Abstract
Here, we provide a protocol to generate synthetic nanobodies, known as sybodies, against any purified protein or protein complex within a 3-week period. Unlike methods that require animals for antibody generation, sybody selections are carried out entirely in vitro under controlled experimental conditions. This is particularly relevant for the generation of conformation-specific binders against labile membrane proteins or protein complexes and allows selections in the presence of non-covalent ligands. Sybodies are especially suited for cases where binder generation via immune libraries fails due to high sequence conservation, toxicity or insufficient stability of the target protein. The procedure entails a single round of ribosome display using the sybody libraries encoded by mRNA, followed by two rounds of phage display and binder identification by ELISA. The protocol is optimized to avoid undesired reduction in binder diversity and enrichment of non-specific binders to ensure the best possible selection outcome. Using the efficient fragment exchange (FX) cloning method, the sybody sequences are transferred from the phagemid to different expression vectors without the need to amplify them by PCR, which avoids unintentional shuffling of complementary determining regions. Using quantitative PCR (qPCR), the efficiency of each selection round is monitored to provide immediate feedback and guide troubleshooting. Our protocol can be carried out by any trained biochemist or molecular biologist using commercially available reagents and typically gives rise to 10-30 unique sybodies exhibiting binding affinities in the range of 500 pM-500 nM.
Introduction
Binding proteins such as antibodies are widely used affinity reagents for the detection and manipulation of proteins in diverse research fields, including structural biology and bio-imaging1–7. However, as the portfolio of validated antibodies is limited, there is an unmet need for binding proteins tailored to fulfill desired technical and biological requirements concerning affinity and specificity against certain proteins or protein domains. This is particularly relevant for binding proteins targeting specific conformational states, which offer unique opportunities in structural biology and fluorescence microscopy7–9.
In the past decades, a diverse array of natural and synthetic scaffolds has been utilized as binding proteins10. Nanobodies, the variable domain of camelid heavy chain antibodies11, have proven to be a particularly successful scaffold, especially for targeting relatively flexible proteins with limited potential epitopes such as membrane proteins3. Despite their size of only ~110 amino acids, nanobodies exhibit full capacity to bind proteins selectively and with high affinity12. Nanobodies are composed of antiparallel β-strands organized in two sheets and stabilized by an internal disulfide bond, which supports the three complementarity determining regions (CDRs) constituting the binding surface (paratope) (Fig. 1). The robust and simple single-domain architecture, combined with the characteristic variation in the length of CDR3, providing surface-complementarity to diverse antigen surfaces including clefts12–15, forms the basis of the success of nanobodies. Nanobodies are typically generated by the immunization of camelids, followed by isolation of peripheral blood lymphocytes to clone the variable regions of the single-domain antibodies for generating a focused immune library for phage display16,17. While immunization generally leads to a broad variation in the binders and high affinities due to antibody maturation, its undefined and poorly controllable character can impede selections against delicate proteins or protein complexes.
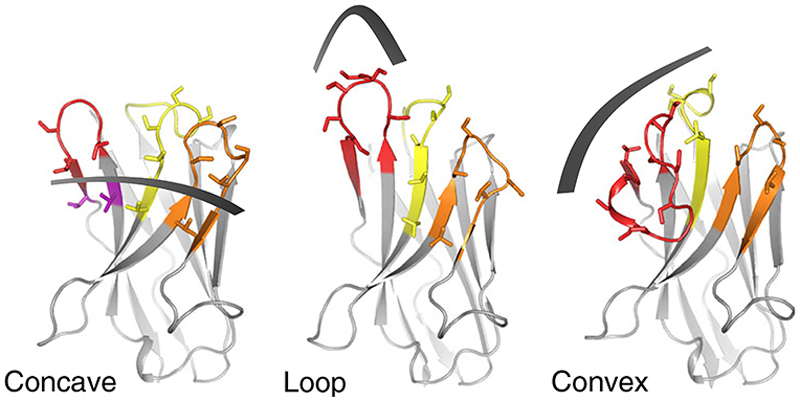
Fig. 1. Sybody libraries.
Shape and randomization scheme of the three sybody libraries: concave, loop and convex. CDR1, CDR2 and CDR3 are colored in yellow, orange and red, respectively. Randomized residues are depicted as sticks. A detailed description of the library design can be found in Zimmermann et al.18.
Synthetic antibody libraries offer an alternative to immune libraries as they allow a higher degree of control. However, until recently their application had been constricted due to limitations regarding library size and selection bias, which need to be appropriately addressed. We recently created three synthetic single-domain antibody (sybody) libraries and established a protocol to select binders against any target protein, including very challenging membrane proteins and rare conformational states stabilized by non-covalent small molecule compounds18. In contrast to immune libraries, where the antibody repertoire is dynamically expanded and optimized in the context of the adaptive immune response, synthetic libraries are rather static. We compensated for this inherent disadvantage by having a larger and more diverse binder repertoire already at the onset of the selections. To account for the exquisite paratope shape variation of natural nanobodies12, we created three different sybody libraries using a structure-based library design approach. The three libraries differ in the length of the main complementarity determining region (CDR3) and thereby exhibit three different interaction surfaces: a concave, a protruding loop and a convex-shaped paratope (Fig. 1). By implementing ribosome display as a first selection step, we start selections with a diversity of 1012 different binder candidates per library, which is two to three orders of magnitude more than approaches relying on transformation, like phage or yeast display19. Finally, our protocol prevents selection bias (i.e., the enrichment of binders unspecific to the target) by combining different display methodologies (ribosome and phage display) and changing the surface chemistry used for target immobilization in every selection round. Using this optimized pipeline, we have overcome several limitations in the use of synthetic antibody libraries, allowing us to successfully select conformation-specific sybodies against highly challenging membrane proteins such as the human solute carriers GlyT1 and ENT1, the bacterial ABC transporters TM287/288 and IrtAB and the intracellular KDEL receptor7,18,20.
Comparison with other approaches
The generation of nanobodies using immunization of camelids is a widely established method with an impressive track record dating back to the mid-1990s17,21. Alternative strategies that do not require camelid immunization rely either on naïve libraries or on synthetic libraries. Naïve libraries are composed of the single-domain antibody repertoire of several (10–20) non-immunized animals22–24. Naïve libraries are typically combined with phage display, limiting their practical library size to 1010 due to the requirement for bacterial transformation. In contrast to synthetic nanobody libraries, naïve libraries represent the full spectrum of the considerable natural variation in CDR3 length and thereby offer more paratope shapes. In addition, as the members of a naïve library have already passed a stringent quality control regarding biophysical characteristics, the overall quality of the binders is expected to be high. These advantages are, however, diminished by the comparably small library size of naïve libraries, which generally result in lower-affinity binders. In addition, the lack of uniformity in the scaffold and the occurrence of additional disulfide bonds required to stabilize long CDR3 loops interfere with standardized approaches and limit their applications. Large and carefully designed synthetic nanobody libraries can compensate for the reduced diversity in paratope shapes of synthetic libraries compared to what is available in a natural immune repertoire. In the case of sybodies, the three libraries account for the natural shape variability of nanobodies. Synthetic nanobodies are inherently more uniform. Therefore, most binders are displayed well and can be tailored to certain applications, such as cytoplasmic expression, allowing for intracellular applications2,25.
Over the last 7 years, our laboratories have generated both natural and synthetic nanobodies (sybodies) against a number of membrane and soluble protein targets, enabling us to directly compare the two methods7,8,18,20,26–28. In our view, both nanobodies and sybodies have their specific advantages and disadvantages. Nanobodies are an excellent choice if the target protein is stable and therefore has an increased likelihood to remain properly folded for prolonged periods (hours to days) at elevated temperatures (37–42 °C) following mixing with (mineral oil–based) adjuvant and injection into the animal. This is likely the case for most soluble proteins, and several prokaryotic α-helical or β-barrel membrane proteins. However, if the target protein or protein complex is labile and/or if one aims at generating conformation-specific binders in the presence of non-covalent ligands, nanobody selection by immunization can be challenging14. For these cases, in vitro generation of sybodies offers unique opportunities. Namely, the entire binder generation process can be carried out under controlled conditions (e.g., at 4 °C and in the presence of a conformation-stabilizing ligand added at saturating concentration18,20). Another advantage of the in vitro selection approach is the possibility of selecting binders against highly conserved proteins that are often non-immunogenic due to self-tolerance. In addition, no ethical considerations restrain the use of toxic compounds, such as the detergent digitonin, during in vitro binder generation. While nanobodies have been successfully generated against toxins29, non-immunogenic targets are a major obstacle to generating nanobodies.
Although both natural and synthetic nanobodies selected using the two approaches have been instrumental in studying structural and functional aspects of our target proteins7,20,26,28, we noted differences in the biophysical properties of the binders. While both procedures generally lead to binders with excellent (<10 nM) binding affinities, the fraction of high-affinity binders is larger for nanobodies obtained from immunizations, due to the affinity maturation by somatic hypermutation. The protein quality control as an inherent part of the antibody maturation process in vivo also results in improved biophysical properties of nanobodies. In a typical selection against integral membrane proteins, ~20–60% of the sybody hits are discontinued due to poor expression, oligomerization or strong interactions with size-exclusion columns. For nanobodies, binders with such adverse properties are already counterselected in vivo, and consequently <10% of the nanobody binder hits are lost due to these problems.
When comparing nanobodies and sybodies, three additional aspects related to the immunization procedure need to be taken into account as well: access to animal facilities, time frame, and sample consumption. Sybody generation does not require access to animal facilities, which allows the immediate ad hoc onset of the selection procedure in one’s own laboratory. While the affinity maturation of nanobodies requires the repetitive immunization during a period of 2 months before the onset of the binder selection17, our sybody selection procedure employs a 1-d ribosome display step to generate a focused library. Of note, the subsequent steps in the nanobody and sybody selection procedure (i.e., two rounds of phage display and ELISA screening) take equally long. Finally, most of the target protein consumption for the generation of nanobodies, amounting to 0.5–1.0 mg, is required for the consecutive immunizations and final boost17,30,31. The subsequent selection procedure requires only ~100 μg, which equals the complete protein consumption required for generating and selecting sybodies.
In addition to our sybodies, there have been three reports on the generation of alternative synthetic nanobody libraries25,32,33. Among them, the McMahon32 and Moutel25 libraries are particularly well described and have been widely applied. Both the McMahon and Moutel libraries were constructed according to a consensus design approach based on a large number of nanobody structures and sequences. Consequently, these libraries feature a single consensus scaffold accommodating three or four length variations of CDR3, respectively. Of special note, the Moutel library was tailored for intracellular expression of their synthetic nanobodies25. Our sybody libraries were constructed according to a structure-based design approach with each CDR3 length variant inspired by a single nanobody structure. As a consequence, our convex sybody library with a long CDR3 length of 16 amino acids possesses an extended hydrophobic core to restrict the flexibility of the CDR3 loop (Fig. 1), as is the case in natural nanobodies34. In contrast, the Moutel and McMahon libraries contain synthetic nanobodies with long CDR3 loops up to 18 amino acids whose flexibility is not restricted, which may decrease binding affinities and protein stability35,36.
Additional differences are found in the methodological approach for selecting high-affinity binders from the three libraries. The Moutel library is available in phage display format and is estimated to have an upper library size of ~1010 members25. The McMahon library is available in a yeast display format and exhibits a reported library size of 108 members32. Although both phage and yeast displays are robust selection procedures compared to ribosome display, the need for transformation to generate these libraries restricts their experimental diversities. In contrast, sybody selections commence with ribosome display, which allows a much larger number (1012) of unique binder molecules to be presented in the initial selection round. Therefore, the average affinity of the final binder hits is generally higher. Regardless of the library size and selection strategy, the affinity of any synthetic nanobody may be readily improved by incorporating an additional affinity maturation step after the selection procedure37. A particular strength of the McMahon library is binder selection by FACS of yeast cells32. However, the standard protocol described in their paper operates by magnetic-based cell sorting and does not foresee FACS, which needs specialized equipment and training.
Overview of the procedure
In this protocol, we outline a general procedure for obtaining sybodies against soluble proteins, membrane proteins and protein complexes. The major steps are summarized in Fig. 2. Our protocol relies on the use of ribosome display and phage display to link genotype (i.e., the open reading frame coding for the binder) and phenotype (i.e., the specificity and affinity of the binder). We employ three selection rounds, one round of ribosome display (Steps 1–22) and two rounds of phage display (Steps 23–113), that lead to an enrichment of target-specific binders. Subsequently, 95 random clones per sybody library are analyzed by ELISA and typically result in 10–30 unique binder hits (Steps 114–152). Unique sybodies meeting the desired specificity are finally purified for further characterization (Steps 153–196) (Fig. 2).
Fig. 2. Sybody selection flowchart.
Sybody selections against target proteins start with one round of ribosome display, followed by two rounds of phage display. Binder hits are identified by ELISA and finally purified. The entire procedure is completed within a period of 3 weeks. PIII, filamentous phage protein III; α-Myc, antibody recognizing Myc-tag; Strep-HRP, streptavidin-horseradish peroxidase conjugate.
Arguably the most important parameter affecting the outcome of our selection procedure is the quality of the target protein, which should be folded in a biologically relevant conformation and stoichiometrically biotinylated. In addition, we emphasize the relevance of carefully planning the selection strategy beforehand by considering the specifications and properties the binder should, or should not, possess in the end. For instance, selections on target proteins holding large fusion proteins or affinity tags may lead to undesired binders targeting these regions. Likewise, stringent initial selections under conditions where only one protein conformation is present will prevent using the resulting enriched pool for selecting binders against other conformations.
Experimental design
Target protein quality and biotinylation
It is crucial that the target protein is pure and well folded, exhibits enzymatic or binding activity (if applicable) and runs as a defined monomer or oligomer on a size-exclusion chromatography (SEC) column. We would recommend assessing the quality of the protein using standard biophysical methods; circular dichroism, multi-angle light scattering and thermal stability are routinely used in our laboratories to assess protein quality. Ideally, the target protein is biophysically characterized in detail before binder selections (e.g., unfolding temperature, optimal buffers, detergents, binding of substrates and inhibitors) to ensure that the target maintains the desired biologically relevant folding state during the selection procedure for ≥1 h. Due to the frequent need for the target protein, it is convenient to freeze aliquots, if freezing is tolerated (Box 1).
Box 1. Requirements for target proteins used for sybody selections.
The success of sybody selections critically depends on the biochemical quality of the target protein. The outcome of the selection is most favorable if the target protein is presented in a well-folded state, as can be demonstrated by its catalytic activity, ability to bind substrates or monodispersity on SEC. Furthermore, the purity of the protein should exceed 95% as assessed by SDS-PAGE.
To allow selective immobilization of the target protein as required for our sybody selection procedure, the target protein needs to be biotinylated38. Ideally, only one biotin group is introduced per protein or protein complex as this allows more flexibility and thus presentation of different protein surfaces. Biotinylation can be achieved chemically or enzymatically. The latter requires the modification of the target protein with an Avi-tag of 15 amino acids. FX cloning vectors for the introduction of N-or C-terminal Avi-tags are available from Addgene (#47069, #47071-5). Subsequent biotinylation of the Avi-tag in vivo or in vitro is straightforward using the E. coli enzyme BirA57. Chemical biotinylation is most conveniently done by targeting the primary amine of surface-exposed lysines using, for example, sulfo-NHS-LC-biotin, but care should be taken to ensure that >90% of the target proteins have at least one biotin attached. Over-biotinylation should be avoided, as this will limit the representation of different protein surfaces and may even directly mask epitopes.
Besides the target protein, it is important to use representative control proteins of high quality. As control protein for a membrane protein target, we recommend using another membrane protein, such as the ABC exporter TM287/288. In case the target is a soluble protein, we recommend MBP as control protein. Both proteins are well expressed and robust and are available as Avi-tagged constructs from Addgene (#132700 and #132701)18. We generally use SEC as a final polishing step after biotinylation. The peak corresponding to the monodisperse target or control protein is then pooled (and may need to be concentrated if the concentration is <100 nM), supplemented with 10% (vol/vol) glycerol, divided into aliquots, snap-frozen in liquid nitrogen and stored at −80 °C. To test whether freezing is tolerated, compare an aliquot before and after a freeze-thawing cycle by SEC. If the target protein elutes again as a monodisperse peak at the expected retention volume, freezing can be regarded as tolerated. If freezing is not tolerated well, freshly purified sample needs to be used.
To perform selections and ELISA with the three sybody libraries, the following protein amounts of biotinylated target proteins are needed: ribosome display and two rounds of phage display: 16 ml at a concentration of 50 nM; ELISA: 30 ml at a concentration of 50 nM; total: ~50 ml at a concentration of 50 nM, which corresponds to 1 ml at a concentration of 2.5 µM. In case of a 50-kDa target protein, as little as 125 µg is required for sybody selections and ELISA.
To perform off-rate selections as part of the second phage display round (Step 98), 700 µl of non-biotinylated target protein at a concentration of 5 µM is needed. In case it is difficult to obtain these protein amounts, the concentration may be reduced to 2.5 µM.
The removal of non-binders during the washing steps requires the selective immobilization of the target protein. Our standard immobilization is mediated by the robust and widely used biotin-streptavidin interaction (Box 1). Consequently, the target protein needs to be biotinylated. We prefer the enzymatic biotinylation of either an N-or ideally C-terminal Avi-tag, as this allows obtaining near equimolar biotin:target ratios38. Chemical biotinylation may be used as well but is less optimal due to the difficulty in obtaining an equimolar ratio. Both under- and over-biotinylation will decrease the overall efficiency of the selection procedure (e.g., by blocking potential epitopes).
In our selection protocol, we use solution panning; that is, the displayed sybody library is first incubated with the biotinylated target in solution before rapid capture of complexes via immobilized streptavidin or neutravidin. Solution panning is preferred over surface panning (i.e., target protein immobilization before incubation with the binder library) as it allows the presentation of additional surfaces of the target protein, limits the time the target protein is in contact with plastic/bead surfaces and generally favors high-affinity interactions and decreases the role of avidity. A direct consequence of solution panning is that a high degree of target biotinylation (>90%) is required, because displayed binders are immobilized only via the biotinylated fraction of target protein. This is particularly relevant for the first ribosome display round, where every binder is present in essentially one copy and where insufficient biotinylation will result in a so-called diversity bottleneck, namely, the undesired reduction in binder diversity.
Ribosome display selection
We distribute our three sybody libraries in the form of mRNA containing all the relevant features required for ribosome display. The mRNA is devoid of a stop codon and contains a stem loop in the 3′-region to induce ribosome stalling. Although ribosome display is generally considered to be difficult to implement39, this mostly concerns establishing the in vitro translation system based on Escherichia coli cell extracts. Instead, we use the commercial in vitro translation kit PUREfrex 2.1 (GeneFrontier) following extensive validation of its predecessor, the PUREfrexSS kit. This kit allowed efficient display of disulfide bond–containing nanobodies and sybodies, and we obtained high recovery efficiency (>80%) of a GFP nanobody18. More recent selections using the new PUREfrex 2.1 indicate a slightly improved performance. The PUREfrex 2.1 reaction mix consists of purified proteins, ribosomes, amino acids and NTPs only and does not require any fine-tuning40. Although the kit, in principle, can use DNA libraries as input, as it contains all components required for in vitro transcription, we add the sybody library in the form of mRNA. Thereby, we have precise control over the amount of mRNA added, namely, 1.6 × 1012 mRNA molecules, which translates to the presence of ~1012 unique binders at the onset of the selection (if we assume a dropout rate of 40% for sybody library members bearing a frameshift mutation or exhibiting poor expression). When using ribosome display, it is important to avoid RNase contaminations. Therefore, we have a dedicated bench to perform ribosome display, which is regularly cleaned with RNase AWAY, a decontamination reagent inactivating RNases. Furthermore, we use filter tips and wear clean gloves during the procedure. Water should be RNAse free, which can be achieved through treatment with diethyl pyrocarbonate.
The expected output of the ribosome display selection round is ~1–5 × 106 purified cDNA molecules. We highly recommend implementing qPCR analyses to experimentally assess the cDNA output quantitatively and qualitatively (Box 2). For the latter, we run two qPCR reactions on the cDNA, one using primers close to the 5′-end and one using primers close to the 3′-end. If the ratio of the two qPCR reactions deviates strongly from 1:1, this is an indicator of mRNA degradation, which starts from the 5′-end. In our experience, capture of the genetic information from mRNA by reverse transcription and PCR amplification is the most delicate part of the protocol. It is therefore important to strictly adhere to the detailed procedures for reverse transcription, cDNA purification and cDNA amplification. Note that for the PCR amplification of the cDNA, a non-proofreading polymerase, such as Taq, should be used. Proofreading polymerases (such as Phusion, Q5 or Pfu) digest single-stranded cDNA due to their 3′–5′-exonuclease activities, which leads to variable outcomes in PCR yields and presents a diversity bottleneck. Despite the use of a non-proofreading polymerase, we thus far have not encountered the problem of excessive mutations in the sybody framework.
Box 2. qPCR analysis of ribosome and phage display outcomes.
We use qPCR to assess the quality of the cDNA output of the ribosome display selection and to monitor the enrichment during the phage display selection steps. The primers (Fig. 3 and Table 2) depend on the selection step, but the composition and cycling conditions stay the same.
Set up the following 10-µl qPCR reactions. We use a 7500 Fast Real-Time PCR System of Applied Biosystems with SYBR select Master Mix. Run a duplicate for every reaction.
| Reagent | Amount (μl) | |
|---|---|---|
| SYBR select Master Mix 2× | 5.0 | |
| DMSO | 0.5 | |
| Primers, each 10 μM | 0.3 (of each) | |
| H2O | 1.9 | |
| Diluted cDNA/phage elution (from Steps 20, 81 and 107) | 2 | |
| Total | 10 | |
| Use the following primer combinations (see also Fig. 3): | ||
| Region to test (library) | Forward qPCR primer | Reverse qPCR primer |
| 5′ region (concave/loop) | qPCR_RD_5′_for | qPCR_RD_S_and_M_5′_rev |
| 5′ region (convex) | qPCR_RD_5′_for | qPCR_ RD_L_5′_rev |
| 3′ region (generic) | qPCR_ RD_tolA_3′_for | qPCR_ RD_tolA_3′_rev |
| Phagemid (generic) | qPCR_PD_pDX_for | qPCR_PD_pDX_rev |
| qPCR program: | ||
| Cycle number | PCR conditions | PCR steps |
| 1 | 95 °C, 2 min | Initial denaturation |
| 2–41 | 95 °C, 10 s; 63 °C, 30 s |
Denaturation anneal/elongate/measure |
| 42 | Perform according to the qPCR machine manual. |
Melt curve |
The Real-Time PCR system needs to be calibrated before first use. A dilution series of a standard construct should be used to determine the PCR efficiency and to quantify the qPCR results. This needs to be performed only once for a specific Real-Time PCR system. Importantly, the threshold cycle must always be the same to be able to compare different runs with the calibration. To generate a standard curve for the ribosome display primers, use pRDV_FX5 vector harboring non-randomized sybodies of the loop and the convex library, respectively (Addgene #132695 and #132696). To obtain a standard curve for the phagemid primers, use pDX_init harboring the non-randomized sybody of the loop library (Addgene #132697).
To analyze the cDNA output of Steps 20, 81 and 107, use the primer pairs specified above in the table. We typically isolate a total of 1–5× 106 cDNA molecules as the output of ribosome display. Usually the ratio of the 5′-region and 3′-region is close to 1. Degradation of mRNA proceeds from 5′ to 3′. We consider a 5′-reaction/3′-reaction ratio of <0.2 as a clear sign of mRNA degradation. In the first phage display round, enrichment factors <2 are fairly normal and do not indicate a failure of the selection process. In the second phage display round, enrichment factors >2 are needed to proceed with ELISA. Enrichment factors measured during selections against three different targets are provided in Table 1.
Generation of phage library
Due to the relatively small number of output mRNAs from ribosome display (~106 molecules), a moderate phage library of ~107 already suffices to maintain full diversity in the binder repertoire. This size of the phage library is similar to the size of a nanobody immune library and can be achieved quite easily17. Our entire pipeline of vectors is compatible with FX cloning (Fig. 3), a facile, inexpensive and robust technology based on the type IIS restriction enzyme SapI or its isoschizomer BspQI41,42. FX cloning allows highly efficient subcloning by restriction and ligation, which is particularly favorable when using synthetic libraries. Subcloning prevents CDR shuffling that cannot be avoided during PCR amplification of the library due to the very high sequence similarity in the non-randomized sybody framework regions. CDR shuffling decreases the selection efficiency due to the formation of new CDR combinations27. Our phagemid pDX_init (Addgene #110101) is an FX-cloning adapted version of the widely used pMESy417. The original amber stop codon in pMESy4 is replaced by a glutamine to allow use of the phage display strain E. coli SS320 that can be efficiently transformed43.
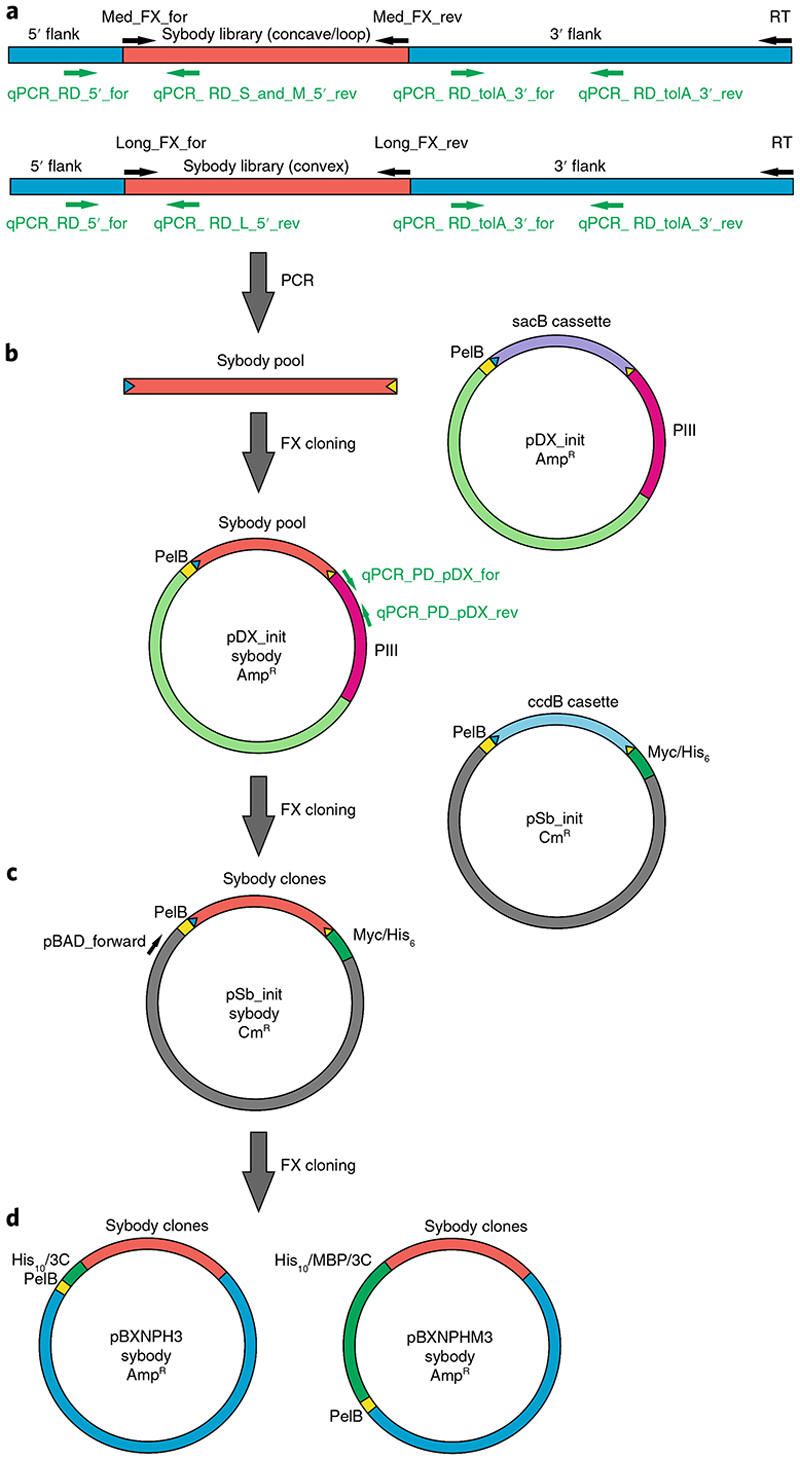
Fig. 3. Overview of genetic constructs and primers.
a, The sybody libraries can be obtained from the authors in the form of mRNA ready for ribosome display. The concave and loop sybodies share the same framework and thus can be amplified with the same set of primers. The convex sybodies have a different framework. Primers used to quantify cDNA after ribosome display by qPCR (green) and to amplify the sybody pools by PCR (black) are indicated. b, Sybody pools amplified in a are cloned into the phagemid vector pDX_init using FX cloning. Note that the BspQI restriction sites (blue and yellow arrowheads) are encoded on the pDX_init backbone, allowing excision of the sybody open reading frames again. Primers used to quantify phages via pDX_init by qPCR are indicated in green. c, For single-clone expression, the output of the second phage display round is sub-cloned into pSb_init using FX cloning. Thereby, a Myc-tag and a His6-tag are attached at the C terminus of the sybodies. Note that the BspQI restriction sites are encoded on the pSb_init backbone. The sequencing primer pBAD_forward is indicated. d, For the production of tag-free proteins, sybodies are sub-cloned into pBXNPH3 or pBXNPHM3. In this cloning step, the BspQI restriction sites are lost.
Phage display selections
Our protocol foresees two rounds of phage display, which are sufficient to obtain highly enriched binder pools. Phage display is robust and widely used44. We realized that the infection rate of M13 phages prepared by the standard precipitation protocol is rather poor (2–5%). It is therefore important that the first phage display round is performed using large panning volumes. This ensures the isolation of sufficient phages to avoid diversity bottlenecks. Hence, for the first phage display selection round, we immobilize the pre-incubated phage-target complexes on 47 wells of a 96-well plate. In the second phage display round, we use magnetic beads and routinely perform an off-rate selection step by washing with an excess of non-biotinylated target45. Thereby, we enrich sybodies with slower off-rates and thus higher affinities. In our protocol, we use qPCR to rapidly quantify the number of phages that are eluted from the target. Unspecific off-target binding is assessed using an unrelated control protein. In case sybodies are raised against a membrane protein, it is important that this control protein is also a membrane protein, to account for unspecific binding to transmembrane helices and/or detergent micelles46. Enrichment is calculated based on qPCR quantification of phages isolated from the target divided by phages isolated from the control panning reaction. In a typical selection, we observe enrichment factors in the range of 2-to 1,000-fold after the second phage display selection round. These numbers vary between different targets. For soluble and highly stable membrane protein targets, enrichment factors >100 are typically observed. For challenging targets and especially for small membrane proteins, enrichment factors in the range of 2–20 can be considered as a good outcome. Furthermore, enrichment factors are typically highest for our loop sybody library (medium-sized CDR3), followed by the concave library (short CDR3) and the convex library (long CDR3).
Binder identification by ELISA
If the final enrichment factors are >5, it is certainly worth proceeding with ELISAs. If the enrichment factor is between 2 and 5, we usually still perform ELISA and often isolate 5–10 binder hits per sybody library (out of 95 tested clones). If the enrichment factor is <2 (i.e., very poor or no enrichment), we recommend either performing a third phage display round or repeating the entire selection with a biochemically improved target protein (e.g., stabilized through mutations) or a different selection condition (e.g., an alternative detergent in the case of a membrane protein).
Using FX cloning, the enriched sybody pool can be subcloned from the phagemid pDX_init into the expression vector pSb_init (Addgene #110100)18. Thereby, we attach a Myc-tag and a His6-tag sequence to the C terminus of the sybody. In our standard protocol, we then perform a small-scale expression in deep-well plates of 95 randomly picked sybody clones, plus one positive control sybody, for each of the three sybody libraries (concave, loop and convex). Using periplasmic extracts of these cultures, we run two 96-well ELISA plates per sybody library (i.e., six ELISA plates in total per target or selection condition). ELISA is performed using the target protein and a control protein side by side. The control must be a membrane protein when sybodies are raised against a membrane protein.
Our ELISA setup operates inverse to the standard nanobody ELISA procedure17 (Fig. 2). It starts with binding of anti-Myc antibodies to an ELISA plate coated with Protein A. Then, the Myc-tagged sybodies are added, followed by the biotinylated target protein that is subsequently detected using a streptavidin–horseradish peroxidase conjugate18,46,47. This setup is less prone to identifying low-affinity and unspecific ‘sticky’ binders. In the case of membrane protein targets, this setup reduces foam formation and costs as (expensive) detergents need to be included only in the last three washing steps. Depending on the enrichment, we typically identify 5–85 target-specific hits per ELISA plate. A specific hit shows binding against the target and no background binding to the control protein. In our experience, the strength of ELISA signals only poorly correlate with binder affinities determined by surface plasmon resonance (SPR). Thus, ELISA signals as small as 1.5-fold above background can indicate a high-affinity binder. It is therefore recommended to pick ELISA hits of different signal intensities.
Sybody sequencing
ELISA hits are subsequently analyzed by Sanger sequencing of purified plasmids. In a typical selection, we sequence between 20 and 80 sybodies in total, ideally originating from all three sybody libraries. The selected sybody hits are typically very diverse. Due to the randomization scheme involving all three CDRs, sybodies cannot be grouped according to the CDR3 alone to find binder families, as is the case for nanobodies17. In particular for the concave sybodies with their short CDR3, we suspect that CDR1 or CDR2 are often involved in providing dominant target interactions, as was the case for some nanobodies selected from immune and semi-synthetic libraries48,49. Therefore, we usually proceed with all identified binders that differ by more than three amino acids.
Sybody production
We usually purify all unique, sequenced sybodies to analyze their biophysical properties. Expression is performed using the pSb_init vector, and the sybodies are purified using high-throughput immobilized metal affinity chromatography (IMAC) from the periplasm of E. coli MC1061 grown in 50 ml of medium. Yields typically range from 0.2 to 1 mg. Purified sybodies are further analyzed by SEC. We generally observe that a significant fraction of sybodies (5–30%, depending on the target) interacts with widely used SEC resins such as Superdex 200. This problem manifests as severely delayed elution peaks of up to four column volumes. Importantly, this delayed elution can be improved using a Sepax SRT-10C SEC100 column and is not correlated to the overall quality of the sybody binder. We generally discontinue binders that show low expression levels or oligomerization. Complete aggregation of sybodies is rarely observed. In a typical binder selection against membrane proteins, up to 30% of the sequenced binders are discontinued due to low expression levels and up to 20% do not show monomeric behavior on SEC. For very difficult targets (e.g., the KDEL receptor), a significant fraction (>60%) of unique, sequenced sybodies may be discontinued due to poor biophysical behavior (Table 1). In contrast, most (>80%) of the selected sybodies are biophysically well behaved for more stable membrane protein targets, such as TM287/288 (Table 1).
Table 1. Sybody selection outcome for three example membrane protein targets.
| Target | Enrichment phage display #1 |
Enrichment phage display #2 |
Number of ELISA hits (out of total analyzed) |
Number of unique binders (out of total sequenced) |
Number of purified sybodies (out of total analyzed) |
|---|---|---|---|---|---|
| TM287/288 | |||||
| Concave | 17 | 170 | 28/95 | 6/6 | 4/6 |
| Loop | 60 | 220 | 84/95 | 23/24 | 22/23 |
| Convex | 3 | 25 | 74/95 | 11/18 | 5/11 |
| UraA | |||||
| Concave | 1.2 | 4.3/6.7a | 25/190b | 5/6 | 3/4 |
| Loop | 0.9 | 11.8/19.6 | 34/190 | 18/27 | 13/15 |
| Convex | 0.9 | 10.2/5.6 | 37/190 | 22/25 | 15/21 |
| KDEL receptor | |||||
| Concave | 1.6 | 7.8 | 27/95 | 21/27 | 5/21 |
| Loop | 1.2 | 4.9 | 8/95 | 3/8 | 3/3 |
| Convex | 0.7 | 3.6 | 33/95 | 7/33 | 4/7 |
The two values represent enrichments obtained in the absence/presence of the ligand uracil.
ELISA hits originating from selections performed in the absence/presence of uracil are summed.
Downstream analyses of sybodies
We routinely use SPR to determine the affinity of sybodies18. Although the absolute affinity values determined by SPR often depend on sample quality and data fitting, SPR is an excellent technique to rank-order a set of sybodies selected against the same target and measured under identical experimental conditions. Importantly, SPR provides valuable information on binder off-rates. Furthermore, SPR allows researchers to perform binding experiments under different conditions (i.e., in the presence or absence of ligands), which can be a very powerful tool to identify conformation-specific sybodies20. Next to SPR, analytical SEC can be used to identify sybodies with affinities better than 500 nM based on their ability to shift the target elution peak and to co-elute. In case only a minor shift of the target peak is expected, we monitor the decrease of the sybody peak as a consequence of co-migration with the target protein (sybody peak observed with versus without the target protein present). We highly recommend analyzing target-sybody complexes isolated by SEC using SDS PAGE to exclude low-affinity binders that can hardly be detected on the gel. Nanobodies as well as sybodies typically bind to three-dimensional epitopes on the folded target protein. To find (rare) binders that recognize linear epitopes (e.g., for western blotting or immunostaining), we refer to published protocols that detail how such analyses can be carried out50.
Production of tag-free sybodies
An important application of sybodies is their use as crystallization chaperones7,20. For the purpose of crystallization, the purified sybody is ideally devoid of flexible tags. To this end, we previously constructed the expression vectors pBXNPH3 (Addgene #110098) and pBXNPHM3 (Addgene #110099)18. Both vectors contain a PelB leader sequence for periplasmic expression. In the case of pBXNPH3, a His10-tag followed by an HRV 3C protease cleavage site is added to the N terminus of the sybody. In the case of pBXNPHM3, an additional maltose binding protein (MBP) fusion is present between the His-tag and the protease site. We generally prefer the pBXNPHM3 construct, because the sybody yields are less variable using this vector. In either case, the sybodies are purified via IMAC, followed by tag cleavage by 3C protease and reverse-IMAC to obtain the compact, tag-free sybody. Expression of tag-free sybodies or nanobodies with these expression vectors generally results in a 10-fold lower expression level compared to constructs containing C-terminal purification tags, as is the case for sybody or nanobody expression using the pSb_init vector (see above). Of note, in the classical nanobody expression vector pMESy4, a non-cleavable His6-tag remains fused to the C terminus17.
Limitations
The outcome of the sybody selections strongly depends on the type and quality of the target protein. Like nanobodies, sybodies recognize three-dimensional epitopes. Thus, the target protein needs to be well folded. At a minimum, the target protein should give rise to a monodisperse peak on SEC at an elution volume corresponding to its expected molecular weight. Elution close to or within the void volume of an SEC column is a strong indication of misfolding and the formation of soluble aggregates. Ideally, the target protein folding state is assessed by an enzymatic or ligand binding assay7,20. Furthermore, intimate knowledge of how the target protein can be stably maintained during the time course of a selection step (~1 h) is beneficial, as our selection procedure is established to allow a high degree of control over the selection conditions.
Although a high degree of adjustment in the sybody selection conditions is tolerated, certain aspects of the system need to be taken into account. The ribosome display buffer contains 50 mM magnesium acetate (MgAc2). Magnesium ions are mandatory to stabilize the ternary mRNA-ribosome-sybody complex that ensures the phenotype-genotype linkage. Therefore, ribosome display in the presence of chelating agents such as EDTA cannot be performed. A further challenge may be the potential destabilization of ribosomal complexes by certain detergents, ligands or buffer conditions. Thus far, we have performed successful selections using the following detergents added at a concentration of 0.1% (wt/vol): Tween-20, n-dodecyl-β-D-maltopyranoside (β-DDM), n-decyl-β-D-maltopyranoside (β-DM), glycol-diosgenin and lauryl maltose neopentyl glycol (LMNG), also in combination with cholesteryl hemisuccinate (CHS). Furthermore, we successfully performed ribosome display at a pH as low as 6.0 (standard pH is 7.4) and in the presence of the reducing agent DTT added at a final concentration of 1 mM. To experimentally validate the stability of ribosomal complexes in alternative conditions, we recommend conducting test experiments in which a single high-affinity sybody or nanobody is displayed on ribosomes, followed by determination of its pull-down efficiency by qPCR. More details on such validation experiments can be found in our original sybody paper18.
The degree of difficulty of selecting sybodies against a protein or a protein complex is inversely related to the fraction of hydrophilic surface, e.g., as observed for compact membrane proteins, and with the fraction of highly dynamic or flexible hydrophilic surfaces, such as found in intrinsically disordered proteins. While these are challenging targets that are still expected to allow sybody selection, the selection of sybodies against linear peptides and/or small molecules is unlikely to result in meaningful binder outcomes. For such cases, we advise generating conventional antibodies via immunization of mice, rats or rabbits.
Materials
Biological materials
M13KO7 Helper Phage (NEB, cat. no. N0315S) ▴CRITICAL We strongly advise determining the helper phage concentration based on its ability to infect E. coli SS320 using either colony forming units or plaque forming units, instead of spectroscopic determinations.
E. coli MC1061 (Lucigen, cat. no. 60514-1)51
E. coli SS320 (Lucigen, cat. no. 60512-1)43
Biotinylated control and target protein (Box 1)
Non-biotinylated target protein for off-rate selection (Box 1)
Reagents
PUREfrex 2.1 (Genefrontier, cat. no. PF213-0.25-EX)
DS supplement (Genefrontier, cat. no. PF005-0.5-EX)
Nuclease-free water (Invitrogen, cat. no. 10977035)
RnaseIn (Promega, cat. no. N2611)
dNTPs (Thermo Scientific, cat. no. R0181)
RNaseAway (Merck, cat. no. 83931)
Trizma base (Merck, cat. no. 93350)
Sodium chloride (Merck, cat. no. 71380)
Magnesium acetate tetrahydrate (Merck, cat. no. M2545)
Magnesium chloride (Merck, cat. no. M8266)
Glacial acetic acid (Carl Roth, cat. no. 3738.4) ! CAUTION Acetic acid is corrosive. Avoid inhalation and exposure to skin and eyes.
Hydrochloric acid (Carl Roth, cat. no. X942.1) ! CAUTION Hydrochloric acid is corrosive. Avoid inhalation and exposure to skin and eyes.
Sodium hydroxide (Carl Roth, cat. no. P031.1) ! CAUTION sodium hydroxide is corrosive. Avoid inhalation and exposure to skin and eyes.
Yeast RNA (Merck, cat. no. R6750)
Rneasy micro (Qiagen, cat. no. 74004) ▴CRITICAL We strongly advise using this product, because it allows elution of the mRNA in small volumes.
AffinityScript Multiple Temperature Reverse Transcriptase (Agilent, cat. no. 600107) ▴CRITICAL We strongly advise using this product, because the reverse transcription conditions need to be optimized for different reverse transcription kits.
NucleoSpin Gel and PCR Clean-up (Macherey-Nagel, cat. no. 740609) ▴ CRITICAL We strongly advise the use of this product, because it allows purification of single-stranded DNA present after cDNA generation.
Heparin sodium salt (Merck, cat. no. H4784)
SYBR select Master Mix (Applied Biosystems, cat. no. 4472908)
GoTaq G2 DNA polymerase (Promega, cat. no. M7841)
Agarose analytical grade (Promega, cat. no. V3125)
HEPES (Carl Roth, cat. no. HN77.3)
Ultrapure glycerol puriss. p.a. (Merck, cat. no. 49770)
BspQI (10 U/µl) (NEB, cat. no. R0712)
T4 DNA ligase (5 U/µl) (Thermo Scientific, cat. no. EL0011)
Bovine serum albumin (Merck, cat. no. A3912)
Sodium hypochlorite solution (Merck, cat. no. 71696)
PEG6000 (Merck, cat. no. 81260)
Neutravidin (Thermo Scientific, cat. no. 31000)
Trypsin (Merck, cat. no. T1426)
4-(2-Aminoethyl)benzenesulfonyl fluoride (AEBSF) (Merck, cat. no. A8456)
QIAprep Spin Miniprep Kit (Qiagen, cat. no. 27106)
L-(+)-Arabinose (Merck, cat. no. A3256)
Sucrose (Merck, cat. no. 84100)
EDTA (Merck, cat. no. EDS)
Lysozyme from chicken egg white (Merck, cat. no. L4919)
Protein A from Staphylococcus aureus (Merck, cat. no. P3838)
Streptavidin-peroxidase polymer, ultrasensitive (Merck, cat. no. S2438)
3,3′,5,5′-Tetramethylbenzidine (TMB) (Merck, cat. no. T2885)
Dimethyl sulfoxide (DMSO) (Merck, cat. no. D8418)
30% (wt/vol) Hydrogen peroxide solution (Merck, cat. no. 216763)
Imidazole (Carl Roth, cat. no. X998.3)
Na2HPO4 (Applichem, cat. no. A3905)
KH2PO4 (Applichem, cat. no. A1043)
Citric acid (Merck, cat. no. 251275)
K2HPO4 (Applichem, cat. no. A1042)
KCl (Merck, cat. no. P3911)
Glucose (Merck, cat. no. G8270)
Tryptone (BD, cat. no. 211699)
Yeast extract (BD, cat. no. 212720)
Ethanol (Merck, cat. no. 51976)
Dynabeads MyOne Streptavidin T1 (Invitrogen, cat. no. 65601)
Dynabeads MyOne Streptavidin C1 (Invitrogen, cat. no. 65001)
His MultiTrap HP (GE Healthcare, cat. no. 28-4009-89)
Ni-NTA Superflow (Qiagen, cat. no. 30430)
Adenosine 5′-triphosphate disodium salt hydrate (Merck, cat. no. A3377)
Primers (Table 2)
Table 2. Primers for PCR, qPCR and sequencing.
| Primer | Sequence (5′to 3′ direction) |
|---|---|
| Med_FX_for* | ATA TGC TCT TCT AGT CAG GTT CAG CTG GTT GAG AGC G |
| Med_FX_rev* | TAT AGC TCT TCA TGC GCT CAC AGT CAC TTG GGT ACC |
| Long_FX_for* | ATA TGC TCT TCT AGT CAA GTC CAG CTG GTG GAA TCG |
| Long_FX_rev* | TAT AGC TCT TCA TGC AGA AAC GGT AAC TTG GGT GCC C |
| RT_Primer | CTT CAG TTG CCG CTT TCT TTC TTG |
| qPCR_RD_5′_for | GGG AGA CCA CAA CGG TTT CCC |
| qPCR_ RD_S_and_M_5′_rev | CAC CGG GAA ACC GCT TGC GGC |
| qPCR_ RD_L_5′_rev | GCC GCT AGC CGC ACA GCT C |
| qPCR_ RD_tolA_3′_for | GCC GAA TTC GGA TCT GGT GGC |
| qPCR_ RD_tolA_3′_rev | CTG CTT CTT CCG CAG CTT TAG C |
| qPCR_PD_pDX_for | GAC GTT CCG GAC TAC GGT TCC |
| qPCR_PD_pDX_rev | CAC AGA CAG CCC TCA TAG TTA GC |
| pBAD_forward | ATG CCA TAG CAT TTT TAT CC |
These primers are ordered in PAGE-purified form.
Antibiotics
Ampicillin (Carl Roth, cat. no. HP62.1)
Kanamycin (Merck, cat. no. K4000)
Tetracycline hydrochloride (Merck, cat. no. T7660)
Chloramphenicol (Carl Roth, cat. no. 3886.1)
Antibodies
Monoclonal anti-c-Myc antibody produced in mouse (Sigma-Aldrich, cat. no. M4439; RRID: AB_439694)
Sybody libraries
The three sybody libraries in the form of mRNA can be obtained from the University of Zurich for academic research through a material transfer agreement ▴ CRITICAL We send the libraries as 10-µl aliquots, which need to be stored at at −80 °C. Repetitive freeze-thawing of the RNA library until the aliquot is empty is well tolerated.
Plasmids
▴CRITICAL Please note that these 14 plasmids are available as a kit called ‘Sybody Generation Toolbox’ (Addgene Kit 1000000160).
pDX_init (Addgene #110101)
pSb_init (Addgene #110100)
pBXNPHM3 (Addgene #110099)
pBXNPH3 (Addgene #110098)
pBXNPHM3 containing Sb_concave (Addgene #110102)
pBXNPHM3 containing Sb_loop (Addgene #110103)
pBXNPHM3 containing Sb_convex (Addgene #110104)
pRDV_FX5 containing Sb_loop (Addgene #132695)
pRDV_FX5 containing Sb_convex (Addgene #132696)
pDX_init containing Sb_loop (Addgene #132697)
pSb_init containing the TM287/288 nanobody Nb_TM#2 (Addgene #132698)
pSb_init containing the MBP sybody Sb_MBP#1 (Addgene #132699)
pBXNH3CA_MBP (Addgene #132700)
pBXNH3LCA_TM287/288 (Addgene #132701)
Equipment
SpectraPor 7 Dialysis Membrane 3.5 kD (Spectrum Labs, cat. no. 132110)
Amicon Ultra 15 Ultracel 3K (Merck, cat. no. UFC900324)
MagnaRac Magnetic Separation Rack (Invitrogen, cat. no. CS15000)
Nunc Maxisorp 96-well immunoplates (Merck, cat. no. M9410)
2-mm electroporation cuvettes (BioRad, cat. no. 1652086)
96-Well deep-well plate PP 2.2 ml (TreffLab, cat. no. 96.09799.9.01)
Gas-permeable foil (Macherey-Nagel, cat. no. 740675)
Adhesive plate seals (Abgene, cat. no. AB-0580)
15-ml tubes (Sarstedt, cat. no. 62.554.002)
50-ml tubes (Sarstedt, cat. no. 62.547.004)
PCR tubes (Sarstedt, cat. no. 72.737.002)
RNase-free/low binding microtubes (Sorenson, cat. no. 39640T (formerly cat. no. 11720) ▴CRITICAL We strongly advise the use of these exact tubes; low binding microtubes from other suppliers may result in less selection efficiency.
Petri dishes, polystyrene, size 60 mm × 15 mm (Merck, cat. no. P5481)
0.2-ml Non-skirted 96-well PCR plate (Abgene, cat. no. AB-0600)
Heraeus Multifuge 4 KR for 15-/50-ml tubes and 96-well plates (VWR, cat. no. 521-1025)
Centrifuge for 1.5-/2.0-ml tubes (Eppendorf, cat. no. 5424R)
Heraeus Primo R Centrifuge for 15-/50-ml tubes (Thermo Scientific, cat. no. 75005440)
Electroporator Gene Pulser Xcell (BioRad, cat. no. 1652662)
SRT-10C SEC100 (Sepax Technologies, cat. no. 239100-10030)
Microfluidizer high-pressure homogenizer (Microfluidics, cat. no. M-110P)
Incubators with orbital shaking platform (Kühner, cat. no. ISF1-X)
Multichannel pipette 10–100 µl (Eppendorf, cat. no. 3125000036)
Multichannel pipette/dispenser 50–1,200 µl (Eppendorf, cat. no. 4861000163)
2-l baffled Erlenmeyer flask (Merck, cat. no. CLS431256)
Spectrophotometer (Eppendorf, cat. no. 6133000001)
Plate reader (Biotek, cat. no. Synergy H1)
qPCR machine 7500fast (Applied Biosystems, cat. no. 4406985)
PCR cycler
Agarose gel casting and electrophoresis equipment (Bio Rad, cat. no. 1704405)
Reagent setup
1 M Tris/acetate stock solution
Dissolve 6.0 g of Tris in 30 ml of ddH2O. Set the pH to 7.4 (at room temperature, i.e., 20–25 °C) with acetic acid. Fill to 50 ml with ddH2O and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
1 M Tris/HCl stock solution
Dissolve 6.0 g of Tris in 30 ml of ddH2O. Set the pH to 8.0 (at room temperature) with HCl. Fill to 50 ml with ddH2O and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
5 M NaCl stock solution
Dissolve 14.6 g of NaCl in a total volume of 50 ml and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
3 M MgAc2 stock solution
Dissolve 32.2 g of MgAc2 tetrahydrate in a total volume of 50 ml and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
1 M MgCl2 stock solution
Dissolve 4.8 g MgCl2 in a final volume of 50 ml of ddH2O and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
1 M KCl stock solution
Dissolve 3.7 g KCl in a final volume of 50 ml of ddH2O and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
1 M HEPES stock solution
Dissolve 11.9 g HEPES in 30 ml of ddH2O. Adjust the pH to 7.4 with NaOH. Fill to 50 ml with ddH2O and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
10% (vol/vol) ultrapure glycerol
Mix 50 ml of ultrapure glycerol with 450 ml of ddH2O and sterilize by autoclaving. Ultrapure glycerol can be stored for ≥6 months at room temperature.
0.5 M Na2HPO4 stock solution
Dissolve 89.0 g of Na2HPO4 in 1 l of ddH20 and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
0.1 M Citric acid stock solution
Dissolve 19.2 g of citric acid in 1 l of ddH20 and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
0.5 M EDTA stock solution
Weigh in 7.5 g of EDTA and add 30 ml of ddH2O. Set the pH to 8.0. EDTA will dissolve only as the pH of the solution turns basic. Fill to 50 ml with ddH2O and sterilize through a 0.22-µm filter. The solution can be stored for ≥6 months at room temperature.
Heparin stock solution
Dissolve 1.0 g of heparin in 5 ml of ddH20. Do not filtrate, because the solution is very viscous. Divide into aliquots and store for ≤2 years at −20 °C.
Yeast RNA stock solution
Dissolve 5.0 mg of yeast RNA in 1 ml of nuclease-free H2O. Prepare 100-µl aliquots and store for ≤2 years at −20 °C.
Neutravidin stock solution
Dissolve 10 mg of neutravidin in 2.5 ml of PBS. This 4-mg/ml solution corresponds to 67 µM. Prepare 25-µl aliquots and snap-freeze in liquid nitrogen. Store for ≤5 years at −80 °C.
10× PBS stock solution
Dissolve 2.4 g of KH2PO4, 14.1 g of Na2HPO4·H2O, 2.0 g of KCl and 80 g of NaCl in 1 l of ddH2O (pH 6.8), and sterilize through a 0.22-µm filter. This corrensponds to a final concentration of 1.37 M NaCl, 100 mM phosphate and 27 mM KCl. This buffer will give a pH of 7.4 when diluted to 1× PBS. The solution can be stored for ≥6 months at room temperature.
10× TBS stock solution
Dissolve 44.2 g Tris and 87.7 g NaCl in 800 ml of ddH2O. Set the pH to 7.4 (at room temperature) using HCl. Fill to 1 l and sterilize through a 0.22-µm filter. This corresponds to 200 mM Tris and 1.5 M NaCl. The solution can be stored for ≥6 months at room temperature.
Lysozyme stock solution
Dissolve 50 mg lysozyme in 1 ml of ddH2O. Store for ≤2 years at −20 °C.
WTB buffer
Use the stock solutions to assemble 50 mM Tris/acetate, 150 mM NaCl and 50 mM MgAc2. Prepare freshly and use immediately.
WTB-BSA buffer
WTB supplemented with 0.5% (wt/vol) BSA. Prepare freshly and use immediately.
WTB-D buffer
WTB supplemented with a detergent of choice added at a final concentration equaling three times the critical micelle concentration (3xCMC) of the respective detergent. Prepare freshly and use immediately.
WTB-BSA-D buffer
WTB supplemented with 0.5% (wt/vol) BSA and 0.1% (wt/vol) or 3xCMC (whichever is higher) of the detergent of choice. At least 0.1% of the detergent is needed to compensate for detergent binding to BSA. Prepare freshly and use immediately.
RD-elution buffer
Use the stock solutions to assemble 50 mM Tris/acetate, 150 mM NaCl, 50 mM EDTA and 100 µg/ml yeast RNA in nuclease-free H2O. Prepare freshly and use immediately.
TBS buffer
10-Fold dilution of 10× TBS. Prepare freshly and use immediately.
TBS-BSA buffer
TBS supplemented with 0.5% (wt/vol) BSA. Prepare freshly and use immediately.
TBS-D buffer
TBS supplemented with 3xCMC of the detergent of choice. Prepare freshly and use immediately.
TBS-BSA-D buffer
TBS supplemented with 0.5% (wt/vol) BSA and 0.1% (wt/vol) of the detergent of choice. Prepare freshly and use immediately.
PD-elution buffer
TBS supplemented with 0.25 mg/ml trypsin, added as powder. Prepare freshly, use the first half immediately and store the rest at –20 °C and use it within 1 week.
AEBSF solution
Dissolve 7.5 mg of AEBSF in 1 ml of ddH2O and store at −20 °C. The solution is stable at −20 °C for ≥2 months. ! CAUTION The serine protease inhibitor is toxic. Minimize dust generation and accumulation.
Periplasmic extraction buffer
Dissolve sucrose to a final concentration of 20% (wt/vol) in half of the final volume of ddH2O. Use the stock solutions to get final concentrations of 50 mM Tris/HCl pH 8.0, 0.5 mM EDTA and 0.5 µg/ml lysozyme. Fill to the final volume with ddH2O. Prepare freshly and use immediately.
Protein A stock solution
Dissolve 5 mg of Protein A in 1 ml of PBS. Prepare 25-µl aliquots and store for ≤2 years at −20 °C.
TMB stock solution
Dissolve 100 mg of TMB in 10 ml of DMSO. Divide into aliquots and store for ≤6 months at −20 °C.
ELISA developing buffer
Mix 10.3 ml of 0.5 M Na2HPO4 stock solution (51.5 mM final concentration), 24.3 ml of 0.1 M citric acid stock solution (24.3 mM final concentration) and 65.4 ml of ddH2O. Before use, add 20 µl of 30% (vol/vol) H2O2 (0.006% (vol/vol) final concentration) and 1 ml of the TMB stock solution (1 mg/ml final concentration). Prepare freshly and use immediately.
20% (wt/vol) glucose stock solution
Weigh out 100 g of glucose and dissolve in a total volume of 500 ml of ddH20 and autoclave to sterilize. The solution can be stored for ≥6 months at room temperature.
LB medium
Weigh out 10 g of tryptone, 5.0 g of yeast extract and 10 g of NaCl, and dissolve in 900 ml of ddH2O. Adjust the pH to 7.0. Fill to 1 l with ddH2O and sterilize by autoclaving. The medium can be stored for ≥6 months at room temperature.
TB medium
Dissolve 12 g of tryptone, 24 g of yeast extract and 4.0 ml of glycerol in a total volume of 900 ml of ddH2O. Sterilize by autoclaving. The medium can be stored for ≥6 months at room temperature. Add 100 ml of 10× TB buffer immediately before use.
10× TB buffer
Dissolve 2.31 g of KH2PO4 (170 mM final concentration) and 12.54 g of K2HPO4 (720 mM final concentration) in a final volume of 100 ml of ddH2O. Sterilize by autoclaving. The buffer can be stored for ≥6 months at room temperature.
2YT medium
Dissolve 16 g of tryptone, 10 g of yeast extract and 5.0 g of NaCl in 900 ml of ddH2O. Set the pH to 7.0 and fill to a final volume of 1l with ddH2O. Sterilize by autoclaving. The medium can be stored for ≥6 months at room temperature.
SOC medium
Dissolve 20 g of tryptone, 5.0 g of yeast extract, 0.50 g of NaCl and 2.5 ml of 1 M KCl in 800 ml of ddH20. Set the pH to 7.0 and fill to a final volume of 970 ml. Sterilize by autoclaving. Immediately before use, add 10 ml of 1 M MgCl2 and 20 ml of 20% (wt/vol) glucose. The medium can be stored for ≥6 months at room temperature.
50× TEA buffer
For agarose gel electrophoresis. Dissolve 242 g of Tris in 500 ml of ddH2O (2 M final concentration). Add 100 ml of 0.5 M Na2EDTA (pH 8.0) (50 mM final concentration) and 57.1 ml of glacial acetic acid (1 M final concentration). Adjust to a final volume of 1 l with ddH2O. The buffer can be stored for ≥6 months at room temperature.
Ampicillin stock solution (100 mg/ml)
Dissolve 1.0 g of ampicillin in 10 ml of H2O and sterilize through a 0.22-µm filter. Divide into aliquots and store at –20 °C for ≤6 months.
Kanamycin stock solution (50 mg/ml)
Dissolve 0.50 g of kanamycin in 10 ml of H2O and sterilize through a 0.22-µm filter. Divide into aliquots and store at –20 °C for ≤6 months.
Tetracycline stock solution (10 mg/ml)
Dissolve 100 mg of tetracycline hydrochloride in 10 ml of H2O and sterilize through a 0.22-µm filter. Divide into aliquots and store at −20 °C for ≤6 months.
Chloramphenicol stock solution (25 mg/ml)
Dissolve 250 mg of chloramphenicol in 10 ml of 100% ethanol. Divide into aliquots and store at −20 °C for ≤6 months.
PEG6000/NaCl solution
Dissolve 100 g of PEG6000 (20% (wt/vol) final concentration) and 73 g of NaCl (2.5 M final concentration) in a total volume of 500 ml. Transfer the solution into an autoclavable flask including the stirrer bar and autoclave. Subsequently, stir the hot solution until it is cooled to room temperature to dissolve the PEG6000. The solution can be stored for ≥6 months at room temperature.
3 M imidazole stock solution
Dissolve 20.4 g of imidazole in 60 ml of ddH2O. Adjust the pH to 8.0 and fill to a total volume of 100 ml. Store the solution protected from light. The solution can be stored for ≥6 months at room temperature.
10 mM ATP stock solution
Dissolve 55 mg of adenosine 5′-triphosphate disodium salt hydrate and 25 mg of MgSO4 heptahydrate in 5 ml of H2O. Measure the pH by spotting 1-μl aliquots on pH paper and adjust the pH to 6.5–7.5 with NaOH. Adjust the volume to 10 ml. Prepare 100-µl aliquots, and store at −20 °C for ≤2 years.
Procedure
▴CRITICAL For the sake of clarity, the protocol is written for binder selections using one sybody library. Typically, selections are performed with the three sybody libraries (concave, loop and convex) in parallel.
Ribosome display ● Timing 3 h
-
Thaw the components of the PUREfrex 2.1 kit and the DS supplement according to the manufacturer’s instructions and keep the solutions on ice. Also, thaw one RNA aliquot of each of the three sybody libraries. Precool the benchtop centrifuge to 4 °C.
! CAUTION Whenever working with RNA, use gloves, RNAse-free plastic ware and filter-tips. Clean the bench regularly with RNase AWAY Decontamination Reagent.
- Assemble three in vitro translation reactions on ice:
Reagent Amount (μl) Nuclease-free water 1.8 Solution I 4.0 10 mM cysteine 0.5 80 mM reduced glutathione 0.5 60 mM oxidized glutathione 0.5 Solution II 0.5 Solution III 1.0 1.875 mg/ml DsbC 0.5 Total 9.3 Mix the components by pipetting and incubate the mixture for 5 min at 37 °C in a PCR cycler.
-
Add 0.7 µl of the RNA library, corresponding to 1.6 × 1012 mRNA strands, to the reaction (Fig. 2). The remaining RNA-library aliquot is immediately frozen again in liquid nitrogen and stored at −80 °C. Repetitive freeze-thawing of the RNA library until the aliquot is empty is well tolerated (we send 10-µl aliquots).
! CAUTION We do not advise amplifying/propagating the RNA library by reverse transcription and PCR amplification, as this results in a loss of library diversity.
Incubate the reactions for 30 min at 37 °C to form ribosomal complexes.
Prepare 10 ml of each buffer (WTB-BSA, WTB-D-BSA and WTB-D) and precool on ice.
Place 12 µl of Dynabeads MyOne Streptavidin T1 in a 1.5-ml RNase-free/low binding microtube. Place the tubes and magnetic rack on ice. Wash the beads twice with 500 µl of WTB-BSA using a magnetic rack to immobilize the beads. The immobilization will be complete in 30 s. Block the beads in 500 µl of WTB-BSA for ≥20 min.
Prepare 100 µl of ribosome display panning solution: WTB-D-BSA supplemented with 2.5 µl of heparin stock solution and 1 µl of RnaseIn.
Dilute the ribosomal complexes from Step 5 in 100 µl of ice-cold panning solution and centrifuge at 20,000g for 5 min at 4 °C.
-
Transfer the supernatant into a fresh 1.5-ml RNase-free/low binding microtube and add the biotinylated target protein (see Box 1) to a final concentration of 50 nM (solution panning). Mix by pipetting and incubate the panning mix for 20 min on ice.
▴ CRITICAL STEP The biophysical quality of the target protein needs to be tested at least by SEC (Box 1). If ligands are available, show that the protein is functional by a ligand binding assay. Only properly folded and non-aggregated target proteins will lead to a successful outcome of the selection. Ideally, the target protein is biotinylated via an Avi-tag to reach an equimolar biotinto-target protein ratio.
Wash the blocked magnetic beads of Step 7 three times with 500 µl of ice-cold WTB-D-BSA.
Add the panning mix of Step 10 to the washed beads of Step 11. Incubate on ice for 5 min to pull down the biotinylated target protein and the bound ribosomal complexes.
-
Wash the beads three times with 500 µl of ice cold WTB-D. Remove the buffer while the beads remain immobilized on the wall of the tube. Then, resuspend the beads with a pipette before pulling them again to the wall of the tube. Make sure to remove as much buffer as possible during the washing rounds. During the last washing step, transfer the beads to a fresh 1.5-ml tube.
▴ CRITICAL STEP Do not wash excessively. Three washing steps without extra incubation time are sufficient. Please note that there is a 30-s waiting time until the beads are completely pulled to the wall of the tube. Excessive washing leads to diversity loss as the sybodies are not displayed redundantly due to the large library size.
Elute the RNA by resuspending the beads in 100 µl of RD-elution buffer. Incubate for 10 min at room temperature. Immobilize the beads on the wall of the tube using the magnetic rack and transfer the supernatant to a fresh 1.5-ml tube.
Purify the eluted RNA with the RNeasy micro kit according to the manufacturer’s instructions. Elute the purified RNA in 15 µl of nuclease-free water. Repeat the elution by loading the eluate to the column again.
Reverse transcription ● Timing 2 h
-
16
Assemble a 40-µl total-volume AffinityScript reverse transcription reaction as outlined in Steps 17 and 18.
-
17Steps 17 and 18 outline denaturation of RNA, primer annealing and reverse transcription. First, mix the components below and incubate for 5 min at 65 °C in a PCR cycler; shock cool on ice afterward.
Reagent Amount (μl) Eluted RNA (Step 15) 14 RT_Primer (Table 2; 100 μM) 2 dNTPs (10 mM) 4 Subtotal 20 -
18Add the components below to the reaction, incubate in a PCR cycler at 37 °C for 1 h and then deactivate the reaction for 5 min at 95 °C.
Reagent Amount (μl) RT-buffer 4 DTT 4 dNTPs (100 mM) 3.2 Nuclease-free H2O 5.8 RNaseln 1 Reverse transcriptase 2 Total 40 ▴CRITICAL STEP It is essential to follow exactly the instructions in Steps 16–18.
-
19
Dilute the 40-µl reaction to 100 µl with nuclease-free H2O and purify the cDNA using the Machery-Nagel NucleoSpin Gel and PCR Clean-up kit using the standard NTI binding buffer according to the manufacturer’s instructions. Elute the cDNA in 30 µl of NE buffer (provided in the kit) and reload the eluate to the column for a second elution step.
-
20
Remove 1 µl of the purified cDNA and dilute it in 9 µl of nuclease-free H2O for qPCR analysis. Use the remaining 29 µl for PCR amplification and immediately proceed to Step 21. Run the qPCR analysis (Box 2).
? TROUBLESHOOTING
PCR amplification of the cDNA recovered from ribosome display ● Timing 3 h
-
21Set up a 100-µl PCR reaction with the components below. The PCR reactions are prepared in a volume of 100 µl and split in 2 × 50 µl. Use primers Med_FX_for and Med_FX_rev for the concave and loop library. Use primers Long_FX_for and Long_FX_rev for the convex library (Fig. 3).
Reagent Amount (μl) 5× GoTaq buffer 20 dNTPs (8 mM) 10 Primers (Table 2; 10 μM) 5 each Purified cDNA (Step 20) 29 Nuclease-free H2O 30.5 GoTaq G2 polymerase (1.25u) 0.5 ▴ CRITICAL STEP It is crucial that non-proofreading DNA polymerase such as GoTaq G2 is used at this step. Proofreading DNA polymerases digest cDNA and thereby result in poor PCR performance and diversity bottlenecks.
Perform the following PCR cycler program with 30 cycles of denaturation, annealing and elongation:Step Temperature (°C) Time (min:sec) Initial denaturation 95 2:00 Denaturation 95 0:30 Annealing 57 0:30 Elongation 72 1:00 Final elongation 72 2:00 -
22
Prepare a 1.5% (wt/vol) agarose gel in 1× TEA buffer, mix the entire PCR reaction with 6× DNA loading dye and load it into several wells. Run the gel. Cut the band at 350–400 bp (Fig. 4) and purify the PCR product using the Machery-Nagel NucleoSpin Gel and PCR Clean-up kit according to the manufacturer’s instructions.
◼PAUSE POINT Purified DNA fragments can be stored at −20 °C until further processing.
? TROUBLESHOOTING
Fig. 4. Exemplary DNA gel of sybody pools.
The entire PCR reaction (100 µl) obtained from amplifying cDNA encoding for enriched pools of the concave, loop and convex sybodies was loaded on a preparative 1.5% (wt/vol) agarose gel in 1× TEA buffer and stained with ethidium bromide (Step 22). The length differences of the three sybody libraries can be distinguished (concave: 342 bp; loop: 360 bp; convex: 372 bp).
Making electrocompetent E. coli SS320 ● Timing 1 d
▴ CRITICAL After growing the cells at 37 °C, all steps are carried out on ice, and centrifugation takes place at 4 °C. Centrifugation buckets must be autoclaved before use.
-
23
Prepare an overnight culture of E. coli SS320 in TB medium supplemented with 10 µg/ml tetracycline, with shaking at 37 °C.
-
24
Inoculate six baffled 2-l flasks containing 900 ml of TB medium (no antibiotic) each with 9 ml of the overnight culture.
-
25
Grow the cultures at 37 °C while shaking at 90 r.p.m. to an OD600 of 0.8.
-
26
Chill the cultures on ice for ≥10 min.
-
27
Centrifuge at 5,000g for 10 min in sterilized buckets.
-
28
Decant the supernatant and resuspend the pellets in 500 ml of ice-cold and sterile 1 mM HEPES pH 7.4.
-
29
Centrifuge at 5,000g for 10 min.
-
30
Decant the supernatant and resuspend the pellets in 500 ml of ice-cold and sterile 1 mM HEPES pH 7.4.
-
31
Centrifuge at 5,000g for 10 min.
-
32
Decant the supernatant and resuspend the pellets in 150 ml of ice-cold and sterile 10% ultrapure glycerol.
-
33
Centrifuge at 5,000g for 10 min.
-
34
Decant the supernatant and add 3 ml of ice-cold and sterile 10% ultrapure glycerol to the first bucket. Resuspend the pellet with a pipette and transfer the suspension to the next tube. Repeat until the six pellets are resuspended.
-
35
Transfer 350-µl aliquots in sterile 1.5-ml tubes and flash-freeze in liquid nitrogen. Store the frozen cells at −80 °C.
◼ PAUSE POINT Competent cells can be kept in the −80 °C freezer for ≥6 months.
Cloning of the phagemid libraries ● Timing 6 h
-
36
Digest 1 µg of the amplified PCR product from Step 22 and 10 µg of the pDX_init vector (Addgene #110101) using BspQI by mixing the components below. Incubate in a PCR cycler for 1.5 h at 50 °C followed by 20 min of BspQI deactivation at 80 °C.
| Digest of PCR product | Amount |
|---|---|
| 10× NEB 3.1 buffer | 10 μl |
| PCR product (Step 22) | Volume corresponding to 1 μg |
| BspQI | 5 μl (50 U) |
| H2O | Fill up to 100 μl |
| Digest of pDX_init | Amount |
| 10× NEB 3.1 buffer | 10 μl |
| PCR product (Step 22) | Volume corresponding to 10 μg |
| BspQI | 5 μl (50 U) |
| H2O | Fill up to 100 μl |
-
37
Purify the digested PCR products directly using the Machery-Nagel NucleoSpin Gel and PCR Clean-up kit according to the manufacturer’s instructions. Elute in 20 µl of NE buffer.
-
38
Prepare a 1% (wt/vol) agarose gel in 1× TEA buffer. Mix the digested pDX_init vector with 6× DNA loading dye and load the complete digest into several wells. Run the gel for ~45 min at a voltage of 120–140 V.
-
39
Digestion with BspQI will have formed two DNA bands. Cut out the larger fragment running at ~5,000 bp and purify it using the Machery-Nagel NucleoSpin Gel and PCR Clean-up kit according to the manufacturer’s instructions. Elute in 30 µl of NE buffer.
-
40
Ligation of the fragments. Mix the following reagents:
| Reagent | Amount |
|---|---|
| 10× T4 ligase buffer | 5 μl |
| Digested pDX_init backbone | Volume corresponding to 1.2 μg |
| Digested PCR product | Volume corresponding to 300 ng |
| H2O | Fill up to 50 μl |
-
41
Heat to 65 °C for 30 s in a PCR cycler, and cool to 37 °C. Add 2.5 µl (12.5 U) of T4 ligase. Incubate at 37 °C for 1.5 h in a PCR cycler. Heat-inactivate the T4 ligase at 65 °C for 10 min.
Electroporation ● Timing 2 h
-
42
Thaw an aliquot (350 µl) of electrocompetent E. coli SS320 cells (Step 35) on ice, and place the ligation reaction and electroporation cuvettes with a 0.2-cm gap on ice as well.
-
43
Mix the 50-µl ligation reaction (Step 41) with the competent cells by pipetting gently up and down.
-
44
Pulse the cells with a Gene Pulser Xcell electroporation system using 2,400 V, 25 µF and 750 Ω.
? TROUBLESHOOTING
-
45
Immediately transfer the electroporated cells to 25 ml of SOC medium.
-
46
Incubate the culture for 30 min while shaking at 37 °C and 160 r.p.m. for recovery.
-
47
Remove 20 µl of the culture and generate a dilution series by diluting three times 10-fold in LB medium. Streak out 5 µl of the dilutions on LB-agar plates supplemented with 120 µg/ml ampicillin to determine the efficiency of the transformation. The total number of transformants needs to be >5 × 106 colonies to recover the diversity of the ribosome display output.
? TROUBLESHOOTING
-
48
Transfer the rest of the recovery culture into 225 ml of 2YT supplemented with 200 µg/ml ampicillin and 2% (wt/vol) glucose. Incubate overnight at 37 °C while shaking at 160 r.p.m.
Phage production ● Timing 1 d
-
49
Make a glycerol stock of the dense overnight culture by mixing 1 ml of the culture with 0.8 ml of sterile 50 % glycerol. Snap-freeze in liquid nitrogen and store at −80 °C.
◼ PAUSE POINT The glycerol stock of the phagemid sublibrary can be stored at −80 °C for ≥1 year.
! CAUTION Use the entire glycerol stock to inoculate a culture to avoid diversity bottlenecks.
-
50
Prepare 50 ml of 2YT supplemented with 200 µg/ml ampicillin and 2% (wt/vol) glucose. Inoculate with 1 ml of the overnight culture and grow the culture at 37 °C while shaking at 160 r.p.m. for 1.5–2 h to an OD600 of 0.6.
-
51
Place 27 µl of the M13KO7 helper phage at 1012 plaque-forming units/ml in a 50-ml tube. Add 10 ml of the culture from Step 50 and swirl to mix well. Incubate at 37 °C without shaking for 30 min.
! CAUTION To prevent phage contaminations, all flasks and tubes in contact with the phages should be soaked in a 1% (wt/vol) sodium hypochlorite solution overnight to inactivate phages. Subsequently, flasks are autoclaved, cleaned and reused.
-
52
Centrifuge at 5,000g for 10 min. Decant the supernatant. Resuspend the pellet in 50 ml of 2YT supplemented with 200 µg/ml ampicillin and 25 µg/ml kanamycin. Grow the culture at 37 °C while shaking overnight at 160 r.p.m. to produce phages.
Phage purification ● Timing 2.5 h
-
53
Centrifuge the overnight culture from Step 52 in a 50-ml tube at 5,000g for 30 min at 4 °C.
-
54
Decant 40 ml of the culture supernatant into a fresh 50-ml tube, add 10 ml of the PEG6000/NaCl solution and mix well by inverting the tube end over end five times.
-
55
Incubate on ice for ≥30 min to precipitate the phages. The solution can turn a bit cloudy.
-
56
Pellet the phages by centrifugation at 5,000g for 30 min at 4 °C.
-
57
Decant and remove any remaining supernatant with a tissue. Resuspend the pellet in 1 ml of PBS with a pipette. Transfer into a fresh 1.5-ml tube.
-
58
Remove cells and aggregates by centrifuging the resuspended phages at 20,000g for 5 min.
-
59
Transfer the supernatant in a fresh 1.5-ml tube.
-
60
Repeat Steps 58 and 59.
-
61
Determine the phage concentration by UV-visible spectroscopy:
Measure the absorption (over a 1-cm pathlength) of the phage solution at 269 nm and 320 nm wavelength in a UV-visible spectrophotometer. To measure in the linear range, dilute the phages 1:10 in PBS. Calculate the phage titer with the following equation:The phagemid titers should be in the range of 1–5 × 1013/ml.
◼ PAUSE POINT Purified phages can be kept at 4 °C for ≥1 month.
? TROUBLESHOOTING
First round of phage display ● Timing 5 h
-
62
On the day before the first phage display round, coat 48 wells of a Nunc Maxisorp 96-well immunoplate with 100 µl per well of 67 nM neutravidin in PBS. Seal the plate using an adhesive plate seal and store the plate at 4 °C overnight.
-
63
Inoculate an overnight culture of E. coli SS320 in 50 ml of 2YT supplemented with 10 µg/ml tetracycline.
▴ CRITICAL STEP All the following steps of the phage display (Steps 64–85) can be performed either at 4 °C (on ice) or at room temperature, if the stability of the protein allows for it.
-
64
Prepare 50 ml of TBS, 50 ml of TBS-BSA, 50 ml of TBS-BSA-D and 150 ml of TBS-D.
-
65
Wash the neutravidin-coated plate once with 250 µl of TBS per well and block the neutravidin with 250 µl of TBS-BSA for 30 min. To reduce carryover of any remaining liquid between the washing steps, beat the plates dry on a paper tissue after decanting the washing buffer.
-
66
Prepare 5 ml of TBS-BSA-D containing 1012 phages/ml.
-
67
Remove 100 µl of this phage solution to a fresh 1.5-ml tube.
-
68
Add the biotinylated target protein to the remaining 4.9 ml of phage solution at a concentration of 50 nM and mix well.
▴ CRITICAL STEP In our hands, only 2–5 % of the phage particles (numbers determined by absorption and qPCR) are infectious and lead to a transfer of the phagemid into E. coli. Due to this low infection rate of M13 phages, it is mandatory to adhere to these large volumes and target protein amounts. Reduction of the volume or target protein concentration will lead to diversity bottlenecks.
-
69
Add a biotinylated negative control protein at 50 nM to the 100 µl of phage solution (Step 67). In case selections are performed against a membrane protein, we recommend using purified and biotinylated TM287/288 (expressed from plasmid Addgene #132701). In case selections are performed against a soluble protein, purified and biotinylated MBP can be used (expressed from plasmid Addgene #132700).
-
70
Incubate for 20 min (solution panning).
-
71
Wash the plates once with 250 µl of TBS-BSA-D per well.
-
72
Transfer the phage-target solution of Step 68 in 47 wells, 100 µl per well.
-
73
Transfer the 100 µl of negative control of Step 69 into the remaining well.
-
74
Incubate for 5 min.
-
75
Freshly prepare 10 ml of the PD elution buffer.
-
76
Wash the plates three times by adding 250 µl of TBS-D per well, discarding the buffer by inverting the plate and beating it dry on a paper tissue. Incubate for 2 min between washing steps.
-
77
Add 100 µl of PD elution buffer per well. Incubate for 10 min at room temperature.
-
78
Immediately freeze the remaining elution buffer and store at −20 °C for use in the next round of phage display.
-
79
Transfer the 100-µl elution of the negative control into a fresh 1.5-ml tube and add 0.8 μl of the AEBSF solution to inhibit the trypsin.
-
80
Pool the 4.7-ml elution of the target-containing wells into a fresh 50-ml tube and add 40 µl of the AEBSF solution to inhibit the trypsin.
-
81
Mix 1 µl of the elutions of Steps 79 and 80 with 9 µl of ddH2O for qPCR (Box 2).
-
82
Inoculate 200 ml of 2YT with 4 ml of E. coli SS320 preculture from Step 63.
-
83
Grow the culture at 37 °C while shaking at 160 r.p.m. to an OD600 of 0.6.
-
84
Add 45 ml of the culture to the 4.7 ml of eluted phages from Step 80. There is no need to infect the negative control as it is only used for qPCR.
-
85
Incubate at 37 °C without shaking for 30 min to infect the E. coli SS320 cells. Add the 50 ml of infected E. coli SS320 cells to 200 ml of 2YT supplemented with 200 µg/ml ampicillin and 2% (wt/vol) glucose. Grow the cultures overnight at 37 °C while shaking at 160 r.p.m.
Second round of phage display ● Timing 2 d
-
86
On the day before the second phage display round, inoculate an overnight culture of E. coli SS320 from a glycerol stock in 50 ml of 2YT supplemented with 10 µg/ml tetracycline.
-
87
Prepare a glycerol stock of the dense overnight culture (from Step 85) by mixing 1 ml of the culture with 0.8 ml of sterile 50% (vol/vol) glycerol. Snap-freeze in liquid nitrogen and store at −80 °C.
◼PAUSE POINT A glycerol stock of the phagemid sublibrary can be stored at −80 °C for ≥1 year.
-
88
Use the remaining overnight culture of the phagemid sublibrary (Step 87) to produce the input phages for the second round of phage display according to Steps 50–61.
-
89
Prepare 10 ml of TBS-BSA, 10 ml of TBS-BSA-D and 10 ml of TBS-D.
-
90
Place 12 µl of Dynabeads MyOne Streptavidin C1 in a 1.5-ml RNase-free/low binding microtube. Wash the beads twice with 500 µl of TBS-BSA using a magnetic rack to immobilize the beads. Block the beads in 500 µl of TBS-BSA for ≥20 min.
-
91
Prepare 250 µl of TBS-BSA-D containing 5 × 1012 phages/ml (from Step 88).
-
92
Place 100 µl of the phage solution in a fresh 1.5-ml RNase-free/low binding microtube and add the biotinylated target protein to a concentration of 50 nM.
-
93
Place another 100 µl of the phage solution in a fresh 1.5-ml RNase-free/low binding microtube and add the biotinylated negative control protein to a concentration of 50 nM. In case selections are performed against a membrane protein, we recommend using purified and biotinylated TM287/288 (expressed from plasmid Addgene #132701). In case selections are performed against a soluble protein, purified and biotinylated MBP can be used (expressed from plasmid Addgene #132700).
-
94
Incubate the mixtures of Steps 92 and 93 for 20 min (this step corresponds to what we call solution panning).
-
95
Wash the beads from Step 90 three times with 500 µl of TBS-BSA-D.
-
96
Resuspend the beads with the 100 µl of panning solutions of Steps 92 and 93.
-
97
Incubate for 5 min.
-
98
Prepare 220 µl of the competition buffer consisting of TBS-BSA-D with 5 µM non-biotinylated target protein.
-
99
Wash the beads once with 500 µl of TBS-BSA-D using a magnetic rack.
-
100
Resuspend the beads in 100 µl of competition buffer. Incubate for 3 min.
-
101
Wash the beads twice with 500 µl of TBS-D. Exchange all tubes with fresh ones during the last washing step.
-
102
Thaw the PD elution buffer remaining from the last round of phage display (Step 78).
-
103
Resuspend the beads in 100 µl of PD elution buffer.
-
104
Incubate for 10 min at room temperature.
-
105
Pull the beads to the wall of the tube using a magnetic rack and transfer the elutions in fresh 1.5-ml tubes.
-
106
Add 0.8 µl of the AEBSF solution to each elution to inhibit the trypsin. Mix by pipetting up and down.
-
107
Mix 1 µl of each elution with 9 µl of ddH2O for quantification of the enrichment by qPCR.
? TROUBLESHOOTING
-
108
Inoculate 50 ml of 2YT with 1 ml of E. coli SS320 of the overnight culture (Step 86).
-
109
Grow cells while shaking at 37 °C to an OD600 of 0.6.
-
110
Add 1.4 ml of the culture to the 100 µl of eluted phages of Step 106. There is no need to infect the negative control as it is only used for qPCR.
-
111
Incubate at 37 °C without shaking for 30 min.
-
112
Add the 1.5-ml phage-infected culture to 10 ml of 2YT supplemented with 200 µg/ml ampicillin and 2% (wt/vol) glucose.
-
113
Grow the cultures overnight while shaking at 37 °C and prepare a glycerol stock of the culture the next day.
◼PAUSE POINT The glycerol stock of the phagemid sublibrary can be stored at −80 °C for ≥1 year.
Cloning of the phage display output into pSB_init ● Timing 4 h
-
114
Harvest 5 ml of the culture of Step 113 by centrifugation at 5,000g and 4 °C for 10 min.
-
115
Use the QIAprep Spin Miniprep Kit to isolate phagemids (pDX_init PD output) according to the manufacturer’s instructions.
-
116
Sub-clone sybodies into pSb_init vector (Addgene #110100) using FX cloning. Mix the following components:
| Component | Amount |
|---|---|
| 10× NEB 3.1 buffer | 1 μl |
| pSb_init | Volume corresponding to 100 ng |
| pDX_init PD output | Volume corresponding to 300 ng |
| BspQI | 1 μl (10 U) |
| H2O | Fill up to 10 μl |
Incubate the reaction in a PCR cycler at 50 °C for 1 h. Deactivate BspQI at 80 °C for 20 min. Cool the reaction to room temperature and add the following components to the reaction:
| Component | Amount |
|---|---|
| 10 mM ATP | 1.2 μl |
| T4 DNA ligase | 1 μl (5 U) |
Incubate the reaction in a PCR cycler at 37 °C for 1 h. Deactivate T4 DNA ligase at 65 °C for 10 min.
-
117
Thaw one 50-µl aliquot of chemically competent E. coli MC1061 on ice.
-
118
Add 5 µl of the cloning reaction to the cells and incubate for 10 min on ice.
-
119
Transform by heat shock at 42 °C for 45 s.
-
120
Place the cells on ice for 5 min.
-
121
Add 500 µl of LB medium to the cells and recover them at 37 °C while shaking at 600 r.p.m. for 30 min.
-
122
Plate 100 µl of cells on LB-agar plates supplemented with 25 µg/ml chloramphenicol. Pellet the rest of the recovery culture, resuspend the pellet in 100 µl and plate it on a second LB-agar plate supplemented with 25 µg/ml chloramphenicol.
-
123
Incubate the plates overnight at 37 °C. A total of ~500–5,000 colonies are expected to appear on both plates.
? TROUBLESHOOTING
Single-clone expression for ELISA ● Timing 1 d
-
124
Prepare one 96-well deep-well plate and add 1.2 ml of TB medium supplemented with 25 µg/ml chloramphenicol per well. Label the plate with ‘preculture’.
-
125
Pick 95 colonies from either the first and/or second plate of Step 123 (depending on the colony density) and inoculate one positive control from a glycerol stock. If sybody selections are performed against a membrane protein, use the TM287/288 selective nanobody Nb_TM#2 (Addgene #132698) as positive control (published in ref. 20). If selections are performed against soluble proteins, use MBP sybody Sb_MBP#1 (Addgene #132699) as positive control (published in ref. 18).
-
126
Seal the plates with a gas-permeable seal and grow the cells at 37 °C while shaking at 300 r.p.m.
-
127
After 4 h of growth, check if the culture has turned turbid. If it is not yet turbid, check in intervals of 30 min for turbidity. Prepare a fresh 96-well deep-well plate with 1 ml of prewarmed TB medium supplemented with 25 µg/ml chloramphenicol per well. Label the plate with ‘expression culture’.
Inoculate the expression culture with 50 µl of the preculture using a multichannel pipet. Seal the plates with a gas-permeable seal.
-
128
Place the ‘preculture’ and ‘expression culture’ plates in the shaker and grow the cells at 37 °C while shaking at 300 r.p.m. for 2 h.
-
129
Change the temperature setting to 22 °C and continue cultivation for ~1.5 h. Following this, the cells will be in the exponential growth phase, and the cultures will have an OD600 of 0.4–0.8.
-
130
Induce the expression culture with 0.02% (wt/vol) L-(+)-arabinose by adding 20 µl of 1% (wt/vol) L-(+)-arabinose. Shake overnight at 22 °C.
-
131
On the next day, collect the cells of all plates by centrifugation at 4,000g and 4 °C for 20 min.
-
132
Store the pellets of the ‘preculture’ plate at −20 °C for plasmid DNA purification and process the plate containing the cell pellets of the ‘expression culture’.
Periplasmic extraction for ELISA ● Timing 2 h
-
133
Prepare 15 ml of the periplasmic extraction buffer.
-
134
Add 100 µl to each cell pellet of the ‘expression culture’ of Step 132 and resuspend the cells by intensive vortexing. Incubate on ice for 30 min.
-
135
Add 900 µl of TBS supplemented with 1 mM MgCl2 to the cells.
-
136
Centrifuge the plate at 4,000g and 4 °C for 20 min.
-
137
Use the supernatant as the periplasmic cell extract for the ELISA in Step 144.
ELISA and sequence analysis ● Timing 2–3d
▴ CRITICAL The ELISA can be performed at room temperature or in a cold room at 4°C, depending on the stability of the target protein.
-
138
On the day before, prepare two 96-well Nunc Maxisorp immunoplates by coating each well with 100 µl of a 1:1,000 dilution of the Protein A stock solution in PBS. Seal the plates using adhesive plate seals and store the plates at 4 °C overnight.
-
139
Wash the plates once with 250 µl of TBS per well.
-
140
Block the plates with TBS-BSA. Incubate for 30 min.
-
140
Wash the plates three times with 250 µl of TBS per well.
-
141
Add 100 µl of a 1:2,000 dilution of the monoclonal anti-c-Myc antibody in TBS-BSA-D to each well. Incubate for 20 min. In case an expensive detergent is used, it can be omitted in the buffer until Step 146.
-
142
Wash the plates three times with 250 µl of TBS-D per well.
-
143
Add 80 µl of TBS-BSA-D to each well and add 20 µl of the periplasmic extract from Step 137 using a multichannel pipette. Add the same periplasmic extract to two wells side by side (e.g., A1 and A2, B1 and B2, etc.) to be able to compare binding to the target and to the negative control. Incubate for 20 min.
-
144
Wash the plates three times with 250 µl of TBS-D per well.
-
145
Add 100 µl of the biotinylated target and negative control at 50 nM in TBS-BSA-D to the respective wells (we place target to wells A1-H1, negative control to wells A2-H2, target to wells A3-H3, etc.). If sybody selections are performed against a membrane protein, the TM287/288 selective nanobody Nb_TM#2 is used as positive control (Step 125), and hence add biotinylated TM287/288 to the respective well (typically well H11 of the second ELISA plate). If selections are performed against soluble proteins, MBP sybody Sb_MBP#1 is used as positive control (Step 125), and hence add biotinylated MBP to the respective well. Incubate for 20 min.
-
146
Wash the plates three times with 250 µl of TBS-D per well.
-
147
Add 100 µl of 1:5,000 diluted streptavidin−peroxidase polymer in TBS-BSA-D. Incubate for 20 min.
-
148
Wash the plates three times with 250 µl of TBS-D per well.
-
150
Add 100 µl of the ELISA developing buffer. Wait until individual wells turn blue, which takes ~5–15 min.
-
151
Measure the absorbance at 650 nm in a plate reader. ELISA signals as small as 1.5-fold above background can indicate a high-affinity binder. It is therefore worthwhile to pick ELISA hits of different signal intensities.
? TROUBLESHOOTING
-
152
Use the QIAprep Spin Miniprep Kit to isolate the pSb_init plasmids from the frozen preculture cell pellets of Step 132 carrying the sybodies corresponding to the positive ELISA hits. Elute plasmids in 30 µl of elution buffer of the QIAprep Spin Miniprep Kit. Typical plasmid yields range from 80 to 120 ng/µl. Send the plasmids for Sanger sequencing using the pBAD_forward primer. Analyze the sequences to determine unique sybodies (Table 3).
! CAUTION Some clones (~1 in 50) may have ambiguous sequencing results in the CDRs caused by transformation with multiple plasmids. We ignore these clones.
Table 3. DNA and protein sequences of sybodies.
| Sybody type | Sequence |
|---|---|
| DNA sequences (5′ to 3′ direction) | |
| Concave |
CAGGTTCAGCTGGTTGAGAGCGGTGGTGGCCTGGTCCAAGCTGGCGGTTCGCTGCGTCTGAGC
TGCGCCGCAAGCGGTTTCCCGGTGNNNNNNNNNNNNATGNNNTGGTATCGTCAGGCACCGGGCA AAGAACGTGAGTGGGTCGCGGCGATTNNNAGCNNNGGTNNNNNNACGNNNTACGCAGATTCTGTTA AGGGCCGCTTTACCATCAGCCGCGACAACGCGAAGAATACGGTCTATTTGCAGATGAATAGCCTGA AACCGGAAGATACCGCGGTTTACTACTGTNNNGTGNNNGTGGGTNNNNNNTACNNNGGCCAAGGTA CCCAAGTGACTGTGAGC |
| Loop |
CAGGTTCAGCTGGTTGAGAGCGGTGGTGGCCTGGTCCAAGCTGGCGGTTCGCTGCGTCTGA
GCTGCGCCGCAAGCGGTTTCCCGGTGNNNNNNNNNNNNATGNNNTGGTATCGTCAGGCACCG GGCAAAGAACGTGAGTGGGTCGCGGCGATTNNNAGCNNNGGTNNNNNNACGNNNTACGCAGAT TCTGTTAAGGGCCGCTTTACCATCAGCCGCGACAACGCGAAGAATACGGTCTATTTGCAGATGA ATAGCCTGAAACCGGAAGATACCGCGGTTTACTACTGTAACGTGAAAGACNNNGGTNN NNNNNNNNNNNNNTACGACTATTGGGGCCAAGGTACCCAAGTGACTGTGAGC |
| Convex |
CAAGTCCAGCTGGTGGAATCGGGTGGTGGTAGCGTCCAGGCGGGTGGTAGCCTGCGTCTGA
GCTGTGCGGCTAGCGGCNNNATTNNNNNNATCNNNTACCTGGGCTGGTTTCGCCAGGC ACCGGGCAAAGAGCGTGAGGGCGTCGCAGCGCTGNNNACCNNNNNNGGTNNNACCTAC TACGCGGACAGCGTTAAGGGTCGTTTCACGGTGAGCCTGGACAACGCCAAGAATA CCGTGTATCTGCAAATGAACAGCTTGAAACCGGAAGATACTGCTTTGTATTACTGC GCGGCAGCCNNNNNNGGCNNNNNNNNNCCGCTGNNNNNNNNNNNNTATNNNTACTGGGGTCAGGGCACCCAAGTTACCGTTTCT |
| Protein sequences | |
| Concave |
QVQLVESGGGLVQAGGSLRLSCAASGFPVXXXXMXWYRQAPGKEREWVAAIXSXGXXTXYADSVK
GRFTISRDNAKNTVYLQMNSLKPEDTAVYYCXVXVGXXYXGQGTQVTVS |
| Loop |
QVQLVESGGGLVQAGGSLRLSCAASGFPVXXXXMXWYRQAPGKEREWVAAIXSXGXX
TXYADSVKGRFTISRDNAKNTVYLQMNSLKPEDTAVYYCNVKDXGXXXXXYDYWGQGTQVTVS |
| Convex |
QVQLVESGGGSVQAGGSLRLSCAASGXIXXIXYLGWFRQAPGKEREGVAALXTXXGXT
YYADSVKGRFTVSLDNAKNTVYLQMNSLKPEDTALYYCAAAXXGXXXPLXXXXYXYWGQGTQVTVS |
For DNA sequences, NNN denotes a randomized position. Randomization was performed by trinucleotide building blocks to achieve defined amino acid compositions as described previously18. For protein sequences, X denotes a randomized position.
Medium scale purification of the unique hits ● Timing 2 d
-
153
Add 1 µl of each unique sybody plasmid into a 96-well PCR plate on ice. Add 10 µl of chemically competent E. coli MC1061 cells to each well containing a plasmid. Heat-shock in a PCR cycler set to 42 °C for 45 s. Cool on ice for 5 min. Add 90 µl of LB medium and recover in a PCR cycler set to 37 °C for 30 min (no shaking required).
-
154
Transfer transformed cells to 1 ml of TB medium supplemented with 25 µg/ml chloramphenicol in a 96-well deep-well plate and grow overnight at 37 °C while shaking at 300 r.p.m.
-
155
Inoculate 50-ml cultures of TB medium supplemented with 25 µg/ml chloramphenicol in 150-ml Erlenmeyer flasks with 500 µl of the precultures.
-
156
Grow the cultures at 37 °C while shaking at 160 r.p.m. for 2 h.
-
157
Change the temperature setting to 22 °C and continue cultivation for ~1.5 h. Following this, the cells will be in the exponential growth phase, and the cultures will have an OD600 of 0.4–0.8.
-
158
Induce expression by adding L-(+)-arabinose to a final concentration of 0.02% (wt/vol).
-
159
Express the sybodies overnight at 22 °C while shaking at 160 r.p.m.
Purification ● Timing 1–3 d
-
160
Transfer the cultures into 50-ml tubes.
-
161
Collect the cells by centrifugation at 4,000g for 20 min.
-
162
Resuspend the cell pellet in 5 ml of periplasmic extraction buffer and incubate at 4 °C for 30 min.
-
163
Add 20 ml of TBS supplemented with 1 mM MgCl2.
-
164
Pellet the cells by centrifugation at 4,000g for 20 min.
-
165
Transfer the supernatant containing the extracted sybodies in a fresh 50-ml tube and add 125 µl of the 3 M imidazole stock solution to a final concentration of 15 mM.
-
166
Pool the entire resin of a His MultiTrap HP plate into a 50-ml tube.
-
167
Add 500 µl of the resin to the supernatants of the periplasmic extracts.
-
168
Incubate for 1 h at room temperature while inverting the tubes occasionally to prevent sedimentation of the resin.
-
169
Collect the resin by centrifugation at 500g for 2 min in a swing-out rotor.
-
170
Decant the supernatant, resuspend pelleted beads with the remaining buffer and transfer to the wells of the His MultiTrap HP plate.
-
171
Wash three times by resuspending the resin with 500 µl of TBS supplemented with 30 mM imidazole followed by centrifugation at 500g for 2 min.
-
172
Place the His MultiTrap HP plate on a fresh 96-well deep-well plate and elute with 500 µl of TBS supplemented with 300 mM imidazole by centrifuging at 500g for 2 min.
-
173
Analyze each sample using a Sepax SRT-10C SEC100 SEC column at a flow rate of 1 ml/min and elute for ≥1.25 column volumes. Collect the peak fractions. Monomeric sybodies elute at a retention volume of 11–12.5 ml from the Sepax SRT-10C SEC100 column. Retention volumes <11 ml indicate oligomerization. Retention volumes in the range of 12.5-14 ml indicate moderate column interaction and retention volumes >14 ml indicate strong column interaction (Fig. 5). We discard sybodies that are not expressed, form oligomers or exhibit strong column interactions. Typical yields are in the range of 200 µg to 1 mg.
! CAUTION We do not advise the use of SEC columns other than Sepax SRT-10C SEC100 due to the fact that 10–30% of the sybodies exhibit column interactions. In particular, column interaction are a problem when using columns made of sugar-based polymers, such as widely used Superdex columns. Sybodies with a retention volume <12.5 ml on the Sepax SRT-10C SEC100 column are safe to use with any other column.
? TROUBLESHOOTING
Fig. 5. SEC analysis of sybodies.
Three exemplary sybodies were purified from medium-scale cultures and analyzed on a Sepax SRT-10C SEC100 column. The sybody giving rise to the blue chromatogram elutes at a retention volume of ~11 ml and is thus a monomer. The red chromatogram with its peak <10 ml represents an oligomerizing sybody. The orange chromatogram with its main peak at ~16 ml represents a sybody exhibiting strong column interaction. AU, arbitrary units.
FX cloning in pBXNPH3 or pBXNPHM3 ● Timing 4 h
▴ CRITICAL Sybodies purified from pSb_init (Step 173) bear a Myc-tag and a His6-tag at the C terminus. They can be further analyzed (e.g., by SPR) in this form. Furthermore, they can be used for cryo-electron microscopy analysis and for microscopy purposes without any problems. However, if the sybodies are used as crystallization chaperones, we highly recommend subcloning them into pBXNPH3 (Addgene #110098) or pBXNPHM3 (Addgene #110099) to obtain tag-free binders. We typically try the pBXNPHM3 construct first and use the pBXNPH3 as the second option.
-
174Set up the following reaction:
Component Amount 10× NEB 3.1 buffer 1 μl pBXNPH3 or pBXNPHM3 Volume corresponding to 100 ng pSB_init Volume corresponding to 300 ng BspQI 1 μl (10 U) H2O Fill up to 10 μl Incubate the reaction in a PCR cycler at 50 °C for 1 h. Deactivate BspQI at 80 °C for 20 min. Cool the reaction to room temperature and add the following components:Component Amount 10 mM ATP 1.2 μl T4 DNA ligase 1 μl (5 U) Incubate the reaction in a PCR cycler at 37 °C for 1 h. Deactivate T4 DNA ligase at 65 °C for 10 min.
-
175
Thaw a 50-µl aliquot of chemically competent E. coli MC1061 on ice.
-
176
Add 5 µl of the cloning reaction to the cells and incubate for 10 min on ice.
-
177
Transform by heat shock at 42 °C for 45 s.
-
178
Place the cells on ice for 5 min.
-
179
Add 500 µl of LB medium to the cells and recover them at 37 °C with shaking at 600 r.p.m. for 30 min.
-
180
Plate 100 µl on an LB-agar plate supplemented with 120 µg/ml ampicillin.
-
181
Incubate the plates overnight at 37 °C.
Large scale expression of sybodies ● Timing 2 d
-
182
Inoculate 50 ml of LB medium supplemented with 100 µg/ml ampicillin with a single colony from Step 181. Grow the culture overnight at 37 °C while shaking.
-
183
Prepare two baffled 2-l flasks with 600 ml of TB medium supplemented with 100 µg/ml ampicillin and inoculate each flask with 6 ml of the overnight preculture.
-
184
Grow the main culture to an OD600 of 0.5, and then set the shaker temperature to 22 °C and continue growth for 1.5 h.
-
185
Induce expression by adding L-(+)-arabinose to a final concentration of 0.02% (wt/vol). Express the sybody overnight.
Purification of tag-free sybodies ● Timing 1–2 d
-
186
Collect the cells by centrifugation at 5,000g and 4 °C for 15 min and resuspend them in 100 ml of ice-cold TBS.
-
187
Lyse the cells by passing them three times through a pre-cooled high-pressure homogenizer at 1–1.5 kbar.
-
188
Centrifuge the lysate at 8,000g and 4 °C for 30 min to pellet cell debris.
-
189
Load the supernatant on a 2-ml Ni-NTA superflow gravity column.
-
190
Wash the column with 20 ml of TBS supplemented with 50 mM imidazole.
-
191
Elute the column with 5 ml of TBS supplemented with 300 mM imidazole.
-
192
Add 400 µg of 3C protease and fill into a dialysis tube with a cutoff of 3 kDa.
-
193
Dialyze against 500 ml of TBS for 3 h at room temperature or overnight at 4 °C.
-
194
Set the imidazole to a final concentration of 30 mM (add imidazole if necessary).
-
195
Load the dialyzed sample again on a 2-ml Ni-NTA superflow column (reverse IMAC). Collect the cleaved, tag-free sybody in the flow-through. Add 5 ml of TBS to the column to collect the remaining sample and pool it with the flow-through.
-
196
Concentrate the sybody eluate to 0.5 ml using an Amicon Ultra 15 concentrator with a molecular weight cutoff of 3 kDa and separate via SEC using a Sepax SRT-10C SEC100 column.
! CAUTION Take care not to overload the column; maximally run 2–4 mg per run. In case the tag-free sybody elutes at a retention volume <12.5 ml, it is safe to run on other columns (e.g., Superdex-200 columns).
? TROUBLESHOOTING
Troubleshooting
Troubleshooting advice can be found in Table 4.
Table 4. Troubleshooting table.
| Step | Problem | Possible reasons | Solution |
|---|---|---|---|
| 20 | Ratio of 5′ qPCR reaction to 3′ qPCR reaction <0.2 | mRNA degradation | Clean bench and pipettes with RNase AWAY and repeat Steps 1–20. Make sure to add RNase inhibitor (RNaseIn) during panning and reverse transcription steps |
| 22 | Weak or absent band on DNA gel | mRNA degradation Failure of reverse transcription reaction Use of proofreading polymerases |
See above for Step 20 Use input mRNA sybody library for a test reverse transcription and cDNA amplification Use a Taq-based polymerase |
| 44 | An arc occurs during electroporation. | Cuvette is wet outside Cells were not washed sufficiently (residual salt) Temperature of electroporation reaction and cuvette is too high |
Make sure the cuvette is dry outside to avoid arcs around the cuvette Repeat the making of electrocompetent cells Make sure the cuvette is ice cold and operate fast enough to avoid warming of cuvette |
| 47 | Phagemid clone number <5 × 106 | Electrocompetent cells are not competent enough | Make sure to use highly pure glycerol for making electrocompetent cells as specified in the protocol. Use covalently closed circular pDX_init plasmid to test the electrocompetency of cells |
| 61 | Phage titer < 5 × 1012 | Cloning or ligation did not work properly Not enough phage produced Phage purification failed |
Repeat cloning reaction. Make sure to use the indicated amounts of BspQI, T4 ligase and DNA. Use freshly purchased enzymes Repeat phage production and purification using the glycerol stock of the sublibrary. Use the complete glycerol stock to inoculate a new culture to rescue the complete sybody repertoire. Make sure to omit glucose in the 2YT medium of Step 52 Make sure that the PEG6000/NaCl solution added in Step 54 is mixed well |
| 107 | Enrichment against target < twofold compared to control protein. | Target protein is not sufficiently biotinylated Target protein is not properly folded |
Make sure that target is sufficiently biotinylated Ensure that target protein runs as a defined species in SEC |
| 123 | Fewer than 100 colonies after cloning into pSb_init | Target protein denatures during selection procedure Digestion or ligation not sufficient DNase contamination in purified phagemid or pSb_init |
Make sure to carry out selections on ice. Consider including ligands that stabilize the protein. Consider using another detergent Repeat cloning reaction. Make sure to use the indicated amounts of BspQI, T4 ligase and DNA. Use freshly purchased enzymes. Prepare new ATP stocks Make new minipreps of phagemid and pSb_init |
| 151 | No specific ELISA hits | No sybody enrichment ELISA failed Cloning of sybodies into pSb_init failed Single-clone expression and/or periplasmic extraction failed Unspecific signals against control protein |
Troubleshoot according to Step 109 Make sure to include a positive control Prepare five randomly picked pSb_init clones and perform Sanger sequencing to ensure that pSb_init carries sybodies as an insert Make sure to include the positive control binders during single-clone expression and periplasmic extraction Make sure to alter immobilization surfaces in each selection round as outlined in the protocol |
| 173 | No protein yield for any purified sybody | Medium-scale expression or purification failed | Establish purification protocol using the MBP sybody Sb_MBP#1 cloned into pSb_init (Addgene #132699) as positive control |
| 196 | No protein yield for any purified tag-free sybody | Large-scale expression or purification failed | Establish purification protocol using a non-randomized sybody cloned into pBXNPHM3 (Addgene #110102, #110103 or #110104) as positive control |
Timing
Steps 1–22, ribosome display, reverse transcription and PCR amplification of cDNA: 1 d Steps 23–35, making of electrocompetent cells: 1 d
Steps 36–48, generation of phagemid libraries: 1 d
Steps 49–85, phage production, phage purification and first round of phage display: 2 d
Steps 86–113, phage production, phage purification and second round of phage display: 2 d
Steps 114–151, sybody cloning into pSb_init, small-scale expression, periplasmic extracts and ELISA: 3 d
Step 152, miniprep and Sanger sequencing of ELISA hits: 2 d
Steps 153–173, medium-scale expression and purification: 3–5 d
Steps 174–196, cloning, large-scale expression and purification of tag-free sybodies: 4–5 d
Anticipated results
We outline here three examples of sybody selections against membrane proteins, all of which were conducted precisely according to this protocol. A special emphasis is put on the selection strategy to obtain conformation-specific binders, which inhibit the membrane protein’s function.
Conformational trapping of an ABC exporter with a sybody
TM287/288 is a bacterial ABC exporter exhibiting drug efflux activity whose structure was solved in two inward-facing states52,53. For many years, we aimed to obtain the outward-facing structure of TM287/288. However, it turned out to be difficult to obtain crystals of the outward-facing transporter for two main reasons: (i) the outward-facing conformation was difficult to populate54, and (ii) the crystal packing of inward-facing TM287/288 was dominant. To further stabilize the outward-facing conformation, we introduced a mutation in the conserved nucleotide-binding domains that blocks ATP hydrolysis without affecting ATP binding (namely the E517A mutation in the Walker B sequence of the consensus ATP binding site) and selected sybodies against TM287/288 in the presence of ATP. This selection regime can be performed only in vitro where ATP is always present at saturating concentrations; upon injection of such a transporter-ATP complex, ATP dissociates and the transporter reverts back to its dominant inward-facing state. There was strong binder enrichment already in the first phage display round (17-, 60- and 3-fold) and 170-fold, 220-fold and 25-fold enrichment in the second phage display round for the concave, loop and convex libraries, respectively (Table 1). The ELISA hit rate ranged from 30% to 89%, and a total of 48 clones giving rise to ELISA signals of different strengths were sequenced. Forty sequences were unique, and 31 sybodies exhibiting good yields and biophysical properties could be purified. We conducted SPR in the presence or absence of ATP to identify conformation-specific binders (Fig. 6b). The affinities obtained using TM287/288_E517A + ATP-Mg ranged from 2–57 nM, 8–1090 nM and 21–422 nM for the concave, loop and convex libraries, respectively18. A total of 11 sybodies were found to be conformation specific, as defined by an affinity difference of ≥10-fold between measurements conducted in the presence or absence of ATP. We then attempted to co-crystallize these 11 state-specific sybodies together with TM287/288_E517A in the presence of ATP-Mg18. Six of them gave crystals. With one convex sybody called Sb_TM#35, we were able to solve the structure of outward-facing TM287/288 at a resolution of 3.2 Å20 (Fig. 6a). Unintended by design, the long 16–amino acid CDR3 was no longer tethered and adopted a stretched conformation. Sb_TM#35 binds to an extracellular wing of TM287/288, and electron spin resonance revealed that it clamps open the extracellular gate and thereby stabilizes the outward-facing conformation of the transporter20.
Fig. 6. A conformation-specific sybody against the ABC transporter TM287/288.
a, The convex sybody Sb_TM#35 (gray with CDR3 in red) served as a crystallization chaperone to solve the structures of outward-facing ABC exporter TM287/288 (TM287 chain in aquamarine, TM288 chain in light pink, bound ATP as spheres) (PDB: 6QUZ). b, SPR analysis showing that the sybody Sb_TM#35 binds only to the outward-facing ABC transporter in the presence of ATP and Mg2+. KD, dissociation constant; RU, response units. The data shown in b were taken from ref. 20.
Sybodies stabilizing distinct conformations of UraA
The SLC23 homolog UraA from E. coli is a secondary active transporter built up of 14 transmembrane helices. The protein is a dimer in the lipid bilayer without solvent-exposed domains. UraA was crystallized with its substrate uracil bound in the inward-facing and occluded conformations55,56. However, low protein stability without bound substrate and conformational flexibility in detergent hamper structural analysis of the full transport cycle. To overcome these limitations, sybodies were selected against detergent-solubilized UraA in the presence and absence of uracil. To enrich binders against uncharacterized conformations, counterselection was performed using a locked variant of UraA based on the published inward-facing conformation55. The enrichment after two rounds of phage display was 4.3-, 11.8- and 10.2-fold without substrate and 6.7-, 19.6- and 5.6-fold in the presence of uracil for the concave, loop and convex libraries, respectively (Table 1). The enrichment was highest for the loop library. While for the concave and loop library the inclusion of uracil during the selection resulted in stronger enrichment factors, the opposite effect was observed for the convex library. ELISA performed with 2 × 95 clones (with or without uracil included during selection) for each of the three sybody libraries gave a total of 96 hits (ELISA hit rate of 17%). Of these 96 hits, 58 were analyzed by sequencing, resulting in 45 unique sybodies. Forty sybodies were purified, yielding 31 binders with a monodisperse size-exclusion profile. Nine sybodies dropped out due to low expression (seven sybodies) or poor elution profile (two sybodies). Further characterization (e.g., conformation specificity and thermostabilization) of these sybodies will be reported elsewhere.
A sybody targeting the peptide binding pocket of the KDEL receptor
The KDEL receptor retrieves endoplasmatic reticulum (ER) luminal proteins from the Golgi by pH-dependent recognition of a carboxyl-terminal Lys-Asp-Glu-Leu (KDEL) signal sequence and thereby fulfils an essential function in eukaryotic cells. To shed light on the structural basis of KDEL peptide binding, sybodies were selected against the purified mouse KDEL receptor. The KDEL receptor unites all features of a challenging membrane protein target. It is a small integral membrane protein with a molecular weight of only 26 kDa, and it consists of seven transmembrane helices devoid of soluble domains on either side of the membrane that could serve as extended paratopes. Furthermore, the purified KDEL receptor needs to be kept at 4 °C to remain stable. Sybody selections were performed at pH 7.5 and on ice in the absence of KDEL peptide. Under these conditions, the receptor is expected to populate mainly its apo state and remains properly folded. In contrast to the selections against the stable and large ABC exporter TM287/288, but similar to the smaller transporter UraA, sybody enrichment was less pronounced, namely 1.6-, 1.2- and 0.7-fold after the first phage display round and 7.8-, 4.9- and 3.6-fold after the second phage display round for the concave, loop and convex libraries, respectively (Table 1). Accordingly, the ELISA hit rate ranged from 8% to 35%. A total of 48 ELISA hits were sequenced, giving rise to 31 unique binder hits. Of these 31 binders, 12 could be purified and were biophysically well behaved on SEC. The other 19 sybodies dropped out due to poor expression (10 sybodies), oligomerization on SEC (7 sybodies) and strong column interaction on SEC (2 sybodies). A qualitative SPR analysis revealed that one of these binders, called ‘Syb37’ and stemming from the loop library, exhibited the slowest off-rate and was cross-reactive with the chicken KDEL receptor. Using biolayer interferometry, Syb37’s affinity for the chicken KDEL receptor was determined to be 15 nM. The KDEL receptor/Syb37 complex was crystallized using lipidic mesophase and crystals diffracted to 2.23 Å7 (Fig. 7a). Due to the small size difference between sybody (15 kDa) and KDEL receptor (26 kDa), it was possible to solve the complex structure by molecular replacement using the MBP sybody (PDB: 5M13) as the search model. Using in turn the built KDEL receptor as the molecular replacement search model, the structures of free apo KDEL receptor as well as of the KDEL receptor with the bound TAEKDEL peptide were solved. The extended CDR3 loop of Syb37 protrudes deep into, and thereby occupies, the peptide-binding pocket (Fig. 7a). In agreement with this observation, Syb37 and TAEKDEL compete for binding to the KDEL receptor7 (Fig. 7b). However, only binding of TAEKDEL peptide leads to a conformational change of the receptor, whereas Syb37 stabilizes the apo conformation. Since sybody selections were selected against the apo receptor in the absence of peptide and at high pH, this result was anticipated. Cell biology experiments finally revealed that the expression of Syb37 with a leader sequence allowing for secretion into the ER interferes with KDEL receptor retrieval from the Golgi to the ER7.
Fig. 7. Structure of the KDEL receptor in complex with a sybody.
a, The loop sybody Syb37 occupies the KDEL peptide-binding pocket of the KDEL receptor via its CDR3 sequence (red) (PDB: 6I6J). b, Peptide-binding assays using tritiated TAEKDEL peptide showing that sybody Syb37 competes with peptide binding. The data shown in panel b were taken from ref. 7. Column heights indicate mean values and the error bars are standard deviations of the individual measurement points shown as circles.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We wish to thank all members of the Seeger and Geertsma laboratoriess for scientific discussions. We acknowledge Jenifer Cuesta Bernal for critical reading. E.R.G. acknowledges financial support from the German Research Foundation via the Cluster of Excellence Frankfurt (Macromolecular Complexes) and the CRC807 (Transport and Communication across Biological Membranes). Work in the Seeger group was supported by an SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823, to M.A.S.), an SNSF NRP 72 grant (407240_177368, to M.A.S.), an SNSF BRIDGE proof-of-concept grant (20B1-1_175192, to P.E.) and a BioEntrepreneur-Fellowship of the University of Zurich (BIOEF-17-002, to I.Z.). R.J.P.D, E.R.G., and M.A.S. acknowledge a grant from the Commission for Technology and Innovation CTI (16003.1 PFLS-LS). Work in the group of S.N. was supported by a Wellcome award (102890/Z/13/Z).
Footnotes
Author contributions
M.A.S., E.R.G. and R.J.P.D. conceived the sybody project. E.R.G. and M.A.S. designed the sybody library. I.Z. and P.E. established the sybody selection platform. C.A.J.H. established the ELISA setup. I.Z., C.A.J.H., B.T.K., P.B. and E.R.G. selected sybodies against protein targets. I.Z., S.N., E.R.G. and M.A.S. supervised students and postdocs. I.Z., B.T.K., E.R.G. and M.A.S. wrote the manuscript. P.E., C.A.J.H., S.N. and R.J.P.D. edited the manuscript.
Competing interests
The authors declare competing financial interests. I.Z., P.E., R.J.P.D. and M.A.S. are co-founders and shareholders of Linkster Therapeutics AG.
Peer review information Nature Protocols thanks Serge Muyldermans, Jamshid Tanha and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
All plasmids have been deposited on Addgene. The sybody libraries can be obtained from the authors via an academic material transfer agreement.
References
- 1.Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- 2.Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 3.Manglik A, Kobilka BK, Steyaert J. Nanobodies to study G protein-coupled receptor structure and function. Annu Rev Pharmacol Toxicol. 2017;57:19–37. doi: 10.1146/annurev-pharmtox-010716-104710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmansa S, Alborelli I, Bieli D, Caussinus E, Affolter M. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. Elife. 2017;6:e22549. doi: 10.7554/eLife.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukowska MA, Grutter MG. New concepts and aids to facilitate crystallization. Curr Opin Struct Biol. 2013;23:409–416. doi: 10.1016/j.sbi.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 7.Bräuer P, et al. Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science. 2019;363:1103–1107. doi: 10.1126/science.aaw2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur H, et al. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J Biomol NMR. 2019;73:375–384. doi: 10.1007/s10858-019-00250-8. [DOI] [PubMed] [Google Scholar]
- 9.Nevoltris D, et al. Conformational nanobodies reveal tethered epidermal growth factor receptor involved in EGFR/ErbB2 predimers. ACS Nano. 2015;9:1388–1399. doi: 10.1021/nn505752u. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez-Lombardi R, et al. Challenges and opportunities for non-antibody scaffold drugs. Drug Discov Today. 2015;20:1271–1283. doi: 10.1016/j.drudis.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 12.De Genst E, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry KA, MacKenzie CR. Antigen recognition by single-domain antibodies: structural latitudes and constraints. MAbs. 2018;10:815–826. doi: 10.1080/19420862.2018.1489633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 17.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann I, et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife. 2018;7:e34317. doi: 10.7554/eLife.34317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradbury ARM, Sidhu S, Dubel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol. 2011;29:245–254. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutter CAJ, et al. The extracellular gate shapes the energy profile of an ABC exporter. Nat Commun. 2019;10:2260. doi: 10.1038/s41467-019-09892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 22.Verheesen P, et al. Reliable and controllable antibody fragment selections from Camelid non-immune libraries for target validation. Biochim Biophys Acta. 2006;1764:1307–1319. doi: 10.1016/j.bbapap.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Olichon A, de Marco A. Preparation of a naïve library of camelid single domain antibodies. Methods Mol Biol. 2012;911:65–78. doi: 10.1007/978-1-61779-968-6_5. [DOI] [PubMed] [Google Scholar]
- 24.Monegal A, et al. Immunological applications of single-domain llama recombinant antibodies isolated from a naïve library. Protein Eng Des Sel. 2009;22:273–280. doi: 10.1093/protein/gzp002. [DOI] [PubMed] [Google Scholar]
- 25.Moutel S, et al. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife. 2016;5:e16228. doi: 10.7554/eLife.16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geertsma ER, et al. Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family. Nat Struct Mol Biol. 2015;22:803–808. doi: 10.1038/nsmb.3091. [DOI] [PubMed] [Google Scholar]
- 27.Egloff P, et al. Engineered peptide barcodes for in-depth analyses of binding protein libraries. Nat Methods. 2019;16:421–428. doi: 10.1038/s41592-019-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrnstorfer IA, Geertsma ER, Pardon E, Steyaert J, Dutzler R. Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat Struct Mol Biol. 2014;21:990–996. doi: 10.1038/nsmb.2904. [DOI] [PubMed] [Google Scholar]
- 29.Prado ND, et al. Inhibition of the myotoxicity induced by Bothrops jararacussu venom and isolated phospholipases A2 by specific camelid single-domain antibody fragments. PLoS One. 2016;11:e0151363. doi: 10.1371/journal.pone.0151363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch K, et al. Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers. Sci Rep. 2017;7:8390. doi: 10.1038/s41598-017-08273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussack G, Arbabi-Ghahroudi M, Mackenzie CR, Tanha J. Isolation and characterization of Clostridium difficile toxin-specific single-domain antibodies. Methods Mol Biol. 2012;911:211–239. doi: 10.1007/978-1-61779-968-6_14. [DOI] [PubMed] [Google Scholar]
- 32.McMahon C, et al. Yeast surface display platform for rapid discovery of conformationally selective nano-bodies. Nat Struct Mol Biol. 2018;25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan JR, Li GH, Hu YH, Ou WJ, Wan YK. Construction of a synthetic phage-displayed Nanobody library with CDR3 regions randomized by trinucleotide cassettes for diagnostic applications. J Transl Med. 2014;12:343. doi: 10.1186/s12967-014-0343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sircar A, Sanni KA, Shi J, Gray JJ. Analysis and modeling of the variable region of camelid single-domain antibodies. J Immunol. 2011;186:6357–6367. doi: 10.4049/jimmunol.1100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumoulin M, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govaert J, et al. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J Biol Chem. 2012;287:1970–1979. doi: 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ring AM, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn BT, et al. Biotinylation of membrane proteins for binder selections. Methods Mol Biol. 2020;2127:151–165. doi: 10.1007/978-1-0716-0373-4_11. [DOI] [PubMed] [Google Scholar]
- 39.Zahnd C, Amstutz P, Plückthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 41.Geertsma ER, Dutzler R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry. 2011;50:3272–3278. doi: 10.1021/bi200178z. [DOI] [PubMed] [Google Scholar]
- 42.Geertsma ER. FX cloning: a simple and robust high-throughput cloning method for protein expression. Methods Mol Biol. 2014;1116:153–164. doi: 10.1007/978-1-62703-764-8_11. [DOI] [PubMed] [Google Scholar]
- 43.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 44.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 45.Zahnd C, Sarkar CA, Pluckthun A. Computational analysis of off-rate selection experiments to optimize affinity maturation by directed evolution. Protein Eng Des Sel. 2010;23:175–184. doi: 10.1093/protein/gzp087. [DOI] [PubMed] [Google Scholar]
- 46.Seeger MA, et al. Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity. Protein Sci. 2013;22:1239–1257. doi: 10.1002/pro.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber T, Steiner D, Röthlisberger D, Plückthun A. In vitro selection and characterization of DARPins and Fab fragments for the co-crystallization of membrane proteins: the Na+-citrate symporter CitS as an example. J Struct Biol. 2007;159:206–221. doi: 10.1016/j.jsb.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Garza JA, Taylor AB, Sherwood LJ, Hart PJ, Hayhurst A. Unveiling a drift resistant cryptotope within Marburgvirus nucleoprotein recognized by llama single-domain antibodies. Front Immunol. 2017;8:1234. doi: 10.3389/fimmu.2017.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zabetakis D, Anderson GP, Bayya N, Goldman ER. Contributions of the complementarity determining regions to the thermal stability of a single-domain antibody. PLoS One. 2013;8:e77678. doi: 10.1371/journal.pone.0077678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussack G, et al. Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain. J Biol Chem. 2011;286:8961–8976. doi: 10.1074/jbc.M110.198754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 52.Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- 53.Hohl M, et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci USA. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timachi MH, et al. Exploring conformational equilibria of a heterodimeric ABC transporter. Elife. 2017;6:e20236. doi: 10.7554/eLife.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu F, et al. Structure and mechanism of the uracil transporter UraA. Nature. 2011;472:243–246. doi: 10.1038/nature09885. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, et al. Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters. Cell Res. 2017;27:1020–1033. doi: 10.1038/cr.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids have been deposited on Addgene. The sybody libraries can be obtained from the authors via an academic material transfer agreement.