Abstract
Homeostasis of hematopoietic stem and progenitor cells is a tightly regulated process. The disturbance of the balance in the hematopoietic progenitor pool can result in favorable conditions for development of diseases such as myelodysplastic syndromes and leukemia. It has been shown recently that mice lacking p15Ink4b have skewed differentiation of common myeloid progenitors toward the myeloid lineage at the expense of erythroid progenitors. The lack of p15INK4B expression in human leukemic blasts has been linked to poor prognosis and increased risk of myelodysplastic syndromes transformation to acute myeloid leukemia. However, the role of p15Ink4b in disease development is just beginning to be elucidated. This study examines the collaboration of the loss of p15Ink4b with Nup98-HoxD13 translocation in the development of hematological malignancies in a mouse model. Here, we report that loss of p15Ink4b collaborates with Nup98-HoxD13 transgene in the development of predominantly myeloid neoplasms, namely acute myeloid leukemia, myeloproliferative disease, and myelodysplastic syndromes. This mouse model could be a very valuable tool for studying p15Ink4b function in tumorigenesis as well as preclinical drug testing.
Keywords: Acute myelogenous leukemia, Myelodysplastic syndromes, Myeloproliferative disease, Nup98-HoxD13, p15Ink4b, Transgenic mouse
Introduction
The myelodysplastic syndromes (MDS) are conditions characterized by ineffective production of white blood cells, red blood cells, and platelets and often progress to acute myeloid leukemia (AML). In the U.S. alone, approximately 20,000 people are diagnosed with MDS yearly [1]. Because of limited therapeutic options and lack of understanding of the disease etiology, it is estimated that ~70% of patients die from MDS-related complications including AML.
A striking 80% of human leukemia and MDS cases have silenced expression of the CDKN2B locus that encodes the tumor suppressor p15Ink4b [2–7]. Nonetheless, the function of p15Ink4b in the disease development remains to be elucidated [8, 9]. We have previously generated a conditional p15Ink4b knockout mouse model that expresses Cre recombinase from the lysozyme M promoter, p15Ink4bfl/fl-LysMCre. Using this model, we demonstrated that myeloid-specific inactivation of p15Ink4b in mice results in the development of nonreactive monocytosis manifested by increased numbers of monocytes in the peripheral blood, expansion of monocytes, myelomonocytes (the type of cell observed in chronic myelomonocytic leukemia), and increased numbers of early granulocyte-macrophage progenitors in the bone marrow [10–12]. The p15Ink4b knockout mice were also found to be more susceptible to MOL4070LTR virus-induced myeloid leukemia [10, 13].
A number of MDS associated oncogenes/translocations have been reported. Many of them have been used to generate chimeric or transgenic animals in an attempt to model this disease [14, 15]. The Nup98-HoxD13 (NHD13) transgenic animals were found to recapitulate many of the features of human MDS, like anemia, neutropenia, lymphocytopenia, and multilineage dysplasia. In this mouse model, the NHD13 expression was directed to hematopoietic tissue by the use of the Vav1 promoter. The MDS was evident at 5 months of age and approximately half of animals with MDS transformed to leukemia. Unlike human MDS, where the AML transformation is predominant, the NHD13 mice developed a variety of leukemias including myeloid, erythroid, lymphoid, and undifferentiated.
It was of our interest to assess the collaboration of loss of p15Ink4b with over-expression of NHD13 in development of MDS and leukemic transformation. For this purpose, we crossed the two animal models and generated the p15Ink4bfl/flNHD13 mouse strain. Here, we report that myeloid-specific loss of p15Ink4b accelerates the disease development in the NHD13 transgenic animals and results predominantly in the development of the myeloid neoplasms.
Materials and Methods
Animals
The p15Ink4bfl/fl-LysMCre mice and the Vav1-NHD13 transgenic animals used in this study were previously described and were maintained on a C57BL/6–129SvJ and FVB background, respectively [10, 15]. The two lines were crossed to generate mice with the following genotype: C57BL6/FVB/129-Tg (Vav1-NUP98/HOXD13)/p15Ink4bfl/fl-LysMCre+ (p15Ink4bfl/flNHD13). All animals were housed at the NCI-SAIC-Fredrick facility. Experiments were carried out according to the protocols approved by The Institutional Animal Care Committee at The National Cancer Institute, NIH. The control animals were obtained from the same cross as experimental groups and wild-type (WT) refers to animals expressing WT p15Ink4b allele and LysM Cre gene (p15Ink4bwt/wtLysM Cre) for accurate comparison.
Genotyping
Mouse tail DNA was used for genotyping. The following primers and PCR condition were used for genotyping. For the floxed allele of p15Ink4b (p15Ink4bfl/fl): FP: ATACCAAGGAAACAAGATCCTC, RP: AGGACAGCCAGGGCTATACAG. Product size: 480 bp (floxed allele) and 360 bp (WT allele). For Cre: FP: CGATGCAACGAGTGATGAGG, RP: GCATTGCTGTCACTTGGTCCT. Product size is 300 bp. For the NHD13 genotyping, a quantitative assay was designed: FP: GGGCCCCTGGATTTAATACTACG, RP: GCACATGTCCGGCTGATTTAGA, FAM-NFQ-Probe: CTCCAAAG CCCAAAGTGGCTGT. The primers and probe designed for the mouse p15Ink4b were used as an internal control for NHD13 genotyping: FP: GGTCCCCAGGCCTTACC, RP: GGTGCCCAGTTAC AACTCACA, FAM-NFQ-Probe: CTCGTGCGCCCTCTGCC.
Blood Collection and Analysis
Blood samples were harvested every other month and at the time of sacrifice by mandibular bleeding technique. Samples were analyzed using a CDC Hemavet blood counter at the Pathology/Histology Laboratory, LASP, Frederick. For differential analysis, blood smears were stained with Diff-Quick (Siemens HealthCare Diagnostics, Tarrytown, NY, usa.healthcare.siemens.com) and scored using an Olympus BH2 and DP70 light microscopes.
Classification of Hematological Neoplasms
Animals were monitored daily for the signs of disease. Typical signs included low activity level, ruffled fur, hunched posture, labored breathing, palpable masses, abdominal swelling, and bleeding. Moribund animals were sacrificed and preliminary necropsy performed noting organomegaly, discoloration of organs, and tumor masses. One femur/tibia was fixed in 10% neutral buffered formalin (NBF). Pieces of spleen, liver, enlarged lymph nodes, and enlarged thymus were fixed in Telly’s, as well as, frozen in O.C.T. blocks in liquid nitrogen. Fresh femur/tibia and a piece of spleen were immunophenotyped for the following antigens: Mac1/Gr1, CD4/CD8, B220/IgM, ckit/Sca1, Ter119/CD71, CD41/CD61, and CD3/CD19. The enlarged thymus was immunophenotyped for CD4/CD8. The following dye-conjugated anti-mouse antibodies were used for immunophenotyping: FITC-CD11b (M1/70, BD Pharmingen, San Jose, CA, www.bdbiosciences.com), PE-Ly6G and Ly6C (RB6–8C5, BD Pharmingen), FITC-CD4 (L3T4, eBiosciences, San Diego, CA, www.ebioscience.com), PE-CD8a (53–6.7, eBiosciences), FITC-B220 (RA3–6B2, eBiosciences), PE-IgM (II/41, eBiosciences), FITC-cKit (2B8, eBioscience), PE-Sca-1 (D7, eBiosciences), FITC-Ter119 (TER-119, eBiosciences), PE-CD71 (C2, BD Pharmingen), FITC-CD41 (eBioMWReg30, eBiosciences), PE-CD61 (2C9.G3, eBiosciences), FITC-CD3e (145–2C11, eBiosciences), and PE-CD19 (eBio1D3, eBiosciences). Data were acquired using BD FACSCanto II equipped with High Throughput Sampler option (Becton Dickinson). Data were analyzed using FlowJo (Tree Star, Inc.). Diagnoses were performed based on the combination of the flow cytometric analysis of fresh tissues, complete blood count (CBCs), morphology, necropsy findings, histology, and immunohistochemistry. Leukemia classification was according to the Bethesda proposal for nonlymphoid and lymphoid neoplasms in mice [16, 17].
Immunohistochemistry
Tissue sections and staining for CD3 (AbD Serotec, Oxford, UK, www.abdserotec.com), myeloperoxidase (MPO) (DAKO, Carpinteria, CA, www.dako.com), F4/80 (eBioscience) antigens, and hematoxylin and eosin (H&E) were performed by the Pathology/Histotechnology Laboratory (Frederick National Laboratory for Cancer Research) using conventional staining techniques of O.C.T. blocks.
Statistical Analysis
Statistical analysis was done using Microsoft Excel and Graph Pad Prism software. The unpaired two-tailed Student’s t test, Mann-Whitney U test, and log-rank tests were used to calculate p values.
Results and Discussion
The Loss of p15Ink4b Tumor Suppressor Collaborates with the NHD13 Transgene
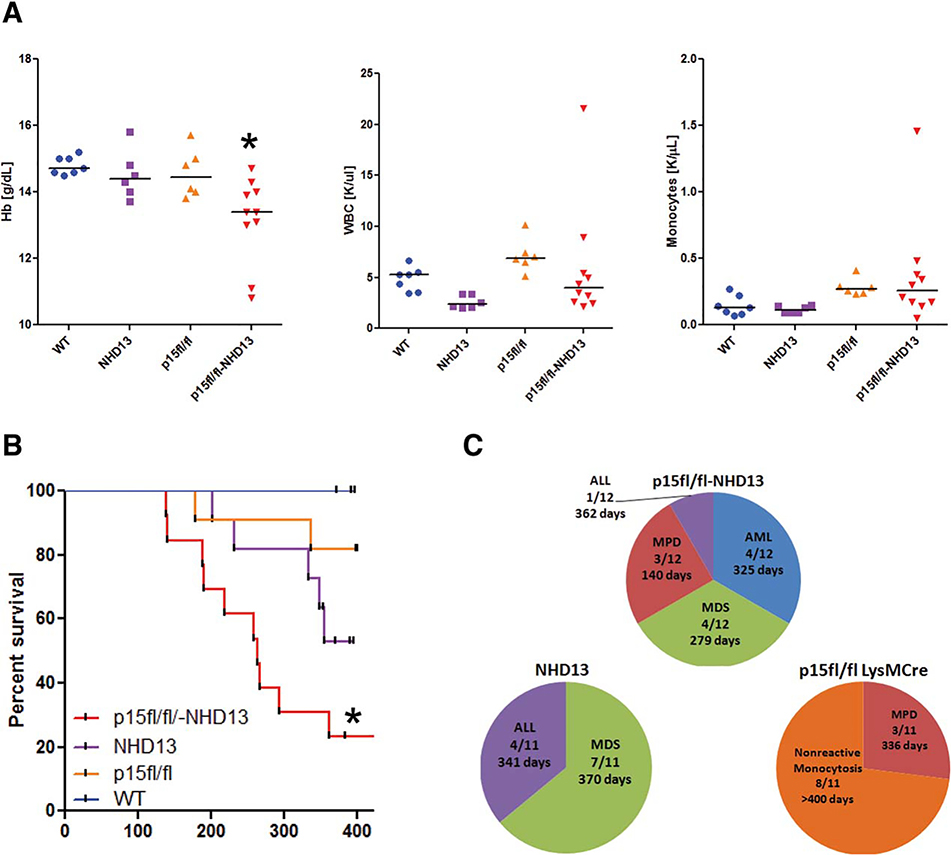
Examination of blood of 3–4-month-old p15Ink4bfl/flNHD13 mice revealed a statistically significant decrease in hemoglobin content as compared to either NHD13 or p15Ink4bfl/fl-LysMCre group alone (Fig. 1A). The total white blood cell counts of p15Ink4bfl/fNHD13 animals were not as elevated as in p15Ink4b knockout animals but were higher than in NHD13 alone animals (Fig. 1A). The increase in white blood cell counts in the p15Ink4bfl/fNHD13 as well as p15Ink4bfl/fl-LysMCre group was due to a higher monocyte count (Fig. 1A). The observed increase in the white blood cell counts in the p15Ink4bfl/fl-LysMCre mice reached statistical significance, as compared to having only an upward tendency in the previous report [10]. This might be due to examination of animals at a younger age and closer age-matching (3–4 months, instead 5–7, or 8–10). Alternatively, it may be a strain-specific phenomenon. Over time the p15Ink4bfl/flNHD13 animals had a tendency toward increased neutrophil and monocyte counts and decreased red blood cell, hemoglobin, and hematocrit levels (data not shown). The survival of p15Ink4bfl/flNHD13 animals was significantly lower as compared to the NHD13 group (Fig. 1B). The 50% survival was 264 days for this group. Approximately 150 days shorter compared to NHD13 transgenic animals. The combination of these two genes accelerates the development of hematological neoplasms as compared to either one of the genes alone. These data provide an additional argument supporting the important function of p15Ink4b as a tumor suppressor.
Figure 1.
Loss of p15Ink4b accelerates development of myeloid neoplasms in mice. Complete blood counts from p15Ink4bfl/flNHD13 mice aged 3–4 months. (A): Complete blood counts from p15Ink4bfl/flNHD13 mice aged 3–4 months. Hb levels were significantly decreased in the p15Ink4bfl/flNHD13 group as compared to WT animals (p15Ink4bwt/wt-LysMCre), *, p = .0024. The WBCs were significantly decreased in the NHD13 group (*, p = .001) and significantly increased in the p15Ink4bfl/fl-LysMCre animals, *, p = .01, as compared to WT. The monocyte count was significantly increased in the p15Ink4bfl/fl-LysMCre group as compared to WT, *, p = .008. (B): Survival of p15Ink4bfl/flNHD13 animals is significantly shorter as compared to the animals in NHD13 group, *, p = .0007. (C): Classification of hematologic malignancies in p15Ink4bfl/flNHD13 mice. The number of animals and the median survival among the animals diagnosed with the same malignancy are shown. Abbreviations: ALL, acute lymphoid leukemia; AML, acute myeloid leukemia; Hb, hemoglobin; MDS, myelodysplastic syndromes; MPD, myeloproliferative disease; NHD13, Nup98-HoxD13; WBC, white blood cell count; WT, wild type.
Increased Incidence of Myeloid Neoplasms in p15Ink4bfl/fl NHD13 Animals
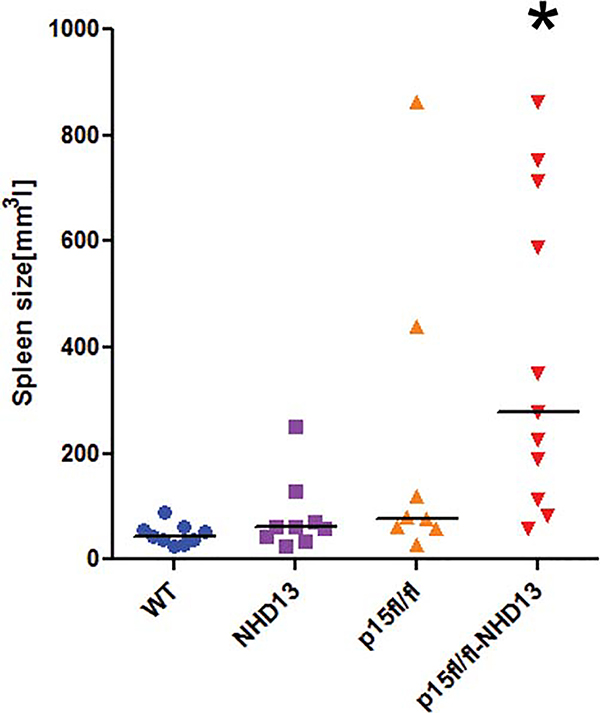
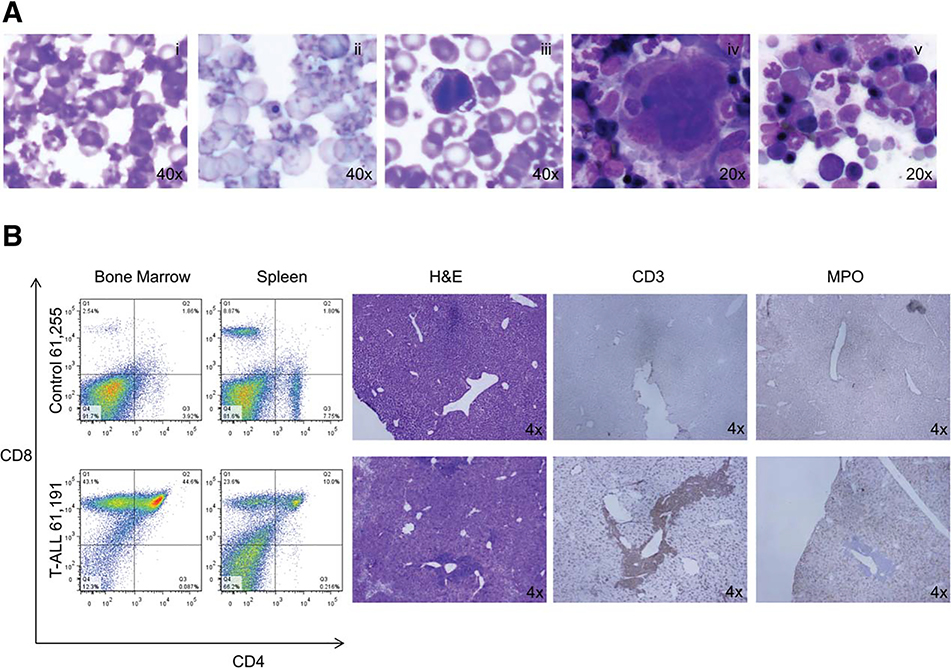
The moribund animals as well as animals that survived until the end of study were sacrificed, autopsied, and tissues analyzed for hematological malignancies as described in Materials and Methods. We found that one-third of p15Ink4bfl/flNHD13 animals develop acute myeloid leukaemia (AML) with evidence of splenomegaly, and dissemination into the blood (Figs. 1C, 2, 3). Examination of bone marrow and spleen of those animals for expression of various lineage-specific markers showed an increase in the number of Mac-1/Gr1-positive cells (Fig. 3, middle panel). Morphological analyses demonstrated the presence of blast cells of myeloid origin in the blood, spleen, and bone marrow (Fig. 3, middle panel). Another third of p15Ink4bfl/flNHD13 animals developed myeloproliferative disease (MPD) as evidenced by an increase in the number of white blood cells, mostly neutrophils and eosinophils, in the bone marrow, spleen, and peripheral blood (Fig. 3, lower panel). The incidence of MPD in the p15Ink4bfl/flNHD13 group was similar to the p15Ink4bfl/fl-LysMCre group but the disease was more aggressive and resulted in a decreased survival rate (Fig. 1C) (140 days for p15Ink4bfl/flNHD13 and 336 days for p15Ink4bfl/fl-LysMCre). The rest of the animals in this group, except for one that had lymphoid leukemia, died of MDS. The median survival of p15Ink4bfl/flNHD13 animals with MDS was 91 days shorter as compared to NHD13 animals. Dysplastic cells of both myeloid and erythroid origins were noted and abnormalities included poikilocytosis, Howell-Jolly bodies, cells with multiple nuclei, and abnormal nuclear maturation (Fig. 4A). Consistent with the previous report, the majority of p15Ink4bfl/fl-LysMCre animals survived until the end of the study (>400 days) and show signs of nonreactive monocytosis [10]. The NHD13 transgenic animals in this study developed MDS with the majority of animals experiencing progressive leukocytopenia, lymphocytopenia, and dyspoiesis in those blood lineages. Some of the animals progressed to acute leukemia. Interestingly, all the leukemias were of lymphoid origin (Figs. 1C, 4B). Although this may be due to the fact that p15Ink4b is not absent in these animals, two other explanations are possible. First, the experiment was terminated at 400 days and there is a possibility that some of the remaining MDS cases would eventually transform to myeloid leukaemia. Second, the ratio of myeloid to lymphoid malignancies may be strain specific. All the genotypes in this study are on the mixed background (50% C57Bl/6–129SvJ and 50% FVB) and the NHD13 transgenic animals on this background developed predominantly lymphoid malignancies. This is similar to the spectrum of hematological malignancies developed by the NHD13 animals on the pure FVB background that has been described previously [15].
Figure 2.
Increased myeloid proliferation in the spleens of p15Ink4bfl/flNHD13 mice. The spleen size of p15Ink4bfl/flNHD13 mice at the time of sacrifice, *, p = .0005. Abbreviations: NHD13, Nup98-HoxD13; WT, wild type.
Figure 3.
AML and MPD in the p15Ink4bfl/flNHD13 mice. Increased numbers of Mac1/Gr-1 double positive cells in the bone marrow and spleen of the p15Ink4bfl/flNHD13 mice with AML (61,010), and MPD (61,131). Diff-Quick-stained smears of peripheral blood, spleen, and bone marrow. In the case of AML, myeloid blasts are present in the peripheral blood, spleen, and bone marrow. In the case of MPD, the bone marrow is comprised of mostly neutrophilic metamyelocytes and mature neutrophils that disseminated to the spleen and peripheral blood. Abbreviations: AML, acute myeloid leukemia; MPD, myeloproliferative disease.
Figure 4.
Myelodysplastic syndromes in p15Ink4bfl/flNHD13 mice and acute T-ALL in Nup98-HoxD13 (NHD13) mice. (A): Diff-Quick-stained smears of peripheral blood of p15Ink4bfl/flNHD13 mice showing: (i) poikilocytosis, (ii) Howell-Jolly bodies, and (iii) cells with multiple nuclei. Diff-Quick-stained smears of bone marrow showing: (iv) hypolobulated megakaryocyte and (v) cells with abnormal nuclear maturation. (B): Increased numbers of CD4/CD8 double positive cells in the bone marrow and spleen of NHD13 mice. H&E staining of liver sections showing metastasis adjacent to large blood vessels. Staining of the liver sections for CD3 and MPO. Abbreviation: T-ALL, acute T lymphoblastic leukemia.
Conclusion
Our data demonstrate that the lack of p15Ink4b results in skewing of hematological neoplasms to those of myeloid origin and in combination with NHD13 a large proportion of animals progress to AML. The result is consistent with the recently described function of p15Ink4b in the regulation of cellular commitment of hematopoietic progenitor cells. Loss of p15Ink4b in the myeloid compartment allows for expansion of granulocyte-macrophage progenitors [8, 9, 11, 12]. We hypothesize that the expanded myeloid cells are the precursors of malignant neoplasms. In summary, we have developed a mouse model that closely recapitulates features of human diseases like MDS, MPD, and AML. The data shed new light on the contribution of p15Ink4b to the development of hematological neoplasms and might be a useful tool in drug discovery for myeloid neoplasms.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Cogle CR, Craig BM, Rollison DE, et al. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: High number of uncaptured cases by cancer registries. Blood 2011;117:7121–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drexler HG. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia 1998;12: 845–859. [DOI] [PubMed] [Google Scholar]
- 3.Herman JG, Civin CI, Isso CI, et al. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res 1997;57:837–841. [PubMed] [Google Scholar]
- 4.Herman JG, Jen J, Merlo A, et al. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res 1996;56:722–727. [PubMed] [Google Scholar]
- 5.Markus J, Garin MT, Bies J, et al. Methylation-independent silencing of the tumor suppressor INK4b (p15) by CBFbeta-SMMHC in acute myelogenous leukemia with inv(16). Cancer Res 2007;67:992–1000. [DOI] [PubMed] [Google Scholar]
- 6.Paul TA, Bies J, Small D, et al. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood 2010;115:3098–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tien HF, Tang JH, Tsay W, et al. Methylation of the p15(INK4B) gene in myelodysplastic syndrome: It can be detected early at diagnosis or during disease progression and is highly associated with leukaemic transformation. Br J Haematol 2001;112:148–154. [DOI] [PubMed] [Google Scholar]
- 8.Wolff L, Bies J. p15Ink4b Functions in determining hematopoietic cell fates: Implications for its role as a tumor suppressor. Blood Cells Mol Dis 2013;50:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolff L, Humeniuk R. Erythroid versus myeloid lineage commitment: Regulating the master regulators. Stem Cells 2013;31:1237–1244. [DOI] [PubMed] [Google Scholar]
- 10.Bies J, Sramko M, Fares J, et al. Myeloid-specific inactivation of p15Ink4b results in monocytosis and predisposition to myeloid leukemia. Blood 2010;116:979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humeniuk R, Rosu-Myles M, Fares J, et al. The role of tumor suppressor p15Ink4b in the regulation of hematopoietic progenitor cell fate. Blood Cancer J 2013;2:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosu-Myles M, Taylor BJ, Wolff L, et al. Loss of the tumor suppressor p15Ink4b enhances myeloid progenitor formation from common myeloid progenitors. Exp Hematol 2007;35:394–406. [DOI] [PubMed] [Google Scholar]
- 13.Wolff L, Garin MT, Koller R, et al. Hypermethylation of the Ink4b locus in murine myeloid leukemia and increased susceptibility to leukemia in p15(Ink4b)-deficient mice. Oncogene 2003;22:9265–9274. [DOI] [PubMed] [Google Scholar]
- 14.Beachy SH, Aplan PD. Mouse models of myelodysplastic syndromes. Hematol Oncol Clin North Am 2010;24:361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin YW, Slape C, Zhang Z, et al. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 2005;106:287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 2002;100:238–245. [DOI] [PubMed] [Google Scholar]
- 17.Morse HC 3rd, Anver MR, Frederickson TN, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 2002;100:246–258. [DOI] [PubMed] [Google Scholar]