Abstract
Glycans are the major carbon sources available to the human colonic microbiota. Numerous N-glycosylated proteins are found in the human gut, from both dietary and host sources, including immunoglobulins such as IgA which are secreted into the intestine at high levels. Here we show that many mutualistic gut Bacteroides spp. have the capacity to utilise complex N-glycans (CNGs) as nutrients, including those from immunoglobulins. Detailed mechanistic studies using transcriptomic, biochemical, structural and genetic techniques reveal the pathway employed by B. thetaiotaomicron (Bt) for CNG degradation. The breakdown process involves an extensive enzymatic apparatus encoded by multiple non-adjacent loci and comprises 19 different carbohydrate-active enzymes (CAZymes) from different families, including a CNG specific endo-glycosidase activity. Furthermore, CNG degradation involves the activity of CAZymes that have previously been implicated in the degradation of other classes of glycan. This complex and diverse apparatus provides Bt with the capacity to access the myriad different structural variants of CNGs likely to be found in the intestinal niche.
Introduction
The composition of the human gut microbiota is closely associated with many aspects of health, development and disease1–4. The major nutrients available to the colonic microbiota are complex carbohydrates and glycan preferences shape the composition of this microbial community5. While dietary plant polysaccharides likely make up the bulk of glycans reaching the large intestine, host glycans are also significant nutrient source and access to these host molecules appears to be important for gut colonisation and survival6–9.
N-linked glycans are commonly present within the gut as almost all secreted eukaryotic proteins are N-glycosylated to some extent. N-glycans are composed of a conserved pentasaccharide core capped with a diverse range of different monosaccharides to give three main types; complex, high mannose and hybrid, although there is significant heterogeneity within these structures (Fig. 1a)10. N-glycans found in the gut come from a range of sources, including dietary animal products and host molecules such as mucins and antibodies11,12. For example, secretory IgA, the major immunoglobulin produced in the intestine, is heavily decorated with mainly CNG structures13,14.
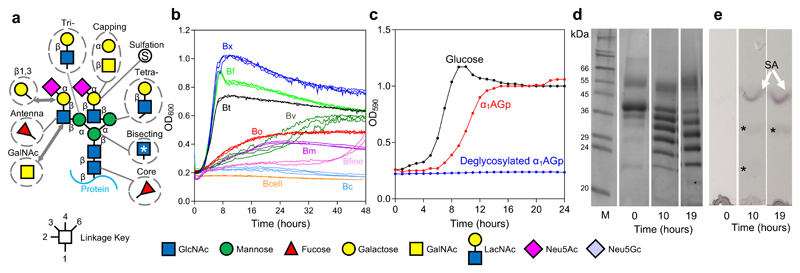
Figure 1. Complex N-glycans as a nutrient source in Bacteroides species.
a, Structure of a biantennary complex N-glycan (CNG) with other possible decorations and variations. The linkage is indicated by the key, however the sialic acid sugars can be attached through α2,3 or 2,6 linkages to the galactose and also to the antennary GlcNAc. Biantennary CNG have both antennae linked through β1,2 bonds and additional antenna have either a β1,4-linkage (on the α1,3 arm) or a β1,6-linkage (on the α1,6 arm) to produce tri- and tetra-antennary CNG, respectively. Common modifications to this core model are shown in dotted circles. b, Growth on native α1AGp of different Bacteroides spp. - B. thetaiotaomicron (Bt, black), B. xylanisolvans (Bx, dark blue), B. ovatus (Bo, red), B. vulgatus (Bv, dark green), B. finegoldii (Bfine, pink), B. massiliensis (Bm, magneta), B. fragilis (Bf, light green), B. caccae (Bc, light blue) and B. cellulolyticus (Bcell, orange). Growth curves were carried out at least twice for each species. c, Growth of B. thetaiotaomicron on glucose (5 mg/ml), α1AGp and deglycosylated α1AGp (both 20mg/ml) as the sole carbon source. d, Supernatant samples were taken at the start, 10 hour and 19 hour time points from the growth of Bt on α1AGp show in panel c and were analysed by SDS-PAGE. Fully glycosylated α1AGp is shown at T0 and fully deglycosylated α1AGp is the bottom 23.5 kDa protein band in the 10 h and 19 h time point lanes. e, TLC of the cell free spent media at the 10 h and 19 h time points to analyse glycans present. The top band is sialic acid (white arrow and “SA”) and two other faint glycan bands are indicated to (*). The results are representative of at least three independent replicates. Full versions of the gels and TLCs can be found in Supplementary Figure 1 and 23.
Despite the prominence of N-glycans in the human intestine, there are only a few studies describing their use as nutrients by the microbiota and the pathways of N-glycan breakdown employed by mutualistic microbes. Some strains of Bifidobacteria have been found to utilise N-glycans on lactoferrin from milk15, while Bt has been shown to express a single PUL to access high mannose N-glycans (HMNG)8. While there is a paucity of data on N-glycan use by the normal gut microbiota, especially of the complex type, these molecules are well known to be key nutrients for some pathogens, as the ability to access N-glycans facilitates survival of these microbes within the extra-intestinal niche16–21. Although several studies describe aspects of N-glycan degradation by enteric pathogens, most of these focus on only a single part of the degradation pathway or the role of this process in virulence21–24.
Bacteroides are one of the dominant genera of the human gut25 and are prominent glycan degraders, with many species encoding hundreds of different CAZymes to facilitate access to the diverse array of glycans available6–8,26–32. In Bacteroides, the genes encoding the glycan degrading apparatus are usually co-localised in discrete clusters known as polysaccharide utilisation loci (PULs)33. PULs are typically defined by their composition as containing SusC-like and SusD-like gene pairs that encode the outer membrane glycan import machinery34, as well as a sensor-regulator and multiple CAZymes. Often a single PUL encodes all of the apparatus required to access a specific glycan structure, although multiple PULs have been shown to be involved in the degradation of some highly complex glycans 5 35.
In this study we set out to further our understanding of CNG breakdown by the normal gut microbiota, focusing on prominent Bacteroides spp. The data reveal that the ability to utilise CNGs is commonplace amongst gut Bacteroides and that growth of Bt on CNG activates an extensive enzymatic apparatus encoded by multiple loci, including a single core PUL that likely controls expression of the other non-adjacent loci. Detailed characterisation of the Bt CNG breakdown pathway provides significant insights into the mechanism of N-glycan degradation by key members of the microbiota and provides knowledge for future studies examining the role of this process in gut survival.
Results
Growth of gut Bacteroides on CNGs
Prominent gut Bacteroides spp. were tested for their ability to grow on native Bovine α1 acid glycoprotein (α1AGp) as the sole carbon source (Fig. 1a). Of the 9 species tested, most could grow to some extent on α1AGp, with only B. caccae and B. celluloslyticus showing no growth over 48h (Fig. 1b). Analysis of supernatant after growths revealed that no protein degradation had occurred and protein remaining was almost completely deglycosylated, indicating that growth was supported by the glycan component of α1AGp alone (Supplementary Fig. 1a&b). Growth on other types of CNG-containing glycoproteins was also assessed (Supplementary Fig. 1f) and described in Supplementary Results and Discussion.
RNAseq to identify CNG degrading apparatus
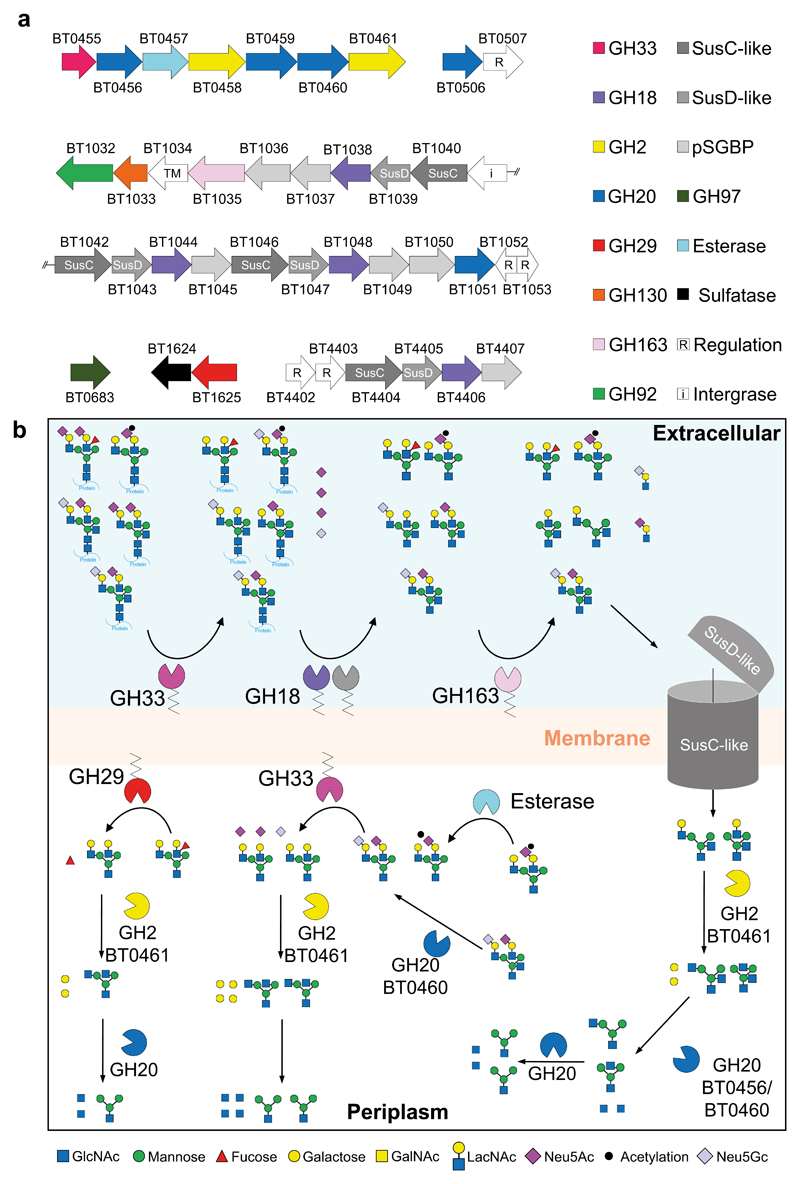
RNAseq was used to identify the predicted CAZyme and associated genes upregulated during growth of Bt on α1AGp. Six discrete loci were identified from this analysis that were composed of in total 19 predicted CAZymes, 4 SusC/D pairs, and several ORFs of unknown function (Fig. 2, Supplementary Table 1&2, Supplementary Results and Discussion, Supplementary Fig. 2). Only two of the loci are classed as PULs in that they encode predicted CAZymes, SusC/D homologues and sensor-regulators33. These data indicate that breakdown of CNGs by Bt requires the cooperative action of a complex multi-locus encoded machinery. To understand the degradative pathway for CNGs in Bt we carried out biochemical characterisation of the CAZymes upregulated on α1AGp.
Figure 2. Genes upregulated in Bt genes during growth on CNG.
a, A schematic of the 6 CAZyme-containing loci upregulated in Bt during growth on α1AGp. TM: transmembrane protein; pSGBP: putative surface glycan binding protein. b, A model of the degradation of CNGs by Bt. BT1625GH29 is placed in the periplasm but could in fact be facing the outside of the cell. The degradation of the common core tetrasaccharide likely occurs through the activity of previously characterised enzymes located in the periplasm and cytoplasm (Supplementary Table 2, Supplemental Results and Discussion).
BT0455GH33 sialidase is required for growth on CNGs and is localised both on the cell surface and inside the cell
Mammalian CNGs are often capped with sialic acid (SA), with N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) being the dominant forms, however only Neu5Ac is found in humans (Fig. 1)36. Neu5Gc would therefore only be available to the microbiota from eukaryotic dietary sources, while Neu5Ac would be present in both dietary and host glycans. BT0455GH33 is the only sialidase encoded by the Bt genome and has been previously characterised as having broad specificity, acting on a range of SA capped structures including CNGs37.
Bt cannot metabolise SA, but requires its removal to enable access to underlying glycan 38 and as expected SA build-up can be seen in the supernatant over the course of growth of Bt on α1AGp (Fig. 1e and Supplementary Fig. 1&3). BT0455GH33 was found to be critical for the utilisation of CNGs by Bt as a BT0455 deletion strain (ΔBT0455) was unable to grow and the defect could be rescued by addition of recombinant BT0455 to the media (Supplementary Fig. 1).
We next examined the cellular location BT0455GH33 using a whole cell activity assays and protease protection assays (Supplementary Fig. 4-7 and Supplementary Results and Discussion). Unexpectedly, the data revealed that BT0455GH33 is localised both inside the cell and on the cell surface. In the same locus as BT0455GH33 is an ORF (BT0457) which shares 70% identity with the Tannerella forsythia SA esterase, NanS39. BT0457 was shown to have similar activity to NanS against differentially acetylated forms of SA (Supplementary Fig. 8) and is predicted to be periplasmic, providing a likely explanation for the unusual dual surface and periplasmic localisation of the sialidase BT0455GH33. This is discussed in more detail in Supplementary Results and Discussion.
CNGs are cleaved from native glycoproteins at the cell surface by GH18 endo-glucosaminidases
Previously characterised members of the GH18 family include endo-β-N-acetylglucosaminidases with a range of different specificities against different types of N-glycans (Supplementary Table 3). The RNAseq data revealed four GH18 family members upregulated during growth on α1AGp (Fig. 2), which were tested for activity against a variety of N-glycosylated proteins including those with different CNG, high mannose, plant-type and hybrid structures (Supplementary Fig. 9a&b). Surprisingly, only BT1044GH18 displayed activity against any of the glycoproteins tested, and of the substrates tested this was limited to release of glycans from α1AGp, IgA and IgG from human serum, and IgA from human colostrum (IgAs, IgGs and IgAc, respectively). None of the four GH18 family members were active on chitin or chito-oligosaccharides (Supplementary Fig. 9c).
To investigate the specificity of BT1044GH18 in more detail, products from digested glycoproteins were labelled with the fluorophore procainamide and analysed by liquid chromatography-fluorescence detection-electrospray-mass spectrometry (LC-FLR-ESI-MS) (Fig. 3a&b, Fig. 4a-c). The structures of the various CNGs were determined using MS/MS and showed predominantly bi-antennary CNG structures released, including species with bisecting GlcNAcs, antenna fucosylation and polyLacNAc antenna structures. Tri-antennary N-glycan can also be removed, but only if the third antenna is limited to a single GlcNAc (e.g. Fig. 4, glycan 5). In comparison, PNGaseF treatment (removes all N-glycan structures) revealed tri-antennary CNG structures were present on IgAs and bovine fetuin, but these were not removed by BT1044GH18 (Supplementary Fig. 10 & 11a-c).
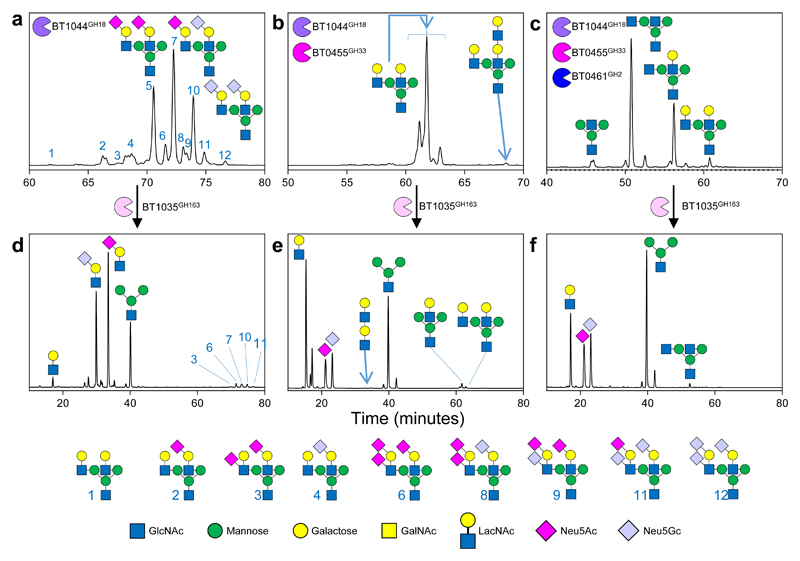
Figure 3. The degradation of biantennary CNG by recombinant enzymes from Bt.
a, α1AGp digested with BT1044GH18 endo-GlcNAc’ase only and b, then in combination with BT0455GH33 sialidase, and c, then also BT0461GH2 β-galactosidase. d-f, Each of the assays shown in a-c were stopped after a 24 h incubation using heat denaturation and BT1035GH163 then added. The time shown for the different chromatograms varies between panels to provide clarity of the main peaks. The glycan products were labelled with procainamide and analysed by LC-FLD-ESI-MS (see Materials and Methods). The most abundant glycan products are annotated on the chromatograms and the minor products are shown in the key at the bottom. The same glycan species detected in multiple peaks is likely due to different linkages and glycosylation on different arms, which cannot be determined by the analytical methods employed. Neu5Ac and Neu5Gc are pink and light blue, respectively. The linkages they are attached through could not be determined using the techniques employed here and also likely vary between glycans and glycoproteins. The results are representative of at least three independent replicates. A sugar key is included in Figure 1.
Figure 4. The activity of BT1044GH18 on immunoglobulin substrates and structural of the enzyme.
Activity of BT1044GH18 against a, human serum IgG b, human serum IgA and c, human colostrum IgA is shown in the presence of BT0455GH33 sialidase. The products with a β1,4 bisecting GlcNAc are indicated to by a white asterisk. The results are representative of at least three independent replicates. d, Cartoon depiction of the predicted orientation of BT1044GH18 on the cell surface of Bt showing the attachment to the cell surface through a lipid anchor. Loops 1-7 on the surface of the enzyme with the active site are coloured yellow, green, orange, blue, magenta, purple and dark green, respectively. e, The surface of the BT1044GH18 crystal structure is shown again with the same loop colouring and the two catalytic residues are in red. For comparison, the previously published crystal structure of the CNG-active enzyme EndoF3 (1EOM) from Elizabethkingia meningoseptica is shown (cyan). The conserved residues from the DxxDxDxE motif have been coloured orange and the approximate +1 (GlcNAc) and +1’ (fucose) subsites are indicated to by white and red dashed lines, respectively. BT1044GH18 has more of a groove (white dashed box) in the equivalent section. The area where a bisecting GlcNAc would be predicted to sit is indicated to by an asterisk. This space is occupied in Endo F3, whereas in BT1044GH18 there is more space to accommodate this sugar, reflecting their respective activities (see Supplementary Fig. 21 for more details). The results are representative of at least three independent replicates.
Assays against sialylated and desialylated forms of α1AGp with BT1044GH18 showed the enzyme can act on both, but with a preference for the latter, suggesting removal of CNGs from the protein mainly occurs after the action of the surface sialidase (Supplementary Fig. 9d). The GH18 EndoE from Enterococcus faecalis also has a preference for desialylated N-glycans 40, whereas the GH18 EndoS2 from Streptococcus pyogenes can remove either type without preference 22 (Supplementary Table 3). BT1044GH18 was also shown to tolerate fucosylation of the core GlcNAc (Supplementary Fig. 12a).
A ΔBT1044 strain displayed only a minor growth defect on α1AGp compared to the wild type strain (Supplementary Fig. 1g). Furthermore, analysis of surface enzyme activity of ΔBT1044 vs wild type cells against α1AGp showed similar product profiles (Supplementary Fig. 7a) and treatment of α1AGp using both BT0455GH33 and BT1044GH18 revealed the level of deglycosylation appeared less than after growth on this glycoprotein (Supplementary Fig. 1a&c). Together, these data suggest that in addition to BT1044GH18, one or more of the other three upregulated GH18 members or a currently unknown enzyme, are also involved in deglycosylating CNGs in vivo. This apparent redundancy in GH18 activity has been observed previously in Bacteroides spp; a B. fragilis mutant strain lacking the main GH18 upregulated during growth on rat transferrin retained the ability to grow on the serum glycoprotein, albeit at a slower rate20. Deletion of the other three Bt GH18 genes upregulated on CNGs was attempted to explore their role further, but we were unable to obtain these mutants.
The structure of BT1044GH18 endo-glucosaminidase
To investigate the structural basis for substrate specificity displayed by BT1044GH18, the crystal structure of the enzyme was solved to 2.4 Å resolution (Fig. 4). BT1044GH18 adopts the canonical (β/α)8 barrel (TIM barrel) fold common to many GH structures.
Notably, the N-terminal region forms a discrete extended structure that would orientate the lipoprotein on the cell surface with the active site facing the extracellular environment (Fig. 4d & Supplementary Results and Discussion). This is likely necessary to prevent spatial restrictions in accessing bulky native glycoprotein substrates. Furthermore, there are ~20 amino acids after the N-terminal lipid anchoring Cys that are not visible in the structure. It appears likely therefore that this region acts as a relatively unstructured linker between the lipid anchor and the folded N-terminal domain to further extend the distance between the cell surface and the enzyme’s active site. To understand substrate specificity in more detail, the structure of BT1044GH18 was compared to that of Elizabethkingia meningoseptica GH18 (EndoF3) in complex with a biantennary CNG product (Fig. 4e & see Supplementary Results and Discussion)41.
CNG structures are predominantly degalactosylated in the periplasm by BT0461GH2
SA removal from CNGs exposes the predominantly β1,4-linked galactose (although occasionally this can be β1,3-linked) (Fig. 1)42. Inspection of the CAZymes upregulated on α1AGp revealed only two possible β-galactosidase candidates; BT0458GH2 and BT0461GH2. As BT0458 has previously been shown to be a β-mannosidase43, BT0461GH2 alone was screened against a variety of defined β-galactose substrates and found to have a preference for the structures and linkages found in CNGs (Supplementary Table 4).
BT0461GH2 was tested on BT1044GH18-liberated CNG from α1AGp pre-treated with BT0455GH33 and the products were predominantly fully degalactosylated (Fig. 3c). PNGaseF-liberated and desialylated CNG from IgAs, IgGs and IgAc substrates were also tested. All of the galactose was removed from the IgGs and IgAs glycans, but there was a minor population of galactose-containing structures remaining from the IgAc sample (Supplementary Fig. 11d-f). The IgAc glycans with antenna fucosylation on the +1 sugar (GlcNAc) were not degalactosylated, indicating the fucose was blocking access of BT0461GH2. A lack of BT0461GH2 activity against fucosylated LacNAc and LNB (Lewis X and A, respectively) supported the need for antennary fucose removal prior to galactose release (Fig. 1, Supplementary Fig. 11k & Supplementary Results and Discussion). The antennary fucose, which often decorates CNG structures, is removed by BT1625GH29 fucosidase (Supplementary Fig. 11d and Supplementary Table 7). Details of BT1625GH29 activity are described in Supplementary Results and Discussion.
The multiple GH20 genes upregulated on α1AGp display complementary but overlapping activities against CNG
Degalactosylation of CNGs reveals β-linked GlcNAcs which are β1,2 linked in biantennary, but β1,4 (on the α1,3 arm) and β1,6 (on α1,6 arm) in tri- and tetra-antennary CNG, respectively (Fig. 1). The core mannose can also have a β1,4 GlcNAc (termed ‘bisecting’). GH20 family members are known to be predominantly exo-acting β-hexosaminidases and four of these were expressed during growth on α1AGp (Fig. 2). Localisation studies indicate all were most likely periplasmic (Supplementary Fig. 7c). Notably, two of the enzymes contain a Type II signal sequence suggesting they are membrane associated but are not trafficked to the cell surface (Supplementary Table 2).
To define the specificity of the four GH20 enzymes they were tested against CNGs from α1AGp, IgGs, IgAs and IgAc liberated with PNGaseF and pre-treated with BT0455GH33 and BT0461GH2 (Fig. 5 and Supplementary Table 6 & Supplementary Fig. 11e-g). Activity of the GH20 family members was also tested against CNGs released by PNGase from IgGs with no pre-treatment with BT0455GH33 and BT0461GH2 (Supplementary Fig. 13).
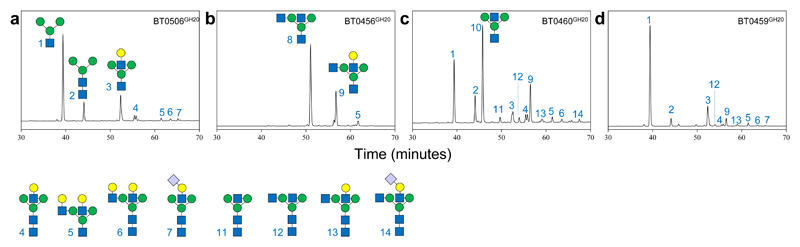
Figure 5. GH20 activity on α1AGp.
Activity was assessed for a, BT0506GH20, b, BT0456GH20 c, BT0460GH20 and d, BT0459GH20. Procainamide labelled products were analysed as described previously for other assays and the samples were pre-digested with BT0455GH33, BT1044GH18 and BT0461GH2. The results are representative of at least three independent replicates.
BT0506GH20 is specific and highly active against the antenna GlcNAcs, but failed to have any activity against the bisecting β1,4-GlcNAc. BT0459GH20 is also predominantly specific for a range of antenna GlcNAcs, however, trace activity on bisecting structures could be detected. The difference between these enzymes is BT0459GH20 struggles to remove both antenna GlcNAcs, usually when a bisecting GlcNAc is present, indicative of a steric block (Supplementary Fig 11 e.g. glycans 28 & 31).
In contrast, BT0456GH20 and BT0460GH20 were both able to remove the bisecting GlcNAc when samples had SA and Gal removed. Any further antenna decoration meant BT0456GH20 was no longer active on bisecting structures. BT0460GH20, however, could remove bisecting GlcNAc even when both antennae had Gal and SA present (Supplementary Fig. 13 & Supplementary Table 6).
In terms of antennary deglycosylation, BT0460GH20 was able to remove all GlcNAc in the substrates tested, while products from the BT0456GH20 had a high proportion with only one uncapped GlcNAc remaining indicating a preference for one arm. Furthermore, if the one antenna has a Gal (or SA as well) cap, then BT0456GH20 cannot not access the GlcNAc on the other antenna, again indicating a preference for one arm or a steric block (Supplementary Fig. 13, glycans 4, 6 and 11 remaining). Overall these data reveal that the four the Bt GH20 family members characterised here display complementary but overlapping activities that facilitate access to the diverse GlcNAc linkages present on CNGs.
Crystal structure of BT0459GH20 GlcNAc’ase
The structure of BT0459GH20 was solvedto 2.4 Å with GlcNAc product present in the -1 subsite. The enzyme is made up of four domains – a small N-terminal domain, a (β/α)8 barrel catalytic domain, an FN3 domain and a C-terminal F5/F8 type C domain that are oriented overall to form a hook-like shape with the C-terminal domain facing the active site (Fig. 6). The potential role of the C-terminal F5/F8 type C domain in substrate binding is discussed in Supplementary Results and Discussion.
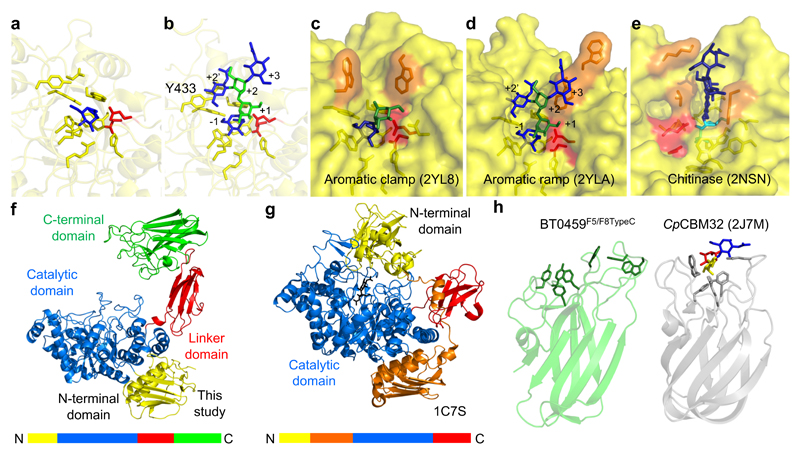
Figure 6. Crystal structure of BT0459GH20.
a, A close up of the active site of BT0459GH20 with the residues forming the -1 subsite shown as sticks and a GlcNAc (blue) product in the active site. b, A close up of the active site of BT0459GH20 overlaid with a CNG structure (from 2YLA). Mannose and GlcNAc are green and blue, respectively, with the antennary GlcNAc in the active site. The α1,3 mannose arm is in the +1 position and the core mannose and GlcNAc occupy the +2 and +3 subsites, respectively. A clash can be seen between the Y433 and the bisecting GlcNAc in the +2’ position. The active sites of the S. pneumonie c, SpGH20A and d, SpGH20B, and e, a GH20 active on chitooligosaccharides from O. furnacalis. The inhibitor in e is TMG-chitotriomycin (TMG and GlcNAcs shown in cyan and dark blue, respectively). Catalytic residues are shown in red and those residues interacting with sugars in the positive subsites are in orange. f, The full length structure of BT0459GH20 and g, the S. marcescens GH20 (1C7S). The order of the modules are shown as coloured bars. h, The C-terminal F5/F8 type C domain of BT0459GH20 is shown (BT0459F5/F8TypeC, left) and aromatic residues on the potential glycan binding surface are shown as sticks. A structural homologue of BT0459F5/F8TypeC is shown for comparison (right) and is a CBM32 from a C. perfringens GH84 (CpCBM32). This has a trisaccharide ligand of fucose, galactose and GlcNAc (red, yellow and blue, respectively) bound and the aromatic residues involved in binding are shown as sticks. The core folds between BT0459F5/F8TypeC and CpCBM32 are very similar but the potential glycan binding surfaces vary greatly (see Supplementary Fig. 12 for further structural homologues).
Compared to other known GH20 structures, BT0459GH20 has a very open active site structure with only the -1 site, indicating little interaction with the +1 sugar and potentially providing a rationale for the broad substrate specificity displayed by this enzyme (Fig. 6a&b and Supplementary Fig. 14-16). A comparison of this active site with CNG-active SpGH20A and SpGH20B from S. pneumoniae (Fig. 6c&d)44 and also a GH20 from Ostrinia furnacalis (Fig. 6e) specific for chitooligosaccharides45 highlights the unusual openness of the BT0459GH20 active site (Supplementary Results and Discussion).
BT1035GH163 is a surface located enzyme with CNG-specific endo GlcNAc’ase activity
Within the large CNG-upregulated locus is an ORF (BT1035) of unknown function that sequence analysis suggests is a glycoside hydrolase. It is predicted to have an N-terminal DUF4838 domain that has distant homology to GH20 and a C-terminal F5/F8 type C domain. Incubation of recombinant BT1035GH163 against BT1044GH18-liberated CNG from α1AGp showed it was able to release sialylated LacNAc (both Neu5Ac and Neu5Gc) (Fig. 3a&d). BT1035GH163 was able to act on the CNG while still attached to the protein, but displayed a preference for the GH18 released glycan (Supplementary Fig. 17c). The enzyme was also capable hydrolysing the same GlcNAc-β1,2-Mannose linkage in desialylated α1AGp (Fig. 3b&e). Comparison of BT1035GH163 activity against sialylated and desialylated CNGs showed that desialylated CNG was found to be a more favourable substrate, possibly indicating that SA attached through particular linkages cannot be accommodated by the enzyme (Supplementary Fig. 18). CNG structures with SA linked to the GlcNAc were not hydrolysed by BT1035GH163, indicating the enzyme has no -1’ subsite (Fig. 3d, no SA-GlcNAc products). BT1035GH163 was also able to remove larger diLacNAc structures from CNG antenna, however the enzyme could not degrade the diLacNAc product further, suggesting that either the +1 subsite cannot tolerate Gal, or has a requirement for Man i.e. is CNG specific, or the enzyme is linkage-specific (Fig. 3b&e).
Using CNGs pre-treated with BT0455GH33 and BT0461GH2 revealed BT1035GH163 was also able to hydrolyse the GlcNAc-β1,2-Mannose linkage to almost totally remove both the remaining GlcNAc and LacNAc, showing that the galactose in the -2 subsite is not an absolute requirement for activity (Fig. 3c&f). Notably, however, BT1035GH163 was unable to cleave the GlcNAc-β1,2-Mannose disaccharide (Supplementary Fig. 17a), indicating interactions with a +2 subsite are required for activity (i.e. with the core mannose), although the context of this sugar would vary depending on the specific arm targeted. Activity against IgA and IgG showed that bisecting GlcNAc does not interfere with BT1035GH163 hydrolysis (Supplementary Fig. 11h-j).
The capacity of to remove LacNAc, sialyl LacNAc and diLacNAc from CNGs and lack of activity against the GlcNAc-Man disaccharide indicate BT1035GH163 displays an endo-like activity that targets the β-GlcNAc-Man found in these structures.
Analysis of the cellular location of BT1035GH163 revealed the enzyme is on the cell surface, providing an explanation for the release of a sialylated-LacNAc structure (predominantly Neu5Gc) outside the cell during growth on α1AGp (Supplementary Figs. 1, 3 & 4). Growth data suggest that BT0455GH33 and BT1035GH163 are both active in vivo from early to mid-exponential onwards, with SA released slightly earlier (Supplementary Fig. 1). Whole cell assay data also show BT0455GH33 and BT1035GH163 both act rapidly on a1AGp in vitro, but the sialylated-LacNAc structures produced by BT1035GH163 remain sialylated. This suggests that BT1035GH163 is accessing glycan structures that BT0455GH33 cannot in vivo and a biological rationale for this activity on the cell surface of BT1035GH163 may be to increase the proportion of CNG structures that can be accessed.
It may also be that CNGs with particular structures are poor substrates for the SusCD glycan import apparatus and thus removal of some trisaccharides by BT1035GH163 facilitates more efficient uptake of these glycans (see Supplementary Results and Discussion).
Discussion
This study characterises the pathway for CNG breakdown by a prominent member of the normal human gut microbiota. CNGs are highly variable in terms of sugar composition and linkages, displaying extensive heterogeneity even within a single glycoprotein46. For example, human IgG is decorated with 18 different N-glycan structures, few of which are shared with the other glycoproteins tested here.
Here we describe the extensive enzyme apparatus Bt uses to deal with this heterogeneity. We show that significant processing of predominantly biantennary forms of CNG occurs at the cell surface prior to import, including removal of glycan from the protein, desialylation and the action of BT1035GH163. The exact biological advantage BT1035GH163 provides could not be fully elucidated in this study, but one role may be to increase the variety of CNGs that Bt can access. After removal of galactose and antennary fucose, the complementary but overlapping activities of the four different GH20 enzymes allow access to a diverse range of antennary and bisecting GlcNAc structures.
Notably, the broad specificity of the exo-acting enzymes could be a reflection of their role in the degradation of multiple types of glycan with conserved structural features in common. Thus, many of the enzymes involved in CNG breakdown in Bt also appear to play a role in the degradation of O-glycans; both the BT0455-BT0461 locus and BT1624-BT1625 have been shown to be expressed during growth of Bt on mucin 26,47. It is possible that the shared structures contained within O-glycans and CNGs (i.e. β-linked Gal and GlcNAcs, as well as capping SA and fucose) targeted by these enzymes and the consistent presence of these glycans from host sources in the gut (i.e. less variable that dietary glycans) have driven the overlapping regulation of these enzymes in Bt.
Related to this, a previous study indicates that the anti-sigma regulator of the core BT1032-BT1053 CNG PUL, BT1053, directly controls expression of the whole multi-locus CNG degradation apparatus48. Thus, when BT1053 was deleted to derepress the genes under its control, the same loci were upregulated as were seen here to be activated by CNGs. As the sensor-regulator for the core BT1032-BT1053 PUL is an ECF-sigma/anti-sigma system, the activating signal for expression of the whole Bt multi-locus CNG apparatus will be import of discrete CNG structures by the core PUL encoded SusCD.
The ability of Bacteroides spp. to deglycosylate secretory IgA is an interesting finding of this work which raises the possibility that this process plays a role in modulating the function of this important gut immune molecule and is an intriguing avenue of research to pursue in the future11,49,50.
Methods
Sources of glycans and glycoproteins
Glycoproteins bovin α1acid glycoprotein, bovine fetuin, bovine RNaseB, chicken egg white ovalbumin, horseradish peroxidase, human serum IgG, human serum IgA, human colostrum IgA and p-Nitrophenyl (PNP) monosaccharides were obtained from Sigma. Purified di- and oligo-saccharides were obtained from Carbosynth. The sialic acid reference panel was obtained from Ludger. Squid chitin was a gift from Prof. Gideon Davies (Univ. of York, UK).
Bacterial strains
Bacteroides strains used were as follows: B. thetaiotaomicron VPI-5482, B. fragilis NCTC9342, B. caccae ATCC43185, B. cellulosilyticus DSM14838, B. massiliensis DSM17679, B. finegoldii DSM17565, B. vulgatus ATCC8483, B. ovatus ATCC8482 and B xylanisolvans XBA1.
Cloning, expression and purification of recombinant proteins
DNA encoding the appropriate genes (excluding the signal sequences) were amplified from genomic DNA using appropriate primers and cloned into pET28b (Novagen) using NheI-XhoI restriction sites. His-tags were located at the N-terminus. Recombinant plasmids were transformed into TUNER (Novagen) cells in LB broth containing 10 μg/ml kanamycin at 37 °C shaking at 180 rpm. One litre cultures were grown to mid-exponential phase in 2 litre baffled flasks, cooled to 16 °C and isopropyl β-D-thiogalactopyranoside (IPTG) added to a final concentration of 0.2 mM. These cells were then incubated for 16 hours at 16 °C in an orbital shaker at 150 rpm. Recombinant His-tagged protein was purified from cell-free extracts using immobilised metal affinity chromatography (IMAC using Talon resin; Clontech) as described previously 51.
The purity and size of the proteins were checked using SDS-PAGE and their concentrations determined using absorbance at 280 nm (NanoDrop 2000c; Thermo Scientific) and their molar extinction coefficients52.
Purification of proteins for crystallisation
IMAC-purified proteins were further purified by SEC using a HiLoad™ Superdex 200 pg on an AKTA Pure FPLC system (GE Healthcare Life Sciences). The purity of the fractions were determined using SDS-PAGE and those of high enough purity were pooled and concentrated to ~ 10 mg/ml.
Crystallisation
We used the vapour diffusion sitting drop method to screen crystallisation conditions. Seleno-methionine (SeMet) crystals for BT1044GH18 were obtained in 100 mM imidazole pH 8.0 and 10 % polyethylene glycol (PEG) 8000. The sample were cryo-protected with paratone-N oil. Additional SeMet BT1044GH18 crystals were obtained in 200 mM sodium fluoride, 100 mM bis-tris propane pH 6.5 and 20 % PEG 3350. These samples were cryo-protected with the addition of 20% PEG 400 to the reservoir solution. BT0459GH20 crystals were obtained in 100 mM sodium acetate, 100 mM bis-tris propane pH 7.5 and 10 % PEG3350.
Data collection, structure solution, model building, refinement and validation
A 3-wavelength interleaved MAD experiment using SeMet derivatives cryo-protected with oil was collected at the beamline I03, Diamond Light Source (UK). The data were indexed and integrated with XDS53. Space group determination was confirmed with Pointless and the data were scaled with Aimless54. The phase problem for BT1044GH18 was solved by autoSHARP 55. ShelxD56 found the requested 8 selenium sites with a final correlation coefficient (E) of 0.59. Subsequent density modification with Parrot57 and automated model building with Buccaneer58 placed 292 sequenced residues out of 364. Once the phase problem was solved, higher resolution datasets were obtained on the BT1044GH18 SeMet samples cryo-protected with PEG 400 (see above). The data were processed as above and the initial experimental phasing model was refined using Refmac559 and manually built using Coot60. The data processing and refinement statistics are reported in Supplementary Table 8; PDB accession 6Q64. The phase problem for BT0459GH20 was solved by using MrBump61. Briefly, the search model 3RCN was prep with Molrep62 and used in Phaser63 all automated through MrBump61. The initial phases obtained from MrBump were improved by density modification using Parrot 57 and the model was automatically build using Buccaneer58. For all models, iterative cycles of model building with Coot60 and refinement with Refmac559 were stopped when the validation with Coot60 and Molprobity64 gave acceptable values. 5 % of the data were randomly selected for Rfree calculation. Structural figures were made using Pymol65 and all other programs used were from the CCP4 suite66. The data processing and refinement statistics are reported in Supplementary Table 8; PDB accession 6Q63.
Growth of Bacteroides species
Starter cultures for most Bacteroides spp. were grown in tryptone-yeast-extract-glucose (TYG) medium with the addition of hematin and inoculated from glycerol stocks32. Bacteroides massiliensis and xylanisolvans were grown on chopped meat broth (CMB) as described previously9,67. Cells were typically grown in 5 ml cultures in glass test tubes in an anaerobic cabinet (Whitley A35 Workstation; Don Whitley, UK) and were monitored at OD600nm using a Biochrom WPA cell density meter. Growth tests using 20 mg/ml glycoprotein as the sole carbon source were carried out in minimal media (MM) 33 in either 5 ml cultures (to allow for taking samples) or 600 μl cultures in a 96 well plate using a Biotek Epoch plate reader. The removal of glycan from peptide was monitored by SDS-PAGE with either Coomassie staining or Pierce™ Glycoprotein staining kit (ThermoFisher Scientific), to detect just protein or glycoprotein, respectively. Replicates are presented individually in the figures and not as averages.
RNA extraction, sequencing and data processing
A starter culture was used to inoculate minimal media (5 ml) of either 5 mg/ml glucose or 10 mg/ml α1AGp, which was then grown to OD580nm 0.4-0.6. Cells were harvested and stored in RNAProtect (QIAGEN) and the RNA extracted using RNeasy Mini Kit (Qiagen). Sample processing, library prep and sequencing took place at Oxford Genomics Centre (University of Oxford, Oxford, UK). Data processing was done by The Bioinformatics Support Unit at Newcastle University using Bowtie2 for alignments68 and genome annotation completed using the Ensembl database69. The read counts aligning to the genomic features were obtained and differential expression analysis was done using R packages70. This data is presented in Supplementary Table 1 has been submitted to https://www.ncbi.nlm.nih.gov/geo/ and has the accession number GSE129572.
Genetic manipulation
Gene deletion mutations were produced by allelic exchange using the pExchange vector6.
Cellular localisation
Cells (15 ml) were grown on minimal media with α1AGp (10 mg/ml final) to mid-exponential growth, harvested by centrifugation, washed in PBS and resuspended in 2 ml PBS. The cells were split into two. One half had Proteinase K (2 mg/ml final) added and was incubated at 37 °C for 2 hours and one half was the control. The Proteinase K sample was harvested by centrifugation, washed in PBS and resuspended in 1 ml. Trichloroacetic acid (200 μl) was added to all samples to precipitate out the protein, incubated for 30 mins on ice, washed four times with ice-cold acetone (1ml) and cell pellets resuspended in 0.5 ml PBS. Samples (15 μl) were run on SDS-PAGE gels with MagicMark XP Western Protein Standard (Thermo Fisher Scientific) and then transferred to Whatman Protran BA 85 nitrocellulose membrane. Specific proteins were detected using anti-sera raised against recombinant versions in rabbit (Eurogentec). The secondary antibody used was a Goat Anti-rabbit HRP congugate (SC-2004, Santa Cruz). Antibodies were detected by chemi-luminescence using Biorad Clarity Western ECL Substrate.
Whole cell assays to analyse cell surface enzyme activity
15 ml cultures grown on minimal media with α1AGp (20 mg/ml final) to mid-exponential and harvested by centrifugation. Cells were washed twice in PBS and resuspended in 1.75 ml PBS. To assess the cell surface activity, 250 μl of the resuspended cells were mixed with 250 μl of fresh α1AGp added (20 mg/ml final) and a 50 μl aliquot was taken at T=0, 0.25, 0.5, 1, 2, 3, 4 and ~16 h. One aliquot was pelleted again and resuspended in 250 μl of BugBuster® (Merck Millipore) to lyse the cells and then mixed with 250 μl of fresh α1AGp (20 mg/ml final). Several controls were also included. Cells (250 μl) were mixed with 250 μl PBS, 250 μl of the spent media from the growth was also mixed with 250 μl of fresh α1AGp (20 mg/ml final) and 500 μl of cells were mixed with 500 μl of PBS. For the latter control, two 50 μl aliquots were taken at every time point and one was checked for activity and one had the cells were removed by centrifugation and the supernatant kept on ice. The supernatant was tested for overnight activity against fresh α1AGp (20 mg/ml final) to check there was no cell lysis during the time course. Cells (750 μl) were then treated with Proteinase K (2 mg/ml final) for two hours and again harvested by centrifugation, washed twice and resuspended back to 750 μl. 250 μl of these cells were mixed with α1AGp (20 mg/ml final) and the same time course run. To check all the Proteinase K had been removed, 250 μl of the treated cells were mixed with 250 μl of bovine serum albumin (20 mg/ml final). The treated cells were also lysed using BugBuster® and assayed against α1AGp (20 mg/ml final).
Thin layer chromatography
Between 3-12 μl of samples were spotted in 3 μl aliquots on to silica plates and resolved in butanol:acetic acid:water (2:1:1). Sugars were visualised using diphenylamine-aniline-phosphoric acid stain71.
Recombinant enzyme assays
Generation of kinetic data was carried out with either assays on PNP-linked sugars (monitored at 400 nm) or using galactose, mannose or fucose detection kits (Megazyme International), as described previously8. To assay the activity of enzymes on CNG, reactions were left overnight (~16h) unless otherwise stated. The activities of the recombinant enzymes were typically assessed in 20 mM MOPS pH 7, at 37 °C, with a final glycoprotein concentration of 20 mg/ml and a final enzyme concentration of 1 μM. Exceptions to this were the final concentrations of IgG, serum IgA and colostrum IgA that were 5 mg/ml, 0.5 mg/ml and 2.5 mg/ml, respectively. All kinetic assays were performed at least in triplicate on multiple occasions and data presented are means and standard deviations. Total N-glycans were released from glycoproteins using PNGaseF (sigma), purified by centrifugation through a 10 kDa protein concentrator, freeze dried and resuspended in the required volume.
Esterase activity
A sialic acid reference panel containing differently esterified sialic acid species was digested with 1 μM of BT0457 overnight at 37 °C, derivatised with DMB (Ludger, Ltd) and analysed using reversed phase HPLC. Here, 25 μl of sample was injected into the LudgerSep R1 column (4.6 x 150 mm, 3 μm particle size) at 30 °C on a Waters Alliance HPLC instrument with fluorescence detection (λex = 373 nm, λem = 448 nm). Mobile phase A was a methanol: acetonitrile: water solution (7:9:84) and mobile phase B was acetonitrile. Analytes were eluted using an isocratic flow at 100 % mobile phase A running at a flow rate of 0.5 mL/min for 19 minutes. Column wash was accomplished by increasing proportion of mobile phase B to 90 %.
High-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD)
HPAEC-PAD was used to analyse BT1035GH163 activity against sialylated and desialylated CNGs. Samples were separated using a CARBOPAC™ PA-100 anion exchange column with a CARBOPAC™ PA-100 guard column (Thermo Fisher Scientific). Flow was 1 ml/min and elution conditions were 0-20 min, 20 mM NaOH; 20-80 min 100 mM NaOH with a 0-500 mM sodium acetate gradient. LacNAc was quantified using a standard curve generated using a range of LacNAc standards between 0.1 and 0.01 μM.
Procainamide labelling
N-glycans were fluorescently labelled by reductive amination using a procainamide labelling kit containing sodium cyanoborohydride as reductant (Ludger, Ltd). Before analysis, derivative glycans were cleaned-up from excess reagents using SPE clean-up plates (Ludger, Ltd).
Analysis of Procainamide labelled glycans
Procainamide labelled glycans were analysed by LC-FLR-ESI-MS. Here, 25 µl of each sample was injected into a Waters ACQUITY UPLC Glycan BEH Amide column (2.1 x 150 mm, 1.7 µm particle size, 130 Å pore size) at 40°C on a Dionex Ultimate 3000 UHPLC instrument with a fluorescence detector (λex = 310 nm, λem = 370 nm) attached to a Bruker Amazon Speed ETD. Mobile phase A was a 50 mM ammonium formate solution (pH 4.4) and mobile phase B was neat acetonitrile. Analyte separation was accomplished by a gradient running from 85-57% mobile phase B over 105 minutes at a flow rate of 0.4 mL/min, with exception of IgG CNG used for screening of GH20 enzymes, where separation was accomplished by a gradient running from 76-58% mobile phase B over 70 minutes. The Amazon Speed was operated in the positive sensitivity mode using following settings: source temperature, 180 C; gas flow, 4 L/min; capillary voltage, 4500 V; ICC target, 200,000; maximum accumulation time, 50.00 ms; rolling average, 2; number of precursor ions selected, 3; scan mode, enhanced resolution; mass range scanned, 400 to 1700.
Analysis of mass spectrometry data
Procainamide labelled glycans were analysed using Bruker Compass Data Analysis software and GlycoWorkbench72. Glycan compositions were elucidated based on MS2 fragmentation and literature knowledge.
Bioinformatics
Putative signal sequences were identified using LipoP73. Alignments and sequence identities were determined using Clustal Omega using the full length protein sequences, not individual modules74. The IMG database was used to analyse genomic locations and synteny75. The CAZy database (www.cazy.org) was used as the main reference for carbohydrate-active enzyme activity76. Dali77 and PDBefold78 were used to carry out structural homology searches of protein modules. Phylogenetic trees were constructed using Phylogeny.fr79,80. Pfam81 and SMART82,83 were used to determine module boundaries and look at the prevalence of modules in other proteins.
Supplementary Material
Acknowledgments
We thank Carl Morland (Newcastle University, UK) for his expert technical assistance, Fiona Cuskin (Newcastle University, UK) and Louise Royle (Ludger Ltd, UK) for insightful conversations about the data. We would like to thank Diamond Light Source (Oxfordshire, UK) for beamtime (proposal mx13587 and mx18598) and the staff of beamline I03 and 104-1 for assistance with crystal testing and data collection. We thank John Casement (Bioinformatics Support Unit, Newcastle University, UK) for analysing the raw RNAseq data. We thank Justin Sonnenberg (Stanford, USA) for the ΔBT0455 Bt strain, Rick Lewis (Newcastle University, UK) for his guidance and advice in looking for structural homologues and Robert Hirt for his advice on phylogenetics. The work was funded by BBSRC/Innovate UK IB catalyst award ‘Glycoenzymes for Bioindustries’ (BB/M029018/1).
Footnotes
Data availability. The full RNAseq data is provided in Supplementary Table 1 and is also submitted to https://www.ncbi.nlm.nih.gov/geo/ with the accession number GSE129572. The crystal structures are deposited in the Protein Data Bank under the accession numbers 6Q63 and 6Q64. The other data that support the findings in this paper are available upon request from the corresponding authors.
Author contributions
J.B., A.S.L. and L.I.C carried out enzyme kinetics. J.B., P.A.U., A.S.L., O.R. and L.I.C. carried out enzymes assays. J.H. made substrates. J.B. and L.I.C. carried out Bacteroides growths. L.I.C carried out the whole cell assays. P.A.U. and O.R. carried out the LC-MS. P.A.U. and L.I.C. analysed the LC-MS data. J.B. and L.I.C. purified proteins and set up crystal trays. A.B. harvested crystals and collected data. A.B. and N.P. solved crystal structures. J.B., D.N. and L.I.C. produced Bacteroides gene deletion strains. L.I.C. carried out the bioinformatic analysis. L.I.C., D.N.B., P.A.U., J.B., A.S.L., D.I.R.S. and E.C.M designed experiments. L.I.C., E.C.L and D.N.B wrote the manuscript.
Competing interests
The authors declare no competing interests
References
- 1.McNeil NI. The contribution of the large intestine to energy supplies in man. The American journal of clinical nutrition. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 3.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. Journal of clinical gastroenterology. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 4.O'Keefe SJ, et al. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. The Journal of nutrition. 2009;139:2044–2048. doi: 10.3945/jn.109.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature reviews. Microbiology. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure (London, England : 1993) 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luis AS, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nature microbiology. 2018;3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai MS, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353.e1321. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung CY, Majewska NI, Wang Q, Paul JT, Betenbaugh MJ. SnapShot: N-Glycosylation Processing Pathways across Kingdoms. Cell. 2017;171:258–258.e251. doi: 10.1016/j.cell.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Mathias A, Corthesy B. N-Glycans on secretory component: mediators of the interaction between secretory IgA and gram-positive commensals sustaining intestinal homeostasis. Gut microbes. 2011;2:287–293. doi: 10.4161/gmic.2.5.18269. [DOI] [PubMed] [Google Scholar]
- 12.Corfield AP. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms. 2018;6 doi: 10.3390/microorganisms6030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mestecky J, Russell MW, Jackson S, Brown TA. The human IgA system: a reassessment. Clinical immunology and immunopathology. 1986;40:105–114. doi: 10.1016/0090-1229(86)90073-5. [DOI] [PubMed] [Google Scholar]
- 14.Hughes GJ, Reason AJ, Savoy L, Jaton J, Frutiger-Hughes S. Carbohydrate moieties in human secretory component. Biochimica et biophysica acta. 1999;1434:86–93. doi: 10.1016/s0167-4838(99)00168-5. [DOI] [PubMed] [Google Scholar]
- 15.Garrido D, et al. Endo-beta-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Molecular & cellular proteomics : MCP. 2012;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebaihia M, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature genetics. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 17.Bohle LA, Mathiesen G, Vaaje-Kolstad G, Eijsink VG. An endo-beta-N-acetylglucosaminidase from Enterococcus faecalis V583 responsible for the hydrolysis of high-mannose and hybrid-type N-linked glycans. FEMS microbiology letters. 2011;325:123–129. doi: 10.1111/j.1574-6968.2011.02419.x. [DOI] [PubMed] [Google Scholar]
- 18.Renzi F, et al. The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLoS pathogens. 2011;7:e1002118. doi: 10.1371/journal.ppat.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collin M, Fischetti VA. A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. The Journal of biological chemistry. 2004;279:22558–22570. doi: 10.1074/jbc.M402156200. [DOI] [PubMed] [Google Scholar]
- 20.Cao Y, Rocha ER, Smith CJ. Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12901–12906. doi: 10.1073/pnas.1407344111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robb M, et al. Molecular Characterization of N-glycan Degradation and Transport in Streptococcus pneumoniae and Its Contribution to Virulence. PLoS pathogens. 2017;13:e1006090. doi: 10.1371/journal.ppat.1006090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sjogren J, et al. EndoS2 is a unique and conserved enzyme of serotype M49 group A Streptococcus that hydrolyses N-linked glycans on IgG and alpha1-acid glycoprotein. The Biochemical journal. 2013;455:107–118. doi: 10.1042/BJ20130126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collin M, Olsen A. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. The EMBO journal. 2001;20:3046–3055. doi: 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupoiron S, et al. The N-Glycan cluster from Xanthomonas campestris pv. campestris: a toolbox for sequential plant N-glycan processing. The Journal of biological chemistry. 2015;290:6022–6036. doi: 10.1074/jbc.M114.624593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forster SC, et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nature biotechnology. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martens EC, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS biology. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogowski A, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nature communications. 2015;6 doi: 10.1038/ncomms8481. 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ndeh D, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagenholm V, et al. Galactomannan Catabolism Conferred by a Polysaccharide Utilization Locus of Bacteroides ovatus: ENZYME SYNERGY AND CRYSTAL STRUCTURE OF A beta-MANNANASE. The Journal of biological chemistry. 2017;292:229–243. doi: 10.1074/jbc.M116.746438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, et al. Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides. Cell reports. 2017;21:417–430. doi: 10.1016/j.celrep.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Temple MJ, et al. A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase. The Journal of biological chemistry. 2017;292:10639–10650. doi: 10.1074/jbc.M117.787606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsbrink J, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. The Journal of biological chemistry. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. Journal of bacteriology. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas F, et al. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environmental microbiology. 2012;14:2379–2394. doi: 10.1111/j.1462-2920.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 36.Muchmore EA, Diaz S, Varki A. A structural difference between the cell surfaces of humans and the great apes. American journal of physical anthropology. 1998;107:187–198. doi: 10.1002/(SICI)1096-8644(199810)107:2<187::AID-AJPA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 37.Park KH, et al. Structural and biochemical characterization of the broad substrate specificity of Bacteroides thetaiotaomicron commensal sialidase. Biochimica et biophysica acta. 2013;1834:1510–1519. doi: 10.1016/j.bbapap.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 38.Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC evolutionary biology. 2009;9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phansopa C, et al. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. The Biochemical journal. 2015;472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- 40.Garbe J, et al. EndoE from Enterococcus faecalis hydrolyzes the glycans of the biofilm inhibiting protein lactoferrin and mediates growth. PloS one. 2014;9:e91035. doi: 10.1371/journal.pone.0091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waddling CA, Plummer TH, Jr, Tarentino AL, Van Roey P. Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3) Biochemistry. 2000;39:7878–7885. doi: 10.1021/bi0001731. [DOI] [PubMed] [Google Scholar]
- 42.Green ED, Adelt G, Baenziger JU, Wilson S, Van Halbeek H. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. The Journal of biological chemistry. 1988;263:18253–18268. [PubMed] [Google Scholar]
- 43.Tailford LE, et al. Mannose foraging by Bacteroides thetaiotaomicron: structure and specificity of the beta-mannosidase, BtMan2A. The Journal of biological chemistry. 2007;282:11291–11299. doi: 10.1074/jbc.M610964200. [DOI] [PubMed] [Google Scholar]
- 44.Pluvinage B, et al. Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis. Structure (London, England : 1993) 2011;19:1603–1614. doi: 10.1016/j.str.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Liu T, et al. Structural determinants of an insect beta-N-Acetyl-D-hexosaminidase specialized as a chitinolytic enzyme. The Journal of biological chemistry. 2011;286:4049–4058. doi: 10.1074/jbc.M110.184796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theodoratou E, et al. Glycosylation of plasma IgG in colorectal cancer prognosis. Scientific reports. 2016;6 doi: 10.1038/srep28098. 28098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. The Journal of biological chemistry. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell host & microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence RM, Pane CA. Human breast milk: current concepts of immunology and infectious diseases. Current problems in pediatric and adolescent health care. 2007;37:7–36. doi: 10.1016/j.cppeds.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Charnock SJ, et al. Key residues in subsite F play a critical role in the activity of Pseudomonas fluorescens subspecies cellulosa xylanase A against xylooligosaccharides but not against highly polymeric substrates such as xylan. The Journal of biological chemistry. 1997;272:2942–2951. doi: 10.1074/jbc.272.5.2942. [DOI] [PubMed] [Google Scholar]
- 52.Gasteiger E, H C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook. :571–607. [Google Scholar]
- 53.Kabsch W. XDS. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans P. Scaling and assessment of data quality. Acta crystallographica. Section D, Biological crystallography. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 55.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods in molecular biology (Clifton, N.J.) 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 56.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta crystallographica. Section D, Biological crystallography. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cowtan K. Recent developments in classical density modification. Acta crystallographica. Section D, Biological crystallography. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta crystallographica. Section D, Biological crystallography. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keegan RM, Winn MD. Automated search-model discovery and preparation for structure solution by molecular replacement. Acta crystallographica. Section D, Biological crystallography. 2007;63:447–457. doi: 10.1107/S0907444907002661. [DOI] [PubMed] [Google Scholar]
- 62.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta crystallographica. Section D, Biological crystallography. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 63.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The PyMOL Molecular Graphics System, Version 2.0. Schrödinger, LLC; [Google Scholar]
- 66.The CCP4 suite: programs for protein crystallography. Acta crystallographica. Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 67.Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zerbino DR, et al. Ensembl 2018. Nucleic acids research. 2018;46:D754–d761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R: A language and environment for statistical computing. 2013.
- 71.Zhang Z, Xie J, Zhang F, Linhardt RJ. Thin-layer chromatography for the analysis of glycosaminoglycan oligosaccharides. Analytical biochemistry. 2007;371:118–120. doi: 10.1016/j.ab.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ceroni A, et al. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. Journal of proteome research. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 73.Juncker AS, et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein science : a publication of the Protein Society. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Markowitz VM, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic acids research. 2012;40:D115–122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic acids research. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holm L, Laakso LM. Dali server update. Nucleic acids research. 2016;44:W351–355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Protein structure comparison service PDBeFold at European Bioinformatics Institute
- 79.Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC evolutionary biology. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic acids research. 2018 doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic acids research. 2018;46:D493–d496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic acids research. 2015;43:D257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.