Abstract
MicroRNA-7 (miR-7) has been identified as a tumor suppressor in non-small cell lung cancer (NSCLC) and a radiosensitivity regulator. Numerous studies have revealed that specific protein 1 (SP1) plays a critical role in the tumorigenesis of various types of cancers and regulates radiosensitivity and tumor suppressor p53-binding protein 1 (TP53BP1), which plays an essential role in DNA repair. However, it is not clear whether miR-7 has a regulatory effect on SP1 and TP53BP1 in NSCLC. In the present study it was revealed that miR-7 directly binds to the 3′UTR of SP1, thereby suppressing SP1 expression to regulate radiosensitivity. Overexpression of miR-7 and SP1 and knockdown of miR-7 and SP1 were performed using lentiviral transfection. Protein and mRNA abundance of SP1 and TP53BP1 were determined using western blotting and RT-qPCR, respectively, while miR-7 binding to SP1 was validated using a luciferase reporter assay. Biological function analysis indicated that miR-7 negatively regulated SP1 and inhibited cell proliferation, migration, and invasion when combined with radiation. It was also revealed that the expression of TP53BP1 was positively regulated by SP1 or negatively regulated by miR-7. In conclusion, SP1 was a target of miR-7, and the decreased expression of SP1 resulting from miR-7 overexpression in NSCLC was vital for improving radiosensitivity in NSCLC cells. Moreover, SP1 expression was detected in 95 paired NSCLC and adjacent normal tissues, and it was determined that SP1 was significantly upregulated in NSCLC tissues and that its upregulation was correlated with the degree of tissue differentiation. Thus, SP1 and/or miR-7 may be potential molecular targets in NSCLC radiotherapy.
Keywords: specific protein 1, microRNA-7, radiosensitivity, non-small cell lung cancer
Introduction
Lung cancer accounts for nearly 20% of cancer-related deaths worldwide, and the high incidence and mortality of lung cancer, with 2.1 million new cases and 1.8 million deaths in 2018, indicate the importance of etiological and therapeutic research (1). In all lung cancer cases, non-small cell lung cancer (NSCLC) accounts for approximately 80% of the cases (2). Radiotherapy, as a critical treatment for inoperable and postsurgical NSCLC patients, has been the main focus in the past decades (3).
MicroRNA (miR) functions as either a tumor suppressor or oncogene due to its direct involvement in cancer initiation and progression through regulation of the expression of essential cancer-related genes (4–6). MicroRNA-7 (miR-7) has been identified as a tumor suppressor in several human cancers. In studies with 41 and 128 pairs of cancer and adjacent tissues, Xiong et al (7) and Su et al (8), respectively, revealed that miR-7 was decreased in NSCLC. miR-7 also increased the radiosensitivity of human cancer cells by enhancing the epidermal growth factor receptor (EGFR) signaling pathway. Lee et al revealed that targeting the miR-7-EGFR network overcame the resistance of X-ray radiotherapy (9). EGFR is expressed in various normal tissues, such as the skin, lung, liver and colon, and the potential pan-effects of EGFR targets require the identification of additional methods by which to regulate radiosensitivity (10). Thus, it is highly valuable to identify more miR-7 target genes as therapeutic targets. However, the regulatory mechanisms of miR-7 and its target genes remain unclear in the radiosensitivity of NSCLC.
Specific protein 1 (SP1) has been revealed to play critical roles in tumorigenesis, promote the repair of DNA double-strand breaks (DSBs) and regulate the radiosensitivity of tumors (11). Following SP1 silencing, cells were revealed to be more sensitive to DNA damage, and the foci degradation of γ-H2AX was delayed (12). In addition, SP1 silencing has been revealed to result in abnormal chromosome accumulation, suggesting that SP1 plays a vital role in DSB repair because SP1 often accumulates at damaged sites of DNA (13). Multiple genes involved in the cell cycle, differentiation, and oncogenesis are regulated by the SP1 protein (14). In lung cancer cells, SP1 overexpression influenced the chemotherapeutic resistance by upregulating the expression of the ATP-binding cassette transporter G2 (ABCG2) (15). Tumor suppressor p53-binding protein 1 (TP53BP1) participates in the cellular response to DNA damage and plays an essential role in DNA repair (16,17) Ward created a TP53BP1-deficient mouse model and found that mice lacking TP53BP1 were sensitive to radiation. Yang et al confirmed that the suppression of BMI-1 (B-cell-specific Moloney murine leukaemia virus integration site-1) increased the radiosensitivity of esophageal carcinoma through regulation of TP53BP1 (18). As a transcription regulator, SP1 is involved in the regulation of downstream gene expression, and the binding sites of SP1 were predicted to be within the promoter region of TP53BP1 (http://genexplain.com/transfac/) (19). However, SP1 has not been investigated in the radioresistance of NSCLC. Additionally, it remains unclear whether miR-7 has a regulatory effect on SP1 and TP53BP1 in NSCLC.
Herein, it was hypothesized that miR-7 increased radiosensitivity in NSCLC by downregulating SP1 and TP53BP1.
Materials and methods
Cell lines
A549 and SK-MES-1 cells were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, China. In addition, 293T cells were obtained from Dr Yu Tao, a researcher from China Medical University. A549 (adenocarcinoma) cells were grown in F-12 medium, and SK-MES-1 (squamous carcinoma) cells were grown in MEM medium. Additionally, 293T cells were grown in DMEM medium. All cells were grown at 37°C in a humidified atmosphere of 5% CO2. All the media contained 10% fetal bovine serum (FBS), and were obtained from Gibco; Thermo Fisher Scientific, Inc.
Tissue collection
The Biomedical Ethics Committee of the Fourth Affiliated Hospital of China Medical University approved the present study (Institutional Review Board-approved protocol no. 2014-039). All relevant patients provided written informed consent, and the collection of specimens was performed according to the Declaration of Helsinki. In total, 95 matched paraffin sections and 8 pairs of NSCLC tissues and adjacent normal tissues (≥5 cm from the NSCLC tissue) were obtained from patients who received surgical resection at The Fourth Affiliated Hospital to China Medical University from May 2008 to June 2018. All patients provided informed consent and did not undergo radiotherapy or chemotherapy before the operation. The age of the patients ranged from 31–76 years old, with a median age of 63±8.96 years, including 24 females and 71 males. Staging was performed according to the UICC/AJCC (8th edition) TNM staging system for lung cancer (20). Organizational credit types were determined according to the standards provided by the World Health Organization (2015) (21). The selection criteria were as follows: i) all paraffin specimens were NSCLC with a definite pathological diagnosis after operation; and ii) all samples had complete clinical data.
RNA extraction, reverse transcription-quantitative PCR (RT-qPCR) for miRNA expression, and mRNA expression
Total RNA from tissues and cells was extracted with the Eastep Super Total RNA Extraction Kit (Promega Corporation). Every step was performed on ice and according to the protocol of the kit. The 175 mg/ml tissue lysate was prepared from the tissue preserved in liquid nitrogen. We collected 3–5×106 cells or 150 µl tissue lysate from the logarithmic growth period into an EP tube. Then, 300 µl RNA lysate was added to the EP tube, which was followed by centrifugation at 12,000 × g for 5 min at 4°C. The upper liquid was absorbed and added to a new EP tube, followed by the addition of anhydrous ethanol, mixing, and the addition of the mixture to the centrifugal column/collection tube. The sample was set aside and centrifuged at 12,000 × g, for 5 min at 4°C, according to the described steps, the filtrate was discarded, and 50 µl DNA enzyme I incubation liquid was added to the centrifugal column. The sample was incubated on ice for 15 min, followed by the addition of 600 µl RNA to the centrifugal column and centrifugation twice. The centrifugation column was moved to the elution tube, and 10–30 µl of nuclease-free water was added to the center of the centrifugal column membrane and allowed to sit on ice for 2 min. It was then centrifuged at 12,000 × g for 1 min at 4 °C to obtain RNA. The purity and concentration of the A260/A280 ratio of RNA detected using a spectrophotometer was between 1.9–2.1, and the concentration was higher than 0.1 µg/µl. For miRNA expression, total RNA (1–3 µg) was converted to cDNA using the Hairpin-it microRNA and U6 snRNA Normalization RT-PCR Quantitation Kit (Shanghai GenePharma Co., Ltd.). For each sample, quantitative PCR (qPCR) was performed in triplicate on an ABI 7500 thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.) after reverse transcription. For mRNA expression, the total RNA (0.5 µg) was converted to cDNA for each sample using the Primer Script RT Reagent Kit (Takara Biotechnology Co., Ltd). In each sample, RT-qPCR was performed in triplicate with SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd). The sequences of the oligonucleotide primers are summarized in Table I. The primer oligonucleotides were purchased from Takara Biotechnology Co., Ltd. qPCR for miRNA, thermocycling conditions were as follows: 95°C for 3 min and 40 cycles at 95°C for 12 sec, and 62°C for 40 sec. qPCR for mRNA thermocycling conditions were as follows: 95°C for 30 sec and 40 cycles at 95°C for 5 sec, and 60°C for 34 sec. All data were analyzed using 2−ΔΔCq method (22)
Table I.
The sequences of oligonucleotide primers.
| Gene | Oligonucleotide primers |
|---|---|
| miR-7 | F: 5′-ACGTTGGAAGACTAGTGATTT-3′ |
| R: 5′-TATGGTTGTTCTGCTCTCTGTCTC-3′ | |
| U6 | F: 5′-ATTGGAACGATACAGAGAAGATT-3′ |
| R: 5′-GGAACGCTTCACGAATTTG-3′ | |
| SP1 | F: 5′-GACTAGGGCTGCAAGTAGTGAGGA-3′ |
| R: 5′-GGAAACTGGAGCACTGGGTAGAC-3′ | |
| TP53BP1 | F: 5′-AGCAGGAGCTGGCTATATCCTTGA-3′ |
| R: 5′-GACAATGCTGATCCGCAATTAGAA-3′ | |
| GAPDH | F: 5′-TCCTGTGGCATCCACGAAACT-3′ |
| R: 5′-GAAGCATTTGCGGTGGACGAT-3′ |
miR-7, microRNA-7; SP1, specific protein 1; TP53BP1, tumor suppressor p53-binding protein 1; F, forward; R, reverse.
SP1 expression and the correlation with TP53BP1
UALCAN (23) (http://ualcan.path.uab.edu/) was used to obtain SP1 transcript and protein expression data of lung adenocarcinoma (LUADC) and lung squamous cell cancer (LUSCC). The GEPIA database (24) (http://gepia.cancer-pku.cn) was used to collect correlation data between SP1 and TP53BP1 in LUADC, LUSCC, and lung tissues.
miRNA target prediction
Putative target genes of miR-7 were identified using five miRNA databases: TargetScan (25) (http://www.targetscan.org/mamm_31/); PicTar (26) (https://pictar.mdc-berlin.de); RNAhybrid (27) (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/); miRanda (28) (http://www.microrna.org/microrna/home.do); and miRbase (29) (http://www.mirbase.org).
Luciferase assay
The pGL3-conSP1-3′-UTR-Wt1 (1152–1159), pGL3-conSP1-3′-UTR-Wt2 (4319–4325), pGL3-con-SP1-3′-UTR-Mut1, and pGL3-con-SP1-3′-UTR-Mut2 reporter plasmids, as well as the control Renilla luciferase pRL-TK vector were purchased from Genomeditech. The LV-hsa-miR-7 and negative control CON238 (Ubi-MCS-SV40-EGFP-IRES-puromycin) were used to establish miR-7-overexpressing and control 293T cells. The reporter plasmids were transfected into 293T cells with Lipofectamine 2000 (Thermo Fisher Scientific, Inc). After 48 h, the cells were collected, lysed and centrifuged in 12,000 × g, and the supernatant was collected. The Dual-Luciferase Reporter Assay System (Promega Corporation) was used to perform the luminescence analysis.
Lentivirus transfection
LV-hsa-miR-7 (miR-7), negative control CON238 (Ubi-MCS-SV40-EGFP-IRES-puromycin), LV-hsa-miR-7-inhibitor (miR-7-inhibitor) and negative control CON137 (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) were designed and synthesized by Shanghai GenePharma Co., Ltd. The LV-shRNA against SP1 mRNA (sh-SP1), control pGMLV-SC5 RNAi (U6-CMV-EGFP-PGK-Puromycin), LV-SP1-overexpression, and overexpression control PGMLV-4931 (CMV-MCS-3Flag-PGK-Puromycin) were designed and synthesized by Genomeditech. The manufacturer's protocol was followed to transfect the cells when the confluence rate was 30–50%. The transfection efficiency was detected using fluorescence microscopy. Western blotting or real-time PCR were used to measure the expression efficiency. RNA was extracted 24 h later and total proteins 48 or 72 h later after transfection. The target sequence of short hairpin RNA for miR-7 was: 5′-TTCTCCGAACGTGTCACGT-3′ Furthermore, the sequences of the oligonucleotide primers for hsa-miR-7 were as follows: Forward (F), 5′-GAGGATCCCCGGGTACCGGCCAAGCAAACTTCTCATTTCTC-3′ and reverse (R), 5′-CACACATTCCACAGGCTAGGGAATTGAAAGTTGTTAATATTTG-3′. The target sequence of short hairpin RNA for SP1 was: 5′-GCGTTTCTGCAGCTACCTTGA-3′ and the sequences of the oligonucleotide primers for SP1 were as follows: F, 5′-CCGCTCGAGGCCACCATGAGCG-3′ and R, 5′-CCGGAATTCgtGAAGCCATTGCCACTGATATTAATGG-3′
Cell proliferation assay
Cells were seeded in a 96-well plate (2,000 cells/well). Two groups were established, namely the irradiated and non-irradiated groups, and each group had five replicates. After adherence, cells were subjected to irradiation with 2 Gy, as well as at 0, 24, 48, and 72 h after irradiation, or with 2, 4, 6, and 8 Gy at 24 h after irradiation, after which 10 µl of the Cell Counting Kit-8 (CCK-8) reagent (Dojindo Molecular Technologies, Inc.) was added to each well followed by a 1- to 4-h incubation and determination of the absorbance at 450 nm on a microplate reader. Cell survival was calculated according to the following formula: Cell viability = (OD value in the irradiation group - OD of blank)/(OD in the non-irradiation group - OD of blank). A cell proliferation curve was generated based on the results.
Clonogenic assay
Exponential growth phase A549 cells were seeded into six-well plates at 300, 500, 800, 1,200, and 2,000 cells/well in triplicate. The cells were irradiated with a single dose of radiation (0, 2, 4, 6, and 8 Gy) (Varian 2300EX at a dose rate of 5 Gy/min; 6 MVX; SSD, 100 cm; Varian Medical Systems) separately after 24 h. Then, the incubated cells were allowed to undergo colony formation at 37°C in a 5% CO2 atmosphere. After incubation for ~14 days, 20% methanol and 0.5% (v/v) crystal violet were added to the plates for 20 min at room temperature to fix and stain the colonies. The cells were observed under a dissecting light microscope (magnification, ×100), and the colonies containing >50 cells were counted. The survival fraction (SF) were calculated as follows: SF = (number of colonies/number of cells plated)irradiated/(number of colonies/number of cells plated)non-irradiated (30) The SF in each group was also analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc.) as previously described (31).
Wound-healing assay
Cell migration was measured using the wound-healing assay. Cells (0.5×106 cells/well) were seeded into six-well plates, and when the confluency of the cells was optimal, a line was generated through the entire center of the well by scraping the cells with a 10-µl tip. The cells were then cultured in medium with 2% FBS for 48 h. Light microscopic images (magnification, ×100) of the cultures were obtained at 0, 24, and 48 h, and then the open wound area was assessed using ImageJ software (version 1.52q; National Institutes of Health). The open wound areas at 0 h in each group were used as the reference point, and the wound area at 24 and 48 h was divided by the area at 0 h, and the resulting relative wound areas were compared.
Transwell invasion assay
Cells were seeded into 6-well plates (0.5×106 cells/well), and two groups, namely the irradiation group and non-irradiation group, were established according to the experimental requirements. After adherence, the cells were irradiated with 0 and 2 Gy, and the culture medium was replaced with serum-free medium for 12 h. Cells were then digested, washed, and suspended in complete medium, and the cell density was adjusted to 1×104/ml. Matrigel (50 µl; BD Bioesciences) was added to the upper chambers of the 24-well 8-µm Transwell plates (Corning-Costar; Corning, Inc.). Suspended cells (200 µl/well) were then added to the upper chamber, and 700 µl of medium containing 10% FBS was added to the lower chamber. Cells were incubated at 5% CO2 and 37°C. After 48 h of culture, the medium was discarded, and the cells were fixed with 20% methanol and stained with crystal violet 0.5% (v/v) for 20 min at room temperature. After washing the cells, images were obtained using a light microscope (magnification, ×400), and the results were analyzed.
Western blot analysis
Protein was extracted from the cells. Cells were lysed in RIPA buffer (Thermo Fisher Scientific, Inc.) containing a protease and phosphatase inhibitor cocktail and incubated at 4°C for 30 min. Cells (5×106) and RIPA buffer were added to the EP tube and mixed evenly; the mixed solution was placed on ice for 5 min to lyse the cells, followed by centrifugation at 13,000 × g for 10 min at 4°C. The supernatant was removed to a new EP tube. To extract the protein from the tissue, first the tissue was cut into small pieces, the RIPA buffer was added, and the tissue was disrupted with an ultrasonic homogenizer. The mixture was placed on ice for 15 min for complete lysis and then centrifuged at 13,000 × g for 10 min at 4°C and removal of the supernatant to a new EP tube. A BCA kit (Thermo Fisher Scientific, Inc) was used for protein quantification. Additionally, 50 µg protein was subjected to 10% SDS-PAGE at a constant 120 V, 120 mA current and transferred to a polyvinylidene fluoride (PVDF) membrane (EMD Millipore). The membranes were blocked in 5% (w/v) bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature and incubated at 4°C overnight with primary antibodies against SP1 (1:2,000 dilution; cat. no. H00006667-M05; Novus Biologicals, Ltd.), TP53BP1 (1:1,000 dilution; product code ab172580; Abcam) and GAPDH (1:2,000 dilution; product code ab8245; Abcam) On the following day, membranes were incubated with anti-mouse (SP1 and GAPDH) or anti-rabbit (TP53BP1) IgG HRP-labeled secondary antibodies (1:10,000 dilution; product codes ab205719 and ab205718, respectively; Abcam) for 1 h at room temperature. Chemiluminescent detection was performed using the SuperSignal West Pico Chemiluminescent Substrate (product no. NCI5079; Thermo Scientific, Inc.). ImageJ software (version 1.52q; National Institutes of Health) was used for densitometric analysis.
Immunohistochemical (IHC) staining
All paraffin specimens were cut into 4-µm serial sections to generate 3 pieces per sample. Sections were attached to the anti-slipping slides and processed in a thermostat (60°C) for 60 min. Two sections were used for immunohistochemical staining, and one section was used as a negative control. IHC staining was performed by a two-step procedure, the paraffin-embedded sections were deparaffinized using xylene and rehydrated using a series of solutions of decreasing alcohol concentrations. Sections were placed in boiling citrate buffer for antigen retrieval, and 3% H2O2 was used to block endogenous peroxidase activity. The primary antibody against SP1 (1:200 dilution; product code ab124804; Abcam) was used to incubate sections overnight at 4°C. The following day, the sections were washed and incubated with rabbit specific IHC polymer detection kit HRP/DAB (product code ab209101; Abcam) at room temperature for 10 min, followed by hematoxylin counter-staining for 30 sec. Images were obtained using a light microscope (magnification, ×100). Pathologists performed a blinded assessment of the IHC staining. Brownish-yellow granular precipitates in the nuclei were considered positive. The following scoring system was used: 3, >60% of tumor cells were stained as positive; 2, 41 to 60% of tumor cells were stained positive; 1, 11 to 40% of tumor cells were stained positive; and 0, <10% of tumor cells were stained positive. If the score was equal to 3, the grade of specimen was classified as overexpression of SP1 (32).
Statistical analysis
The mean ± SEM from two or three independent experiments was used to express the data, and GraphPad Prism 5.0 software (GraphPad Software, Inc.) and SPSS 20.0 software (IBM Corp.) were used to perform statistical analyses. ANOVA was used for multigroup comparisons, and multiple comparisons between two groups were performed using analysis of variance and least significant difference (LSD) post hoc tests (Figs. 4D and E and 6D and E). Multiple comparison between two groups were performed using Tukey's test (Figs. 2C, 3A and 5A). To determine the SP1 expression differences between tumor tissue and non-tumor tissue, McNemar's test was used to analyze the data in Table II. To determine the associations between SP1 and clinicopathological features, Fisher's exact test was used to analyze the data in Table III. A paired t-test was used to compare tumor vs. adjacent normal tissue (Fig. 1). An unpaired Student's t-test was conducted to compare two groups (Figs. 2B and D, 3B, 4A-C, 5B and C, 6A-C and 7, except 7E). Spearman's test was used to analyze the correlation coefficient (Fig. 7E). A P-value <0.05 was considered to indicate a statistically significant difference
Figure 4.
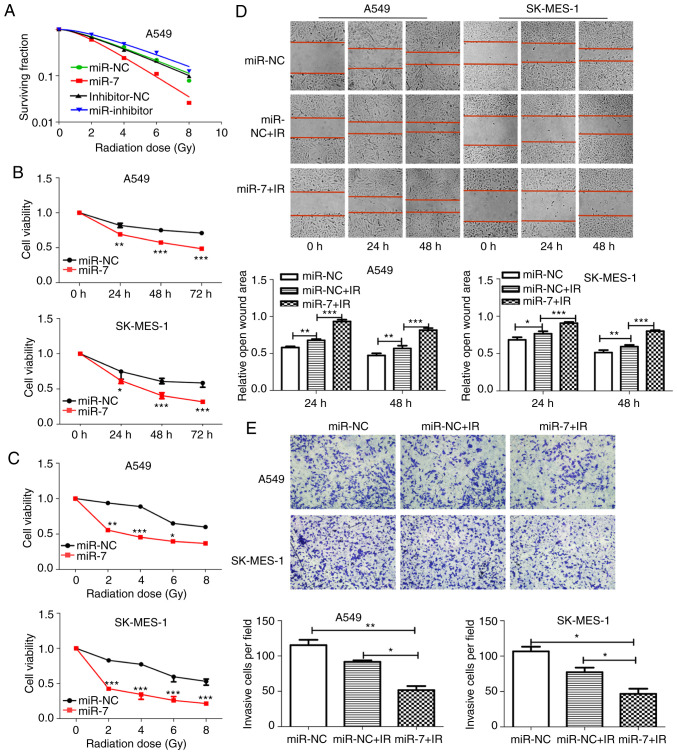
miR-7 enhances radiosensitivity of NSCLC cells. (A) Clonogenic cell survival curves were generated. miR-7 overexpression decreased survival fractions of A549 cells after irradiation (SER=1.54), and inhibition of miR-7 increased the survival fraction (SER=0.89). (B and C) Cell proliferation assays revealed that miR-7-overexpressing cells had decreased viability with (B) time course (0, 24, 48 and 72 h) and (C) increasing radiation dose (0, 2, 4, 6 and 8 Gy) after 2 Gy irradiation. The 24-h time-point was selected as the representative time-point in C. (D) Wound healing (magnification, ×100) and (E) Transwell invasion (magnification, ×400) assays demonstrated that miR-7 combined with irradiation significantly inhibited the migration and invasion of NSCLC cells. *P<0.05, **P<0.01 and ***P<0.001. miR-7, microRNA-7; NSCLC, non-small cell lung cancer; SER, sensitivity enhancement ratio.
Figure 6.
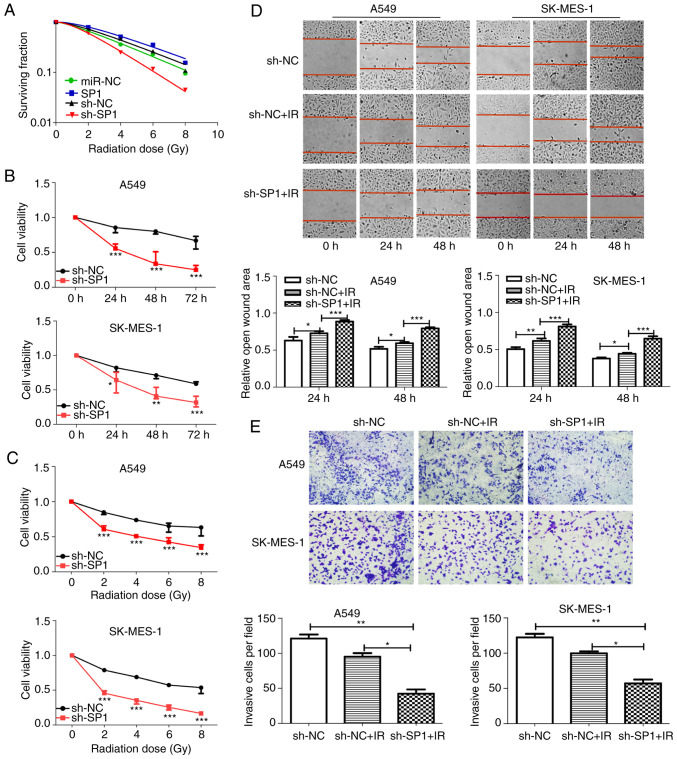
sh-SP1 enhances radiosensitivity of NSCLC cells. (A) Clonogenic cell survival curves were generated. The SP1-knockdown A549 cells decreased survival fractions after irradiation (SER=1.59), and overexpression of SP1 increased the survival fraction (SER=0.88). (B and C) Cell proliferation assays revealed that SP1-knockdown cells had decreased viability with B) time course (0, 24, 48 and 72 h) and (C) increasing radiation dose (0, 2, 4, 6 and 8 Gy) after 2 Gy irradiation. The 24-h time-point was selected as the representative time-point in C. (D) Wound healing (magnification, ×100) and (E) Transwell invasion (magnification, ×400) assays demonstrated that knockdown of SP1 combined with irradiation significantly inhibited the migration and invasion of NSCLC cells. *P<0.05, **P<0.01 and ***P<0.001. SP1, specific protein 1; sh, shRNA; NSCLC, non-small cell lung cancer; SER, sensitivity enhancement ratio.
Figure 2.
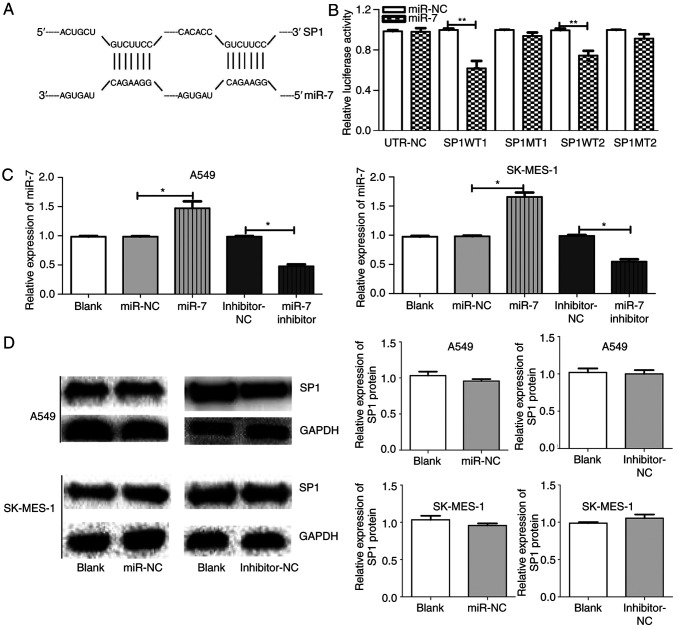
SP1 is a direct target of miR-7 in NSCLC. (A) Prediction of TargetScan; the binding sites of SP1 and miR-7 (1152–1159 and 4319–4325). (B) Luciferase reporter assays indicated that miR-7 targeted SP1 3′-UTR in A549 and SK-MES-1 cells. (C) The miR-7 expression between groups in A549 and SK-MES-1 cells after transfection with blank, miR-NC, miR-7, inhibitor-NC, or miR-7 inhibitor. (D) SP1 expression between groups in A549 and SK-MES-1 cells after transfection with miR-NC, inhibitor-NC, or blank. *P<0.05, **P<0.01 and. SP1, specific protein 1; miR-7, microRNA-7; NSCLC, non-small cell lung cancer; NC, negative control.
Figure 3.
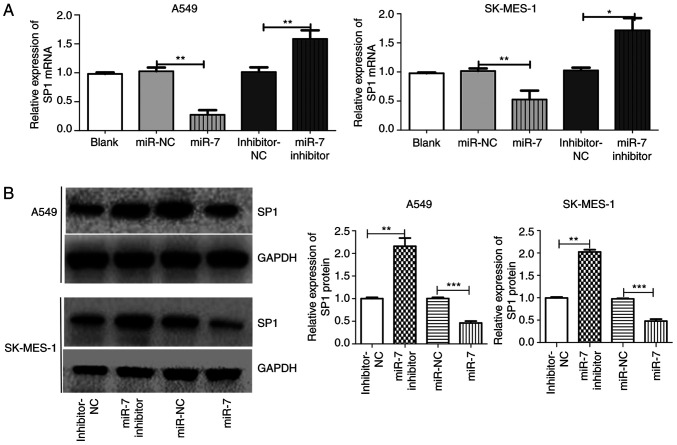
SP1 is a direct target of miR-7 in NSCLC. (A) SP1 mRNA expression between groups in A549 and SK-MES-1 cells after transfection with blank, miR-NC, miR-7, inhibition-NC or miR-7 inhibition. (B) SP1 protein expression between groups in A549 and SK-MES-1 cells after transfection with inhibitor-NC, miR-7 inhibitor, miR-NC or miR-7. *P<0.05, **P<0.01 and ***P<0.001. SP1, specific protein 1; miR-7, microRNA-7; NSCLC, non-small cell lung cancer; NC, negative control.
Figure 5.
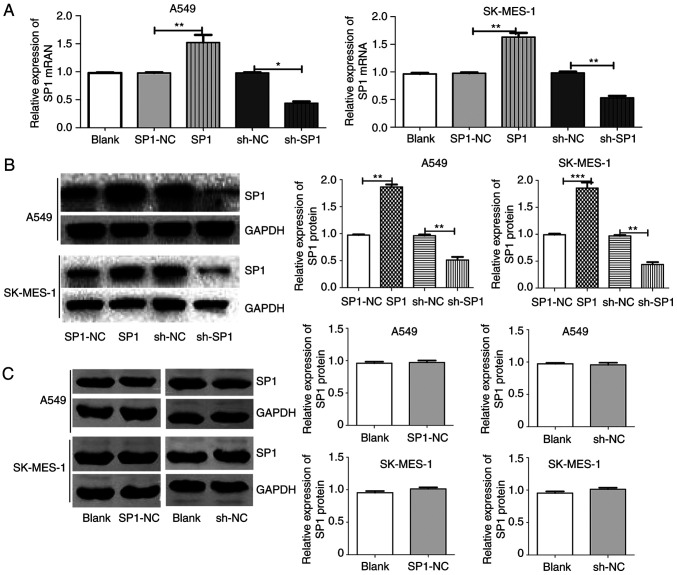
SP1 expression after SP1 overexpression and knockdown transfection. (A) SP1 mRNA expression between groups in A549 and SK-MES-1 cells after transfection with blank, SP1-NC, SP1, sh-NC or sh-SP1. (B) SP1 protein expression between groups in A549 and SK-MES-1 cells after transfection with SP1-NC, SP1, sh-NC or sh-SP1. (C) SP1 protein expression between groups in A549 and SK-MES-1 cells after transfection with blank, SP1-NC or sh-NC. *P<0.05, **P<0.01 and ***P<0.001. SP1, specific protein 1; NC, negative control; sh, shRNA.
Table II.
Expression of SP1 protein in carcinoma and adjacent tissues.
| Cancer | Adjacent normal | |||||
|---|---|---|---|---|---|---|
| Cancer type | N | Low | High | Low | High | P-valuea |
| NSCLCb | 95 | 36 | 59 | 67 | 28 | <0.001 |
| LUADb | 49 | 17 | 32 | 32 | 17 | <0.001 |
| LUSCc | 46 | 19 | 27 | 35 | 11 | 0.002 |
McNemar test.
The approximate P-value of the McNemar test was calculated according to Chi square distribution.
The exact P-value of the McNemar test was calculated according to binomial distribution. Bold indicates statistical significance. SP1, specific protein 1; NSCLC, non-small cell lung cancer; LUADC, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Table III.
Associations between SP1 expression and clinicopathologic parameters of NSCLC patients.
| SP1 expression | ||||
|---|---|---|---|---|
| Parameters | No. of cases | Low | High | P-valuea |
| Sex | Male (n=71) | 27 | 44 | 0.963 |
| Female (n=24) | 9 | 15 | ||
| Age (years) | <60 (n=28) | 10 | 18 | 0.821 |
| >60 (n=67) | 26 | 41 | ||
| Smoking | No (n=29) | 9 | 20 | 0.441 |
| NSCLC | Yes (n=66) | 27 | 39 | |
| No (n=24) | 9 | 15 | 0.769 | |
| LUAD | Yes (n=25) | 8 | 17 | |
| No (n=5) | 0 | 5 | 0.041 | |
| LUSC | Yes (n=41) | 19 | 22 | |
| Pathology | LUAD (n=49) | 17 | 32 | 0.533 |
| LUSC (n=46) | 19 | 27 | ||
| Differentiation | Low (n=32) | 5 | 27 | 0.002 |
| NSCLC | High (n=63) | 31 | 32 | |
| LUAD | Low (n=21) | 4 | 17 | 0.040 |
| High (n=28) | 13 | 15 | ||
| LUSC | Low (n=11) | 1 | 10 | 0.016 |
| High (n=35) | 8 | 17 | ||
| T stage | T1+T2 (n=71) | 28 | 43 | 0.635 |
| T3+T4 (n=24) | 8 | 16 | ||
| Lymph node | Negative (n=52) | 17 | 35 | 0.292 |
| Positive (n=43) | 19 | 24 | ||
| TNM stage | I+II (N=67) | 24 | 43 | 0.643 |
| III (N=28) | 12 | 16 | ||
Fisher's exact probability. Bold indicates statistical significance. SP1, specific protein 1; NSCLC, non-small cell lung cancer; LUADC, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Figure 1.
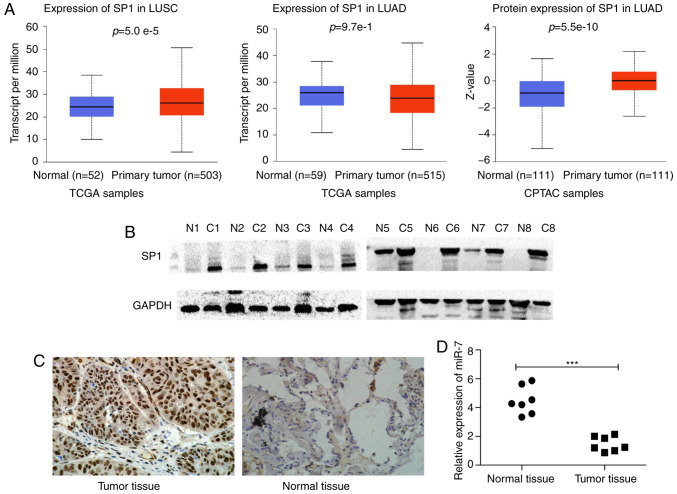
SP1 is upregulated and miR-7 is downregulated in NSCLC tissues. (A) SP1 transcript expression in LUSC, LUAD, and normal tissues and SP1 protein expression in LUAD tissues and normal tissues. (B) SP1 expression was examined in 8 cases of NSCLC tissues and adjacent normal tissues by western blotting. N indicates normal tissues and C indicates cancer tissues. (C) SP1 expression was examined in 95 cases of NSCLC tissues and adjacent non-cancer tissues by IHC staining. The brown granules in the image are strongly positive in the nucleus (magnification, ×100). (D) miR-7 expression in primary NSCLC tissues and matched adjacent lung tissues. ***P<0.001. SP1, specific protein 1; miR-7, microRNA-7; NSCLC, non-small cell lung cancer; LUADC, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; IHC, immunohistochemical.
Figure 7.
miR-7 regulates TP53BP1 through SP1. The (A and B) mRNA and (C and D) protein levels of TP53BP1 in A549 and SK-MES-1 cells are higher with upregulation of SP1 or downregulation of miR-7, and are lower with downregulation of SP1 or upregulation of miR-7 compared with the negative control. (E) The correlation coefficient (Spearman's test) between SP1 and TP53BP1 in LUAD, LUSC, and lung tissue from GEPIA, revealed that SP1 was correlated with the TP53BP1 in lung cancer tissue. *P<0.05, **P<0.01. miR-7, microRNA-7; TP53BP1, tumor suppressor p53-binding protein 1; SP1, specific protein 1; LUADC, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Results
Expression and clinical characteristics of SP1 in NSCLC
SP1 expression information obtained from UALCAN revealed that SP1 was highly expressed in NSCLC tissues compared with normal samples. (Fig. 1A). We detected SP1 expression in 8 pairs of matched NSCLC and paracancerous tissues by western blotting. SP1 expression in cancer tissues was markedly higher than in paracancerous normal tissues (Fig. 1B). Furthermore, SP1 expression in 95 pairs of paraffin-embedded NSCLC tissues was detected by IHC and the results revealed that the expression of SP1 in NSCLC was significantly higher than that in normal tissues (Fig. 1C). The expression of SP1 in NSCLC (LUAD, LUSC or together) was significantly higher than that in paracancerous tissues (Table II).
According to SP1 expression, 95 patients with NSCLC were divided into low and high groups. The results revealed that the expression of SP1 was correlated with the degree of tumor differentiation (LUAD P=0.040, LUSC P=0.016 and NSCLC P=0.002) and with smoking the status in LUSC (P=0.041) (Table III).
SP1 emerges as a target of miR-7
Through bioinformatics analysis, SP1 was predicted as a target of miR-7 in all five databases (miRanda, TargetScan, PicTar, miRbase and RNAhybrid). It was also determined that miR-7 expression was low (Fig. 1D) and SP1 expression was high (Fig. 1A-C) in NSCLC tumor tissue compared to normal tissue. The predicted binding site between miR-7 and the 3′UTR of SP1 included the following bases: 1152–1159 and 4319–4325 (Fig. 2A). The wild-type (WT-3′UTR) and mutant (MT-3′UTR) sequences of the SP1 3′UTR were cloned into a luciferase plasmid, and the generated plasmids were transfected into 293T cells to investigate whether the 3′UTR of SP1 was a functional target of miR-7. As anticipated, miR-7 suppressed the luciferase activity of SP1 WT-3′UTR, but it did not affect the luciferase activity of SP1 MUT-3′UTR. These results indicated that miR-7 directly acted on the 3′UTR of SP1, thereby playing a negative role in regulating the expression of SP1 (Fig. 2B). At the first transfection, the cells of the transfected NC group were compared with the blank cells, and the expression levels of miR-7 (Fig. 2C) and SP1 (Figs. 2D and 3A) were examined. No statistical differences in the expression of miR-7 were found compared with the control blank, and no difference in the expression of SP1 in terms of both mRNA and protein levels was identified. A549 and SK-MES-1 cells were transfected with miR-7 and miR-7 inhibitor, and the transfection efficiency was assessed by qPCR (Fig. 2C). The SP1 expression was assessed by qPCR (Fig. 3A) and western blotting (Fig. 3B). Western blotting and qPCR analyses revealed increased expression of SP1 in miR-7-depleted cells and decreased expression of SP1 in miR-7-overexpressing cells at the protein and mRNA levels. All data indicated that SP1 is a downstream target of miR-7 in NSCLC cells.
miR-7 increases the radiosensitivity of NSCLC by suppressing tumor cell growth, migration and invasion
The role of miR-7 in the radiosensitivity of NSCLC cells was explored. miR-7-overexpressing A549 cells had decreased clone formation compared with control cells after irradiation (Fig. S1). The survival fractions of miR-7-overexpressing A549 cells were markedly decreased after irradiation (Fig. 4A, SER=1.54). In contrast, inhibition of miR-7 increased the survival fraction (Fig. 4A, SER=0.89). The CCK-8 assay (Fig. 4B and C), wound-healing assay (Fig. 4D), and Transwell invasion (Fig. 4E) assays revealed that miR-7-overexpressing cells had decreased viability, migration ability, and invasion ability compared with control cells after irradiation. These findings indicated that miR-7 enhanced radiosensitivity in NSCLC cells.
Knockdown of SP1 by shRNA increases the radiosensitivity of NSCLC by suppressing tumor cell growth, migration, and invasion
The role of SP1 in the radiosensitivity of NSCLC cells was investigated. SP1-expressing and sh-SP1 lentiviral vectors were transfected into A549 and SK-MES-1 cells, and the transfection efficiency was assessed by qPCR (Fig. 5A) and western blotting (Fig. 5B). At the first transfection, the cells of the transfected NC group were compared with the blank cells, the expression levels of SP1 (Fig. 5A and C) were examined. No statistical differences in the expression of SP1 in terms of mRNA and protein levels were found. SP1-knockdown A549 cells had decreased clone formation compared with control cells after irradiation (Fig. S2). The survival fractions of SP1-knockdown A549 cells were markedly decreased after irradiation (Fig. 6A, SER=1.59). In contrast, overexpression of SP1 significantly increased the survival fraction (Fig. 6A, SER=0.88). The CCK-8 (Fig. 6B and C), wound-healing (Fig. 6D), and Transwell invasion (Fig. 6E) assays revealed that sh-SP1 cells had decreased viability, migration ability, and invasion ability compared with control cells after irradiation. The aforementioned findings indicated that the depletion of SP1 enhanced radiosensitivity in NSCLC cells.
miR-7 suppresses the expression of the DNA repair protein TP53BP1 by downregulating SP1
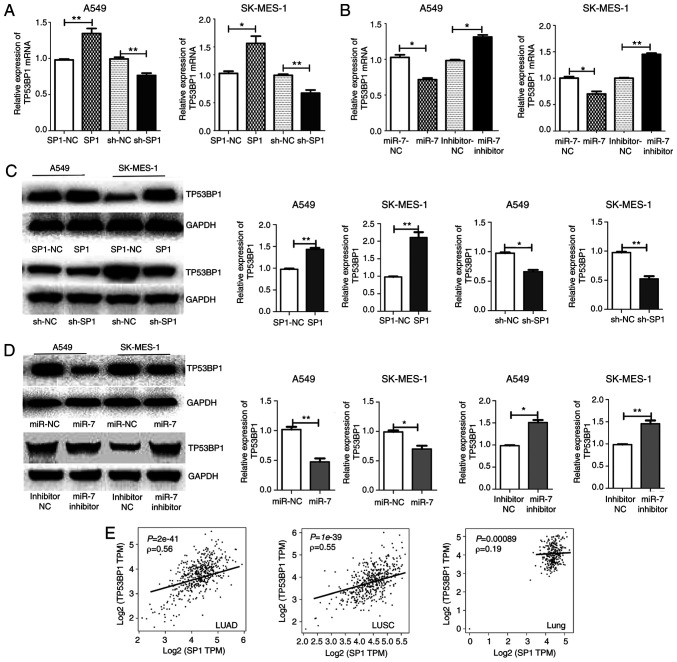
A549 and SK-MES-1 cells were transfected with miR-7, miR-7 inhibitor, SP1, and sh-SP1, and TP53BP1 expression levels were then assessed. The mRNA and protein expression levels of TP53BP1 were decreased after overexpression of miR-7 or downregulation of SP1 in both A549 and SK-MES-1 cells compared with control cells. TP53BP1 expression increased with the inhibition of miR-7 or the overexpression of SP1 (Fig. 7A-D). The correlation between SP1 and TP53BP1 in LUAD, LUSC, and lung tissues from GEPIA revealed that SP1 was correlated with TP53BP1 in lung cancer tissue (Fig. 7E). These finding indicated that the miR-7-induced increase in radiosensitivity may occur, in part, through the inhibition of SP1 and subsequent reduction of TP53BP1 expression.
Discussion
Radioresistance is a major issue in radiotherapy, which is a primary method for treating malignant tumors, however, the underlying reasons remain poorly understood. Numerous studies supported that miRNAs mediate the radiosensitivity of cancer cells by regulating target genes. In nasopharyngeal carcinoma cells, miR-210 has been revealed to negatively regulate radiosensitivity (33). In human cervical carcinoma cells, miR-218 was revealed to promote radiation-induced apoptosis, increasing the radiosensitivity (34). miR-214 promoted radiosensitivity by inhibiting autophagy that was mediated by autophagy-related gene 12 (ATG12) in colorectal cancer (35). In recent years, several studies have demonstrated that miR-7 plays an antitumor role in tumorigenesis, including pancreatic (36), lung (7), thyroid (37), and gastric cancer (38). Concurrently, miR-7 has been revealed to reduce the levels of EGFR and AKT by activating the EGFR/PI3K/AKT signaling pathway, which delays radiation-induced DNA DSBs and increases the radiosensitivity of A549 cells, laryngeal squamous cell carcinoma SQ20B cells, breast cancer MDA-MB-468 cells and malignant glioma U251 and U87 cells (9).
The present results revealed that miR-7 combined with radiation had a significant inhibitory effect on NSCLC cell proliferation, migration and invasion. miR-7 increased radiosensitivity in NSCLC, suggesting that miR-7 may be a potential target for assisting NSCLC radiotherapy. Notably, it was revealed that miR-7 directly targeted SP1, which led to changes in the expression of SP1 downstream genes, such as TP53BP1, in NSCLC cells.
SP1 has been revealed to be aberrantly expressed and activated in tumor tissues, and to participate in the regulation of cancer cell biological functions, such as proliferation, invasion, and angiogenesis (39). SP1 may promote cancer progression by altering the expression of other genes. For instance, Liu et al reported that SP1 binds to the promoter region of lncRNA SNHG14, resulting in the overexpression of lncRNA SNHG14 in clear cell renal carcinoma (40). Zhang et al revealed that the SP1-induced upregulation of the lncRNA LUCAT1 promoted cervical cancer cell proliferation, migration and invasion (41). SP1 is also a microRNA target gene involved in the regulation of radiosensitivity. Kang et al validated SP1 as a target gene of miR-24 and found that the miR-24/SP1 pathway was involved in the regulation of cell viability and radiosensitivity of nasopharyngeal carcinoma cells (42). In their study, however, the expression of SP1 in human nasopharyngeal carcinoma was not studied, and they used transient transfection to overexpress the microRNA and knock down SP1. In the present study, SP1 expression was detected in NSCLC tissue and transfected cells with lentivirus to obtain stably transfected cell lines. In the present study, SP1 was upregulated in NSCLC tissue and was related to the differentiation degree of tumor tissue. Additionally, SP1 has been revealed as a target of the DNA damage response pathway and may play a role in modulating the cellular response to DNA damage (11). It has been reported that SP1 is involved in DSB repair through a mechanism that is independent of its sequence-directed transcriptional effects. SP1 is rapidly recruited to the region immediately adjacent to sites of DNA DSBs. Cells are more sensitive to DNA damage when SP1 is depleted as they show a delayed resolution of γ-H2AX foci with an accumulation of chromosomal abnormalities, suggesting that SP1 plays a role in DSB repair (12).
Proliferation, migration and invasion are important for tumorigenesis, and play a vital role in influencing tumor recurrence and metastasis (43–45). Radiation affects tumor cells in numerous aspects, including cell proliferation, colony formation, apoptosis rate, cell invasion, as well as other biological responses (46,47). The inhibition of tumor cell proliferation, migration and invasion by radiotherapy is of great value for controlling the progression of tumor cells (47,48). Therefore, to investigate the role of miR-7 and SP1 combined with radiation in cell proliferation, migration and invasion, cell proliferation, clonogenic, scratch wound healing and Transwell invasion assays were performed. Overexpression of miR-7 or depletion of SP1 inhibited the proliferation, migration, and invasion of NSCLC, which was enhanced by combination with irradiation.
Overexpression of miR-7 and silencing of SP1 had the same radiosensitization effect. Therefore, miR-7 may increase the X-ray sensitivity of NSCLC cells through the negative regulation of SP1.
DSB repair is an essential protective pathway for the genome to maintain stability after external injury, and it is also the main pathway for the restoration of cancer cells after radiotherapy and the main factor affecting the radiosensitivity of tumor cells (49,50). TP53BP1 is one of the critical regulatory proteins in DNA DSB repair, and it plays an essential role in regulating damage repair balance and maintaining genomic stability (51,52).
In the present study it was demonstrated that the binding sites of miR-7 were in the 3′UTR of SP1 and that SP1 was directly downregulated by miR-7. Upregulated/downregulated miR-7 and SP1 were evaluated to ascertain whether they could regulate radiosensitivity. It was revealed that the radiosensitivity was increased by upregulation of miR-7 or downregulation of SP1. The results illustrated the relationship of miR-7 and SP1 with radiosensitivity. Especially after miR-7 was upregulated/downregulated, the mRNA and protein expression of SP1 inversely changed following miR-7 expression. The consequential results, such as clonogenic formation, proliferation, migration and invasion, were all altered following the transfection. However, we did not perform rescue experiments, which is a limitation of our study.
Concurrently, the binding sites of SP1 were predicted in the promoter region of TP53BP1, and PCR and western blot experiments confirmed the positive regulation of SP1 on TP53BP1. Therefore, it was concluded that the overexpression of miR-7 decreases the expression of SP1, downregulates the transcription of TP53BP1, reduces TP53BP1 protein, and increases the radiosensitivity of NSCLC.
miR-7 was revealed to increase the radiosensitivity of human cancer cells. The present study focused not only on miR-7 but also on SP1. As a transcription factor, SP1 alters the expression of downstream genes. The analysis of SP1 and TP53BP1 in LUAD and LUSC from GEPIA revealed that SP1 was correlated with TP53BP1 in lung cancer tissue. SP1, as a member of the SP transcription factor family, contains a special zinc finger DNA-binding domain. This zinc finger DNA-binding domain binds to the promoter and enhancer regions at the sites with GC boxes, thereby regulating numerous housekeeping genes (53). SP1 may regulate TP53BP1 by binding to the zinc finger domain. Binding sites of SP1 were predicted in the promoter region of TP53BP1, and PCR and western blot experiments confirmed the positive regulation of TP53BP1 by SP1. However, we did not perform a luciferase reporter assay to verify the SP1-binding sites on TP53BP1, which is a limitation of the present study.
In conclusion, it was revealed that miR-7 directly targeted SP1 to increase radiosensitivity in NSCLC. Moreover, SP1 reduced radiosensitivity, in part, through the regulation of TP53BP1. The results revealed that the miR-7/SP1/TP53BP1 axis may play a pivotal role in NSCLC radiosensitivity.
Supplementary Material
Acknowledgements
The authors thank all participating patients. We would also like to thank Dr Zhou Yuan from the Department of Thoracic Surgery at The Fourth Affiliated Hospital of China Medical University for helping to collect the specimens as well as Dr Li Hong and Dr Xu Huihui at the Department of Pathology of The Fourth Affiliated Hospital of China Medical University for their help in preparing and evaluating the specimens.
Funding
The present study was supported by grants from the Clinical Capability Construction Project for Liaoning Provincial Hospital (grant no. LNCCC-B08-2014), and the Youth Backbone Support Program of China Medical University (grant no. QGZ2018064).
Availability of data and materials
Upon reasonable request, the datasets used in this study can be obtained from the corresponding author.
Authors' contributions
YZ, YY, GG, and LL guided the study design, performed analysis of data, interpreted data, and supervised of the study. GG and GS performed most of the experiments and statistical analysis. GG drafted the manuscript. LL, GS, and JW carried out the experiments and sample collection. Each author revised the article critically for important intellectual content. The final manuscript was approved by all authors.
Ethics approval and consent to participate
The Biomedical Ethics Committee of the Fourth Affiliated Hospital of China Medical University approved the present study (Institutional Review Board-approved protocol no. 2014-039). All relevant patients provided written informed consent. The authors are responsible for all aspects of the work. It is ensured that a proper investigation was performed and that issues were resolved to ensure the accuracy and completeness of this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Koh PK, Faivre-Finn C, Blackhall FH, De Ruysscher D. Targeted agents in non-small cell lung cancer (NSCLC): Clinical developments and rationale for the combination with thoracic radiotherapy. Cancer Treat Rev. 2012;38:626–640. doi: 10.1016/j.ctrv.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 5.Redis RS, Berindan-Neagoe I, Pop VI, Calin GA. Non-coding RNAs as theranostics in human cancers. J Cell Biochem. 2012;113:1451–1459. doi: 10.1002/jcb.24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherni I, Weiss GJ. miRNAs in lung cancer: Large roles for small players. Future Oncol. 2011;7:1045–1055. doi: 10.2217/fon.11.74. [DOI] [PubMed] [Google Scholar]
- 7.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Qian J, Gu J, Chang L, Ge D, Chu Y. PA28gamma emerges as a novel functional target of tumour suppressor microRNA-7 in non-small-cell lung cancer. Br J Cancer. 2014;110:353–362. doi: 10.1038/bjc.2013.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KM, Choi EJ, Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol. 2011;101:171–176. doi: 10.1016/j.radonc.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 10.Yano S, Kondo K, Yamaguchi M, Richmond G, Hutchison M, Wakeling A, Averbuch S, Wadsworth P. Distribution and function of EGFR in human tissue and the effect of EGFR tyrosine kinase inhibition. Anticancer Res. 2003;23:3639–3650. [PubMed] [Google Scholar]
- 11.Beishline K, Kelly CM, Olofsson BA, Koduri S, Emrich J, Greenberg RA, Azizkhan-Clifford J. Sp1 facilitates DNA double-strand break repair through a nontranscriptional mechanism. Mol Cell Biol. 2012;32:3790–3799. doi: 10.1128/MCB.00049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olofsson BA, Kelly CM, Kim J, Hornsby SM, Azizkhan-Clifford J. Phosphorylation of Sp1 in response to DNA damage by ataxia telangiectasia-mutated kinase. Mol Cancer Res. 2007;5:1319–1330. doi: 10.1158/1541-7786.MCR-07-0374. [DOI] [PubMed] [Google Scholar]
- 13.Iwahori S, Yasui Y, Kudoh A, Sato Y, Nakayama S, Murata T, Isomura H, Tsurumi T. Identification of phosphorylation sites on transcription factor Sp1 in response to DNA damage and its accumulation at damaged sites. Cell Signal. 2008;20:1795–1803. doi: 10.1016/j.cellsig.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, Wang X, Xia Z, Yang L, Ding Z, Chen S, Lai B, Zhang N. Transcriptional factor specificity protein 1 (SP1) promotes the proliferation of glioma cells by up-regulating midkine (MDK) Mol Biol Cell. 2015;26:430–439. doi: 10.1091/mbc.E14-10-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang WJ, Song MJ, Park EY, Lee JJ, Park JH, Park K, Park JH, Kim HP. Transcription factors Sp1 and Sp3 regulate expression of human ABCG2 gene and chemoresistance phenotype. Mol Cells. 2013;36:368–375. doi: 10.1007/s10059-013-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. P53 binding protein 1 (53bp1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XX, Ma M, Sang MX, Zhang XY, Liu ZK, Song H, Zhu SC. BMI-1 suppression increases the radiosensitivity of oesophageal carcinoma via the PI3K/Akt signaling pathway. Oncol Rep. 2018;39:667–678. doi: 10.3892/or.2017.6136. [DOI] [PubMed] [Google Scholar]
- 19.Wingender E, Schoeps T, Haubrock M, Krull M, Dönitz J. TFClass: Expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018;46:D343–D347. doi: 10.1093/nar/gkx987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kay FU, Kandathil A, Batra K, Saboo SS, Abbara S, Rajiah P. Revisions to the tumor, node, metastasis staging of lung cancer (8th edition): Rationale, radiologic findings and clinical implications. World J Radiol. 2017;9:269–279. doi: 10.4329/wjr.v9.i6.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travis WD. The 2015 WHO classification of lung tumors. Pathologe. 2014;35(Suppl 2):188. doi: 10.1007/s00292-014-1974-3. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 27.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui X, Xiao D, Cui Y, Wang X. Exosomes-derived long non-coding RNA HOTAIR reduces laryngeal cancer radiosensitivity by regulating microRNA-454-3p/E2F2 axis. Onco Targets Ther. 2019;12:10827–10839. doi: 10.2147/OTT.S224881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du M, Wang J, Chen H, Wang S, Chen L, Xu Y, Su F, Lu X. MicroRNA-200a suppresses migration and invasion and enhances the radiosensitivity of NSCLC cells by inhibiting the HGF/c-Met signaling pathway. Oncol Rep. 2019;41:1497–1508. doi: 10.3892/or.2018.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, Lu YY, Tang YA, Yang YC, Yang PC, Wang YC. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–5817. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 33.Li BY, Luo Y, Zhao WS, Zhang L, Zhou HJ, Zou YC, Zhang T. MicroRNA-210 negatively regulates the radiosensitivity of nasopharyngeal carcinoma cells. Mol Med Rep. 2017;16:1401–1408. doi: 10.3892/mmr.2017.6694. [DOI] [PubMed] [Google Scholar]
- 34.Yuan W, Xiaoyun H, Haifeng Q, Jing L, Weixu H, Ruofan D, Jinjin Y, Zongji S. MicroRNA-218 enhances the radiosensitivity of human cervical cancer via promoting radiation induced apoptosis. Int J Med Sci. 2014;11:691–696. doi: 10.7150/ijms.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y, Qian XL, Liao WT, Ding YQ, Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7:16. doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia J, Cao T, Ma C, Shi Y, Sun Y, Wang ZP, Ma J. miR-7 suppresses tumor progression by directly targeting MAP3K9 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:121–132. doi: 10.1016/j.omtn.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue K, Wang X, Wu Y, Zhou X, He Q, Duan Y. microRNA-7 regulates cell growth, migration and invasion via direct targeting of PAK1 in thyroid cancer. Mol Med Rep. 2016;14:2127–2134. doi: 10.3892/mmr.2016.5477. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Dou W, He L, Liang S, Tie J, Liu C, Li T, Lu Y, Mo P, Shi Y, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363–1372. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 39.Chang WC, Hung JJ. Functional role of post-translational modifications of Sp1 in tumorigenesis. J Biomed Sci. 2012;19:94. doi: 10.1186/1423-0127-19-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Ye Z, Zhao X, Ji Z. SP1-induced up-regulation of lncRNA SNHG14 as a ceRNA promotes migration and invasion of clear cell renal cell carcinoma by regulating N-WASP. Am J Cancer Res. 2017;7:2515–2525. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Liu SK, Song L, Yao HR. SP1-induced up-regulation of lncRNA LUCAT1 promotes proliferation, migration and invasion of cervical cancer by sponging miR-181a. Artif Cells Nanomed Biotechnol. 2019;47:556–564. doi: 10.1080/21691401.2019.1575840. [DOI] [PubMed] [Google Scholar]
- 42.Kang M, Xiao J, Wang J, Zhou P, Wei T, Zhao T, Wang R. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 2016;5:1163–1173. doi: 10.1002/cam4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R, Peng C, Zhang X, Wu Y, Pan S, Xiao Y. Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci. 2017;182:80–84. doi: 10.1016/j.lfs.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 45.Banyard J, Chung I, Migliozzi M, Phan DT, Wilson AM, Zetter BR, Bielenberg DR. Identification of genes regulating migration and invasion using a new model of metastatic prostate cancer. BMC Cancer. 2014;14:387. doi: 10.1186/1471-2407-14-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan XM, Liu XN, Li YX, Cao YQ, Silayiding A, Zhang RK, Wang JP. MicroRNA-498 promotes proliferation, migration, and invasion of prostate cancer cells and decreases radiation sensitivity by targeting PTEN. Kaohsiung J Med Sci. 2019;35:659–671. doi: 10.1002/kjm2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pordanjani SM, Hosseinimehr SJ. The role of NF-κB inhibitors in cell response to radiation. Curr Med Chem. 2016;23:3951–3963. doi: 10.2174/0929867323666160824162718. [DOI] [PubMed] [Google Scholar]
- 48.Yan J, Jiang Y, Lu J, Wu J, Zhang M. Inhibiting of proliferation, migration, and invasion in lung cancer induced by silencing interferon-induced transmembrane protein 1 (IFITM1) Biomed Res Int. 2019;2019:9085435. doi: 10.1155/2019/9085435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeggo PA, Löbrich M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem J. 2015;471:1–11. doi: 10.1042/BJ20150582. [DOI] [PubMed] [Google Scholar]
- 51.Iwabuchi K, Li B, Massa HF, Trask BJ, Date T, Fields S. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 52.Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JA, Suh DC, Kang JE, Kim MH, Park H, Lee MN, Kim JM, Jeon BN, Roh HE, Yu MY, et al. Transcriptional activity of Sp1 is regulated by molecular interactions between the zinc finger DNA binding domain and the inhibitory domain with corepressors, and this interaction is modulated by MEK. J Biol Chem. 2005;280:28061–28071. doi: 10.1074/jbc.M414134200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Upon reasonable request, the datasets used in this study can be obtained from the corresponding author.