Abstract
Background
Chitosan (CTS), a natural polysaccharide, exhibits multiple functions of stress adaptation regulation in plants. However, effects and mechanism of CTS on alleviating salt stress damage are still not fully understood. Objectives of this study were to investigate the function of CTS on improving salt tolerance associated with metabolic balance, polyamine (PAs) accumulation, and Na+ transport in creeping bentgrass (Agrostis stolonifera).
Results
CTS pretreatment significantly alleviated declines in relative water content, photosynthesis, photochemical efficiency, and water use efficiency in leaves under salt stress. Exogenous CTS increased endogenous PAs accumulation, antioxidant enzyme (SOD, POD, and CAT) activities, and sucrose accumulation and metabolism through the activation of sucrose synthase and pyruvate kinase activities, and inhibition of invertase activity. The CTS also improved total amino acids, glutamic acid, and γ-aminobutyric acid (GABA) accumulation. In addition, CTS-pretreated plants exhibited significantly higher Na+ content in roots and lower Na+ accumulation in leaves then untreated plants in response to salt stress. However, CTS had no significant effects on K+/Na+ ratio. Importantly, CTS enhanced salt overly sensitive (SOS) pathways and also up-regulated the expression of AsHKT1 and genes (AsNHX4, AsNHX5, and AsNHX6) encoding Na+/H+ exchangers under salt stress.
Conclusions
The application of CTS increased antioxidant enzyme activities, thereby reducing oxidative damage to roots and leaves. CTS-induced increases in sucrose and GABA accumulation and metabolism played important roles in osmotic adjustment and energy metabolism during salt stress. The CTS also enhanced SOS pathway associated with Na+ excretion from cytosol into rhizosphere, increased AsHKT1 expression inhibiting Na+ transport to the photosynthetic tissues, and also up-regulated the expression of AsNHX4, AsNHX5, and AsNHX6 promoting the capacity of Na+ compartmentalization in roots and leaves under salt stress. In addition, CTS-induced PAs accumulation could be an important regulatory mechanism contributing to enhanced salt tolerance. These findings reveal new functions of CTS on regulating Na+ transport, enhancing sugars and amino acids metabolism for osmotic adjustment and energy supply, and increasing PAs accumulation when creeping bentgrass responds to salt stress.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02720-w.
Keywords: Ion transportation, Sugar metabolism, SOS pathway, Amino acids, Osmotic adjustment, Compartmentalization, Gene expression
Background
Crop growth and productivity is severely affected by salt stress all over the world. Plants uptake large amount of sodium ions (Na+) in cells resulting in the ionic imbalance under salt stress [1]. Excessive accumulation of Na+ leads to tissue necrosis and early aging of blades due to inhibition in protein synthesis, enzyme reactions, and photosynthesis [2–4]. Plants mediate Na+ concentration depending on the salt overly sensitive (SOS) pathways, Na+/H+ antiporters (NHX), and high-affinity Na+/K+-permeable transporter (HKT) in vacuoles and plasma membranes [5–7]. SOS1 encodes a plasma membrane Na+/H+ antiporter which extrudes Na+ from the cytosol into the rhizosphere and is also involved in the long-distance transport of Na+ [8, 9]. SOS3 and SOS2 are necessary for the synthesis of SOS1 mRNA and protein under salt stress and jointly participate in the regulation of SOS1 phosphorylation [10]. In addition, the SOS2 and SOS3 may also regulate activities of H+-ATPase, H+-PPase, and NHX in the vacuolar membrane [11]. NHX mediates the exchange of Na+/H+ and K+/H+, thus affecting salt tolerance and potassium ions (K+) nutrition [12, 13]. It has been reported that NHXs overexpression can promote the efflux of excessive Na+ from cytoplasm and separate excess Na+ into vacuoles, resulting in increased salt tolerance in different plant species [14–16]. HKT1 plays an important role in retrieving Na+ from the xylem sap, thereby inhibiting Na+ transport to the photosynthetic tissues [7]. H+-ATPase or H+-PPase hydrolyzes ATP or PPi in the cytoplasm to pump H+ into the vacuole, which generates the electrochemical gradient and a proton motive force for the expulsion and separation of Na+ in cells [16–18]. Improved gene expression encoding H+-PPase could be associated with enhanced salt tolerance in transgenic Lotus corniculatus and alfalfa (Medicago sativa) [19, 20].
Sugars metabolism is regarded as a common response under different abiotic stresses [21]. Previous studies have shown that sucrose metabolism is one of key regulatory systems that confer tolerance to abiotic stresses, such as drought, high temperature and salt stress [22–24]. Chitosan (CTS) is a polycationic polysaccharide obtained by de-acetylation of chitin and also widely exists in plant species. In recent years, the CTS has been widely used in agricultural production as an exogenous additive substance being safe and cheap. The CTS exhibits positive effects on plant growth and tolerance to abiotic and biotic stresses [25, 26]. The study of Zhang et al. [27] found that CTS regulated a range of metabolic pathways including carbon and nitrogen metabolism in wheat (Triticum aestivum) leaves, thereby promoting plant growth. Exogenous CTS application effectively alleviated drought damage in white clover (Trifolium repens) through enhancing sucrose, mannose, and fructose accumulation in leaves [28]. The CTS also plays positive role in regulating salt tolerance in plants. For example, the CTS improved salt tolerance of maize (Zea mays) seedlings associated with enhancement in photosynthesis, glycolysis, and nitrogen assimilation [29]. With increasing salt stress, safflower (Carthamus tinctorius) and sunflower (Helianthus annuus) seed priming with CTS significantly improved seeds germination and antioxidant enzymes catalase (CAT) and peroxidase (POD) activities to mitigate oxidative damage in seedings [30]. Although these previous studies demonstrated that the CTS alleviated salt stress associated with some physiological effects such as antioxidant capacity, photosynthesis, and nitrogen assimilation, CTS-induced salt tolerance in relation to sugar and amino acid metabolism, polyamines (PAs) accumulation, and Na+ transport is still not completely elucidated in plants.
The turfgrass used in the sports industry needs higher frequency of maintenance and management than it is used in other area. A long-term use of chemicals, fertilizers, pesticides, and reclaimed water accelerates the soil salinization leading to the decline in turf quality and the increase in management difficulty and maintenance cost [31–33]. Creeping bentgrass (Agrostis stolonifera) is one of the most important turfgrasses and used extensively in the sports industry including golf courses and tennis lawns because of its high turf quality and creeping growth characteristics, however, salt tolerance of creeping bentgrass has been ranked as moderately sensitive [34]. Objectives of this study were to examine physiological effects of CTS on regulating water balance, antioxidant capacity, and photosynthesis salt stress. This study further hypothesizes that CTS-induced salt tolerance could be related to metabolite accumulation, changes in endogenous PAs, and Na+ transport in leaf and root. Current study provides a new and comprehensive insight into complex mechanism of CTS-regulated salt tolerance in plants.
Results
Effects of CTS on water status and photosynthesis in leaf
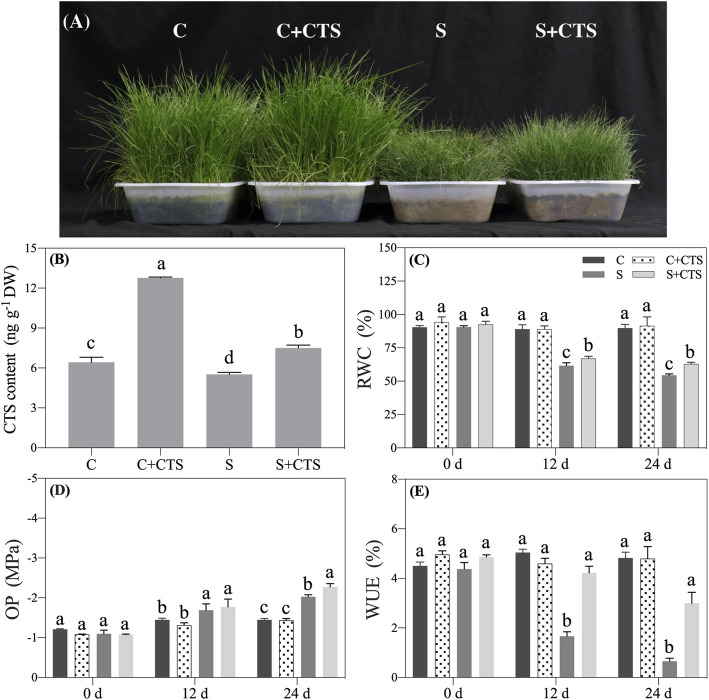
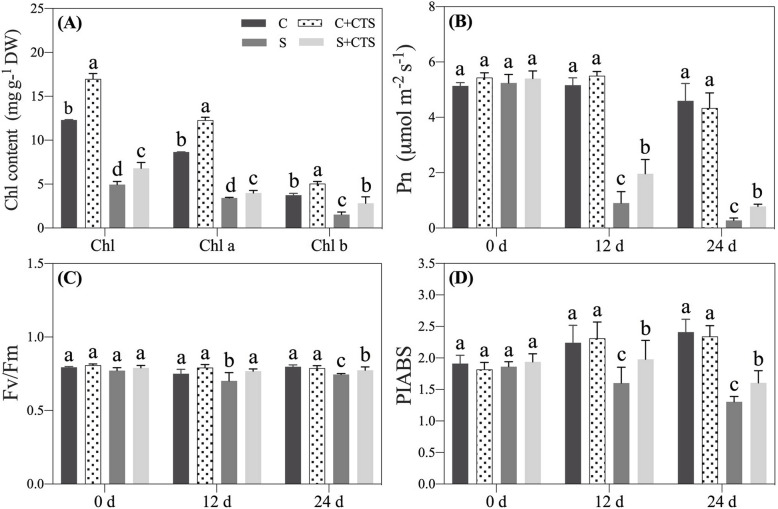
To avoid salt shock and acclimate to salt stress, a successive increasing salt concentration (100 mmol/L NaCl solution for 4 days, 150 mmol/L NaCl solution for another 4 days, and 200 mmol/L NaCl for 16 days.) was applied in this study. Plant growth was inhibited under salt stress, but CTS-treated plants maintained better growth than untreated plants under salt stress (Fig. 1a). Salt stress significantly decreased CTS content, whereas exogenous CTS application improved CTS accumulation in leaves under normal condition and salt stress (Fig. 1b). Salt stress led to 30.70%, 16.81%, or 67.06% decrease in relative water content (RWC), osmotic potential (OP), or water use efficiency (WUE) in leaves at 12 d, respectively (Fig. 1c-e). CTS-treated plants had 15.65% increase of RWC and four times higher WUE than untreated plants at 24 d of salt stress (Fig. 1c and e). The OP indicates the potential of water movement between two regions associated with the water potential in cells. There was a significant difference in OP between CTS-treated and untreated plants at 24 d of salt stress (Fig. 1d). The application of CTS effectively alleviated salt-induced decline in chlorophyll (Chl), Chl a, and Chl b content in leaves (Fig. 2a). CTS-treated plants showed significantly higher net photosynthesis rate (Pn) than untreated plants at 12 or 24 d of salt stress (Fig. 2b). The performance index on absorption basis (PIABS) is a comprehensive photosynthetic index indicating the health status of leaves. For changes in photochemical efficiency (Fv/Fm) and PIABS, exogenous CTS did not affect these two parameters at 0, 12, and 24 d of normal condition, but significantly improved Fv/Fm and PIABS in leaves at 12 and 24 d of salt stress (Fig. 2c and d).
Fig. 1.
Changes in (a) phenotypes at 24 d, (b) CTS content at 24 d, (c) relative water content (RWC), (d) osmotic potential (OP), and (e) water use efficiency (WUE) in leaves of creeping bentgrass affected by the application of chitosan (CTS) under normal and salt stress conditions. Bars represent standard errors. Same letters above columns indicate no significant difference at a given day of treatment (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
Fig. 2.
Changes in (a) chlorophyll (Chl) content at 24 d, (b) net photosynthesis rate (Pn), (c) photochemical efficiency (Fv/Fm), and (d) performance index on absorption basis (PIABS) in leaves of creeping bentgrass affected by the application of chitosan (CTS) under normal and salt stress conditions. Bars represent standard errors. Same letters above columns indicate no significant difference at a given day of treatment (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
Effects of CTS on antioxidant capacity in leaf
Malondialdehyde (MDA) is a key parameter related to lipid peroxidation level and electrolyte leakage (EL) reflects the degree of cell membrane stability. The MDA content increased significantly in leaves under salt stress. CTS pretreatment did not cause significant changes in MDA content under normal condition, but CTS-pretreated plants exhibited 31.37% decrease in MDA content than untreated plants at 24 d of salt stress (Fig. 3a). The EL gradually increased during salt stress. EL level was significantly lower in CTS-treated plants than that in untreated plants at 12 and 24 d of salt stress (Fig. 3b). For antioxidant enzyme activities, salt stress significantly increased superoxide dismutase (SOD), ascorbate peroxidase (APX), and CAT activities, but decreased POD activity in leaves of creeping bentgrass without CTS application (Fig. 3c). CTS-pretreated plants exhibited 17.24%, 18.80%, or 18.62% increase in SOD, POD, or CAT activity than untreated plants under salt stress, respectively. Exogenous CTS had no significant effects on POD activity under normal and stress conditions (Fig. 3c).
Fig. 3.
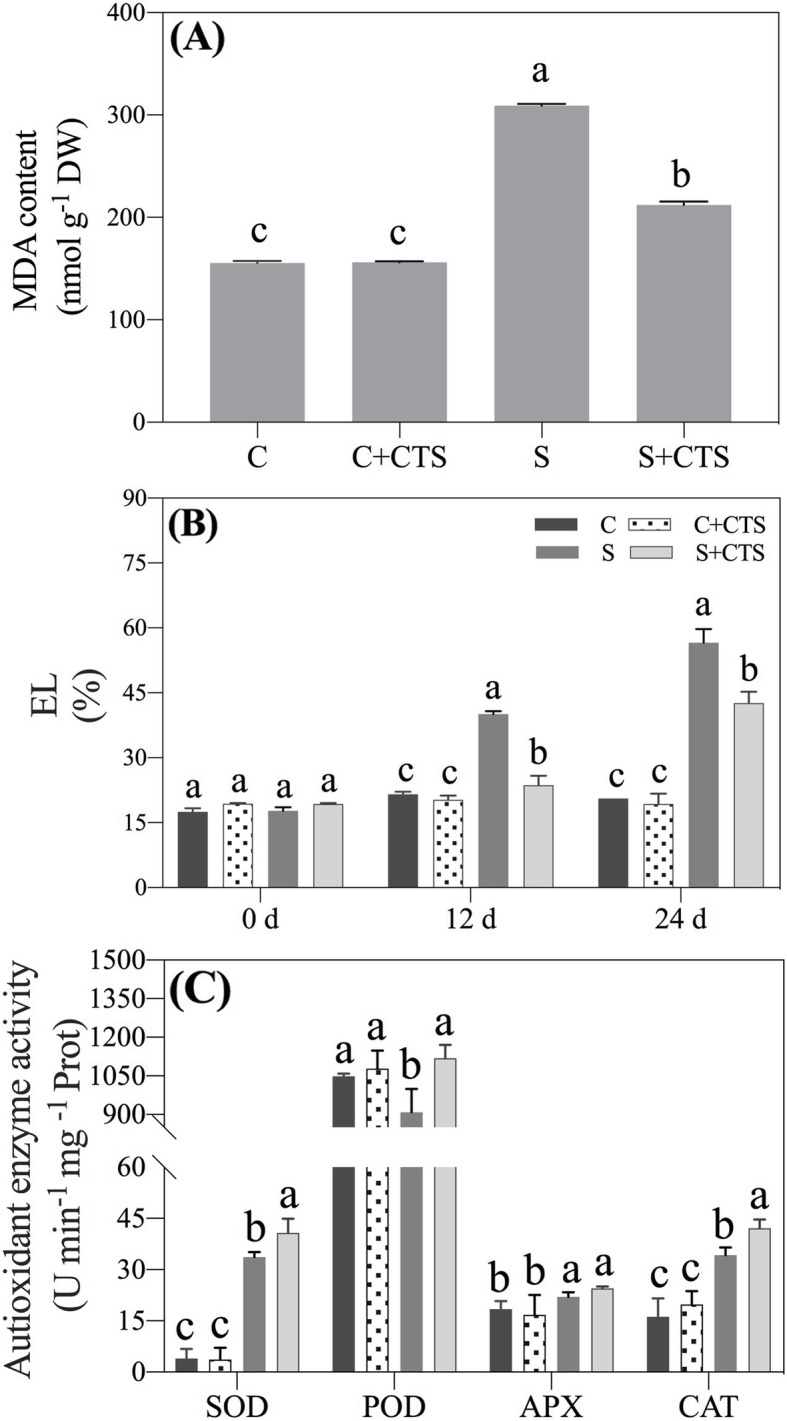
Changes in (a) malondialdehyde (MDA) content at 24 d, (b) electrolyte leakage (EL), and (c) antioxidant enzyme activities (SOD, superoxide dismutase; POD, peroxidase; APX, ascorbate peroxidase; CAT, catalase) in leaves of creeping bentgrass at 24 d affected by the application of chitosan (CTS) under normal and salt stress conditions. Bars represent standard errors. Same letters above columns indicate no significant difference (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
Effect of CTS on sugars, amino acids, and energy metabolism in leaf
Salt stress significantly enhanced sucrose, glucose, and fructose accumulation in plants (Table 1). The ‘S + CTS’ treatment had 75.98% increase in sucrose content than the ‘S’ treatment (Table 1). CTS-pretreated plants exhibited significantly lower glucose and fructose content than untreated plants under salt stress (Table 1). Salt stress activated sucrose synthase (SS) and sucrose phosphate synthase (SPS) activities, but significantly inhibited invertase activity as showed in Table 1. CTS-pretreated plants demonstrated 33.04% increase in SS activity and also had 16.79% and 47.04% decreases in invertase and SPS activities than untreated plants under salt stress, respectively (Table 1). Total amino acids content (TAA), free proline, and glutamic acid (Glu) content increased significantly in leaves under salt stress, however, a significant decrease in γ-aminobutyric acid (GABA) content was observed under salt stress (Table 1). CTS-treated plants exhibited 19.84%, 6.79%, and 29.48% increases in TAA, glutamic acid, and GABA content compared with untreated plants under salt stress (Table 1). Exogenous CTS significantly decreased free proline content in leaves under salt stress (Table 1). Pyruvic acid (PA) content increased in response to salt stress, however, no significant difference in PA content between CTS-treated and untreated plants under normal and salt stress conditions was noticed (Table 1). CTS pretreatment did not cause significant change in pyruvate kinase (PK) activity under normal condition, but significantly increased PK activity under salt stress (Table 1).
Table 1.
Effects of exogenous chitosan (CTS) on changes in sucrose content, fructose content, glucose content, sucrose synthase (SS) activity, sucrose phosphate synthase (SPS) activity, invertase activity, total amino acids (TAA) content, free proline (Pro) content, glutamic acid (Glu) content, γ-aminobutyric acid (GABA) content, pyruvic acid (PA) content, and pyruvate kinase (PK) activity in leaves of creeping bentgrass under normal and salt stress conditions at 24 d. Same letters indicate no significant difference (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
| Parameters | C | C + CTS | S | S + CTS |
|---|---|---|---|---|
| Sucrose content (mg g− 1 DW) | 12.45 + 1.09 d | 21.88 + 4.51 c | 36.05 + 3.22 b | 63.43 + 8.58 a |
| Fructose content (mg g− 1 DW) | 9.51 + 1.55 c | 3.46 + 0.65 d | 28.66 + 0.29 a | 23.71 + 0.70 b |
| Glucose content (μmol g− 1 DW) | 7.79 + 0.85 c | 5.44 + 0.48 c | 41.27 + 1.60 a | 25.83 + 2.27 b |
| SS activity (μg min−1 mg− 1 Prot) | 0.89 + 0.06 c | 1.01 + 0.06 bc | 1.16 + 0.05 b | 1.54 + 0.01 a |
| SPS activity (μg min−1 mg− 1 Prot) | 29.08 + 1.82 c | 18.67 + 1.51 c | 127.26 + 7.28 a | 67.40 + 3.85 b |
| Invertase activity (μg min−1 mg− 1 Prot) | 627.50 + 12.71 a | 643.37 + 12.70 a | 425.33 + 10.28 b | 353.93 + 9.64 c |
| TAA content (μmol mg−1 Prot) | 54.19 + 6.54 c | 62.27 + 5.99 c | 114.75 + 7.09 b | 137.53 + 3.06 a |
| Free Pro content (μg g−1 DW) | 1.06 + 0.31 c | 0.85 + 0.09 c | 4.87 + 0.31 a | 3.15 + 0.07 b |
| Glu content (μg g−1 DW) | 374.00 + 5.19 b | 332.85 + 5.46 c | 386.48 + 9.42 b | 412.71 + 3.08 a |
| GABA content (μmol mg−1 DW) | 821.47 + 18.74 b | 895.28 + 10.92 a | 414.45 + 9.18 d | 536.65 + 20.73 c |
| PA content (μg mg−1) | 155.99 + 13.40 b | 158.34 + 17.79 b | 207.39 + 11.26 a | 208.77 + 5.50 a |
| PK activity (nmol−1 min − 1 mg Prot) | 299.15 + 7.17 c | 280.86 + 6.46 c | 635.5 + 19.61 b | 689.03 + 12.33 a |
Effects of CTS on physiological changes in root
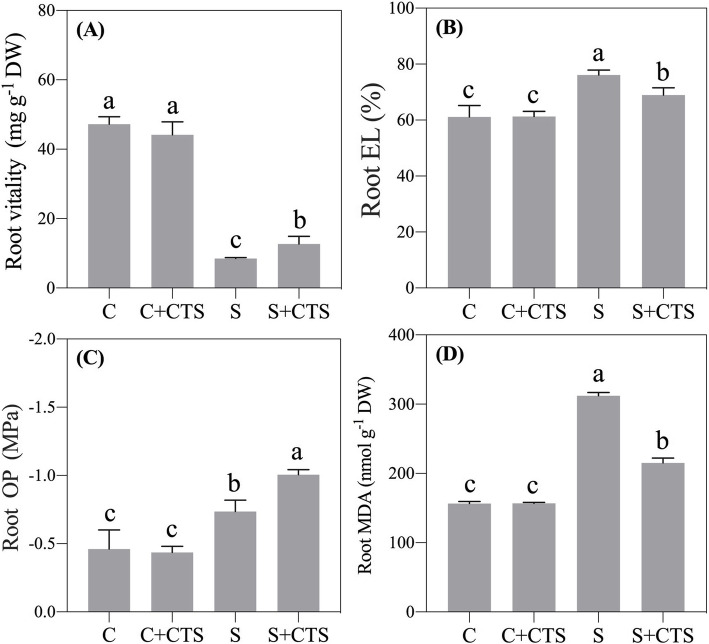
Under normal condition, exogenous CTS did not significantly affect root vitality, EL, OP, and MDA content in roots (Fig. 4a-d). The root vitality reflects the health status of roots. Successive increasing salt concentration decreased root vitality, but the CTS application significantly alleviated the decline in root vitality(Fig. 4a). Salt stress also significantly decreased OP and increased EL and MDA content in roots (Fig. 4b-d). CTS-pretreated plants showed 13.15% or 31.07% decrease in EL or MDA content in roots than untreated plants, respectively (Fig. 4b and d). For OP in roots, CTS-pretreated plants had lower OP than untreated plants under salt stress (Fig. 4c).
Fig. 4.
Changes in (a) root vitality, (b) electrolyte leakage (EL), (c) osmotic potential (OP), and (d) malondialdehyde (MDA) content in roots of creeping bentgrass affected by the application of chitosan (CTS) under normal and salt stress conditions at 24 d. Bars represent standard errors. Same letters above columns indicate no significant difference (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
Effects of CTS on changes of endogenous PAs, Na+ content, K+ content, and transcript levels of genes involved in Na+ transportation in leaf and root
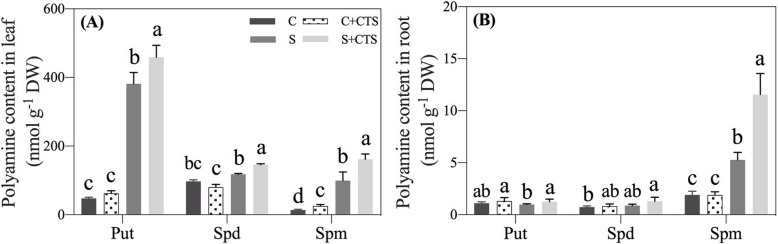
Under normal condition, the level of endogenous putrescine (Put), spermidine (Spd) and spermine (Spm) content in leaves were relatively lower, whereas salt stress significantly increased Put, Spd, and Spm accumulation in leaves (Fig. 5a). CTS-pretreated plants exhibited 20.32%, 23.39%, or 62.78% increase in Put, Spd, and Spm content compared with untreated plants in leaves under salt stress (Fig. 5a). For changes of PAs in roots, CTS-pretreated plants had significantly higher Put content than untreated plants in roots under salt stress (Fig. 5b). There was no significant difference in Spd content between CTS-pretreated and untreated plants in roots under normal condition or salt stress. Spd content in roots only significantly increased in CTS-treated plants in response to salt stress. Salt stress significantly induced Spm accumulation in roots and CTS-pretreated plants had 100% increase in Spm content than untreated plants in roots under salt stress (Fig. 5b).
Fig. 5.
Changes in polyamines content in (a) leaves and (b) roots of creeping bentgrass affected by theapplication of chitosan (CTS) under normal and salt stress conditions at 24 d. Bars represent standard errors. Same letters above columns indicate no significant difference (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
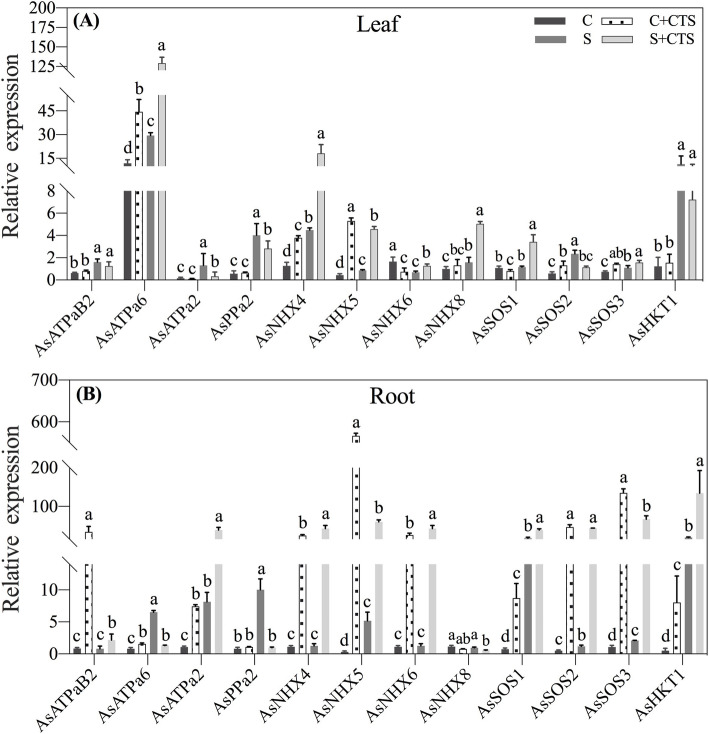
Under normal condition, the Na+ content was maintained at a very low level in leaves and roots, and CTS pretreatment had no effect on the Na+ content (Fig. 6a). Salt stress significantly increased Na+ content in roots and leaves. CTS-pretreated plants accumulated significantly higher Na+ in roots, but lower Na+ in leaves than untreated plants under salt stress (Fig. 6a). The K+ content and K+/Na+ ratio were significantly higher in CTS-untreated plants than those in CTS-pretreated plants in leaves and roots under normal condition (Fig. 6b and c). Exogenous CTS pretreatment had significant effects on K+ content in leaves under salt stress but no significant effects on K+ content in roots and K+/Na+ ratio in roots and leaves under salt stress (Fig. 6b and c). For changes of genes expression involved in Na+ transport in leaves, exogenous CTS had no significant effects on genes encoding AsATPaB2, AsATPa2, AsPPa2, AsNHX8, and AsHKT1, but significantly up-regulated AsATPa6, AsNHX4, AsNHX5, and AsSOS2 under normal condition (Fig. 7a). Under salt stress, CTS-pretreated plants showed significantly higher expression levels of AsATPa6, AsNHX4, AsNHX5, AsNHX6, AsNHX8, AsSOS1, and AsSOS3 than untreated plants in leaves (Fig. 7a). Salt stress significantly up-regulated AsATPa6, AsATPa2, AsPPa2, AsNHX5, AsSOS1, AsSOS3, and AsHKT1 in roots (Fig. 7b). Under normal condition, exogenous CTS application induced significant increases in AsATPaB2, AsATPa6, AsATPa2, AsNHX4, AsNHX5, AsNHX6, AsSOS1, AsSOS2, AsSOS3, and AsHKT1 in roots. Under salt stress, AsATPaB2, AsATPa6, AsATPa2, AsNHX4, AsNHX5, AsNHX6, AsSOS1, AsSOS2, AsSOS3, and AsHKT1 were significantly up-regulated by exogenous CTS in roots (Fig. 7b). Figure 8 showed proposed key pathways involved in immanent and CTS-regulated adaptive response to salt stress in perennial creeping bentgrass.
Fig. 6.
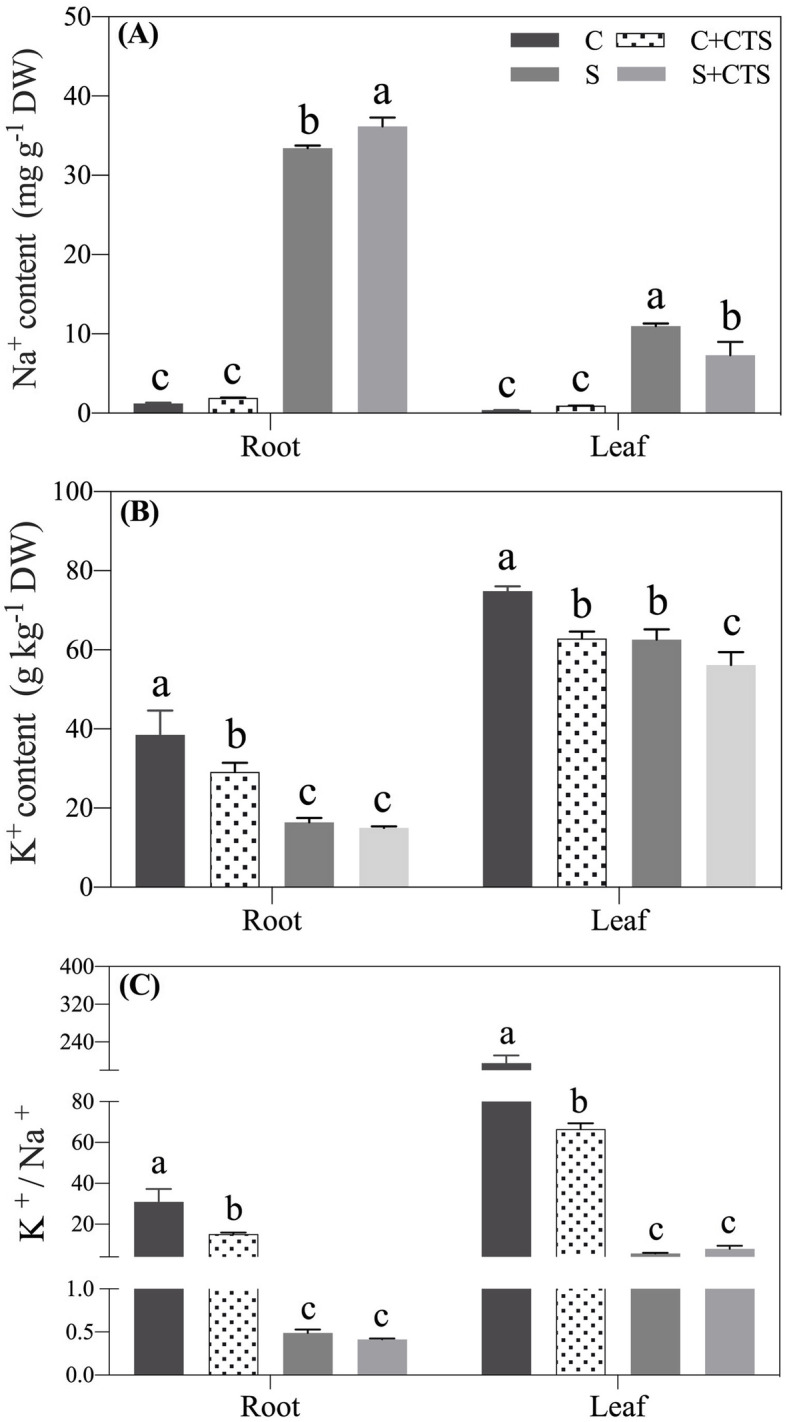
Changes in (a) Sodium (Na+) content, (b) potassium (K+) content, and (c) K+/Na+ affected by theapplication of chitosan (CTS) in leaves and roots of creeping bentgrass under normal and salt stress conditions at 24 d. Bars represent standard errors. Same letters above columns indicate no significant difference (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
Fig. 7.
Changes in genes relative expression in (a) leaves and (b) roots of creeping bentgrass affected by the application of chitosan (CTS) under normal and salt stress conditions at 12 d. Bars represent standard errors. Same letters above columns indicate no significant difference (n = 4, and p ≤ 0.05). C, control; C + CTS, control pretreated with CTS; S, salt stress; S + CTS, salt-stressed plants pretreated with CTS
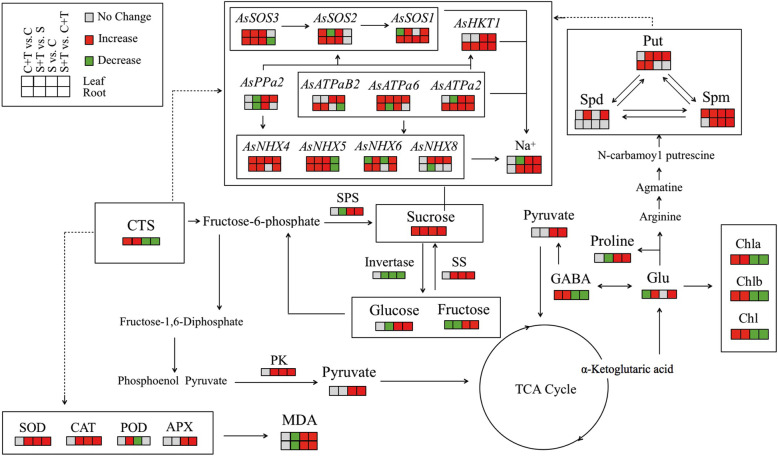
Fig. 8.
Proposed key pathways involved in immanent and CTS-regulated adaptive response to salt stress in perennial creeping bentgrass
Discussion
High soil salinity severely reduces water uptake leading to physiological drought, thereby inhibiting plant growth [35]. It has been well documented that maintenance of higher osmotic adjustment (OA) capacity and WUE contributed to better water status in plants under salt stress [36]. In the current study, salt stress significantly decreased OP in leaves (Fig. 1d) and roots (Fig. 4c) that could be a positive response for acclimating to salt damage in creeping bentgrass. CTS-pretreated creeping bentgrass maintained significantly higher WUE and lower OP in leaves (Fig. 1d-e), which could be associated with better water status in plants under salt stress. Our recent study also found that exogenous GABA application effectively alleviated salt-induced damage through enhancing OA and WUE in creeping bentgrass [37]. When plants are exposed to salt stress, accelerated oxidative damage and water deficit will cause photoinhibition [38]. Successive increasing salt concentration significantly decreased Chl content, Pn, Fv/Fm, and PIABS in leaves of creeping bentgrass (Fig. 2). However, CTS-pretreated creeping bentgrass had significantly higher Chl content, Pn, PIABS, and Fv/Fm than untreated plants, which reflected better photosynthetic capacity in CTS-pretreated plants under salt stress in this study. In addition, the application of CTS also improved plant growth and Chl content under normal condition. It has been reported that the CTS could be used as an exogenous plant growth regulator for improving growth and quality of crops [27]. Thus, dual positive functions of CTS in regulating growth under normal condition and stress tolerance highlight its importance in agricultural cultivation and production.
Salt stress increases accumulation of reactive oxygen species (ROS) such as superoxide radical and hydrogen peroxide (H2O2) that are responsible for oxidative damage and ultimately cell death [5]. Improvement or maintenance of higher antioxidant enzyme activities such as SOD, CAT, POD, and APX is important for reducing oxidative damage in different plant species under salt stress [39–41]. Current results found that exogenous CTS played an important role in reducing lipid peroxidation in association with significant increase in antioxidant enzyme (SOD, CAT, and POD) activities in creeping bentgrass under salt stress (Fig. 3), which was consistent with previous study about the effect of CTS on improving the salt tolerance in sunflower seedlings through enhancing CAT and POD activities. As the primary organ for water and nutrition intake, root system is easily damaged by high soil Na+ [42]. Successive increasing salt concentration seriously reduced root vitality (Fig. 4a), whereas CTS application effectively alleviated the negative effect in creeping bentgrass. These phenotypic and physiological changes in roots and leaves indicated positive effects of CTS on alleviating salt stress damage in creeping bentgrass.
Regulation of sugar metabolism plays an essential role for plants acclimation to environmental stress [43]. Exogenous application of CTS significantly increased endogenous CTS content in leaves of creeping bentgrass (Fig. 1b). Based on metabolic pathways, the CTS could be converted directly to other sugars and pyruvate that participates in the tricarboxylic acid cycle and the biosynthesis of glutamic acid and other amino acids (Fig. 8). When plants are subjected to abiotic stress, soluble sugars such as fructose, glucose, and sucrose accumulate in cells as important signaling molecules for stress signal transduction, osmolytes for OA, and energy substances for energy supply [44, 45]. Sucrose is a major photoassimilate and can be transferred from source leaves to roots. Invertase catalyzes sucrose degradation into glucose and fructose for cell biosynthesis and metabolism [46]. Sucrose synthesis can be catalyzed by SPS from fructose-6-phosphate or transformed from glucose and fructose by SS. In this study, salt stress significantly improved sucrose synthesis through activating SPS and SS activities and reducing invertase activity (Table 1). Interestingly, exogenous CTS application further increased salt-induced sucrose accumulation that could be associated with higher SS activity and lower invertase in leaves of creeping bentgrass. In an earlier study, it has been found that sucrose accumulation was conducive to PSII protection in plants under salt stress [47]. The study of Schwender et al. [48] demonstrated that 90% glucose could be converted to pyruvate by glycolysis in Brassica napus. PK is one of main rate-limiting enzymes in the glycolysis process for energy production [49, 50]. CTS-improved PK activity might enhance conversion of CTS, glucose, and fructose into pyruvate for energy metabolism in creeping bentgrass (Table 1), but the mechanism underlying changes of sugars and metabolic enzymes still need to be further investigated during salt stress.
Amino acids accumulation and metabolism are important for plants to deal with stress damage [51]. Proline has been extensively studied as a stress-responsive amino acid and its function is involved in OA and redox balance in plants during abiotic stress [52, 53]. Glu is a main metabolite of nitrogen metabolism in plants associated with the maintenance of carbon-nitrogen balance, other amino acid metabolism, Chl biosynthesis during normal growth and stress response [54]. Increasing evidences indicate that GABA is involved in regulating tolerance to abiotic stress through enhancing OA, antioxidant defense, and metabolic balance in plants [55]. Previous study has shown that amino acid synthesis pathway was significantly enhanced in barley (Hordeum vulgare) leaf under salt stress, and amino acids such as Glu and GABA were key metabolites involved in salt stress response [56]. Exogenous GABA induced salt tolerance associated with increase in sugars (glucose, fructose, maltose, and trehalose) and amino acids (Glu, alanine, leucine, valine, asparagine, lysine, threonine, and cysteine) content in leaves of creeping bentgrass [37]. In the current study, CTS-pretreated creeping bentgrass exhibited significantly higher TAA, Glu, and GABA content than untreated plants under salt stress (Table 1), which could explain why CTS-pretreated plants had better OA capacity, water status, and metabolic homeostasis as compared to untreated plants. However, the application of exogenous CTS alleviated salt stress-induced increase in proline accumulation in leaves (Table 1). The proline accumulation is also often regarded as an indicator of stress damage. More severe stress damage, more proline accumulation in plants under salt or other abiotic stress [37, 57, 58].
Many studies have reported that increased PAs level is an important adaptive response when plants are subjected to salt stress [59–61]. PAs-regulated salt tolerance could be associated with activation of SOS signaling pathways, nitrogen metabolism, and antioxidant defense system to maintain ions and metabolic balance in plants under salt stress [62]. A previous study found that the salt-tolerant barley cultivar 'J4′ accumulated significantly higher endogenous Spd and Spm than salt-sensitive 'KP7′ in response to successive increasing salt concentration [60]. The CTS can be converted into pyruvate involved in the tricarboxylic acid cycle for the production of glutamic acid which can be metabolized to arginine for the synthesis of PAs (Fig. 8). It has been found that exogenous CTS significantly improved endogenous Put, Spd, and Spm accumulation contributing to enhanced drought tolerance in white clover (Trifolium repens) [28, 63]. In this study, Put, Spd, and Spm accumulated significantly in leaves and Spm increased considerably in roots of creeping bentgrass under salt stress (Fig. 5). Exogenous CTS application further promoted salt-induced PAs accumulation in leaves and roots of creeping bentgrass. These findings suggested that CTS-regulated salt tolerance could be involved in PAs synthesis in creeping bentgrass.
High Na+ levels are generally considered to be the primary factor in salt toxicity in plants [64]. In a saline environment, a possible survival strategy for plants is to effectively isolate excess Na+ in roots and to inhibit the transfer of Na+ from roots to shoots [65]. Our results showed that the creeping bentgrass with CTS application had significantly higher Na+ content in roots, but lower Na+ accumulation in leaves as compared to the plants without CTS pretreatment (Fig. 6). Analyses of genes expression also found that salt stress significantly up-regulated the AsHKT1 expression in leaves and roots. More importantly, exogenous CTS could further enhance the AsHKT1 expression in roots of creeping bentgrass, which indicated that CTS could regulate AsHKT1 to isolate Na+ in roots and to inhibit Na+ transport to leaves during a long period of salt stress. The CTS pretreatment also significantly activated the SOS pathway in roots under salt stress (Fig. 7). The study of Wang et al. (2014) found out that main function of AtSOS1 is to extrude Na+ from the cytosol into the rhizosphere in Arabidopsis thaliana under the normal K+ plus salt stress. However, the AtSOS1 regulated the long-distance transport of Na+ from roots to leaves in Arabidopsis thaliana under the low K+ plus salt stress [66]. In current study, the CTS-activated SOS pathway could be involved in Na+ excretion, since the creeping bentgrass was subjected to salt stress with enough K+ supply in Hoagland’ solution. In addition, the separation of Na+ into vacuoles is considered as a key mechanism to avoid toxic effects of Na+ in the cytoplasm. The Na+ compartmentalization also can provide additional osmotic adjustment for water maintenance under salt stress [17, 67]. The CTS-treated creeping bentgrass (S + CTS) had significantly higher expression levels of AsNHX4, AsNHX5, and AsNHX6 than untreated plants (S) in roots and leaves under salt stress (Fig. 7), suggesting that the CTS could enhance the capacity of Na+ compartmentalization in cells. Interestingly, the CTS significantly up-regulated AsATPaB2 and AsATPa2 in roots and AsATPa6 in leaves, which could be related to enhanced proton motive force in creeping bentgrass under salt stress. A previous study has proved that the inhibition of Spd and Spm biosynthesis decreased tonoplast H+-ATPase activities resulting in significant decrease in salt tolerance of barley seedlings [60]. It is worth further study whether PAs are involved in CTS-regulated Na+ transport in plants during salt stress.
Conclusion
The addition of exogenous CTS is a cheap and effective measure to alleviate growth inhibition and stress damage caused by salt stress in creeping bentgrass. In response to salinity, exogenous CTS increased antioxidant enzyme activities, thereby reducing oxidative damage to roots and leaves. CTS-induced increase in sucrose and GABA accumulation and metabolism played important role in OA and energy metabolism during stress. The CTS also regulated Na+ transport though increasing AsHKT1 expression for inhibiting Na+ transport to photosynthetic tissues, enhancing SOS pathway associated with Na+ excretion from cytosol into rhizosphere, and up-regulating the expression of AsNHX4, AsNHX5, and AsNHX6 involved in Na+ compartmentalization from cytoplasm into vacuoles in roots and leaves under salt stress. In addition, CTS-induced PAs accumulation could be an important regulatory mechanism contributing to enhanced salt tolerance in creeping bentgrass. These findings reveal important functions of CTS on regulating Na+ transport, enhancing sugars and amino acids metabolism, and increasing PAs accumulation, which contributes to the alleviation of salt stress in perennial creeping bentgrass. To further understand the CTS-induced salt tolerance in plants, future studies will focus on investigating changes in global metabolites based on metabolomics and analyzing possible signal transduction pathways involved in GABA-regulated PAs accumulation, antioxidant defense, and Na+ transport under salt stress.
Methods
Plant materials and treatments
Seeds of creeping bentgrass (cv. PA-1) were purchased from Tee-2-Green Company (Oregon, USA) and used as a plant material. Seeds (3.8 g/m2) were sown in seedling pots filled with sterilized quartz sand and germinated in a plant growth chamber (photoperiod cycle of 10/14 h light/dark, 21/18 °C day/night, 65% relative humidity, and 700 μmol m− 2·s− 1 PAR) for 8 days. Seedlings were then grown in Hoagland’ solution for 20 days [68]. 28-day-old plants were pretreated with or without 0.5 g/L CTS for 4 days. The CTS or NaCl was dissolved in Hoagland’ solution. The effective dose of CTS was selected based on a preliminary test with a range of concentrations (0.1, 0.2, 0.5, 1, and 2 g/L) for the most effective dose on phenotypic changes. The CTS-pretreated and untreated plants were subjected to NaCl-induced salt stress for 24 days. For salt stress, plants were grown in 100 mmol/L NaCl solution for 4 days, 150 mmol/L NaCl solution for another 4 days, and 200 mmol/L NaCl for 16 days. All solutions were refreshed every day. Four biological replicates were set for each treatment. Plants were sampled at 0, 12, and 24 d of salt stress, respectively, and 4 biological replicates were used to estimate all parameters.
Physiological measurements
For the estimation of EL, the method of Blum and Ebercon was used [69]. RWC was determined according to the method of Barrs and Weatherley [70]. For OP of leaf and root, fresh leaves or roots were immediately immerged in distilled water for 12 h at 4 °C. After being blotted dry, leaves or roots were frozen in liquid nitrogen for 10 min, and then thawed for 30 min at 4 °C. Leaves or roots were pressed to get cell fluid. The osmolality of cell fluid was measured by using a vapor pressure osmometer (Wescor, Logan, UT, USA), and the OP was converted based on -c × 2.58 × 10− 3 [71]. For root viability, the method of McMichael and Burke was used [72]. The determination of Chl content was performed according to the method of Arnon et al. [73]. The Fv/Fm and PIABS were recorded by a Chl fluorescence system (Pocket PEA, Hansatech, the United Kingdom). Leaves were placed in dark for 30 min with leaf clips before analysis. Pn and WUE were measured using a portable photosynthetic system (CIRAS-3, PP Systems, USA) that provided 400 μl L− 1 CO2 and 800 μmol photon m− 2 red and blue light.
Measurements of total antioxidant capacity and antioxidant enzyme activities
Fresh leaves (0.2 g) were ground on ice with 1.5 mL of 50 mM cold phosphate buffer (pH 7.8) and centrifuged at 12000 g for 30 min at 4 °C. The supernatant was used for the determination of antioxidant enzyme activities and MDA content. Activity of SOD, CAT, POD, or APX was measured by recording changes in absorbance at 560, 240, 470, or 290 mm, respectively [74–76]. Protein content was measured according to the method of Bradford [77]. For MDA content, 0.5 mL of the supernatant was mixed with 1 mL of reaction solution containing 20% trichloroacetic acid and 0.5% thiobarbituric acid (TBA). The mixture was heated in a water bath at 95 °C for 15 min and then rapidly cooled in an ice bath. The solution was centrifuged at 8000 g for 10 min at 4 °C. The absorbance of supernatant was recorded at 532, 600 and 450 nm [28].
Measurements of sugars, amino acids, and enzyme activities related to sugars and energy metabolism
The CTS content (Art. No. ml020132) was determined by using a Test Kit (mlbio Good elisakit producers, Shanghai, China) according to the manufacturer’s instructions. Pyruvate kinase (PK) activity (Art. No. PK-2-Y), pyruvic acid content (Art. No. PA-1-Y), sucrose synthase (SS) activity (Art. No. SSII-1-Y), sucrose phosphate synthase (SPS) activity (Art. No. SPS-1-Y), hexokinase activity (Art. No. HK-2-Y), invertase activity (Art. No. ZTM-1-Y), sucrose content (Art. No. ZHT-2-Y), glucose content (Art. No. PT-2-Y), and fructose content (Art. No. GT-2-Y)were determined by using Test Kits (Suzhou Comin Biotechnology, Suzhou, China) according to manufacturer’s instructions. For free proline content, fresh leaves (0.2 g) were ground with 3% aqueous sulfosalicylic acid and then centrifuged at 10000 g for 10 min to obtain supernatant. The reaction mixture (1 mL of the supernatant, 1.5 mL of acid ninhydrin, and 1.5 mL of glacial acetic acid) was boiled for 30 min. After being cooled at room temperature, the reaction mixture was extracted with five mL of benzene and the absorbance was recorded at 520 nm [78].
Measurement of endogenous polyamine and Na+ / K+ content
For the measurement of endogenous PAs content, the methods of Duan [79] and Li [80] were used. The Na+ or K+ content was detected by using a flame atomic absorption spectrophotometer (Analytik Jena AG, Jena, Germany) and the assay method has been recorded in our previous study [37].
Genes expression analyses
For determining effects of CTS on changes in selected gene transcript levels, real-time quantitative polymerase chain reaction (qRT-PCR) was used. The Rneasy Mini Kit (Qiagen, Duesseldorf, Germany) was used for extracting total RNA in fresh leaves or roots. The RNA was reverse-transcribed to cDNA using a revert Aid First Stand cDNA Synthesis Kit (Fermentas, Lithuania). Primers of sodium-hydrogen antiporter genes (AsNHX4, AsNHX5, AsNHX6, and AsNHX8), H+ transporter genes (AsATPaB6, AsATPa6, AsATPa2, and AsPPa2), salt overly sensitive genes (AsSOS1, AsSOS2, and AsSOS3), high-affinity Na+/K+-permeable transporter (AsHKT1), and reference gene (β-actin) were given in Table S1. For all genes, PCR conditions (iCycler iQ qRT-PCR detection system with SYBR Green Supermix, Bio-Rad, USA) were as follows: 5 min at 94 °C and 30 s at 95 °C (45 repeats of denaturation), annealing and extending 45 s at 62 °C, and amplicon from 60 to 95 °C to obtain the melting curve. Transcript levels of all genes were calculated according to the formula 2−∆∆Ct described by Livak and Schmittgen [81].
Statistic
The SPSS 23 (IBM, Armonk, NY, USA) was used for analyzing all data. Significant differences among C, C + NaCl, CTS, and CTS + NaCl treatments were tested by using the least significant difference (LSD) at p ≤ 0.05.
Supplementary Information
Additional file 1 Table S1. Primer sequences of gene.
Acknowledgments
Not applicable.
Abbreviations
- CTS
Chitosan
- Na+
Sodium ions
- ROS
Reactive oxygen species
- H2O2
Hydrogen peroxide
- SOS
Salt overly sensitive pathways
- NHX
Na+/H+ antiporters
- K+
Potassium ions
- PAs
Polyamines
- RWC
Relative water content
- FW
Fresh weight
- SW
Saturated weight
- DW
Dry weight
- WUE
Water use efficiency
- OP
Osmotic potential
- Chl
Total chlorophyll
- Fv/Fm
Photochemical efficiency
- PIABS
Performance index on absorption basis
- Pn
Net photosynthetic rate
- MDA
Malondialdehyde
- EL
electrolyte leakage
- SOD
Superoxide dismutase
- CAT
Catalase
- POD
Peroxidase
- APX
Ascorbate peroxidase
- SS
Sucrose synthase
- SPS
Sucrose phosphate synthase
- TAA
Total amino acids
- Pro
Free proline
- Glu
Glutamic acid
- ROS
Reactive oxygen species
- GABA
γ-Aminobutyric acid
- PA
Pyruvic acid
- PK
Pyruvate kinase
- Put
Putrescine
- Spd
Spermidine
- Spm
Spermine
- OA
Osmotic adjustment
Authors’ contributions
ZL conceived the research and designed experiments; WG performed the experiments and analyzed the data; WG and ZL wrote the manuscript; ZL, MJH, and YP reviewed the manuscript. All authors have approved the final version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31702182). The funding agency had no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Niu ML, Xie JJ, Chen C, Cao HS, Sun JY, Kong QS, Shabala S, Shabala L, Huang Y, Bie ZL. An early ABA-induced stomatal closure, Na+ sequestration in leaf vein and K+ retention in mesophyll confer salt tissue tolerance in cucurbita species. J Exp Bot. 2018;69(20):4945–4960. doi: 10.1093/jxb/ery251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy SJ, Negrão S, Tester M. Salt resistant crop plants. Curr Opin Biotechnol. 2014;26(4):115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Yuan S, Zhao J, Li Z, Hu Q, Yuan N, Zhou M, Xia X, Noorai R, Saski C, Li S, et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Horticulture Res. 2019;6(1):48. doi: 10.1038/s41438-019-0130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asha K, Paromita D, Kumar PA, Agarwal PK. Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front Plant Sci. 2015;6:537. doi: 10.3389/fpls.2015.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsubasa S, Koya S, Tatsuya A, Yayoi K. Huazhong, Shi HZ, Zhang JK, Aua R, Hasegawa PM, Hashimoto T: salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol. 2006;47(8):1158–1168. doi: 10.1093/pcp/pcj090. [DOI] [PubMed] [Google Scholar]
- 6.Wu GQ, Xi JJ, Wang Q, Bao AK, Ma Q, Zhang JL, Wang SM. The ZxNHX gene encoding tonoplast Na+/H+ antiporter from the xerophyte Zygophyllum xanthoxylum plays important roles in response to salt and drought. J Plant Physiol. 2011;168(8):758–767. doi: 10.1016/j.jplph.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Almeida DM, Oliveira MM, Saibo NJM. Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol. 2017;40:326–345. doi: 10.1590/1678-4685-gmb-2016-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F, Zhang HR, Zhao HS, Gao TW, Song AP, Jiang JF, Chen FD, Chen SM. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol J. 2018;16(7):1311–1321. doi: 10.1111/pbi.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi HZ, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14(2):465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi HZ, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A. 2000;12(97):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J-K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2000;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36(2):229–239. doi: 10.1046/j.1365-313X.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- 13.Leidi EO, Verónica B, Rubio L, El-Hamdaoui A, Pardo JM. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2009;61(3):495–506. doi: 10.1111/j.1365-313X.2009.04073.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Wang QQ, Yu MM, Zhang YY, Wu YB, Zhang HX. Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively expressing an Arabidopsis thaliana vacuolar Na+ /H+ antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots. Plant Cell Environ. 2008;31(9):1325–1334. doi: 10.1111/j.1365-3040.2008.01838.x. [DOI] [PubMed] [Google Scholar]
- 15.Huertas R, Rubio L, Cagnac O, García-Sánchez MJ. JD Alché, Venema K: the K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant Cell Environ. 2013;36(12):2135–2149. doi: 10.1111/pce.12109. [DOI] [PubMed] [Google Scholar]
- 16.Bassil E, Blumwald E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr Opin Plant Biol. 2014;22:1–6. doi: 10.1016/j.pbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Leidi EO, Pardo JM. How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav. 2010;5(7):792–795. doi: 10.4161/psb.5.7.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465(2):140–151. doi: 10.1016/S0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 19.Bao AK, Du BQ, Touil L, Kang P, Wang QL, Wang SM. Co-expression of tonoplast Cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol J. 2016;14(3):964–975. doi: 10.1111/pbi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao AK, Wang YW, Xi JJ, Liu C, Zhang JL, Wang SM. Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances salt and drought tolerance in transgenic Lotus corniculatus by increasing cations accumulation. Funct Plant Biol. 2014;41(2):203–214. doi: 10.1071/FP13106. [DOI] [PubMed] [Google Scholar]
- 21.Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J Biosci. 2005;30(5):761–776. doi: 10.1007/BF02703574. [DOI] [PubMed] [Google Scholar]
- 22.Yl R, Jin Y, Yang YJ, Li GJ. Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant. 2010;6(3):943–955. doi: 10.1093/mp/ssq044. [DOI] [PubMed] [Google Scholar]
- 23.Sturm A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121(1):1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen XY, Liu HQ, Li FL, Su LP. Effects of exogenous sucrose and Ca2+ on salt tolerance of buckwheat seedlings. Zhiwu Shengli Xuebao/Plant Physiol J. 2012;48(12):1187–1192. [Google Scholar]
- 25.Kurita K. Chitin and chitosan: functional biopolymers from marine crustaceans. Mar Biotechnol. 2006;8(3):203–226. doi: 10.1007/s10126-005-0097-5. [DOI] [PubMed] [Google Scholar]
- 26.Massimo M, Raffaella C. Chitosan effects on plant systems. Int J Mol Sci. 2016;17(7):996. doi: 10.3390/ijms17070996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang XQ, Li KC, Xing R, Liu S, Li P. Metabolite profiling of wheat seedlings induced by chitosan: revelation of the enhanced carbon and nitrogen metabolism. Front Plant Sci. 2017;8:2017–2029. doi: 10.3389/fpls.2017.02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Zhang Y, Zhang XQ, Merewitz E, Peng Y, Ma X, Huang LK, Yan YH. Metabolic pathways regulated by chitosan contributing to drought resistance in white clover. J Proteome Res. 2017;16(8):3039–3052. doi: 10.1021/acs.jproteome.7b00334. [DOI] [PubMed] [Google Scholar]
- 29.Turk H. Chitosan-induced enhanced expression and activation of alternative oxidase confer tolerance to salt stress in maize seedlings. Plant Physiol Biochem. 2019;141:415–422. doi: 10.1016/j.plaphy.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Jabeen N, Ahmad R. The activity of antioxidant enzymes in response to salt stress in safflower (Carthamus tinctorius L.) and sunflower (Helianthus annuus L.) seedlings raised from seed treated with chitosan. J Sci Food Agric. 2013;93(7):1699–1705. doi: 10.1002/jsfa.5953. [DOI] [PubMed] [Google Scholar]
- 31.Qian YL, Mecham B. Long-term effects of recycled wastewater irrigation on soil chemical properties on golf course fairways. Agron J. 2005;97(3):717–721. doi: 10.2134/agronj2004.0140. [DOI] [Google Scholar]
- 32.Mantell A. Effect of irrigation frequency and nitrogen fertilization on growth and water use of a kikuyugrass lawn (Pennisetum clandestinum Hochst.) Agron J. 1966;58(6):559–561. doi: 10.2134/agronj1966.00021962005800060001x. [DOI] [Google Scholar]
- 33.Kunkel BA, Held DW, Potter DA. Lethal and sublethal effects of bendiocarb, halofenozide, and imidacloprid on harpalus pennsylvanicus (Coleoptera: Carabidae) following different modes of exposure in turfgrass. J Econ Entomol. 2001;94(1):60–67. doi: 10.1603/0022-0493-94.1.60. [DOI] [PubMed] [Google Scholar]
- 34.Fu JM, Qian YL. Responses of creeping bentgrass to salinity and mowing management: growth and turf quality. HortScience. 2005;40(2):463–467. doi: 10.21273/HORTSCI.40.2.463. [DOI] [Google Scholar]
- 35.Patel AD, Pandey AN. Effect of soil salinity on growth, water status and nutrient accumulation in seedlings of suaeda nudiflora (Chenopodiaceae) An Biol. 2009;31:61–70. [Google Scholar]
- 36.Strogonov BP. Physiological basis of salt tolerance of plants. Basic Life Sci. 1965;99(5):356. [Google Scholar]
- 37.Li Z, Cheng BZ, Zeng WH, Zhang XQ, Peng Y. Proteomic and metabolomic profilings reveal crucial function of γ-aminobutyric acid (GABA) on regulating ionic, water, and metabolic homeostasis in creeping bentgrass under salt stress. J Proteome Res. 2020;19:769–780. doi: 10.1021/acs.jproteome.9b00627. [DOI] [PubMed] [Google Scholar]
- 38.Mostofa MG, Hossain MA, Fujita M. Trehalose pretreatment induces salt tolerance in rice (Oryza sativa L.) seedlings: oxidative damage and co-induction of antioxidant defense and glyoxalase systems. Protoplasma. 2015;252(2):461–475. doi: 10.1007/s00709-014-0691-3. [DOI] [PubMed] [Google Scholar]
- 39.Mandhania S, Madan S, Sawhney V. Antioxidant defense mechanism under salt stress in wheat seedlings. Biol Plant. 2006;50(2):227–231. doi: 10.1007/s10535-006-0011-7. [DOI] [Google Scholar]
- 40.Hu LX, Li HY, Pang HC, Fu JM. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne L.) differing in salt tolerance. J Plant Physiol. 2012;169(2):146–156. doi: 10.1016/j.jplph.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Chojak-Koźniewska J, Linkiewicz A, Sowa S, Radzioch MA, Kuźniak E. Interactive effects of salt stress and Pseudomonas syringae pv. Lachrymans infection in cucumber: involvement of antioxidant enzymes, abscisic acid and salicylic acid. Environ Exp Bot. 2017;136:9–20. doi: 10.1016/j.envexpbot.2017.01.004. [DOI] [Google Scholar]
- 42.Ahmad P, Azooz MM, Prasad MNV: Ecophysiology and responses of plants under salt stress || Adaptive plasticity of salt-stressed root systems. 2013, 4(6):169–201.
- 43.Sellami S, Le HR, Thorpe MR, Vilaine F, Wolff N, Brini F, Dinant S. Salinity effects on sugar homeostasis and vascular anatomy in the stem of the Arabidopsis thaliana inflorescence. Int J Mol Sci. 2019;20(13):3167–3186. doi: 10.3390/ijms20133167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofius D, Börnke FAJ. Photosynthesis, carbohydrate metabolism and sourcesink relations. Potato Biol Biotechnol. 2007:257–85.
- 45.Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63(4):1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang SY, Chi YH, Wang JZ, Zhou JX, Cheng YS, Zhang BL, Ma AL. Sucrose metabolism gene families and their biological functions. Sci Rep. 2015;5:17583–17607. doi: 10.1038/srep17583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peter H. Homann: stabilization of the water oxidizing polypeptide assembly on photosystem II membranes by osmolytes and other solutes. Photosynth Res. 1992;33(1):29–36. doi: 10.1007/BF00032980. [DOI] [PubMed] [Google Scholar]
- 48.Schwender J. Probing in vivo metabolism by stableisotope labeling of storage lipids and proteins in developing brassica napus embryos. Plant Physiol. 2002;130(1):347–361. doi: 10.1104/pp.004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carl A, Christoph B. Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol. 2007;145(4):1670–1680. doi: 10.1104/pp.107.108340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knowles VL, McHugh SG, Hu Z-Y, Dennis DT, Miki BL, William C. Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol. 1998;116(1):45–51. doi: 10.1104/pp.116.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rai VK. Role of amino acids in plant responses to stresses. Biol Plant. 2002;45(4):481–487. doi: 10.1023/A:1022308229759. [DOI] [Google Scholar]
- 52.Per TS, Khan NA, Reddy PS, Masood A, Hasanuzzaman M, Khan MIR, Anjum NA. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: phytohormones, mineral nutrients and transgenics. Russ J Plant Physiol. 2017;115:126–140. doi: 10.1016/j.plaphy.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Lehmann S, Funck D, László S, Rentsch D. Proline metabolism and transport in plant development. Amino Acids. 2010;39(4):949–962. doi: 10.1007/s00726-010-0525-3. [DOI] [PubMed] [Google Scholar]
- 54.Majumdar R, Minocha R, Minocha SC, D'Mello JPF: Ornithine: at the crossroads of multiple paths to amino acids and polyamines. In: Amino acids in higher plants Edited by D’Mello J; 2015:156–176.
- 55.Ham T, Chu S, Han SJ, Ryu SN. γ-Aminobutyric acid metabolism in plant under environment stressses. Korean J Crop Sci. 2012;57(2):144–150. doi: 10.7740/kjcs.2012.57.2.144. [DOI] [Google Scholar]
- 56.Wu DZ, Shen QF, Cai SG, Chen ZH, Dai F, Zhang GP. Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol. 2013;54(12):1976–1988. doi: 10.1093/pcp/pct134. [DOI] [PubMed] [Google Scholar]
- 57.Du H, Wang Z, Yu W, Liu Y, Huang BR. Differential metabolic responses of perennial grass Cynodon transvaalensis × Cynodon dactylon (C4) and Poa Pratensis (C3) to heat stress. Physiol Plant. 2011;141(3):251–264. doi: 10.1111/j.1399-3054.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, Fu JY, Shi DW, Peng Y. Myo-inositol enhances drought tolerance in creeping bentgrass through alteration of osmotic adjustment, photosynthesis, and antioxidant defense. Crop Sci. 2020;60(4):2149–2158. doi: 10.1002/csc2.20186. [DOI] [Google Scholar]
- 59.Li SC, Cui LL, Zhang YJ, Wang YW, Mao PS. The variation tendency of polyamines forms and components of polyamine metabolism in Zoysiagrass (Zoysia japonica Steud.) to salt stress with exogenous spermidine application. Front Physiol. 2017;8:208. doi: 10.3389/fphys.2017.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Yu BK, Liu YL. Effects of spermidine and spermine levels on salt tolerance associated with tonoplast H+-ATPase and H+-PPase activities in barley roots. Plant Growth Regul. 2006;49(3):119–126. doi: 10.1007/s10725-006-9001-1. [DOI] [Google Scholar]
- 61.Krishnamurthy R, Bhagwat KA. Polyamines as modulators of salt tolerance in rice cultivars. Plant Physiol. 1989;91(2):500–504. doi: 10.1104/pp.91.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L, Liu YP, Wu GW, Zhang N, Shen QR, Zhang RF. Beneficial rhizobacterium bacillus amyloliquefaciens SQR9 induces plant salt tolerance through spermidine production. Mol Plant-Microbe Interact. 2017;30(5):423–432. doi: 10.1094/MPMI-02-17-0027-R. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Li Z, Li YP, Zhang XQ, Ma X, Huang LK, Yan YH, Peng Y. Chitosan and spermine enhance drought resistance in white clover, associated with changes in endogenous phytohormones and polyamines, and antioxidant metabolism. Funct Plant Biol. 2018;45(12):1205–1222. doi: 10.1071/FP18012. [DOI] [PubMed] [Google Scholar]
- 64.Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109(3):735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H, Ding GH, Fang K, Zhao FG, Qin P. New perspective on the mechanism of alleviating salt stress by spermidine in barley seedlings. Plant Growth Regul. 2006;49(3):147–156. doi: 10.1007/s10725-006-9004-y. [DOI] [Google Scholar]
- 66.Wang Q, Guan C, Wang SM. Coordination of AtHKT1;1 and AtSOS1 facilitates Na+ and K+ homeostasis in Arabidopsis thaliana under salt stress. J Plant Biol. 2014;57(5):282–290. doi: 10.1007/s12374-014-0222-y. [DOI] [Google Scholar]
- 67.Apse MP, Blumwald E. Na+ transport in plants. FEBS Lett. 2007;581(12):2247–2254. doi: 10.1016/j.febslet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 68.Hogland CR, Arnon DI. The solution-culture method for growing plants without soil. Calif Agric Exp Circ. 1950;247:16–21. [Google Scholar]
- 69.Blum A, Ebercon A. Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci. 1981;21(1):43–47. doi: 10.2135/cropsci1981.0011183X002100010013x. [DOI] [Google Scholar]
- 70.Barrs HD, Weatherley PE. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci. 1962;15(3):413–428. doi: 10.1071/BI9620413. [DOI] [Google Scholar]
- 71.Blum A. Osmotic adjustment and growth of barley genotypes under drought stress. Crop Sci. 1989;29(1):230–233. doi: 10.2135/cropsci1989.0011183X002900010052x. [DOI] [Google Scholar]
- 72.McMichael BL, Burke JJ. Metabolic activity of cotton roots in response to temperature. Environ Exp Bot. 1994;34(2):201–206. doi: 10.1016/0098-8472(94)90039-6. [DOI] [Google Scholar]
- 73.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Cell. 1949;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chance B, Maehly AC. Assay of catalases and peroxidases. Methods Enzymol. 1955;55(2):764–775. doi: 10.1016/S0076-6879(55)02300-8. [DOI] [PubMed] [Google Scholar]
- 75.Yoshiyuki N, Kozi A. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1980;22(5):867–880. [Google Scholar]
- 76.Agarwal S, Pandey V. Antioxidant enzyme responses to NaCl stress in Cassia angustifolia. Biol Plant. 2004;48(4):555–560. doi: 10.1023/B:BIOP.0000047152.07878.e7. [DOI] [Google Scholar]
- 77.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 78.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39(1):205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 79.Duan JJ, Li J, Guo SR, Kang YY. Exogenous spermidine affects polyamine metabolism in salinity-stressed cucumis sativus roots and enhances short-term salinity tolerance. J Plant Physiol. 2008;165(15):1620–1635. doi: 10.1016/j.jplph.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 80.Li Z, Li YP, Zhang Y, Cheng BZ, Peng Y, Zhang XQ, Ma X, Huang LK, Yan YH. Indole-3-acetic acid modulates phytohormones and polyamines metabolism associated with the tolerance to water stress in white clover. Plant Physiol Biochem. 2018;129:251–263. doi: 10.1016/j.plaphy.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 81.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. Primer sequences of gene.
Data Availability Statement
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.