Abstract
Alterations in the expression of microRNA (miR)-138 have been demonstrated to result in the development of several malignant tumours. However, the possible function of miR-138 in human glioma cells remains unclear. The present study demonstrated that miR-138 was significantly downregulated in 48 human glioma specimens by quantitative PCR analysis. The upregulation of miR-138 exerted significant antiproliferative and anti-invasive effects on glioma cells and promoted their apoptosis. In addition, cAMP response element-binding protein 1 (CREB1) was confirmed as a direct target gene of miR-138 by luciferase gene reporter assay, and the antitumour effect of miR-138 on glioma cells was significantly reversed by CREB1 overexpression. Moreover, the molecular mechanisms underlying the tumour-suppressive role of miR-138 in malignant glioma may be associated with the dephosphorylation of AKT/mTOR caused by the miR-138 upregulation-induced decrease in CREB1 expression in glioma cells. The results of the present study indicated that miR-138 may affect CREB1/AKT/mTOR signalling to regulate the proliferation, apoptosis and invasion of glioma cells and the malignant progression of glioma, thereby suggesting that miR-138 may be a potential target for the treatment of gliomas.
Keywords: microRNA-138, glioma, cAMP response element-binding protein 1, apoptosis, proliferation, invasion
Introduction
Gliomas are the most frequent primary tumours of the central nervous system and a major cause of death among patients with intracranial tumours (1). Aggressive interventions, including surgery, chemotherapy and radiotherapy, are not always curative, particularly for glioblastoma (GBM) (2). In recent years, increasing attention has been focused on the molecular genetics of GBM in order to improve the prognosis and treatment of patients with GBM (3). However, no definite conclusions have been reached regarding the mechanisms underlying GBM tumourigenesis. Therefore, it is urgent to identify new molecular biomarkers that regulate the malignant biological behaviour of GBM.
MicroRNAs (miRNAs) constitute a class of single-stranded non-coding RNAs that play an important role in the translation of tumour genes and the regulation of downstream proteins. Accumulating evidence indicates that miR-138 may act as a tumour suppressor via its interaction with critical signalling pathways in tumourigenesis, such as the hypoxia-inducible factor-1α, extracellular-signal regulated protein kinase and nuclear factor-κB pathways (4,5). Specifically, aberrantly low expression levels of miR-138 have been reported in several malignant tumours, such as gallbladder carcinoma, anaplastic thyroid carcinoma (ATC), non-small cell lung cancer (NSCLC) and oral squamous cell carcinoma (OSCC) (6–9). Recent studies have demonstrated that the upregulation of miR-138 in tumour cells may cause reversion of the malignant phenotype (10,11). Moreover, Stojcheva et al observed that miR-138 promoted acquired alkylator resistance in GBM by targeting BIM, highlighting the importance of this miRNA in GBM (12). However, the molecular mechanisms underlying the function of miR-138 in glioma remain unclear.
The cAMP response element-binding protein 1 (CREB1) is a multifunctional molecule that mediates transcriptional responses to various growth factors and stress signals involved in tumour progression (13). Typically, CREB1 is overexpressed in a series of human neoplasms, and high expression of CREB1 in several cancers, such as astrocytoma, hepatocellular carcinoma and NSCLC, has been associated with an unfavourable overall survival (OS) outcome (14–16). Furthermore, Rodon et al revealed that CREB1 may promote a malignant transforming growth factor-β2 autocrine loop in GBM (17). However, current research cannot fully explain the molecular mechanism of action of CREB1 in GBM, and the presence of an interactive network mainly involving CREB1 has not been established. Therefore, the role of CREB1 in GBM cell proliferation, apoptosis and invasion and the underlying regulatory mechanism require further investigation.
The aim of the present study was to investigate miR-138 expression in glioma specimens and evaluate miR-138 as a candidate biomarker. In addition, the expression of miR-138 was upregulated in U87 and U251 cells to observe the effect of miR-138 on cell proliferation, invasion and apoptosis. Furthermore, it was investigated whether CREB1 is a direct target miR-138 in order to determine whether the miR-138/CREB1/mTOR pathway may play a critical role in the tumourigenic behaviour of glioma cells.
Materials and methods
Cell culture
The 293 cell line and the malignant glioma cell lines U87 (glioblastoma of unknown origin, ATCC HTB-14™) and U251 were obtained from the Chinese Academy of Sciences (Shanghai, China). All glioma cell lines were grown in DMEM supplemented with 10% FBS in a humidified atmosphere containing 5% CO2 at 37°C.
Tissue samples
The study protocol complied with the National Regulations on the Use of Clinical Samples in China. The Specialty Committee on Ethics of Biomedicine Research, Navy Medical University, approved the use of human specimens in this study (PJ2011-012-03). Written informed consent was obtained from all participants prior to the present study. The glioma cancer specimens were obtained from patients who underwent surgery at Changzheng Hospital and Shanghai Tongji Hospital between July 2012 and July 2018.
Cell viability assay
In the present study, MTT was used to monitor cell proliferation. An MTT assay was conducted as previously described to measure cell viability (18). Cell viability was analysed with a Cell Counting Kit (cat. no. 11465007001, Roche Diagnostics GmbH). The optical density at 570 nm was measured 24, 48, 72, 96 and 120 h after transfection.
Cell transfections
The miR-138 mimics and miRNA-negative control (NC) mimics were obtained from Qiagen GmbH. U87 and U251 cells were transfected with miR-138 mimics or miRNA-NC by Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.). The target sequences of miR-138 mimics were as previously described (19). The scrambled sequence (5′-UUCUCCGAACGUGUCACGUTT-3′) was used to create non-targeting miR-NC and GFP-siRNA. CREB1 siRNA (sc-35111) and siRNA Reagent System (sc-45064) were purchased from Santa Cruz Biotechnology, Inc. and all transfections were conducted according to the manufacturer's instructions. CREB1 overexpression vectors were purchased from GenePharma and successfully transfected into the corresponding cells according to the manufacturer's instructions in the presence of Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.). A full-length human CREB1 complementary DNA containing the entire coding sequence tagged with GFP or GFP alone (Lenti-GFP) was cloned into the lentiviral vector pLenti6/V5-DEST (Invitrogen; Thermo Fisher Scientific, Inc.) to create the complete functional overexpression vector Lenti-CREB1. The efficacy of the transfection was tested by using western blotting.
Invasion assay
The invasion assay was conducted as previously described (20). Equal numbers (1×105) of cells stably transfected with miR-NC, miR-138 mimics, GFP-siRNA or CREB1-siRNA were plated in separate 24-well Matrigel-coated cell culture inserts with an 8-µm pore size. The invasion rate was measured in three independent experiments.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from glioma tissues and cells lines by using TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized using miRNA RT assay (TaqMan; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and miRNA expression levels were analysed using the 7900 fast RT-PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using miR-138-specific primers. The 25-µl reaction mixture contained 14 µl 2X SYBR Green Master Mix, 1 µl forward primer (10 µM), 1 µl reverse primer (10 µM), 3 µl cDNA template and 6 µl ddH2O was set up. The thermocycling parameters were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec, 60°C for 1 min, and a detection step at 72°C for 30 sec. The sequences for the primers of miR-138 and U6 were as previously described (21) and were as follows: miR-138, forward, 5′-CCCAGGGTCTGGTGCGGAGA-3′ and reverse, 5′-CAGGGGCTGAGCGGTGAGGG-3′; and U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. U6 probe was used as an endogenous control. Relative fold expression of the target genes was calculated according to the 2−∆∆Cq method (22).
Apoptosis assay
Apoptosis was assessed by dual staining using Annexin V-FITC and propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.). All cells were analysed by a FACSCalibur System (BD Biosciences). The apoptosis assays were performed using U87 and U251 cell lines with or without miR-138 overexpression. The analyses were performed using FlowJo software (v10.6.2; FlowJo LLC).
Dual luciferase reporter assay
A total of 100 ng CREB1 wild-type (WT) 3′-untranslated region (UTR) or CREB1 mutant (MT) 3′-UTR luciferase reporter plasmids were co-transfected into 293 cells. At 48 h after transfection, the luciferase activity in the cells was measured with a Dual Luciferase Reporter Assay System (Promega Corporation). The transfections were performed three times.
Statistical analysis
All data are expressed as the mean ± SD values of data from triplicate experiments and paired groups were compared by Student's t-test, whereas three or more groups were compared by ANOVA followed by Tukey's test for multiple pairwise comparisons. The Kruskal-Wallis with Dunn multiple comparisons test was used to compare the immunolabeling results between the glioma tissue grades. The correlation between miR-138 and CREB1 expression was examined by Spearman's correlation coefficient. OS was investigated by conducting a Kaplan-Meier analysis, and the predictors of survival were analysed by a Cox proportional hazards regression analysis. Statistical analysis was performed with SPSS 13.0 software (SPSS Inc.) and P<0.05 was considered to indicate statistically significant differences.
Results
miR-138 is frequently downregulated in clinical glioma tissue samples
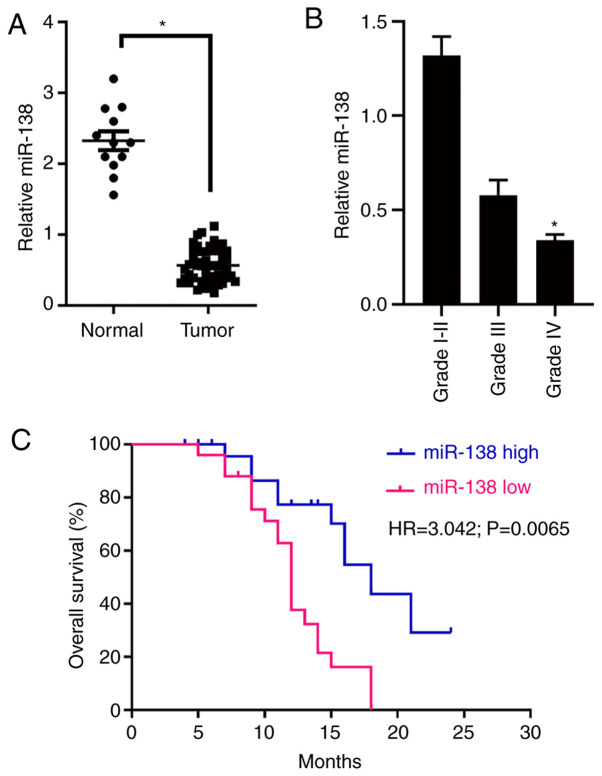
RT-qPCR was first used to measure miR-138 expression in 48 human glioma specimens and 12 normal brain tissue specimens. miR-138 expression in the glioma specimens was significantly lower compared with that in the normal brain specimens (Fig. 1A). In addition, high-grade glioma specimens (WHO grades III–IV) exhibited a lower expression of miR-138 compared with the low-grade glioma specimens (WHO grades I–II; Fig. 1B). Subsequently, the association between miR-138 expression and clinicopathological characteristics was investigated. According to the miR-138 expression level in the specimens, the 48 patients with glioma were divided into the low expression (<4.12, n=22) and high expression (≥4.12, n=26) groups, and a statistical analysis was conducted (Table I). The expression of miR-138 was found to be significantly associated with histological grade (P=0.02). Furthermore, it was demonstrated that patients with high expression levels of miR-138 had significantly longer OS compared with those with low miR-138 expression levels by a Kaplan-Meier survival analysis (Fig. 1C). Collectively, these results indicate that miR-138 expression may be a key prognostic index of glioma patient survival.
Figure 1.
(A) miR-138 expression was measured by RT-qPCR analysis in normal brain tissues and glioma specimens. All expression levels were normalized to those of U6 small nuclear RNA. *P<0.05. (B) miR-138 expression was measured by RT-qPCR analysis in different-grade gliomas. *P<0.05. (C) Kaplan-Meier survival curves according to miR-138 expression. The expression of miR-138 in patient tumours was analysed by RT-qPCR, and the patients were divided into the high (blue line) and low (red line) expression groups, as described in Materials and methods (P=0.002). RT-qPCR, reverse transcription-quantitative PCR.
Table I.
Association between miR-138 expression and clinicopathological variables of glioma patients.
| miR-138 expression | ||||
|---|---|---|---|---|
| Characteristics | Value | High | Low | P-value |
| No. of patients | 48 | 25 | 23 | |
| Age, years | 0.28 | |||
| <50 | 26 | 15 | 11 | |
| ≥50 | 22 | 10 | 12 | |
| Sex | 0.35 | |||
| Male | 28 | 13 | 15 | |
| Female | 20 | 12 | 8 | |
| Mean tumour diameter, cm | 0.29 | |||
| <3 | 29 | 14 | 15 | |
| ≥3 | 19 | 11 | 8 | |
| Predominant location | 0.17 | |||
| Frontal lobe | 12 | 8 | 4 | |
| Temporal lobe | 16 | 9 | 7 | |
| Parietal lobe | 11 | 5 | 6 | |
| Cerebellum | 9 | 3 | 6 | |
| Stage | 0.013 | |||
| I/II | 19 | 14 | 5 | |
| III/IV | 29 | 11 | 18 | |
miR-138 regulates cell viability, invasion and apoptosis in gliomas
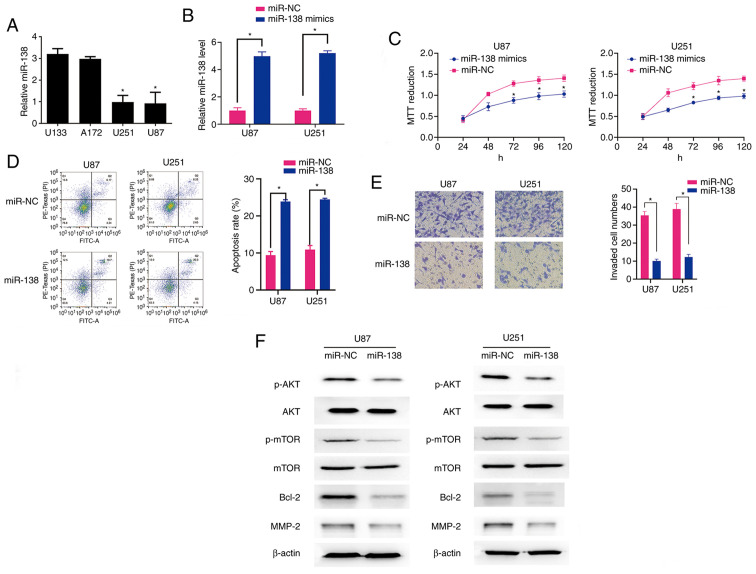
Subsequently, RT-qPCR was used to measure the miR-138 expression level in four glioma cell lines (U87, U251, A172, SW1088 and U133) (Fig. 2A). The U87 and U251 cells exhibited significantly higher miR-138 expression compared with the A172 and U133 cells (Fig. 2A). Therefore, the U87 and U251 cells were selected for the subsequent in vitro experiments. The U87 and U251 cells transfected with miR-138 mimics exhibited a significant increase in miR-138 expression (Fig. 2B). Then, an MTT assay was used to assess the effect of miR-138 expression on U87 and U251 cell viability. Compared with the miR-NC cells, the transfection of the U87 and U251 cells with the miR-138 mimics resulted in a significant reduction in cell viability (Fig. 2C). The upregulation of miR-138 in the U87 and U251 cells was correlated with enhanced apoptosis (Fig. 2D). Furthermore, compared with the miR-NC cells, the glioma cells transfected with the miR-138 mimics exhibited significantly decreased invasion ability (Fig. 2E). These data suggest that miR-138 can increase the proliferation, promote invasion and inhibit apoptosis of glioma cells.
Figure 2.
(A) RT-qPCR analysis demonstrated that the expression level of miR-138 in U87 and U251 glioma cells was significantly higher compared with that in U133 and A172 cells. *P<0.05. (B) RT-qPCR analysis demonstrated that miR-138 expression was significantly upregulated in U87 and U251 cells transfected with miR-138 mimics compared to that in cells transfected with miR-NC. *P<0.05. (C) Effect of miR-138 overexpression on U87 and U251 cell viability, as measured by the MTT assay. *P<0.05. (D) Left panel: Effect of miR-138 overexpression on U87 and U251 cell apoptosis rates. Right panel: The number of apoptotic cells among U87 and U251 cells overexpressing miR-138 was significantly higher compared with that in miR-NC-transfected cells, as measured by flow cytometry. *P<0.05. (E) Left panel: Increased miR-138 expression inhibited glioma cell invasion in the Matrigel assay. Right panel: Data show the mean number of migrated cells 24 h after the cells were seeded in the upper compartment of the chambers. *P<0.05. (F) The association of miR-138 overexpression with alterations in AKT/mTOR signalling and induction of cell apoptosis was evaluated by detection of apoptosis- and invasion-associated protein expression in U87 and U251 cells by western blotting. RT-qPCR, reverse transcription-quantitative PCR.
The antitumour effect of miR-138 is associated with the dephosphorylation of AKT/mTOR
To explore the underlying mechanism through which miR-138 acts as a tumour suppressor, the expression levels of signalling molecules involved in the AKT/mTOR pathway were measured. A marked decrease in the levels of p-AKT and p-mTOR was observed in the U87 and U251 cells transfected with the miR-138 mimics, while the levels of total AKT and total mTOR did not differ between the mimic-transfected and miR-NC-transfected groups (Fig. 2F). In addition, using RT-qPCR analysis, no change was observed in the expression levels of miR-138 following treatment with the AKT inhibitor SH-5 inU87 and U251 cells (Fig. S1), suggesting that miR-138 is an upstream regulator of the AKT/mTOR pathway. Moreover, the levels of the anti-apoptotic protein Bcl-2 and the invasion-associated protein MMP-2 were decreased in the miR-138-transfected U87 and U251 cells (Fig. 2F). These data suggest that the dephosphorylation of AKT/mTOR may mediate the function of miR-138 in glioma cells. Activating miR-138 in glioma cells may promote the dephosphorylation of AKT and mTOR, thereby enhancing apoptosis and inhibiting invasion in these cells by decreasing the levels of anti-apoptotic proteins and invasion-promoting factors.
CREB1 is a novel target of miR-138
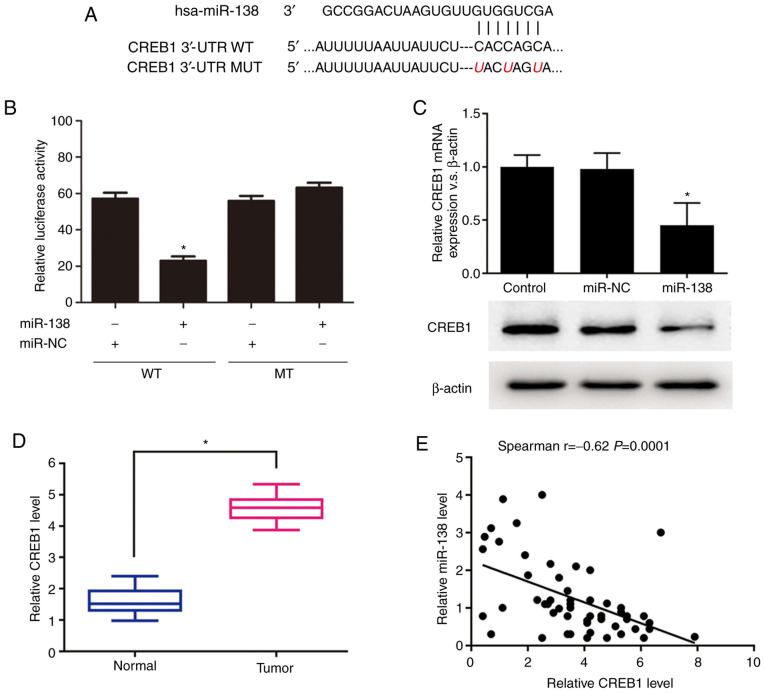
As CREB1 was predicted by TargetScan (http://www.targetscan.org/vert_72/) to contain a putative miR-138 target site in its 3′-UTR, it was hypothesized that CREB1 is a potential target of miR-138. The WT or MUT target site was inserted into identical luciferase reporter vectors (Fig. 3A). The transfection of the reporter vectors containing the putative target sequence resulted in a significant decrease in the relative luciferase activity compared to that in the 293 cells cotransfected with the MUT sequence and miR-138 (Fig. 3B). In addition, the miR-138 transfection significantly decreased both the mRNA and protein expression levels of CREB1 (Fig. 3C). The expression levels of CREB1 and miR-138 were further explored in 48 glioma tissues to investigate their clinical relevance in vivo. The RT-qPCR results revealed that the mRNA expression level of CREB1 was significantly increased in the glioma samples compared to that in the normal brain samples (Fig. 3D). Furthermore, the Spearman's correlation analysis of the mRNA expression levels demonstrated that CREB1 expression was inversely associated with miR-138 expression (r=−0.62, P=0.0001; Fig. 3E). Taken together, these results suggest that CREB1 is a novel target of miR-138.
Figure 3.
(A) Putative miR-138-binding sequences in the CREB1 3′-UTR. (B) A significant decrease in relative luciferase activity was observed following co-transfection of the CREB1 3′-UTR and miR-138 mimics. *P<0.05. (C) RT-qPCR and western blot analyses of CREB1 expression levels after the introduction of the mimic control and miR-138 mimics into 293 cells. *P<0.05. (D) RT-qPCR analysis was used to measure the miRNA expression levels of CREB1 in human glioma and normal samples. *P<0.05. (E) Correlation between CREB1 mRNA expression levels and miR-138 expression levels in human glioma tissues. P=0.00001. CREB1, cAMP response element-binding protein 1; UTR, untranslated region; RT-qPCR, reverse transcription-quantitative PCR.
CREB1 functions as an oncogene in glioma
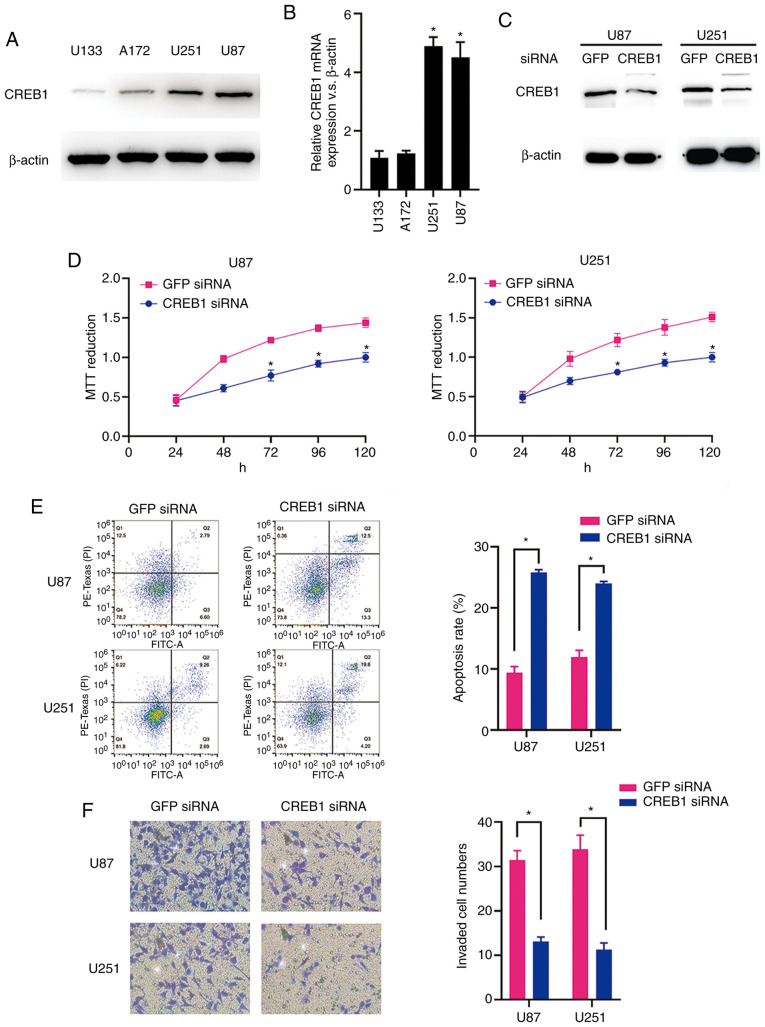
The CREB1 mRNA and protein expression levels in the glioma cell lines (U87, U251, A172 and U133) were measured by RT-qPCR analysis and western blotting. As predicted, the U87 and U251 cells exhibited significantly higher expression levels of CREB1 mRNA and protein compared with the A172 and U133 cells (Fig. 4A and B). The western blot analysis results revealed that CREB1 protein expression was significantly suppressed in both cell types following transfection of siRNA targeting CREB1 (Fig. 4C). The MTT assay results demonstrated that the downregulation of CREB1 significantly suppressed the viability of U87 and U251 cells (Fig. 4D). In addition, the downregulation of CREB1 promoted apoptosis of U87 and U251 cells, as demonstrated by the flow cytometric analysis (Fig. 4E). Furthermore, compared to the cells transfected with control siRNA, the knockdown of CREB1 significantly inhibited the invasion of U87 and U251 cells transfected with CREB1 siRNA, as indicated by the Transwell assay results (Fig. 4F). These data suggest that CREB1 acts as a critical oncogene in glioma cells in vitro.
Figure 4.
(A) CREB1 protein expression levels were measured in the U87, U251, U133 and A172 cell lines by western blotting. (B) CREB1 mRNA expression levels in the four glioma cell lines were measured by RT-qPCR analysis. *P<0.05. (C) CREB1 expression was significantly decreased in U87 and U251 cells transfected with CREB1-siRNA, as assessed by western blotting. (D) Effect of CREB1 knockdown on U87 and U251 cell viability, as measured by the MTT assay. *P<0.05. (E) The number of apoptotic U87 and U251 cells transfected with CREB1-siRNA was significantly higher compared with that of U87 and U251 cells transfected with control siRNA, as measured by flow cytometry. *P<0.05. (F) CREB1 inhibition in U87 and U251 cells by transfection of CREB1-siRNA decreased cell invasion compared with that in GFP-siRNA-transfected cells, as assessed by Matrigel assay. *P<0.05. CREB1, cAMP response element-binding protein 1; RT-qPCR, reverse transcription-quantitative PCR.
Rescue of CREB1 expression overcomes the antitumour effects of miR-138 through the AKT/mTOR signalling pathway
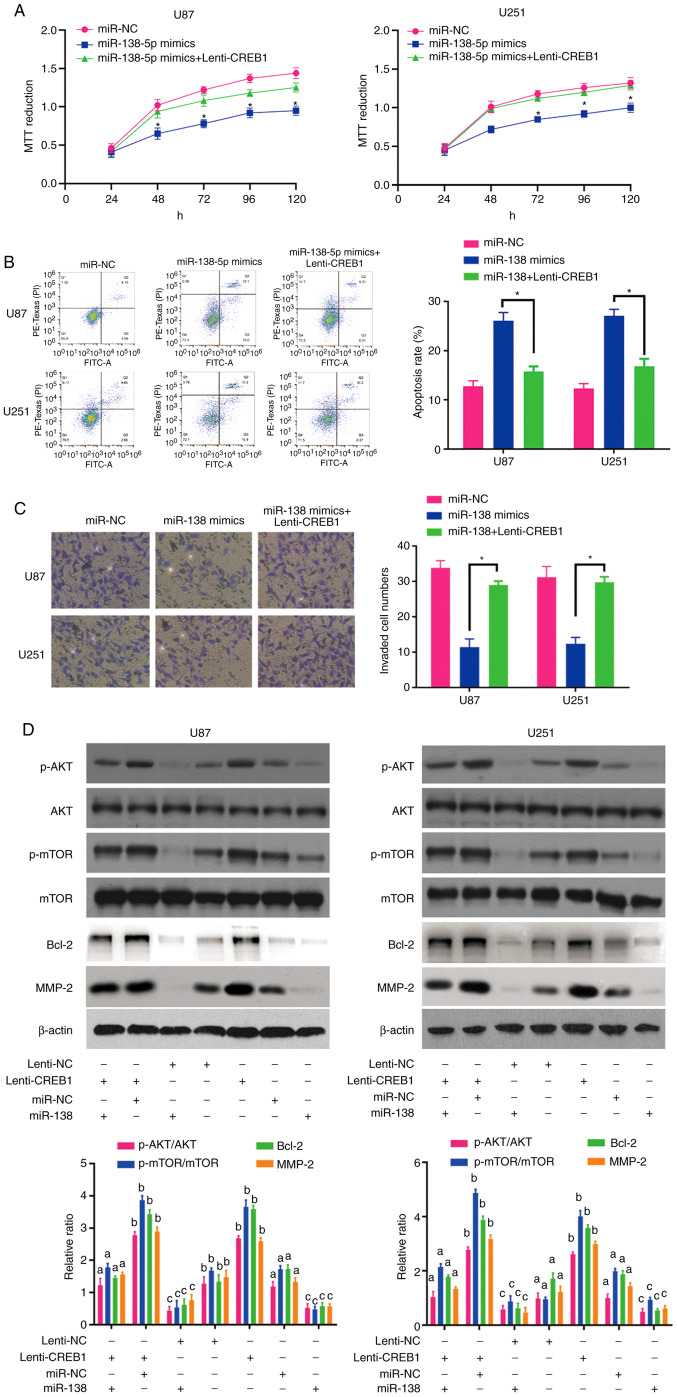
To confirm that CREB1 functions as a critical downstream target of miR-138, rescue experiments were performed by culturing miR-138 mimic-transfected U87 and U251 cells in the presence of purified CREB1. The results of the MTT, apoptosis and invasion assays revealed that the upregulation of CREB1 restored the viability (Fig. 5A), apoptosis (Fig. 5B) and invasion (Fig. 5C) of the miR-138 mimic-transfected glioma cells, suggesting that CREB1 plays an important role in mediating the antitumour effects of miR-138 in glioma cells.
Figure 5.
(A) MTT assay of U87 and U251 cells co-transfected with miR-138 mimics and CREB1 expression vector or control vector. *P<0.05. (B) Apoptosis assay of U87 and U251 cells co-transfected with miR-138 mimics and CREB1 expression vector or control vector. *P<0.05. (C) Invasion assay of U87 and U251 cells co-transfected with miR-138 mimics and CREB1 expression vector or control vector. *P<0.05. (D) CREB1 overexpression abolished the effect of miR-138 on AKT/mTOR dephosphorylation without affecting the levels of total AKT and mTOR. miR-138 suppressed Bcl-2 and MMP-2 protein expression through CREB1. Mean ± SD (n=3) values marked with different letters are significantly different. *P<0.05. CREB1, cAMP response element-binding protein 1.
The association between miR-138 and CREB1 in glioma cells prompted us to explore the activation of the AKT/mTOR signalling pathway and the expression of Bcl-2 and MMP-2 in response to changes in CREB1 expression. After increasing the expression of CREB1 in U87 and U251 cells via Lenti-CREB1, the expression levels of MMP-2 were increased in these cells (Fig. 5D). In addition, CREB1 overexpression activated the AKT/mTOR signalling pathway and enhanced Bcl-2 expression by increasing the levels of p-AKT and p-mTOR, without altering the levels of total AKT and mTOR (Fig. 5D). In particular, the upregulation of CREB1 drastically abrogated the decreases in AKT/mTOR activity and Bcl-2 expression in cells overexpressing miR-138 (Fig. 5D). Furthermore, CREB1 transfection rescued the inhibitory effect of miR-138 on MMP-2 expression in U87 and U251 cells (Fig. 5D). These data indicate that miR-138 likely mediates apoptosis and invasion of glioma cells by targeting CREB1, thus suppressing MMP-2 expression and AKT/mTOR signalling pathway activation.
Discussion
miRNAs have been reported to participate in several malignant biological behaviours, including cell proliferation, metabolism and differentiation, at the post-transcriptional level (23,24). Mutations affecting miRNAs or their functional interactions with oncogenes and tumour suppressors may mediate tumourigenesis (25). Recent studies have indicated that miR-138 may act as a tumour suppressor in various cancers, such as ATC, NSCLC and OSCC (7,9,10). Notably, certain studies demonstrated that miR-138 inhibits GBM cell proliferation by suppressing the EZH2-CDK4/6-pRb-E2F1 signalling loop and suppresses low-grade glioma development and metastasis by regulating IGF2BP2 (19,26). However, Chan et al demonstrated that miR-138 acts as an oncogene in glioma stem cells (GSCs) (27). These contradictory results suggest that the molecular function of miR-138 in glioma remains elusive. A preliminary miRNA microarray analysis of 5 high-grade glioma specimens and 5 normal brain tissue specimens was recently conducted and revealed a markedly lower level of miR-138 expression in high-grade glioma samples (data not shown). These results led to the hypothesis that miR-138 may be used as a prognostic marker of GBM. The present study demonstrated that miR-138 expression was lower in the GBM samples and was inversely associated with advanced pathological stage. These results are consistent with those reported by previous studies, indicating that miR-138 may play an important role in inhibiting the proliferation and malignant behaviour of tumour cells. In addition, the statistical analysis revealed that glioma patients with low expression levels of miR-138 had a markedly longer OS, indicating that miR-138 expression may induce less aggressive tumour behaviour. Thus, the results of the present study indicate that miR-138 expression is inversely associated with pathological grade and that high miR-138 expression is a prognostic factor for a more favourable outcome.
The cellular origin of gliomas remains a controversial topic in cancer research. Recent studies involving animal models indicate that the two different lineages of tumour-initiating cells may contribute to the different biological and genomic phenotypes of GBM (28). Notably, certain miRNAs, such as miR-92a-3p, miR-221, miR-222 and miR-21, exert opposite effects on glioma cells and GSCs (29,30). This obvious paradox may be attributed to the fact that miRNAs may target several important downstream signal molecules, some of which may have opposite functions, which may partially explain why miR-138 could act as either an onco-miR in GSCs (27) or a tumour suppressor in glioma cells (19,26). The present study confirmed that miR-138 exerts an antitumour effect in glioma cells through the dephosphorylation of AKT/mTOR by targeting CREB1, which should be further investigated in future studies.
Recent studies have reported that miR-138 functions as a tumour suppressor by targeting various downstream genes related to cell growth, autophagy, invasion and apoptosis (5,6,10). Moreover, other recent studies have suggested that miR-138 may play an even broader role. Wang et al revealed that miR-138 participates in the process of DNA damage, which affects cell sensitivity to DNA-damaging agents (21). In addition, miR-138 has been confirmed to participate in crosstalk with other pathways (31,32). Hrdlickova et al observed that miR-138 can inhibit TCF7, MSI1 and PAX5, which are crucial target genes of the Wnt signalling pathway involved in the regulation of cell proliferation (33). As miR-138 expression has been demonstrated to be inversely correlated with aggressive behaviour in other tumours, a similar correlation may be expected in GBM. Consistent with this hypothesis, cultured U87 and U251 cells were used to demonstrate the tumour-suppressive role of miR-138, which attenuated the aggressive nature of GBM. Treatment with miR-138 resulted in significant decreases in the levels of p-AKT2 and p-mTOR, without affecting the levels of total AKT2 and mTOR, which is consistent with the results of a previous study suggesting that miR-138 can decrease cell proliferation ability by attenuating the phosphorylation of its target gene. In addition, the AKT inhibitor SH-5 did not affect the expression levels of miR-138 in U87 and U251 glioma cells. Given the importance of miR-138 in AKT/mTOR signalling, it is reasonable to hypothesise that agents specifically targeting miR-138 may be beneficial in GBM patients, particularly those with GBMs exhibiting a low expression of miR-138.
CREB1 produces oncogenic effects by enhancing several signalling pathways in human cancer cells (13). Notably, a recent study revealed that the knockdown of CREB1 decreased the phosphorylation of IRK1/2 and AKT in U251 glioma cells (34). AKT serves as a critical downstream effector of the PI3K pathway. Approximately 88% of gliomas have somatic alterations in RTK/PI3K pathway genes, highlighting the importance of AKT in glioma (35). The findings of the present study indicated that CREB1 acts as an oncogene in glioma cells and that the overexpression of CREB1 can reverse the antitumour effects of miR-138 treatment on glioma cells. To address the molecular mechanisms underlying miR-138-mediated cell proliferation, CREB1, AKT and mTOR expression was further explored and it was observed that miR-138 can directly bind the 3′-UTR of CREB1 and inhibit the phosphorylation of AKT and mTOR without affecting the total expression levels of AKT and mTOR, forming a miR-138/CREB1/AKT/mTOR interactive network that plays an important role in promoting proliferation and suppressing apoptosis of GBM cells. The aforementioned results are also consistent with those of a previous study demonstrating that the C/EBPβ responsive element is essential for miR-138 to participate in the development of gliomas (36). The present study strongly suggests that the significant suppression of glioma cell proliferation induced by miR-138 treatment may be attributed to an enhancement of AKT/mTOR-induced apoptosis.
A hallmark of malignant glioma cells is their highly invasive nature (37). The disruption of the extracellular matrix (ECM) has been identified as an initial step in the penetration of glioma cells into adjacent normal brain tissue (38). Emerging evidence indicates that MMP-2, which is an important member of the MMP family, is required for the degradation of the physical ECM barrier at tumour invasion fronts during this process (39,40). Notably, the expression level of MMP-2 increases as the pathological stage advances and is the highest in malignant gliomas (39). In particular, MMP-2 is a well-documented candidate predictive biomarker of glioma invasion (41). These results indicate that MMP-2 may be strongly associated with tumour invasion. Therefore, the expression level of MMP-2 was measured after miR-138 treatment to elucidate the mechanisms underlying GBM cell invasion. It was demonstrated that the upregulation of miR-138 expression was correlated with a decrease in MMP-2 expression, suggesting a functional interaction between miR-138 and MMP-2.
In summary, the results of the present study indicate that miR-138 mimic transfection into U87 and U251 cells induced apoptosis via the AKT/mTOR signalling pathway and decreased the invasiveness of GBM cells by decreasing MMP-2 expression. However, the mechanisms through which miR-138 regulates GBM cell apoptosis and invasion require further investigation.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Science Foundation of China (grant no. 81802489), the Shanghai Natural Science Foundation (grant nos. 18ZR1434500, 19ZR1448900 and 18411962500), the Scientific Research Initial Funding of Shanghai Tongji Hospital (grant no. RCQD1704), the Featured Clinical Discipline Project of Shanghai Pudong (grant no. PWYst2018-01) and the Key Discipline Group Construction Project of Shanghai Pudong (grant no. PWZxq2017-02).
Availability of data and materials
The datasets generated and analysed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CZ and QW were responsible for designing the study and writing the manuscript. XZ, LZ and HC were responsible for the data analysis. JQ and QB performed the experiments and revised the manuscript. CL and GH contributed to the design and critically revised the manuscript for important intellectual content YY and JG were involved in acquisition and interpretation of RT-qPCR data. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Fearon C, Loftus T, Byrne AL, Heffernan J, Cooney M, Heeney C, Walsh A, Lorigan J, Beausang A, Cryan J, et al. Impact of the 2016 world health organization classification of tumours of the central nervous system: An Irish experience. Ir J Med Sci. 2020;189:799–803. doi: 10.1007/s11845-019-02144-y. [DOI] [PubMed] [Google Scholar]
- 2.Siegelin MD, Schneider E, Westhoff MA, Wirtz CR, arpel-Massler G. Current state and future perspective of drug repurposing in malignant glioma. Semin Cancer Biol. 2019 Nov 14; doi: 10.1016/j.semcancer.2019.10.018. (Epub ahead of print). doi: 10.1016/j.semcancer.2019.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Tamura R, Miyoshi H, Yoshida K, Okano H, Toda M. Recent progress in the research of suicide gene therapy for malignant glioma. Neurosurg Rev. 2019 Nov 28; doi: 10.1007/s10143-019-01203-3. (Epub ahead of print). doi: 10.1007/s10143-019-01203-3. [DOI] [PubMed] [Google Scholar]
- 4.Song T, Zhang X, Wang C, Wu Y, Cai W, Gao J, Hong B. MiR-138 suppresses expression of hypoxia-inducible factor 1α (HIF-1α) in clear cell renal cell carcinoma 786-O cells. Asian Pac J Cancer Prev. 2011;12:1307–1311. [PubMed] [Google Scholar]
- 5.Islam M, Datta J, Lang JC, Teknos TN. Down regulation of RhoC by microRNA-138 results in de-activation of FAK, Src and Erk1/2 signaling pathway in head and neck squamous cell carcinoma. Oral Oncol. 2014;50:448–456. doi: 10.1016/j.oraloncology.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One. 2016;11:e0150026. doi: 10.1371/journal.pone.0150026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao L, Zhou H, Li XP, Chen J, Fang C, Mao CX, Cui JJ, Zhang W, Zhou HH, Yin JY, Liu ZQ. MicroRNA-138 acts as a tumor suppressor in non small cell lung cancer via targeting YAP1. Oncotarget. 2016;7:40038–40046. doi: 10.18632/oncotarget.9480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Ma F, Zhang M, Gong W, Weng M, Quan Z. MiR-138 suppresses cell proliferation by targeting Bag-1 in gallbladder carcinoma. PLoS One. 2015;10:e0126499. doi: 10.1371/journal.pone.0126499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang B, Mu W, Wang J, Lu J, Jiang S, Li L, Xu H, Tian H. MicroRNA-138 functions as a tumor suppressor in osteosarcoma by targeting differentiated embryonic chondrocyte gene 2. J Exp Clin Cancer Res. 2016;35:69. doi: 10.1186/s13046-016-0348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Pan ZG, Shu L, Li QJ. Podocalyxin-like, targeted by miR-138, promotes colorectal cancer cell proliferation, migration, invasion and EMT. Eur Rev Med Pharmacol Sci. 2018;22:8664–8674. doi: 10.26355/eurrev_201812_16631. [DOI] [PubMed] [Google Scholar]
- 11.Nama S, Muhuri M, Di Pascale F, Quah S, Aswad L, Fullwood M, Sampath P. MicroRNA-138 is a prognostic biomarker for triple-negative breast cancer and promotes tumorigenesis via TUSC2 repression. Sci Rep. 2019;9:12718. doi: 10.1038/s41598-019-49155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stojcheva N, Schechtmann G, Sass S, Roth P, Florea AM, Stefanski A, Stühler K, Wolter M, Müller NS, Theis FJ, et al. MicroRNA-138 promotes acquired alkylator resistance in glioblastoma by targeting the Bcl-2-interacting mediator BIM. Oncotarget. 2016;7:12937–12950. doi: 10.18632/oncotarget.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: Implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009;15:2583–2587. doi: 10.1158/1078-0432.CCR-08-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JQ, Yao QH, Kuang YQ, Ma Y, Yang LB, Huang HD, Cheng JM, Yang T, Liu EY, Liang L, et al. Prognostic value of coexistence of abnormal expression of micro-RNA-200b and cyclic adenosine monophosphate-responsive element-binding protein 1 in human astrocytoma. Hum Pathol. 2014;45:2154–2161. doi: 10.1016/j.humpath.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, Guo X, Zhang P, Qi R, Li Z, Zhang S. Cyclic adenosine monophosphate-responsive element-binding protein activation predicts an unfavorable prognosis in patients with hepatocellular carcinoma. Onco Targets Ther. 2014;7:873–879. doi: 10.2147/OTT.S63594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo HS, Liu DD, Bekele BN, Kim MK, Pisters K, Lippman SM, Wistuba II, Koo JS. Cyclic AMP response element-binding protein overexpression: A feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res. 2008;68:6065–6073. doi: 10.1158/0008-5472.CAN-07-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodon L, Gonzàlez-Juncà A, Inda Mdel M, Sala-Hojman A, Martínez-Sáez E, Seoane J. Active CREB1 promotes a malignant TGFβ2 autocrine loop in glioblastoma. Cancer Discov. 2014;4:1230–1241. doi: 10.1158/2159-8290.CD-14-0275. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Zhang L, Cui Y, Zhang C, Chen H, Gu J, Qian J, Luo C. Increased RLIP76 expression in IDH1 wildtype glioblastoma multiforme is associated with worse prognosis. Oncol Rep. 2020;43:188–200. doi: 10.3892/or.2019.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, Peng Y. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta. 2013;1832:1697–1707. doi: 10.1016/j.bbadis.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Qian J, Wang J, Luo C, Chen J, Hu G, Lu Y. Knockdown of RLIP76 expression by RNA interference inhibits invasion, induces cell cycle arrest, and increases chemosensitivity to the anticancer drug temozolomide in glioma cells. J Neurooncol. 2013;112:73–82. doi: 10.1007/s11060-013-1045-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res. 2011;9:1100–1111. doi: 10.1158/1541-7786.MCR-11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Syed SN, Brune B. MicroRNAs as emerging regulators of signaling in the tumor microenvironment. Cancers (Basel) 2020;12:911. doi: 10.3390/cancers12040911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SiamiGorji S, Jorjani I, Tahamtan A, Moradi A. Effects of microRNAs polymorphism in cancer progression. Med J Islam Repub Iran. 2020;34:3. doi: 10.34171/mjiri.34.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandiford OA, Moore CA, Du J, Boulad M, Gergues M, Eltouky H, Rameshwar P. Human aging and cancer: Role of miRNA in tumor microenvironment. Adv Exp Med Biol. 2018;1056:137–152. doi: 10.1007/978-3-319-74470-4_9. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Liu X, Cheng L, Li L, Wei Z, Wang Z, Han G, Wan X, Wang Z, Zhang J, Chen C. Tumor suppressor microRNA-138 suppresses low-grade glioma development and metastasis via regulating IGF2BP2. Onco Targets Ther. 2020;13:2247–2260. doi: 10.2147/OTT.S232795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan XH, Nama S, Gopal F, Rizk P, Ramasamy S, Sundaram G, Ow GS, Ivshina AV, Tanavde V, Haybaeck J, et al. Targeting glioma stem cells by functional inhibition of a prosurvival oncomiR-138 in malignant gliomas. Cell Rep. 2012;2:591–602. doi: 10.1016/j.celrep.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Alcantara Llaguno SR, Xie X, Parada LF. Cell of origin and cancer stem cells in tumor suppressor mouse models of glioblastoma. Cold Spring Harb Symp Quant Biol. 2016;81:31–36. doi: 10.1101/sqb.2016.81.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aldaz B, Sagardoy A, Nogueira L, Guruceaga E, Grande L, Huse JT, Aznar MA, Díez-Valle R, Tejada-Solís S, Alonso MM, et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS One. 2013;8:e77098. doi: 10.1371/journal.pone.0077098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song H, Zhang Y, Liu N, Zhao S, Kong Y, Yuan L. MiR-92a-3p exerts various effects in glioma and glioma stem-like cells specifically targeting CDH1/β-catenin and Notch-1/Akt signaling pathways. Int J Mol Sci. 2016;17:1799. doi: 10.3390/ijms17111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sha HH, Wang DD, Chen D, Liu SW, Wang Z, Yan DL, Dong SC, Feng JF. MiR-138: A promising therapeutic target for cancer. Tumour Biol. 2017;39:1010428317697575. doi: 10.1177/1010428317697575. [DOI] [PubMed] [Google Scholar]
- 32.Huang H, Xiong Y, Wu Z, He Y, Gao X, Zhou Z, Wang T. MIR-138-5P inhibits the progression of prostate cancer by targeting FOXC1. Mol Genet Genomic Med. 2020;8:e1193. doi: 10.1002/mgg3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hrdlickova R, Nehyba J, Bargmann W, Bose HR., Jr Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS One. 2014;9:e86990. doi: 10.1371/journal.pone.0086990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng KB, Xie J, Li YT, Yuan Y, Wang Y, Li C, Shi YF. Knockdown of CERB expression inhibits proliferation and migration of glioma cells line U251. Bratisl Lek Listy. 2019;120:309–315. doi: 10.4149/BLL_2019_049. [DOI] [PubMed] [Google Scholar]
- 35.Robison NJ, Kieran MW. Identification of novel biologic targets in the treatment of newly diagnosed diffuse intrinsic pontine glioma. Am Soc Clin Oncol Educ Book. 2012;2012:625–628. doi: 10.14694/EdBook_AM.2012.32.190. [DOI] [PubMed] [Google Scholar]
- 36.Di Pascale F, Nama S, Muhuri M, Quah S, Ismail HM, Chan XH, Sundaram GM, Ramalingam R, Burke B, Sampath P. C/EBPβ mediates RNA polymerase III-driven transcription of oncomiR-138 in malignant gliomas. Nucleic Acids Res. 2018;46:336–349. doi: 10.1093/nar/gkx1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vollmann-Zwerenz A, Leidgens V, Feliciello G, Klein CA, Hau P. Tumor cell invasion in glioblastoma. Int J Mol Sci. 2020;21:1932. doi: 10.3390/ijms21061932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrer VP, Moura Neto V, Mentlein R. Glioma infiltration and extracellular matrix: Key players and modulators. Glia. 2018;66:1542–1565. doi: 10.1002/glia.23309. [DOI] [PubMed] [Google Scholar]
- 39.Zhou W, Yu X, Sun S, Zhang X, Yang W, Zhang J, Zhang X, Jiang Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed Pharmacother. 2019;118:109369. doi: 10.1016/j.biopha.2019.109369. [DOI] [PubMed] [Google Scholar]
- 40.Kast RE, Halatsch ME. Matrix metalloproteinase-2 and −9 in glioblastoma: A trio of old drugs-captopril, disulfiram and nelfinavir-are inhibitors with potential as adjunctive treatments in glioblastoma. Arch Med Res. 2012;43:243–247. doi: 10.1016/j.arcmed.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Chintala SK, Tonn JC, Rao JS. Matrix metalloproteinases and their biological function in human gliomas. Int J Dev Neurosci. 1999;17:495–502. doi: 10.1016/S0736-5748(99)00010-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the present study are available from the corresponding author on reasonable request.