Abstract
A primary calcium (Ca2+) entry pathway into non-excitable cells is through the store-operated Ca2+ release activated Ca2+ (CRAC) channel. Ca2+ entry into cells is responsible for the initiation of diverse signalling cascades that affect essential cellular processes like gene regulation, cell growth and death, secretion and gene transcription. Upon depletion of intracellular Ca2+ stores within the endoplasmic reticulum (ER), the CRAC channel opens to refill depleted stores. The two key limiting molecular players of the CRAC channel are the stromal interaction molecule (STIM1) embedded in the ER-membrane and Orai1, residing in the plasma membrane (PM), respectively. Together, they form a highly Ca2+ selective ion channel complex. STIM1 senses the Ca2+ content of the ER and confers Ca2+ store-depletion into the opening of Orai1 channels in the PM for triggering Ca2+-dependent gene transcription, T-cell activation or mast cell degranulation. The interplay of Orai and STIM proteins in the CRAC channel signalling cascade has been the main focus of research for more than twelve years. This chapter focuses on current knowledge and main experimental advances in the understanding of Orai1 activation by STIM1, thereby portraying key mechanistic steps in the CRAC channel signalling cascade.
Keywords: CRAC, Orai, STIM, Ca2+ signaling
1. Introduction
As diverse signalling pathways depend on Ca2+ levels within cells, the refilling of the intracellular stores in the ER is a crucial step in Ca2+ signalling. More than 30 years ago Putney et al proposed a concept of a Ca2+ entry pathway that is activated upon store-depletion of these internal Ca2+ reservoirs [1]. Today this Ca2+ influx pathway is referred to as store-operated Ca2+ entry (SOCE), which can be observed in a variety of cell types including lymphocytes, pancreatic acinar, vascular endothelial and smooth and skeletal muscle cells [2–6]. Under physiological conditions, SOCE is initiated upon Ca2+ release from the ER lumen into the cytoplasm after receptor activation. Inositol-1,4,5-triphosphate (IP3) is produced upon activation of receptors that are coupled to phospholipase C (PLC) stimulation which hydrolyses phosphatidylinositol-4, 5-bisphosphate (PIP2) [7,8]. Subsequently, IP3 binds to its receptor in the ER membrane, thereby initiating Ca2+ release [9,10]. The Ca2+ release activated Ca2+ (CRAC) channel is the best-characterized store-operated channel (SOC) and its currents are defined by a set of characteristic features [5,11].
Decades after Putney’s concept of store-operated Ca2+ entry, the two key players of the CRAC channel complex have been identified: STIM1 and Orai1 [12–16]. Genetic screens by RNAi have revealed that STIM1 constitutes a functional component of the CRAC channel [12,13] and senses the Ca2+ content of the ER. Orai1 has been shown to play an important role in T-cell effector functions due to the fact that a single missense mutation (R91 W) abrogates CRAC channel function, which leads to a severe immunodeficient phenotype (SCID – severe combined immunodeficiency) [16].
Ca2+ ions enter the cell upon physical interaction of STIM1 with Orai1 and act as second messengers to sustain Ca2+ oscillations, to maintain Ca2+ homeostasis and to provide long-term Ca2+ signals. These long-term Ca2+ signals regulate a plethora of signalling cascades including cytokine secretion, mast cell degranulation, T-cell differentiation into effector subsets, B-cell differentiation into plasma cells, lysis of cancerous or infected target cells by cytolytic T-cells, cell proliferation or gene transcription [14–25].
In the following sections we will specify CRAC channel proteins and describe key findings that have established our current knowledge of the Orai1 activation process by STIM1.
2. Key players in the CRAC signalling cascade
The STIM1 protein is uniformly distributed and shows constitutive movement along microtubules within the ER when Ca2+ stores are full. During store-depletion, bound Ca2+ ions dissociate from the luminal EF hand of STIM1, a signal that triggers STIM1 to oligomerize and translocate to ER-PM junctions, leading to puncta at sites near the PM [18,26–28]. The pore-forming subunit of the CRAC channel in the PM, Orai1, accumulates at sites opposite to STIM1. Thus, high proximity between these two proteins allows for direct interaction and leads to CRAC channel activation and subsequent refilling of internal Ca2+ stores [18,28,29].
3. Characteristics of Orai proteins
There are three members of the Orai family (Orai1 – 3) in mammals, each within selected tissues exhibiting elevated expression [19,30] and each of them forming a hexameric channel complex with a highly Ca2+ selective pore in the PM [31–33].
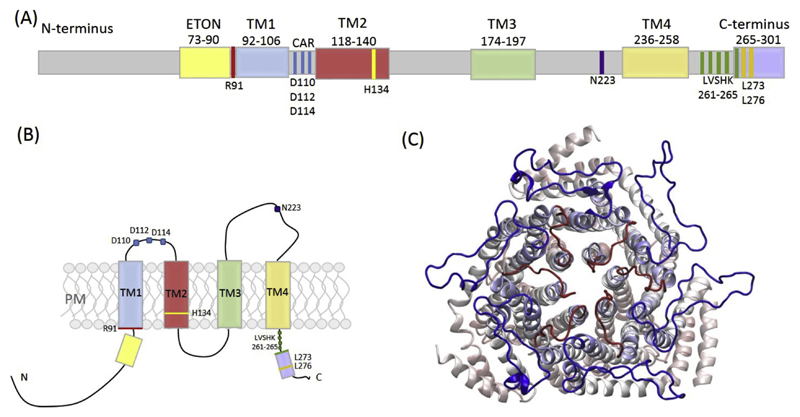
Orai1 is a 33 kDa protein (composed of 301 amino acids) containing four transmembrane domains (TM) with both the N-terminus and the C-terminus residing in the cytosol [16] (Fig. 1A, B). The hexameric assembly of the closed Orai channel of Drosophila melanogaster, which shows 73% sequence identity to hOrai1, has been resolved by X-ray crystallography in 2012 [32]. This structure, achieved with truncated NH2 and COOH termini (residues 132–341 in dOrai1 (corresponding residues 60–296 in hOrai1) used for the crystal structure), the mutation of two non-conserved cysteine residues (dOrai C224,283 T; hOrai1 S153,T240) and a mutated loop3 (dOrai1 P276,277R; corresponding residues in hOrai1 K204,Q205), has revealed that six Orai monomers are arranged around a central axis, in which the TM1 of each subunit forms the inner ring of the pore. The remaining three TMs are arranged in concentric rings around the ion conducting pore (Fig. 1C).
Fig. 1.
(A) Cartoon of full-length hOrai1 depicting the overall structure with important regions highlighted. (B) Schematic representation of a single hOrai1 monomer with the 4 transmembrane (TM) regions, N- and C-terminus and highlighted important residues like in (A). (C) Top view of a modelled hOrai1 hexameric structure showing TM1 (inner ring-pore), TM2-4, loop1 (red) and loop3 (blue) representing a closed channel.
The cytosolic C-termini of the TM4 helices have been described as arranged in pairs, with the helices bending in the opposite directions to form coiled-coil interactions with other Orai subunits. It has been shown that mutations in the coiled coil regions of hOrai1 (L273 and L276) lower the coiled-coil probability and inhibit STIM1 binding and channel activation [28,34,35]. Quite recently, the Long lab managed to crystallize the Drosophila melanogaster Orai channel in its open conformation [36]. By using a gain-of-function (GOF) mutation in Orai (dOrai H206 A corresponding to hOrai1 H134 A, which switches the channel into a constitutively open conformation as shown by Frischauf et al in 2017 [37]) they have been able to identify that the pore dramatically dilates on the cytosolic end being ~10 Å apart at K159 (corresponding to K87 in hOrai1) upon channel opening. This opening requires the release of cytosolic latches with P288 (P245 in hOrai1) and the SHK motif (aa263,264,265 in hOrai1) as hinge points. A straightening of TM4 and TM4ext leads to new TM4ext-TM4ext interactions from different channel subunits and a continuous α-helix that extends 45 Å into the cytosol.
Although it appears in the crystal that pore lining amino acids closer to the extracellular side stay locally constant in closed and open conformations, the low resolution limits to observe if the opening of the channel involves a slight rotation of the TM1 helix as has been proposed by Yamashita et al in 2016 [38]. Hou et al. have also found that the unlatching of cytosolic regions alone is not responsible for inducing the open conformation of the channel, rather it seems to expose cytosolic docking sites for STIM1. The subsequent interaction with STIM1 potentially stabilizes the unlatched conformation and a widening of the pore, which allows for Ca2+ influx. Taken together, the open dOrai channel conformation at a 6.7 Å resolution exhibits a dilated pore within the basic regions and changes in cytosolic latch regions, an outward rotation of TM1-TM4 and subtle changes towards the extracellular side [36].
In earlier studies it has been hypothesized that upon STIM1 binding, the Orai1 C-termini straighten, which breaks their coiled-coil interaction and allows binding of STIM1 [32]. However, an NMR structure suggests a different model where Orai1 C-termini keep their conformation, with some minor alterations only in the angles of the Orai1 helices [39]. These findings may also reflect earlier, transient steps in the activation cascade of the CRAC channel. The closed Drosophila melanogaster crystal structure also reveals that the narrowest part of the pore (~6 Å) is formed by a ring of glutamates at position E106 in hOrai1 (E178 in dOrai), constituting the selectivity filter of the Orai channel, which has a highly negative electrostatic potential [32]. The identification of E106 to be the selectivity filter for Ca2+ is in line with the high-affinity of this residue to Ca2+ [40–42]. Other hOrai1 residues facing the inner side of the pore in the closed as well as in the open channel configuration are V102, G98, L95 and R91, all of them discussed to have a specific role in gating of the channel [32]. Mutational analysis has shown that the polar substitution of V102 yielded constitutively open channels [43] while G98 has been suggested to function as gating hinge [44]. Also R91, together with the formation of a water layer, may act as gate of the channel [37]. V102 and F99 have been proposed to work in concert to form a hydrophobic gate, as mutation to more polar amino acids leads to leaky gates [38].
Recently, Frischauf et al. have identified a cluster of Orai1 mutations, located in TM2, to be a trigger site for Orai1 gating [37]. Functional analysis of constitutively open Orai1 mutants, derived from large-scale cancer genomics data sets and introduced in fluorescence-labelled Orai1 constructs, has revealed two fundamental gates regulating Ca2+ influx: Arginine side-chains were displaced so they no longer blocked the pore and a chain of water molecules formed in the hydrophobic pore region. In molecular dynamics (MD) simulations residue E190 within TM3 has also been shown to contribute to selectivity and gating by reducing the number of water molecules in this region when mutated to glutamine [45]. Another important region of human Orai1 was described by Frischauf et al in the extracellular loop region, in close proximity to the selectivity filter. This Ca2+-accumulating region (CAR) enhances Ca2+ permeation at low Ca2+ concentrations via accumulating Ca2+ at the outer mouth of the channel thereby increasing the local Ca2+ concentration. Mutational analysis has revealed two aspartate residues (D110, D112) within the first extracellular loop of Orai1 channels to function as Ca2+ accumulating sites (Fig. 1A, B) [46].
4. Characteristics of STIM proteins and their activation
There are two mammalian homologs of STIM, i.e. STIM1 and STIM2. These proteins are type I, single-pass ER membrane proteins, which interlink Ca2+ store-depletion and Ca2+ influx through the PM.
STIM1 displays constitutive movement along microtubules in resting cells. Via a tandem EF hand like sequence on its luminal side, STIM1 senses a drop in [Ca2+]ER that triggers its translocation into defined ER-PM junctions [12,13,24,26,47]. Mutations of the acidic residues within the EF hand mimicked the store-depleted state as STIM1 was no longer able to bind Ca2+ [12,26].
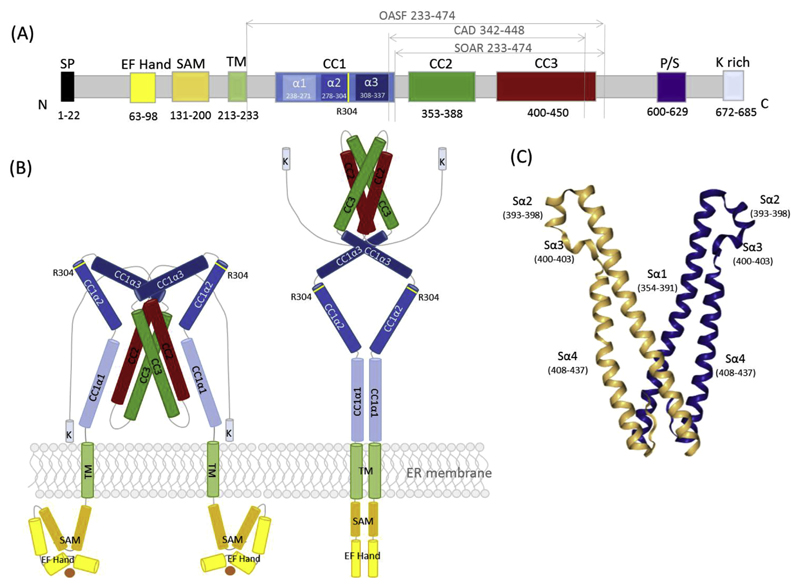
STIM1 is a 75 kDa protein (built up of 685aa) with its N-terminus located in the ER lumen, followed by a single TM spanning domain and a cytosolic C-terminus (Fig. 2).
Fig. 2.
(A) Schematic representation of full-length human STIM1 depicting the essential regions for interaction with Orai together with the overall structure. (B) Model of STIM1 in resting (left) and activated (right) state. (C) Model of the STIM1 SOAR domain depicting Sα1-4 regions [53,72].
STIM2 is a 84 kDa protein (composed of 746aa) that mediates Orai1 channel activation in the absence of agonist stimulation under resting conditions [48] attributed to a weaker Ca2+ binding affinity of STIM2 compared to STIM1 [12,49]. Alternative splicing of STIM2 results in the STIM2β isoform (also named STIM2.1) that contains an 8aa insertion near the end of CC2 [50,51]. The STIM2.1 protein has just recently been shown to directly preclude Orai1 channel activation by inducing structural perturbations in the STIM1 CC region [52].
Compared to STIM1, the STIM2 SOAR induces a weaker activation of Orai1 although it binds to the channel efficiently [53,54]. The C-terminal polybasic region of STIM2 shows a stronger affinity for PIP2 than that of STIM1 and a lack of this specific domain eliminates STIM2 clustering [55,56]. Upon Ca2+ dissociation from the luminal EF hand of STIM2, conformational changes within the N-terminus show different temporal dynamics compared to STIM2 [57]. Overexpressed STIM2 in cells exhibits pre-clustering near the PM and causes constitutive activation of Orai1 [48,50,51,53,58].
As this review is predominantly focused on STIM1 activation of Orai1, we will not further discuss other STIM or Orai isoforms in detail.
4.1. Luminal domains of STIM1
Studies on the activation and homo-/oligo-merization process of STIM1 all have in common that the initial trigger for homo-/oligomerization of the luminal STIM1 part is a decrease in [Ca2+]ER, a signal that is consecutively transduced through the TM segment, reaching cytosolic coiled-coil (CC) domains which results in higher order STIM1 oligomerization [12,13,18,27,35,47,59–61].
Essential domains within the luminal part are the canonical and hidden EF hand (cEF and hEF, respectively) and the sterile alpha motif (SAM) (Fig. 2A) [24,62,63]. Structurally, the EF hand exhibits a helix-loop-helix motif with negatively charged aspartates and glutamates that interact with Ca2+ as long as [Ca2+]ER is full. The binding affinity of the EF hand is adequate to [Ca2+]ER ranging from 400 to 800 μM to ensure an accurate response to varying Ca2+ concentrations. Dissociation of Ca2+ from the EF hand destabilizes the entire EF-SAM entity [62]. Stathopulos et al have examined the EF-SAM complex in detail [24], showing that the EF-Ca2+-SAM holoform is monomeric whereas the apoform is at least a dimer [24,62]. STIM2 proteins also contain a luminal EF-SAM domain which has been shown to have different cellular functions [48].
4.2. The transmembrane domain of STIM1
Within the TM domain a single point-mutation has been identified (C227 W) that switches the CRAC channel constitutively open [64]. This GOF mutant has been shown to resemble an activated state of full-length STIM1, forming puncta at ER-PM junctions. This is of advantage when examining the ER lumen-to-cytosol signal transduction, as it represents an activated STIM1 protein without the necessity of store-depletion, thereby bypassing physiological steps of activation. In resting cell conditions, the C227 W mutant shows proximity of the N-terminal TM parts similar to EF-SAM domains upon store-depletion. FRET measurements point to a decreased interaction of CC1 with SOAR in case of C227 W, consistent with formation of the activated STIM1 form that is physiologically obtained by Ca2+ depletion-induced homo-/oligo-merization [64]. Three glycines within the TM domain (G223, G225 and G226) have been suggested to grant flexibility needed for conformational changes to transduce the activation signal from the ER lumen to cytosolic STIM1 domains [64,65].
Further mechanistic aspects for the transduction of the signal from the luminal domain of STIM1 to the cytoplasmic SOAR region have recently been published [66]. It has been shown that the TM domains are not in close proximity in the resting state of STIM1, but the loss of Ca2+ from the luminal EF-hand actually brings them close. By this mechanism the signal of reduced Ca2+ content is transmitted from the luminal part of the TM region to the STIM1 cytoplasmic region at amino acids 230–233. Hirve et al. stated that the physiological signal propagation is directional, which means that the signal is originating in the ER lumen and is propagated to the cytoplasm. In contrast, the conformational change of the STIM1 protein is cooperative and therefore non-directional [66].
4.3. Cytosolic domains of STIM1
Successive to the α-helical TM domain, STIM1 contains a cytosolic strand including three CC regions, a CRAC modulatory domain (CMD), a serine/proline and a lysine-rich region (Fig. 2A) [21,67]. Independent studies [68,69] have shown that the STIM1 C-terminus alone is sufficient to activate CRAC currents through Orai1. Several cytosolic STIM1 fragments capable to activate Orai1 have been identified thereafter: OASF (Orai activating small fragment, aa233-450), CAD (CRAC activation domain, aa342-448), SOAR (STIM1-Orai1 activation region, aa344-442) and CCb9 (aa339-444) [54,69–71]. All these Orai-activating STIM1 fragments have the CC2 (aa363-389) and CC3 (aa399-423) region with additional 19 amino acids in common. Together with CC2 and CC3, a STIM1 homomerization domain (SHD, aa421-450) has been identified for mediating homomerization and is required in the coupling to and activation of the CRAC channel. An SHD deletion within the OASF fragment decreases drastically interaction with Orai1 and abrogates CRAC channel activation in patch-clamp experiments [69].
In 2013 Stathopulos et al published an NMR structure of a SOAR overlapping fragment [39]. The X-ray crystallographic structure of a cytosolic portion of STIM1 has been published in 2012 [72] revealing a human SOAR protein (aa345-444 with mutations L374 M, V419 A and C437 T) in a dimeric assembly with intra- as well as intermolecular interactions (Fig. 2C). The dimeric SOAR structure exhibits an R-shaped structure where an antiparallel arrangement of CC2 and CC3 with two inter-adjacent short α-helices is visualized. This dimeric assembly is generated by coiled-coil interactions whereby C-terminal residues of one monomer (R429, W430, I433 and L436) interact with N-terminal residues of the other (T354, L351, W350 and L347), forming an overall V-shaped structure of the SOAR dimer which is its physiological conformation. The CC2 domains cross each other at position Y361, forming a stacked interaction. Mutation of this binding interface in SOAR or full-length STIM1 abrogated co-localization with and activation of Orai1 channels [72]. A basic amino acid cluster (K382, K384, K385, K386 and R387) is located at the tips of the V-shaped SOAR dimer [72,73].
The polybasic cluster at the C-terminal end of STIM1 together with the SOAR/CAD domain have been shown to be crucial for puncta formation through a diffusion-trap mechanism via interaction with PM-resident phospholipids and Orai channels. In contrast, it has been shown that this K-rich region is not necessary for the oligomerization of STIM1 [60,68,70,74,75]. At resting state, the C-terminal STIM1 domains, including the CC1, SOAR/CAD and the polybasic region are inactive through a folded-back configuration. Upon ER Ca2+ store-depletion, the STIM1 C-terminus adopts a more extended configuration by exposing the SOAR/CAD as well as the polybasic domain. At ER-PM contact sites, the polybasic tail can interact with PIP2 in the inner leaflet of the PM [64,76].
Within STIM1 fragment structures available, the CC1 domain is not or only partially visible. This STIM1 region comprises three alpha helical segments (CC1α1 aa238-271, CC1α2 aa278-304 and CC1α3 aa308-337 also known as inhibitory helix) [21,72,76]. The role of STIM1 CC1 is on the one hand to bridge the distance at ER-PM junctions allowing for interaction with Orai1 and on the other hand to keep STIM1 in a tight and compact conformation at high [Ca2+]ER levels [64,76–78]. The structural rearrangements upon Ca2+ store-depletion range from luminal oligomerisation and rearrangement of TM domains to a conformational change in cytosolic STIM1 regions and SOAR exposure. The most prominent change there is the switch from the inactive, tight state to the activated, extended state [64,76,78]. Through this switch SOAR is exposed upon the release of an intramolecular CC1-CC3 clamp.
To monitor STIM1 C-terminal conformational changes, a double-labelled conformational sensor construct has been developed by Muik et al. [77] based on STIM1 OASF. FRET data show that OASF folds into a rather compact structure which likely corresponds to the quiescent state of STIM1. Point mutations within OASF induce a conformational change which results in different intramolecular FRET signals. Mutations in CC1 (L251S) or CC3 (L416S, L423S) for example lead to an extended OASF structure. Introducing these mutations in full-length STIM1 lead to constitutive STIM1 activation and CRAC channel activation [76,77]. These findings suggest the existence of an intramolecular CC clamp within the quiescent STIM1 protein. These inhibitions are most likely released by either CC1α1 or CC3 mutations or physiologically upon store-depletion [77]. Likewise, Zhou et al. have observed an extended STIM1 conformation induced by artificial cross-linking of CC1 domains to promote dimerization or by introducing the L251S STIM1 activating mutation [78]. Taken together, these results reveal the impact of CC1α1 on the transition of STIM1 from a quiescent, tight state to an activated, extended state. Fahrner et al have identified a direct CC1α1-CC3 interaction as key molecular determinant in setting the quiescent state of STIM1 [76], by use of a FRET-based method called FIRE (FRET Interaction in a Restricted Environment). Additionally, among the three α helices of CC1, CC1α2 also contributes to the activation of STIM1.
Constitutive cluster formation and CRAC channel activation are characteristics of the Stormorken Syndrome [79–81]. This syndrome is associated with the STIM1 R304 W mutant carrying the mutation at the end of CC1α2, in which the CC1α2 inherent function is enhanced by this mutation resulting in permanently oligomerized and activated STIM1 proteins. A more recent study by Fahrner et al. [82] proposes a dual mechanism that switches the STIM1 R304 W Stormorken mutant into its activated state. The identified R304 has been proven to be a crucial position for the extension of STIM1 and the exposure of the CAD/SOAR domain. Mutation from arginine to tryptophane (R304 W) enforces CC1 homomerization together with an elongation of the helicity of CC1, thereby preventing CC1 – CC3 clamp formation and locking STIM1 in the active state [82].
Covington et al have studied the homomerization of STIM1 C-terminal truncation mutants using FRET [83] to investigate the impact of the cytosolic CC domains. They came to the conclusion that the mere presence of CC1 leads to store-independent but unstable oligomerization. While longer constructs including CC3 and SHD lead to strong and store-dependent oligomerization. Besides this important role of CC1 it has been shown that it also plays a regulatory role in the activation cascade upon store-depletion. Several studies focused on how CC1 governs the switch between the quiescent and the activated state of the STIM1 protein. Cui et al have managed the structural resolution of CC1, revealing a long helix which dimerizes in an antiparallel manner [84]. Korzeniowski et al have first addressed the role of CC1 in the control of STIM1 activation by an autoinhibitory clamp between a glutamate-rich segment within CC1α3 (aa308-337) and a lysine-rich region within CC2 (aa382-387) [73]. As evident from the CC1-SOAR crystal structure of Caenorhabditis elegans, the CC1α3 (inhibitory helix) and the lysine-rich region are not in the proximity to form the suggested electrostatic interactions [72]. Based on these findings Yang et al propose residues within CC1α3 which interact with residues at the beginning of CC2 as well as at the end of CC3. Their proposed model states that in resting conditions STIM1 forms a dimer with the SOAR domain responsible for dimerization and CC1α3 stabilizing the inactive state of STIM1. Yu et al presented an intramolecular shielding model that keeps STIM1 inactive [85]. They suggest that the contribution of electrostatic or coiled-coil interactions is rather unlikely, and consequently present a model in which the amphipathic properties of CC1α3 regulate the STIM1 activation state.
In summary, the STIM1 conformation (Fig. 2B) is pivotal for the function of CRAC channel activation. The activation process of STIM1 is dynamic and is initiated by low [Ca2+]ER. In resting cell conditions when [Ca2+]ER is full, inactive STIM1 proteins exhibit a tightly packed structure of their C-termini. The dimeric, tight state is stabilized by intramolecular CC1-CC3 clamp formation [76,77]. The spark for inducing a STIM1 luminal conformational change originates upon Ca2+ flux from the ER lumen into the cytosol [62,63]. This signal is further transmitted through the transmembrane domain to the cytosolic STIM1 domains [64]. As a result, the clamp of CC1-CC3 is released and SOAR exposed [39,64,76–78]. Rearrangement of CC2 and CC3 results in the formation of SOAP (STIM1 Orai activation pocket, see below) and may trigger exposure of CC3 for homo-/oligomerization and clustering [39]. Finally, higher order oligomers are formed via CC3 of STIM1 [76,77].
5. Coupling of STIM1 and activation of the Orai channel
Conformational rearrangements of STIM1 upon store-depletion are thought to facilitate spanning the distance between ER and PM [77]. Within ER-PM junctions, clusters of STIM1 and Orai proteins form, representing hotspots of local Ca2+ entry [86]. Several studies have suggested a direct interaction of these two proteins [28,35,87]. STIM1 coupling to the Orai1 channel involves the cytosolic strand of STIM1 and both, N- and C-termini of Orai1. It has been shown that the CAD fragment directly binds to both strands of Orai1 [54,70] and that the Orai1 C-terminus is indispensable for proper coupling to STIM1. As can be seen in the closed dOrai crystal structure [32], the TM4 of Orai extends into the cytosol. In the open dOrai structure the TM4 regions straighten and extend 45 Å into the cytosol granting accessibility for the STIM1 protein [36]. All three mammalian Orai homologs show certain probabilities for the formation of C-terminal CC domains. Frischauf et al have suggested a weak coiled-coil probability for Orai1 whereas the one of Orai2 and Orai3 has been shown to be 15–17 fold higher. We suppose that the affinity towards STIM1 increases with the higher CC probability of Orai C-terminal domains [88]. Within Orai2 and Orai3, mutations of at least two hydrophobic core positions within the CC region are required to fully interrupt interaction with STIM1, whereas in Orai1 single mutations are sufficient [88]. It has been shown that hydrophobic residues L273 and L276 in Orai1 play an important role in coupling to STIM1 and in the C-terminal dimerization process within the hexameric channel complex [32] (Fig. 3).
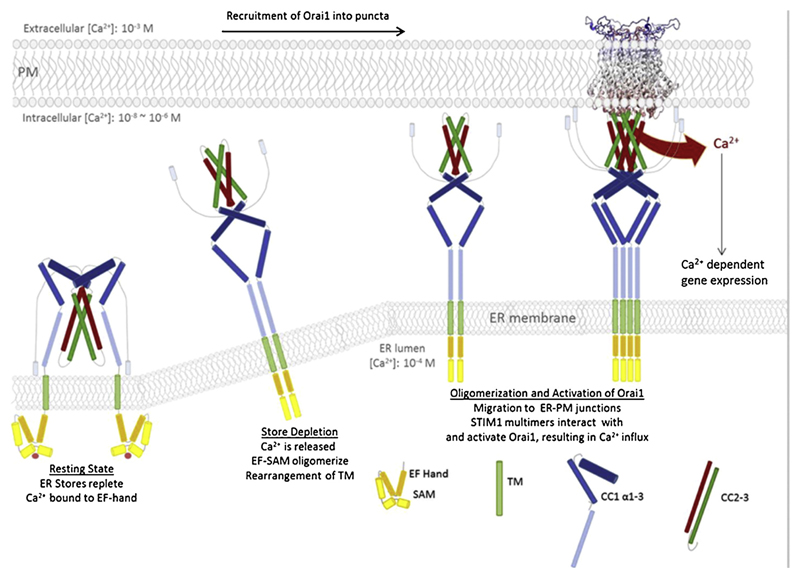
Fig. 3.
Simplified model of the CRAC activation process. Starting with STIM1 at resting state with Ca2+ bound at the luminal EF-hand which remains as monomer together with SAM, followed by the activation of STIM1 due to Ca2+ dissociation off the EF hand. Subsequent interaction of the TM and the EF-SAM domains and further signal propagation via a dynamic rearrangement of the cytosolic portion of STIM1. Undocking of the SOAR/CAD domain from CC1 allows interaction with the Orai1 protein in the PM, upon multimerization of STIM1. Ca2+ influx, achieved by the gating of the Orai1 channel via STIM1 activates downstream effector functions like Ca2+ dependent gene expression. (This model does not take stoichiometry issues, uni- or bimolecular coupling models or rearrangements within the SOAR molecule into account).
Besides the important role of CC structures in the interaction event, Calloway et al have proposed an electrostatic interaction model between STIM1 and Orai [89,90]. Acidic residues within the Orai1 C-terminus and positively charged amino acids in STIM1 (KIKKKR aa382-387) were hypothesized to form salt bridges, however, mutating these residues in Orai1 still allowed for coupling to STIM1 and retained channel function, suggesting that the coupling process involves other structural components [39,90]. Taken together, all these data strongly suggest a direct coupling of STIM1 to Orai via CC domains. Conclusive evidence was provided in 2013 by Stathopulos et al [39] with the NMR structure of a STIM1 fragment including the rear part of CC1 as well as the whole CC2 domain (aa 312–387) together with an Orai1 C-terminal domain ranging from aa 272 to 292. These data have been the first to prove a C-terminal interaction of STIM1 and Orai1 on a structural basis, although it has to be kept in mind that the STIM1 fragment lacks the important CC3 domain. The SOAP involves the Orai1 leucines L273 and L276 as well as negatively charged aspartates D284, D287 and D291 interacting with basic STIM1 residues K382, K384, K385 and K386. Furthermore, several other STIM1 residues (L347, L351, Y362, L373 and A376) are highlighted that are in close proximity to the Orai1 C-terminus. Summarized, the coupling process mediated between Orai1 C-terminus and STIM1 CC2 involves hydrophobic as well as electrostatic interactions.
The closed dOrai hexameric crystal structure pinpoints to an antiparallel crossing of Orai1 C-termini within three Orai1 dimers which is strongly supported by hydrophobic interactions of L273 and L276 [32]. Mutation of these residues to less hydrophobic ones abolishes Orai1 activation [91]. Via cysteine-crosslinking analysis of L273C, L276C mutants, STIM1 binding is inhibited and only reversed upon disulphide-bond break [92].
A flexible linker region connects the TM4 and the C-terminus of Orai1 which is crucial for the orientation of the C-termini and in turn for coupling to STIM1 (Fig. 1A) [91–93]. The binding of STIM1 to Orai1 mutants within the linker region is significantly reduced compared to wild type proteins, yet only completely destructed by additional C-terminal Orai1 mutations [91,93]. These data suggest that this Orai1 region is necessary for establishing the optimal position of the C-termini required for STIM1 binding. Besides a couple of Orai1 linker mutants that do lose function [93,94] [92], there were some constitutively active mutants characterized which exhibit biophysical characteristics comparable to wild type Orai1 [93]. It has been concluded that these mutants induce conformational changes within the Orai1 protein similar to those occurring through STIM1 binding to accomplish signal propagation elementary for channel opening. This is in strong agreement with the crystal structure of open dOrai1, which proposes conformational changes of the TM4 and the extended TM4 region upon channel opening. While the closed structure shows an interaction of TM4 and TM3, it is disrupted in the open conformation. This unlatching is apparently necessary for STIM1 binding, but is not sufficient open the channel [36].
Another region identified to be a crucial segment for C-terminal STIM1 binding is the helical extension of TM4. Especially a segment of five amino acids (aa 261–265, LVSHK), termed “nexus”, was recently proven to transmit information from the STIM1-binding site to the core of the Orai1 channel (Fig. 1A). Mutation of four amino acids in this region (LVSHK to ANSGA) led to a constitutively active channel, which appears to have the same properties like a channel activated by STIM1. It seems that L261 and V262 are hydrophobically attached to the TM3, position L174, respectively [93]. The SHK region of Orai has also been shown to be a hinge point for channel opening [36] as its unlatching is required to expose docking sites for STIM1, a prerequisite for channel opening.
These functional and structural data indicate a predominant role of the Orai1 C-terminus in coupling to STIM1. Nevertheless, the N-terminus of Orai1 is also a prerequisite for correct channel gating as it has been shown that N-terminal deletions or point mutations lead to loss of function [35,93,95]. The interaction of STIM1 with the N-terminus of Orai1 involves a conserved sequence (aa 73–90) close to the TM1 region, termed extended transmembrane Orai1 N-terminal (ETON) region [70,95]. Orai1 mutants that lack the ETON region, completely lose function and the introduction of mutations at positions L74 and W76 results in highly reduced interaction with STIM1, suggesting this N-terminal Orai1 region contributes to a STIM1 binding interface [95]. The three Orai homologs, although fully conserved among the ETON region, are differently affected by N-terminal deletions, for example Orai3 retains activity upon deletion of a larger N-terminal fragment. It has been recently shown that the fully conserved ETON region is needed either in Orai1 as well as in Orai3 to restore CRAC channel activity together with STIM1 in case of constitutive active TM3/TM4 mutants as well as in store-operated channel configuration. The domains of the conserved N-terminal region seem to fulfil different functions, as the latter stretch (Orai1 aa 80–90; Orai3 aa 55–65) is involved in general channel activity, the other part (Orai1 aa 73–79; Orai3 aa 47–54) appears to fine-tune Ca2+ entry [96,97]. In addition, Zheng et al have examined progressive N-terminal deletions yielding the result that the Orai1 N-terminus is essential for binding to STIM1 [98]. They suggest that STIM1 first binds to the Orai1 C-terminus followed by binding to the N-terminus. A recent study [53] hypothesized that a specific region within STIM1 between CC2 and CC3 (aa 393–403) functions as binding site to Orai1. Furthermore, residue F394 is proposed to interact with hydrophobic residues in the N-terminus of Orai1 (aa73-85), thereby probably constituting part of the STIM-Orai binding site, because mutation to hydrophilic amino acids abolishes the activation of Orai1 [53]. In addition, the loop2 of Orai1, which connects TM2 and TM3, has been moved in the focus of interest. Not only the Orai1 N-terminal region alone, but also the interaction of the N-terminus with loop 2 seems to have impact on STIM1 coupling. This fine-tuning of Orai1 channel function seems to take place due to predicted interactions of residues Y80 in the N-terminus and N156 in loop2 [99]. Palty et al proposed a sequential Orai1-activation model by SOAR. The first step is SOAR binding to Orai1, that partially activates the channel and leads to structural rearrangements. This change in structure permits sequential steps of SOAR binding to fully activate the channel [100].
STIM1 and Orai1 proteins have been proven sufficient to reconstitute CRAC currents. Nevertheless, various proteins and lipids have been identified that modulate the STIM1-Orai1 signalling cascade: STIMATE [101], Septin [102], SARAF [103,104], CRACR2 A [105] and cholesterol [106,107].
In summary, besides the Orai1 C-terminus, the N-terminal ETON region contributes to the STIM1 binding interface and provides electrostatic components that impact the structure of the elongated pore. The created bridge upon STIM1-Orai1 binding may generate a force that induces a conformational change of the ETON and TM1 regions, in line with the open dOrai structure – a signal that is propagated to the channels gate and results in Ca2+ influx.
Although for a long time believed to be a tetramer, the hexameric dOrai structure is now widely accepted [31,32,108–111]. Still, the exact stoichiometry of a functional STIM1-Orai1 complex remains elusive. Several studies hypothesized that CRAC channel activation depends on the number of STIM1 molecules bound [34,112,113]. For Ca2+ dependent inactivation (CDI) a higher amount of STIM1 to Orai1 molecules leads to more pronounced inactivation [114]. Other studies with Orai1 proteins linked to tandem CAD fragments indicated that eight STIM1 molecules are required for maximal activation and inactivation [34]. The selectivity for Ca2+ is also enhanced by an increasing amount of coupled STIM1 fragments, which has been shown by McNally et al who used Orai1 and Orai1 V102 A proteins linked to single and tandem CAD/SOAR domains [115]. The NMR structure of the SOAP region suggests a 1:1 ratio of STIM1 to Orai1 proteins in an active channel complex, which probably denotes that six STIM1 proteins attach to six Orai1 subunits [39]. Having the dOrai crystal structure in view, it is better conceivable that six but not eight STIM1 proteins bind to a hexamer of Orai subunits.
Based on the SOAP structure, it has been hypothesized that Orai1 channel activation occurs upon interaction of a SOAR dimer within STIM1 with a dimer of adjacent Orai1 subunits in the hexameric Orai1 channel – a model termed bimolecular interaction [39]. Alternatively, a unimolecular coupling model has been suggested by Zhou et al [53,93,116]. By the use of SOAR1/SOAR2 chimeras, F394 in SOAR1 has been identified as critical residue that determines differences in binding to and activation of Orai1 by STIM1 and STIM2. The introduction of F394 point mutations (to L, A or H) in wild type STIM1 decreases or fully abolishes SOCE, with F394H as most effective in disrupting STIM1 interaction with Orai1. As a SOAR concatemeric dimer with one functional SOAR monomer linked to one F394H containing SOAR monomer has a similar effect on Orai1 binding and activation as a SOAR concatemer with two functional SOAR monomers, a unimolecular interaction model has been suggested with each Orai1 channel subunit in the hexameric complex bound to one STIM1 of a STIM1 dimer. This model further leads to speculate that the second, uncoupled STIM1 within a STIM1 dimer interacts with an adjacent Orai1 subunit of another hexameric channel complex thereby enabling clustering of Orai1 channels in the PM [117].
To finally prove a bi- or unimolecular coupling model, clearly more experiments are necessary, as is it conceivable that the observed clustering of Orai1 channels may also occur via a bimolecular STIM1-Orai1 binding, as clustering between STIM1 proteins may be able via their CC3 regions in the active state [76].
Summing up, STIM1 to Orai1 coupling predominantly occurs via the C-terminus of Orai1. The coupling process highly likely involves a conformational rearrangement of Orai1 subdomains, a process that is followed by a bridging to the Orai1 N-terminus that results in CRAC channel activation. Clearly, further functional and structural data of human Orai1 proteins in complex with STIM1 are needed to throw light on the underlying mechanism of STIM1/Orai1 activation.
5.1. Open Orai channel structure
Since the discovery of the CRAC channels key players, STIM1 and Orai1, in 2005 and 2006 respectively [12,13,15,16,118], extensive progress has been made and parts of the CRAC channel system have been resolved with 3D atomic resolution.
Just recently, the open dOrai structure has complemented our knowledge on structural rearrangements that occur in the open channel. Both Drosophila melanogaster Orai crystal structures have experimentally been designed with dOrai protein deletions (aa 133–341) and mutations (P276R, P277R, C224S and C283 T). It is of note that the open dOrai structure is based on the usage of a constitutively open channel (H106 A corresponding to H134 A in hOrai1 proteins) that does not require STIM binding for channel opening. Physiologically, upon Ca2+ store-depletion STIM1 couples to the cytosolic strands of Orai1 that gates the channel into the open state. While the coupling of STIM1 to the C-terminus of Orai1 is widely accepted, the role of the N-terminus needs to be clarified in more detail. Deductively, there is no structural knowledge about the N-terminal regions in closed and open Orai proteins available. Binding of STIM1 to the Orai1 C-terminus allosterically affects the whole Orai1 protein. A recently published study by Frischauf et al suggests that a patch of amino acids in the TM2 acts as trigger for hOrai1 channel gating [37]. This is in line with the findings in the crystal structure of dOrai, which shows that the amino acids in TM1 hardly change their position except of a dilation of the pore itself. This indicates that the gating trigger has to be located elsewhere than in TM1. We have also shown in this study that residue H134 in hOrai1 forms a hydrogen bond with residue S93 which further stabilizes the channel and suggests an effect of the TM2 domain on the pore during gating [36,37]. Further experiments and experimental data are needed that should be orientated towards elucidating the physiological coupling of STIM1 to Orai1 to gain a complete description of gating in the wild type CRAC channel.
6. Perspectives/open questions
The conformational rearrangements within Orai1 that are induced by STIM1 binding are only partially resolved. Constitutively active Orai1 mutants which act independently of STIM1 (H134 A, P245 L, ANSGA) [37,93] (Fig. 4) have the potential to provide structural resolution of further open hOrai1 channel conformations, as it has been recently shown with the new crystal structure of dOrai [36]. These mutations can help to provide hints how the transition from the closed to the open state(s) may take place in detail. Nevertheless, one has to keep in mind that dOrai shares only 73% sequence homology with hOrai1 [32] and differences in these two protein sequences could exhibit unique structural and functional characteristics, in addition to the fact that STIM1 as physiological activator is still missing in these structures. Just recently it has been shown that the gating mechanism of Orai channels radically differs between mammalian and C. elegans forms [119]. In contrast to STIM1 binding to the N- and C-termini of Orai1 in mammals, it interacts with the intracellular loop2 of Orai in C. elegans. This previously unknown mechanism suggests that different gating mechanism have evolved and diverged between invertebrates and mammals.
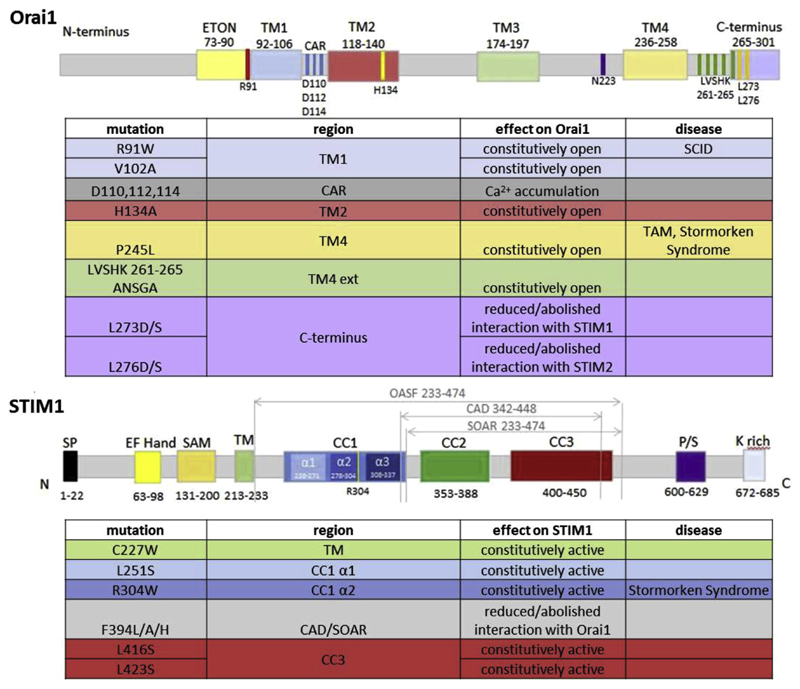
Fig. 4.
Overview of important functional mutants within Orai1 and STIM1 proteins mentioned in this review. Diseases linked to specific mutations are indicated (SCID: severe combined immunodeficiency; TAM: tubular aggregate myopathy).
It is likely that STIM1 binding to one or both Orai1 termini induces a signal propagation by exerting a force on the four TM regions. It needs to be clarified whether it is a unidirectional signal propagation from Orai1 C-terminus to the TM1 pore or whether additional sites in TM regions exist that have the ability to control TM1 pore opening. Recently the group of Prakriya [120] has proposed a rotation of the TM1 helix that is linked with conformational changes in the Orai1 C-terminus upon STIM1 binding and channel opening. Alternatively, Frischauf et al have shown that a small local widening of the pore occurs upon channel opening [37]. The open dOrai crystal structure [36] reveals a widening of the basic region in the channels pore and has shown that pore-lining amino acids keep almost the same position during channel opening. Nevertheless, the limited resolution may have prevented identification of a slight rotation of the pore helix. Clearly, this mechanism needs further clarification.
For STIM1, 3D atomic structures are available only for STIM1 C-terminal fragments and exhibit substantial differences. To resolve the mechanisms that switch STIM1 from its inactive, tight conformation to the active, extended one, larger crystallized portions of the STIM1 protein, at least including the CC1 domain, are required.
Furthermore, optogenetic engineering approaches are emerging. Studies focus on the generation and testing of a photo-activated CRAC channel and clearly will shed light on the regulatory mechanisms of STIM1-Orai1 downstream signalling pathways [121,122].
Despite all considerable advances in the knowledge of CRAC channel function, a thorough understanding of this fascinating coupling machinery clearly requires further studies, with results from the powerful cryo EM technique so far still missing. The additional proteins, only shortly mentioned in this review, that modulate STIM1/Orai1 function even enhance the complexity of the native CRAC channel system and have to be taken into account for their physiological impact on STIM1 and Orai1 protein function.
Acknowledgment
This work was supported by the Austrian Science Fund (FWF) grants No. P28872 (to I.F.) and No. P27263 (to C.R.).
References
- [1].Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- [2].Abdullaev IF, Bisaillon JM, Potier M, Gonzalez JC, Motiani RK, Trebak M. Stim1 and Orai1 mediate CRAC currents and store-operated calcium entry important for endothelial cell proliferation. Circ Res. 2008;103:1289–1299. doi: 10.1161/01.RES.0000338496.95579.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Albert AP, Saleh SN, Peppiatt-Wildman CM, Large WA. Multiple activation mechanisms of store-operated TRPC channels in smooth muscle cells. J Physiol. 2007;583:25–36. doi: 10.1113/jphysiol.2007.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Prakriya M. The molecular physiology of CRAC channels. Immunol Rev. 2009;231:88–98. doi: 10.1111/j.1600-065X.2009.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- [6].Mogami H, Lloyd Mills C, Gallacher DV. Phospholipase C inhibitor, U73122, releases intracellular Ca2+, potentiates Ins(1,4,5)P3-mediated Ca2+ + release and directly activates ion channels in mouse pancreatic acinar cells. Biochem J. 1997;324(Pt 2):645–651. doi: 10.1042/bj3240645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Parekh AB, Putney JW., Jr Store-operated calcium channels. Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- [8].Lewis RS. Calcium signaling mechanisms in T lymphocytes. Annu Rev Immunol. 2001;19:497–521. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- [9].Berridge MJ. Capacitative calcium entry. Biochem J. 1995;312(Pt 1):1–11. doi: 10.1042/bj3120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kuno M, Gardner P. Ion channels activated by inositol 1,4,5-trisphosphate in plasma membrane of human T-lymphocytes. Nature. 1987;326:301–304. doi: 10.1038/326301a0. [DOI] [PubMed] [Google Scholar]
- [11].Zweifach A, Lewis RS. Mitogen-regulated Ca2+ + current of T lymphocytes is activated by depletion of intracellular Ca2+ + stores. Proc Natl Acad Sci U S A. 1993;90:6295–6299. doi: 10.1073/pnas.90.13.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang SL, Yeromin AV, Zhang XHF, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc Natl Acad Sci U S A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- [17].Hogan PG, Rao A. Dissecting I(CRAC), a store-operated calcium current. Trends Biochem Sci. 2007;32:235–245. doi: 10.1016/j.tibs.2007.03.009. [DOI] [PubMed] [Google Scholar]
- [18].Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Feske S, Skolnik EY, Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat Rev Immunol. 2012;12:532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cahalan MD. STIMulating store-operated Ca(2+) entry. Nat Cell Biol. 2009;11:669–677. doi: 10.1038/ncb0609-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kar P, Bakowski D, Di Capite J, Nelson C, Parekh AB. Different agonists recruit different stromal interaction molecule proteins to support cytoplasmic Ca2+ oscillations and gene expression. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1201204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Di Capite J, Parekh AB. CRAC channels and Ca2+ signaling in mast cells. Immunol Rev. 2009;231:45–58. doi: 10.1111/j.1600-065X.2009.00808.x. [DOI] [PubMed] [Google Scholar]
- [24].Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- [25].Pores-Fernando AT, Zweifach A. Calcium influx and signaling in cytotoxic t-lymphocyte lytic granule exocytosis. Immunol Rev. 2009;231:160–173. doi: 10.1111/j.1600-065X.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- [26].Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Baba Y, Hayashit K, Fujii Y, Mizushima A, Watarai H, Wakamori M, Numaga T, Mori Y, Iino M, Hikida M, Kurosaki T. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Navarro-Borelly L, Somasundaram A, Yamashita M, Ren D, Miller RJ, Prakriya M. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu P, Lu J, Li Z, Yu X, Chen L, Xu T. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- [30].Shaw PJ, Qu B, Hoth M, Feske S. Molecular regulation of CRAC channels and their role in lymphocyte function. Cell Mol Life Sci: CMLS. 2013;70:2637–2656. doi: 10.1007/s00018-012-1175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cai X, Zhou Y, Nwokonko RM, Loktionova NA, Wang X, Xin P, Trebak M, Wang Y, Gill DL. The Orai1 store-operated calcium channel functions as a hexamer. J Biol Chem. 2016;291:25764–25775. doi: 10.1074/jbc.M116.758813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yen M, Lokteva LA, Lewis RS. Functional analysis of Orai1 concatemers supports a hexameric stoichiometry for the CRAC channel. Biophys J. 2016;111:1897–1907. doi: 10.1016/j.bpj.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li Z, Liu L, Deng Y, Ji W, Du W, Xu P, Chen L, Xu T. Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 2011;21:305–315. doi: 10.1038/cr.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Muik M, Frischauf I, Derler I, Fahrner M, Bergsmann J, Eder P, Schindl R, Hesch C, Polzinger B, Fritsch R, Kahr H, et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- [36].Hou X, Burstein SR, Long SB. Structures reveal opening of the store-operated calcium channel Orai. Elife. 2018;7 doi: 10.7554/eLife.36758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Frischauf I, Litvinukova M, Schober R, Zayats V, Svobodova B, Bonhenry D, Lunz V, Cappello S, Tociu L, Reha D, Stallinger A, et al. Transmembrane helix connectivity in Orai1 controls two gates for calcium-dependent transcription. Sci Signal. 2017;10 doi: 10.1126/scisignal.aao0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yamashita M, Yeung PS, Ing CE, McNally BA, Pomes R, Prakriya M. STIM1 activates CRAC channels through rotation of the pore helix to open a hydrophobic gate. Nat Commun. 2017;8 doi: 10.1038/ncomms14512. 14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun. 2013;4 doi: 10.1038/ncomms3963. 2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- [41].Yamashita M, Somasundaram A, Prakriya M. Competitive modulation of CRAC channel gating by STIM1 and 2-aminoethyldiphenyl borate (2-APB) J Biol Chem. 2010;286:9429–9442. doi: 10.1074/jbc.M110.189035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yamashita M, Prakriya M. Divergence of Ca(2+) selectivity and equilibrium Ca(2+) blockade in a Ca(2+) release-activated Ca(2+) channel. J Gen Physiol. 2014;143:325–343. doi: 10.1085/jgp.201311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McNally BA, Prakriya M. Permeation, selectivity and gating in store-operated CRAC channels. J Physiol. 2012;590:4179–4191. doi: 10.1113/jphysiol.2012.233098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang SL, Yeromin AV, Hu J, Amcheslavsky A, Zheng H, Cahalan MD. Mutations in Orai1 transmembrane segment 1 cause STIM1-independent activation of Orai1 channels at glycine 98 and channel closure at arginine 91. Proc Natl Acad Sci U S A. 2011;108:17838–17843. doi: 10.1073/pnas.1114821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Alavizargar A, Berti C, Ejtehadi MR, Furini S. Molecular dynamics simulations of Orai reveal how the third transmembrane segment contributes to hydration and Ca(2+) selectivity in calcium release-activated calcium channels. J Phys Chem B. 2018;122:4407–4417. doi: 10.1021/acs.jpcb.7b12453. [DOI] [PubMed] [Google Scholar]
- [46].Frischauf I, Zayats V, Deix M, Hochreiter A, Jardin I, Muik M, Lackner B, Svobodova B, Pammer T, Litvinukova M, Sridhar AA, et al. A calcium-accumulating region, CAR, in the channel Orai1 enhances Ca(2+) permeation and SOCE-induced gene transcription. Sci Signal. 2015;8:ra131. doi: 10.1126/scisignal.aab1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. %R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca(2+) levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zheng L, Stathopulos PB, Schindl R, Li GY, Romanin C, Ikura M. Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry. Proc Natl Acad Sci U S A. 2011;108:1337–1342. doi: 10.1073/pnas.1015125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Miederer AM, Alansary D, Schwar G, Lee PH, Jung M, Helms V, Niemeyer BA. A STIM2 splice variant negatively regulates store-operated calcium entry. Nat Commun. 2015;6 doi: 10.1038/ncomms7899. 6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rana A, Yen M, Sadaghiani AM, Malmersjo S, Park CY, Dolmetsch RE, Lewis RS. Alternative splicing converts STIM2 from an activator to an inhibitor of store-operated calcium channels. J Cell Biol. 2015;209:653–669. doi: 10.1083/jcb.201412060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chung S, Zhang M, Stathopulos PB. The 2beta splice variation alters the structure and function of the stromal interaction molecule coiled-coil domains. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19113316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang X, Wang Y, Zhou Y, Hendron E, Mancarella S, Andrake MD, Rothberg BS, Soboloff J, Gill DL. Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat Commun. 2014;5 doi: 10.1038/ncomms4183. 3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ercan E, Momburg F, Engel U, Temmerman K, Nickel W, Seedorf M. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 2009;10:1802–1818. doi: 10.1111/j.1600-0854.2009.00995.x. [DOI] [PubMed] [Google Scholar]
- [56].Bhardwaj R, Muller HM, Nickel W, Seedorf M. Oligomerization and Ca2+/calmodulin control binding of the ER Ca2+-sensors STIM1 and STIM2 to plasma membrane lipids. Biosci Rep. 2013;33 doi: 10.1042/BSR20130089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Stathopulos PB, Ikura M. Structure and function of endoplasmic reticulum STIM calcium sensors. Curr Top Membr. 2013;71:59–93. doi: 10.1016/B978-0-12-407870-3.00003-2. [DOI] [PubMed] [Google Scholar]
- [58].Soboloff J, Spassova MA, Hewavitharana T, He LP, Xu W, Johnstone LS, Dziadek MA, Gill DL. STIM2 is an inhibitor of STIM1-mediated store-operated Ca(2+) entry. Curr Biol. 2006;16:1465–1470. doi: 10.1016/j.cub.2006.05.051. [DOI] [PubMed] [Google Scholar]
- [59].Wang Y, Deng X, Hewavitharana T, Soboloff J, Gill DL. Stim, orai and trpc channels in the control of calcium entry signals in smooth muscle. Clin Exp Pharmacol Physiol. 2008;35:1127–1133. doi: 10.1111/j.1440-1681.2008.05018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Malli R, Naghdi S, Romanin C, Graier WF. Cytosolic Ca2+ prevents the subplasmalemmal clustering of STIM1: an intrinsic mechanism to avoid Ca2+ overload. J Cell Sci. 2008;121:3133–3139. doi: 10.1242/jcs.034496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Stathopulos PB, Li G-Y, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ + depletion-induced oligomerization of STIM1 via the EF-SAM region: an initiation mechanism for capacitive Ca2+ + entry. J Biol Chem. 2006 doi: 10.1074/jbc.M608247200. M608247200, %R. [DOI] [PubMed] [Google Scholar]
- [63].Stathopulos PB, Ikura M. Partial unfolding and oligomerization of stromal interaction molecules as an initiation mechanism of store operated calcium entry. Biochem Cell Biol. 2010;88:175–183. doi: 10.1139/o09-125. [DOI] [PubMed] [Google Scholar]
- [64].Ma G, Wei M, He L, Liu C, Wu B, Zhang SL, Jing J, Liang X, Senes A, Tan P, Li S, et al. Inside-out Ca signalling prompted by STIM1 conformational switch. Nat Commun. 2015;6 doi: 10.1038/ncomms8826. 7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dong H, Fiorin G, Carnevale V, Treptow W, Klein ML. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proc Natl Acad Sci U S A. 2013;110:17332–17337. doi: 10.1073/pnas.1316969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hirve N, Rajanikanth V, Hogan PG, Gudlur A. Coiled-coil formation conveys a STIM1 signal from ER lumen to cytoplasm. Cell Rep. 2018;22:72–83. doi: 10.1016/j.celrep.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Fahrner M, Muik M, Derler I, Schindl R, Fritsch R, Frischauf I, Romanin C. Mechanistic view on domains mediating STIM1-Orai coupling. Immunol Rev. 2009;231:99–112. doi: 10.1111/j.1600-065X.2009.00815.x. [DOI] [PubMed] [Google Scholar]
- [68].Huang GN, Zeng W, Kim JY, Yuan JP, Han L, Muallem S, Worley PF. STIM1 carboxyl-terminus activates native SOC, i(crac) and TRPC1 channels. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- [69].Muik M, Fahrner M, Derler I, Schindl R, Bergsmann J, Frischauf I, Groschner K, Romanin C. A cytosolic homomerization and a modulatory domain within STIM1 c terminus determine coupling to ORAI1 channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yang X, Jin H, Cai X, Li S, Shen Y. Structural and mechanistic insights into the activation of stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci U S A. 2012;109:5657–5662. doi: 10.1073/pnas.1118947109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Korzeniowski MK, Manjarres IM, Varnai P, Balla T. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci Signal. 2010;3:ra82. doi: 10.1126/scisignal.2001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zheng S, Zhou L, Ma G, Zhang T, Liu J, Li J, Nguyen NT, Zhang X, Li W, Nwokonko R, Zhou Y, et al. Calcium store refilling and STIM activation in STIM- and Orai-deficient cell lines. Pflug Arch. 2018 doi: 10.1007/s00424-018-2165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wu MM, Covington ED, Lewis RS. Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol Biol Cell. 2014;25:3672–3685. doi: 10.1091/mbc.E14-06-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fahrner M, Muik M, Schindl R, Butorac C, Stathopulos P, Zheng L, Jardin I, Ikura M, Romanin C. A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1) J Biol Chem. 2014;289:33231–33244. doi: 10.1074/jbc.M114.610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Muik M, Fahrner M, Schindl R, Stathopulos P, Frischauf I, Derler I, Plenk P, Lackner B, Groschner K, Ikura M, Romanin C. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 2011;30:1678–1689. doi: 10.1038/emboj.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat Struct Mol Biol. 2013;20:973–981. doi: 10.1038/nsmb.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Misceo D, Holmgren A, Louch WE, Holme PA, Mizobuchi M, Morales RJ, De Paula AM, Stray-Pedersen A, Lyle R, Dalhus B, Christensen G, et al. A dominant STIM1 mutation causes Stormorken syndrome. Hum Mutat. 2014;35:556–564. doi: 10.1002/humu.22544. [DOI] [PubMed] [Google Scholar]
- [80].Morin G, Bruechle NO, Singh AR, Knopp C, Jedraszak G, Elbracht M, Bremond-Gignac D, Hartmann K, Sevestre H, Deutz P, Herent D, et al. Gain-of-function mutation in STIM1 (p.R304W) is associated with stormorken syndrome. Hum Mutat. 2014;35:1221–1232. doi: 10.1002/humu.22621. [DOI] [PubMed] [Google Scholar]
- [81].Nesin V, Wiley G, Kousi M, Ong EC, Lehmann T, Nicholl DJ, Suri M, Shahrizaila N, Katsanis N, Gaffney PM, Wierenga KJ, et al. Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis. Proc Natl Acad Sci U S A. 2014;111:4197–4202. doi: 10.1073/pnas.1312520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Fahrner M, Stadlbauer M, Muik M, Rathner P, Stathopulos P, Ikura M, Muller N, Romanin C. A dual mechanism promotes switching of the Stormorken STIM1 R304W mutant into the activated state. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03062-w. 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol Biol Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cui B, Yang X, Li S, Lin Z, Wang Z, Dong C, Shen Y. The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1. PLoS One. 2013;8:e74735. doi: 10.1371/journal.pone.0074735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Yu F, Sun L, Hubrack S, Selvaraj S, Machaca K. Intramolecular shielding maintains the ER Ca(2)(+) sensor STIM1 in an inactive conformation. J Cell Sci. 2013;126:2401–2410. doi: 10.1242/jcs.117200. [DOI] [PubMed] [Google Scholar]
- [86].Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Barr VA, Bernot KM, Srikanth S, Gwack Y, Balagopalan L, Regan CK, Helman DJ, Sommers CL, Oh-Hora M, Rao A, Samelson LE. Dynamic movement of the calcium sensor STIM1 and the calcium channel Orai1 in activated t-cells: puncta and distal caps. Mol Biol Cell. 2008;19:2802–2817. doi: 10.1091/mbc.E08-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Frischauf I, Muik M, Derler I, Bergsmann J, Fahrner M, Schindl R, Groschner K, Romanin C. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant. J Biol Chem. 2009;284:21696–21706. doi: 10.1074/jbc.M109.018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Calloway N, Vig M, Kinet JP, Holowka D, Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol Biol Cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Calloway NT, Holowka DA, Baird BA. A basic sequence in STIM1 promotes Ca2+ + influx by interacting with the C-terminal acidic coiled-coil of Orai1. Biochemistry. 2010;49:1067–1071. doi: 10.1021/bi901936q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Palty R, Stanley C, Isacoff EY. Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res. 2015;25:963–980. doi: 10.1038/cr.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tirado-Lee L, Yamashita M, Prakriya M. Conformational changes in the Orai1 c-terminus evoked by STIM1 binding. PLoS One. 2015;10:e0128622. doi: 10.1371/journal.pone.0128622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhou Y, Cai X, Loktionova NA, Wang X, Nwokonko RM, Wang X, Wang Y, Rothberg BS, Trebak M, Gill DL. The STIM1-binding site nexus remotely controls Orai1 channel gating. Nat Commun. 2016;7 doi: 10.1038/ncomms13725. 13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Palty R, Isacoff EY. Cooperative binding of stromal interaction molecule 1 (STIM1) to the N and C termini of calcium release-activated calcium modulator 1 (Orai1) J Biol Chem. 2016;291:334–341. doi: 10.1074/jbc.M115.685289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Derler I, Plenk P, Fahrner M, Muik M, Jardin I, Schindl R, Gruber HJ, Groschner K, Romanin C. The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J Biol Chem. 2013;288:29025–29034. doi: 10.1074/jbc.M113.501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fahrner M, Schindl R, Muik M, Derler I, Romanin C. The STIM-Orai pathway: the interactions between STIM and Orai. Adv Exp Med Biol. 2017;993:59–81. doi: 10.1007/978-3-319-57732-6_4. [DOI] [PubMed] [Google Scholar]
- [97].Derler I, Butorac C, Krizova A, Stadlbauer M, Muik M, Fahrner M, Frischauf I, Romanin C. Authentic CRAC channel activity requires STIM1 and the conserved portion of the Orai N terminus. J Biol Chem. 2018;293:1259–1270. doi: 10.1074/jbc.M117.812206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zheng H, Zhou MH, Hu C, Kuo E, Peng X, Hu J, Kuo L, Zhang SL. Differential roles of the C and N termini of Orai1 protein in interacting with stromal interaction molecule 1 (STIM1) for Ca2+ + release-activated Ca2+ + (CRAC) channel activation. J Biol Chem. 2013;288:11263–11272. doi: 10.1074/jbc.M113.450254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fahrner M, Pandey SK, Muik M, Traxler L, Butorac C, Stadlbauer M, Zayats V, Krizova A, Plenk P, Frischauf I, Schindl R, et al. Communication between N terminus and loop2 tunes Orai activation. J Biol Chem. 2018;293:1271–1285. doi: 10.1074/jbc.M117.812693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Palty R, Fu Z, Isacoff EY. Sequential steps of CRAC channel activation. Cell Rep. 2017;19:1929–1939. doi: 10.1016/j.celrep.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, Shi X, et al. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx. Nat Cell Biol. 2015;17:1339–1347. doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 2012;149:425–438. doi: 10.1016/j.cell.2012.01.055. [DOI] [PubMed] [Google Scholar]
- [104].Jardin I, Albarran L, Salido GM, Lopez JJ, Sage SO, Rosado JA. Fine-tuning of store-operated calcium entry by fast and slow Ca(2+)-dependent inactivation: involvement of SARAF. Biochim Biophys Acta. 2018;1865:463–469. doi: 10.1016/j.bbamcr.2017.12.001. [DOI] [PubMed] [Google Scholar]
- [105].Srikanth S, Jung HJ, Kim KD, Souda P, Whitelegge J, Gwack Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol. 2010;12:436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Derler I, Jardin I, Stathopulos PB, Muik M, Fahrner M, Zayats V, Pandey SK, Poteser M, Lackner B, Absolonova M, Schindl R, et al. Cholesterol modulates Orai1 channel function. Sci Signal. 2016;9:ra10. doi: 10.1126/scisignal.aad7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Pacheco J, Dominguez L, Bohorquez-Hernandez A, Asanov A, Vaca L. A cholesterol-binding domain in STIM1 modulates STIM1-Orai1 physical and functional interactions. Sci Rep. 2016;6 doi: 10.1038/srep29634. 29634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Demuro A, Penna A, Safrina O, Yeromin AV, Amcheslavsky A, Cahalan MD, Parker I. Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc Natl Acad Sci U S A. 2011;108:17832–17837. doi: 10.1073/pnas.1114814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Maruyama Y, Ogura T, Mio K, Kato K, Kaneko T, Kiyonaka S, Mori Y, Sato C. Tetrameric Orai1 is a teardrop-shaped molecule with a long, tapered cytoplasmic domain. J Biol Chem. 2009;284:13676–13685. doi: 10.1074/jbc.M900812200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Mignen O, Thompson JL, Shuttleworth TJ. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J Physiol. 2008;586:419–425. doi: 10.1113/jphysiol.2007.147249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Penna A, Demuro A, Yeromin AV, Zhang SL, Safrina O, Parker I, Cahalan MD. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hoover PJ, Lewis RS. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci U S A. 2011;108:13299–13304. doi: 10.1073/pnas.1101664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Scrimgeour N, Litjens T, Ma L, Barritt GJ, Rychkov GY. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J Physiol. 2009;587:2903–2918. doi: 10.1113/jphysiol.2009.170662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Scrimgeour NR, Wilson DP, Barritt GJ, Rychkov GY. Structural and stoichiometric determinants of Ca release-activated Ca (CRAC) channel Ca-dependent inactivation. Biochim Biophys Acta. 2014;1838:1281–1287. doi: 10.1016/j.bbamem.2014.01.019. [DOI] [PubMed] [Google Scholar]
- [115].McNally BA, Somasundaram A, Yamashita M, Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 2012;482:241–245. doi: 10.1038/nature10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhou Y, Wang X, Wang X, Loktionova NA, Cai X, Nwokonko RM, Vrana E, Wang Y, Rothberg BS, Gill DL. STIM1 dimers undergo unimolecular coupling to activate Orai1 channels. Nat Commun. 2015;6 doi: 10.1038/ncomms9395. 8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhou Y, Nwokonko RM, Cai X, Loktionova NA, Abdulqadir R, Xin P, Niemeyer BA, Wang Y, Trebak M, Gill DL. Cross-linking of Orai1 channels by STIM proteins. Proc Natl Acad Sci U S A. 2018;115:3398–3407. doi: 10.1073/pnas.1720810115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Vig M, Beck A, Billingsley JM, Lis A, Parvez S, Peinelt C, Koomoa DL, Soboloff J, Gill DL, Fleig A, Kinet JP, et al. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kim KM, Wijerathne T, Hur JH, Kang UJ, Kim IH, Kweon YC, Lee AR, Jeong SJ, Lee SK, Lee YY, Sim BW, et al. Distinct gating mechanism of SOC channel involving STIM-Orai coupling and an intramolecular interaction of Orai in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2018;115:4623–4632. doi: 10.1073/pnas.1714986115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Yeung PS, Yamashita M, Prakriya M. Pore opening mechanism of CRAC channels. Cell Calcium. 2016;63:14–19. doi: 10.1016/j.ceca.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].He L, Zhang Y, Ma G, Tan P, Li Z, Zang S, Wu X, Jing J, Fang S, Zhou L, Wang Y, et al. Near-infrared photoactivatable control of Ca(2+) signaling and optogenetic immunomodulation. Elife. 2015;4 doi: 10.7554/eLife.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ishii T, Sato K, Kakumoto T, Miura S, Touhara K, Takeuchi S, Nakata T. Light generation of intracellular Ca(2+) signals by a genetically encoded protein BACCS. Nat Commun. 2015;6 doi: 10.1038/ncomms9021. 8021. [DOI] [PMC free article] [PubMed] [Google Scholar]