Abstract
The targeting and modulation of G-protein-coupled receptors (GPCRs) has immense therapeutic potential. A study in Nature now reports on the successful targeting of intracellular allosteric sites that effectively bias GPCR signaling, which has opened new opportunities to develop safer therapeutic agents.
GPCRs have a pivotal role in regulating various physiological processes through their interactions with a huge diversity of ligands. Their importance is evident, as approximately 35% of US Food and Drug Administration (FDA)-approved drugs target GPCRs, as agonists, antagonists, biased agonists, allosteric modulators or ago-allosteric modulators1,2. However, the traditional approach of developing small molecules that bind to the orthosteric site presents challenges in receptor-specific targeting owing to the high conservation of this site across GPCRs, resulting in potential side effects3. To address this limitation, the exploration of allosteric sites has emerged as a promising strategy for drug design. By targeting allosteric sites, drugs can modulate downstream signaling without directly competing with the endogenous ligand4,5. Allosteric ligands identified so far predominantly include antagonists, positive allosteric modulators (PAMs), negative allosteric modulators (NAMs), and ago-PAMs1,2,4 (ago-PAM can activate the receptor without the orthosteric ligand). However, whether biased agonist binding can occur at allosteric sites within the intracellular cavity remains unclear.
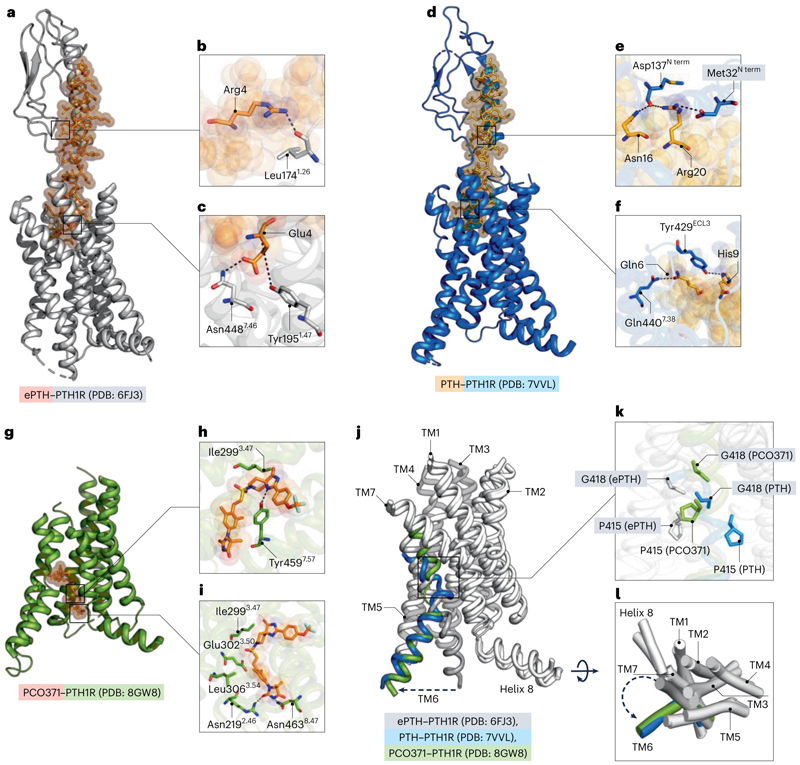
In a recent study, Kobayashi et al.6 identified an intracellular ligand-binding pocket in the human parathyroid hormone type 1 receptor (PTH1R), which diverges from the conventional extracellular orthosteric sites typically observed in class B1 GPCR structures7 (Fig. 1a–f). This discovery sheds light on new mechanisms of GPCR activation and presents opportunities for future drug discovery endeavors, combining intracellular and biased approaches. The work demonstrated that the PTH1R agonist PCO371 causes a distinct active state in PTH1R that combines an inactive extracellular conformation with an intracellular conformation that is similar to an active state (PTH-bound conformation) (Fig. 1g–i). The unique mode of action of PCO371 contributes to a greater knowledge of ligand–GPCR interactions and has created opportunities for investigating pharmacological therapies. By examining three key motifs —the PYQ active motif, the PXXG switch, and the HETY inactive motif — the authors found that endogenous ligand binding causes rearrangement in the PYQ motif, but in the case of mutation, no conformational change was observed; in addition, Pro4156.47 (in which superscripts denote relative positions in the receptor according to the Wootten class B1 GPCR numbering) is a specific residue in the intracellular allosteric motif of the receptor. Mutation of Pro4156.47 to Leu resulted in loss of activity of PCO371 (Fig. 1j,k). The TM6 conformation is altered when PCO371 binds to the intracellular allosteric site of the receptor (Fig. 1j–l), causing it to solely interact with the Gs protein. This biased signal is demonstrated by the absence of β-arrestin recruitment. Thus, a pocket generated by highly conserved residues on class B1 GPCRs interacts with the versatile agonist PCO371, activating 7 of the 15 class B1 receptors. The proline residue at position 6.47 seems to be important for the ability of PCO371 to recognize PTH1R, which suggests a connection between the activity of this compound and this residue (Fig. 1d–f). The failure of the molecule to activate PTH2R is thought to be caused by a distinct amino acid at position 6.47 (Fig. 1k). According to the findings, PCO371 stimulates receptors that may mobilize the intracellular and extracellular halves of TM6 separately. This research highlights the importance of certain residues and transmembrane structures in affecting the receptor activation and selectivity of substances such as PCO371. However, although both the synthetically made PCO371 and the endogenous ligand PTH activate the receptor, they do not have any overlapping residue contact. In addition, PTH binding generates a kink on TM6 at the Gly418 residue. By contrast, PCO371 generates the kink at the Pro415 residue but ultimately leads the receptor to an active state (Fig. 1j–l). As a result, Kobayashi et al.6 have identified an intracellular agonistic region in PTH1R where PCO371 functions as a molecular wedge to trigger a special activation pathway. In contrast to other ligands, PCO371 immediately binds to the intracellular transducer pocket and activates the receptor, which indicates that it penetrates the cell membrane and enters the intracellular or intramembrane area. PCO371 is an allosteric activator with a broad target base among class B1 GPCRs and this lack of specificity limits its usability. Nevertheless, the work provides a template for designing an allosteric modulator that provides opportunities for biased GPCR drug development.
Fig. 1. Structural details of PTH1R–ligand interactions.
a, Overview of engineered PTH (ePTH)-bound PTH1R structure viewed from the membrane plane (PTH1R depicted in gray, ePTH in orange). The ePTH-bound structure represents an inactive-like conformation of PTHR1. ePTH is shown in stick representation, with spheres of high transparency. b, Magnified view of binding sites of N terminus of PTH1R, showing Leu1741.26–Arg4 (nitrogen and oxygen atoms are shown in blue and red, respectively). c, Magnified view of the orthosteric site showing an interaction between Tyr1951.47–Glu4 and Asn4487.46–Glu4. d, Overview of PTH–PTH1R complex structure viewed from membrane plane (PTH1R in marine blue, PTH in light orange). PTH is shown in stick representation, with spheres of high transparency. e, Magnified view of binding sites of N terminus of PTH1R, showing Asp137–Asn16, Arg20, and Met32–Arg20 (nitrogen and oxygen atoms are shown in blue and red, respectively). f, Magnified view of the orthosteric site showing the interaction between Tyr429ECL3–His9 and Gln4407.38. ECL3, extracellular loop 3. g, Overview of PCO371–PTH1R complex structure viewed from membrane plane (PTH1R depicted in split-pea, and PCO371 in orange). PCO371 is shown in stick representation with high transparency. h, Magnified view of Tyr4597.57–PCO371 interaction. Ile2993.47 is shown in the cleft. i, Magnified view of Asn2192.46–PCO371 and Asn4638.47–PCO371 interactions. Leu3063.54, Glu3023.50 and Ile2993.47 are shown in the cleft. j, Overlapping view of PCO371–PTH1R, PTH–PTH1R and ePTH–PTH1R (TM6 is in split-pea, marine blue and grey, respectively, and the remaining helices are white). k, Stick representation of the movement of Pro415 and Gly418. l, Cylindrical helices representation of overlapped structures with all transmembrane domains colored white except TM6. Dotted arrow shows the direction of helix 6 movements away from the receptor core. Figure created with The PyMOL Molecular Graphics System, v.2.5.5, Schrödinger, LLC.
Although biased drugs for GPCRs hold much promise in reducing side effects, limited success in clinical trials has been a major bottleneck to their commercial success. Achieving biasing by targeting allosteric sites has the potential to revolutionize the next generation of safer drugs for several pathophysiological conditions across a range of therapeutic areas. Until then, understanding these intricate relationships between orthosteric and allosteric sites, and their functions in canonical and non-canonical GPCR activation7–9 that lead to biased and unbiased signaling10 will remain an important area of focus.
Acknowledgements
The research program in M.B.’s laboratory is supported by the India Alliance DBT Wellcome Trust (IA/E/20/1/505691) and the Institute Seed Grant (SGT-100081) Indian Institute of Technology Jammu.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz Zacarías NV, Lenselink EB, IJzerman AP, Handel TM, Heitman LH. Trends Pharmacol Sci. 2018;39:547–559. doi: 10.1016/j.tips.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powers AS, et al. Nat Chem Biol. 2023;19:805–814. doi: 10.1038/s41589-022-01247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan HCS, Li Y, Dahoun T, Vogel H, Yuan S. Trends Biochem Sci. 2019;44:312–330. doi: 10.1016/j.tibs.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Katritch V, Cherezov V, Stevens RC. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi K, et al. Nature. 2023;618:1085–1093. doi: 10.1038/s41586-023-06169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baidya M, Dwivedi H, Shukla AK. Nat Struct Mol Biol. 2017;24:500–502. doi: 10.1038/nsmb.3418. [DOI] [PubMed] [Google Scholar]
- 8.Kumari P, Inoue A, Chapman K, Lian P, Rosenbaum DM. Proc Natl Acad Sci USA. 2023;120:e2219569120. doi: 10.1073/pnas.2219569120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullard A. Nat Rev Drug Discov. 2023;22:347–348. doi: 10.1038/d41573-023-00064-2. [DOI] [PubMed] [Google Scholar]
- 10.Smith JS, Lefkowitz RJ, Rajagopal S. Nat Rev Drug Discov. 2018;17:243–260. doi: 10.1038/nrd.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]