Abstract
During X-chromosome inactivation (XCI) one of the two X-inactivation centers (Xic) up-regulates the non-coding Xist RNA to initiate chromosomal silencing in cis. How one Xic is chosen to up-regulate Xist remains unclear. Models proposed include localization of one Xic at the nuclear envelope, or transient homologous Xic pairing followed by asymmetric transcription factor distribution at Xist’s antisense Xite/Tsix locus. Here we use a TetO/TetR system that can inducibly relocate one or both Xics to the nuclear lamina in differentiating embryonic stem cells (ESCs). We find that neither nuclear lamina localization, nor reduction of Xic homologous pairing influence monoallelic Xist up-regulation or choice-making. We also show that transient pairing is associated with biallelic expression, not only at Xist/Tsix but at other X-linked loci that can escape XCI. Finally we show that Xic pairing occurs in wave-like patterns, coinciding with genome dynamics and the onset of global regulatory programmes during early differentiation.
Introduction
X-chromosome inactivation (XCI) is the mechanism ensuring dosage compensation in female mammals. The X-inactivation center (Xic) is required in at least two copies in a diploid genome to trigger the initiation of random XCI1 (for reviews see2–4) in epiblast cells of the mouse embryo. The Xic harbors the main regulator of XCI, the X-inactive specific transcript (Xist) gene in eutherian mammals5–10. Robust Xist up-regulation is triggered only in cells with more than one X chromosome and in diploid cells just one of the two X chromosomes becomes inactivated during random XCI. How this choice of only one of the two Xics to up-regulate Xist occurs, has remained enigmatic. Failure to establish monoallelic Xist up-regulation can potentially result in both X chromosomes remaining active or being silenced, leading to embryonic lethality11–13. Random XCI can be recapitulated in vitro in differentiating mouse embryonic stem cells (mESCs). In undifferentiated mESCs, both X chromosomes are active and Xist is repressed by several mechanisms including pluripotency factors, and cis-regulatory elements including Tsix, which is transcribed antisense through the Xist gene and its promoter. Several lines of evidence have led to the proposal that Tsix is involved in choice-making and monoallelic Xist expression at the onset of random XCI14–20: Tsix becomes down-regulated on one Xic during a similar time-window as Xist up-regulation21,22; deletion of Tsix’s promoter results in non-random Xist up-regulation from the mutated allele23; deletion of both Tsix promoters in female ESCs leads to chaotic XCI with increased proportions of cells showing no Xist up-regulation or biallelic up-regulation during differentiation14.
How random monoallelic Xist expression is achieved and the role of Tsix have been debated. Both Xist and Tsix are regulated by pluripotency factors and Xist is controlled by trans-acting factors such as Rnf12/RLIM 24–29(see 30 for review). However, both Xics are exposed to the same trans-acting factors and the same or similar cis-regulatory elements within the nucleus. Numerous models have been evoked to explain how two genetically identical Xics become oppositely expressed during differentiation, but few have been directly tested.
One model for initiating asymmetric Xist and Tsix expression involves transient homologous Xic pairing during initiation of XCI31–34. Pairing at Xite, a putative Tsix enhancer, was proposed to facilitate choice-making and monoallelic Tsix expression31,35. Another region, Xpr, located several hundred kilobases away was proposed to facilitate Xite pairing but also to sense the presence of two Xics and help promote initiation of Xist expression35 (see 2 for review), although Xpr’s precise role remains unclear36. Studies involving heterokaryons suggested that Xist can be activated even without two X chromosomes within the same nucleus29. Nevertheless, Xic pairing may still be required for symmetry breaking in XX cells. Indeed, live-cell imaging using Xic TetO-tagged female ESCs to visualize Tsix/Xite pairing, combined with FISH to visualize nascent expression of Tsix after pairing events, suggested that the outcome of pairing might be transient monoallelic Tsix down-regulation, which could favour monoallelic Xist up-regulation in cis34. These studies and others, evoking Oct4 and CTCF as potential asymmetrically distributed factors following pairing33,37, all point to a potential role for Xic pairing in Tsix/Xist symmetry-breaking and Xic choice. However, the role of Xic pairing in choice-making has never been directly tested, something that we undertake here.
Another long-standing hypothesis for differential treatment of two X chromosomes during random XCI is nuclear compartmentalization. For example association of one X chromosome with the nuclear periphery or another nuclear compartment might facilitate asymmetric access to transcription or replication factors38,39. Indeed, the Xic lying closest to the nuclear periphery may be more prone to show initial Xist RNA accumulation in early differentiating XX ESCs32. More recently, it was proposed that association of the X chromosome with the lamina and the lamin B receptor (LBR), could facilitate Xist RNA–mediated gene silencing40.
Similar mechanisms have been evoked for other examples of monoallelic regulation, such as the immunoglobulin (Ig) and Tcr loci. Homologous pairing and asymmetric association with pericentric heterochromatin (so called “chromocenters”), as well as locus repositioning into the interior of the nucleus, have been proposed to play roles in ensuring faithful recombination and monoallelic expression during lymphocyte maturation41–45. Roles for homologous pairing have also been proposed in Drosophila, for both positive and negative transcriptional regulation in trans (transvection). Examples include the Ultrabithorax (Ubx), yellow (y), decapentaplegic (dpp), eyes absent (eya) loci and the brown (bw) gene) (reviewed in 46,47). Transvection has also been visualized by live-cell imaging in Drosophila embryos48.
Although a large body of circumstantial evidence links nuclear positioning with transcriptional regulation, including monoallelic expression, few studies have directly tested such hypotheses. Indeed, the design of experiments that specifically affect nuclear positioning, as opposed to affecting cis-acting elements that might have other effects on gene expression is challenging. In this paper we established direct functional tests that seek to assess whether subnuclear positioning of the Xic or homologous Xic pairing at the level of the Tsix region, actually contribute to monoallelic Xist regulation and choice-making during initiation of random XCI in early differentiating female ESCs. We use a TetO/TetR system, previously used for Xic pairing visualization, to reversibly relocate one or both Xic loci to the nuclear lamina via a TetR-EGFP-LaminB1 fusion protein. Similar strategies have been used to relocate LacO array-tagged autosomal loci to the nuclear lamina in mammalian cell lines and Drosophila upon expression of various LacI fusion proteins49–52. We assess the impact of Xic tethering to the lamina on (i) gene expression; (ii) Xic pairing; (iii) initiation of monoallelic Xist up-regulation and random XCI. We find that neither localization at the nuclear periphery, nor Xic pairing are major determinants for the initiation of XCI, at least during early ESC differentiation. Rather, we show that Xic pairing seems to be a consequence of biallelic Xist/Tsix transcription (sense or antisense) in the context of mESC differentiation.
Results
Repositioning of the Xic to the nuclear periphery
To explore the impact of relocating the Xic to the nuclear envelope, we used a female mESC line (PGK12.1) that carries a TetO array (224 repeats) 40 kb upstream of Tsix34,53. This line displays random XCI and has been used to visualize Xic dynamics in living cells via a TetR-mCherry or GFP protein fusion34,53(Figure 1A). In order to relocate the single Xic-TetO (in XXTetO ESCs) or both Xic-TetOs, (XTetOXTetO ESC) to the nuclear lamina, a transgene for TetR-EGFP-LaminB1 (T-E-LaminB1) fusion protein was stably introduced (Figure 1B,C; see Online Methods; Supplementary Figure 1A). In this system, association with the lamina is inducible, as TetR binding is doxycycline (dox) sensitive and thus reversible. In cells grown without dox, TetR binds to the TetO array; addition of dox prevents binding (Figure 1D). The TetR-EGFP-LaminB1 expressing ESCs were thus generated in dox-containing medium, to exclude effects of TetR binding (and Xic lamina targeting) during their derivation. TetR binding could then be induced (removing dox from the medium) at specific time points. For each experiment cells were split into two populations: one in dox-containing medium (referred to as ‘control’), the other in medium without dox (referred to as ‘bound’) (Figure 1D,E).
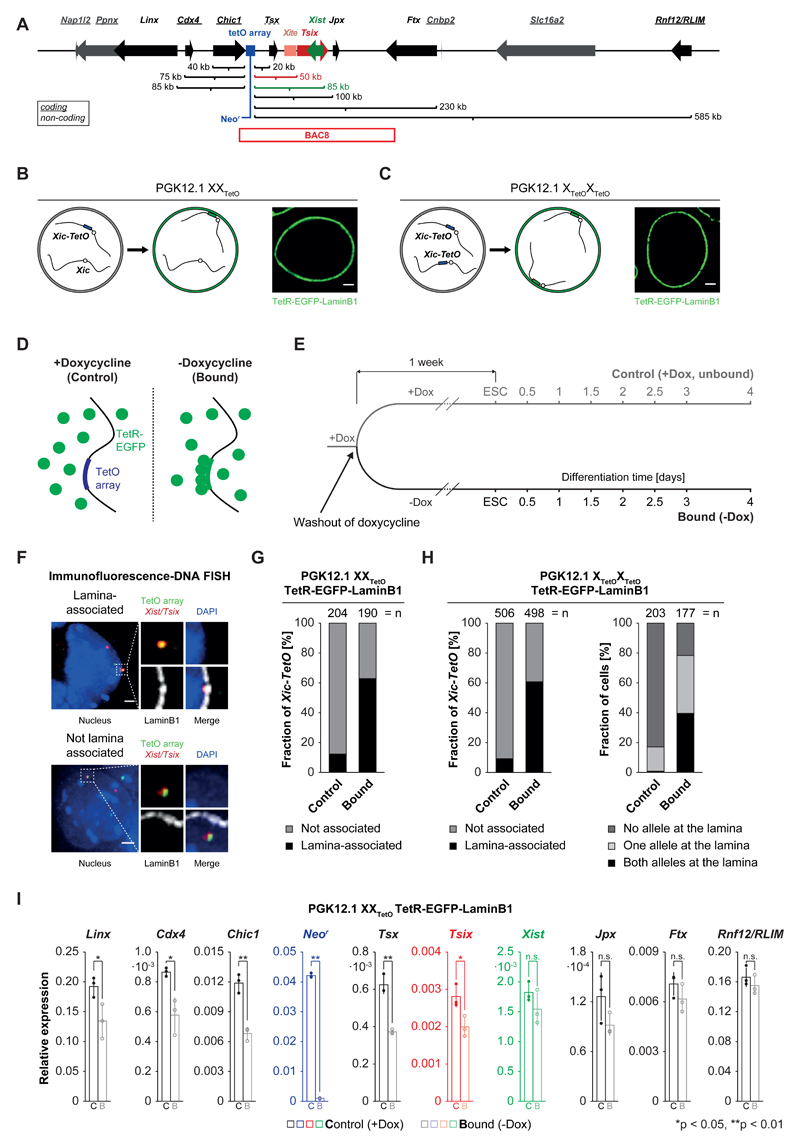
Figure 1. Expression of TetR-EGFP-LaminB1 in PGK12.1 XXTetO/XTetOXTetO cells induces relocalization of the Xic-TetO and gene repression in the relocalized Xic.
A) Schematic representation of the murine X-inactivation center (Xic) with an insertion of a TetO array (blue box, 224 repeats, 11.2 kb). Indicated are the linear genomic distances of the transcriptional start sites of genes in the Xic with respect to the insertion site of the TetO array. Genes giving rise to non-coding transcripts are underlined. The position of BAC8 is indicated below the scheme. B) Schematic representation of the experimental approach for the relocalization of the single Xic-TetO to the nuclear lamina upon expression of a TetR-EGFP-LaminB1 fusion protein in heterozygous PGK12.1 XXTetO cells. The EGFP fluorescence and localization of the fusion protein are detectable in PGK12.1 XXTetO stably expressing the transgene. C) Like B) for PGK12.1 XTetOXTetO cells. D) Schematic representation of binding of TetR fusion proteins to the TetO array in the absence (bound) or presence (control) of doxycycline. E) Scheme of the experimental set up. One population of ‘control’ cells (+Dox) was separated into one population of ‘control’ cells and one population of ‘bound’ cells one week prior to initiation of differentiation. F) Immunofluorescence (IF) DNA fluorescence in situ hybridization (FISH) for the nuclear lamina (anti-LaminB1 IF) and for the TetO array locus and the Tsix/Xist region in the Xic (DNA FISH). Depicted are two representative cells with association (upper panel) or no association (lower panel) of the TetO array locus (Xic-TetO) with the nuclear lamina. Depicted is the entire nucleus as a projection (without the lamina signal) and the region of interest magnified on the right of the nucleus (with the lamina signal). Scored as association was only a direct overlap between the TetO array locus signal and the signal of the nuclear lamina as depicted in the example. Depicted are representative cells for each condition. G) Quantification of association of the single Xic-TetO with the nuclear lamina in PGK12.1 XXTetO ESCs based on IF DNA FISH as depicted in D). (n = number of scored Xic-TetO) H) Quantification of the association of the two Xic-TetO in homozygous PGK12.1 XTetOXTetO for all the Xic-TetO in a population of cells (left panel) and quantification of the association of the two Xic-TetO in single cells (right panel). (n (left) = number of scored Xic-TetO, n(right) = number of cells). I) Mean relative expression of Xic-linked genes in control (empty bars) and bound (empty bars, lighter shade) PGK12.1 XXTetO TetR-EGFP-LaminB1 cells (n = 3), assessed by qRT-PCR relative to the expression level of the Arp0 gene. Individual data points are depicted as filled and empty circles. Significant differences in gene expression are marked (asterisk: p < 0.05; double asterisk: p < 0.01; t-test (unpaired, two-tailed); error bars indicate standard deviation; p-values for genes with p<0.05: Linx 0.037, Cdx4 0.015, Chic1 0.0016, Neor 9x10-8, Tsx: 0.0011, Tsix: 0.028).
Xic localization at the nuclear lamina in control versus bound conditions was assessed by scoring direct overlap of TetO DNA FISH and LaminB1 immunofluorescence in heterozygous XXTetO ESCs (Figure 1F). In unbound (control) conditions, the single Xic-TetO locus co-localizes with the nuclear lamina in 13% (n>200) of XXTetO T-E-LaminB1 ESCs (Figure 1G). As a comparison, frequency of localization of the Xic-TetO to DAPI-dense pericentric heterochromatin or “chromocenters” in control XXTetO T-E-LaminB1 cells was ~10% (Supplementary Figure 1B,C). Following one week of dox washout, over 60% (n=190) of Xic-TetO loci co-localized with the nuclear lamina in bound XXTetO T-E-LaminB1 ESCs (Figure 1G). Thus, in absence of dox, binding of T-E-LaminB1 to the Xic-TetO results in efficient relocalization of the single Xic-TetO to the nuclear lamina of heterozygous XXTetO ESCs. This was also accompanied by a decrease in Xic-TetO chromocenter colocalization (Supplementary Figure 1C) demonstrating that Xic-TetO localization to the lamina also reduced the probability of the locus visiting other subnuclear compartments (Supplementary Figure 1B,C).
We next analyzed the impact of T-E-LaminB1 binding at both Xic-TetO loci in homozygous XTetOXTetO T-E-LaminB1 ESCs by IF-DNA FISH (as above). The total fraction of Xic-TetO loci that co-localized with the nuclear lamina in control (+dox) conditions was similar in XXTetO and XTetOXTetO T-E-LaminB1 ESCs (compare Figure 1G and 1H, n>400). Simultaneous nuclear lamina association of both Xic-TetO loci was rarely observed (1%, n=203) in control (XTetOXTetO T-E-LaminB1) ESCs. However in bound (-dox) XTetOXTetO T-E-LaminB1 ESCs, the two Xic-TetO loci were both at the lamina in 40% of cells (Figure 1H, n=177).
Thus, binding of TetR-EGFP-LaminB1 in ESCs containing either one or two Xic-TetO loci, leads to efficient Xic relocalization to the nuclear lamina in ESCs, allowing us to address the impact of lamina tethering on gene expression and the early steps of random XCI.
Relocalization of the Xic-TetO to the nuclear lamina does not affect Xist’s regulation during differentiation
We first assessed whether relocalization of one Xic-TetO to the nuclear lamina, a supposedly repressive compartment, affects nearby gene expression using qPCR and nascent RNA FISH (see Supplementary Note). Although genes closest to the TetO array (Tsix, Chic1, Tsx) showed some degree of repression upon TetR-EGFP-LaminB1 binding, the effects on other Xic genes (including Xist) were minor and not linearly proportional to the distance from the TetO array (Figure 1A,I). To determine the impact of relocating the Xic-TetO to the nuclear lamina on the initiation of random XCI, we induced differentiation of control and bound XXTetO T-E-LaminB1 ESCs by LIF withdrawal. Xist up-regulation was assayed by FISH for nascent RNA (Xist intron 1 probes, see Online Methods), and spliced, accumulating Xist RNA (Xist exon pool probes)(Figure 2A). Xist RNA steady state levels were also analyzed by qPCR (Supplementary Figure 2). Samples were collected each day for 4 days and several hundred cells counted at each time point (Figure 1E). No differences in the fraction of cells showing monoallelic or biallelic Xist transcription or accumulation could be found in bound versus control XXTetO T-E-LaminB1 cells (Figure 2B,C, n>200). Furthermore, Xist RNA domains were normal in appearance in bound cells (Figure 2A). Expression levels of Xist and pluripotency factors (by qPCR) were similar between bound and control cells at each timepoint (Supplementary Figure 2). In conclusion, neither time of onset, kinetics nor levels of Xist up-regulation were affected when one of the two Xics was relocalized to the nuclear lamina during early ESC differentiation.
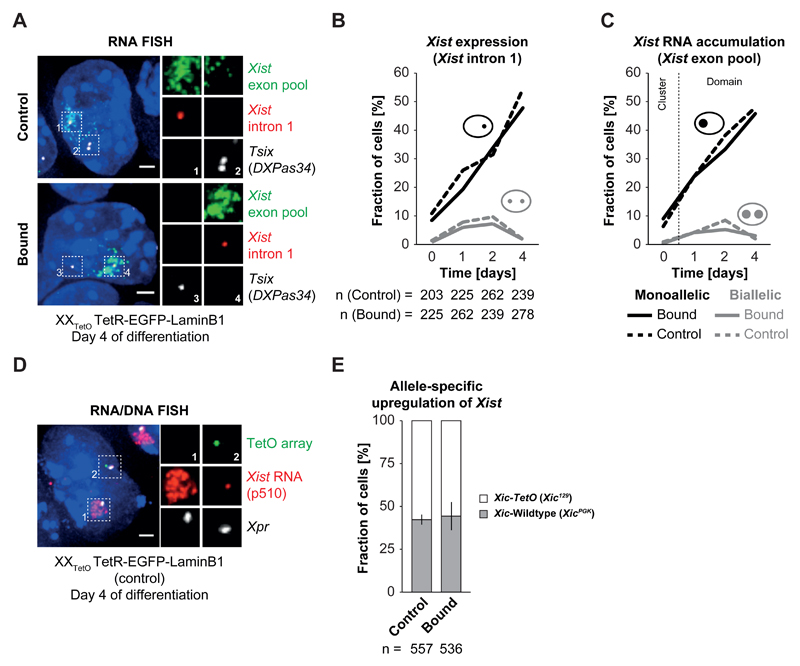
Figure 2. Xist upregulation from the Xic-TetO remains unaffected upon relocalization to the nuclear lamina.
A) RNA FISH for Xist (red) and Tsix (white) nascent transcripts as well as for processed, accumulating Xist RNA (green) in control and bound PGK12.1 XXTetO TetR-EGFP-LaminB1 cells on day 4 of differentiation by LIF withdrawal. Depicted are representative cells for each condition. B) Quantification of Xist expression by presence of nascent Xist RNA as focal signal as depicted in A) in control and bound PGK12.1 XXTetO TetR-EGFP-LaminB1 cells in ESCs and throughout differentiation by LIF withdrawal (n = number of cells). C) Quantification of Xist RNA accumulation by presence of Xist RNA clouds/domains as depicted in A) in control and bound PGK12.1 XXTetO TetR-EGFP-LaminB1 cells in ESCs and throughout differentiation by LIF withdrawal. In ESCs no Xist RNA domains could be detected and only the presence of small clusters of Xist RNA was scored (n = number of cells). D) Simultaneous RNA/DNA FISH for Xist RNA (red) and Xpr (X pairing region, BAC 5 (474E04), white) and the TetO array locus (green) performed on control PGK12.1 XXTetO TetR-EGFP-LaminB1 cells on day 4 of differentiation by LIF withdrawal. Xist RNA is stable during denaturation and can be detected after performing DNA FISH. Depicted is a representative example in which Xist RNA became up-regulated from the wildtype Xic and started coating the wildtype X chromosome. E) Quantification of the distribution of up-regulation of Xist from the Xic-TetO or the wildtype Xic in control and bound PGK12.1 XXTetO TetR-EGFP-LaminB1 cells on day 4 of differentiation by LIF withdrawal based on simultaneous RNA/DNA FISH as depicted in D) (n = total number of cells; mean results of three experiments; error bars indicate standard deviation; for B,C: one representative experiment is depicted in the quantification).
We also assessed whether choice-making was affected in this context, using Xist RNA FISH and Xic/TetO DNA FISH to distinguish the two alleles (Figure 2D) at day 4 of differentiation (see Supplementary Note for controls). By this stage, XCI is established and non-random XCI should be apparent in the overall cell population. Upon binding of TetR-EGFP-LaminB1 to the single Xic-TetO in heterozygous XXTetO T-E-LaminB1 cells, the average ratio of Xist up-regulation from the Xic-TetO versus wildtype Xic was not significantly altered (p > 0.5, Student’s t-test) when compared to control cells (XistXic-TetO : XistXic = 55% : 45%, n>500, Figure 2E), although the variance between the performed experiments was slightly higher for the bound population of cells.
In summary, enforced localization of the Xic-TetO locus to the nuclear periphery neither alters the general capacity of the Xic to up-regulate Xist, nor impacts primary choice-making between the two Xics at the onset of random XCI. This argues against a role for subnuclear localization as a deterministic factor in choice-making during initiation of random XCI.
Relocalization of both Xics at the nuclear lamina reduces the probability of homologous Xic pairing events but does not impact on the onset of XCI
Given these results, we next assessed whether relocalization of the Xic-TetO to the nuclear lamina would affect homologous Xic pairing. We reasoned that moving the Xic to a more peripheral position and restricting its diffusion within the nucleoplasm might influence the frequency with which the two Xics come into close proximity (pair). To test this, we performed DNA FISH for the Xist/Tsix region, which undergoes homologous pairing during early ESC differentiation31–35,37. We measured 3D distances between the two Xics in control and bound XXTetO and XTetOXTetO T-E-LaminB1 ESCs (Supplementary Figure 3A-F). Relocalization of just one Xic to the nuclear lamina, in heterozygous bound XXTetO T-E-LaminB1 ESCs, resulted in a slight, but significant, increase in overall Xic-Xic distances (Supplementary Figure 3B and 3C). This was even more pronounced in homozygous bound XTetOXTetO T-E-LaminB1 ESCs (Supplementary Figure 3E and 3F), shifting to even greater Xic-Xic distance distributions.
We went on to assess whether the frequency of homologous pairing events was affected upon relocalization of one or both Xic-TetO loci to the nuclear lamina using DNA FISH for the Tsix/Xist region in differentiating control and bound XTetOXTetO TetR-EGFP-LaminB1 cells (Figure 3A-D). We measured 3D distances between Xist/Tsix loci at high temporal resolution (0, 0.5, 1, 1.5, 2, 2.5, 3 and 4 days of differentiation) and we counted several hundred (usually n>600) cells at each time point, to ensure that our detection of any differences in Xic pairing frequencies would be robust.
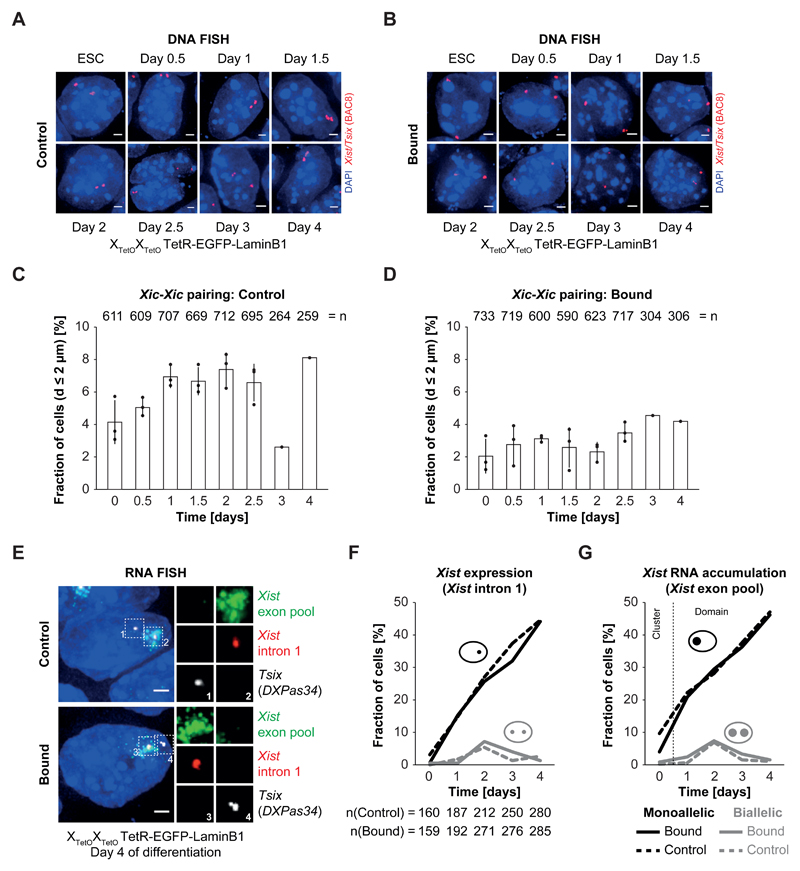
Figure 3. Relocalization and tethering of both Xic in PGK12.1 XTetOXTetO reduces the relative number of Xic trans associations but has no impact on initiation of XCI.
A) + B) DNA FISH for the Xist/Tsix region (red, BAC 8) in control (A) and bound (B) differentiating PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 cells. Depicted are representative cells for each condition. C) + D) Quantification of the mean relative amount of cells with Xic at pairing distance of d < 2 µm in populations of differentiating control (C) and bound (D) PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 cells. The distance between the centers of mass of intensity of the DNA FISH signals was determined in 3D. The displayed relative amount represents the ratio of cells with d < 2 µm and cells with d > 2 µm (n = number of total cells analyzed; mean pairing frequency of three independent experiments for day 0 – day 2.5 of differentiation; error bars indicate standard deviation; one experiment was performed for day 3 and day 4 of differentiation; individual data points are depicted as filled circles). E) RNA FISH for nascent Tsix (white) and Xist (red) transcripts as well as for processed, accumulating Xist RNA (green) in control and bound PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 cells at day 4 of differentiation by LIF withdrawal. Depicted are representative cells for each condition. F) Quantification of Xist expression by presence of nascent Xist RNA as focal signal as depicted in E) in control and bound PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 cells in ESCs and throughout differentiation by LIF withdrawal (n = number of cells). G) Quantification of Xist RNA accumulation by presence of Xist RNA clouds/domains as depicted in E) in control and bound PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 cells in ESCs and throughout differentiation by LIF withdrawal. In ESCs no Xist RNA domains could be detected and only the presence of small clusters of Xist RNA was scored (n = number of cells; for F,G: one representative experiment is depicted in the quantification).
In control XTetOXTetO T-E-LaminB1 cells (+dox), the fraction of cells with Xic-Xic distances of d < 2 µm (defined as Xic pairing35) increased during early differentiation, especially between day 1 and 2.5 and at day 4 of differentiation (Figure 3C). These patterns are similar to those previously seen in wildtype PGK12.1 cells35 (also Panel in Supplementary Figure 3G). We noted that Xic pairing occurs at several time points in a continuous pattern rather than at single discrete time points during differentiation, possibly due to the absence of retinoic acid during differentiation, unlike previous studies. Also, as reported before31 the Xic-Xic distance distribution in the entire population decreased during the first day of differentiation, increased between day 1 and day 1.5/2 and decreased again after day 1.5/2 in a wave-like pattern (Supplementary Figure 3H-J).
When the Xic-TetO loci were relocalized to the nuclear lamina (-dox) in XTetOXTetO T-E-LaminB1 cells, a substantial reduction in Xic-Xic pairing events, compared to unbound controls, was found throughout early differentiation (Figure 3D). We assessed whether this reduction in Xic pairing had an impact on Xist RNA patterns during differentiation. We first analyzed Xist expression (intronic) and Xist RNA accumulation (mRNA) by RNA FISH in early differentiating bound and control XTetOXTetO T-E-LaminB1 cells (Figure 3E). Frequencies of Xist RNA expression or accumulation from one of the two Xics were similar between bound and control XTetOXTetO T-E-LaminB1 cells at all time points (Figure 3F,G, n>150). The appearance of Xist RNA clouds was also similar (Figure 3E). The fraction of cells with aberrant initiation of XCI (biallelic Xist RNA accumulation, no Xist up-regulation) was also comparable between bound and control cells (Figure 3F,G, n>150). Expression of Xist and pluripotency factors (by qPCR) were also comparable between bound and control cells (Supplementary Figure 3K).
Altogether, these results demonstrate that substantial reduction in Xic pairing during ESC differentiation has little impact on Xist up-regulation patterns, frequency and timing in cell populations. Thus, monoallelic Xist up-regulation and Xist/Tsix expression symmetry-breaking are unlikely to result directly from homologous Xic pairing at the Xite/Tsix locus, at least in this system of early ESC differentiation.
Homologous pairing of the Xic is not correlated with a specific transcription event in the Xic but rather biallelic activity of the Xist/Tsix unit
Although the above results indicate that Xic pairing is not a cause of monoallelic Xist/Tsix expression, several lines of evidence have linked Xic pairing with the capacity to initiate XCI and up-regulate Xist31,32. To better understand the basis for this connection, we investigated Tsix and Xist expression during Xic pairing events in differentiating XTetOXTetO cells stably expressing TetR-EGFP53. TetR-EGFP binding to the Xic-TetO loci allowed us to simultaneously assess pairing events and ongoing transcription of Tsix and Xist (intronic probe) by RNA FISH (not previously possible using TetR-mCherry34, see Online Methods for further details).
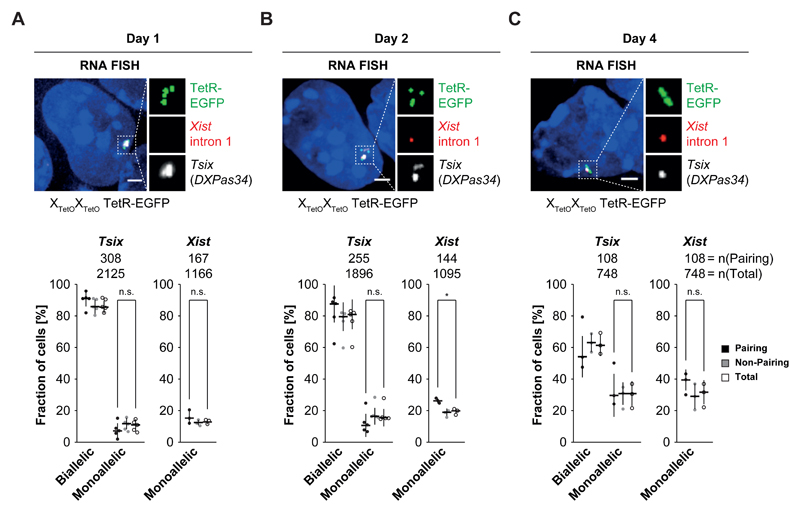
Xic pairing was not specifically associated with a unique expression pattern of Tsix and/or Xist during early differentiation (Figure 4A-C, see Supplementary Note for a more detailed description). Although Xic pairing events usually had biallelic Tsix transcription, different combinations of Xist and Tsix expression were seen, e.g. monoallelic Tsix & biallelic Xist; no Tsix & mono/biallelic Xist. Thus Xic pairing does not seem to be facilitated by a unique Xist/Tsix expression state and pairing does not induce a specific expression pattern. Overall, this data nevertheless indicated that transcription from both Xics (whether sense or antisense, Xist and/or Tsix) occurs at the majority of Xic pairing events suggesting that transcription in the Xic may indeed be linked to its pairing33.
Figure 4. Pairing of the Xic is associated with biallelic Tsix expression or expression of Tsix and Xist.
A) + B) + C) RNA FISH for nascent Xist (red) and Tsix (white) transcripts in differentiating PGK12.1 XTetOXTetO TetR-EGFP cells at day 1 (A), day 2 (B) and day 4 (C) of differentiation. Binding of TetR-EGFP can be detected after RNA FISH as focal green fluorescent signal. Two focal signals per allele indicate the presence of sister chromatids after replication of the Xic. Depicted are three examples of cells with pairing Xic (d < 2 µm) at day 1, day 2 and day 4 after LIF withdrawal. Note the different expression patterns of Tsix and Xist in these cells, demonstrating that no single expression pattern is associated with homologous Xic pairing. The bottom part of panels A), B) and C) depicts the quantification of Tsix and Xist expression in the total population of cells (empty circles), the fraction of cells with non-pairing Xic (grey circles) and cells with pairing Xic (black circles) at day 1, day 2 and day 4 of differentiation based on the presence of focal RNA FISH signals (lines indicate the median). (n = number of cells; * = p < 0.05; t-test (unpaired, two-tailed); scale bar = 2 µm; error bars indicate standard deviation of at least three independent experiments)
To explore this further, we examined Xic pairing in female mESCs harboring a doxycycline-inducible Xist promoter on one of the two X chromosomes (TX1072, Figure 5 and Supplementary Note) and another line carrying a homozygous Xist deletion (ΔXist DKO, Figure 6 and Supplementary Note). Inducing Xist resulted in almost exclusive monoallelic Xist expression, with monoallelic Tsix expression from the other allele (Figure 5C,D and Supplementary Figure 4A). Deletion of Xist from both Xics resulted in mainly biallelic Tsix expression throughout differentiation of ΔXist DKO mESCs (Figure 6B,C). In both situations, Xic pairing frequencies were similar to wildtype cells during early differentiation (Figure 5E,F and Figure 6D,E). Thus, exclusive monoallelic Tsix and Xist expression on the two Xics, or no Xist (only biallelic Tsix) expression, are both still compatible with pairing. This indicates that neither biallelic Tsix expression, nor Xist expression are required per se for Xic pairing to occur; and Xist RNA accumulation has no effect on Xic pairing during early ESC differentiation. Furthermore, Xic pairing in differentiating XXTetO Xist DKO cells does not enable monoallelic Tsix expression as the fraction of cells with monoallelic Tsix is similar at timepoints with high and low frequencies of Xic pairing (Figure 6C,E). In conclusion, Xic pairing was unaffected by any of the XCI initiation events tested: monoallelic Xist up-regulation, monoallelic Tsix down-regulation or no Xist expression and biallelic Tsix. Rather, expression at both Xic loci – whether of Xist and/or of Tsix – seems to be sufficient for Xic pairing to be seen during ESC differentiation.
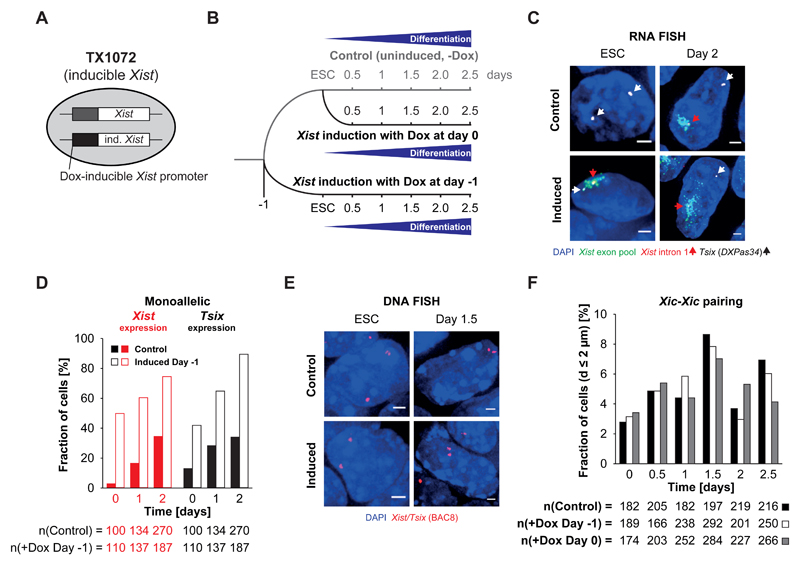
Figure 5. Homologous Xic pairing can occur in the absence of biallelic Tsix expression.
A) Schematic representation of a female ESC line with a doxycycline-inducible promoter regulating the expression of one endogenous Xist allele (TX1072). B) Schematic representation of the initiation of differentiation and induction of Xist expression by addition of doxycycline in TX1072 cells. C) RNA FISH for nascent Xist (red, red arrowheads) and Tsix (white, white arrowheads) transcripts as well as processed and accumulating Xist RNA (green) in undifferentiated TX1072 or TX1072 cells differentiated for 2 days by LIF withdrawal either in the presence or absence of doxycycline in the culture medium. Depicted are representative cells for each condition. D) Quantification of the fraction of cells with monoallelic Tsix expression and monoallelic Xist (intron 1) expression in differentiating TX1072 either not induced with doxycycline or with Xist expression induced one day prior to the initiation of differentiation. E) DNA FISH for the Xist/Tsix region (red, BAC 8) in induced and uninduced TX1072 ESCs and at day 1.5 of differentiation after LIF withdrawal. Depicted are representative cells for each condition. F) Fraction of cells with Xic at pairing distance in differentiating induced and uninduced TX1072 cells. 3D distance was determined after DNA FISH for the Xist/Tsix region (BAC 8) as depicted in D). (n = number of cells, scale bar = 2 µm)
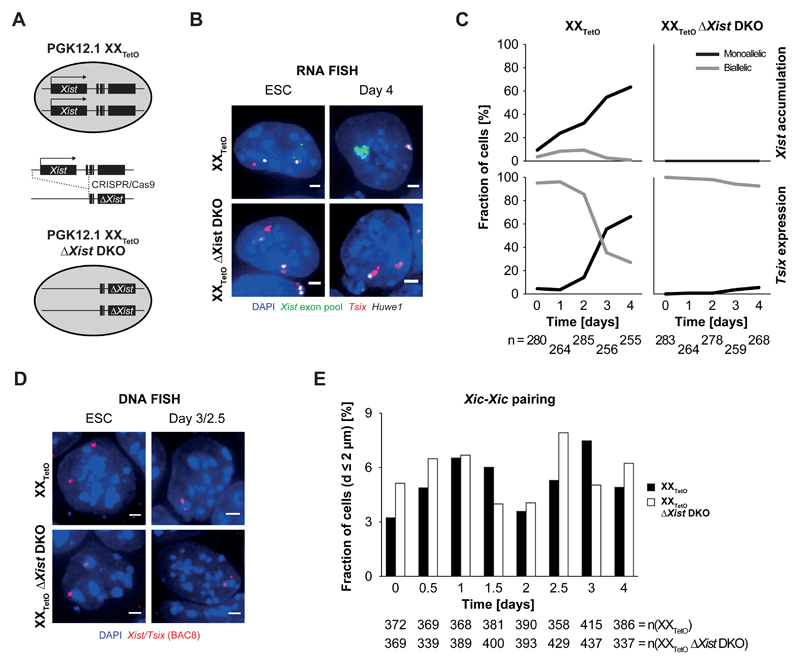
Figure 6. Xist expression is not necessary for homologous Xic pairing.
A) Schematic representation of the derivation of a homozygous Xist double knock-out (DKO) cell line in PGK12.1 XXTetO cells by CRISPR/Cas9-mediated genetic engineering. B) RNA FISH for nascent Tsix (red) and Huwe1 (white) transcripts as well as processed and accumulating Xist RNA (green) performed in PGK12.1 XXTetO (control) and PGK12.1 XXTetO ΔXist DKO in ESCs and at day 4 of differentiation after LIF withdrawal (depicted are representative cells for each condition). C) Quantification of monoallelic and biallelic Xist RNA accumulation as well as monoallelic and biallelic Tsix expression determined by presence of Xist RNA domains/clusters as well as focal signals for Tsix in RNA FISH. D) DNA FISH for the Xist/Tsix region (BAC 8, red) performed in PGK12.1 XXTetO (control) and PGK12.1 XXTetO ΔXist DKO in ESCs and at day 4 of differentiation after LIF withdrawal. Depicted are representative cells for each condition. E) Fraction of cells with Xic at pairing distance in differentiating PGK12.1 XXTetO (control) and PGK12.1 XXTetO ΔXist DKO cells. 3D distance was determined after DNA FISH for the Xist/Tsix region (BAC 8) as depicted in D). (n = number of cells, scale bar = 2 µm)
Loci escaping X-inactivation also undergo homologous pairing during ESC differentiation
Given the above results, we decided to investigate whether biallelic expression of other X-linked loci might be linked to homologous trans associations during early differentiation, as has been suggested for some autosomal loci54. We analyzed 3D distances between homologues of several different X-linked loci by DNA FISH in differentiating female control PGK XTetOXTetO TetR-EGFP-LaminB1 cells. We chose biallelically expressed loci55 (escape from XCI) eg Utx (Kdm6a) and Jarid1c (Kdm5c), and loci that are not expressed (parts of the Rhox cluster), or only at low levels (Dach2) (Figure 7A,B).
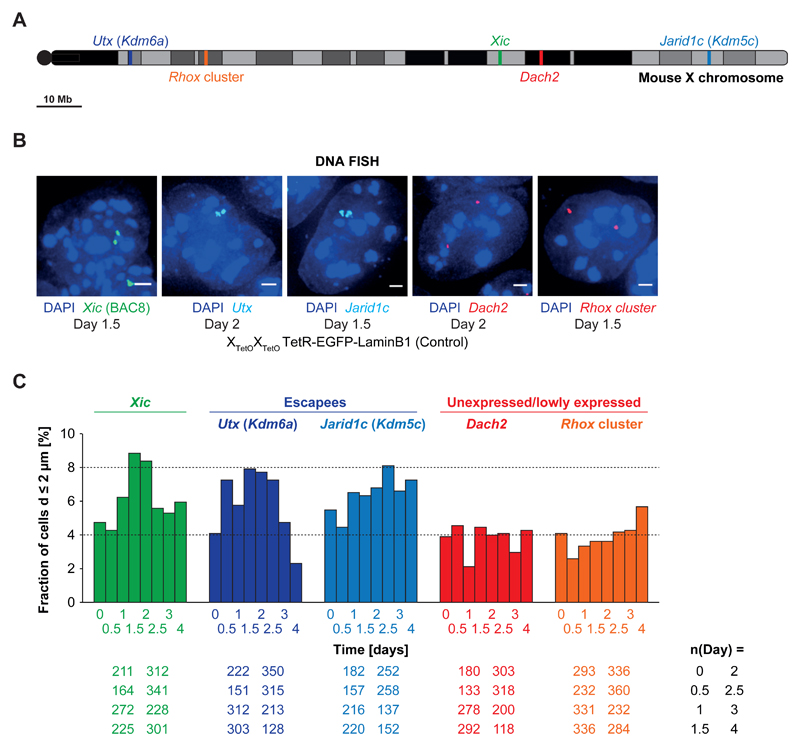
Figure 7. Other biallelically active loci on the X chromosome show homologous pairing.
A) Schematic overview of the position of the analyzed X-linked loci Xic (green), Utx (dark blue), Jarid1c (light blue), Dach2 (red) and the Rhox cluster (orange) on the X chromosome. Dach2 and Rhox7/8 (spanned by the BACs used for the FISH analysis) expression is very low but detectable in undifferentiated female ESCs. Rhox6 and Rhox9 (also spanned by the BAC used for FISH analysis) are expressed at higher levels than Rhox 7/8 (comparable to the lowly expressed Xic gene Chic1). Utx and Jarid1c are well expressed at levels similar to the house-keeping genes Dhfr and G6pdx. B) DNA FISH for the Xic (BAC 8), Utx, Jarid1c, Dach2 and the Rhox cluster in differentiating PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 (control) cells. Depicted are representative cells for each condition. C) Fraction of cells with Xic, Utx, Jarid1c, Dach2 or Rhox loci at pairing distance in differentiating PGK12.1 XTetOXTetO TetR-EGFP-LaminB1 (control) cells. 3D distance was determined after DNA FISH (n = number of cells, scale bar = 2 µm).
As expected, homologous pairing (d < 2µm) at the Xic (Xist/Tsix region) increased during differentiation and peaked at day 1.5/2 (8-9% of the cells, n>200) (Figure 7C). The two escapee loci also displayed homologous pairing that increased during differentiation and peaked at day 1.5/2 for Utx (8%, n>300) and at day 2.5 for Jarid1c (8%, n>250)(Figure 7C). On the other hand, the Rhox cluster and Dach2 showed low frequencies of d < 2µm distances (≤ 4%) (Figure 7C). Furthermore, a locus containing genes which become silenced early on during XCI was previously found to show no significant pairing during female mESC differentiation35. Together, these results suggest that pairing is not restricted to the Xic but can also occur at X-linked loci that are biallelically expressed during early differentiation. Thus homologous trans-associations may be a feature of some biallelically transcribed loci in early differentiating ESCs.
Waves of Xic homologous pairing during early differentiation coincide with the onset of cyclical changes in circadian clock and metabolic genes
We noted that the Xic and escapee loci are expressed biallelically in undifferentiated female ESCs as well as throughout early differentiation, and yet homologous Xic pairing and Xic-Xic distance distributions only occur during differentiation and in a non-linear, rather wave-like pattern (Figures 3C, 5F, 6E, 7C and Supplementary Figure 3J; also see31,34,35). These wave-like patterns did not correlate with patterns of pluripotency factor expression (Oct4, Nanog, Rex1, Esrrb), assayed (by qPCR) at 12h intervals during differentiation (Supplementary Figure 6). However Lamin B (Lmnb), which is implicated in nuclear organization, displayed wave-like changes in mRNA levels (Supplementary Figure 6), very similar in rhythm to Xic-Xic distance distributions (Supplementary Figure 3J). Given these cyclical changes in locus dynamics and gene expression, we analyzed expression of circadian clock genes (Cry1, Cry2, Per1, Per2, Bmal1) and the metabolic Glut8 gene, previously proposed to initiate rhythmic gene expression in differentiating ESCs56,57. Only Per1 and Glut8 showed cyclic fluctuations in expression, and only from day 2 of differentiation (Supplementary Figure 6). Thus the wave-like patterns in Xic pairing and genome dynamics observed during early differentiation of ESCs precede or coincide with, but do not follow, the onset of circadian rhythm patterns.
Discussion
Nuclear localization has been proposed as a mechanism for establishing or maintaining random monoallelic gene expression in processes such as XCI, allelic exclusion of Ig and Tcr genes and olfactory receptor genes31–35,37,41,42,44,45,58,59. Proof of causality between gene positioning and expression has been challenging to obtain. Genetic manipulations (deletions or ectopically inserted transgenes) that point to links between nuclear positioning and gene expression, cannot readily prove causality, as DNA sequence alterations can act through other mechanisms (factor binding, chromatin changes). The TetO/TetR ESC system we used here enabled us to change the subnuclear localization of one or both Xics in a reversible manner without deleting or perturbing regulatory sequences in the Xic. This allowed us to address the causal relationships between subnuclear localization of the Xic, gene expression, homologous Xic pairing and the initiation of random XCI.
By moving the Xic to the nuclear periphery, we found that the nuclear lamina in ESC did not induce complete or stable silencing of nearby endogenous genes of the Xic. Thus, the repressive effect of the nuclear periphery of ESCs and their differentiating derivatives, is less potent than in somatic cells49–52. Although Tsix, Xist’s antisense regulator, was modestly repressed upon relocalization to the nuclear lamina in ESCs, this had no significant effect on Xist regulation, presumably because antisense transcription across Xist continued albeit at a slightly reduced rate. These findings are consistent with previously published data showing that high transcription factor concentrations can override gene silencing at the nuclear lamina49. Furthermore, mechanisms of gene repression at the nuclear periphery in ESCs might differ from somatic cells60.
In this study we also show that Xist up-regulation does not require association with the nuclear lamina and that subnuclear localization at a repressive compartment has no major role in the choice of which Xic will up-regulate Xist and initiate random XCI. Our results suggest that nuclear localization in general is unlikely to be a major determinant in initiation of monoallelic Xic regulation, as the forced relocalization of the Xic to the nuclear lamina simultaneously reduced the likelihood that the Xic would occupy any other parts of the nucleus. It was previously hypothesized that the observed peripheral location of the Xic32, or indeed of the whole X chromosome61, might facilitate the onset of XCI39,40. In fact, positioning at the nuclear periphery (or at the nucleolus) may simply be a consequence of the repetitive nature of the X chromosome. Such positioning may nevertheless facilitate downstream events in XCI, such as maintenance of Xist RNA-mediated gene silencing40 although causality of nuclear position in the context of XCI maintenance remains to be demonstrated.
Pairing of homologous loci has been observed in several experimental systems and many hypotheses about the causes and consequences of this phenomenon exist (reviewed in 62). Our system enabling relocalization of both Xics to the nuclear lamina allowed us to show that reducing Xic pairing had no impact on the monoallelic regulation of Xist during differentiation. Thus homologous Xic pairing at the level of the Xite/Tsix region, does not play an important role in random XCI, at least during in vitro ESC differentiation. The fact that Xic pairing events occur reproducibly during the exact time window of random XCI initiation and the previous studies involving Xic transgenes and deletions31,33,35 that linked pairing with Xic choice-making, nevertheless pointed to an intimate connection between this phenomenon and the capacity to induce normal, random XCI. Our data suggests that expression from both Xist/Tsix alleles during early ESC differentiation appears to be one of the criteria underlying Xic transient homologous trans-association events. The notion that trans-associations might be facilitated by gene expression (or factors involved in it), previously proposed for some biallelically expressed autosomal loci54,63–65, is further supported here by our discovery that loci escaping XCI, but not monoallelic or lowly expressed loci can associate in trans.
Another intriguing question is why Xic pairing events occur only in early differentiating cells, but not in undifferentiated ESCs or in somatic cells. Live cell imaging previously revealed that during early differentiation, genomic loci (not just the Xic) can be particularly dynamic, even more so than in undifferentiated ESCs34, and homologues might be more likely to encounter each other. Transcription of two homologous alleles, with similar binding of transcription and chromatin factors, might favor prolonged associations following chance encounters in a dynamic nuclear context. A role for transcription factors and transcripts was also recently proposed in the context of PAR and Xic pairing66. Indeed, transcription and CTCF binding to Tsix have been proposed to facilitate pairing of the Xic67. Whatever the molecular mediators of trans associations, our data suggests that trans associations might be stabilized when the two regions involved are transcribed, whilst upon loss of expression, loci might dissociate. This would be in line with our previous observations involving live-imaging followed by RNA FISH, where Tsix was found to be monoallelic in a high fraction of cells shortly after homologous pairing had occurred34.
A final intriguing feature of Xic pairing is that Xic-Xic distance distributions change in a wave-like pattern during differentiation, with more frequent pairing at discrete time points (days 1-2 and 4 of differentiation)31,34,35. We find that expression of some genes related to nuclear architecture (Lmnb), circadian rhythm (Per2) and metabolism (Glut8) show similar temporal dynamics to pairing (Supplementary Figure 6). Although circadian rhythms do not exist in ESCs57, circadian rhythm genes may become up-regulated during ESC differentiation. In light of these considerations, the observed changes in chromosome dynamics and the waves of pairing at transcribed loci during early ESC differentiation may occur in the context of a more global (non-linear) regulatory framework being set up in development. Indeed, it has previously been shown that contact maps in trans can change with circadian rhythms68 (eg clock-dependent gene Dbp in mouse embryonic fibroblasts). Homologous pairing events of the X chromosomes and other loci might thus be driven by these general changes in cell physiology that occur in a wave-like pattern upon the initiation of differentiation.
In conclusion, we provide functional evidence that positioning at the lamina and homologous pairing between the Xic loci are not critical determinants of XCI during early ESC differentiation. We do not exclude that homologous pairing has a role at later stages of XCI. Indeed the fact that Xic pairing occurs in multiple waves during differentiation indicates that it may provide a fail-safe strategy to ensure XCI is accurately under way. We also make the unexpected discovery that during early differentiation, cyclical chromosome-wide and local DNA dynamics, including Xic pairing, are accompanied by wave-like patterns of circadian and metabolic gene expression. Although causality has so far not been found, this opens up the exciting possibility that chromosome and nuclear dynamics during early development may be linked to the progressive onset of the circadian clock and to appropriate metabolic programmes.
Online Methods
Cell lines
The female PGK12.1 ESC line was a gift from the laboratory of N. Brockdorff69. PGK12.1 XXTetO and PGK12.1 XTetOXTetO were previously described34. TX1072 cells, which carry a doxycycline responsive promoter upstream the Xist gene transcriptional start site on one X chromosome were previously described70. A homozygous deletion of Xist has also been introduced into PGK12.1 XXTetO cells by CRISPR/Cas9 mediated genetic engineering (see below).
Embryonic stem cell culture and differentiation
Feeder-free embryonic stem cells were essentially cultured as described previously53,70. In brief, ESCs were grown in flasks or dishes pre-coated with 0.1% (w/v) gelatin in PBS in serum-containing ESC medium (15% fetal bovine serum (FBS, Gibco), 0.1 mM 2-mercaptoethanol (Gibco), 1000 U/ml leukemia inhibitory factor (LIF, Chemicon) in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma)). The culture medium was supplemented or not with the following components: doxycycline (1 µg/ml doxycycline (dox, Sigma), 2i components (3µM GSK-3 inhibitor XVI CT-99021 (R&D System) and 1 µM MEK inhibitor PD0325901 (Cliniscience)) and antibiotics (250 µg/ml hygromycin B (Invitrogen)). ESCs were differentiated by LIF withdrawal. Prior to the initiation of differentiation, cells were seeded at a density of 1-1.5x104 cells/cm2 into the wells of 6-well plates. For the initiation of differentiation cells were first washed three times with PBS without resuspension and then cultured in differentiation medium (10% FBS (Eurobio), 0.1 mM 2-mercaptoethanol in DMEM). For imaging purposes cells were seeded and cultured on glass coverslips, which were sterilized by ethanol-flaming and pre-coated with gelatin.
Cell lines stably expressing TetR-EGFP-LaminB1 were cultured in doxycycline containing medium. For each experiment involving a ‘control’ and ‘bound’ population, cells were trypsinized, washed three times in PBS in suspension and split into a population cultured in doxycycline containing medium (referred to as ‘control’) and a population cultured in medium without doxycycline (‘bound’). In order to compromise between incomplete TetR binding after doxycycline washout and avoidance of putative long-term effects of TetR binding as well as to avoid differences by keeping the two ESC populations separate for too long, we performed fixations and initiations of differentiation (see above) one week after doxycycline washout.
Gene expression constructs
Gene expression constructs for the expression of TetR-EGFP fusion proteins are based on the pBROAD3 expression vector (Invivogen) containing a mouse ROSA26 promoter. Plasmid maps can be found in the supplementary information accompanying this publication. Plasmid sequences are available on request. TetR fusion protein constructs were generated based on the TetR fusion protein construct previously generated by Osamu Masui34. The pBROAD3-TetR-EGFP fusion plasmid was generated by exchange of the mCherry coding sequence in pBROAD3-TetR-mCherry34 with the EGFP coding sequence amplified by PCR with respective overhangs (BamHI/EcoRI) for cloning from pEGFP-N1 (Clontech). The pBROAD3-TetR-EGFP-LaminB1 fusion plasmid was generated from a hybrid plasmid containing the coding sequence of a TetR-EGFP-mCherry by insertion of the EGFP coding sequence amplified from pEGFP-N1 with the respective overhangs for cloning into the original vector (BglII/BamHI). The LaminB1 coding sequence was amplified by PCR with the respective overhangs for cloning (BglII/NheI) from pTRE-EYFP-LaminB1 (gift from Andrew Belmont) and exchanged in frame against the coding sequence of mCherry in pBROAD3-TetR-EGFP-mCherry. The plasmid p10L7 (same as 34) provided the hygromycine resistance for the generation of stable ESC lines.
Generation of transgenic ESC cell lines
PGK12.1 XXTetO and XTetOXTetO ESC lines stably expressing TetR-EGFP fusion proteins were generated as described before53. Female ESCs were seeded into the wells of a 6-well plate pre-coated with 0.1% gelatin and grown to a confluence of 70-80%. The plasmids containing the coding sequence for the respective fusion protein were mixed with plasmids containing the respective selectable marker at a ratio of 15:1 and 3 µg of the plasmid mix used for the lipofection of one well of a 6-well plate. Lipofection of the plasmids was performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. One day after transfection different dilutions of the cells were split into 10-cm dishes pre-coated with 0.1% gelatin. Selection with the respective antibiotic was started 1 day after transfer to 10-cm dishes. Approximately 1 week after transfer into 10-cm dishes and selection single colonies could be picked and transferred into 96-well plates. Upon reaching optimal density in 96-well plates, cells were split into a new 96-well plate and a 96-well plate with glass bottom. The selected clones were screened by live-cell imaging in 96-well plates with glass bottom for expression of the respective TetR-EGFP fusion protein. Clones were selected based on expression level, localization of the TetR-EGFP fusion protein and expression mosaicism in the population of cells. Selected clones were amplified and analyzed by RNA and DNA FISH for presence of two X chromosomes. Cells expressing TetR-EGFP-LaminB1 were cultured in the presence of hygromycin B and doxycycline. During differentiation no antibiotics were supplemented to the differentiation medium.
RNA and DNA fluorescence in situ hybridization
RNA and DNA FISH, imaging and FISH signal distance and intensity measurements were performed essentially as described before35,53,71,72.
ESCs and their differentiating derivatives were grown on glass coverslips. The cells were washed once with PBS and fixed in 3% paraformaldehyde (Panreac) in PBS for 10 minutes at room temperature. Cells were washed three times with PBS and permeabilized for 5 minutes on ice in ice-cold permeabilization buffer (0.5% (v/v) Triton-X-100 (Euromedex), 2 mM vanadylribonucleoside complex (New England Biolabs) in PBS), washed 3 times in 70% (v/v) ethanol and stored at -20°C in 70% ethanol.
A list of the RNA and DNA FISH probes used for this study can be found in Supplementary Table 2. Plasmid, fosmid and BAC-derived probes were labeled by nick translation (Abbot) following the manufacturer’s instructions. The probes were either ethanol-precipitated or dried and resuspended in an appropriate amount of formamide while shaking at 37°C in a Thermomixer (Eppendorf). BAC- and fosmid-derived probes were co-precipitated with mouse Cot-1 DNA (Invitrogen) and competition to block repetitive sequences was performed after denaturation at 75°C for 10 minutes for at least 1 hour at 37°C, before mixing the probes with 1 volume of 2x hybridization buffer (40% dextran sulfate (w/v), 2 µg/µl bovine serum albumin, 20 mM vanadyl ribonucleoside complex in 4x SSC; after mix with probes in formamide: 50% formamide, 20% dextran sulfate, 1 µg/µl bovine serum albumin, 10 mM vanadyl ribonucleoside complex in 2x SSC). Probes that did not require competition were denatured at 75°C for 10 minutes and stored on ice until mixing with 1 volume of 2x hybridization buffer.
Prior to RNA and DNA FISH the stored coverslips were desiccated by sequential incubation in 70%, 80%, 95% and 2x 100% ethanol for 5 minutes shaking at room temperature. The coverslips were air-dried completely and lowered onto a drop of the probe/hybridization buffer mix and incubated over night at 37°C for RNA FISH. For DNA FISH the coverslips were washed 3 times in 2x SSC and incubated for 1 hour at 37°C in 2x SSC supplemented with 0.1 mg/ml RNase A (Fermentas) and 10 U/ml RNase H (New England Biolabs). After the RNase treatment the coverslips were desiccated again as described above. Prior to the hybridization the cells on coverslips were denatured for 36-38 minutes (denaturation time was established through a series of prior tests as described in 53) at 80°C in 50% formamide in 2x SSC (pH7.4 at room temperature), washed 3 times in ice-cold 2x SSC and then lowered onto a drop of the probe/hybridization buffer mix and incubated over night at 42°C. The next day the coverslips were washed 3 times at 42°C/45°C in 50% formamide in 2x SSC (pH7.4 at room temperature) and 3 times at 42°C/45°C in 2x SSC. Nuclei were counterstained with DAPI and the coverslips mounted and cells imaged.
For immunofluorescence DNA FISH, coverslips were not dessicated but transferred and washed three times in PBS. The coverslips were incubated for at least 15 minutes in blocking buffer (1% BSA in PBS) and then lowered on a drop of primary antibody solution (antibody in blocking buffer, see list of used antibodies in Supplementary Table 3) and incubated at room temperature for 1 hour. The coverslips were then washed 3 times for 5 minutes at room temperature and incubated for 1 hour with the secondary antibody in the dark (see list with secondary antibodies in Supplementary Table 3). The coverslips were washed three times in PBS at room temperature and the cells fixed for 2 minutes in 3% PFA. After the post-fixation the cells were washed 3 times in PBS and 3 times in 2x SSC and denatured as described above for DNA FISH.
Cells were imaged using a Deltavision Core fluorescent microscope equipped with a CoolSNAP HQ2 camera and a 60x or 100x PlanApo oil immersion objective. 3D image stacks were analyzed using custom-made ImageJ routines as described elsewhere35,71,73.
Quantitative real-time PCR
To isolate RNA, 1 ml Trizol was added to the cells in the well of a 6-well plate after washing the cells once in PBS. The Trizol suspension was transferred to a reaction tube and 200 µl chloroform were added, the mix vortexed for 10-20 seconds and centrifuged for 15 minutes at 4°C and 12,000xg. The upper aqueous solution was transferred to a new reaction tube and mixed with 1 volume 70% ethanol. RNA was isolated from the solution by using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instruction including an on-column DNase digestion (Qiagen) step.
Reverse transcription of isolated RNA was carried out using the SuperScript III Reverse Transcriptase Kit (Invitrogen) using random primers (Invitrogen) according to the manufacturer’s instruction.
Quantitative real-time PCR was carried out using the SYBR green PCR Master Mix (Applied Biosystems) following the manufacturer’s instructions. Reactions were carried out using a ViiA7 real time PCR system (Applied Biosystems). Relative expression levels were normalized as indicated based on the expression level of Rplp0 (Arp0), Rrm2 and beta-Actin (a list of primers can be found in Supplementary Table 1).
Generation of the Xist double knock-out using the CRISPR/Cas9 approach
An approximately 18 kb large deletion was introduced into both alleles of PGK12.1 XXTetO cells using the CRISPR/Cas9 system74.
The pairs of oligonucleotides containing the target sequences for the guide RNAs were cloned into pSpCas9(BB)-2A-puro (pSpCas9(BB)-2A-Puro (PX459) was a gift from Feng Zhang (Addgene plasmids #48139 & #62988)). In brief, the vector was digested with BbsI and the linearized vector gel purified using the Qiagen gel extraction kit following the manufacturer’s instructions. 0.1 µmol of each oligonucleotide (see primer list) containing the target sequence were phosphorylated in a 10 µl reaction using T4 polynucleotide kinase (NEB) in the respective buffer for 30 minutes at 37°C in a Thermocycler (Eppendorf). Oligonucleotide pairs were then annealed after denaturation at 95°C for 5 minutes by cooling down at 5 K per minute to 25°C. 1 µl of a 1:200 dilution of annealed and phosphorylated oligonucleotides was used in a 11 µl ligation reaction containing 50 ng of BbsI digested vector, 1x Quick Ligation Buffer (NEB) and 1 µl Quick Ligase (NEB). The ligation reaction was incubated for 10 minutes at room temperature and subjected to standard transformation into chemically competent bacteria. Ligation of the target sequence into pSpCas9(BB)-2A-puro was confirmed by sequencing.
pSpCas9(BB)-2A-puro plasmids containing the target sequences as well as a Cas9 expression cassette were delivered into mESCs using nucleofection. In brief, mESCs were trypsinized and counted. 5x106 cells were pelleted in a 1.5-ml reaction tube, resuspended in nucleofection mix (Lonza) containing nucleofector solution, supplement 3 and 2.5 µg of each plasmid (5 µg total) and transferred to a nucleofection cuvette. Nucleofection was performed using the mESC settings on a 4D-Nucleofector system (Lonza). The cells in the nucleofection mix were resuspended in 1 ml pre-warmed ESC culture medium and transferred into 9 ml of pre-warmed ESC culture medium in a 15-ml polypropylene tube, mixed and seeded onto 10-cm tissue culture dishes in various dilutions. Cells were grown for one day in ESC culture medium. After one day the medium was exchanged against ESC culture medium containing puromycin (1 µg/ml, Sigma). Cells were grown for one day in selection medium before the medium was changed to regular ESC culture medium without puromycin. Cells were cultured until single ESC colonies were detectable by eye, which were then picked and transferred into 96-well plates (similar to what has been described above for the generation of stable cell lines).
Single clones were screened for deletion by PCR after extraction of genomic DNA using genotyping primer indicating the presence of wildtype alleles or deleted alleles (see primer list). A clone homozygous for the 18-kb deletion in Xist was selected (PGK N-TetO #106 ΔXist TSS-intron 3 clone A9), amplified and subjected to further analysis. Deletion of the Xist allele was confirmed by sequencing. Presence of both X chromosomes was confirmed using DNA FISH. Loss of both Xist alleles was further confirmed by the lack of Xist expression/up-regulation during differentiation using RNA FISH and qPCR.
Supplementary Material
Acknowledgements
We would like to thank the Heard lab for helpful discussions and input. This work was supported by a Institut Curie PhD student fellowship and a fellowship from Fondation ARC (Aides individuelles jeunes chercheurs) to T. Pollex; E. Heard is supported by ERC Advanced Investigator award (ERC-2014-AdG no. 671027), Labelisation La Ligue, FRM (grant DEI20151234398), ANR DoseX 2017, Labex DEEP (ANR-11-LBX-0044), part of the IDEX Idex PSL (ANR-10-IDEX-0001-02 PSL), and ABS4NGS (ANR-11-BINF-0001).
Footnotes
Data availability statement
All data generated or analyzed during this study are included in this published article (and its supplementary information). The raw datasets generated during and or analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
T.P. and E.H. conceived the project and designed the experiments. T.P. carried out all experiments and analyzed the data. T.P. and E.H. wrote the manuscript.
Competing interests
The authors declare no competing financial interest.
References
- 1.Rastan S, Robertson EJ. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- 2.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 3.Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Pollex T, Heard E. Recent advances in X-chromosome inactivation research. Curr Opin Cell Biol. 2012;24:825–832. doi: 10.1016/j.ceb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 7.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 8.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 9.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto I, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 11.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 12.Borensztein M, et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol. 2017;24:226–233. doi: 10.1038/nsmb.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi N, Abe K. Detrimental effects of two active X chromosomes on early mouse development. Development. 1990;109:189–201. doi: 10.1242/dev.109.1.189. [DOI] [PubMed] [Google Scholar]
- 14.Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 15.Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21:8512–8520. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci USA. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Navarro P, Page DR, Avner P, Rougeulle C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev. 2006;20:2787–2792. doi: 10.1101/gad.389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 21.Debrand E, Chureau C, Arnaud D, Avner P, Heard E. Functional analysis of the DXPas34 locus, a 3' regulator of Xist expression. Mol Cell Biol. 1999;19:8513–8525. doi: 10.1128/mcb.19.12.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 23.Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 24.Navarro P, et al. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 25.Navarro P, et al. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 26.Jonkers I, et al. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Barakat TS, et al. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 2011;7:e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gontan C, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 29.Barakat TS, et al. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014;53:965–978. doi: 10.1016/j.molcel.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Schulz EG, Heard E. Role and control of X chromosome dosage in mammalian development. Curr Opin Genet Dev. 2013;23:109–115. doi: 10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Xu N, Tsai C-L, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 32.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 33.Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 34.Masui O, et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Augui S, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 36.Sun S, Fukue Y, Nolen L, Sadreyev R, Lee JT. Characterization of Xpr (Xpct) reveals instability but no effects on X-chromosome pairing or Xist expression. Transcription. 2010;1:46–56. doi: 10.4161/trns.1.1.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comings DE. The rationale for an ordered arrangement of chromatin in the interphase nucleus. Am J Hum Genet. 1968;20:440–460. [PMC free article] [PubMed] [Google Scholar]
- 39.Heard E, Chaumeil J, Masui O, Okamoto I. Mammalian X-chromosome inactivation: an epigenetics paradigm. Cold Spring Harb Symp Quant Biol. 2004;69:89–102. doi: 10.1101/sqb.2004.69.89. [DOI] [PubMed] [Google Scholar]
- 40.Chen C-K, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 41.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 42.Skok JA, et al. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 43.Brown KE, et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 44.Skok JA, et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 45.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 47.Fukaya T, Levine M. Transvection. Curr Biol. 2017;27:R1047–R1049. doi: 10.1016/j.cub.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim B, Heist T, Levine M, Fukaya T. Visualization of Transvection in Living Drosophila Embryos. Mol Cell. 2018;70:287–296.e6. doi: 10.1016/j.molcel.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 52.Dialynas G, Speese S, Budnik V, Geyer PK, Wallrath LL. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–3077. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollex T, Piolot T, Heard E. Live-Cell Imaging Combined with Immunofluorescence, RNA, or DNA FISH to Study the Nuclear Dynamics and Expression of the X-Inactivation Center. Methods Mol Biol. 2013;1042:13–31. doi: 10.1007/978-1-62703-526-2_2. [DOI] [PubMed] [Google Scholar]
- 54.Krueger C, et al. Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PLoS ONE. 2012;7:e38983. doi: 10.1371/journal.pone.0038983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chow JC, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 56.Paulose JK, Rucker EB, Cassone VM. Toward the Beginning of Time: Circadian Rhythms in Metabolism Precede Rhythms in Clock Gene Expression in Mouse Embryonic Stem Cells. PLoS ONE. 2012;7:e49555. doi: 10.1371/journal.pone.0049555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yagita K, et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci USA. 2010;107:3846–3851. doi: 10.1073/pnas.0913256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armelin-Correa LM, Gutiyama LM, Brandt DYC, Malnic B. Nuclear compartmentalization of odo…. Proceedings of the National Academy of Sciences. 2014;111:2782–2787. doi: 10.1073/pnas.1317036111. [Proc Natl Acad Sci U S A. 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo L, et al. The nuclear periphery of embryonic stem cells is a transcriptionally permissive and repressive compartment. J Cell Sci. 2009;122:3729–3737. doi: 10.1242/jcs.052555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyle S, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 62.Joyce EF, Erceg J, Wu C-T. Pairing and anti-pairing: a balancing act in the diploid genome. Curr Opin Genet Dev. 2016;37:119–128. doi: 10.1016/j.gde.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 64.Brown JM, et al. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu H-P, et al. PAR-TERRA directs homologous sex chromosome pairing. Nat Struct Mol Biol. 2017;24:620–631. doi: 10.1038/nsmb.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kung JT, et al. Locus-Specific Targeting to the X Chromosome Revealed by the RNA Interactome of CTCF. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguilar-Arnal L, et al. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norris DP, et al. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 70.Schulz EG, et al. The Two Active X Chromosomes in Female ESCs Block Exit from the Pluripotent State by Modulating the ESC Signaling Network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 71.Giorgetti L, Piolot T, Heard E. High-Resolution 3D DNA FISH Using Plasmid Probes and Computational Correction of Optical Aberrations to Study Chromatin Structure at the Sub-megabase Scale. Methods Mol Biol. 2015;1262:37–53. doi: 10.1007/978-1-4939-2253-6_3. [DOI] [PubMed] [Google Scholar]
- 72.Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol. 2008;463:297–308. doi: 10.1007/978-1-59745-406-3_18. [DOI] [PubMed] [Google Scholar]
- 73.Giorgetti L, et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.