Abstract
Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shedding was observed from the upper respiratory tract of a female immunocompromised individual with chronic lymphocytic leukemia and acquired hypogammaglobulinemia. Shedding of infectious SARS-CoV-2 was observed up to 70 days, and of genomic and subgenomic RNA up to 105 days, after initial diagnosis. The infection was not cleared after the first treatment with convalescent plasma, suggesting a limited effect on SARS-CoV-2 in the upper respiratory tract of this individual. Several weeks after a second convalescent plasma transfusion, SARS-CoV-2 RNA was no longer detected. We observed marked within-host genomic evolution of SARS-CoV-2 with continuous turnover of dominant viral variants. However, replication kinetics in Vero E6 cells and primary human alveolar epithelial tissues were not affected. Our data indicate that certain immunocompromised individuals may shed infectious virus longer than previously recognized. Detection of subgenomic RNA is recommended in persistently SARS-CoV-2-positive individuals as a proxy for shedding of infectious virus.
Keywords: SARS-CoV-2, COVID-19, immunocompromised, asymptometic, long-term shedding, infectious virus, within host evolution, convalescent plasma, chronic lymphocytic leukemia
Graphical Abstract
Highlights
-
•
Persistent SARS-CoV-2 infection and shedding in immunocompromised individual
-
•
Infectious SARS-CoV-2 isolated up to 70 days after diagnosis
-
•
Observed within-host genetic variation with continuous turnover of viral variants
-
•
SARS-CoV-2 isolates from the individual do not display altered replication
This case study describes a female immunocompromised individual with chronic lymphocytic leukemia and acquired hypogammaglobulinemia who became persistently infected with SARS-CoV-2. Although asymptomatic throughout the course of infection, she demonstrated prolonged shedding of infectious SARS-CoV-2 virus and RNA. This study demonstrates that certain individuals may remain infectious for prolonged periods of time and highlights the need for further studies to understand risk factors for prolonged infectious SARS-CoV-2 shedding.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA can be detected at various sites, including samples obtained from the nares, nasopharynx, pharynx, bronchoalveolar lavage (BAL) fluid, feces, and blood (Wang et al., 2020a; Sun et al., 2020; Judson and Munster, 2020). The duration of SARS-CoV-2 RNA shedding is generally between 3 and 46 days after symptom onset (Fu et al., 2020; Qian et al., 2020; Liu et al., 2020c). Asymptomatic individuals shed SARS-CoV-2 RNA comparably with symptomatic individuals regarding duration and viral load (Lee et al., 2020; Long et al., 2020; Zou et al., 2020). Persistent SARS-CoV-2 RNA shedding has been documented, with patients remaining qRT-PCR-positive for up to 63 days (Li et al., 2020; Liu et al., 2020b). In addition, there are reports of symptomatic and asymptomatic individuals testing positive again after a period of negative testing (Lan et al., 2020; Hu et al., 2020). Because qRT-PCR detects viral RNA but does not confirm the presence of infectious SARS-CoV-2, these observations raise questions about the duration of infectious SARS-CoV-2 shedding and transmission potential for symptomatic and asymptomatic individuals.
Estimates suggest that infectiousness begins 2.3 days prior to symptom onset and declines within 7 days of symptom onset (He et al., 2020b). Consistent with this, infectious SARS-CoV-2 has been isolated from patient samples taken up to 8 days after symptom onset but typically not thereafter (Wölfel et al., 2020; Bullard et al., 2020). In contrast to prolonged shedding of SARS-CoV-2 RNA, the longest detected shedding of infectious SARS-CoV-2 virus is up to 20 days after the initial positive test result (van Kampen et al., 2020; Liu et al., 2020b). The probability of isolating SARS-CoV-2 decreases with a lower viral load, when the duration of symptoms exceeds 15 days, and upon generation of detectable neutralizing antibodies (van Kampen et al., 2020).
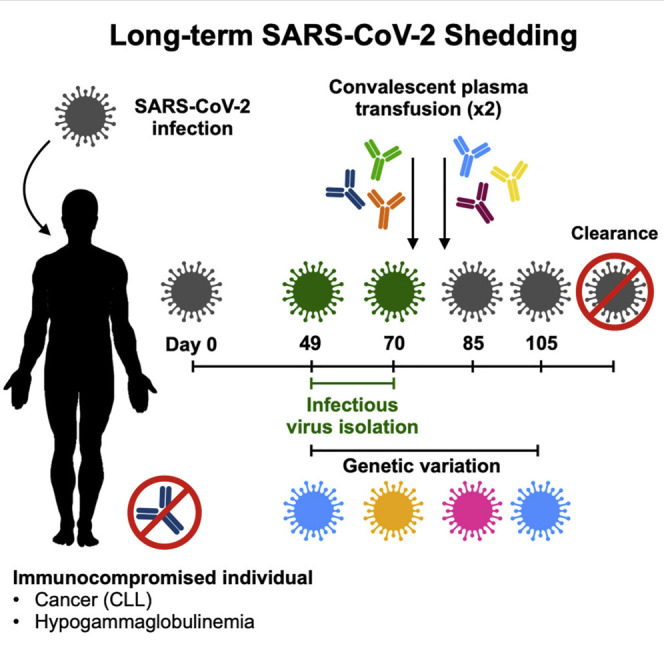
On January 19, 2020, the first case of coronavirus disease 2019 (COVID-19) was identified in the United States, in Snohomish County, Washington, in a traveler returning from Wuhan, China. Community spread in the Seattle region became evident in late February of 2020 (Bhatraju et al., 2020), with extensive spread in a long-term care facility (McMichael et al., 2020a). Here we describe an asymptomatic, immunocompromised individual persistently testing positive for SARS-CoV-2 by qRT-PCR who was infected during the early phase of SARS-CoV-2 spread in the United States. Infectious SARS-CoV-2 was successfully isolated from nasopharyngeal swabs 49 days and 70 days after the initial positive qRT-PCR test. Convalescent plasma treatment was not immediately successful in clearing the infection, but evidence of SARS-CoV-2 RNA was eventually cleared after 105 days.
Results
Clinical Presentation of an Immunocompromised Individual Persistently Infected with SARS-CoV-2
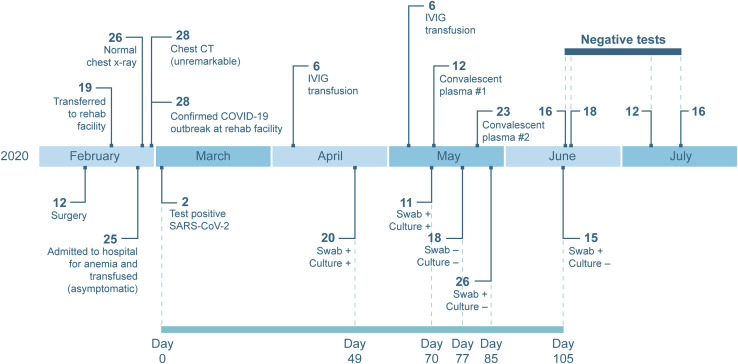
On February 12, 2020, a 71-year-old woman with a 10-year history of chronic lymphocytic leukemia (CLL), acquired hypogammaglobulinemia, anemia, and chronic leukocytosis presented to the emergency department with low back and lower extremity pain. She underwent surgery for a spinal fracture and stenosis related to her cancer on February 14, 2020 (biopsy results in Table S1) and was subsequently transferred to a rehabilitation facility on February 19, 2020. On February 25, 2020, she was re-hospitalized for anemia and underwent a chest X-ray the following day, which was normal. She could not return to her rehabilitation center because of a confirmed outbreak of COVID-19 at the facility (McMichael et al., 2020a, 2020b). Chest computed tomography (CT), performed on February 28, 2020, was unremarkable. The patient had no respiratory or systemic symptoms during this time. Because she was residing in the rehabilitation facility around the time of the COVID-19 outbreak, she was tested and found positive for SARS-CoV-2 on March 2, 2020 (Figure 1 ). After the initial SARS-CoV-2 diagnosis, she was kept in an isolation ward in a single room with negative airflow. Attending medical staff were using full personal protective equipment comprised of powered air-purifying respirators (PAPR) or N95 respirators with goggles, gowns, and gloves. Over the course of the next 15 weeks, she was tested for SARS-CoV-2 another 14 times by several diagnostic companies and remained positive through June 15, 2020, 105 days since the initial positive test. Subsequently, she tested negative on four consecutive swabs from June 16 to July 16, indicating that her infection had cleared.
Figure 1.
Timeline of Clinical Presentation, Diagnostic Tests, and Treatments of an Immunocompromised Individual with Long-Term Shedding of SARS-CoV-2
Dates of relevant clinical events, such as surgeries, therapies, and outcome of diagnostic tests, are shown. Diagnostic qRT-PCR-positive nasopharyngeal and oropharyngeal swabs taken 49, 70, 77, 85, and 105 days after the initial positive sample were sent to Rocky Mountain Laboratories, NIH, for further analysis. Serum and plasma samples pre- and post-transfusion as well as a sample from the donor plasma were also provided. See also Tables S1–S3 for additional laboratory values and clinical information.
Because of acquired hypogammaglobulinemia caused by her CLL, the individual received intravenous immunoglobulin (IVIG) every 4–6 weeks as part of her treatment regimen. She received IVIG treatment on April 6 and May 6, 2020. The manufacture date of her specific lot of IVIG preceded January 1, 2020, the beginning of the COVID-19 pandemic, and therefore did not contribute to any SARS-CoV-2 serology results (Table S2). Because of the persistence of her SARS-CoV-2 infection, serum samples were tested for antibodies against the spike glycoprotein through a study at the NIH Clinical Center, and no spike-specific antibodies were detected (Burbelo et al., 2020). On May 12, 2020, she was transfused with 200 mL of SARS-CoV-2 convalescent plasma provided by Bloodworks Northwest under a US Food and Drug Administration (FDA) emergency investigational new drug (eIND) protocol with a virus-neutralizing (VN) titer of 60 (Table 1 ). Her infection persisted, and on May 23, 2020, she received another 200-mL dose of convalescent plasma from a different donor with a VN titer of 160 under the same protocol (Table 1). Additional laboratory values are available in Table S3.
Table 1.
Virus Neutralization Titers in Pre- and Post-transfusion Sera from the Individual and Convalescent Plasma Used for Transfusion
| Serum | USA/WA1/2020 | Day 49 Isolate | Day 70 Isolate | |
|---|---|---|---|---|
| Day 49 | <10 | <10 | <10 | |
| Day 71 | <10 | <10 | <10 | |
| Day 71 after transfusion | <10 | <10 | <10 | |
| Day 77 | <10 | <10 | <10 | |
| Day 82 | < 10 | 10 | <10 | |
| Day 82 after transfusion | 10 | 10 | 15 | |
| Day 105 | 10 | <10 | <10 | |
| Convalescent plasma 1 | 60 | 40 | 40 | |
| Convalescent plasma 2 | 160 | 160 | 60 | |
Virus neutralization assays were performed for all serum and plasma samples with SARS-CoV-2 strains USA/WA1/2020 and the day 49 and day 70 isolates from the individual. Each serum/plasma sample was tested in duplicate.
Long-Term Shedding of Genomic RNA, Subgenomic RNA, and Infectious SARS-CoV-2
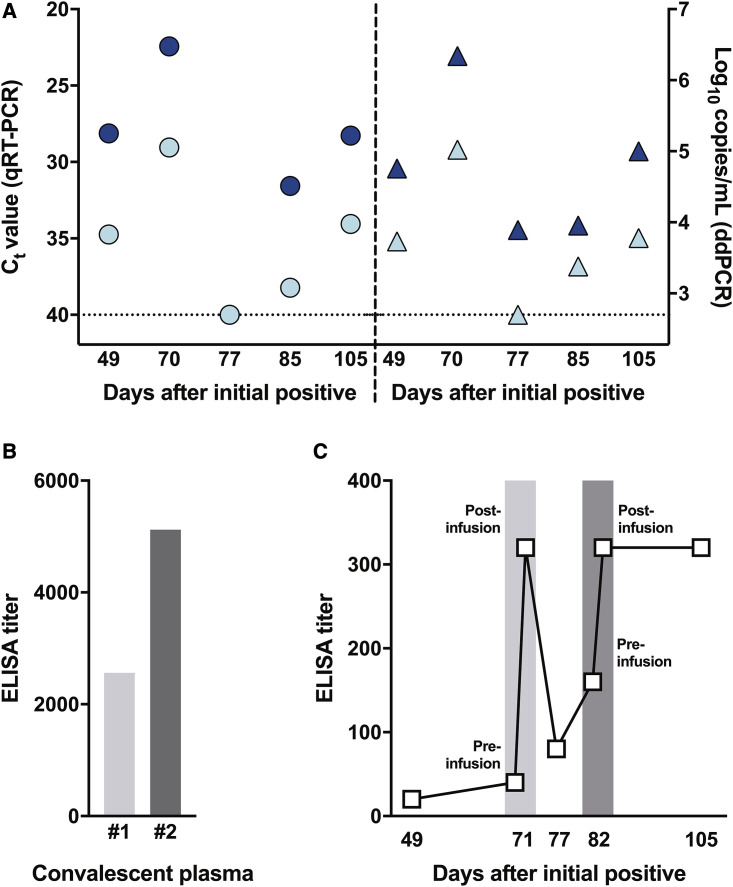
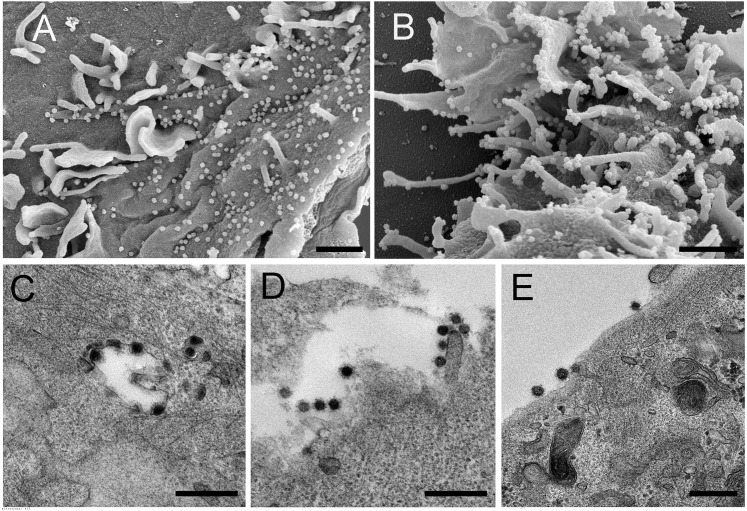
SARS-CoV-2 shedding kinetics in the individual were monitored using detection of genomic RNA (gRNA), subgenomic RNA (sgRNA), and infectious SARS-CoV-2. RNA was extracted from nasopharyngeal and oropharyngeal swabs collected 49, 70, 77, 85, 105, and 136 days after the initial diagnosis and evaluated for the presence of viral gRNA (Corman et al., 2020) and sgRNA (Wölfel et al., 2020). gRNA and sgRNA were detected in nasopharyngeal swabs out to day 105, except for the swab taken on day 77 (Figure 2 A), although the test through EvergreenHealth was positive at this time. None of the oropharyngeal swabs were positive for gRNA or sgRNA, suggesting that the infection was confined to the nasopharynx. Absolute quantification of gRNA and sgRNA on positive swabs was performed by droplet digital PCR (ddPCR) (Figure 2A). The highest viral load was detected in the day 70 swab, at 2.2 × 106 gRNA copies/mL (cycle threshold [Ct] 22.44) and 1.1 × 105 sgRNA copies/mL (Ct 29.05). Detection of sgRNA in swabs is indicative of active SARS-CoV-2 replication because only actively replicating SARS-CoV-2 initiates RNA synthesis, resulting in replication and transcription of sgRNAs (Wang et al., 2020b; Kim et al., 2020), and sgRNA, unlike gRNA, does not persist in the nasal cavity in the absence of virus replication (Speranza et al., 2020). Virus isolation was attempted on all qRT-PCR-positive samples. Infectious SARS-CoV-2 was successfully cultured from the nasopharyngeal swabs collected on day 49 and day 70. Scanning and transmission electron microscopy on SARS-CoV-2 cultured from the nasopharyngeal swabs collected on days 49 and 70 showed viral particles consistent with coronavirus morphology, supporting persistent SARS-CoV-2 infection with shedding of infectious virus in this individual (Figure 3 ).
Figure 2.
Assessment of Viral Load and Seroconversion in an Individual Persistently Infected with SARS-CoV-2 and Treated with Convalescent Plasma
(A) Viral loads were in nasopharyngeal swabs collected at different time points after the initial SARS-CoV-2 diagnosis. Viral RNA extracted from a nasopharyngeal swab was analyzed for the presence of genomic RNA (gRNA; dark blue) and subgenomic RNA (sgRNA; light blue symbols) by qRT-PCR and reported as a cycle threshold (Ct) value (circles, left panel) and in ddPCR and reported as copy numbers (triangles, right panel).
(B) IgG titers against the full-length recombinant SARS-CoV-2 spike ectodomain were determined by ELISA in convalescent plasma used for transfusion. The light gray bar represents the IgG titer of the first donor (convalescent plasma 1), and the dark gray bar represents the second donor (convalescent plasma 2).
(C) IgG titers against the full-length recombinant SARS-CoV-2 spike ectodomain were determined by ELISA in patient serum collected at several time points, including immediately before and after transfusion with convalescent plasma on days 71 (light gray) and 82 (dark gray). Each serum/plasma sample was tested in duplicate.
See also Figure S1 for IgG titers against the SARS-CoV-2 receptor binding domain (RBD).
Figure 3.
Electron Microscopy Confirms Isolation of Coronavirus from the Individual’s Nasopharyngeal Swabs
SARS-CoV-2 cultured from the individual’s nasopharyngeal swabs was used to inoculate Vero E6 cells for imaging by scanning and transmission electron microscopy (SEM and TEM, respectively).
(A and B) SEM images of the day 49 (A) and day 70 (B) isolates.
(C–E) TEM images of the day 49 (C) and day 70 (D and E) isolates.
SEM scale bars, 1 μM; TEM scale bars, 0.5 μM.
Convalescent Plasma Treatment Did Not Clear SARS-CoV-2 from the Upper Respiratory Tract
In an attempt to treat the persistent SARS-CoV-2 infection, the individual received two doses of convalescent plasma therapy on days 71 and 82. Pre- and post-transfusion serum samples and the transfusion convalescent plasma samples were analyzed for the presence of full-length spike and spike receptor binding domain (RBD) antibodies by ELISA assay and of SARS-CoV-2-neutralizing antibodies in a VN assay (Figures 2B and 2C; Figures S1 A and S1B; Table 1; Amanat et al., 2020; Wrapp et al., 2020). The first dose of convalescent plasma (convalescent plasma 1) had an immunoglobulin G (IgG) spike titer of 2,560, RBD titer of 3,840, and VN titer of 60. The second dose of convalescent plasma (convalescent plasma 2) had an IgG spike titer of 5,120, RBD titer of 5,120, and VN titer of 160 (Figure 2B; Figure S1A; Table 1). Prior to the first dose of plasma given on day 71, detectable spike and RBD IgG antibody titers were very low in serum collected from the individual, with IgG titers between 1:10 and 1:40 on days 49 and 71 pre transfusion; no VN titers were detected in these samples. Immediately after the first transfusion on day 71, the spike and RBD IgG antibody titers rose to 1:320 and then decreased to 1:80 and 1:160, respectively, on day 77. No VN titers were detected on days 71 and 77 (Figure 2C; Figure S1B; Table 1). Immediately after the second transfusion on day 82, the spike and RBD IgG titers increased to 1:320 and 1:640, respectively, and remained elevated by day 105 (Figure 2C; Figure S1B). Low neutralizing titers of 1:10 were observed on day 82 and 105 (Table 1).
Figure S1.
ELISA Titers against the SARS-CoV-2 RBD, Related to Figure 2
(A) IgG titers against SARS-CoV-2 receptor binding domain (RBD) were determined in ELISA on convalescent plasma used for transfusion. The light gray bar is the IgG titer of the fist donor (convalescent plasma 1) and the dark gray is the second donor (convalescent plasma 2). (B) IgG titers against SARS-CoV-2 (RBD) were determined in ELISA on patient serum collected on several time points, including immediately before and after transfusion with convalescent plasma at day 71 (light gray) and day 82 (dark gray). Each serum/plasma sample was tested in duplicate.
Despite two transfusions of convalescent plasma, nasopharyngeal swabs on days 85 and 105 remained positive for gRNA and sgRNA, suggesting that the convalescent plasma therapy was not successful in rapidly clearing the infection from the upper respiratory tract in this individual. Although the presence of sgRNA at these time points suggests active viral replication, infectious SARS-CoV-2 could not be cultured after day 70.
Genetic Analysis of Patient Swab Samples Links Infection to the Primary Washington State Outbreak
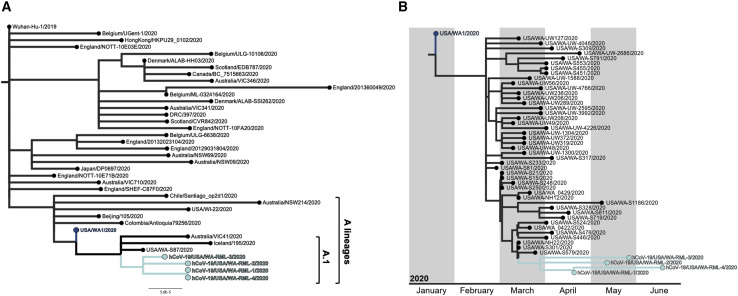
SARS-CoV-2 full genome sequences were obtained from nasopharyngeal swabs collected on days 49, 70, 85, and 105 (Table S4). Full genomes were obtained by sequencing using the ARTIC primer set (https://artic.network/) and assembling reads to MN985325.1 (USA/WA1/2020) as the reference genome (Harcourt et al., 2020). The SARS-CoV-2 lineage was determined using Pangolin software (https://pangolin.cog-uk.io/), which placed the individual’s viral genomes in lineage A.1, which consists of genomes originating from the primary outbreak in Washington state (Rambaut et al., 2020). A maximum-likelihood tree was generated using representative SARS-CoV-2 genomes from previously described lineages (Rambaut et al., 2020) obtained from the GISAID database (https://www.gisaid.org/; Shu and McCauley, 2017). The individual’s SARS-CoV-2 full-length genomes cluster together within lineage A.1 (Figure 4 A). This suggests that she was infected with a virus from the SARS-CoV-2 A.1 lineage, which circulated after the initial import from China, followed by exponential growth and local transmission in Washington state.
Figure 4.
Phylogenomic Analyses of Described SARS-CoV-2 Strains in a Persistently Infected Individual
(A) Full-genome SARS-CoV-2 sequences representing previously described lineages (Rambaut et al., 2020) were downloaded from GISAID (Shu and McCauley, 2017). Lineages were then assigned using Pangolin v.2.0.3 (https://pangolin.cog-uk.io/). Using a representative genome from the assigned lineages and the four SARS-CoV-2 sequences from the individual, a maximum-likelihood tree was inferred using PhyML v.3.3.20180621 (Guindon et al., 2010) implemented in Geneious Prime v.2020.1.2 (https://www.geneious.com/) with a general time-reversible model of nucleotide substitution and rooted at the Wuhan-Hu-1/2019 SARS-CoV-2 strain. Sequences from the A and A.1 lineages are labeled, and the individual’s SARS-CoV-2 sequences are shown in cyan. hCoV-19/USA/WA-RML-1, -2, -3, and -4 are the genome sequences derived from the individual from day 49, 70, 85, and 105 nasopharyngeal swabs, respectively.
(B) Full SARS-CoV-2 genomes were subsampled from Washington state, representing NextStrain clade 19B, including the four full-genome sequences recovered from the individual and the Wuhan-Hu-1/2019 sequence and aligned using MAFFT v.1.4 (Katoh and Standley, 2013; Katoh et al., 2002) implemented in Geneious Prime v.2020.1.2 (https://www.geneious.com/). A maximum-likelihood tree was then reconstructed with PhyML v.3.1 (Guindon et al., 2010), and a tree showing temporal divergence was inferred in TreeTime v.0.7.6 (Hadfield et al., 2018). The individual’s SARS-CoV-2 sequences are shown in cyan, and hCoV-19/USA/WA-RML-1, -2, -3, and -4 are the genome sequences derived from the individual from day 49, 70, 85, and 105 nasopharyngeal swabs, respectively.
See also Figure S2.
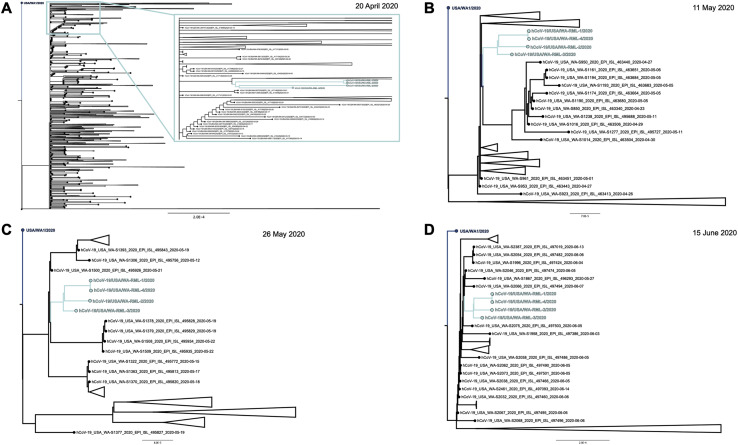
To visualize the temporal relationships of the patient isolates, 44 full SARS-CoV-2 genome sequences from Washington state belonging to NextStrain clade 19B (http://clades.nextstrain.org/) were subsampled from the GISAID database (https://www.gisaid.org/; Shu and McCauley, 2017) representing strains collected in Washington state from February to May 2020. A full genome alignment was performed with four of the full genome sequences recovered from the persistently infected individual, the USA/WA1/2020 genome sequence, and the Wuhan-Hu-1/2019 genome sequence with MAFFT v.1.4 (Katoh and Standley, 2013; Katoh et al., 2002) implemented in Geneious Prime v.2020.1.2 (https://www.geneious.com/). A maximum-likelihood tree was reconstructed with PhyML v.3.1 (Guindon et al., 2010), and a tree showing temporal divergence (Figure 4B) was inferred in TreeTime v.0.7.6 (Sagulenko et al., 2018; Hadfield et al., 2018) using the HKY85 model of nucleotide substitution and a fixed molecular clock at 8e−4 with a standard deviation of 4e−4, as implemented in the NextStrain pipeline (https://nextstrain.org/sars-cov-2/). Divergence dating estimates place the patient isolates sharing a most recent common ancestor between February 27 and March 31, 2020, within 90% of the marginal probability distribution. This is consistent with the timing of the individual’s first positive test on March 2, 2020. To further evaluate the relationship between the SARS-CoV-2 genomes recovered from the patient swabs and other SARS-CoV-2 genomes circulating in Washington state at the times of sampling (April 20, May 11, May 26, and June 15, 2020), Washington SARS-CoV-2 genomes were downloaded from the GISAID database (Shu and McCauley, 2017). The quality of the sequences was determined by Nextclade v.0.7.5 (https://nextstrain.org/ncov/global), and 1,789 sequences on April 20, 385 sequences between April 20 and May 11, 268 sequences between May 11 and May 26, and 709 sequences between May 26 and June 15 were kept for further phylogenetic analysis. Maximum likelihood trees using the curated sets of sequences, the four patient genomes, and the USA/WA1/2020 genome were inferred using ModelFinder (Kalyaanamoorthy et al., 2017) and ultrafast bootstrap (Hoang et al., 2018) implemented in IQ-TREE v.1.6.12 (Nguyen et al., 2015). The phylogenetic trees show that the patient genomes in this study cluster as a monophyletic clade consistent with infection in late February/early March, followed by virus persistence (Figure S2 ).
Figure S2.
Maximum-Likelihood Trees of the Individual with SARS-CoV-2 with Other SARS-CoV-2 Genomes Circulating in Washington State at the Times of Sampling (April 20, May 11, May 26, and June 15, 2020), Related to Figure 4 and Table 2
(A) Maximum likelihood tree using 1789 full genome SARS-CoV-2 sequences deposited to GISAID until 20 April 2020. Inset shows a close up of the monophyletic clade of the genomes directly obtained from the patient samples (cyan). (B) Maximum likelihood tree using 385 full genome SARS-CoV-2 sequences deposited to GISAID between 20 April and 11 May, 2020. The monophyletic clade of the genomes directly obtained from the patient samples is shown in cyan. (C) Maximum likelihood tree using 268 full genome SARS-CoV-2 sequences deposited to GISAID between 11 May and 26 May, 2020. The monophyletic clade of the genomes directly obtained from the patient samples is shown in cyan. (D) Maximum likelihood tree using 709 full genome SARS-CoV-2 sequences deposited to GISAID between 26 May and 15 June, 2020. The monophyletic clade of the genomes directly obtained from the patient samples is shown in cyan.
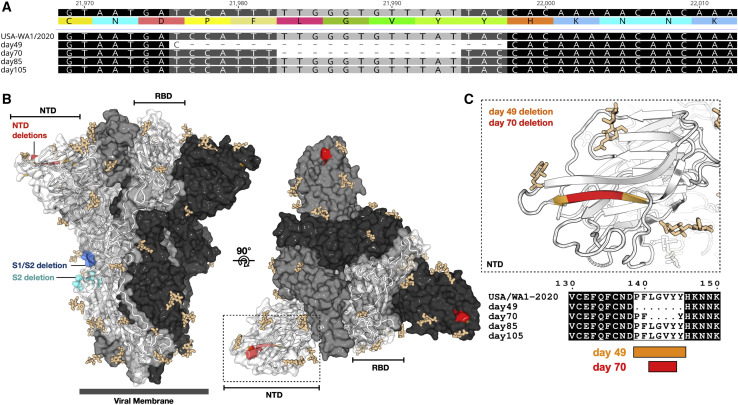
Next, full genome sequences from the two SARS-CoV-2 isolates were obtained (Table S4), and the consensus level variants in the sequences obtained from nasopharyngeal swabs and SARS-CoV-2 isolates cultured from those swabs were compared with the reference strain USA/WA1/2020 (MN985325.1) (Harcourt et al., 2020). Several single-nucleotide (nt) substitutions were observed within the ORF1ab, spike, M, and ORF8 coding sequence in the full-genome sequences obtained directly from the individual’s swabs and the SARS-CoV-2 isolates. In addition, a 3-nt deletion leading to loss of a methionine residue was observed in nsp1 in day 49 and day 70 samples (Table 2 ). Within the genomes of the two SARS-CoV-2 isolates, two in-frame deletions were observed in the spike glycoprotein coding region. A 21-nt in-frame deletion (residues 21,975–21,995) was found in the N-terminal domain (NTD) of S1, leading to a 7-amino-acid deletion (amino acids [aa] 139–145) in the spike glycoprotein of the SARS-CoV-2 day 49 isolate. A smaller, 12-nt deletion (residues 21,982–21,993) was detected in the day 70 isolate, leading to a 4-aa deletion (aa 141–144) in the NTD, which falls within the 7-aa deletion found in the day 49 isolate (Figure 5 A). These observed deletions in the spike glycoprotein map to a region in the NTD that is partially solvent exposed and forms a β strand in a compact conformation of the spike (Wrobel et al., 2020; Figures 5B and 5C). This region is unmodelled in other structures representing additional conformational states of the spike and, thus, is likely flexible (Wrapp et al., 2020; Walls et al., 2020). It is possible that the apparent plasticity in this region of the molecule may contribute to the structural permissibility of the identified deletions. The position of these deletions is distinct from those observed in other SARS-CoV-2 isolates, which locate to the S1/S2 and S2′ cleavage sites (Andrés et al., 2020; Lau et al., 2020; Liu et al., 2020d).
Table 2.
Consensus Sequence Variants in Clinical Samples from the Individual and SARS-CoV-2 Isolates Compared with Reference USA/WA1/2020 (MN985325.1)
| Position | Gene | Nucleotide Change | Protein Change | Day 49 Individual | Day 49 Isolate | Day 70 Individual | Day 70 Isolate | Day 85 Individual | Day 105 Individual |
|---|---|---|---|---|---|---|---|---|---|
| 518–520 | orf1ab | 3-bp deletion | M → del | 22%a | 100% | 100% | 100% | – | – |
| 2,113 | orf1ab | C → T | none | – | – | 100% | 100% | – | – |
| 4,084 | orf1ab | C → T | none | 87.5% | 100% | – | – | – | 97% |
| 17,747 | orf1ab | C → T | P → L | 100% | 100% | 100% | 100% | 100% | 100% |
| 17,858 | orf1ab | A → G | Y → C | 100% | 100% | 100% | 100% | 100% | 100% |
| 19,420 | orf1ab | T → C | S → P | 72% | 98% | – | – | – | 92% |
| 21,975–21,995 | spike | 21-bp deletion | DPFLGVYY → D | 1%a | 100% | – | – | – | – |
| 21,982–21,993 | spike | 12-bp deletion | FLGVY → F | – | – | 100% | 100% | – | – |
| 23,010 | spike | T → C | V → A | 100% | 100% | 100% | 100% | 100% | 99% |
| 23,616 | spike | G → A | R → Q | – | – | – | 95% | – | – |
| 23,617 | spike | T → A | – | – | – | 95% | – | – | |
| 26,526 | M | G → T | A → S | – | – | 16%a | 100% | – | – |
| 27,899 | orf8 | A → T | K → N | – | – | 100% | 100% | – | – |
| 29,308 | N | T → A | N → K | – | – | – | – | 56% | – |
| 29,854 | – | C → T | – | – | – | – | 100% | – | – |
Minor variants present in less than 50% of the reads were not included in the consensus, but these minor variants were included in the table to demonstrate their presence in clinical samples as well as the isolate.
Figure 5.
Deletions in the NTD of S1 of the Spike Protein
(A) Nucleotide and amino acid sequence alignment of the region of the spike gene of the four sequences from the individual and the reference USA/WA1/2020 genome sequence containing the deletions observed in the day 49 and day 70 samples. Alignment was generated with MAFFT v.1.4 (Katoh and Standley, 2013; Katoh et al., 2002) implemented in Geneious Prime 2020.1.2 (https://www.geneious.com).
(B) Amino acid residues removed by the day 49 (orange) and day 70 (red) spike deletions are highlighted on a SARS-CoV-2 spike trimer (PDB: 6zge; Wrobel et al., 2020). Each protomer of the trimer is shown in surface representation, colored in shades of gray. A single protomer is annotated, and its secondary structure is shown in cartoon representation. Glycans are shown as beige sticks. Previously reported spike deletions observed at the S1/S2 and S2′ cleavage sites (Andrés et al., 2020; Lau et al., 2020; Liu et al., 2020d) are colored blue and cyan, respectively.
(C) Close-up view of the indicated region of (B) (dotted box) with the protein surface removed for clarity and accompanying amino acid sequence alignment, generated using Multalin (Corpet, 1988) and plotted with ESPript (Robert and Gouet, 2014).
Comparison of the full genome sequences obtained directly from the individual’s samples with the genome data obtained from the two SARS-CoV-2 isolates showed that the 21-nt deletion was present in a minority of sequencing reads (1%) in the genome obtained from the individual’s sample from day 49 (Table 2) and was selected for upon passage in cell culture. The 12-nt deletion on day 70 was present in 100% of the reads in the clinical sample and tissue culture isolate. Notably, neither spike deletion was detected in the genome sequences from the day 85 and day 105 swabs (Table 2). It is possible that other minor variants exist at low levels that were undetected by the depth of sequencing coverage or were not reflected in the sampling at that time point. The variation observed between the different full-length genomes obtained at various time points during the course of infection points to a quasispecies complex with continuous turnover of dominant viral species.
Growth Kinetics of SARS-CoV-2 Patient Isolates
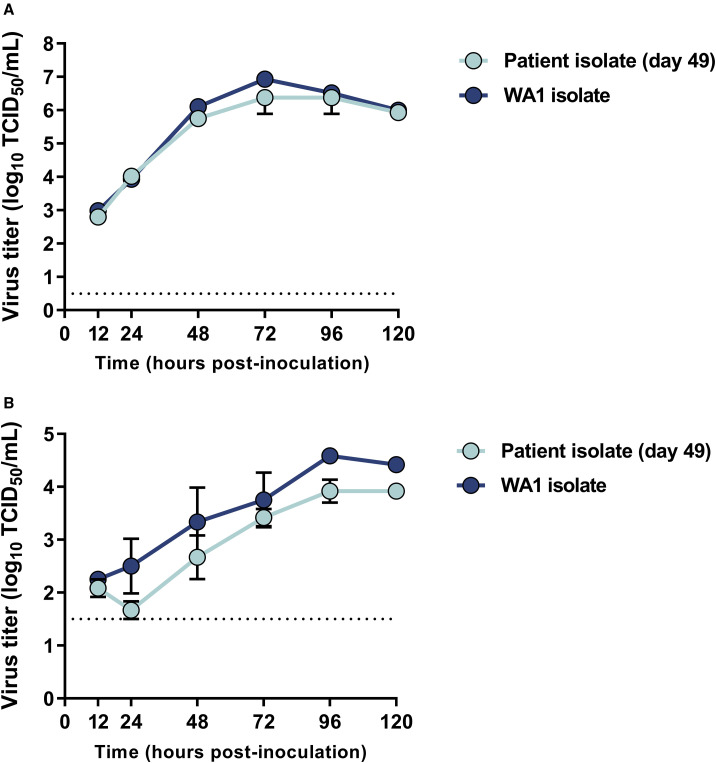
The replication kinetics of the day 49 isolate SARS-CoV-2 were compared with those of the reference strain USA/WA1/2020 in Vero E6 cells. Despite the observed mutations in the day 49 isolate, no difference in replication kinetics were observed between the day 49 isolate and the reference strain (Figure 6 A). To determine growth kinetics in a more functionally relevant cell type, growth curves were also performed on primary human alveolar epithelial tissues (EpiAlveolar; MatTek, Ashland, MA, USA). No significant differences were observed between the individual’s isolate and the reference strain in these cells (Figure 6B).
Figure 6.
Growth Kinetics of the Day 49 Isolate from the Individual in Vero E6 Cells and Primary Human Alveolar Epithelial Tissues
(A) Vero E6 cells were inoculated with the day 49 patient isolate and the reference USA/WA1/2020 strain at a MOI of 0.01 in triplicate.
(B) Primary 3D human alveolar epithelial tissues grown in 3D Transwell culture were inoculated with the same isolates at a MOI of 0.1. Supernatant was harvested at designated time points for assessment of viable virus using endpoint titration.
Data shown are the mean and the standard error of the mean for three independent replicates. Statistical analysis using a 2-way ANOVA in GraphPad Prism shows no significant difference between the isolates at any of the time points.
Discussion
In this report, we describe long-term SARS-CoV-2 shedding in an immunocompromised individual with CLL and acquired hypogammaglobulinemia out to 105 days after the initial positive test. Although the exact time point when the individual acquired SARS-CoV-2 is unknown, it is likely that the exposure occurred in the long-term care facility where she resided between February 19–25, 2020, shortly before a large COVID-19 outbreak was identified in that facility on February 28, 2020. The individual remained asymptomatic throughout the course of infection despite isolation of infectious SARS-CoV-2 49 and 70 days after the initial diagnosis, much longer than shedding of infectious virus up to day 20, as reported previously (van Kampen et al., 2020). The information available to date about SARS-CoV-2 infection in immunocompromised individuals, including those with cancers such as CLL, is limited and mostly focuses on disease severity and outcome (He et al., 2020a; Paneesha et al., 2020; Baumann et al., 2020; Fürstenau et al., 2020; Jin et al., 2020; Soresina et al., 2020; Zhu et al., 2020; Fill et al., 2020). Although it is difficult to extrapolate from a single individual, our data suggest that long-term shedding of infectious virus may be a concern in certain immunocompromised people. Given that immunocompromised individuals could have prolonged shedding and may not have typical symptoms of COVID-19, symptom-based strategies for testing and discontinuing transmission-based precautions, as recommended by the Centers for Disease Control and Prevention (CDC) (CDC, 2020b), may fail to detect whether certain individuals are shedding infectious virus.
The individual eventually cleared the SARS-CoV-2 infection from the upper respiratory tract after developing low neutralizing antibody titers. How the virus was cleared and the effect of convalescent plasma on clearance of the virus is unknown. The initial administration of convalescent plasma was followed by a decreased viral load in nasal swabs, but viral loads subsequently increased, despite administration of a second dose of convalescent plasma comprising higher antibody titers. Therapeutic administration of convalescent plasma is focused on treatment of severe or life-threatening COVID-19. Several clinical trials are investigating the efficacy of convalescent plasma, but currently the effect of convalescent plasma therapy on COVID-19 outcome remains equivocal (Mira et al., 2020; Salazar et al., 2020). The limited effect of convalescent plasma treatment on clearance of SARS-CoV-2 could be due to the fact that intravenously (i.v.) administered antibodies do not distribute well to the nasal epithelium (Ikegami et al., 2020) compared with the lower respiratory tract (Mira et al., 2020).
Throughout the course of infection, there was marked within-host genomic evolution of SARS-CoV-2. Deep sequencing revealed a continuously changing virus population structure with turnover in the relative frequency of the observed genotypes over the course of infection. With SARS-CoV-2, there is generally relatively limited within-host variation reported, and over the course of infection, the major SARS-CoV-2 population remains identical (Jary et al., 2020; Shen et al., 2020; Capobianchi et al., 2020). Potential factors contributing to the observed within-host evolution is prolonged infection and the compromised immune status of the host, possibly resulting in a different set of selective pressures compared with an immune-competent host. These differential selective pressures may have allowed a larger genetic diversity with continuous turnover of dominant viral species throughout the course of infection. Although some sequence variants remain consistent throughout the duration of infection, we also observed variants unique to individual time points, such as the spike deletions observed on day 49 and day 70. Previously reported spike deletions, distinct from those reported here, were observed at relatively low frequency in clinical samples but were enriched upon virus isolation (Andrés et al., 2020; Liu et al., 2020d). Similar to these reports, the spike deletion in the isolate on day 49 was observed as a minor variant in the individual’s sample but was also selected for during passage upon virus isolation.
In contrast to the previously reported deletions at the cleavage sites, both spike deletions observed on day 49 and 70 in the individual are located in the NTD of S1, a region distal from the receptor binding site. These deleted residues are not modeled in a number of spike structures (Wrapp et al., 2020; Walls et al., 2020), suggesting that this region is conformationally labile. Although the NTD has been identified as an antigenic target (Brouwer et al., 2020; Chi et al., 2020; Liu et al., 2020a), no clear difference in virus neutralization was observed between the two patient isolates and the prototype USA/WA1/2020 SARS-CoV-2 isolate.
Despite genetic changes in the SARS-CoV-2 isolated from the individual, the replication kinetics did not change significantly compared with the USA/WA1/2020 virus in Vero E6 cells and primary human alveolar epithelial tissues. This indicates that, most likely, the infectious virus shed by the individual would still be able to establish productive infection in contacts upon transmission, assuming that viral growth kinetics in vitro are a suitable surrogate for virus fitness in vivo. Moreover, despite prolonged replication exclusively in the upper respiratory tract, the virus was still able to replicate in epithelial cells derived from the lower respiratory tract, suggesting that it could still cause pneumonia.
Many current infection control guidelines assume that persistently PCR-positive individuals are shedding residual RNA and not infectious virus, with immunocompromised people thought to remain infectious for no longer than 20 days after symptom onset (CDC, 2020a). Here we show that certain individuals may shed infectious, replication-competent virus for much longer than previously recognized (van Kampen et al., 2020). Although infectious virus could be detected up to day 70, sgRNA, a molecular marker for active SARS-CoV-2 replication (Speranza et al., 2020), could be detected up until day 105. An immunocompromised state has been identified as a risk factor for development of severe disease and complications from COVID-19 (CDC, 2020b). A wide variety of conditions and treatments can alter the immune system and cause immunodeficiency, creating opportunities for prolonged viral replication and shedding of infectious SARS-CoV-2. Although this report focuses on long-term shedding of one immunocompromised individual, an estimated 3 million people in the United States have some form of immunocompromising condition, including individuals with HIV infection, solid organ transplant recipients, hematopoietic stem cell transplant recipients, and individuals receiving chemotherapy and corticosteroids (Kunisaki and Janoff, 2009). This transient or chronic immunocompromised population is at higher risk of respiratory disease complications with respiratory infections such as influenza A virus and SARS-CoV-2 (Kunisaki and Janoff, 2009). Prolonged shedding of pH1N1 shedding was observed in immunocompromised individuals with a variety of immunocompromising conditions during the previous pandemic in 2009, such as people with cancer on chemotherapy and solid organ transplant recipients (van der Vries et al., 2013). For the SARS-CoV-2 related Middle East respiratory syndrome CoV (MERS-CoV), prolonged shedding up to 38 days was observed in individuals with myelodysplastic syndrome, autologous peripheral blood stem cell transplantation for treatment of large B cell lymphoma, and an individual with peripheral T cell lymphoma (Kim et al., 2017). MERS-CoV shedding was higher and longer in experimentally infected non-human primates immunosuppressed with cyclophosphamide and dexamethasone, providing experimental support for the effect of immunosuppression on virus-host dynamics observed here (Prescott et al., 2018).
Limitations of Study
A limitation of the present study is that it comprises only a single case, making it difficult to draw general conclusions regarding use of convalescent plasma for clearance of the virus, potential alternative mechanisms involved in virus clearance, and the frequency of persistent SARS-CoV-2 infection and shedding in individuals with other immunocompromising conditions. Identification of additional cases of persistent infection and long-term shedding of infectious virus are needed so the infection dynamics can be studied in more detail in this diverse population. Understanding the mechanism of virus persistence and eventual clearance will be essential for providing appropriate treatment and preventing transmission of SARS-CoV-2 because persistent infection and prolonged shedding of infectious SARS-CoV-2 might occur more frequently. Because immunocompromised individuals are often cohorted in hospital settings, a more nuanced approach to testing these individuals is warranted, and the presence of persistently positive people by performing SARS-CoV-2 gRNA and sgRNA analyses on clinical samples should be investigated.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Human IgG Fc fragment Secondary Antibody (HRP) | Novus biologicals | Cat# NBP1-73529; Lot 34900 |
| Bacterial and Virus Strains | ||
| hCoV-19/USA/WA1/2020 | CDC, Atlanta, USA (Harcourt et al., 2020) | GenBank: MN985325.1 |
| hCoV-19/USA/WA-RML-5/2020 (SARS-CoV-2 patient genome d49 isolate) | This paper | GenBank: MT982401 |
| hCoV-19/USA/WA-RML-6/2020 (SARS-CoV-2 patient genome d70 isolate) | This paper | GenBank: MT982404 |
| Biological Samples | ||
| Patient nasopharyngeal swabs | This paper | EvergreenHealth |
| Patient oropharyngeal swabs | This paper | EvergreenHealth |
| Patient serum and plasma | This paper | EvergreenHealth |
| Donor convalescent plasma | This paper | EvergreenHealth |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SARS-CoV-2 spike protein | This paper (Wrapp et al., 2020) | N/A |
| SARS-CoV-2 receptor binding domain protein | This paper (Amanat et al., 2020) | N/A |
| PEI transfection reagent | Polysciences | Cat# 23966-1 |
| Blocker Casein in PBS | ThermoFisher | Cat# 37528 |
| TMB 2-Component Microwell Peroxidase Substrate Kit | SeraCare | Cat# 5120-0047 |
| KPL TMB Stop Solution | SeraCare | Cat# 5150-0020 |
| Trizol Reagent | Invitrogen | Cat# 15596026 |
| Karnovsky’s EM fixative | Electron Microscopy Sciences | Cat#15720 |
| Sodium Cacodylate | Sigma | Cat#C4945-10G; CAS#6131-99-3 |
| Osmium Tetroxide | Electron Microscopy Sciences | Cat#19190; CAS#20816-12-0 |
| Potassium Ferrocyanide | Sigma | Cat#P-3289; CAS#14459-95-1 |
| Uranyl Acetate | Ted Pella | Cat#19481; CAS#6159-44-0 |
| Critical Commercial Assays | ||
| PureLink RNA Mini Kit | Invitrogen | Cat# 12183018A |
| SuperScript IV First-Strand Synthesis System | Invitrogen | Cat# 18091050 |
| Quantifast Probe RT-PCR Kit (for Rotorgene) | QIAGEN | Cat# 204556 |
| ddPCR Supermix for Probes (no dUTP) | Biorad | Cat# 1863024 |
| Q5 Hot Start High-Fidelity DNA Polymerase – 100U | New England Biolabs | Cat#M0493S |
| ARTIC nCoV-2019 V3 Panel, 500rxn | Integrated DNA Technologies | Cat#10006788 |
| TruSeq DNA PCR-Free LT Library Prep | Illumina | Cat#20015962 |
| TruSeq DNA Idx Kit Set A | Illumina | Cat#20015960 |
| MiSeq Reagent Nano Kit, v2 (500 cycles) (1M) | Illumina | Cat#MS-103-1003 |
| Deposited Data | ||
| Data used to generate figures | This paper | Mendeley Data at https://dx.doi.org/10.17632/3n377gv8kb. |
| hCoV-19/USA/WA-RML-1/2020 (SARS-CoV-2 patient genome d49 NP swab) | This paper | GenBank: MT982403 |
| hCoV-19/USA/WA-RML-2/2020 (SARS-CoV-2 patient genome d70 NP swab) | This paper | GenBank: MT982402 |
| hCoV-19/USA/WA-RML-3/2020 (SARS-CoV-2 patient genome d85 NP swab) | This paper | GenBank: MT982405 |
| hCoV-19/USA/WA-RML-4/2020 (SARS-CoV-2 patient genome d105 NP swab) | This paper | GenBank: MT982406 |
| hCoV-19/USA/WA-RML-5/2020 (SARS-CoV-2 patient genome d49 isolate) | This paper | GenBank: MT982401 |
| hCoV-19/USA/WA-RML-6/2020 (SARS-CoV-2 patient genome d70 isolate) | This paper | GenBank: MT982404 |
| Experimental Models: Cell Lines | ||
| Freestyle 293-F | ThermoFisher | Cat# R79007; RRID CVCL_D603 |
| VeroE6 | Ralph Baric | ATCC CRL-1586 |
| MatTek EpiAlveolar | MatTek Life Sciences (https://www.mattek.com/products/epialveolar/) | Cat# ALV-100-FT-PE12 |
| Oligonucleotides | ||
| Primer to E genomic (E_Sarbeco_F1) AACAGGTACGTTAATAGTTAATAGCGT | Corman et al., 2020; Integrated DNA Technologies | https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf |
| Primer to E subgenomic (sgLeadSARS2-F)CGATCTCTTGTAGATCTGTTCTC | Wölfel et al., 2020; Integrated DNA Technologies | N/A |
| Reverse primer to E (E_Sarbeco_R2)ATATTGCAGCAGTACGCACACA | Corman et al., 2020; Integrated DNA Technologies | https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf |
| Probe for E (E_Sarbeco_P1) FAM-ACACTAGCCATCCTTACTGCGCTTCG-ZEN-IBHQ | Corman et al., 2020; Integrated DNA Technologies | https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf |
| Recombinant DNA | ||
| pαH SARS-CoV-2 spike plasmid | Kizzemekia Corrbett and Barney GrahamVaccine Research Center, NIH, Bethesda, USA (Wrapp et al., 2020) | N/A |
| pCAGGS SARS-CoV-2 receptor binding domain plasmid | Florian KrammerIcahn School of Medicine at Mt. Sinai, New York, USA (Amanat et al., 2020) | N/A |
| Software and Algorithms | ||
| MAFFT align | (Katoh and Standley, 2013, Katoh et al., 2002) | Geneious Prime 2020.1.2 plugin |
| Multalin sequence alignment | (Corpet, 1988) | http://multalin.toulouse.inra.fr/multalin/ |
| ESPript 3.0 | (Robert and Gouet, 2014) | http://espript.ibcp.fr/ESPript/ESPript/ |
| Geneious Prime 2020.1.2 | Geneious | https://www.geneious.com |
| PhyML 3.320180621 | Guindon et al., 2010 | Geneious Prime 2020.1.2 plugin |
| FigTree v1.4.4 | http://tree.bio.ed.ac.uk/software/figtree/ | https://github.com/rambaut/figtree/ |
| Pangolin COVID-19 Lineage Assigner | Rambaut et al., 2020 | https://pangolin.cog-uk.io |
| Pymol Molecular Graphics System version 2.0.1 | Schrödinger | https://www.schrodinger.com/pymol |
| Prism 8.2.0 | GraphPad | https://www.graphpad.com:443/ |
| NextClade v0.7.5 | https://github.com/nextstrain/nextclade | https://nextstrain.org/ncov/global?c=region |
| ModelFinder | Kalyaanamoorthy et al., 2017 | http://www.iqtree.org/ |
| Ultrafast bootstrap | Hoang et al., 2018 | http://www.iqtree.org/ |
| IQ-TREE v1.6.12 | Nguyen et al., 2015 | http://www.iqtree.org/ |
| TreeTime v.0.7.6 | Sagulenko et al., 2018 | https://github.com/neherlab/treetime |
| BCFtools v1.10.2 | Li et al., 2009 | https://www.htslib.org |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| AdapterRemoval v2.2.2 | Schubert et al., 2016 | https://github.com/MikkelSchubert/adapterremoval |
| Picard 2.18.7 | Broad Institute, 2018 | https://broadinstitute.github.io/picard/ |
| GATK 4 v 4.1.2.0 | McKenna et al., 2010 | https://github.com/broadinstitute/gatk/releases |
| Other | ||
| Ni Sepharose 6 Fast Flow | GE Lifesciences | Cat# 17531802 |
| NiNTA Agarose | QIAGEN | Cat# 30230 |
| Phasemaker Tubes | Invitrogen | Cat# A33248 |
| Thermanox coverslips | Ted Pella | Cat#26028 |
| Silicon Chips | Ted Pella | Cat#16007 |
| Aluminum specimen mounts | Ted Pella | Cat#16111 |
| Double-sided carbon tape | Ted Pella | Cat#16084-1 |
| Spurr’s resin | Ted Pella | Cat#18300-4221 |
| Iridium target | Electron Microscopy Sciences | Cat#3431 |
| Bal-Tec Drier | Balzers, Liechtenstein | Cat#CPD 030 |
| Quorum sputter coater | Electron Microscopy Sciences, Hatfield, PA | Cat#EMS300T D |
| Hitachi field emission scanning electron microscope | Hitachi, Tokyo, Japan | Model#SU-8000 |
| Leica UC7 ultramicrotome | Leica Microsystems | N/A |
| FEI BT Tecnai transmission electron microscope | Thermofisher/FEI | N/A |
| Gatan Rio camera | Gatan | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vincent Munster (Vincent.munster@nih.gov).
Materials Availability
This study did not generate new reagents.
Data Availability
The data and the Supplementary Tables from this study have been deposited to Mendeley Data at https://dx.doi.org/10.17632/3n377gv8kb.
Genome sequences have been deposited to GenBank: MT982403, MT982402, MT982405, MT982406, MT982401 and MT982404.
Experimental Model and Subject Details
Human Patient
The patient described in this case study is a 71 year old female with a 10 year history of chronic lymphocytic leukemia (CLL), acquired hypogammaglobulinemia, anemia, and chronic leukocytosis. The patient tested positive for SARS-CoV-2 on March 2, 2020, and remained positive through June 15, 2020. During the course of the study, the patient was transfused with intravenous immunoglobulin (IVIG, 25 g) on April 6 and May 6, 2020, and convalescent plasma against SARS-CoV-2 on May 12 and May 23, 2020. After the initial SARS-CoV-2 diagnosis, the patient was kept in isolation in an isolation ward in a single room with negative airflow. Anonymized plasma, serum and swabs from a patient at EvergreenHealth, Kirkland, Washington were obtained under an NIH Institutional Review Board exemption. Verbal and signed consent were obtained from the patient to allow analyses of the samples.
Cells
Vero E6 is a female African green monkey kidney epithelial cell line. Vero E6 cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum, 1 mM L-glutamine, 50 U/mL penicillin and 50 μg/mL streptomycin. Vero E6 cells were provided by Dr. Ralph Baric. Cells were authenticated by cytochrome B sequencing. Mycoplasma testing was performed monthly, and no mycoplasma was detected.
FreeStyle 293-F (RRID: CVCL_D603) is a female human embryonic cell line adapted for growth in suspension culture. FreeStyle 293-F cells were grown in Freestyle 293 Expression Medium (GIBCO) at 37°C and 8% CO2, shaking at 130 rpm. Cells were not authenticated in house. Mycoplasma testing was performed monthly, and no mycoplasma was detected.
MatTek EpiAlveolar is a 3D co-culture model of the air-blood barrier produced from primary human alveolar epithelial cells, pulmonary endothelial cells and fibroblasts, and maintained according to manufactures instructions (https://www.mattek.com/products/epialveolar/). Cells were not authenticated in house. Mycoplasma testing was performed monthly, and no mycoplasma was detected.
SARS-CoV-2 Virus
SARS-CoV-2 strain nCoV-WA1-2020 (MN985325.1) (Harcourt et al., 2020) was provided by CDC, Atlanta, USA. SARS-CoV-2 isolates were propagated on Vero E6 cells grown in DMEM supplemented with 2% fetal bovine serum (GIBCO), 1 mM L-glutamine (GIBCO), 50 U/mL penicillin and 50 μg/mL streptomycin (GIBCO) (virus isolation medium), at 37°C and 5% CO2.
Infectious titer of SARS-CoV-2 virus stocks was determined by end-point titration and is reported as log10 50% tissue culture infective dose (TCID50/mL). 1.5 × 104 Vero E6 cells were seeded into each well in 96-well plates in DMEM supplemented with 10% fetal bovine serum, 1 mM L-glutamine, 50 U/mL penicillin and 50 μg/mL streptomycin and incubated overnight at 37°C and 5% CO2. The following morning, when the cells were at approximately 90% confluency, the wells were inoculated with ten-fold serial dilutions of virus stock diluted in virus isolation medium (100 uL per well, with 10 replicate wells for each dilution). The plates were incubated at 37°C and 5% CO2, and the cytopathic effect (CPE) was assessed for each well after 5 days. Wells that demonstrated CPE were counted, and the titer was determined by the method of Spearman and Kärber using 10 replicates as follows:
where X is log10 of the lowest dilution with all wells positive for CPE, d is log10 of the dilution factor (10 in these titrations), and S is the sum of the fraction of wells positive for CPE at all tested dilutions.
Method Details
Clinical Sample RNA Extraction and qRT-PCR
Clinical samples were deidentified as part of their analyses. Nasopharyngeal and oropharyngeal swabs were shipped on wet ice in viral transport medium (VTM) to Rocky Mountain Laboratories (NIH). RNA was extracted using Trizol (Invitrogen), Phasemaker tubes (Invitrogen) and the PureLink RNA Mini Kit (Invitrogen) according to manufacturer’s instructions and eluted in 100 μL RNase-free H2O. First strand cDNA synthesis was performed with the SuperScript IV First Strand Synthesis System (Invitrogen), using 11 μL input RNA and random hexamers. qRT-PCR was performed using 5 μL of cDNA using the QuantiFast Probe kit (QIAGEN) using E gRNA (Corman et al., 2020) and sgRNA specific assays (Wölfel et al., 2020). To quantify viral load within the patient samples, 5 μL of cDNA was analyzed using droplet digital PCR (Biorad) using the same E gRNA and sgRNA assays. The SARS-CoV-2 testing through EvergreenHealth were performed by University of Washington, LabCrop, Cepheid, and GenMark. Kashi clinical laboratories and Magnolia diagnostics performed the negative tests taken at the care facilities.
Virus Isolation
Virus isolation of the clinical specimen was performed on Vero E6 cells in 96 well plates. In brief, media was removed from wells and replaced with 100 μL of undiluted swab sample, or swab sample diluted 1:10 in DMEM supplemented with 2% fetal bovine serum (GIBCO), 1 mM L-glutamine (GIBCO), 50 U/mL penicillin and 50 μg/mL streptomycin (GIBCO) (virus isolation medium). Diluted and undiluted samples were inoculated onto 7 wells. Spin inoculation was performed at 1000 x g for 1 hour at 35°C. Inoculum was removed and wells were washed twice with and replaced with 100 μL of virus isolation medium and incubated at 37°C and 5% CO2. After 5 days, replicate wells were pooled, diluted 10x in virus isolation medium, and used to inoculate T25 flasks of Vero E6 cells in virus isolation medium and incubated at 37°C and 5% CO2. Flasks were observed for cytopathic effect. RNA was extracted, as described above, for confirmation of SARS-CoV-2 by qRT-PCR and next generation sequencing.
Growth kinetics of SARS-COV-2 isolates
Vero E6 cells were seeded in 6 well plates at a density of 4 × 105 cells/well in DMEM supplemented with 2% fetal bovine serum (GIBCO), 1 mM L-glutamine (GIBCO), 50 U/mL penicillin and 50 μg/mL streptomycin (GIBCO) (virus isolation medium) and incubated overnight at 37°C and 5% CO2. The following day, the media was removed from the wells and replaced with 1 mL of virus isolation medium containing virus at a MOI of 0.01. The patient day49 isolate and the USA/WA1/2020 strain were tested in triplicate, with mock control wells in triplicate. After a 1-hour incubation at 37°C and 5% CO2, the inoculum was removed, and wells were washed 3x with PBS and replaced with a fresh 2 mL of virus isolation medium. Supernatant samples were taken at 0, 12, 24, 48, 72, 96, and 120 hours post inoculation. Titer of infectious virus from supernatant was determined by endpoint titration in Vero E6 cells, as described above, but using 4 replicates per sample to determine the TCID50/mL using the Spearman-Karber method. The EpiAlveolar cell growth kinetic experiment was set up similar to the Vero E6 cells but with the following differences. Cells were provided by MatTek with 2.5 × 105 cells/transwell insert. Cells were infected by adding 75 μL of ALI medium containing virus at an MOI of 0.01 to the apical side of the transwell insert. After the above outlined incubation, the inoculum was removed, wells were washed 1x with PBS and replaced with 75 μL of ALI medium upon the apical surface. During sampling of the EpiAlveolar cells, 500 μL of DMEM medium was added to the apical side, gently pipetted to mix, removed, and 75 μL of fresh ALI medium replaced on the apical surface.
Expression and Purification of SARS-CoV-2 Spike and Receptor Binding Domain
Expression plasmids encoding the codon optimized SARS-CoV-2 full length spike and receptor binding domain (RBD) were kindly provided Kizzmekia Corbett and Barney Graham (Vaccine Research Center, Bethesda, USA) and Florian Krammer (Icahn School of Medicine at Mt. Sinai, New York, USA), respectively (Wrapp et al., 2020; Amanat et al., 2020). Both plasmids were expressed in Freestyle 293-F cells (Thermofisher), maintained in Freestyle 293 Expression Medium (GIBCO/ThermoFisher) at 37°C and 8% CO2 in a humidified incubator shaking at 130 rpm. Cultures totaling 500 mL were transfected with PEI at a density of one million cells per mL. Supernatant was harvested 7 days post transfection, clarified by centrifugation and sterile filtered through a 0.22 μM membrane. The protein was purified using Ni-NTA immobilized metal-affinity chromatography (IMAC) using Ni Sepharose 6 Fast Flow Resin (GE Lifesciences) or NiNTA Agarose (QIAGEN) and gravity flow. After elution the protein was buffer exchanged into 10 mM Tris pH8, 150 mM NaCl buffer before further use or frozen at −80°C for storage.
Enzyme-Linked Immunosorbent Assay (ELISA)
Purified SARS-CoV-2 full length spike or RBD protein was diluted to 1 μg/mL in PBS. Maxisorp plates (Nunc) were coated with 100 μL per well (100 ng protein per well) and incubated overnight at 4°C. Plates were washed 3x with PBST (0.1% Tween) and blocked with 100 μL casein in PBS blocking buffer (ThermoFisher) for 1 hour at room temp. Plates were again washed 3x with PBST (0.1% Tween), and 100 μL of serum samples, serially diluted 2 fold in casein in PBS blocking buffer, in duplicate, was added to the wells and incubated at room temperature for 1 hour. Plates were washed 4x with PBST (0.1% Tween), and 100 μL secondary antibody, rabbit anti-human IgG Fc HRP (Novus Biologicals, NBP1-73529) diluted 1:4000 in casein in PBS blocking buffer, was added to the wells and incubated for 1 hour at room temperature. The wells were washed 5x with PBST (0.1% Tween) and developed with the KPL TMP 2-component peroxidase substrate kit (Seracare, 5120-0047). The reaction was stopped with KPL stop solution (Seracare, 5150-0020) and read at 450 nm. The threshold for positivity was calculated as the average plus 3 times the standard deviation of negative control sera. Reported titers are the reciprocal value of the highest dilution at which signal was observed above the calculated threshold.
Virus Neutralization assay
Serum and plasma samples were heat inactivated at 56°C for 30 minutes. Two-fold serial dilutions were prepared in DMEM supplemented with 2% FBS, with each sample diluted in duplicate in 96 well plate format. 100 TCID50 of SARS-CoV-2, in virus isolation medium, was then added to each well. The virus-serum/plasma mixture was incubated at 37°C for 1 hour to allow for neutralization, then 100 uL per well was added to Vero E6 cells in 96 well plates and incubated at 37°C and 5% CO2. After 5 days, wells were observed for cytopathic effect. The virus neutralization titer is displayed as the reciprocal value of the highest dilution of serum/plasma that still inhibited virus replication at which no cytopathic effect was observed.
Next generation sequencing of patient clinical samples and isolates
Clinical Samples - Viral RNA was extracted from patient nasopharyngeal swabs using Trizol (Invitrogen) for use with the ARTIC nCoV-2019 sequencing protocol V.1 (Protocols.io; https://www.protocols.io/view/ncov-2019-sequencing-protocol-bbmuik6w). 30-35 PCR cycles were used to generate tiled-PCR amplicons. Primer pools consisted of the ARTIC nCoV-2019 v3 Panel (Integrated DNA Technologies, Belgium) and were diluted and used in PCR reactions following the instructions. Products from Pool 1 and Pool 2 were combined, AmPure XP cleaned, and quantitated as per the instructions – through step 16.18. Following assessment on a BioAnalyzer DNA Chip (Agilent Technologies, Santa Clara, CA), a volume consisting of 500 ng of product was taken directly into TruSeq DNA PCR-Free Library Preparation Guide, Revision D. (Illumina, San Diego, CA) beginning with the Repair Ends step (q.s. to 50 μL with RSB) and subsequent cleanup consisted of a single 1:1 AmPure XP/reaction ratio. All downstream steps followed the manufacturer’s instructions. Final libraries were visualized on a BioAnalyzer HS chip (Agilent Technologies, Santa Clara, CA) and quantified using KAPA Library Quant Kit (Illumina) Universal qPCR Mix (Kapa Biosystems, Wilmington, MA) on a CFX96 Real-Time System (BioRad, Hercules, CA).
Isolates - Viral RNA was extracted from clarified cell culture supernatant using Trizol (Invitrogen). Extracted RNA was depleted of rRNA using Ribo-Zero Gold H/M/R (Illumina, San Diego, CA) based on manufacturer’s protocols. After Ampure RNAClean XP (Beckman Coulter, Brea, CA) purification, the enriched RNA was eluted in 6 μL of water and assessed on a BioAnalyzer RNA Pico Chip (Agilent Technologies, Santa Clara, CA). Following the Truseq Stranded mRNA Library Preparation Guide, Revision E., (Illumina, San Diego, CA), the remaining RNA was added to Elute-Frag-Prime Buffer and continued through second-strand cDNA synthesis. The resulting double-stranded cDNAs were treated with a combined mixture of RiboShredder RNase Blend (Lucigen, Middleton, WI) and high concentration DNase-free RNase (Roche Diagnostics, Indianapolis, IN). After AMpure XP purification (Beckman Coulter, Brea, CA), samples were analyzed on a RNA Pico chip to confirm no remaining RNA. Library preparation continued with adenylation of ends following manufacturer’s recommendations. All downstream steps followed the manufacturer’s instructions. Final libraries were visualized on a BioAnalyzer DNA1000 chip (Agilent Technologies, Santa Clara, CA) and quantified using KAPA Library Quant Kit (Illumina) Universal qPCR Mix (Kapa Biosystems, Wilmington, MA) on a CFX96 Real-Time System (BioRad, Hercules, CA).
Sequencing and bioinformatics
Libraries were diluted to 2 nM stock, pooled together as needed in equimolar concentrations and sequenced on the MiSeq (Illumina, Inc, San Diego, CA) using on-board cluster generation and 2 × 150 paired-end sequencing. Raw image files were converted to fastq files using bcl2fastq (v2.20.0.422, Illumina, Inc. San Diego, CA) and trimmed of adaptor sequences using cutadapt version 1.12 (Martin, 2011). Adaptor-trimmed reads were trimmed and filtered to remove low quality sequence using fastq_quality_trimmer and fastq_quality_filter tools from the FASTX Toolkit, v 0.0.14 (Gordon, 2018). Singletons were removed and quality filtered reads were coordinate-order sorted using a custom perl script.
Reads were filtered for repeat sequence, rRNA, and PhiX contaminants and then mapped to the SARS-CoV-2 isolate 2019-nCoV/USA_WA1 (MN985325.1) reference genome using bowtie2 with –no-mixed –no-unal -X 1500 options (Langmead and Salzberg, 2012). Aligned SAM files were converted to BAM format, then sorted and indexed using SAMtools (Li et al., 2009). Duplicate reads were removed from the mapped reads using picard’s MarkDuplicates tool (Broad Institute, 2018)
To process the ARTIC data a custom pipeline was developed. Fastq read pairs were first compared to a database of ARTIC primer pairs to identify read pairs that had correct, matching primers on each end. Once identified, the ARTIC primer sequence was trimmed off. Read pairs that did not have the correct ARTIC primer pairs were discarded. Remaining read pairs were collapsed into one sequence using AdapterRemoval (Schubert et al., 2016), requiring a minimum 25 base overlap and 300 base minimum length, generating ARTIC amplicon sequences. Identical amplicon sequences were removed and the unique amplicon sequences were then mapped to the SARS-CoV-2 genome (MN985325.1) using Bowtie2 (Langmead and Salzberg, 2012). Aligned SAM files were converted to BAM format, then sorted and indexed using SAMtools (Li et al., 2009).
Variant calling was performed using Genome Analysis Toolkit (GATK, version 4.1.2) HaplotypeCaller with ploidy set to 2 (McKenna et al., 2010). Single nucleotide polymorphic variants were filtered for QUAL > 200 and quality by depth (QD) > 20 and indels were filtered for QUAL > 500 and QD > 20 using the filter tool in bcftools, v1.9 (Li et al., 2009). The accuracy of the filtered variant calls was manually inspected in Broad’s Integrative Genomics Viewer (IGV) (Robinson et al., 2017). Consensus sequences were generated using bcftools consensus (Li et al., 2009) and subsequently aligned using MAFFT (Katoh and Standley, 2013; Katoh et al., 2002) with 2,434 GISAID Washington SARS2 reference sequences in addition to the 2019-nCoV/USA_WA1 genome used for mapping.
Phylogenomic Analysis
Available SARS-CoV-2 full genome sequences were downloaded from the GISAID database (https://gisaid.org/; Shu and McCauley, 2017). The sequences were then assigned to previously described lineages (Rambaut et al., 2020) using Pangolin v2.0.3 (https://pangolin.cog-uk.io/), and aligned using MAFFT v. 1.4 (Katoh and Standley, 2013; Katoh et al., 2002). A maximum likelihood tree with the patient SARS-CoV-2 genomes, the Wuhan-Hu-1/2019 genome sequence, the USA/WA-1/2020 genome, and a representative genome from the assigned lineages was inferred using PhyML v.3.3.20180621 (Guindon et al., 2010) implemented in Geneious Prime v.2020.1.2 (https://www.geneious.com/) with a general time reversible model of nucleotide substitution and rooted at the Wuhan-Hu-1/2019 SARS-CoV-2 strain. The final figure was made using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). For the time tree, full SARS-CoV-2 genomes were subsampled from Washington state representing NextStrain clade 19B, including the four patient genomes sequences and the Wuhan-Hu-1/2019 genome sequence. The sequences were aligned using MAFFT v. 1.4 (Katoh and Standley, 2013; Katoh et al., 2002) implemented in Geneious Prime v. 2020.1.2 (https://www.geneious.com/), a maximum likelihood tree reconstructed with PhyML v.3.1 (Guindon et al., 2010), and the time tree showing temporal divergence inferred in TreeTime v.0.7.6 (Hadfield et al., 2018) using the HKY85 model of nucleotide substitution and a fixed molecular clock at 8e-4 with a standard deviation of 4e-4 as implemented in the NextStrain pipeline (https://nextstrain.org/sars-cov-2/).
To evaluate the relationship between the SARS-CoV-2 genomes recovered from the patient swabs with other SARS-CoV-2 genomes from Washington state, genomes at the times of sampling (April 20, May 11, May 26, and June 15, 2020) from Washington state were downloaded from the GISAID database (https://gisaid.org/; Shu and McCauley, 2017). The sequences were aligned by MAFFT (Katoh and Standley, 2013; Katoh et al., 2002). The sequences were analyzed by the Nextclade server v0.7.5 (https://clades.nextstrain.org/) for quality and sequences that were not of sufficient quality were discarded. 1,789 sequences at April 20, 385 sequences between April 20 and May 11, 268 sequences between May 11 and May 26, and 709 sequences between May 26 and June 15 were kept for further phylogenetic analysis. Maximum likelihood trees using the curated sets of genomes, the four patient genomes, and the USA/WA1/2020 genome, were inferred using ModelFinder (Kalyaanamoorthy et al., 2017) and ultrafast bootstrap (Hoang et al., 2018) implemented in IQ-TREE (Nguyen et al., 2015), and rooted at USA/WA1/2020. Final figures were made using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). A table of acknowledgments for the GISAID genome sequences used to within this work is available at Mendeley Data at https://dx.doi.org/10.17632/3n377gv8kb.
Electron Microscopy
Vero E6 cells cultured in DMEM supplemented with 10% fetal bovine serum, 1 mM L-glutamine, 50 U/mL penicillin and 50 μg/mL streptomycin were plated at 5 × 104 cells/well in 24 well plates containing Thermanox coverslips (Ted Pella, Redding, CA) for transmission electron microscopy or silicon chips (Ted Pella, Redding, CA) for scanning electron microscopy in the wells, and incubated overnight at 37°C and 5% CO2. The next day, media was carefully aspirated from the wells and replaced with 1 mL of virus isolation medium containing SARS-CoV-2 virus at a MOI of 1 and incubated for 1 hour at 37°C and 5% CO2. Wells were washed three times with PBS, then replaced with 1 mL fresh virus isolation medium and incubated at 37°C and 5% CO2. At 24 and 48 hours post-infection, wells were washed three times with PBS, then fixed as described below.
Scanning electron microscopy
Cells were fixed with Karnovsky’s formulation of 2% paraformaldehyde/2.5% glutaraldehyde in 0.1 M Sorenson’s phosphate buffer, and then post-fixed with 1.0% osmium tetroxide/0.8% potassium ferrocyanide in 0.1 M sodium cacodylate buffer washed with 0.1 M sodium cacodylate buffer then stained with 1% tannic acid in dH2O. After additional buffer washes, the samples were further osmicated with 2% osmium tetroxide in 0.1M sodium cacodylate, then washed with dH2O. Specimens were dehydrated with a graded ethanol series from 50%, 75%, 100% x 3 for 5 minutes each, critical point dried under CO2 in a Bal-Tec model CPD 030 Drier (Balzers, Liechtenstein), mounted with double sided carbon tape on aluminum specimen mounts (Ted Pella), and sputter coated with 35 Å of iridium in a Quorum EMS300T D sputter coater (Electron Microscopy Sciences, Hatfield, PA) prior to viewing at 5 kV in a Hitachi SU-8000 field emission scanning electron microscope (Hitachi, Tokyo, Japan).
Transmission electron microscopy
Specimens were fixed as described above for scanning electron microscopy and additionally stained overnight with 1% uranyl acetate at 4°C after the second osmium staining and then dehydrated with the same graded ethanol series and embedded in Spurr’s resin. Thin sections were cut with a Leica UC7 ultramicrotome (Buffalo Grove, IL) prior to viewing at 120 kV on a FEI BT Tecnai transmission electron microscope (Thermofisher/FEI, Hillsboro, OR). Digital images were acquired with a Gatan Rio camera (Gatan, Pleasanton, CA).
Structure Mapping
The Pymol Molecular Graphics System (https://www.schrodinger.com/pymol) was used to map the location of the observed deletions onto a SARS-CoV-2 spike structure (PDB: 6ZGE; Wrobel et al., 2020). Nucleotide sequence alignments were generated using MAAFT align (Katoh and Standley, 2013; Katoh et al., 2002) implemented in Geneious Prime v.2020.1.2 (https://www.geneious.com) and amino acid sequence alignments were generated with Multalin (Corpet, 1988) and plotted with ESPript (Robert and Gouet, 2014).
Quantification and Statistical Analysis
Data and statistical analysis was performed using GraphPad Prism 8.2.0. Replicates and statistical details can also be found in the methods and figure legends. For ELISA and virus neutralization assays, the serum/plasma samples were diluted and tested in duplicate. For the growth curves, both virus isolates (day 49 patient isolate and USA/WA1/2020) were tested in three replicate wells for both Vero E6 cells and the primary human alveolar epithelial cells. The growth curve data shown are the mean and standard error of the mean for the three independent replicates. The statistical analysis was performed using a 2-way ANOVA in Graphpad Prism 8.2.0. Further methods to determine whether the data met assumptions of the statistical approach were not relevant for these analyses.
Acknowledgments
We would like to thank Neeltje van Doremalen, Jonathan Schulz, Myndi Holbrook, Anita Mora, and Rose Perry for excellent technical assistance. We would like to thank MatTek for providing the alveolar tissue culture system and technical support as well as all the originating and submitting laboratories and authors who deposited SARS-CoV-2 genomes to GISAID. We would also like to thank the health care workers and laboratorians at EvergreenHealth for selfless service to patients. Finally, we thank the individual described in this report for her gracious willingness to participate and contribute to these studies as we sought to understand this enigmatic infection. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID). T.A.B. is supported by the Medical Research Council UK (MR/S007555/1). The Wellcome Centre for Human Genetics is supported by Wellcome Centre grant 203141/Z/16Z.
Author Contributions
Conceptualization, F.X.R. and V.J.M.; Resources, E.R.F., C.M., and F.X.R.; Methodology, V.A.A., M.J.M., S.N.S., B.N.W., E.R.F., C.M., T.A.B., and E.D.W.; Investigation, V.A.A., M.J.M., S.N.S., R.P., B.N.W., S.L.A., K.B., E.R.F., and E.d.W.; Writing – Original Draft, V.A.A., M.J.M., F.X.R., and V.J.M; Writing –Review & Editing, S.N.S., S.D.J., C.M., T.A.B., and E.d.W.; Data Curation, C.M.; Supervision, T.A.B., E.d.W., F.X.R., and V.J.M.
Declaration of Interests
The authors declare no competing interests.
Published: November 4, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.10.049.
Supplemental Information
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés C., Garcia-Cehic D., Gregori J., Piñana M., Rodriguez-Frias F., Guerrero-Murillo M., Esperalba J., Rando A., Goterris L., Codina M.G. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg. Microbes Infect. 2020;9:1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann T., Delgado J., Montserrat E. CLL and COVID-19 at the Hospital Clinic of Barcelona: an interim report. Leukemia. 2020;34:1954–1956. doi: 10.1038/s41375-020-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., Greninger A.L., Pipavath S., Wurfel M.M., Evans L. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. N. Engl. J. Med. 2020;382:2012–2022. doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020:ciaa638. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Jr., Cohen J.I. Detection of Nucleocapsid Antibody to SARS-CoV-2 is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. medRxiv. 2020 2020.04.20.20071423. [Google Scholar]
- Capobianchi M.R., Rueca M., Messina F., Giombini E., Carletti F., Colavita F., Castilletti C., Lalle E., Bordi L., Vairo F. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin. Microbiol. Infect. 2020;26:954–956. doi: 10.1016/j.cmi.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. Duration of Isolation and Precautions for Adults with COVID-19.https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html [Google Scholar]
- CDC . 2020. People with Certain Medical Conditions.https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fill L., Hadney L., Graven K., Persaud R., Hostoffer R. The clinical observation of a patient with common variable immunodeficiency diagnosed as having coronavirus disease 2019. Ann. Allergy Asthma Immunol. 2020;125:112–114. doi: 10.1016/j.anai.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Han P., Zhu R., Bai T., Yi J., Zhao X., Tao M., Quan R., Chen C., Zhang Y. Risk factors for viral RNA shedding in COVID-19 patients. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.01190-2020. 2001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstenau M., Langerbeins P., De Silva N., Fink A.M., Robrecht S., von Tresckow J., Simon F., Hohloch K., Droogendijk J., van der Klift M. COVID-19 among fit patients with CLL treated with venetoclax-based combinations. Leukemia. 2020;34:2225–2229. doi: 10.1038/s41375-020-0941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. 2018. http://hannonlab.cshl.edu/fastx_toolkit/ FASTX-Toolkit.
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Chen L., Chen L., Yuan G., Fang Y., Chen W., Wu D., Liang B., Lu X., Ma Y. COVID-19 in persons with haematological cancers. Leukemia. 2020;34:1637–1645. doi: 10.1038/s41375-020-0836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X., Ma H., Chen W., Lin Y., Zheng Y. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami S., Benirschke R., Flanagan T., Tanna N., Klein T., Elue R., Debosz P., Mallek J., Wright G., Guariglia P. Persistence of SARS-CoV-2 nasopharyngeal swab PCR positivity in COVID-19 convalescent plasma donors. Transfusion. 2020 doi: 10.1111/trf.16015. Published online August 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad Institute 2018. http://broadinstitute.github.io/picard Picard Toolkit.
- Jary A., Leducq V., Malet I., Marot S., Klement-Frutos E., Teyssou E., Soulié C., Abdi B., Wirden M., Pourcher V. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.07.032. S1198-743X(20)30440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X.H., Zheng K.I., Pan K.H., Xie Y.P., Zheng M.H. COVID-19 in a patient with chronic lymphocytic leukaemia. Lancet Haematol. 2020;7:e351–e352. doi: 10.1016/S2352-3026(20)30074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson S.D., Munster V.J. A framework for nosocomial transmission of emerging coronaviruses. Infect. Control Hosp. Epidemiol. 2020:1–2. doi: 10.1017/ice.2020.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Ko J.H., Park G.E., Cho S.Y., Ha Y.E., Kang J.M., Kim Y.J., Huh H.J., Ki C.S., Jeong B.H. Atypical presentations of MERS-CoV infection in immunocompromised hosts. J. Infect. Chemother. 2017;23:769–773. doi: 10.1016/j.jiac.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki K.M., Janoff E.N. Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect. Dis. 2009;9:493–504. doi: 10.1016/S1473-3099(09)70175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR Test Results in Patients Recovered From COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.Y., Wang P., Mok B.W., Zhang A.J., Chu H., Lee A.C., Deng S., Chen P., Chan K.H., Song W. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., Park S.Y., Son H.J., Yu S., Park J.W. Clinical Course and Molecular Viral Shedding Among Asymptomatic and Symptomatic Patients With SARS-CoV-2 Infection in a Community Treatment Center in the Republic of Korea. JAMA Intern Med. 2020;180:1–6. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang L., Liu B., Song D. Case Report: Viral Shedding for 60 Days in a Woman with COVID-19. Am. J. Trop. Med. Hyg. 2020;102:1210–1213. doi: 10.4269/ajtmh.20-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent Neutralizing Monoclonal Antibodies Directed to Multiple Epitopes on the SARS-CoV-2 Spike. bioRxiv. 2020 doi: 10.1101/2020.06.17.153486. [DOI] [PubMed] [Google Scholar]
- Liu W.D., Chang S.Y., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L., Chang S.C. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J. Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen X., Zou X., Luo H. A severe-type COVID-19 case with prolonged virus shedding. J. Formos. Med. Assoc. 2020;119:1555–1557. doi: 10.1016/j.jfma.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J. Identification of common deletions in the spike protein of SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.00790-20. e00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael T.M., Clark S., Pogosjans S., Kay M., Lewis J., Baer A., Kawakami V., Lukoff M.D., Ferro J., Brostrom-Smith C. COVID-19 in a Long-Term Care Facility - King County, Washington, February 27-March 9, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:339–342. doi: 10.15585/mmwr.mm6912e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael T.M., Currie D.W., Clark S., Pogosjans S., Kay M., Schwartz N.G., Lewis J., Baer A., Kawakami V., Lukoff M.D., Public Health–Seattle and King County, EvergreenHealth, and CDC COVID-19 Investigation Team Epidemiology of Covid-19 in a Long-Term Care Facility in King County, Washington. N. Engl. J. Med. 2020;382:2005–2011. doi: 10.1056/NEJMoa2005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira E., Yarce O.A., Ortega C., Fernández S., Pascual N.M., Gómez C., Alvarez M.A., Molina I.J., Lama R., Santamaria M. Rapid recovery of a SARS-CoV-2-infected X-linked agammaglobulinemia patient after infusion of COVID-19 convalescent plasma. J. Allergy Clin. Immunol. Pract. 2020;8:2793–2795. doi: 10.1016/j.jaip.2020.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paneesha S., Pratt G., Parry H., Moss P. Covid-19 infection in therapy-naive patients with B-cell chronic lymphocytic leukemia. Leuk. Res. 2020;93:106366. doi: 10.1016/j.leukres.2020.106366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J., Falzarano D., de Wit E., Hardcastle K., Feldmann F., Haddock E., Scott D., Feldmann H., Munster V.J. Pathogenicity and Viral Shedding of MERS-CoV in Immunocompromised Rhesus Macaques. Front. Immunol. 2018;9:205. doi: 10.3389/fimmu.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G.Q., Chen X.Q., Lv D.F., Ma A.H.Y., Wang L.P., Yang N.B., Chen X.M. Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect. Dis. (Lond.) 2020;52:511–512. doi: 10.1080/23744235.2020.1748705. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku316. W320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., Mesirov J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017;77:e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko P., Puller V., Neher R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E., Perez K.K., Ashraf M., Chen J., Castillo B., Christensen P.A., Eubank T., Bernard D.W., Eagar T.N., Long S.W. Treatment of Coronavirus Disease 2019 (COVID-19) Patients with Convalescent Plasma. Am. J. Pathol. 2020;190:1680–1690. doi: 10.1016/j.ajpath.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Pérez-Pérez L., Mead-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J. SARS-CoV-2 infection dynamics in lungs of African green monkeys. bioRxiv. 2020 doi: 10.1101/2020.08.20.258087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Xiao J., Sun R., Tang X., Liang C., Lin H., Zeng L., Hu J., Yuan R., Zhou P. Prolonged Persistence of SARS-CoV-2 RNA in Body Fluids. Emerg. Infect. Dis. 2020;26:1834–1838. doi: 10.3201/eid2608.201097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vries E., Stittelaar K.J., van Amerongen G., Veldhuis Kroeze E.J., de Waal L., Fraaij P.L., Meesters R.J., Luider T.M., van der Nagel B., Koch B. Prolonged influenza virus shedding and emergence of antiviral resistance in immunocompromised patients and ferrets. PLoS Pathog. 2013;9:e1003343. doi: 10.1371/journal.ppat.1003343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020 doi: 10.1101/2020.06.08.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Grunewald M., Perlman S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2020;2203:1–29. doi: 10.1007/978-1-0716-0900-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Xu X., Ma K., Yang J., Guan H., Chen S., Chen Z., Chen G. Successful recovery of COVID-19 pneumonia in a renal transplant recipient with long-term immunosuppression. Am. J. Transplant. 2020;20:1859–1863. doi: 10.1111/ajt.15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and the Supplementary Tables from this study have been deposited to Mendeley Data at https://dx.doi.org/10.17632/3n377gv8kb.
Genome sequences have been deposited to GenBank: MT982403, MT982402, MT982405, MT982406, MT982401 and MT982404.