Abstract
The 24h cycle of activity and sleep provides perhaps the most familiar example of circadian rhythms. In mammals, circadian activity rhythms are generated by a master biological clock located in the hypothalamic suprachiasmatic nuclei (SCN). This clock is synchronized (entrained) to the external light environment via light input from retinal photoreceptors. However, sleep is not a simple circadian output and is also regulated by a homeostatic process whereby extended wakefulness increases the need for subsequent sleep. As such, the amount and distribution of sleep depends upon the interaction between both circadian and homeostatic processes. The study of circadian activity and sleep is not only confined to these specialised fields. Sleep and circadian rhythm disruption is common in many conditions, ranging from neurological and metabolic disorders to aging. Such disruption is associated with a range of negative consequences including cognitive impairment, mood disorders, as well as immune and metabolic dysfunction. As circadian activity and sleep are hallmarks of normal healthy physiology, they also provide valuable welfare indicators. However traditional methods for the monitoring of circadian rhythms and sleep in mice can require separate specialised resources as well as significant expertise. Here we outline a low-cost, non-invasive and open source method for the simultaneous assessment of circadian activity and sleep in mice. This protocol describes both the assembly of the hardware used and the capture and analysis of data without the need for expertise in electronics or data processing.
Keywords: Circadian, sleep, home-cage monitoring, welfare, refinement
Introduction
Circadian rhythms are approximately 24h cycles of physiology and behaviour that optimise physiology to the demands of the day/night cycle. In mammals, the master circadian clock is located in the suprachiasmatic nuclei (SCN) in the anterior hypothalamus (Reppert and Weaver, 2002). The SCN is synchronised (entrained) to the external light/dark cycle by light input from specialised photoreceptors in the retina and regulates physiological and behavioural outputs such as the timing of locomotor activity and the entrainment of peripheral circadian clocks found in cells and tissues throughout the body (Dibner et al., 2010). However, the timing of sleep and wake is not a simple circadian output. Whilst the SCN clock undoubtedly plays a role in the regulation of the timing of sleep and wake (Borbely et al., 2016; Mistleberger 2005), sleep is also dependent upon the duration of preceding wakefulness in a homeostatic manner. As such, the amount and distribution of sleep depends upon the interaction of both circadian and homeostatic processes (Borbely 1982; Borbely et al., 2016). Although specific hypothalamic circuits control the switch from sleep to wake (Scammell et al 2017), the homeostat may depend upon cellular mechanisms rather than specific neuronal circuits (Wang et al 2018).
In healthy individuals cycles of activity and rest are very robust with similar recurring patterns across multiple days. Disruption of these regular activity/sleep patterns contributes to impaired cognitive performance, metabolic and cardiovascular disorders and even the risk of developing cancer (Bechtold et al., 2010; Hastings et al, 2003; Jagannath et al., 2013; West and Bechtold, 2015; Wulff et al., 2010). In addition, circadian disruption has also been characterised in many different disease states as well as a result of our modern 24h society, with jet-lag, shift work and light at night all perturbing the normal activity/rest cycle (Brown et al., 2019). As a result, the ability to measure changes in activity and sleep cycles in model systems such as the mouse is of value not only to the circadian and sleep community but also to researchers studying a wide range of disorders. In addition, animals display characteristic changes in physiology and behaviour in response to illness, including reduced activity and increased sleep (Tizard et al., 2008). As such, home cage activity and sleep provides a valuable potential indicator of healthy physiology as well as disease progression.
Measuring activity and sleep in mice typically requires specialised resources. In the case of circadian activity, usually cages equipped with running wheels are housed in light-tight environmentally-controlled chambers with independent lighting and ventilation. Wheel revolutions are recorded via micro-switches to computer systems running specialist software (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015). By contrast, sleep is traditionally recorded using electroencephalography (EEG) combined with electromyography (EMG), which involve invasive surgery and collection of high resolution electrophysiological data. The detailed characterisation of sleep critically depends upon such EEG/EMG data, in particular the analysis of specific vigilance states and spectral analysis which require specialist software and time-consuming sleep-scoring. However, behavioural correlates of sleep provide a valuable approach to studying the amount and distribution of sleep in a high throughput manner, prior to focused EEG/EMG studies or as part of a larger screening process. Both video and piezoelectric methods have been used in this manner (Pack et al., 2007; Flores et al., 2007; Fisher et al., 2012). Continuous Open Mouse Phenotyping of Activity and Sleep Status (COMPASS) was developed as a low cost, accessible technique for measuring simultaneous activity and sleep in mice. This system is based upon passive infrared (PIR) sensors that continuously monitor home cage movement and store data onto a local PC via an Arduino microprocessor (Figure 1). Validation against EEG/EMG data shows that accurate sleep data can be obtained by monitoring periods of extended immobility using calibrated positioning of the PIR sensors (Brown et al 2017). Here we provide a step-by-step protocol for the assembly of the COMPASS system, the collection of PIR activity data and the analysis of this data for circadian activity and sleep phenotyping.
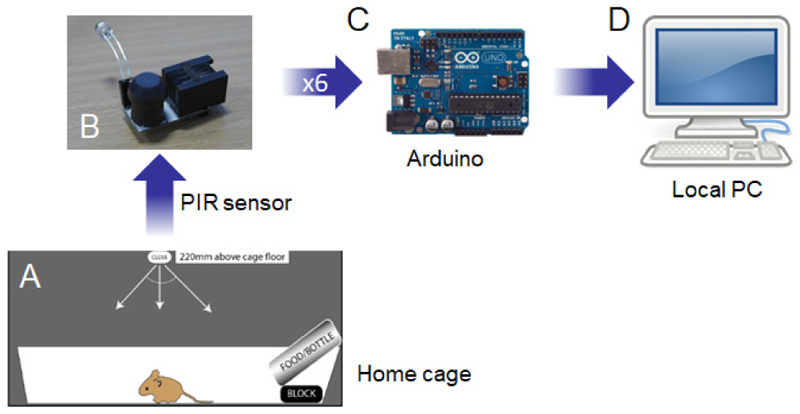
Figure 1. COMPASS system overview.
(A) Singly-housed animal in home cage. To prevent the animal moving out of the range of the passive infrared (PIR) sensor, a block is placed under the food hopper ensuring the animal still has sufficient cage space. (B) The PIR sensor is placed 220mm above the cage floor for optimum sensitivity for the assessment of immobility defined sleep (Brown et al 2017). The sensor may also have an integrated light dependent resistor (LDR) to measure light levels. Typically only one LDR is required for all 6 channels if these are in the same light environment. (C) Up to six PIR sensors are then connected to the main board, which is designed to interface with the Arduino microprocessor. (D) The Arduino is then connected to a local PC to store data. Alternatively, a Raspberry Pi microprocessor may be used instead of a PC, particularly when multiple different systems are being run in parallel.
Strategic Planning
Activity and sleep in mice are highly sensitive to light exposure, with light suppressing activity and promoting sleep. Moreover, mistimed light exposure can also disrupt the circadian system by producing circadian phase shifts or fragmentation of activity rhythms (Peirson et al 2018; Brown et al., 2019). Light history may also influence sleep via its action upon the circadian system or by directly regulating sleep. PIR activity and sleep recordings should therefore ideally be performed under controlled light environments. For detailed circadian analysis, the use of light-tight environmentally controlled chambers with inbuilt timer-controlled lighting is strongly recommended. The construction of such chambers has been described in detail elsewhere (see appendix of Albrecht and Foster 2002). See also Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015). The type of lighting used, as well as the light intensity should also always be reported. Ideally, the spectral power distribution of the light source should also be measured and included in any publication (Lucas et al., 2014). If light-tight chambers are not available, PIR recordings can be performed in normal cage racks or procedure rooms. This may be more appropriate for monitoring health status or disease progression in animals under long-term study. However, in such situations it is important to maintain a stable lighting environment. While 12h light/12h dark cycles are commonly used in animal facilities, unexpected light changes will affect the data obtained (e.g. lights being accidently turned on or off in a room or light exposure during the dark phase due to light from external corridors). Clear signage warning users of the need for care in the light environment may be necessary. Additional precautions such as blackout curtains on room entrances may also help provide a light-controlled environment. Some animal facilities also use ramped dawn/dusk lighting, and users should be aware that this may result in different activity and sleep data in comparison with mice housed under a simple lights on/off condition (Comas and Hut, 2009). To ensure that the light environment is consistent during data collection, the build below includes a light-dependent resistor (LDR) to monitor the light environment throughout data collection.
Basic Protocol
Assembly of a PIR system for basic activity and sleep recordings
This protocol describes the assembly of the PIR sensors and Arduino microprocessor, and their connection to the PC-based recording software. The recording and analysis of home cage activity and sleep data is also described.
NOTE: It is recommended that the COMPASS system is built and tested prior to any home cage animal activity study. Exact part codes may be discontinued, though supplier sites will typically suggest alternatives. For the recording computer, the system has been successfully run on old computers (< 512Mb RAM, and <1GHz Pentium III processors), and operating systems from Windows XP- Windows 10 (Microsoft), MacOS X (Apple Inc.) and Ubuntu Linux 14.04 LTS and above (Canonical Ltd.).
NOTE: The technique outlined below is based upon the PIR sensor being positioned above the cage. The mouse should therefore be prevented from reaching areas of the cage not covered by the sensor field. To this end, the underside of the food hopper should be made inaccessible and home cage bedding may need to be reduced. We have found that Nestlets work well. Please note that PIR sensors are unable to penetrate IVC cage lids, so recordings need to be made without such lids in place.
Materials
For construction of PIR sensors
PIR sensors (Panasonic AMN32111) (http://uk.mouser.com/ cat. no 769-AMN32111); one for each cage to be monitored.1 k Ohm Resistor (0.25W, 1%) (Farnell cat. no 9341102); one for each cage to be monitored.
10 k Ohm Resistor (0.25W, 1%) Farnell cat. no 2329855); one for each cage to be monitored (for PIR sensors that require a LDR).
Printed PIR circuit boards (designed and manufactured using Fritzing (http://fritzing.org/home/, versions up to 0.9.3b); one for each cage to be monitored. The circuit board design can be found here: https://aisler.net/LABrown/compass/1pir_ldr.
Light-dependant resistor (LDR) (Farnell cat. no 3168335); one for each lighting environment (e.g. light-tight cabinet or room) to be monitored.
RJ12 receptacle for PIR board (JACK, 6/4 LOW PROFILE) (Farnell cat. no 1560166).
OPTIONAL: Small PIR case (BOX, 18×40×28MM) (Farnell cat. no 1871051). Alternatively, 3D printing .stl files can be found at: http://users.ox.ac.uk/˜anat0144/COMPASS_updates.html
For construction of the main unit
Printed main circuit board (designed and manufactured using Fritzing (http://fritzing.org/home/, versions up to 0.9.3b). The circuit board design can be found here: https://aisler.net/LABrown/compass/6pir_board.
Arduino microcontroller (AVR ARDUINO MEGA2560 REV 3) (http://uk.mouser.com/ cat. no 782-A000067)
Arduino case (Farnell cat. no 1848692)
RJ12 sockets (VERTICAL, 6WAY, 4WIRE) (Farnell cat. no 1097915).
Jumper Wires (MALE TO MALE 15CM 10PK) (http://uk.mouser.com/ cat. no 932-MIKROE-513)
2-pin header (2.54MM, SINGLE, 2WAY) Farnell cat. no 1667508)
24-pin header (2.54MM, 2X12WAY) (Farnell cat no 1804656)
Cables
Cable reel (FCC68, 4CORE, BLACK, 100M) (Farnell cat. no 1202607)
Crimp tool (4P/4C, 6P/4C, 6P/6C) (Farnell cat. no 106321)
Crimp plugs (RJ12, 6P4C, FLAT) (Farnell cat. no 1712385)
USB cable (2.0 A TO B 3M) (Farnell cat. no 2444242)
Miscellaneous
Standard soldering equipment (soldering iron, solder, safety mat). NB: Ensure appropriate ventilation.
For activity recording and analysis
Individual cages
Recording PC. Recording PC (minimal computational requirements), with Arduino integrated development environment (IDE, versions 1.0.1-1.0.5 and 1.5.6- 1.6.11 tested, available at http://arduino.cc/en/Main/Software) and Processing (version 3.4 tested, available at http://processing.org/) installed.
Analysis PC (for Python scripts). Running Python (2.7.11: https://www.python.org/downloads/) and Pycharms (https://www.jetbrains.com/pycharm/). In Pycharms, install the following packages: Pandas 0.18.0; Numpy 1.10.4; Matplotlib 1.5.1; Seaborn 0.7.0.
Data analysis software: e.g. ClockLab (Actimetrics), Actogram J (Schmid et al 2011), Rethomics (Geissmann et al 2020), or a spreadsheet or data analysis package (e.g. Microsoft Excel, MatLab or R).
OPTIONAL: Light-tight environmentally-controlled chambers may be required for more controlled lighting conditions for circadian studies (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015).
PIR sensor assembly
Please note that all circuit board diagrams are available (Brown et al 2017). Alternatively, circuit board details can also be found at: http://users.ox.ac.uk/˜anat0144/COMPASS_updates.html
-
Insert the resistors, the RJ12 jack and the PIR module through the PIR board (Figure 2A) in the marked locations and pull tight by twisting the wire ends or bending the wire legs apart on the other side (Figures 2B-E).
RJ11 and RJ12 connections use the same size plug, but RJ11 uses 4 channels whereas RJ12 uses 6. This system requires only 4 channels (RJ11) but RJ12 plugs and sockets can be used interchangeably. As the cable is 6 channels, RJ12 is used throughout the protocol.
-
Carefully solder all holes where connections come through. Check there are no loose components or connections (check both sides of the board and fix with more solder if needed). Tidy by removing loose wires with tin snips, just beyond the soldered points.
CAUTION: Ensure that appropriate ventilation is available during soldering
Insert a 10 k resistor and 2-pin header to any board that will have and LDR mounted (Figure 2F-G).
Solder resistor/header into place.
-
Run final checks for loose connections or missing components and add LDRs if needed. Ensure the wire legs of the LDR do not cross using heat-shrink tubing or tape.
OPTIONAL: If using a case to house the PIR unit, it can be placed inside, ensuring that the sensor is unobscured.
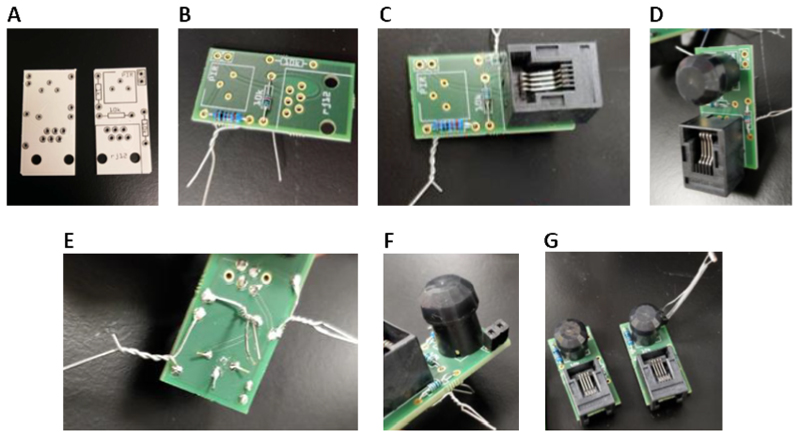
Figure 2. Assembly of the PIR sensor units.
(A) Image of the PIR circuit board prior to addition of components. (B) Addition of the 1 k and 10 k Ohm Resistors to the board. (C) Addition of the RJ12 jack to the board. (D) Addition of the PIR module to the board. (E) Twisting wires and bending of legs will secure components prior to soldering. (F) Addition of a 2-pin header to the board for LDR connection. (G) Final assembly of PIR sensor units both with (right) and without (left) an LDR.
Main board assembly
The main board involves a simple circuit board that collects inputs from up to 6 PIR sensors to the Arduino microprocessor.
-
6.
Mount the headers to the underside of the main board (Figure 3A) and carefully solder in place (Figures 3B-C).
These headers allow the connection of the individual PIR sensors via standard RJ12 connector plugs.
-
7.
Mount the RJ12 connectors to the other side of the board and solder these in place (Figures 3D-E).
-
8.
If using the Arduino case, fit the Arduino Uno microcontroller into the lower half of the case (a small section of the case either side of the existing hole may need to be removed to mount the board flat to the case).
-
9.
Insert the jumper cables into the correct pins of the microcontroller (start with GND (black) and 5V (red)) (Figure 3F).
Additional information regarding these connections can be found in the original publication (Brown et al., 2017).
-
10.
Mount the remaining jumper wires with A2 for the LDR and digital pins 2 to 8 for the 6 PIR sensors.
Pin 4 is not used because this setup was designed to allow that to be used for other add-ons
-
11.
Run the other ends of the jumper wires through the hole in the case and into the headers on the main board (PIR 1-6 from one end, + and – for power in the middle, then LDR pins. LDR pin for PIR sensor 1 is closest to the power pins) (Figure 3F).
-
12.
Close the case and secure the main board to the front of the Arduino case. This can be achieved with any suitable adhesive (Figure 3G).
For clarity, the connections between the main board and Arduino are shown in more detail in Figure 4.
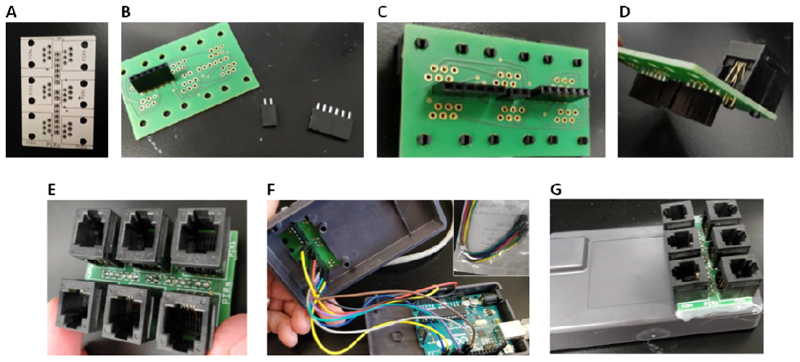
Figure 3. Assembly of the main board.
(A) Image of the main circuit board prior to addition of components. (B) Headers to be mounted to the underside of the main board. (C) Headers mounted to the underside of the board. (D) RJ12 connectors to be mounted to the main board. (E) Addition of RJ12 connectors to the main board. (F) Addition wiring connections between the Arduino Uno microcontroller (lower case section) and the headers on the underside of the main board (upper case section). (G) Final assembly of the main board on the Arduino case.
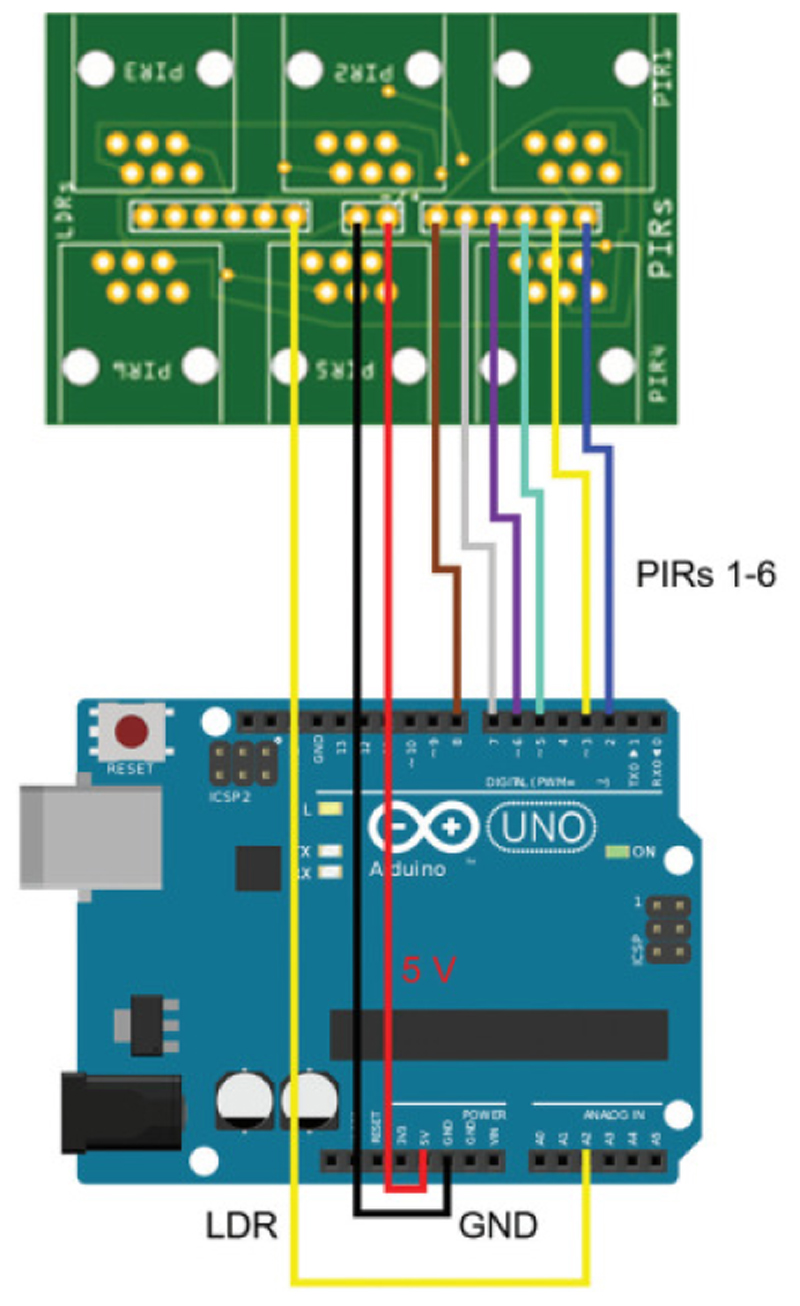
Figure 4. Schematic of connections of main board to Arduino.
The power (5V, red) and ground (GND, black) channels are connected to the two central pins. PIR channels 1-6 are connected to ports 2 – 8. In the example above a single LDR channel is used on PIR sensor 1, and this is connected to analogue port A2. For clarity, the same colour scheme for wiring is used as in image in Figure 3F.
Arduino box setup
-
13.
Connect each of the PIR sensors to the main board using RJ12 cables. Please note that the PIR sensor attached to the LDR needs to go into the ‘PIR1’ port. Connect the main board to the recording PC via the USB connection.
-
14.
Download and install the Arduino integrated development environment (IDE) on the data collection PC (www.arduino.cc/en/main/software). Open the IDE once installed.
-
15.
Load the 6chan_arduino.ino configuration sketch (sketch can be found in the repository here: https://zenodo.org/record/3838998#.Xsei3ndFwjY) in the IDE.
-
16.
In the tools tab of the IDE, confirm that the correct board and port are selected.
-
17.
Test the PIR sensors using the ‘Serial Monitor’ found in the tools tab of the IDE. Check the Serial Baud Rate (bottom right of Serial Monitor window) matches that in the Sketch, so that they are communicating at the same speed. This Output from the Serial Monitor will confirm that readings are being taken from the sensors every 10 seconds.
-
18.
Close the Arduino IDE before trying to access serial data through Processing.
Processing setup
-
19.
Close the IDE and open Processing.
-
20.
The activity_monitor_P3_update.pde sketch used by Processing can be found in the repository here: https://zenodo.org/record/3838998#.Xsei3ndFwjY. Load the .pde sketch in Processing. Within the sketch confirm that the ‘port index’ is to the COM port specified by the IDE.
-
21.
Press the ‘play’ icon on the Processing sketch to begin a test recording. While recording, Processing opens a new window showing the activity data recorded by each sensor for both the previous hour and the previous 24 hours.
-
22.
Prior to commencing actual experiments, the Processing window read out should be used to test that the field of each PIR sensor covers the entire base of the cage and that there is no cross contamination between sensors. The Arduino IDE ‘Serial Monitor’ can also be used for this purpose.
-
23.
The recording can be stopped by either pressing ‘X’ or by closing the window showing the individual PIR channels.
-
24.
Once the system has been built and tested, it can then be transferred to the animal facility ready to use.
Recording animal activity
-
25.Position each PIR sensor 220mm above the cage floor.If there are no convenient fittings from which the sensors can be suspended, frames can be constructed from which the sensors can be mounted. However the sensors are mounted, care should be taken that they are stable, as any movement from cage changes or mechanical vibrations will be detected as mouse movement during analysis. Be careful to ensure that the PIR sensor does not obscure the cage lighting.
-
26.
Prepare animal cages for recording (minimal bedding, locations in which the PIR sensor will be obscured are blocked to animal entry).
-
27.When ready to commence the experimental recording, singly house each animal in a cage with a PIR sensor positioned above (as described above).Ensure that there are no areas of the cage where the animal will be out of range of the sensor. For example, in many cages, the area under the food hopper may need to be blocked off. We normally use Perspex blocks, which are cheap and easy to clean (always make sure there are no sharp edges). Always ensure that animals have sufficient cage space, as set out by local regulations. We normally use larger cages for long-term circadian and sleep studies. In addition, nesting material may need to be reduced to prevent the animal being obscured. We have found that Nestlets work well. Other environmental enrichment may be included if it does not obscure the animal. For example, using wooden chew blocks or forage mix would not pose a problem, but Perspex mouse houses or cardboard tubes would not be suitable.
-
28.
When ready, open Processing and begin the recording by pressing the play icon.
OPTIONAL: PIR recordings can also be performed in group housed conditions. Such recordings can be used to analyse activity patterns at a whole cage level, but cannot distinguish individuals within the cage. Care must therefore be taken in the interpretation of group housed data.
Experimental recording and data collection
-
29.
Record data for at least 5 days under a 12h light/12h dark cycle.
During this time, perform daily visual checks of mice and top up food and water as necessary (in accordance with local regulations). During these checks, minimise any disruption of animals and movement of the cage. If cages are moved following animal checks, this can lead to inconsistent data. If cages must be moved to enable animal checks, care should be taken that the cages are returned to the same position as before. If necessary guides or runners can be installed to assist cage return in the same location, though this is usually less of a problem when normal animal cage racks are used.
-
30.
Once the screening period had ended, if necessary remove mice from cages (and cabinets if used) and end the recording on the computer.
-
31.
The data will be saved as a .csv file in the same folder as the Processing file.
The activity data will be recorded in 10 second bins in the following the columns: column 1: time bin; column 2: Arduino ID; column 3 onwards: data from individual PIR sensors. Note that the .csv is updated in real time. It is therefore possible to take a copy of the .csv data file for analysis at any time while leaving the screen running. In this way initial analyses can be performed and the ongoing screening conditions updated based upon the results of the initial analysis.
Data analysis
Data analysis can be performed using the raw data found in the .csv file. Basic data analysis can be performed using any spreadsheet or data analysis package (such as Microsoft Excel, MatLab or R), or using commercially available circadian analysis tools such as ClockLab (Actimetrics) or freely-available alternatives such as ActogramJ (Schmid et al 2011). We have provided several analytical tools for Python, which we describe the use of below.
-
32.
On the analysis computer, open the Pycharm program and open the following Python scripts in Pycharm: settings.py; binning.py; sleep_bout_lengths.py; sleep_counting.py (https://github.com/MRC-Harwell/sleep-tracking/tree/master/scripts).
-
33.
In the ‘settings’ script, set the following options: set where the output files are to be stored in the ‘outdir’ line (line 8); set the .csv data file location and name in the ‘data path’ line (line 11); set the date and time period across which the analysis is to be performed using the ‘time_start’ and ‘time_end’ lines (time and date format = “YYYY-MM-DD HH:MM:SS”) (lines 25 and 26).
The Python scripts provided can be used for analyses as described below.
-
34.
In all cases an output .csv file is generated in which the first column is the specified time bin and the subsequent columns are the re-binned data from each PIR sensor. In the case of the binning.py script the final column shows the light data from the LDR.
Users are encouraged to generate analysis techniques best suited to their specific needs. Additionally, please see the STATISTICAL ANALYSIS section below regarding how best to fully understand and analyse this data.-
a)Change the bin size of the activity data: While the .csv data file gives activity data, it is in 10 second time bins. This can produce a very large number of data points that can be very unwieldy to analyse. The binning.py script allows the user to change the size of the time bins to alternative bin sizes (e.g. 60 mins). The output bin size (in minutes) is specified the settings.py script (line 16).
-
b)Convert the activity data into sleep data: When the PIR senor is positioned as described above, immobility bouts of 40 seconds or more are highly correlated with time spent asleep (Brown et al 2017). The sleep_counting.py script extracts periods of zero movement for 40 seconds or more to give a measure of the time spent in immobility defined sleep. The time bins in which this data is returned is specified in the sleep_counting.py script (line 13).
-
c)Change the activity data into sleep bout data: The period between the onset of a sleep episode and the subsequent waking is defined as a sleep bout. Sleep bout analysis will give a measure of sleep fragmentation, which is usually apparent as an increase in the number of short bouts. The sleep_bout_lengths.py script will produce two different outputs. The sleep_bout_length .csv file returns the mean length of the sleep bouts in time bins specified in the settings.py script (line 22). The sleep_bin_counts_by_pir folder returns .csv files of the total number of sleep bouts of different lengths in bins of one minute, up to 15 minutes. There will be a different .csv for each 12 hour period analysed.OPTIONAL: All three scripts can be run as one combined script. The run_sleep_scripts.py script will run the three scripts described above, with the options for this running set in the sleep.toml script (both scripts can be found at https://github.com/MRC-Harwell/sleep-tracking/).
-
a)
Alternate Protocol
Data collection using Raspberry Pi
An obvious limitation of the system outlined in the basic protocol above is the requirement of one laptop for every 6 PIR channels. Whilst this may not be a consideration for screening small numbers of animals, for large-scale screening involving tens or even hundreds of cages, using one laptop for every 6 cages is very cumbersome and often impractical. As such, an alternative to using a laptop is the use of a low cost single-board computer such as a Raspberry Pi. Raspberry Pi boards are much cheaper and smaller than laptops and can easily be integrated into the PIR system to continuously upload data to online resources such as Dropbox. There are a wealth of online resources for working with Raspberry Pi units (https://www.raspberrypi.org/) and Processing is also an option here (https://pi.processing.org/). As such, the authors recommend that users consider such alternatives depending upon their experimental requirements. This alternate protocol uses Node-RED (https://nodered.org/), a simple and low-code system.
Materials
Raspberry Pi kit (including board, case and power supply; Raspbian operating system) (Farnell cat no RPI3-MODBP-STARTER).
HDMI-capable monitor (or adapter), keyboard and mouse for initial set up of the Pi system. If required/preferred, these can be removed following set up.
Sample Data (incorporated into the protocol steps)
With the Raspberry Pi connected to a monitor, keyboard and mouse, install and set up Node-RED on the Raspberry Pi (details for this can be found here: https://nodered.org/docs/getting-started/raspberrypi).
Once the web interface for Node RED is available install the ‘thethingbox-node-timestamp’ node via the 'manage pallete' option on the web page.
If remote backup of data to Dropbox is required, install the ‘node-red-node-dropbox.’
Import the ‘COMPASS_Arduino string to .csv’ flow onto the Pi (the flow can be found at https://flows.nodered.org/flow/70424972bc835ead155e3264f7d100b1).
If remote backup of data to Dropbox is required, install the ‘COMPASS_backup to Dropbox’ flow (https://flows.nodered.org/flow/5d825ee3fd1194398cc3bb57fdc79745).
Assemble and set up the PIR sensors as described in steps 1 to 4 of the basic protocol above.
Connect the main board to the Pi unit and find the connected unit by opening the serial node in the web interface. Data from the serial port can be checked using a debug node and reading the messages.
The flow can be duplicated for more than one Arduino base unit and Node-RED provides a wealth of other options for local and remote storage as well as other sensors.
Support Protocol
Circadian analysis using PIR sensors
The protocol described below provides a standard circadian phenotyping screen. We and others have previously published detailed guidelines for circadian analysis of mice (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015; Hughes et al., 2015; Brown et al., 2019). Please refer to these protocols for more comprehensive guidance of circadian protocols and materials. Be aware that a detailed circadian screen using different light/dark protocols will require the use of light-tight chambers with independent lighting control.
Materials
See Basic Protocol
Follow the steps outlined in the basic protocol above, with PIR sensors (one above each cage) in a light-tight environmentally controlled chamber with independent lighting control.
Singly house mice and place cages into the chamber, ensuring that there are no regions of the cage blocked from the PIR sensor that the mouse can access (see Basic Protocol Steps 25 to 27).
Once all cages are in position, close the cabinet and start the PIR recording, as described in the basic protocol step 28.
-
Set the lighting timers of the circadian cabinets to those required by the experiment.
For a basic circadian analysis we recommend 7 to 14 days in a 12h light/12h dark cycle, followed by 10 to 14 days in constant darkness to assess free running period.
Alternatively, more detailed protocols including phase shifting light pulses or conditions known to result in circadian disruption may be used (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015; Hughes et al., 2015; Brown et al., 2019).
During recording ensure a light-controlled environment is maintained and that cage disturbance is kept to a minimum (see Basic Protocol).
Always include an LDR on at least one PIR sensor to monitor the light environment throughout the study.
After the completion of the recording, remove the mice from the circadian cabinets and stop and close the recording software.
Convert the .csv data file produced by the recording into an .awd file which can be read by the Clocklab circadian analysis software (Actimetrics). This can be achieved by running the binning.py script, setting the output bin size to 1 minute. The re-binned data file will display the data for each sensor in a different column in one minute bins.
Open the .awd template file (https://github.com/MRC-Harwell/sleep-tracking/tree/master/data) in Notebook (note that a new .awd file must be created for each animal to be analysed). Fill in the additional fields as noted in lines 1,2,3,5 and 7. Paste the data for the specified animal starting from line 8 (below the ‘sex’ field), with each data point on a new line. Save this with an .awd extension. Note that the repository link above also includes an example .awd file that can be compared to the template and used as a guide for data entry.
-
Alternatively, using a text editor such as Notebook, simply add the following 7 line header before the data:
- NAME
- START DATE (dd-mon-yyyy), where the month uses stardard 3-letter abbreviations (lower case)
- START TIME (24 hour) Always is 2 digits for both hour and minute, as in 04:20
- INTERVAL (sample interval, or the time between data points, in minutes multiplied by 4)
- AGE (fill in any number just as a place holder, it will be ignored)
- ID (also ignored, put in something random as a place holder)
- SEX (ignored)
Following this header, just put the data points one after another, one number per line. Finally, give the file the extension ".AWD". The file can now be read by Clocklab and analysed following program instructions.
An example is provided below:- Taconic2
- 21-nov-2000
- 00:00
- 24
- 0
- 9wild
- M
-
253 (First data point)OPTIONAL: Open source circadian analysis software such as ActogramJ (Schmid et al 2011) or Rethomics (Geissmann et al 2020) can also be used for circadian analysis. The activity data generated by protocols described here can be used directly in such programs following the program instructions. However it may still be advantageous to re-bin the raw data for ease of use.
Commentary
Background information
Circadian rhythms are approximately 24h cycles of physiology and behaviour that optimise internal biology to the varying demands of day and night. Whilst gene expression, cognition, cardiovascular function, digestion, hormone synthesis and body temperature all show robust circadian rhythms (Rijo-Ferreira and Takahashi 2019), perhaps the most recognisable circadian rhythm is that of activity and rest. In mammals, circadian rhythms in activity are regulated by a master circadian pacemaker located in the suprachiasmatic nuclei (SCN) of the anterior hypothalamus. The SCN receives light input from the retina, entraining the endogenous biological clock to the external light/dark cycle (Reppert and Weaver, 2002). However, sleep is not a simple output of the circadian system. Whilst the circadian system contributes to the regulation of the timing of sleep and wake, sleep is also regulated by a homeostatic process which increases the propensity for sleep with extended wakefulness. These circadian and homeostatic processes interact to determine the amount and distribution of sleep across the 24h cycle (Borbely 1982; Borbely et al., 2016). There is an increasing appreciation that circadian rhythms and sleep are essential for normal healthy physiology. Sleep and circadian rhythm disruption results in numerous deleterious effects, and is also associated with a range of conditions, including neurological and metabolic disease as well as aging (Bechtold et al., 2010; Hastings et al, 2003; Jagannath et al., 2013; West and Bechtold, 2015; Wulff et al., 2010; Banks et al., 2015).
Laboratory mice provide a versatile animal model for the study of circadian rhythms and sleep. Both the genetic and neuronal circuits regulating these processes are conserved between mice and other mammals, including humans (Lo et al 2004; Weiergraber et al 2005). EEG studies of sleep have also demonstrated that the structure of sleep and the composition of characteristic frequencies of neuronal activity are also largely conserved between mice and humans (Fuller et al., 2006). Additionally, mice show the same associations between sleep and circadian rhythm disturbances and metabolic or cognitive disruptions as have been shown to occur in humans (Fisk et al., 2018). While such evidence demonstrates the power of the mouse as a model for understanding conserved processes, a number of practical considerations also make the mouse a useful model in the study of circadian and sleep function. Studies of long term sleep and circadian rhythm disruption (such as repeated jetlag or sleep deprivation) which would be practically and ethically challenging to perform in humans can readily be studied in mice. Critically, genetically modified mice provide an ever-expanding resource to understanding the molecular mechanisms underlying these biological processes. Repositories of genetically modified mice have been successfully utilised for large scale screens for sleep and circadian phenotypes (Parsons et al 2015; Banks et al 2018; Potter et al 2016; Funato et al 2016; Zhang et al 2020). In addition, methods such as gene editing allow the role of specific human mutations to be studied in mice to establish their role in sleep or circadian rhythms (Kurien et al 2019). Finally, techniques such as conditional and inducible gene targeting as well as optogenetics allow researchers to generate increasingly specific spatial and temporal genetic models as well as enabling the study of specific neuronal circuits in sleep and circadian biology (Wilcox et al 2017; Yang et al 2016).
Historically, a range of methods have been used to analyse circadian activity and sleep in mice. Of the two parameters, activity is the easiest to measure over long time periods. The most common activity measure has been to use cages equipped with running wheels. Mice, when housed with free access to a wheel, will spend the majority of the active phase of their daily cycle running on the wheel. Running wheel activity can be monitored via a micro-switch attached to the wheel, and saved to a local computer to give a measure of activity over time (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015). While wheel running screens are robust and relatively inexpensive, they only give a measure of one aspect of home cage activity. If the animal is feeding, grooming or in a state of quiet wakefulness, no activity will be detectable even though the animal is clearly still awake and active. For this reason, running wheel data alone does not provide a reliable indicator of sleep. Instead, the gold standard methodology for the assessment of sleep is electroencephalography (EEG) combined with electromyography (EMG) (Lo et al 2004; Weiergraber et al 2005). EEG/EMG provides a detailed breakdown of sleep structure, enabling rapid eye movement (REM) and non-rapid eye movement (NREM) – also known as slow-wave sleep (SWS) - to be measured. Moreover, it also allows spectral analysis of the specific frequencies of neuronal activity to be assessed. However, EEG/EMG requires the surgical implantation of electrodes. As such, non-invasive behavioural phenotyping methods can provide a valuable preliminary screen prior to focused EEG/EMG studies, as well as providing an ideal tool for high-throughput genetic screens. Sleep is also frequently disrupted in response to injury of infection, but as measuring sleep has typically necessitated invasive measures, sleep is rarely used as a biomarker of animal health or disease progression.
In mice, periods of extended immobility of 40 seconds or more are highly correlated with sleep (Pack et al 2007). This enables researchers to conduct non-invasive behavioural phenotyping of both sleep and activity in the same animals (Fisher et al 2012). While behavioural sleep scoring cannot distinguish between REM and NREM sleep, it can be used to rapidly measure the amount and distribution of sleep over multiple cycles (Banks et al 2015; Heise et al 2015). However, video tracking generates very large data files which presents issues for data storage and sharing – particularly for large scale screens. This data also requires specialist software for analysis and can be computationally intensive. The COMPASS technique we describe here relies upon the same behaviourally-defined sleep criteria as video tracking, and provides a comparable accuracy versus EEG/EMG scored sleep. A key difference is that the output data files are small and can be easily analysed in real time (Brown et al 2017). This enables days, weeks or even months of data to be easily collected, archived and shared. Additionally, the relative simplicity of the components means that PIR screening is both low-cost and can be assembled and run with minimal expertise in electronics or data processing. It is important to note that the COMPASS system was never intended to be a static or finalised methodology. The protocol outlined here should allow users to set up the basic equipment and make recordings in an animal house environment, but the system is extremely flexible and amenable to customisation. For example, temperature, noise or other environmental sensors can easily be incorporated. As such, we recommend that users adapt and modify the system to suit their own specific research requirements. Updates and modifications to the system - as well as additional guidance can be found online at: http://users.ox.ac.uk/˜anat0144/COMPASS_updates.html and individual files are also available from: https://zenodo.org/record/3838998#.Xsei3ndFwjY.
Critical Parameters:
Electrical components
If unfamiliar with simple electronics, it is recommended that users familiarise themselves with the basics of soldering and assembly of electrical components. Guides for soldering are freely available online and should be consulted if the user is inexperienced with such techniques. It is also recommended that spare components are ordered to allow for practice and accidental damage. All components are relatively cheap, and this also provides a ready reserve should components need replacing. Following the construction of each circuit board, it is vital that each solder point is tested to ensure there are no loose connections. Additionally, care must be taken that soldered connections or wires do no cross (heat-shrink tubing or tape can be used if necessary). Finally, good crimp connections on cables or wires are essential for reliable recordings. Again, if necessary, additional components should be ordered to establish good practice. The inbuilt Arduino IDE Serial Monitor tool provides an ideal way of testing PIR sensors and light sensors are working properly prior to use in experiments and enables crosstalk between PIR channels to be easily detected during testing. These are most likely to occur due to soldered points being accidentally joined or crimp connectors being incorrectly attached (see Troubleshooting below).
Mouse age, sex and strain
Strain specific differences in both circadian rhythms and sleep have been demonstrated in a number of studies (Banks et al 2015; Hasan et al 2012). Therefore, when comparing cohorts of mice, the genetic background of the cohorts should be as closely matched as possible. The best comparison is between littermates, so if possible, use siblings for experimental and control groups. A number of studies provide background on the genetic and behavioural differences between mouse strains (Simon et al 2013). Such studies may also provide guidance of the most appropriate strain to use for particular studies. Age-related changes have also been reported for both activity rhythms and sleep (Banks et al 2015; Hasan et al 2012). In general, older animals show reduced activity and increased sleep with increasing fragmentation and variability. While older mice can still be used for analysis - and age-related phenotypes may be of interest - the increased variability in data may require larger cohort numbers or the use of within-subject longitudinal experimental designs. Finally, sex specific differences have been demonstrated in both circadian rhythms and sleep (Bailey and Silver 2014). Therefore, comparisons should be based upon mice of the same sex, and where possible, both male and female mice should be analysed to test for sex-dependent differences.
Disturbance
The measurment of both circadian rhythms and sleep demands as little disturbance from external sources as possible. Disturbance to the cage as well as noise, light or changes in temperature will all disturb animals, altering their behaviour and potentially resulting in stress which will influence both circadian activity and sleep. As mice are nocturnal, they are typically inactive and asleep during the day, when both technicians and scientists will be working. As such, daily animal checks should be performed with minimal disruption and cage changes should be minimised wherever possible within the necessary limits of local regulations. Where checks are conducted in the dark phase, use of dim red light is recommended to minimise potential circadian disruption (>600nm wavelength and ideally <1 lux or <11.8 log quanta/cm2/s, Peirson et al., 2018). Unless required by the study (e.g. scheduled feeding), food and water should be accessible ad libitum to prevent entrainment to feeding regimes. The best method of minimising external disruption is the use of light-tight environmentally controlled chambers, as described elsewhere (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015).
Light source
Both circadian activity rhythms and sleep are extremely sensitive to light. A cool white light source generating a light intensity of 100-300 lux is standard for circadian entrainment, though mice are capable of responding to far lower light levels (Peirson et al., 2018). The use of light sources such as fluorescent or LED bulbs in dedicated circadian cabinets has been discussed elsewhere (Albrecht and Foster 2002, Jud et al 2005, Banks and Nolan 2011, Eckel-Mahan and Sassone-Corsi 2015). However, be aware that white light sources are not all equivalent. Despite their similar appearance to the human eye, different white light sources can be spectrally very different, and may exert differing visual and non-visual effects as well as appearing different to mice. As such, it is always recommended that the spectral power distribution of the light source is properly characterised in any circadian or sleep study (Lucas et al., 2014). Researchers should also be aware that if studies are conducted in procedure rooms or cage racks, care must be taken that the light intensity is the same across all cages being recorded. This is a particular issue when using standard laboratory mouse cage racks where the light intensity can vary dramatically due to cage position. However, even in light-tight environmentally controlled chambers light intensity can potentially vary between chambers and cage positions, which can be a source of systematic experimental bias.
Troubleshooting
Common technical problems and likely solutions are provided below:
| Steps | Problem | Possible solutions |
|---|---|---|
| 13-18 | Ardunio microcontroller board not recognized when plugged in | Check Arduino IDE is open Attach board by USB cable Under Tools in IDE, check for correct board and port entries Unplug microcontroller and plug in again |
| 19-21 | No readout or saved file after running Processing sketch | Close Arduino IDE Re-run Processing sketch Change the ‘portindex1 =’ value towards the top of the Processing sketch to match the designated port |
| 1-12 | Output values are random values | Check cable and sensor (if possible test replacements) Check all soldering connections |
| 1-5 and 13 | Constant readings of 1 or 100 | If on all sensors, ensure the RJ12 cable does not cross over If on a single sensor replace cable or sensor and check connections to main board and microcontroller |
| 3, 10 and 11 | Failure to read environmental light via the LDR | Ensure the face of the LDR faces the light source Ensure the legs of the LDR are not crossed and touching |
| 27-29 | Changes of read quality following cage change or animal check | Confirm that the PIR sensor has not been moved during the cage change Ensure that the cage has been returned to the same location relative to the sensor. If necessary, runners or guides can be installed to maintain correct cage placement |
| 32-34 | Failure to read .csv data file by Python scripts | Confirm that the file location designated in the settings.py script includes the .csv extension in the filename Confirm that all data in the .csv is numerical. Any non-numerical data should be removed from the file If being read and saved by Windows, the .csv file can initially be saved in such a way that it cannot be read by python scripts. If so, open the .csv file in excel and resave before reading in Python |
Statistical Analysis
The statistical methods used to analyse the data generated in PIR experiments will depend upon the specific hypothesis under investigation and the experimental design. For example, where gross differences in activity or sleep under a 12h light/12h dark cycle are of interest, a simple comparison between total day-time and night-time activity and sleep counts may be sufficient. Such data can be analysed by simple ANOVA or Students t-test, with 2-way ANOVAs where control vs treated or wildtype vs knockout designs are used. However, as PIR recordings allow longitudinal data to be analysed over time, the distribution of activity or sleep over one or more days may provide considerably more data. Such data are amenable to analysis using repeated measures ANOVA. In many cases, the primary circadian and sleep literature provide a rich resource to help plan experiments, as well as to obtain measurements of variance to allow power calculations to determine the necessary sample size.
As described above, light exerts profound effects on both activity and sleep in mice. The lighting environment of the cage should therefore be taken into consideration when conducting any form of analysis. Light and dark phase data should be considered separately, as the behaviour of animals will differ markedly between these conditions. Additionally, if unexpected light changes are evident on the lighting channel data may need to be excluded from the analysis. The response of mice to different lighting conditions provides additional critical data regarding different aspects of circadian function, for which the reader is referred elsewhere (Hughes et al., 2015).
Circadian researchers have developed a wide range of detailed methods for analysing activity rhythm data, particularly for the measurement of circadian period (Refinetti 1993; Refinetti et al 2007; Zielinski et al., 2014). Commercial analysis programs such as ClockLab (Actimetrics) provide user-friendly methods of analysing circadian data, including period estimates, activity onsets and phase shifting. Alternatively, similar analyses can be performed using open source software. For example, ActogramJ can perform a range of periodogram analyses (Schmid et al 2011), as can online resources such as the Circadian Rhythm Laboratory (https://www.circadian.org/softwar.html) or BioDare 2 (https://biodare2.ed.ac.uk/, Zielinski et al., 2014). Finally, disruption of circadian rhythms and sleep can be assessed using a range of different measures (Brown et al., 2019), including interdaily stability (IS) and intradaily variability (IV) (Van Someren et al 1999), implementations of which are also available as online R scripts (Blume et al 2016).
Understanding Results
“It seems that many researchers think that circadian biology involves simply placing a mouse in a cage with a running wheel and watching papers fall out the other end”
- Anonymous
Although measuring the levels of home cage activity and sleep using the PIR system described herein is relatively straightforward, as in all research areas the interpretation of data often depends upon a detailed understanding of the underlying biology. The interpretation of results from different types of circadian analyses have been described in detail elsewhere (Albrecht and Foster 2002; Jud et al 2005; Banks and Nolan 2011; Eckel-Mahan and Sassone-Corsi 2015; Hughes et al., 2015). While PIR circadian recordings will generate comparable data to wheel running screens, users should also be aware that some differences will exist. As PIR sensors detect all animal movement rather than just voluntary running wheel activity, in our experience greater activity will be observed during the inactive light phase, activity is often less consolidated and there may be a greater day-to-day variability in PIR data when compared to wheel running data.
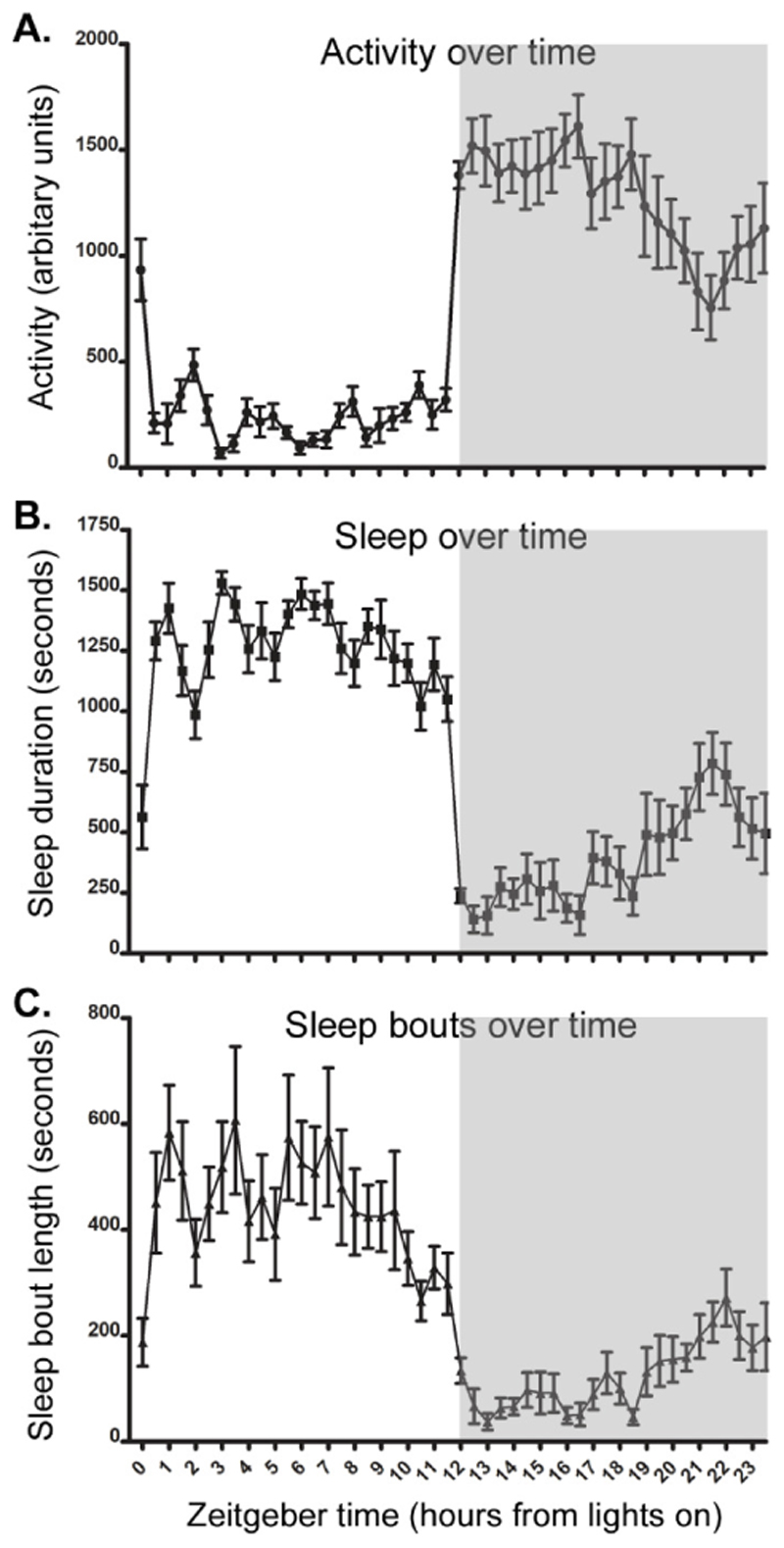
An example of activity, sleep duration and sleep bout data generated from a wildtype C57BL/6N animal in a standard 12h light/12h dark cycle is shown in Figure 5. In the light phase, mice spend ˜70% of their time asleep. While the animal will show some activity during the light phase, this will be much lower. The length of the sleep bouts of an animal also shows high variability across a day. However, in general sleep in the light phase consists of fewer, longer bouts than sleep in the dark phase. Notably, previous data has demonstrated that patterns of activity and sleep are relatively stable across different mouse strains and ages in the light phase (Banks et al 2015). However, in the dark phase strain- and age-dependent patterns of activity and sleep are more apparent. Furthermore, conspecific differences are also apparent, and even genetically identical mice may show quite different patterns of activity and sleep, though these patterns are very stable within any individual (Brown et al., 2017).
Figure 5. Typical activity, sleep and sleep bout length in C57BL/6N animals (females, 34 weeks old, n=9).
Data given in 30-min time bins. Grey section of the graph represents the dark phase of the light-dark cycle. (A) Activity over 24h. (B) Time spent in immobility defined sleep over 24h. (C) Average length of sleep bouts over 24h.
Time Considerations
Users should allow sufficient time to assemble all components and test the PIR system prior to starting any animal work. Once assembled, the system can easily be tested by simply running the Arduino IDE ‘Serial Monitor’ tool, which allows the user to see the signal generated in real time by each sensor channel – typically 6 separate PIR sensors and a single light channel. This enables crosstalk between channels or non-functioning channels to be easily detected. Following this, running the system using Processing or using Node red on a Raspberry Pi will allow the system to be tested over several days to ensure everything works properly before starting experiments. We have often left a system running in the lab to measure human traffic and light levels as a simple way of testing that the system works prior to animal studies.
The time taken for activity and sleep analysis is dependent upon the experiment or condition the measures are being taken for. However, it should be noted that once running the system is fully automated, requiring little input from the investigator throughout recording. PIR analysis of C57BL/6J animals has demonstrated that individual animals show highly stable day to day patterns of activity and sleep (Brown et al 2017). It is therefore possible to establish baseline levels of activity and sleep from 3-5 days of data. However, it is worth noting that this represents a minimum, and circadian screens often require 7-10 days under each lighting condition to obtain sufficient data for detailed analysis. For both baseline activity and sleep analysis and detailed circadian screening, it is worth noting that the first 1-2 days an animal is housed in a new cage or lighting condition their activity and sleep may be more variable. For this reason, cage changes for husbandry purposes should be planned in advance so as not to coincide with critical periods of data recording.
Finally, if the PIR system is for monitoring health or disease progression, recordings may be necessary for extended periods. However, establishing a healthy baseline is critical, as deviations from this baseline are likely to provide the most reliable welfare indicators. For example, if assessing the effects of surgical interventions on activity and sleep, a comparison of at least a week pre- and post-surgery would be required. By contrast, models of neurodegenerative disease may require monitoring over a period of months as the condition progresses. In our experience the PIR system can make stable continuous measurements for at least a year, so the protocols we describe here can easily be used for such longitudinal measurements associated with biological processes such as aging. In all cases, it is recommended that experiments are discussed with local animal care and veterinary staff to address welfare considerations prior to commencing such a study. For example, some neurodegenerative models may not thrive under single housing conditions, which may limit the monitoring of activity and sleep in individual animals. Alternatively, separating male animals to record activity and sleep and then returning animals to cohoused conditions can result in fighting.
Acknowledgements
P.M.N. is supported by the MRC (grant codes MC_U142684173). S.N.P is supported by the BBSRC (BB/I021086/1 and BB/S015817/1), Wellcome (098461/Z/12/Z) and the NC3Rs (NC/V000977/1). S.L.W. is funded by the NC3Rs (NC/S001689/1). We would like to thank Rhys Hancocks and Angus Fisk for their assistance in establishing and running the PIR systems in our institutes and the biomedical staff at the Mary Lyon Centre, MRC Harwell Institute. Thanks to Robert Dallmann (University of Warwick) for advice on how to convert data for ClockLab.
Literature Cited
- Albrecht U, Foster RG. Placing ocular mutants into a functional context: a chronobiological approach. Methods. 2002;28(4):465–77. doi: 10.1016/s1046-2023(02)00266-9. [DOI] [PubMed] [Google Scholar]
- Bailey M, Silver R. Sex differences in circadian timing systems: Implications for disease. Front Neuroendocrinol. 2014;35(1):111–39. doi: 10.1016/j.yfrne.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks GT, Nolan PN. Assessment of Circadian and Light-Entrainable Paramters in Mice Using Wheel-Running Activity. Curr Protoc Mouse Biol. 2011;1(3):369–81. doi: 10.1002/9780470942390.mo110123. [DOI] [PubMed] [Google Scholar]
- Banks G, Heise I, Starbuck B, Osborne T, Wisby L, Potter P, Jackson IJ, Foster RG, Peirson SN, Nolan PN. Genetic background influences age-related decline in visual and nonvisual retinal responses, circadian rhythms, and sleep. Neurobiol Aging. 2015;36(1):380–93. doi: 10.1016/j.neurobiolaging.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks G, Lassi G, Hoerder-Suabedissen A, Tinarelli F, Simon MM, Wilcox A, Lau P, Lawson TN, Johnson S, Rutman A, Sweeting M, et al. A Missense Mutation in Katnal1 Underlies Behavioural, Neurological and Ciliary Anomalies. Mol Psychiatry. 2018;23(3):713–722. doi: 10.1038/mp.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold DA, Gibbs JE, Loudon AS. Circadian dysfunction in disease. Trends Pharmacol Sci. 2010;31(5):191–8. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Blume C, Santhi N, Schabus M. 'nparACT' Package for R: A Free Software Tool for the Non-Parametric Analysis of Actigraphy Data. MethodsX. 2016;3:430–5. doi: 10.1016/j.mex.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1(3):195–204. [PubMed] [Google Scholar]
- Borbély AA, Daan S, Wirz-Justice A, Deboer T. The two-process model of sleep regulation: a reappraisal. J Sleep Res. 2016;25(2):131–43. doi: 10.1111/jsr.12371. [DOI] [PubMed] [Google Scholar]
- Brown LA, Hasan S, Foster RG, Peirson SN. COMPASS: Continuous Open Mouse Phenotyping of Activity and Sleep Status. Wellcome Open Research 2017. 2017;1:2. doi: 10.12688/wellcomeopenres.9892.2. [version 2; peer review: 4 approved] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LA, Fisk AS, Pothecary CA, Peirson SN. Telling the Time with a Broken Clock: Quantifying Circadian Disruption in Animal Models. Biology (Basel) 2019;8(1):18. doi: 10.3390/biology8010018. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas M, Hut RA. Twilight and photoperiod affect behavioral entrainment in the house mouse (Mus musculus. J Biol Rhythms. 2009;24(5):403–12. doi: 10.1177/0748730409343873. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. Phenotyping circadian rhythms in mice. Curr Protoc Mouse Biol. 2015;5:271–281. doi: 10.1002/9780470942390.mo140229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SP, Godinho SIH, Pothecary CA, Hankins MW, Foster RG, Peirson SN. Rapid Assessment of Sleep-Wake Behavior in Mice. J Biol Rhythms. 2012;27(1):48–58. doi: 10.1177/0748730411431550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk AS, Tam SKE, Brown LA, Vyazovskiy VV, Bannerman DM, Peirson SN. Light and Cognition: Roles for Circadian Rhythms, Sleep, and Arousal. Front Neurol. 2018 Feb 9;9:56. doi: 10.3389/fneur.2018.00056. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AE, Flores JE, Deshpande H, Picazo JA, Xie XS, Franken P, Heller HC, Grahn DA, O’Hara BF. Pattern recognition of sleep in rodents using piezoelectric signals generated by gross body movements. IEEE Trans Biomed Eng. 2007;54(2):225–33. doi: 10.1109/TBME.2006.886938. [DOI] [PubMed] [Google Scholar]
- Fuller PM, Gooley JJ, Saper CB. Neurobiology of the Sleep-Wake Cycle: Sleep Architecture, Circadian Regulation, and Regulatory Feedback. J Biol Rhythms. 2006;21(6):482–93. doi: 10.1177/0748730406294627. [DOI] [PubMed] [Google Scholar]
- Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z, Ma J, Nakane S, Tomita J, Ikkyu A, Kakizaki M, et al. Forward-genetics Analysis of Sleep in Randomly Mutagenized Mice. Nature. 2016;539(7629):378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann Q, Rodriguez LG, Beckwith EJ, Gilestro GF. Rethomics: An R framework to analyse high-throughput behavioural data. PLoS ONE. 2020;14(1):e0209331. doi: 10.1371/journal.pone.0209331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S, Dauvilliers Y, Mongrain V, Franken P, Tafti M. Age-related changes in sleep in inbred mice are genotype dependent. Neurobiol Aging. 2012;33:195.e13-195.e26. doi: 10.1016/j.neurobiolaging.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4(8):649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Heise I, Fisher SP, Banks GT, Wells S, Peirson SN, Foster RG, Nolan PM. Sleep-like behaviour and 24-h rhythm disruption in the Tc1 mouse model of Down syndrome. Genes Brain Behav. 2015;14:209–216. doi: 10.1111/gbb.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S, Jagannath A, Hankins MW, Foster RG, Peirson SN. Photic regulation of clock systems. Methods Enzymol. 2015;552:125–43. doi: 10.1016/bs.mie.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Jagannath A, Peirson SN, Foster RG. Sleep and circadian rhythm disruption in neuropsychiatric illness. Curr Opin Neurobiol. 2013;23(5):888–94. doi: 10.1016/j.conb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Jud C, Schmutz I, Hampp G, Oster H, Albrecht U. A guideline for analyzing circadian wheel-running behavior in rodents under different lighting conditions. Biol Proced Online. 2005;7:101–16. doi: 10.1251/bpo109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurien P, Hsu P-K, Leon J, Wu D, McMahon T, Shi G, Xu Y, Lipzen A, Pennacchio LA, Jones CR, Fu Y-H, et al. TIMELESS mutation alters phase responsiveness and causes advanced sleep phase. PNAS. 2019;116(24):12045–12053. doi: 10.1073/pnas.1819110116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CC, Chou T, Penzel T, Scammell TE, Strecker RE, Stanley HE, Ivanov PCh. Common scale-invariant patterns of sleep-wake transitions across mammalian species. Proc Natl Acad Sci U S A. 101(50):17545–8. doi: 10.1073/pnas.0408242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas RJ, Peirson SN, Berson DM, Brown TM, Cooper HM, Czeisler CA, Figueiro MG, Gamlin PD, Lockley SW, O’Hagan JB, Price LL, et al. Measuring and using light in the melanopsin age. Trends Neurosci. 2014;37(1):1–9. doi: 10.1016/j.tins.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE. Circadian regulation of sleep in mammals: role of the suprachiasmatic nucleus. Brain Res Brain Res Rev. 2005;49(3):429–54. doi: 10.1016/j.brainresrev.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Pack AI, Galante RJ, Maislin G, Cater J, Metaxas D, Lu S, Zhang L, Von Smith R, Kay T, Lian J, Svenson K, et al. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics. 2007;28:232–238. doi: 10.1152/physiolgenomics.00139.2006. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Brancaccio M, Sethi S, Maywood ES, Satija R, Edwards JK, Jagannath A, Couch Y, Finelli MJ, Smyllie NJ, Esapa C, et al. The Regulatory Factor ZFHX3 Modifies Circadian Function in SCN via an AT Motif-Driven Axis. Cell. 2015;162(3):607–21. doi: 10.1016/j.cell.2015.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson SN, Brown LA, Pothecary CA, Benson LA, Fisk AS. Light and the Laboratory Mouse. J Neurosci Methods. 2018;300:26–36. doi: 10.1016/j.jneumeth.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter PK, Bowl MR, Jeyarajan P, Wisby L, Blease A, Goldsworthy ME, Simon MM, Greenaway S, Michel V, Barnard A, Aguilar C, et al. Novel Gene Function Revealed by Mouse Mutagenesis Screens for Models of Age-Related Disease. Nat Commun. 2016;18(7):12444. doi: 10.1038/ncomms12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R. Laboratory instrumentation and computing: comparison of six methods for the determination of the period of circadian rhythms. Physiol Behav. 1993;54(5):869–75. doi: 10.1016/0031-9384(93)90294-p. [DOI] [PubMed] [Google Scholar]
- Refinetti R, Lissen GC, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 2007;38(4):275–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of Circadian Timing in Mammals. Nature. 2002;418(6901):935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Rijo-Ferreira F, Takahashi JS. Genomics of Circadian Rhythms in Health and Disease. Genome Med. 2019;11(1):82. doi: 10.1186/s13073-019-0704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scammell TE, Arrigoni E, Lipton J. Neuronal Circuitry of Wakefulness of Sleep. Neuron. 2017;93(4):747–765. doi: 10.1016/j.neuron.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Helfrich-Förster C, Yoshii T. A new ImageJ plugin "ActogramJ" for chronobiological analyses. J Biol Rhythms. 2011;26:464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- Simon MM, Greenaway S, White JK, Fuchs H, Gailus-Durner V, Wells S, Sorg T, Wong K, Bedu E, Cartwright EJ, Dacquin R, et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013;14:R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizard I. Sickness behavior, its mechanisms and significance. Anim Health Res Rev. 2008;9(1):87–99. doi: 10.1017/S1466252308001448. [DOI] [PubMed] [Google Scholar]
- Van Someren EJW, Swaab DF, Colenda CC, Cohen W, McCall WV, Rosenquist PB. Bright Light Therapy: Improved sensitivity to its effects on rest-activity rhythms in Alzheimer patients by application of nonparametric methods. Chronobiol Int. 1999;16(4):505–518. doi: 10.3109/07420529908998724. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ma J, Miyoshi C, Li Y, Sato M, Ogawa Y, Lou T, Ma C, Gao X, Lee C, Fujiyama T, et al. Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature. 2018;558(7710):435–439. doi: 10.1038/s41586-018-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiergräber M, Henry M, Hescheler J, Smyth N, Schneider T. Electrocorticographic and deep intracerebral EEG recording in mice using a telemetry system. Brain Res Brain Res Protoc. 2005;14(3):154–64. doi: 10.1016/j.brainresprot.2004.12.006. [DOI] [PubMed] [Google Scholar]
- West AC, Bechtold DA. The cost of circadian desynchrony: Evidence, insights and open questions. Bioessays. 2015;37(7):777–88. doi: 10.1002/bies.201400173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AG, Vizor L, Parsons MJ, Banks G, Nolan PM. Inducible Knockout of Mouse Zfhx3 Emphasizes Its Key Role in Setting the Pace and Amplitude of the Adult Circadian Clock. J Biol Rhythms. 2017;32(5):433–443. doi: 10.1177/0748730417722631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci. 2010;11(8):589–99. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- Yang G, Chen L, Grant GR, Paschos G, Song W-L, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, FitzGerald GA. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. 2016;8:324ra16. doi: 10.1126/scitranslmed.aad3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Xie P, Dong Y, Liu Z, Zhou F, Pan D, Huang Z, Zhai Q, Gu Y, Wu Q, Tanaka N, et al. High-throughput Discovery of Genetic Determinants of Circadian Misalignment. PLoS Genet. 2020;16(1):e1008577. doi: 10.1371/journal.pgen.1008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ. Strengths and limitations of period estimation methods for circadian data. PLoS One. 2014;9(5):e96462. doi: 10.1371/journal.pone.0096462. [DOI] [PMC free article] [PubMed] [Google Scholar]