Abstract
Various studies have revealed that the Hedgehog (Hh) signaling pathway promotes ovarian cancer invasion, migration and drug resistance. Previous studies by our group have identified a set of genes, including multidrug resistance gene 1 (MDR1), that are regulated by Hh signaling in ovarian cancer. However, the association between Hh signaling activation and MDR1 expression requires further validation. In the present study, reverse transcription-quantitative PCR or western blot assays were used to evaluate the mRNA and protein expression levels of MDR1, Sonic Hh (Shh), glioma-associated oncogene 2 (Gli2), Gli1 and γ-phosphorylated H2A.X variant histone (γ-H2AX). MTT and colony-formation assays were performed to determine the effect of cisplatin (DDP) after inhibiting the Hh pathway in ovarian cancer cells. The results indicated that MDR1, Gli2 and Shh levels were much higher in SK-OV-3 cells with acquired DDP resistance than in native SK-OV-3 cells. ES-2 cells with overexpression of Gli2 were capable of efficiently forming colonies in the presence of low DDP concentrations. By contrast, Gli2 knockdown in SK-OV-3 cells decreased the colony-forming ability under the same concentration of DDP. As determined by MTT assays, knockdown of Gli2 or targeting of the Hh signaling pathway with either Gli-antagonist 61 (GANT61) or cyclopamine, in combination with DDP treatment, diminished the viability of ES-2 and SK-OV-3 cells, whereas Gli2 overexpression increased the viability of ES-2 cells in the presence of DDP. Knockdown of Gli2 or targeting the Hh signaling pathway with GANT61 also increased γ-H2AX levels but decreased the expression of MDR1 in the presence of DDP. MDR1 expression is regulated by the Hh signaling pathway and is likely a downstream transcription factor of Gli2. In conclusion, targeting the Hh signaling pathway increases the sensitivity of ovarian cancer to DDP. MDR1 is a target gene of the Hh signaling pathway and this pathway may affect chemoresistance of ovarian cancer to DDP via MDR1.
Keywords: hedgehog signaling pathway, ovarian cancer, multidrug resistance protein 1, chemotherapy resistance
Introduction
Ovarian cancer is the most fatal type of tumor of the female genital tract (1). Although surgical techniques and chemotherapy have been improved in recent years, the prognosis for this disease remains poor. According to the GLOBOCAN statistics for 2018, the mortality rate (4.4%) was higher than the rate of new cases (3.4%) in 185 countries (2). This may be due to recurrence and chemotherapy resistance. Therefore, it is important to explore the mechanisms of drug resistance.
The Hedgehog (Hh) gene was first reported in 1980 (3). There are three highly conserved ligand proteins: Sonic Hh (Shh), Indian Hh and Desert Hh (4). The glioma-associated oncogene 1 (Gli) gene family comprises three members: Gli1, Gli2 and Gli3. Gli1 is a target gene of Gli2. The Hh signaling pathway is suppressed in the absence of ligand binding to the transmembrane protein receptor Patched (Ptch). Ligand binds to Ptch, releasing Ptch-mediated suppression of Smoothened (Smo), which translocates into the primary cilium (PC) with Gli, Suppressor of Fused (Sufu) and kinesin family member 7; then, activated Smo induces Sufu to release Gli, which results in Hh signaling pathway activation (5). The Hh signaling pathway has an important role in embryogenesis, as well as in the initiation and development of basal-cell carcinoma (6), lung cancer (7) and ovarian cancer (8).
Several studies have indicated that the Hh signaling pathway correlates with chemotherapy resistance. Song et al (9) revealed widespread expression of Smo, Gli1 and Ptch in ovarian tumors. In addition, the expression of Smo and Gli1 in the cisplatin (DDP)-resistant A2780 cells (A2780/DDP) was significantly higher than that in native A2780 cells. Steg et al (10) suggested that smoothened antagonists are able to reverse taxane resistance in ovarian cancer. LDE-225, an Smo inhibitor, increased the sensitivity of chemotherapy-resistant ovarian cancer cells to paclitaxel by downregulating multidrug resistance protein 1 (MDR1) expression. In addition, Bcl2 (11,12) and forkhead box M1 (13,14), which are target genes of the Hh signaling pathway, are significantly associated with DDP resistance in ovarian cancer and result in poor outcomes. Furthermore, the Hh signaling pathway induces drug resistance via DNA damage repair (15), DNA methylation (16) and epithelial-mesenchymal transition (17). Cancer stem cells (CSCs) and the stem cell signaling pathway are associated with chemotherapy resistance (18). However, the mechanisms of the chemoresistance associated with the Hh signaling pathway have remained elusive. Therefore, it is important to explore the mechanistic roles of the Hh signaling pathway in the drug resistance of ovarian cancer.
MDR1/P-glycoprotein, which is encoded by the ATP-binding cassette (ABC) subfamily B member 1 gene, is a member of the ABC transporter family. This molecular efflux pump eliminates drugs from cancer cells to reduce xenobiotic molecule accumulation, resulting in drug resistance (19). A large number of studies have revealed that MDR1 expression is positively correlated with poor prognosis and drug resistance (20,21). In addition, MDR1 knockdown reverses paclitaxel resistance in ovarian CSCs. Cui et al (22) suggested that blocking the Hh pathway increases glioma cell sensitivity to chemotherapy by downregulating the expression of MDR1, multidrug resistance protein 1, major vault protein, O6-methylguanine-DNA methyltransferase, Bcl-2 and survivin genes.
In the present study, the role of the Hh signaling pathway in DDP resistance in ovarian cancer was investigated. Steg et al (18) revealed that knockdown of Gli2 diminished the viability of ES2 cells. Increased sensitivity to DDP was noted in ES2 cells transfected with Gli2 small interfering (si)RNAs, but not Gli1 siRNAs. This may indicate Gli2 has an important role in drug resistance in ovarian cancer. A complementary (c)DNA microarray revealed that MDR1 may be a target gene. To date, no previous study has assessed the promotion of DDP resistance in ovarian cancer via the Hh pathway based on the transcription factor Gli2 and MDR1, to the best of our knowledge. Thus, the present study focused on Gli2, which has been indicated to influence the proliferation and DNA damage repair and correlate with resistance to chemotherapy in ovarian cancer. The present study explored whether MDR1 is regulated by Gli2 to promote drug resistance in ovarian cancer.
Materials and methods
Reagents and antibodies
MTT, DMSO and protease inhibitor (cat. no. P8340) were purchased from Sigma-Aldrich (Merck KGaA). Puromycin (cat. no. P8230-25) was purchased from Solarbio. Doxycycline was purchased from Sangon Biotech. Lipofectamine 2000 (cat. no. 11668-019), TRIzol (cat. no. 15596016), penicillin/streptomycin (cat. no. 15140122) and a bicinchoninic acid (BCA) protein assay kit (cat. no. 23225) were purchased from Thermo Fisher Scientific, Inc. Cyclopamine (cat. no. S1146) was purchased from Selleck and Gli-antagonist 61 (GANT61; cat. no. HY-13901) was purchased from MedChem Express. Anti-Gli1 primary antibody (cat. no. 2643S) was purchased from Cell Signaling Technology, Inc. (CST) and anti-MDR1 primary antibody (cat. no. ab170904) was obtained from Abcam. Anti-phospho-histone H2A.X (Ser139) primary antibody (cat. no. 2577) was purchased from CST and anti-GAPDH antibody (cat. no. MAB374) was purchased from EMD Millipore. Verapamil, an MDR1 inhibitor, was purchased from Sigma Aldrich (Merck KGaA).
Cell lines and culture
The human ovarian cancer cell lines SK-OV-3 and ES-2 were purchased from the Cell Bank of the Chinese Academy of Sciences between 2010 and 2013. The SK-OV-3 cell line, a hypodiploid cell line, was established from an adenocarcinoma tumor. The ES-2 cell line with a complex hyperdiploid karyotype of 66XX to 88XX was established from a surgical clear-cell carcinoma specimen. The cells were authenticated in December 2017. 293T cells were purchased from the American Type Culture Collection between 2010 and 2015. SK-OV-3 and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; cat. no. C11995500BT) supplemented with 10% FBS (cat. no. 04-001-1A; Biological Industries) and 1X penicillin/streptomycin. ES-2 cells were cultured in McCoy's 5A medium (cat. no. 01-075-1AS; BI) supplemented with 10% FBS and 1X penicillin/streptomycin. All cells were cultured in a 37°C in an atmosphere with 5% CO2. Transient cell transfection was performed with Lipofectamine 2000 or PEI according to the manufacturer's protocol.
Complementary (c)DNA microarray analysis
SKOV3 cells were treated with GANT61 or DMSO. RNA was isolated with TRIzol. Double-stranded cDNA and biotin-labeled cRNA were synthesized from RNA (300 ng) using an IlluminaH TotalPrep RNA Amplification Kit (Ambion Inc.) according to the manufacturer's protocol. Each biotinylated cRNA (750 ng) was hybridized to an Illumina Human HT expression BeadChip V4 (Illumina Inc.). Following standard washing steps and staining with StreptavidinCy3, the arrays were scanned using an IlluminaH BeadArray Reader. Genes with a DiffScore of <200 or >20 (i.e. P<0.01) were considered as the differentially expressed genes (23).
Lentivirus (LV) transfection
A Lenti-X-small hairpin (sh)RNA Tet-On system (pGV307-red fluorescent protein) comprising shRNA-Gli2 targeting the sequence 5′-TCCTGAACATGATGACCTA-3′ of Gli2 and an LV (SL) system (GV356-enhanced green fluorescence protein) comprising LV-activated Gli2 (Gli2A; GenBank accession no. NM_005270) were constructed and packaged by GeneChem. LV-shRNA-control (shControl) targeted the sequence 5′-TTCTCCGAACGTGTCACGT-3′. SK-OV-3 cells seeded in a 24-well dish at 30% density were infected by shGli2 LV [1×108 transfection units (TU)/ml] or shControl LV (1×108 TU/ml), which were diluted by enhanced infection solution (GeneChem), and 5 µg/ml of polybrene (GeneChem) was used to enhance the transfection efficiency. After 12 h of transfection, the medium was replaced with fresh medium. After 48 h, the cells were cultured in a 12-well dish. ES-2 cells seeded in a 24-well dish at 30% density were infected with LV-Gli2A (5×108 TU/ml) or LV-control (8×108 TU/ml), which were diluted by enhanced infection solution (GeneChem), and 5 µg/ml of polybrene (GeneChem) was used to enhance the transfection efficiency. After 12 h of transfection, the medium was replaced by fresh medium. After 48 h, the cells were cultured in a 12-well dish. Cells were selected with puromycin (2 µg/ml; Sangon Biotech) and doxycycline (2 µg/ml; Sangon Biotech). The cell overexpression/knockdown efficiency was confirmed by reverse transcription-quantitative PCR (RT-qPCR) and western blot analyses (Fig. S1A and B) (24).
Cell transfection
pCMV6-Entry-Gli2-myc (cat. no. RC217291) containing human Gli2 mRNA (GenBank accession no. NM_005270) and empty vector (cat. no. PS100001) were obtained from OriGene. The first 984 bases of Gli2 mRNA were deleted to generate an active form of Gli2, Gli2A (25). pUB6/V5-hisB-Gli1 was obtained from Dr Shiwen Luo and Dr Yong Li (Center for Experimental Medicine, the First Affiliated Hospital of Nanchang University, Nanchang, China) who constructed it according to a published protocol (26). pUB6/V5-hisB vector (cat. no. V25020) was obtained from Invitrogen (Thermo Fisher Scientific, Inc.). SK-OV-3 cells were plated in 6-well plates at a confluency of 60% and cultured overnight. 2 µg of pCMV6-Entry-Gli2-myc plasmid, pUB6/V5-hisB-Gli1 plasmid or empty vector was transfected into SK-OV-3 cells using Lipofectamine 2000 (cat. no. 11668-019; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The medium was replaced prior to transfection. After 3–6 h of transfection, the medium containing Lipofectamine was replaced by fresh medium. After 24–48 h, the cells were harvested for further validation and investigations.
Secretory Flag-tagged N-terminal Shh domain (N-Shh)-conditioned medium
An expression plasmid encoding the N-Shh, the secreted segment of the Shh ligand with ligand activity, was constructed as previously described by inserting the amino-terminal signaling domain of a human Shh cDNA (cat. no. aa26-184; GenBank accession no. NM_000193.2) into a pFlag-cytomegalovirus (CMV)1 vector (cat. no. E7273; Sigma-Aldrich; Merck KGaA) and the pCMV-Flag1 vector was used as a control (23). 293T cells at 25% density were transfected with the Flag-N-Shh plasmid in a 10-cm dish; 293T cells at 25% density in another 10-cm dish were transfected with the pCMV-Flag1 vector. After ~12 h, the medium was replaced with DMEM supplemented with 2% FBS. All cells were cultured for an additional 24 h and the medium containing secreted N-Shh was then harvested and stored at 4°C (27). Western blot analysis confirmed the presence of N-Shh in the conditioned medium (Fig. S1C). When culturing ES-2 and SK-OV-3 cells, the conditioned Shh medium or the control medium was mixed with an equivalent volume of fresh culture medium supplemented with 5% FBS (24).
Induction of SK-OV-3/DDP cells
SK-OV-3 cells were cultured with different concentrations of DDP for 72 h and the IC50 of DDP was determined using the MTT assay. The cells were then cultured in the presence of DDP at the IC50 value. Every month, a new IC50 was detected. When the new IC50 value was significantly greater than the previous one, the cells were cultured under DDP at the new IC50 value. After 6 months, SK-OV-3/DDP cells were fully induced by DDP from SK-OV-3 cells. The resistance index was calculated as the IC50 value of the resistant cells divided by the IC50 value of the native cells. The resistance index of the SK-OV-3/DDP cells was 3.0 (28) and the characteristics of resistance were maintained by continuous culture with 1.25 µmol/l DDP.
RT-qPCR
Total RNA was isolated from cultured cells using TRIzol reagent. A total of 2 micrograms of RNA was reverse-transcribed to cDNA using a PrimeScript RT Reagent kit (cat. no. RRO47A; Takara Bio, Inc.) according to the manufacturer's protocols. Gene expression was then analyzed in these samples using the Applied Biosystems Step One Plus™ Real-Time PCR Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR mixture contained the following: SYBR® Premix Ex Taq™ II (2X) 2 µl, PCR forward primer (10 µM) 0.8 µl, PCR reverse primer (10 µM) 0.8 µl, ROX reference dye (50X) 0.4 µl, cDNA (taken from the RT product mixture) 2 µl, deionized H2O 6 µl. The real-time PCR conditions were as follows: Activation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec, primer annealing and extension at 60°C for 30 sec and a ramp up back to 95°C. The quantification cycle (Cq) values for each gene, normalized to the expression levels of GAPDH, were calculated by the 2−∆∆Cq method (29). The respective forward and reverse primer sequences for real-time PCR were as follows: MDR1, 5′-TGCTCAGACAGGATGTGAGTTG-3′ and 5′-TAGCCCCTTTAACTTGAGCAGC-3′; GAPDH, 5′-CAGGGCTGCTTTTAACTCTG-3′ and 5′-GATTTTGGAGGGATCTCGC-3′; Gli2, 5′-CTCAAGGAAGATCTGGACAGG-3′ and 5′-GATGTGCTCGTTGTTGATGTG-3′; and Gli1, 5′-AGCGTGAGCCTGAATCTGTG-3′ and 5′-CAGCATGTACTGGGCTTTGAA-3′.
Western blot analysis
Total protein was extracted using a buffer [3 M NaCl, 10% Nonidet P-40, 0.5 M EDTANa2, 1 M Tris-HCl (pH 7.5) and 1% protease inhibitor] and the protein concentrations were detected by a BCA protein assay kit (cat. no. 23225; Thermo Fisher Scientific, Inc.). The proteins (20–100 µg) were separated by 8–10% SDS-PAGE for 4 h and then transferred to nitrocellulose membranes (HATF-V0010; EMD Millipore). The following primary antibodies were used: Gli1 (1:500 dilution), MDR1 (1:500 dilution), γ-H2AX (1:1,000 dilution) and GAPDH (1:2,000 dilution). Incubation was performed for 12–16 h at 4°C. The blots were then incubated with a goat anti-rabbit secondary antibody (cat. no. 31460; 1:1,000 dilution; Thermo Fisher Scientific, Inc.) to detect MDR1, Gli1 and γ-H2AX, and with a goat anti-mouse secondary antibody (cat. no. 31430; 1:2,000 dilution; Thermo Fisher Scientific, Inc.) to detect GAPDH; incubation was performed for 4 h at 4°C. The secondary antibodies were conjugated with horseradish peroxidase. Images of the immunoblot bands were captured on X-ray film (Kodak) and then scanned with an Epson scanner (Epson Perfection V700 Photo). Quantified results obtained by densitometric analysis using ImageJ software (v1.48; National Institutes of Health).
MTT and colony formation assays
SK-OV-3 cells were seeded in 96-well plates at 2,000 cells per well. Subsequently, they were treated with different concentrations of DDP (0, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 and 50 µmol/l) with or without cyclopamine (30 µmol/l) for 72 h. Next, the cell viability was assessed using an MTT assay according to the manufacturer's protocol with the optical density measured at 570 mm. SK-OV-3 cells were transfected with shRNA-Gli2 or shControl. After determining stable expression via detection of red fluorescence, the cells were replated in 96-well plates at 2,000 cells per well. The cells were then treated with different concentrations of DDP (0, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 and 50 µmol/l) for 72 h and subsequently, cell viability was assessed using an MTT assay according to the manufacturer's protocol with the optical density measured at 570 mm. Next, the cells were plated in 6-well plates at 500 cells per well. ES-2 cells were treated with 0, 0.1 and 0.2 µmol/l DDP, and SK-OV-3 cells were treated with 0, 0.2, 0.5, 2 and 5 µmol/l DDP; all cells were then cultured for 10–14 days and colonies were counted.
Statistical analysis
All experiments were performed three times. Statistical analysis and figure plotting were performed with GraphPad Prism 8 (GraphPad Software, Inc.). Values are expressed as the mean ± the standard deviation. The results of the MTT assay were analyzed by analysis of variance for repeated measurements. Multi-group comparisons were performed by analysis of variance and post-hoc tests (least-significant differences test). Two different groups of quantitative data were compared by an unpaired t-test.
Results
MDR1, Gli2 and Shh expression is elevated in resistant cells
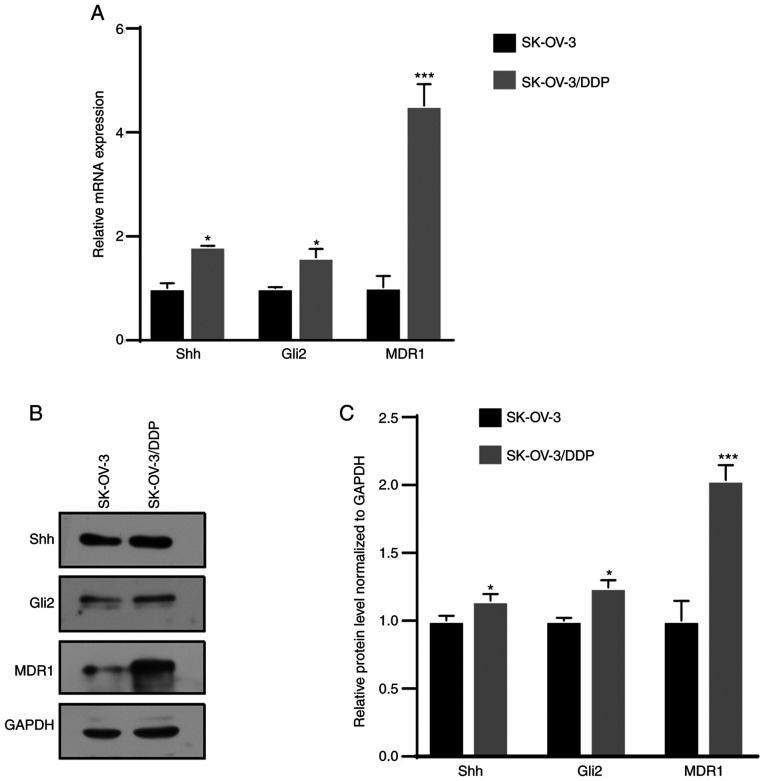
The cDNA microarray revealed that MDR1 was downregulated by GANT61, a Gli inhibitor, in SK-OV-3 cells (Fig. S1D). MDR1 is a well-reported drug resistance gene in ovarian cancer (20). Next, it was attempted to determine whether there was any association between Hh signaling and drug resistance in ovarian cancer. Western blot and RT-qPCR assays revealed that Shh, Gli2 and MDR1 expression was significantly higher in SK-OV-3/DDP cells than in SK-OV-3 cells (Fig. 1). This result indicated that the Hh signaling pathway was aberrantly activated in chemoresistant cells.
Figure 1.
The Hedgehog signaling pathway is aberrantly activated in chemotherapy-resistant cells. (A) Reverse transcription-quantitative PCR assay revealed upregulation of Shh, Gli2 and MDR1 mRNA in SK-OV-3/DDP cells. (B and C) Western blot assay revealed upregulation Shh, Gli2 and MDR1 protein in SK-OV-3/DDP cells. (B) Representative western blot image and (C) quantified results obtained by densitometric analysis using ImageJ software (v1.48). Values are expressed as the mean ± standard deviation (n=3). Data were analyzed by unpaired t-tests. *P<0.05, ***P<0.001 vs. SK-OV-3. SK-OV-3/DDP, SK-OV-3 cell line with acquired cisplatin resistance; Shh, Sonic Hedgehog; Gli2, glioma-associated oncogene 2; MDR1, multidrug resistance protein 1.
Inhibition of Hh signaling or knockdown of Gli2 enhances DDP sensitivity of ovarian cancer cells
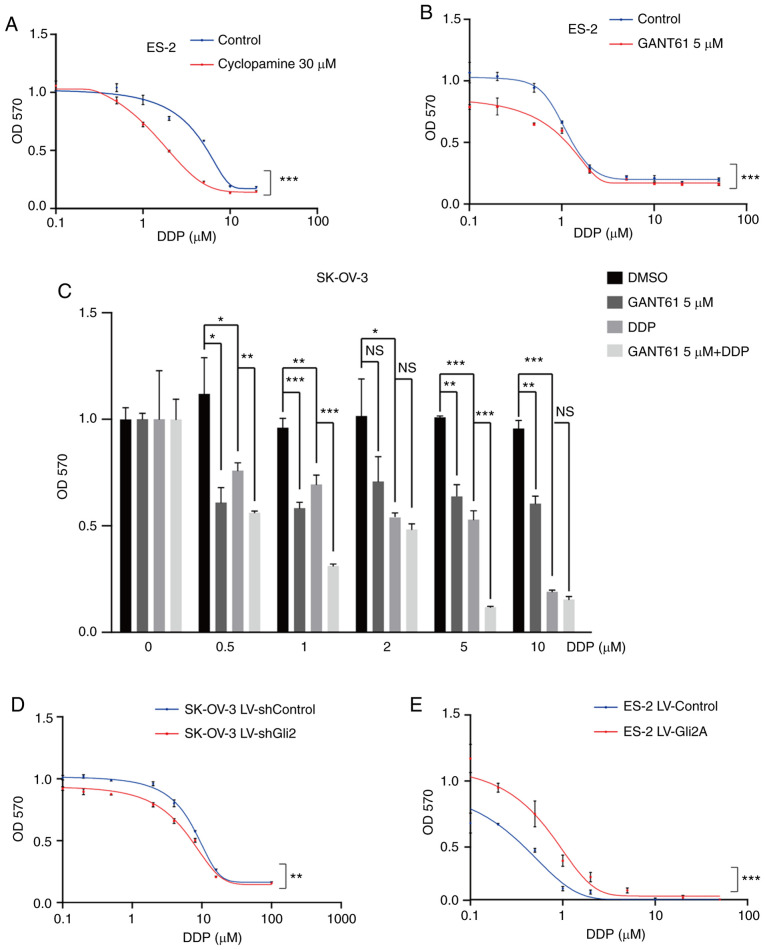
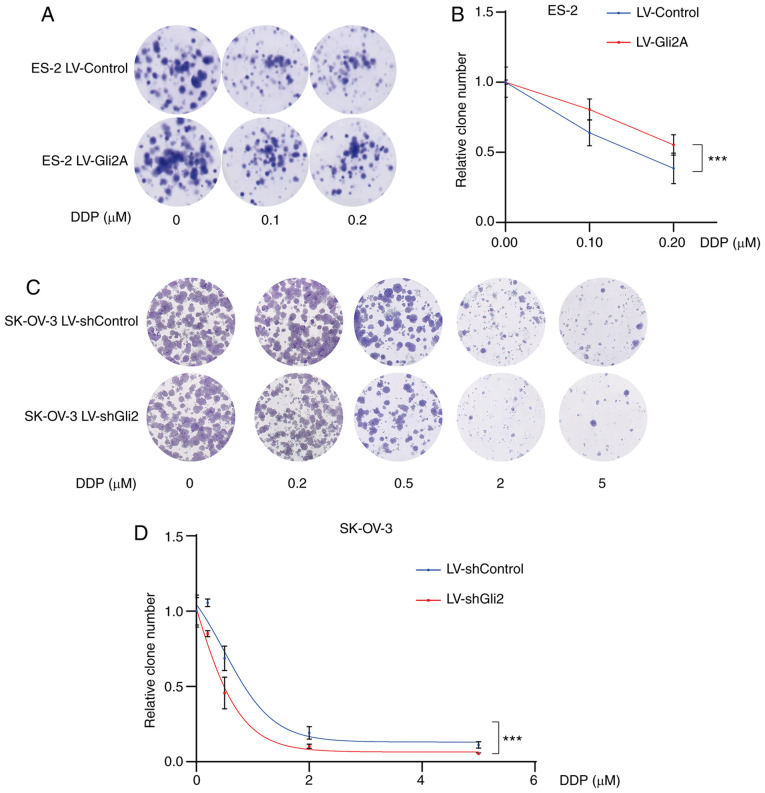
In clinical treatment, most patients are sensitive to DDP during the first chemotherapy, while drug resistance usually occurs after several treatments. Furthermore, SK-OV-3/DDP cells are artificial cells induced by SK-OV-3. Various resistance studies have been performed with native cells (18,30,31). Therefore, native cells were chosen to explore the roles and mechanisms of the Hh pathway in the effectiveness of DDP in ovarian cancer. To explore the association between the Hh signaling pathway and DDP resistance in ovarian cancer, ES-2 cells were treated with cyclopamine (30 µmol/l), an Smo inhibitor, or DMSO (30 µmol/l) and the cell viability was detected using the MTT assay after increasing the DDP concentration. Cells were also treated with GANT61 (5 µmol/l), a Gli antagonist, or DMSO (5 µmol/l). The combination of DDP and cyclopamine or GANT61 resulted in sensitivity to chemotherapy (P<0.001; Fig. 2A and B). Furthermore, treatment of SK-OV-3 cells with GANT61 in combination with DDP was more effective than treatment with either of the drugs alone (Fig. 2C), particularly at 0.5, 1 and 5 µmol/l DDP. These results indicated that targeting the Hh signaling pathway may increase the sensitivity of ovarian cancer cells to DDP. Gli2 expression was much higher in SK-OV-3/DDP cells than in SK-OV-3 cells. As Gli2 is one of the most important transcription factors, its role in DDP resistance was explored. As presented in Fig. 2D, a significant increase in cell death under DDP treatment was observed after knocking down Gli2 in SK-OV-3 cells (P<0.01). By contrast, DDP resistance was observed in ES-2 cells overexpressing Gli2 (P<0.001; Fig. 2E). Similar results were also obtained in the colony formation assays. As compared to the control conditions, Gli2 overexpression in ES-2 cells resulted in a significantly greater number of colonies under the same DDP concentration (Fig. 3A and B), while knockdown of Gli2 in SK-OV-3 cells resulted in fewer colonies under the same DDP concentration (Fig. 3C and D). These results indicated that the Hh signaling pathway has a significant role in DDP resistance.
Figure 2.
Hh signaling pathway inhibition or Gli2 knockdown enhance the effect of DDP on ovarian cancer cells. (A and B) MTT assays revealed that inhibiting the Hh signaling pathway with (A) cyclopamine (30 µmol/l) and (B) GAT61 (5 µmol/l) increased cell death induced by cisplatin in ES-2 cells. (C) MTT assays indicated that treatment with GANT61 in combination with DDP was more effective than treatment with each drug alone in SK-OV-3 cells. (D) MTT assays indicated that Gli2 knockdown decreased the proliferation of SK-OV-3 cells under DDP treatment. (E) Gli2 overexpression increased ES-2 cell viability under cisplatin treatment, as determined by the MTT assay. GraphPad Prism 8 software was used for nonlinear regression (curve fit) measurements. Experiments were performed three times. Data were analyzed by repeated-measures two-factor ANOVA in A, B, D and E, and by ANOVA and post-hoc tests (least-significant differences) in C. *P<0.05, **P<0.01, ***P<0.001. NS, no significance; ANOVA, analysis of variance; OD570, optical density at 570 nm; Hh, Hedgehog; DDP, cisplatin; SK-OV-3 LV-shControl, SK-OV-3 cells transfected with LV plasmid expressing control small hairpin RNA; SK-OV-3 LV-shGli2, SK-OV-3 cells transfected with LV plasmid expressing small hairpin RNA targeting Gli2; Gli2, glioma-associated oncogene 2; LV, lentivirus; GANT61, Gli-antagonist 61.
Figure 3.
Targeting Gli2 affects DDP sensitivity in ovarian cancer cells. (A and B) Colony-formation assays demonstrated a significantly increased number of colonies of ES-2 LV-Gli2 cells compared to control ES-2 cells under cisplatin treatment (0.1, 0.2 µmol/l). (A) Representative images of colonies and (B) quantification of colonies using ImageJ software. There was a statistically significant difference in the number of colonies formed between the Gli2 overexpression and control groups (F=45.995, P<0.05) and between the different doses of DDP (F=62.158, P<0.0001), while the interaction between these terms was not significant. A post-hoc test revealed significant pairwise differences between shControl vs. shGli2, between cisplatin 0 and 0.1 µM, between cisplatin 0 and 0.2 µM, and between cisplatin 0.1 and 0.2 µM. (C and D) Colony formation assays indicated significantly decreased numbers of colonies of SK-OV-3 LV-shGli2 cells compared to control SK-OV-3 cells. (C) Representative images of colonies and (D) quantification of colonies using ImageJ software. There was a statistically significant difference in the number of colonies formed between the Gli2 knockdown and control groups (F=17.653, P<0.001) and between the different doses of DDP (F=139.978, P<0.0001), but the interaction between these terms was not significant. A post-hoc test revealed that all comparisons were significant except for the comparison between DDP 2 and 5 µM. Data were analyzed by two-way analysis of variance. ***P<0.001. DDP, cisplatin; SK-OV-3 LV-shControl, SK-OV-3 cells transfected with LV plasmid expressing control small hairpin RNA; SK-OV-3 LV-shGli2, SK-OV-3 cells transfected with LV plasmid expressing small hairpin RNA targeting Gli2; Gli2, glioma-associated oncogene 2; LV, lentivirus.
Targeting the Hh signaling pathway increases the DNA damage effect of DDP
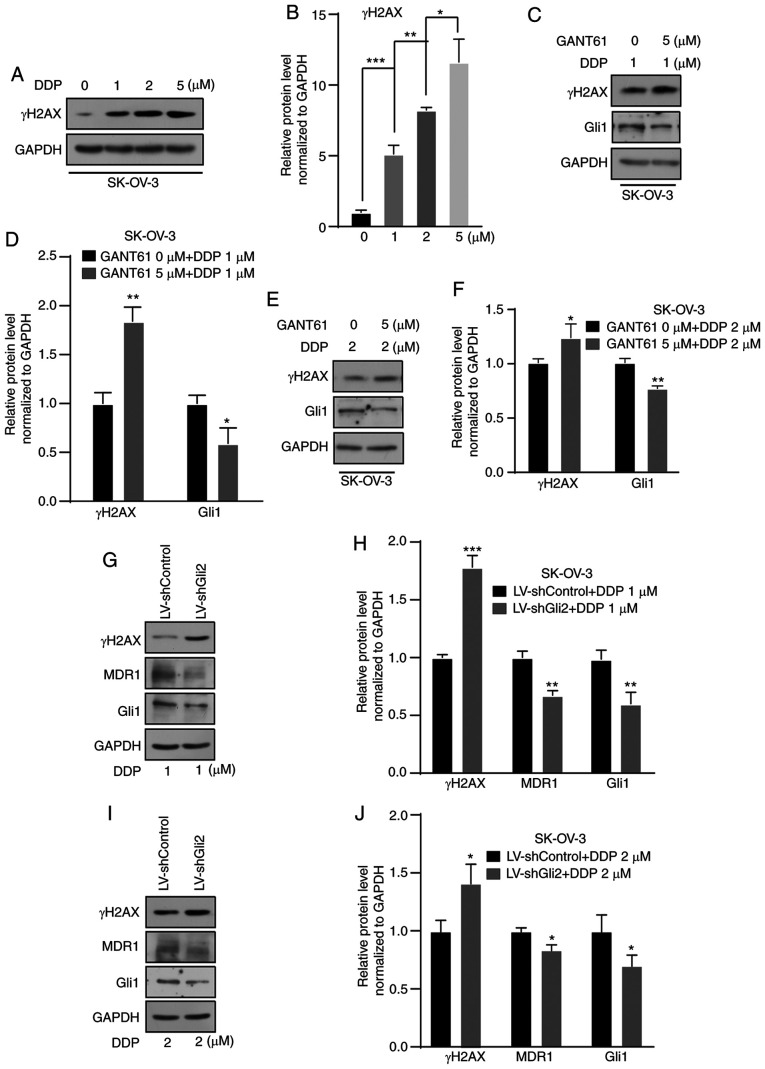
DDP is commonly used to treat ovarian cancer. The mechanism of its anticancer action is cytotoxicity by binding to DNA molecules to form a platinum-DNA adduct (32). γ-H2AX is a DNA damage marker. Therefore, in the present study, changes in the levels of γ-H2AX were detected and the levels were indicated to be increased in a concentration-dependent manner after DDP treatment for 72 h (Fig. 4A and B). An increase in γ-H2AX was observed in SK-OV-3 cells after treatment with GANT61 (5 µmol/l) combined with DDP (1 or 2 µmol/l) for 72 h compared with DDP only (1 or 2 µmol/l) (Fig. 4C-F). After Gli2 was knocked down in SK-OV-3 cells, a further elevation of γ-H2AX under DDP treatment still occurred, but MDR1 expression decreased (Fig. 4G-J). These results indicated that targeting the Hh signaling pathway or Gli2 promoted DNA damage, consequently enhancing the sensitivity of ovarian cancer cells to DDP. Previous studies suggested that MDR1 is a candidate downstream gene of the Hh signaling pathway. In various studies, MDR1 has been significantly correlated with the chemoresistance of ovarian cancer (20,21,33). In the present study, suppression of MDR1 expression was observed (Fig. 4G-J). Therefore, the possible relationship between the Hh signaling pathway and MDR1 expression was further explored.
Figure 4.
Suppression of the Hedgehog signaling pathway increases DNA damage resulting from DDP treatment. (A and B) Western blot analysis indicated an increase in γ-H2AX after treatment with 1, 2 and 5 µmol/l DDP in SK-OV-3 cells. (A) Representative western blot image and (B) quantified results obtained by densitometric analysis using ImageJ software. (C and D) Higher levels of γ-H2AX after treatment with 1 µmol/l DDP in combination with GANT61 (5 µmol/l) was observed in SK-OV-3 cells by western blot analysis. (C) Representative western blot image and (D) quantified results obtained by densitometric analysis. (E and F) Higher levels of γ-H2AX after treatment with 2 µmol/l DDP in combination with GANT61 (5 µmol/l) was observed by western blot assay in SK-OV-3 cells. (E) Representative western blot image and (F) quantified results obtained by densitometric analysis. (G and H) Gli2 knockdown in combination with 1 µmol/l DDP treatment increased γ-H2AX levels, decreased MDR1 levels in SK-OV-3 cells. (G) Representative western blot image and (H) quantified results obtained by densitometric analysis. (I and J) Gli2 knockdown in combination with 2 µmol/l DDP treatment increased γ-H2AX levels, decreased MDR1 levels in SK-OV-3 cells. (I) Representative western blot image and (J) quantified results obtained by densitometric analysis. Gli1 was used as a positive control. Values are expressed as the mean ± standard deviation (n=3). Data were analyzed by unpaired t-tests or analysis of variance and post-hoc tests. *P<0.05, **P<0.01, ***P<0.001. DDP, cisplatin; SK-OV-3 LV-shControl, SK-OV-3 cells transfected with LV plasmid expressing control small hairpin RNA; SK-OV-3 LV-shGli2, SK-OV-3 cells transfected with LV plasmid expressing small hairpin RNA targeting Gli2; Gli2, glioma-associated oncogene 2; γ-H2AX, γ-phosphorylated H2A.X variant histone; MDR1, multidrug resistance protein 1; LV, lentivirus.; GANT61, Gli-antagonist 61.
MDR1 expression is regulated by the Hh signaling pathway via Gli2
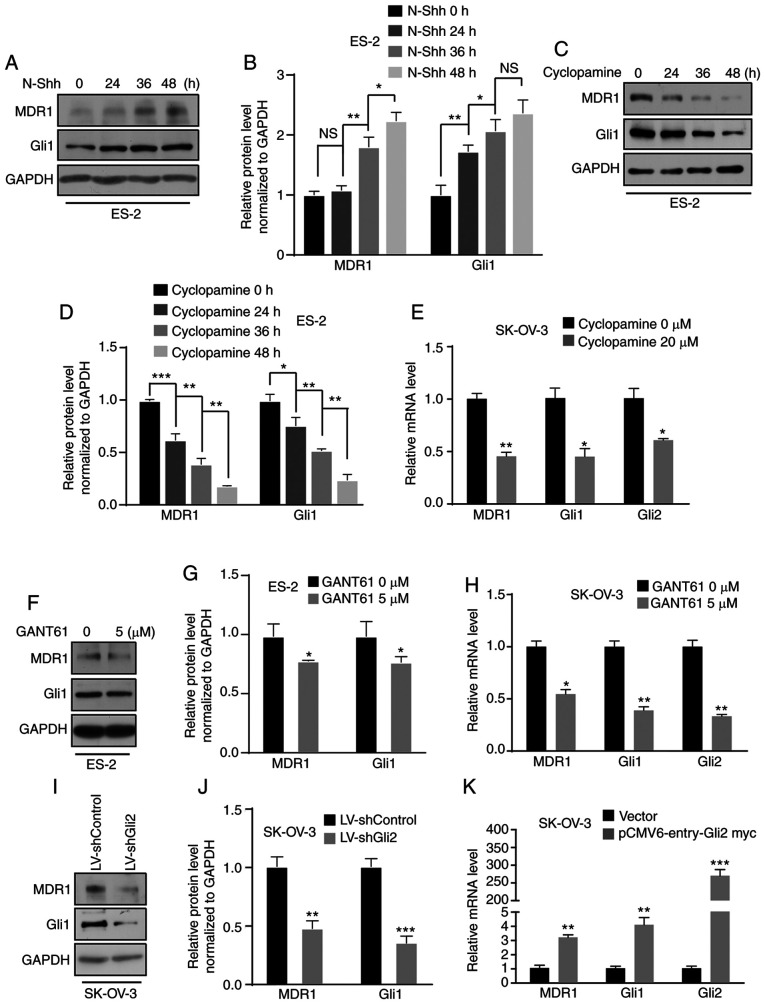
In previous studies by our group, several target genes were explored using a cDNA microarray and it was demonstrated that aberrant activation of the Hh signaling pathway promotes ovarian cancer invasion and migration via target genes such as CD24 (34) and MMP7 (24). The present study focused on MDR1, which is considered a candidate gene. First, ES-2 cells were treated with N-Shh-conditioned media or control media and a time-dependent increase in MDR1 protein levels was observed by western blot analysis (Fig. 5A and B). Furthermore, a time-dependent increase in MDR1 protein expression in SK-OV-3 cells was observed after N-Shh treatment (Fig. S2A and B). ES-2 and SK-OV-3 cells were then treated with cyclopamine and the results indicated that the protein expression of MDR1 was suppressed in a time-dependent manner in ES-2 cells (Fig. 5C and D), and the mRNA level was decreased in SK-OV-3 cells (Fig. 5E). Furthermore, the protein and mRNA levels of MDR1 were decreased in ES-2 and SK-OV-3 cells after GANT61 treatment (Fig. 5F-H; Fig. S2C). Simultaneously, MDR1 expression was downregulated in SK-OV-3 cells after Gli2 knockdown (Fig. 5I and J). By contrast, there was an increase in the level of MDR1 mRNA after transfecting SK-OV-3 cells with the Gli2 overexpression plasmids (Fig. 5K). Regardless, there was no increase in the level of MDR1 mRNA after transfecting SK-OV-3 cells with Gli1 overexpression plasmid (Fig. S2D). These results revealed that MDR1 is regulated by the Hh signaling pathway via Gli2. To define the mechanism by which the Hh pathway promotes DDP resistance via MDR1, ES-2 cells with or without overexpression of Gli2 were treated with verapamil (5 µmol/l), an MDR1 inhibitor, and the cell viability was detected by the MTT assay after increasing the DDP concentration. An increase in cell viability was observed in ES-2 cells overexpressing Gli2 under DDP treatment, but there was no increase in the viability of Gli2-overexpressing ES-2 cells after treatment with verapamil (Fig. S3A). Thus, the Hh signaling pathway promotes chemotherapy resistance via MDR1.
Figure 5.
MDR1 is regulated by the Hedgehog signaling pathway. (A and B) ES-2 cells were incubated with N-Shh for 0, 24, 36 and 48 h. Western blot analysis revealed that MDR1 expression was upregulated. Gli1 was used as a positive control. (A) Representative western blot image and (B) quantified results obtained by densitometric analysis using ImageJ software. (C and D) ES-2 cells were treated with cyclopamine (20 µmol/l) for 0, 24, 36 and 48 h. Western blot assays indicated that MDR1 expression was downregulated. (C) Representative western blot image and (D) quantified results obtained by densitometric analysis. (E) RT-qPCR indicated a reduction in the mRNA level of MDR1 in SK-OV-3 cells treated with cyclopamine (20 µmol/l). Gli1 and Gli2 were used as positive controls. (F and G) ES-2 cells were treated with GANT61 (5 µmol/l). Western blot assays indicated that MDR1 expression was downregulated. (F) Representative western blot image and (G) quantified results obtained by densitometric analysis. (H) RT-qPCR revealed a decrease in the mRNA level of MDR1 in SK-OV-3 cells treated with GANT61 (5 µmol/l). Gli1 and Gli2 were used as positive controls. (I and J) Gli2 knockdown in SK-OV-3 cells resulted in a decrease in MDR1 protein expression. (I) Representative western blot image and (J) quantified results obtained by densitometric analysis. (K) RT-qPCR revealed that Gli2 overexpression in SK-OV-3 cells induced an increase in MDR1 mRNA levels. Values are expressed as the mean ± standard deviation (n=3). Data were analyzed by unpaired t-tests or analysis of variance and post-hoc tests (least-significant difference). *P<0.05, **P<0.01, ***P<0.001 vs. control. NS, no significance; DDP, cisplatin; SK-OV-3 LV-shControl, SK-OV-3 cells transfected with LV plasmid expressing control small hairpin RNA; SK-OV-3 LV-shGli2, SK-OV-3 cells transfected with LV plasmid expressing small hairpin RNA targeting Gli2; Gli2, glioma-associated oncogene 2; N-Shh, N-terminal ‘Hedge’ domain; MDR1, multidrug resistance protein 1; RT-qPCR, reverse transcription-quantitative PCR; LV, lentivirus; GANT61, Gli-antagonist 61.
Discussion
Ovarian cancer has been indicated to be the leading cause of cancer-related death among cancers of the female genital tract (1). DDP and other chemotherapeutic drugs are commonly used to treat ovarian cancer, but the prognosis is poor due to chemotherapy resistance (35). The present study sought to determine ways to reverse DDP resistance.
Hh was first reported in Drosophila melanogaster in 1980 and has an important role in embryogenesis (36). The Hh signaling pathway participates in the invasion, metastasis and drug resistance of a variety of tumor types, including ovarian cancer (37), hepatoma (38), cervical cancer (39) gliomas (22) and gastric cancer (40). Zahreddine et al (41) suggested that Gli1 alone should be sufficient to facilitate UGT1A-dependent glucuronidation of cytarabine and ribavirin and ultimately drug resistance. A Gli1 inhibitor was able to restore drug sensitivity and thereby provide therapeutic benefits. Steg et al (37) indicated that proteasome inhibition, through alteration of microtubule dynamics and Hh signaling, was able to reverse taxane-mediated chemoresistance. Another study revealed that Gli1, Gli2 or Smo knockdown enhances taxane sensitivity (10), which was consistent with the results of the present study. The protein and RNA levels of Gli2, Shh and MDR1 are higher in chemoresistant cell lines than in wild-type ovarian cancer cells, indicating that the Hh signaling pathway may be activated in chemotherapy-resistant ovarian cancer. MTT and colony formation assays confirmed that treatment with a Gli inhibitor (GANT61), treatment with an Smo antagonist (cyclopamine) or Gli2 knockdown increased the sensitivity of ovarian cancer cell lines to DDP. By contrast, Gli2 overexpression promoted the resistance of ES-2 cells to DDP. However, after inhibiting MDR1, overexpression of Gli2 in ES-2 cells did not increase cell viability under DDP. Therefore, it is indicated that the Hh signaling pathway is abnormally activated in chemoresistant ovarian cancer cell lines and that targeting the Hh signaling pathway, Gli2 or MDR1 is able to reverse chemoresistance to DDP.
The mechanisms of DDP resistance are associated with DNA repair alterations and cellular accumulation and drug inactivation (42). Platinum compounds induce cytotoxicity by binding to DNA molecules to form a platinum-DNA adduct, which may be removed by nucleotide excision repair (NER). ERCC excision repair 1, endonuclease non-catalytic subunit (ERCC1), which is regarded as one of the most potent biomarkers for DDP resistance, is associated with the removal of DNA-platinum adducts and results in DDP resistance (43). Targeting Gli1 alters the expression of NER-associated genes (e.g., c-JUN, ERCC1, and X-ray repair cross-complementing 1) and increases DDP sensitivity (44–46). Therefore, it may be speculated that the Hh signaling pathway is involved in DNA damage repair to promote chemotherapy resistance in ovarian cancer. γ-H2AX is a DNA damage marker. GANT61, an inhibitor of Gli, increases γ-H2AX expression in glioma cells in response to temozolomide treatment (47). In the present study, an increase in γ-H2AX protein expression, along with a decrease in MDR1, was detected in ovarian cancer cells at 72 h following DDP treatment combined with GANT61 treatment or Gli2 knockdown. At such a time-point, DNA damage is more common than DNA damage repair (48). It has been confirmed that there is an increase in DNA damage under DDP treatment combined with Hh signaling pathway targeting. Further exploration of the mechanism of the Hh signaling pathway in DNA damage or DNA damage repair may provide strategies to reverse chemotherapy resistance in ovarian cancer.
Targeting the Hh signaling pathway in ovarian cancer alters cell viability and the DNA damage response to treatment with DDP, which indicates that the Hh signaling pathway has an important role in ovarian cancer cell resistance to DDP. However, the mechanisms underlying this process remain to be fully elucidated. As a drug resistance gene, MDR1 has been demonstrated to be a cause of drug resistance. It has been revealed that Hh signaling increases resistance to multiple chemotherapeutic agents in part by promoting drug efflux through regulation of MDR1 expression (19). A gene microarray suggested that MDR1 is a candidate target gene of the Hh signaling pathway after treatment of SK-OV-3 cells with GANT61. Western blot and RT-qPCR assays demonstrated that MDR1 is a target gene of the Hh signaling pathway. There was a decrease in the expression of MDR1 along with an increase in γ-H2AX, increase of DNA damage and reduction of MDR1 expression after DDP treatment combined with targeting of the Hh signaling pathway.
In conclusion, targeting the Hh signaling pathway increases DDP sensitivity in ovarian cancer. MDR1 is a target gene of the Hh signaling pathway. The Hh signaling pathway may affect DDP chemoresistance in ovarian cancer via MDR1.
Supplementary Material
Acknowledgements
The authors thank Dr Shiwen Luo and Dr Yong Li (Center for Experimental Medicine, The First Affiliated Hospital of Nanchang University, Nanchang, China) for producing and providing the plasmid.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (grant no. 81960470) and the Key R&D Project of Jiangxi Science and Technology Department (grant no. 20171BBG70008), as well as the Graduate Innovation Foundation of Nanchang University (grant no. CX2018195).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HZ and LH performed the MTT assays and colony formation assays; HZ analyzed and interpreted the data; LH drafted the initial manuscript; MC revised the manuscript and performed statistical analysis; QW and XH performed the western blot and RT-qPCR analyses; QC provided the experimental design, managed the study, reviewed the manuscript and provided funding support. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 4.Ingham PW, McMahon AP. Hedgehog signaling in animal development_ paradigms and princip. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 5.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 6.Bakshi A, Chaudhary SC, Rana M, Elmets CA, Athar M. Basal cell carcinoma pathogenesis and therapy involving hedgehog signaling and beyond. Mol Carcinog. 2017;56:2543–2557. doi: 10.1002/mc.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, Bernard K, Conklin JF, Szczepny A, Yuan J, et al. A crucial requirement for hedgehog signaling in small cell lung cancer. Nat Med. 2011;17:1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Li J, Feng L. Hedgehog signaling pathway as a therapeutic target for ovarian cancer. Cancer Epidemiol. 2016;40:152–157. doi: 10.1016/j.canep.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Song X, Yan L, Lu C, Zhang C, Zhu F, Yang J, Jing H, Zhang Y, Qiao J, Guo H. Activation of hedgehog signaling and its association with cisplatin resistance in ovarian epithelial tumors. Oncol Lett. 2018;15:5569–5576. doi: 10.3892/ol.2018.8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steg AD, Katre AA, Bevis KS, Ziebarth A, Dobbin ZC, Shah MM, Alvarez RD, Landen CN. Smoothened antagonists reverse taxane resistance in ovarian cancer. Mol Cancer Ther. 2012;11:1587–1597. doi: 10.1158/1535-7163.MCT-11-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao X, Siu MK, Au CW, Wong ES, Chan HY, Ip PP, Ngan HY, Cheung AN. Aberrant activation of hedgehog signaling pathway in ovarian cancers: Effect on prognosis, cell invasion and differentiation. Carcinogenesis. 2009;30:131–140. doi: 10.1093/carcin/bgn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Hamo R, Zilberberg A, Cohen H, Bahar-Shany K, Wachtel C, Korach J, Aviel-Ronen S, Barshack I, Barash D, Levanon K, Efroni S. Resistance to paclitaxel is associated with a variant of the gene BCL2 in multiple tumor types. NPJ Precis Oncol. 2019;3:12. doi: 10.1038/s41698-019-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Li ZY, Duan XJ, Fan XM, Liu WN, Li YH. Clinical significance of FOXM1 and gli-1 protein expression in high-grade ovarian serous carcinoma. Zhonghua Zhong Liu Za Zhi. 2016;38:904–908. doi: 10.3760/cma.j.issn.0253-3766.2016.12.005. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 14.Tassi RA, Todeschini P, Siegel ER, Calza S, Cappella P, Ardighieri L, Cadei M, Bugatti M, Romani C, Bandiera E, et al. FOXM1 expression is significantly associated with chemotherapy resistance and adverse prognosis in non-serous epithelial ovarian cancer patients. J Exp Clin Cancer Res. 2017;36:63. doi: 10.1186/s13046-017-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng E, Hanna A, Samant RS, Shevde LA. The impact of hedgehog signaling pathway on DNA repair mechanisms in human cancer. Cancers (Basel) 2015;7:1333–1348. doi: 10.3390/cancers7030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang RL, Gu F, Kirma NB, Ruan J, Chen CL, Wang HC, Liao YP, Chang CC, Yu MH, Pilrose JM, et al. Comprehensive methylome analysis of ovarian tumors reveals hedgehog signaling pathway regulators as prognostic DNA methylation biomarkers. Epigenetics. 2014;8:624–634. doi: 10.4161/epi.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke Z, Caiping S, Qing Z, Xiaojing W. Sonic hedgehog-gli1 signals promote epithelial-mesenchymal transition in ovarian cancer by mediating PI3K/AKT pathway. Med Oncol. 2015;32:368. doi: 10.1007/s12032-014-0368-y. [DOI] [PubMed] [Google Scholar]
- 18.Steg AD, Bevis KS, Katre AA, Ziebarth A, Dobbin ZC, Alvarez RD, Zhang K, Conner M, Landen CN. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin Cancer Res. 2012;18:869–881. doi: 10.1158/1078-0432.CCR-11-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bossennec M, Di Roio A, Caux C, Ménétrier-Caux C. MDR1 in immunity: Friend or foe? Oncoimmunology. 2018;7:e1499388. doi: 10.1080/2162402X.2018.1499388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Katsaros D, Wiley A, de la Longrais IA, Puopolo M, Yu H. Expression of MDR1 in epithelial ovarian cancer and its association with disease progression. Oncol Res. 2007;16:395–403. doi: 10.3727/000000006783980892. [DOI] [PubMed] [Google Scholar]
- 21.Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE, Ferguson MJ, Smith G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. 2016;115:431–441. doi: 10.1038/bjc.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288:156–166. doi: 10.1016/j.jns.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q, Xu R, Zeng C, Lu Q, Huang D, Shi C, Zhang W, Deng L, Yan R, Rao H, et al. Down-Regulation of gli transcription factor leads to the inhibition of migration and invasion of ovarian cancer cells via integrin beta4-mediated FAK signaling. PLoS One. 2014;9:e88386. doi: 10.1371/journal.pone.0088386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Wang Y, Chen T, Zhang Y, Xu R, Wang W, Cheng M, Chen Q. Aberrant activation of hedgehog signalling promotes cell migration and invasion via matrix metalloproteinase-7 in ovarian cancer cells. J Cancer. 2019;10:990–1003. doi: 10.7150/jca.26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- 26.Wang D, Hu G, Du Y, Zhang C, Lu Q, Lv N, Luo S. Aberrant activation of hedgehog signaling promotes cell proliferation via the transcriptional activation of forkhead Box M1 in colorectal cancer cells. J Exp Clin Cancer Res. 2017;36:23. doi: 10.1186/s13046-017-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan R, Peng X, Yuan X, Huang D, Chen J, Lu Q, Lv N, Luo S. Suppression of growth and migration by blocking the hedgehog signaling pathway in gastric cancer cells. Cell Oncol (Dordr) 2013;36:421–435. doi: 10.1007/s13402-013-0149-1. [DOI] [PubMed] [Google Scholar]
- 28.Guo P, Peng D, Xiong X, Zhang S. Expression of microRNA-100 and its correlation with drug resistance in human ovarian cancer SKOV3/DDP cells. Nan Fang Yi Ke Da Xue Xue Bao. 2015;35:1624–1627. (In Chinese) [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Shi YL, Zhou K, Wang LL, Yan ZX, Liu YL, Xu LL, Zhao SW, Chu HL, Shi TT, et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018;9:739. doi: 10.1038/s41419-018-0776-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao ZJ, Yuan WD, Yuan JQ, Yuan K, Wang Y. Downregulation of HIF-2alpha reverse the chemotherapy resistance of lung adenocarcinoma A549 cells to cisplatin. Med Sci Monit. 2018;24:1104–1111. doi: 10.12659/MSM.906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed E, Larkins TL, Chau CH, Figg WD. DNA repair: ERCC1, nucleotide excision repair, and platinum resistance. In: Rudek MA, et al., editors. Handbook of Anticancer Pharmacokinetics and Pharmacodynamics. Cancer Drug Discovery and Development. Springer Science and Business Media; New York: 2014. pp. 333–349. DOI 10.1007/978-1-4614-9135-4_18. [DOI] [Google Scholar]
- 33.Ding Y, Niu W, Zhang T, Wang J, Cao J, Chen H, Wang R, An H. Levistolide A synergistically enhances doxorubicininduced apoptosis of k562/dox cells by decreasing MDR1 expression through the ubiquitin pathway. Oncol Rep. 2018;41:1198–1208. doi: 10.3892/or.2018.6889. [DOI] [PubMed] [Google Scholar]
- 34.Zeng C, Chen T, Zhang Y, Chen Q. Hedgehog signaling pathway regulates ovarian cancer invasion and migration via adhesion molecule CD24. J Cancer. 2017;8:786–792. doi: 10.7150/jca.17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiechert A, Saygin C, Thiagarajan PS, Rao VS, Hale JS, Gupta N, Hitomi M, Nagaraj AB, DiFeo A, Lathia JD, Reizes O. Cisplatin induces stemness in ovarian cancer. Oncotarget. 2016;7:30511–30522. doi: 10.18632/oncotarget.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barakat MT, Humke EW, Scott MP. Learning from jekyll to control hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steg AD, Burke MR, Amm HM, Katre AA, Dobbin ZC, Jeong DH, Landen CN. Proteasome inhibition reverses hedgehog inhibitor and taxane resistance in ovarian cancer. Oncotarget. 2014;5:7065–7080. doi: 10.18632/oncotarget.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding J, Zhou XT, Zou HY, Wu J. Hedgehog signaling pathway affects the sensitivity of hepatoma cells to drug therapy through the ABCC1 transporter. Lab Invest. 2017;97:819–832. doi: 10.1038/labinvest.2017.34. [DOI] [PubMed] [Google Scholar]
- 39.Chaudary N, Pintilie M, Hedley D, Hill RP, Milosevic M, Mackay H. Hedgehog inhibition enhances efficacy of radiation and cisplatin in orthotopic cervical cancer xenografts. Br J Cancer. 2017;116:50–57. doi: 10.1038/bjc.2016.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu B, Gu D, Zhang X, Li J, Liu B, Xie J. GLI1-Mediated regulation of side population is responsible for drug resistance in gastric cancer. Oncotarget. 2017;8:27412–27427. doi: 10.18632/oncotarget.16174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, Cormack G, Jaquith JB, Cerchietti L, Cocolakis E, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511:90–93. doi: 10.1038/nature13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol Res. 2016;106:27–36. doi: 10.1016/j.phrs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Yu JJ, Mu C, Yunmbam MK, Slavsky D, Cross CL, Bostick-Bruton F, Reed E. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res. 2000;20:645–652. [PubMed] [Google Scholar]
- 44.Laner-Plamberger S, Kaser A, Paulischta M, Hauser-Kronberger C, Eichberger T, Frischauf AM. Cooperation between GLI and JUN enhances transcription of JUN and selected GLI target genes. Oncogene. 2009;28:1639–1651. doi: 10.1038/onc.2009.10. [DOI] [PubMed] [Google Scholar]
- 45.Kudo K, Gavin E, Das S, Amable L, Shevde LA, Reed E. Inhibition of gli1 results in altered c-Jun activation, inhibition of cisplatin-induced upregulation of ERCC1, XPD and XRCC1, and inhibition of platinum-DNA adduct repair. Oncogene. 2012;31:4718–4724. doi: 10.1038/onc.2011.610. [DOI] [PubMed] [Google Scholar]
- 46.Agyeman A, Mazumdar T, Houghton JA. Regulation of DNA damage following termination of hedgehog (HH) survival signaling at the level of the GLI genes in human colon cancer. Oncotarget. 2012;3:854–868. doi: 10.18632/oncotarget.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Cai J, Zhao S, Yao K, Sun Y, Li Y, Chen L, Li R, Zhai X, Zhang J, Jiang C. GANT61, a GLI inhibitor, sensitizes glioma cells to the temozolomide treatment. J Exp Clin Cancer Res. 2016;35:184. doi: 10.1186/s13046-016-0463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo LJ, Yang LX. γ-H2AX-a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–310. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.