S. Use of early tumor shrinkage to predict long-term outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2013;31:3764–3775. doi: 10.1200/JCO.2012.42.8532. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HE, Krishnamurthy Radhakrishna V, Siravegna G, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu CH, Thurley K, Wei S, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song C, Piva M, Sun L, Hong A, Moriceau G, Kong X, Zhang H, Lomeli S, Qian J, Yu CC, Damoiseaux R, et al. Recurrent tumor cell-intrinsic and -extrinsic alterations during MAPKi-induced melanoma regression and early adaptation. Cancer Discov. 2017;7:1248–1265. doi: 10.1158/2159-8290.CD-17-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, Karras P, Brown D, Chang YH, Debiec-Rychter M, Adriaens C, et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell. 2018;174:843–855. doi: 10.1016/j.cell.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, Hemmati G, Martins MM, Maynard A, Kuhn J, Galeas J, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25:111–118. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, Wolf M, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 13.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 14.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 15.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 16.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C, Ribero D, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 17.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, Ercan D, Rogers A, Roncalli M, Takeda M, Fujisaka Y, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, Apicella M, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658–673. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, Searleman AC, Shen W, Monsey J, Trusolino L, Jacobs SA, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5:832–541. doi: 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, Sausen M, Phallen J, Hruban CA, Tokheim C, Niknafs N, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanella ER, Galimi F, Sassi F, Migliardi G, Cottino F, Leto SM, Lupo B, Erriquez J, Isella C, Comoglio PM, Medico E, et al. IGF2 is an actionable target that identifies a distinct subpopulation of colorectal cancer patients with marginal response to anti-EGFR therapies. Sci Transl Med. 2015;7:272ra12. doi: 10.1126/scitranslmed.3010445. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Brenes IA, Komarova NL, Wodarz D. Tumor growth dynamics: insights into evolutionary processes. Trends Ecol Evol. 2013;28:597–604. doi: 10.1016/j.tree.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 23.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basak O, Beumer J, Wiebrands K, Seno H, van Oudenaarden A, Clevers H. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 2017;20:177–190. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Clevers H. Stem cells: a unifying theory for the crypt. Nature. 2013;495:53–54. doi: 10.1038/nature11958. [DOI] [PubMed] [Google Scholar]
- 26.Barker N, van Oudenaarden A, Clevers H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell. 2012;11:452–460. doi: 10.1016/j.stem.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Budiyanto A, Bito T, Kunisada M, Ashida M, Ichihashi M, Ueda M. Inhibition of the epidermal growth factor receptor suppresses telomerase activity in HSC-1 human cutaneous squamous cell carcinoma cells. J Invest Dermatol. 2003;121:1088–1094. doi: 10.1046/j.1523-1747.2003.12529.x. [DOI] [PubMed] [Google Scholar]
- 28.Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 29.Barriga FM, Montagni E, Mana M, Mendez-Lago M, Hernando-Momblona X, Sevillano M, Guillaumet-Adkins A, Rodriguez-Esteban G, Buczacki SJA, Gut M, Heyn H, et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;20:801–816. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki N, Sachs N, Wiebrands K, Ellenbroek SI, Fumagalli A, Lyubimova A, Begthel H, van den Born M, van Es JH, Karthaus WR, Li VS, et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci USA. 2016;113:E5399-5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 33.Isella C, Brundu F, Bellomo SE, Galimi F, Zanella E, Porporato R, Petti C, Fiori A, Orzan F, Senetta R, Boccaccio C, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun. 2017;8:15107. doi: 10.1038/ncomms15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 35.Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 36.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 37.Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell. 2013;24:459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, Asthana S, Neel D, Yan JJ, Lu X, Pham L, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Montminy T, Azad T, Lightbody E, Hao Y, SenGupta S, Asselin E, Nicol C, Yang X. PI3K positively regulates YAP and TAZ in mammary tumorigenesis through multiple signaling pathways. Mol Cancer Res. 2018;16:1046–1058. doi: 10.1158/1541-7786.MCR-17-0593. [DOI] [PubMed] [Google Scholar]
- 40.Yarden Y, Pines YG. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- 41.Pinkas-Kramarski R, Lenferink AE, Bacus SS, Lyass L, van de Poll ML, Klapper LN, Tzahar E, Sela M, van Zoelen EJ, Yarden Y. The oncogenic ErbB-2/ErbB-3 heterodimer is a surrogate receptor of the epidermal growth factor and betacellulin. Oncogene. 1998;16:1249–1258. doi: 10.1038/sj.onc.1201642. Y. [DOI] [PubMed] [Google Scholar]
- 42.Alimandi M, Wang LM, Bottaro D, Lee CC, Kuo A, Frankel M, Fedi P, Tang C, Lippman M, Pierce JH. Epidermal growth factor and betacellulin mediate signal transduction through co-expressed ErbB2 and ErbB3 receptors. EMBO J. 1997;16:5609–5617. doi: 10.1093/emboj/16.18.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S, Lightfoot H, Cokelaer T, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, Okimoto RA, Lin L, Neel DS, Sabnis S, Flanagan J, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21:1038–1047. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leto SM, Sassi F, Catalano I, Torri V, Migliardi G, Zanella ER, Throsby M, Bertotti A, Trusolino L. Sustained Inhibition of HER3 and EGFR Is Necessary to Induce Regression of HER2-Amplified Gastrointestinal Carcinomas. Clin Cancer Res. 2015;21:5519–5531. doi: 10.1158/1078-0432.CCR-14-3066. [DOI] [PubMed] [Google Scholar]
- 46.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, Russo M, Cancelliere C, Zecchin D, Mazzucchelli L, Sasazuki T, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–2866. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, Sproesser K, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jani TS, DeVecchio J, Mazumdar T, Agyeman A, Houghton JA. Inhibition of NF-kappaB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin. J Biol Chem. 2010;285:19162–19172. doi: 10.1074/jbc.M109.091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sathe A, Chalaud G, Oppolzer I, Wong KY, von Busch M, Schmid SC, Tong Z, Retz M, Gschwend JE, Schulz WA, Nawroth R. Parallel PI3K, AKT and mTOR inhibition is required to control feedback loops that limit tumor therapy. PLoS One. 2018;13:e0190854. doi: 10.1371/journal.pone.0190854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mancini M, Gaborit N, Lindzen M, Salame TM, Dall’Ora M, Sevilla-Sharon M, Abdul-Hai A, Downward J, Yarden Y. Combining three antibodies nullifies feedback-mediated resistance to erlotinib in lung cancer. Sci Signal. 2015;8:ra53. doi: 10.1126/scisignal.aaa0725. [DOI] [PubMed] [Google Scholar]
- 51.Bosch-Vilaró A, Jacobs B, Pomella V, Abbasi Asbagh L, Kirkland R, Michel J, Singh S, Liu X, Kim P, Weitsman G, Barber PR, et al. Feedback activation of HER3 attenuates response to EGFR inhibitors in colon cancer cells. Oncotarget. 2017;8:4277–4288. doi: 10.18632/oncotarget.13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gala K, Chandralapaty S. Molecular pathways: HER3 targeted therapy. Clin Cancer Res. 2014;20:1410–16. doi: 10.1158/1078-0432.CCR-13-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dienstmann R, Patnaik A, Garcia-Carbonero R, Cervantes A, Benavent M, Susana Roselló S, Tops BB, van der Post R, Argilés G, Skartved NJ, Hansen UH, et al. Safety and activity of the first-in-class Sym004 anti-EGFR antibody mixture in patients with refractory colorectal cancer. Cancer Discov. 2015;5:598–609. doi: 10.1158/2159-8290.CD-14-1432. [DOI] [PubMed] [Google Scholar]
- 56.Jacobsen HJ, Poulsen TT, Dahlman A, Kjær I, Koefoed K, Sen JW, Weilguny D, Bjerregaard B, Andersen CR, Horak ID, Pedersen MW, et al. Pan-HER, an antibody mixture simultaneously targeting EGFR, HER2, and HER3, effectively overcomes tumor heterogeneity and plasticity. Clin Cancer Res. 2015;21:4110–4122. doi: 10.1158/1078-0432.CCR-14-3312. [DOI] [PubMed] [Google Scholar]
- 57.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, Schlicker A, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 58.Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, Baldi F, Lin KH, Truini M, Trusolino L, Bertotti A, Di Nicolantonio F, et al. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun. 2015;6:8305. doi: 10.1038/ncomms9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, Le Magnen C, Chester D, Mostaghel EA, Califano A, Rubin MA, et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7:736–749. doi: 10.1158/2159-8290.CD-16-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baralis E, Bertotti A, Fiori A, Grand A. LAS: a software platform to support oncological data management. J Med Syst. 2012;36(Suppl. 1):S81-90. doi: 10.1007/s10916-012-9891-6. [DOI] [PubMed] [Google Scholar]
- 62.Jammula S, Pasini D. EpiMINE, a computational program for mining epigenomic data. Epigenetics Chromatin. 2016;9:42. doi: 10.1186/s13072-016-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luraghi P, Reato G, Cipriano E, Sassi F, Orzan F, Bigatto V, De Bacco F, Menietti E, Han M, Rideout WM, 3rd, Perera T, et al. MET signaling in colon cancer stem-like cells blunts the therapeutic response to EGFR inhibitors. Cancer Res. 2014;74:1857–1869. doi: 10.1158/0008-5472.CAN-13-2340-T. [DOI] [PubMed] [Google Scholar]
- 64.Pupo E, Ducano N, Lupo B, Vigna E, Avanzato D, Perera T, Trusolino L, Lanzetti L, Comoglio PM. Rebound Effects Caused by Withdrawal of MET Kinase Inhibitor Are Quenched by a MET Therapeutic Antibody. Cancer Res. 2016;76:5019–5029. doi: 10.1158/0008-5472.CAN-15-3107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data have been deposited in GEO with accession code GSE108277. RNA-seq data have been deposited in the Sequence Read Archive (SRA) with accession code EGAS00001003765. WES data have been deposited in SRA with accession code EGAS00001003764. PDX models and derivatives thereof are available for solely academic purposes from L.T. or A.B. under a material transfer agreement with the University of Torino. All data associated with this study are present in the paper or the Supplementary Materials.