Abstract
Conformational dynamics of RNA molecules play a critical role in governing their biological functions. Measurements of RNA dynamic behavior sheds important light on sites that interact with their binding partners or cellular stimulators. However, such measurements using solution-state NMR are difficult for large RNA molecules (> 70 nt; nt = nucleotides) owing to severe spectral overlap, homonuclear 13C scalar couplings, and line broadening. Herein, a strategic combination of solid-phase synthesis, site-specific isotopic labeled phosphoramidites, and enzymatic ligation is introduced. This approach allowed the position-specific insertion of isotopic probes into a 96 nt CCR5 RNA fragment. Accurate measurements of functional dynamics using the Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion (RD) experiments enabled extraction of the exchange rates and populations of this RNA. NMR chemical shift perturbation analysis of the RNA/microRNA-1224 complex indicated that A90-C1′ of the pseudoknot exhibits similar changes in chemical shift observed in the excited state. This work demonstrates the general applicability of a NMR-labeling strategy to probe functional RNA structural dynamics.
Keywords: CCR5 mRNA pseudoknot, dynamics, microRNA, NMR spectroscopy, RNA solid-phase synthesis
The C-C chemokine receptor type 5 (CCR-5) is a coreceptor involved in membrane fusion of human immunodeficiency type 1 virus (HIV-1) particle and plays a critical role in facilitating viral entry into human cells.[1] Significant efforts have been made to develop therapeutic agents or strategies either targeting the receptor itself[2] or manipulating its gene[3] for treatment of HIV-1 infection. Recently, a highly-conserved pseudoknot motif was identified in the CCR-5 encoding mRNA that stimulates programmed −1 ribosomal frameshifting (−1 PRF) and that acts as an mRNA destabilizing element by directing translating ribosomes to a −1 frame termination codon.[4] Interaction between this element and a microRNA was proposed to stimulate −1 PRF, thus fine-tuning gene expression by controlling mRNA abundance, suggesting that this element may be novel for controlling HIV-1 viral replication.[4] Unlike other non-coding functional RNA elements currently being pursued as targets for various diseases,[5] the pseudoknot motif is within the coding region of CCR5 mRNA.[4] Therefore, any information about the CCR5 pseudoknot structure and interaction with miRNA molecules will provide insight into gene regulation connected with −1 PRF mechanism and foundation for RNA/miRNA-based drug design and development.
Although structures are desirable, not all RNA molecules are amenable to high-resolution structural characterization by X-ray crystallization and nuclear magnetic resonance (NMR) spectroscopy, mostly because RNA molecules are highly flexible. Their inherent plasticity renders them capable of sampling a vast ensemble of conformations[6] RNA structures also typically undergo conformational changes in response to ligand binding, cellular signals or environmental fluctuations; indeed, interconversions among different conformational states are crucial for RNA molecules to fulfil their diverse biological functions.[7] Therefore, to better understand how CCR5 functions in −1 PRF and gene regulation, we need to not only determine high resolution unbound structures and complexed with miRNA-1224, but to also understand internal motions and dynamic behavior.
NMR, as a proven and powerful approach, has been applied extensively to characterize RNA dynamics over a wide range of timescales.[8] However, these applications have mainly been limited to small RNA systems (< 50 nt; nt = nucleotides) owing to severe spectral overlap and line broadening, in addition to conformational heterogeneity in larger RNA species.[8a, 9] To bypass these problems, ″divide-and-conquer″ strategies have been employed for structural characterization of sub-domains derived from larger functional RNA molecules. However, these approaches have led to questions regarding whether the isolated sub-domains adopt the same structures, behave with the same dynamic properties, and carry out the same functions as those encoded by the full-length RNA molecules.[10] A hybrid solid-liquid phase transcription method allowing region-specific isotope labeling for RNA molecules larger than 50 nt was previously reported.[11] This, however, was found to be highly sequence restrictive and generated very low yields for large RNA molecules.[11] Recently, coupling of site-specific isotope-labeling techniques with enzymatic ligation has expanded the size limitation of RNA NMR.[12] Of particular promise are approaches combining site-specific isotope-labeling, solid-phase synthesis, and enzymatic ligation in investigating RNA structure and dynamics. Indeed, this chemo-enzymatic approach significantly simplifies the NMR spectra, allows unambiguous resonance assignment and identification of key residues involved in molecular recognition, and enables accurate measurements of dynamic features at atomic resolution that are typically difficult to obtain for large RNA molecules.[8b,e]
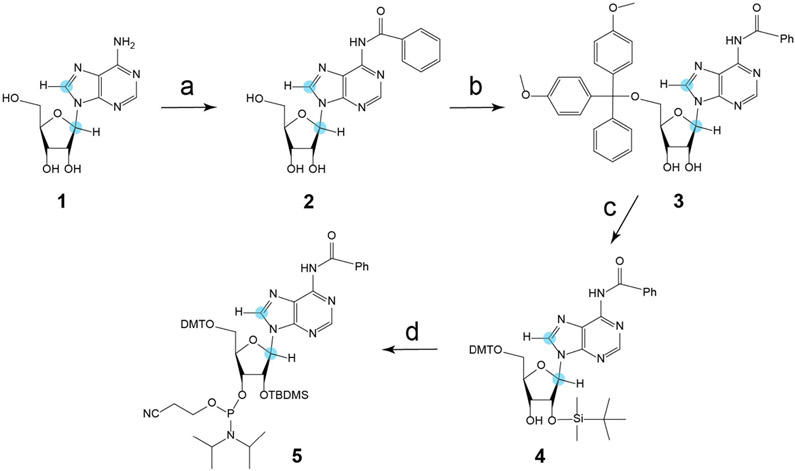
We report herein the characterization of site-specific motions of a 96 nt CCR5 RNA −1 PRF pseudoknot element that are correlated to interaction with miR-1224 based on the strategy described above using NMR spectroscopy. We first prepared a key precursor (8,1′-13C)-modified ATP by coupling (8-13C)-adenine with (1′-13C)-ribose using chemo-enzymatic synthesis with an excellent yield above 90%,[13] followed by dephosphorylation with recombinant shrimp alkaline phosphatase to afford (8,1′-13C)-adenosine (Figure S1, Supporting Information; compound 1 in Scheme 1). The exocyclic amino N6 group of (8,1′-13C)-adenosine (1) was then benzoylated to yield 2. Selective protection of the 2′-hydroxyl group with tert-butyldimethylsilyl chloride through transient blocking of the 5′- and 3′-hydroxy groups using di-tert-butylsilandiylditriflate gave intermediate 3. Selective removal of the 3′,5′-di-tert-butylsilanediyl group and subsequent tritylation gave N6-Bz-5′-O-DMT-(8,1′-13C) adenosine (4). Lastly, the (8,1′-13C)-adenosine phosphoramidite building block 5, which is not commercially available, was obtained for the first time by treatment with 2′-cyanoethyl N,N-diisopropylchlorophosphoramidite (Scheme 1). This building block is crucial for insertion of isotopic probes into CCR5 RNA residue-specifically to facilitate subsequent NMR studies.
Scheme 1.
Synthesis of N6-benzoyl-8,1′-13C-2′-O-TBDMS-adenosine phosphoramidite 5. Reaction conditions: a) Benzoylchloride, trimethylsilyl chloride, pyridine, 2.5 h, rt, then water, 28% NH3, 39%. b) Di-tert-butylsilyl bis(trifluoromethanesulfonate) in DMF, 0 °C, 1 h, then tert-butyldimethylsilyl chloride, 3 h, 60 °C, 40%. c) HF•Py in CH2Cl2, 0 °C, 1 h, then DMT-Cl in pyridine, rt, 3 h, 91%. d) CEP-Cl, DiPEA in THF, rt, 3 h, 80%. Orange dot = 13C.
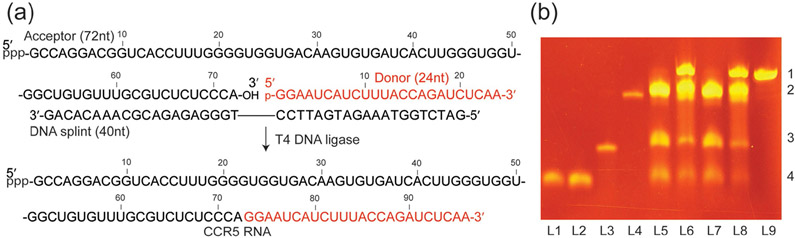
Based on previous mutational studies that suggested Stem 2 of CCR5 pseudoknot motif interacts with miR-1224,[4] the RNA was split into two fragments such that a 24 nt donor fragment containing eight adenosine residues spans the proposed interacting sequence (Figure 1 a). The donor fragments bearing (8,1′-13C)-modified adenosine at each of five positions were synthesized using RNA solid-phase synthesis (Table S1 and Figure S2). Unlabeled 2′-O-TBDMS-protected nucleoside phosphoramidite building blocks were first utilized to optimize the conditions for synthesizing the 24 nt CCR5 donor fragments. A prolonged coupling time (6 min) was employed to guarantee yields greater than 98%. Next, the synthesized 2′-O-TBDMS-(8,1′-13C)-adenosine phosphoramidite was introduced into the donor fragments at different locations of the RNA in combination with three other unlabeled 2′-O-TBDMS-protected nucleoside phosphoramidite building blocks. After successful incorporation of the (8,1′-13C)-adenosine label, we deprotected these donor fragments using methylamine/ammonium hydroxide followed by treatment with 3 m triethylamine trihydrofluoride (TEA•3HF) in anhydrous DMSO.[14] This desilylation agent was very efficient even in the presence of low moisture. After purification to homogeneity by denaturing polyacrylamide gel electrophoresis (PAGE), we obtained 0.3 μmol of the donor fragments typically from a 1 μmol synthesis-scale column (Table S1).
Figure 1.
Ligation of the CCR5 pseudoknot element. a) Scheme for preparation of segmentally and selectively 13C-labeled CCR5 RNA. The unlabeled 72 nt acceptor fragment was synthesized by in vitro transcription. The donor was prepared by either in vitro transcription followed by enzymatic dephosphorylation using RNA 5′ polyphosphatase or solid-phase synthesis followed by phosphorylation with T4 polynucleotide kinase to obtain the desired monophosphorylated group at the 5′-terminus. A 40 mer DNA splint was used to facilitate RNA ligation. b) A 12% denaturing PAGE showing the ligation results. L1: de-phosphorylated donor fragment obtained from in vitro transcription; L2: phosphorylated donor fragment from solid-phase synthesis; L3: DNA splint; L4: acceptor fragment ; L5: ligation reaction at 0 h using the donor from L1; L6: after 3 h reaction of L5; L7: ligation reaction at 0 h using the donor from L2; L8: after 3 h reaction of L7; L9: CCR5 mRNA pseudoknot prepared by in vitro transcription. Similar to prior observations,[8f] addition of the DNA splint to the reaction system resulted in changes of the migration pattern of all components in the system. The numbers at the right side of the image (1, 2, 3, and 4) correspond to full-length CCR5, 72 nt acceptor, DNA splint, and 24 nt donor fragment, respectively. Typically, excess amounts of the acceptor fragment and the DNA splint were applied and the ligation yield was >60% based on the conversion of the donor fragment which, in most cases, was prepared with different isotope labels.
Next, we ligated an unlabeled 72 nt acceptor fragment, synthesized with T7 RNA polymerase, to a solid-phase-synthesized residue/site-specific labeled 24 nt donor fragment (Figure 1). The unlabeled acceptor fragment had a protective 5′-triphosphate group and a reactive 3′-OH group by virtue of using T7 RNA polymerase for in vitro transcription. The synthesized residue/site-specific labeled 24 nt donor fragment was phosphorylated by T4 polynucleotide kinase to obtain the desired monophosphorylated group at the 5′-terminus. A donor fragment with different labeling patterns could also be prepared by in vitro transcription with T7 RNA polymerase and dephosphorylated using RNA 5′-polyphosphatase prior to ligation (Figure 1 b). In general (detailed in the Supporting Information), excess amounts of the acceptor fragment and the DNA splint were used in the ligation reaction to achieve maximum conversion of the donor fragment into the ligated residue/site-specific isotope labeled product with a typical ligation yield (based on the donor fragment) of > 60% (Figure 1 b).
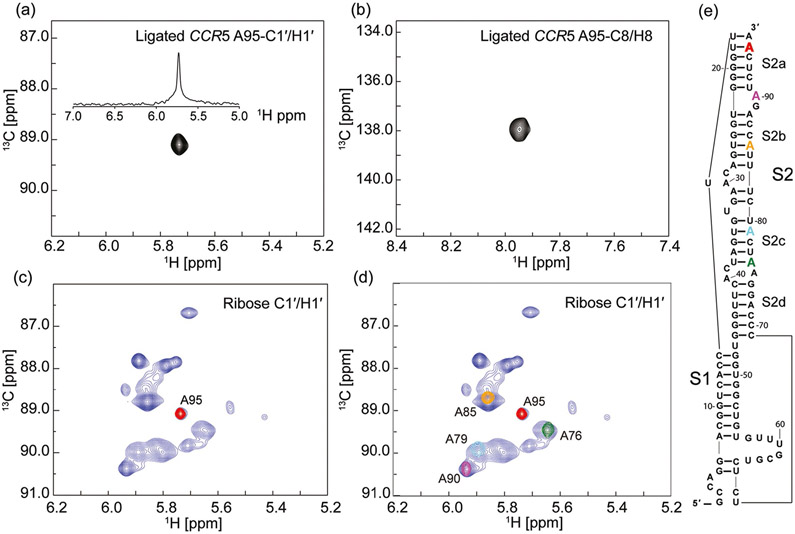
The residue/site-specific insertion of 13C probes into CCR5 RNA was confirmed by NMR spectroscopy. 2D 1H–13C heteronuclear multiple quantum coherence (HMQC) spectra obtained for the ligated and residue/site-specific labeled CCR5 RNA in the absence of miR-1224 exhibited a single cross peak, corresponding to either ribose C1′ or base C8 of the residue A95 (Figure 2a,b). Unambiguous assignments of A95 C1′/H1′ was readily achieved by overlaying the ribose C1′ HMQC spectrum with that of the CCR5 RNA containing all seventeen adenosine residues prepared from in vitro transcription with atom-specific-labeled (8,1′-13C)-ATP and unlabeled CTP/GTP/UTP (Figure 2c). Continued incorporation of 13C probes at different positions of the RNA resulted in identification of ribose C1′/H1′ atoms of A76, A79, A85, and A90 (Figure 2d, e) and base C8/H8 atoms of these residues (Figure S3 a, b).
Figure 2.
Heteronuclear multiple quantum coherence (HMQC) spectra of the CCR5 pseudoknot element. a) Ribose C1′ and b) base C8 HMQC spectra of ligated CCR5 RNA bearing site-specific (8,1′)-13C-labeled probes at residue A95. The donor fragment was prepared by solid-phase synthesis on a DNA synthesizer first using unlabeled RNA amidites followed by incorporation of a selective (8,1′)-13C-labeled adenosine phosphoramidite at different positions. Upon ligation to the unlabeled acceptor fragment, the resulting ligated full length CCR5 pseudoknot element afforded a single peak at either ribose C1′ or base C8 region. The projected 1D 1H spectrum in part (a) displayed a single peak across a broader H1′ region, demonstrating the success in isotope labeling of a specific residue of the full length 96 nt RNA. c) Overlay of ribose HMQC spectra obtained from two CCR5 RNA pseudoknot samples. The spectrum in blue was recorded for a full length CCR5 RNA with all 17 adenosine residues labeled selectively with (8,1′)-13C atoms; the spectrum in red was from the ligated CCR5-A95 RNA. d) Ligation between the unlabeled acceptor and four different residue-specific labeled donor fragments resulted in resonance assignments of four additional residues: A90, A85, A79, and A76 at both ribose C1′ and base C8 regions. The spectrum in blue is the same as that in part (c). This method allowed for straightforward resonance assignments and provided staring points for assignment of the neighboring residues based on 2D/3D NOESY experiments. e) Secondary structure of CCR5 pseudoknot RNA based on previous SHAPE experiments.[4]
To distinguish the three remaining adenosine residues in the donor fragment, we used conventional NMR techniques and prepared a CCR5 RNA sample by ligating the unlabeled acceptor fragment and a donor fragment labeled only with eight site-specific (8,1′-13C)-adenosines. A 2D edited-edited NOESY spectrum obtained for the RNA sample showed inter-residue cross-peaks between A75H1′ and A76H8, and between A95H1′ and A96H8, allowing straightforward and unambiguous assignments of residues A75 and A96 (Figure S3). The last adenosine A88 within the donor fragment region was exclusively determined (Figure S3). The eight adenosine ribose anomeric atoms in the donor fragment of the ligated CCR5 RNA were well-resolved in the 2D HMQC spectrum (Figure S3b).
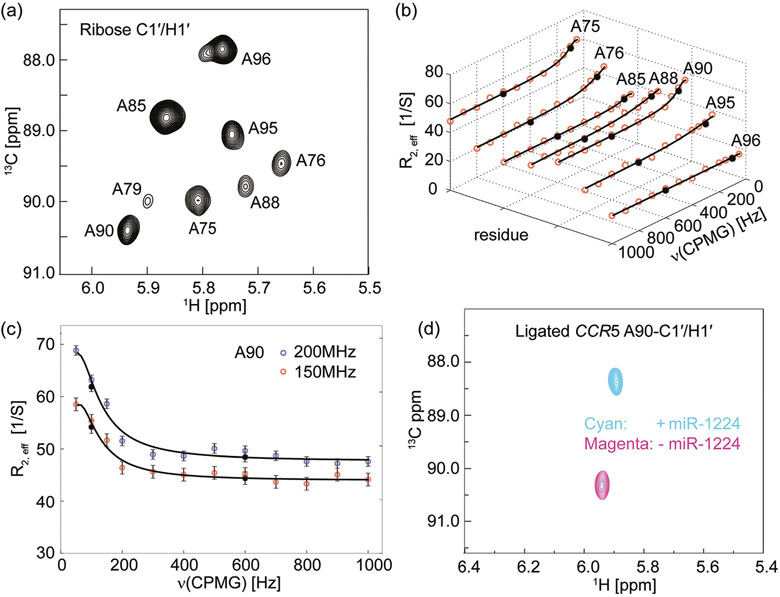
In short, this labeling strategy provides a number of advantages. First, the use of site-specific isotopic probes helps to resolve substantial ambiguities in resonance assignments of RNA molecules of this size. Second, these labels are effective and reliable for quickly assigning large (> 60 nt) RNA molecules. This represents an important advance for RNA structural studies by NMR spectroscopy, currently stuck at a median size of 25 nt in the PDB. Third, these labels remove complicated scalar coupling effects or dipolar interactions with adjacent nuclei, thereby providing an ideal platform for performing, especially, CPMG RD experiments to investigate CCR5 conformational exchanges on micro- to millisecond timescales.[13b, 15] other RD experiments such as R1ρ and CEST will also benefit from this labeling pattern.[8b,f,g] A pseudo-3D 1H–13C HMQC version of CPMG RD experiment was used to minimize relaxation loses and the effect of magnetic field inhomogeneity.[16] The eight well-dispersed 13C probes of the ligated CCR5 RNA in the 2D 1H–13C correlation map with Trelax = 0 (Figure 3a) allowed for accurate measurements of the functional dynamics and subsequent extraction of information about the exchange processes in which the pseudoknot participates. As a result, the 1′ anomeric carbons of adenosines A75, A76, A88, A90, and A95 of the ligated CCR5 RNA at 150 MHz 13C Larmor frequency showed non-flat dispersion profiles whereas that of A85 and A96 exhibited flat dispersion profiles (Figure 3 b). Unfortunately the signal intensity of A79-C1′ rapidly decreases during the CPMG relaxation delay which prevents further characterization and analysis. Additionally A76 and A96 appear to have shoulder peaks suggestive of multiple conformations. More detailed analyses are needed to elucidate the precise nature of these conformations.
Figure 3.
Chemical exchange in CCR5 revealed by 13C Carr–Purcell–Meiboom–Gill relaxation dispersion (CPMG RD) experiments. a) 2D 1H–13C HMQC spectrum of a ligated CCR5 RNA sample labeled only with site-specific (8,1′-13C)-adenosines within the donor fragment. This was recorded in the absence of miR-1224, at 298 K, at 600 MHz proton Larmor frequency, with Trelax=0 ms. The expected eight anomeric H-C cross-peaks are well-resolved. b) 13C CPMG RD profiles of the ligated CCR5 RNA at 150 MHz 13C Larmor frequency. Red dots represent experimental data and black dots represent repeated experiments. Solid lines are best-RD-curve fits of the CPMG profiles. Residues A75, A76, A88, A90, and A95 show non-flat dispersion profiles whereas A85 and A96 show flat dispersion profiles. Note that the A79 resonance was too broad to adequately fit. c) 13C CPMG RD profiles of A90 ribose anomeric carbon atom of the ligated CCR5 RNA at 150 (red dots) and 200 MHz (blue dots) 13C Larmor frequency. Black circles represent repeated experiments and solid lines are the best fits of the CPMG profiles. d) NMR titration of the ligated CCR5-A90 RNA with miR-1224. The 2D HMQC spectrum in magenta recorded in the absence of miR-1224 exhibits a single cross-peak corresponding to ribose C1′ of the residue A90. Upon addition of miR-1224, the ribose A90-C1′ chemical shift changed dramatically from 90.3 ppm to 88.3 ppm (in cyan). The large chemical shift change (up to 2.0 ppm) indicates that the interaction with miR-1224 alters the local chemical environment of the A90 residues. In the presence of an equimolar amount of miR-1224, the CCR5 RNA is not completely converted to its bound form, with ca. 9% remaining in the free conformation. This is consistent with the EMSA result (Figure S6). However, adding 20% extra amount of miR-1224 converts it into the completely bound form (data not shown).
CPMG RD experiments were also conducted at 200 MHz carbon Larmor frequency (magnet field strength: 18.8 T; Figure 3c and Figure S4). Subsequent global fitting resulted in extracting kinetic and thermodynamic parameters of the conformational transition.[13b, 15a] Initially, we expected to obtain the same dynamic parameters because the seven probes report on the same biological exchange process. Surprisingly, fits to a two-state exchange model resulted in residue-specific variations in dynamic behaviors: two probes (A85 and A96) showed no conformational exchange, whereas the other five displayed different thermodynamic parameters (Table 1). A76 samples a minor-populated state of PB ≈ 30%, while residues A75, A88, A90 and A95 sample an invisible-populated state of PB < 10% (Table 1). The exchange rate constants (kex) of A75, A88, and A90 were very close to one another within experimental error, whereas the rate parameters of A76 and A95 were roughly one magnitude higher (Table 1). Longitudinal relaxation rates were also recorded for these sites. Larger longitudinal relaxation rate of residues A85 and A96 indicated the presence of rapid local motion and high flexibility, indicating complicated motions occurring on various time scales (Table S2 and Figure S5). Thus these probes experience local independent complex motions or conformational exchanges.
Table 1.
Kinetic and thermodynamic parameters extracted from CPMG RD experiments.[a]
| Residue (ribose C1′) | kex [S−1] | PB [%] | Δω [ppm] | χ2 |
|---|---|---|---|---|
| A75 | 179±138 | 8±5 | 0.8±0.1 | 1.08 |
| A76 | 1014±188 | 30±12 | 0.3±0.2 | 1.12 |
| A88 | 356±97 | 3±1 | 0.6±0.1 | 0.86 |
| A90 | 165±121 | 9±5 | 0.8±0.1 | 1.46 |
| A95 | 943±197 | 6±2 | 0.5±0.2 | 0.96 |
The CPMG RD experiments were carried out on the ligated CCR5 RNA sample labeled with site-specific (8,1′-13C)-adenosine groups within the donor fragment in the absence of miR-1224 at 298 K. The CPMG data of the ribose C1′ of all five adenosine residues were fitted together to slow exchange two-state models and the results were derived from 1000 Monte Carlo runs (see the Supporting Information). Note that strictly PB and Δω are not separable in the absence of other data.
A 2D 1H–13C HMQC spectrum of the ligated and residue/site-specific labeled CCR5-A90 RNA in the absence of miR-1224 exhibited a cross peak corresponding to the ribose C1′/H1′ (Figure 3d). Upon addition of miR-1224, the ribose H1′ chemical shift was barely perturbed from 5.86 ppm to 5.83 ppm. In contrast, the ribose C1′ chemical shift was strongly perturbed from 90.3 ppm to 88.3 ppm (Figure 3d). A similar result was obtained for the A90-C8/H8 chemical shift changes (data not shown here). The large chemical shift adjustment (up to 2.0 ppm) unambiguously indicated that miR-1224 binding alters the local chemical environment of the A90-C1′ atom. It is notable that the chemical shift (δC = 90.3 ppm) of A90-C1′ in the free-form CCR5 pseudoknot suggests that the A90 residue either base-pairs or is structurally constrained to a fixed conformation. The presence of miR-1224 triggered a highly specific interaction, flipping A90 out to participate in an intermolecular interaction, possibly base stacking with the miR-1224 bases but not base pairing as indicated by its up field chemical shift value (δC = 88.3 ppm). The A90-C1′ chemical shift of the minor-populated invisible state (∣Δω∣ = 0.8±0.1 ppm) likely samples local base-flipping motions to facilitate intermolecular recognition. The base C8 and ribose C1′ of CCR5-G89 also showed large chemical shift changes (data not shown), suggesting that the predicted internal bulge involving residues G89 and A90 is vital to the intermolecular interaction with miR-1224. More detailed analysis using these and other RD measurements will be needed to clearly delineate the sign of chemical shift changes.
Our NMR data indicate that the secondary structure of the CCR5 RNA pseudoknot is slightly different from that derived from SHAPE experiments.[4] We are continuing with resonance assignment using this strategy to obtain a more accurate secondary structure of the CCR5 RNA pseudoknot using NMR spectroscopy.
In summary, herein we have expanded the labeling protocol from previously reported (6,1′-13C) cytidine labels to (8,1′-13C) base and ribose building blocks for preparing residue/site-specifically isotopic labeled 96nt CCR5 RNA to study conformational dynamics.[8f, 15a] The strategic combination of solid-phase synthesis of RNA with labeled phosphoramidites, enzymatic ligation, and NMR spectroscopy allows investigations into the structures and dynamics of large RNA systems, than have been possible until now, with biologically important functions that are otherwise impeded by severe spectral overlap.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Paul Paukstelis for access to the DNA synthesizer and help with setup of DNA/RNA synthesis. This work was funded by grants to J.D.D. from the NIH (R01 GM117177), and to T.K.D. from NSF for NMR instrumentation (DBI1040158) and NIH (U54GM103297).
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/chem.201705948.
Conflict of interest
The authors declare no conflict of interest.
References
- [1].a) Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA, Science 1996, 272, 1955–1958; [DOI] [PubMed] [Google Scholar]; b) Doms RW, Moore JP, J. Cell Biol 2000, 151, 9–14; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Nature 1996, 382, 722–725; [DOI] [PubMed] [Google Scholar]; d) Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy PE, Kumaraswami V, Giorgi JV, Detels R, Hunter J, Chopek M, Berger EA, Fauci AS, Nutman TB, Murphy PM, Mol. Med 1997, 3, 23–36; [PMC free article] [PubMed] [Google Scholar]; e) Gallo SA, Finnegan CM, Viard M, Raviv Y, Dimitrov A, Rawat SS, Puri A, Durell S, Blumenthal R, Biochim. Biophys. Acta Biomembr 2003, 1614, 36–50; [DOI] [PubMed] [Google Scholar]; f) Chan DC, Kim PS, Cell 1998, 93, 681–684. [DOI] [PubMed] [Google Scholar]
- [2].a) Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M, Proc. Natl. Acad. Sci. USA 1999, 96, 5698–5703; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dragic T, Trkola A, Thompson DAD, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying WW, Smith SO, Sakmar TP, Moore JP, Proc. Natl. Acad. Sci. USA 2000, 97, 5639–5644; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Watson C, Jenkinson S, Kazmierski W, Kenakin T, Mol. Pharmacol 2005, 67, 1268–1282; [DOI] [PubMed] [Google Scholar]; d) Maeda K, Nakata H, Koh Y, Miyakawa T, Ogata H, Takaoka Y, Shibayama S, Sagawa K, Fukushima D, Moravek J, Koyanagi Y, Mitsuya H, J. Virol 2004, 78, 8654–8662; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Trkola A, Ketas TJ, Nagashima KA, Zhao L, Cilliers T, Morris L, Moore JP, Maddon PJ, Olson WC, J. Virol 2001, 75, 579–588; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Strizki JM, Treblay C, Xu S, Wojcik L, Wagner N, Gonsiorek W, Hipkin RW, Chou CC, Pugliese-Sivo C, Xiao YS, Tagat JR, Cox K, Priestley T, Sorota S, Huang W, Hirsch M, Reyes GR, Baroudy BM, Anti-microb. Agents Chemother 2005, 49, 4911–4919; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, McCombie S, Baroudy B, Moore JP, Sakmar TP, Dragic T, J. Virol 2003, 77, 5201–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, N. Engl. J. Med 2014, 370, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Belew AT, Meskauskas A, Musalgaonkar S, Advani VM, Sulima SO, Kasprzak WK, Shapiro BA, Dinman JD, Nature 2014, 512, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Matsui M, Corey DR, Nat. Rev. Drug Discovery 2017, 16, 167–179; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ling H, Fabbri M, Calin GA, Nat. Rev. Drug Discovery 2013, 12, 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Al-Hashimi HM, Walter NG, Curr. Opin. Struct. Biol 2008, 18, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM, Nature 2012, 482, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Fürtig B, Richter C, Wöhnert J, Schwalbe H, ChemBioChem 2003, 4, 936–962; [DOI] [PubMed] [Google Scholar]; b) Hansen AL, Al-Hashimi HM, J. Am. Chem. Soc 2007, 129, 16072–16082; [DOI] [PubMed] [Google Scholar]; c) Zhang QQ, Sun X, Watt ED, Al-Hashimi HM, Science 2006, 311, 653–656; [DOI] [PubMed] [Google Scholar]; d) Rinnenthal J, Buck J, Ferner J, Wacker A, Furtig B, Schwalbe H, Acc. Chem. Res 2011, 44, 1292–1301; [DOI] [PubMed] [Google Scholar]; e) Bothe JR, Nikolova EN, Eichhorn CD, Chugh J, Hansen AL, Al-Hashimi HM, Nat. Methods 2011, 8, 919–931; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Chen B, LeBlanc RM, Dayie TK, Angew. Chem. Int. Ed 2016, 55, 2724–2727; [DOI] [PubMed] [Google Scholar]; Angew. Chem 2016, 128, 2774–2777; [Google Scholar]; g) Zhao B, Hansen AL, Zhang Q, J. Am. Chem. Soc 2014, 136, 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scott LG, Hennig M in Methods Mol. Biol, Vol. 452, Humana, Totowa, 2008, pp. 29–61. [DOI] [PubMed] [Google Scholar]
- [10].a) Spriggs S, Garyu L, Connor R, Summers MF, Biochemistry 2008, 47, 13064–13073; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lukavsky PJ, Virus Res. 2009, 139, 166–171; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ganser LR, Al-Hashimi HM, Proc. Natl. Acad. Sci. USA 2016, 113, 13263–13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y, Holmstrom E, Zhang J, Yu P, Wang J, Dyba MA, Chen D, Ying J, Lockett S, Nesbitt DJ, Ferre-D′Amare AR, Sousa R, Stagno JR, Wang Y-X, Nature 2015, 522, 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Thakur CS, Sama JN, Jackson ME, Chen B, Dayie TK, J. Biomol. NMR 2010, 48, 179–192; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Alvarado LJ, LeBlanc RM, Longhini AP, Keane SC, Jain N, Yildiz ZF, Tolbert BS, D′Souza VM, Summers MF, Kreutz C, Dayie TK, ChemBioChem 2014, 15, 1573–1577; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kawahara I, Haruta K, Ashihara Y, Yamanaka D, Kuriyama M, Toki N, Kondo Y, Teruya K, Ishikawa J, Furuta H, Ikawa Y, Kojima C, Tanaka Y, Nucleic Acids Res. 2012, 40, e7; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kim I, Lukavsky PJ, Puglisi JD, J. Am. Chem. Soc 2002, 124, 9338–9339; [DOI] [PubMed] [Google Scholar]; e) D′Souza V, Dey A, Habib D, Summers MF, J. Mol. Biol 2004, 337, 427–442; [DOI] [PubMed] [Google Scholar]; f) Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF, Science 2011, 334, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Longhini AP, LeBlanc RM, Dayie TK, Methods 2016, 103, 11–17; [DOI] [PubMed] [Google Scholar]; b) Longhini AP, LeBlanc RM, Becette O, Salguero C, Wunderlich CH, Johnson BA, D′Souza VM, Kreutz C, Dayie TK, Nucleic Acids Res. 2016, 44, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Westmanu E, Stromberg R, Nucleic Acids Res. 1994, 22, 2430–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Wunderlich CH, Spitzer R, Santner T, Fauster K, Tollinger M, Kreutz C, J. Am. Chem. Soc 2012, 134, 7558–7569; [DOI] [PubMed] [Google Scholar]; b) Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE, Nature 2004, 430, 586–590. [DOI] [PubMed] [Google Scholar]
- [16].Grzesiek S, Bax A, J. Biomol. NMR 1995, 6, 335–339. [DOI] [PubMed] [Google Scholar]
- [17].Wunderlich CH, Spitzer R, Santner T, Fauster K, Tollinger M, Kreutz C, J. Am. Chem. Soc 2012, 134, 7558–7569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.