Abstract
To explore the epigenetic alterations in response to DNA damage following polycyclic aromatic hydrocarbons (PAHs) exposure and the crosstalk between different epigenetic regulations, we examined trimethylated Lys 36 of histone H3 (H3K36me3) and methylation of ‘long interspersed element-1 (LINE-1)’ and ‘O 6-methylguanine-DNA methyltransferase (MGMT)’ in peripheral blood lymphocytes (PBLCs) of 173 coke oven workers (PAH-exposed group) and 94 non-exposed workers (control group). The PAH-exposed group showed higher internal PAH exposure level, enhanced DNA damage and increased MGMT expression (all P < 0.001). Notably, the methylation of LINE-1 and MGMT decreased by 3.9 and 40.8%, respectively, while H3K36me3 level was 1.7 times higher in PBLCs of PAH-exposed group compared to control group (all P < 0.001). These three epigenetic marks were significantly associated with DNA damage degree (all P < 0.001) and PAH exposure level in a dose–response manner (all P < 0.001). LINE-1 hypomethylation is correlated with enhanced H3K36me3 modification (β = −0.198, P = 0.002), indicating a synergistic effect between histone modification and DNA methylation at the whole genome level. In addition, MGMT expression was positively correlated with H3K36me3 modification (r = 0.253, P < 0.001), but not negatively correlated with MGMT methylation (r = 0.202, P < 0.05). The in vitro study using human bronchial epithelial cells treated with the organic extract of coke oven emissions confirmed that H3K36me3 is important for MGMT expression following PAH exposure. In summary, our study indicates that histone modification and DNA methylation might have synergistic effects on DNA damage induced by PAH exposure at the whole genome level and H3K36me3 is more essential for MGMT expression during the course.
Keywords: occupational PAH exposure, DNA damage, H3K36me3, LINE-1 methylation, MGMT methylation
Introduction
Epigenetics alterations could induce heritable phenotypic changes without altering DNA sequence [1, 2]. Epigenetic regulation can be modified by environment, and these changes contribute to the development of human diseases [3–6]. Regulation of gene expression is a result of crosstalk among different epigenetic patterns. Recent studies have revealed that DNA methylation and histone modifications regulate gene expression by constituting a specific ‘epigenetic code’ [7, 8]. Based on the results of studies on environmental epigenetics, numerous epigenetic biomarkers were found to be associated with both environmental chemical exposures and adverse health effects [9–11]. However, most of these studies focus on one type of epigenetic modification and fail to clarify the interactions between different patterns of epigenetic modifications, such as DNA modifications, chromatin modifications, noncoding long RNA, and small RNA. Thus, integrated strategies should be developed to explore epigenetic mechanism and interpret crosstalk between different epigenetic modes, particularly in the context of human studies.
DNA damage is well known to be an important biomarker of polycyclic aromatic hydrocarbons (PAHs) exposure. Upon environmental stimuli, DNA methylation and histone modification are mobilized to accommodate the DNA damage and involved in regulation of DNA damage response [12–14]. It has been reported that PAH exposure induces histone modification and DNA methylation changes [13, 14]. However, it remains unclear whether these epigenetic marks work together in regulation of critical genes in the course of DNA damage response. In this study, we aimed to address the interplay between specific histone modification and DNA methylation in coke oven workers exposed to high levels of PAHs.
O6-methylguanine-DNA methyltransferase (MGMT) encodes a DNA repair protein that can reverse DNA alkylation caused by alkylating agents [15]. It is one of the important DNA damage response genes and its expression can be regulated by DNA methylation [16, 17]. It has been demonstrated that methylation of MGMT was negatively associated with the levels of PAH exposure and the degree of DNA damage in peripheral blood lymphocytes (PBLCs) of PAHs exposed-workers [12]. Our previous study revealed that MGMT gene body physically interacted with H3K36me3 and H3K36me3 modification played an important role in regulation of MGMT expression [14]. Studies conducted by other groups enhanced the notion that MGMT could be regulated by both DNA methylation and histone modification [16–19]. However, it remains unclear how these epigenetic alterations cooperate on regulation of MGMT expression in response to PAH exposure.
In this study, we attempt to address the associations between different epigenetic alterations, PAH exposure, and DNA damage in occupational PAH-exposed workers. DNA methylation (long interspersed element-1 [Line-1] and MGMT) and histone modification (H3K36me3) are used for investigating the interplay between different epigenetic alterations, especially in regulation of MGMT expression. In vitro cell experiments are employed to verify these associations. In addition, mediation analysis is conducted to statistically assess how much PAH exposure-induced epigenetic modification is accounted for by DNA damage. These findings provide new insights into a crosstalk between different epigenetic patterns in regulating DNA damage response upon PAH exposure.
Materials and Methods
Study population
In this study, 173 male coke oven workers and 94 male control workers from the hot rolling workshop were included. Workers suffering from acute or chronic infection or autoimmune diseases or exposed to mutagenic agents (such as X-ray radiation) within 2 months were excluded. All subjects were from the Benxi Iron and Steel Group Corporation in Liaoning Province, China. The coke oven workers are mentioned as PAH-exposed group in this paper and the control workers are mentioned as control group. About 15 ml urine and 3 ml of peripheral blood were collected from each worker for measurement of urinary 1-hydroxypyrene (1-OHP) and isolation of PBLCs, respectively. Genomic DNA of the subjects was extracted using the TIANamp blood DNA kit (Tiangen Biotech, China). Detailed information of the subjects, including demographic data, occupational history, medical history, alcohol consumption, smoking history, and grilled food consumption were acquired using a structured questionnaire. Informed consent in written form was acquired from all subjects. The research protocol was approved by the Ethical Review Committee of Sun Yat-sen University.
Detection of histone modification of H3K36me3 in PBLCs
H3K36me3 modification was determined using the sandwich enzyme-linked immunosorbent assay (ELISA) as reported previously [20, 21]. Briefly, the histones were extracted from PBLCs and concentration was measured using the BCA kit according to the manufacturer’s instructions (Beyotime, China). Histone H3 antibody (1:20 000 dilution, Abcam, Cambridge, MA, USA) was used to coat the microplates. Then, the plates were blocked with 5% milk in PBST (1× PBS, 0.05% Tween-20). The recombinant protein of H3K36me3 (Active Motif, Carlsbad, CA, USA) was added to the plates and followed by addition of PBLC histones at a final concentration of 4 μg ml−1. Primary antibodies including histone H3 (1:10 000 dilution, Sigma-Aldrich, USA) and H3K36me3 (1:5000 dilution, Abnova, Taiwan) were added to each well separately. After washing with PBST, the secondary antibody (1:1000 dilution, Santa Cruz Biotechnology, CA, USA) was added. The optical density was read at 450 nm using a microplate spectrophotometer (BIO-TEC, USA). The analyses were performed in triplicate. The relative percent of histone modification was calculated from the standard curve.
Bisulfite treatment and pyrosequencing
One microgram of genome DNA was subjected to bisulfite conversion using EpiTech bisulfite kit (Qiagen, USA). Pyrosequencing PCR was conducted using the PyroMark PCR Kit (Qiagen, USA). About 5 μl PCR products was subjected to gel electrophoresis for a quality control and sequencing of MGMT and LINE-1 was conducted on a PyroMark Q96 sequencer (Qiagen, USA) according to the protocol reported previously [22, 23]. For LINE-1 the primers 5′-TTTTTTGAGTTAGGTGTGGGATA-3′ and 5′-Biotin-AAAAATCAAAAAATTCCC TTTCC-3′ were used for PCR, primer 5′-GGGTGGGAGTGAT-3′ was used for pyrosequencing [22]. For MGMT, the primers 5′-GGATATGTTGGGATAGTT-3′ and 5′-Biotin-ACCCAAACACTCACCAAATC-3′ were used for PCR, primer 5′-GGATATGTTGGGATAGT-3′ was used for pyrosequencing [23]. Nine CpG sites in MGMT promoter and three sites in LINE-1 were analyzed, and mean percentages of methylation were calculated. In our study, a non-CpG cytosine was used as built-in control to verify bisulfite conversion, which was included in the software. In addition, sentinel quality control was set for each batch of sequencing, and the variability of sentinel samples tested by different batches was less than 10%.
COEs collection and HBE cell treatment
Coke oven emission (COEs) were collected and the organic extracts were prepared according to a protocol described previously [12, 24]. Briefly, COEs were collected using glass fiber filter at the top oven area of a coking plant in Benxi, Liaoning, China, and organic extracts were dissolved in DMSO to a final concentration of 1 mg ml−1 and stored at −20°C. The human bronchial epithelial (HBE) cell line (16HBE14σ, abbreviated as HBE) was kindly provided by Dr D.C. Gruenert (University of California, San Francisco). HBE cells were seeded in triplicate at a density of 2.5 × 105 in 6 cm dishes. After culturing to 50% confluence, the cells were exposed to COEs at concentrations of 1, 5, and 10 μg ml−1 for 48 h. DMSO was used as a vehicle control. After the cells reached ~90% confluence, the cultures were split and subjected to another round of treatment. The cells were treated weekly for four times in total.
Quantitative real-time PCR analysis
Total RNA was isolated using TRIzol Reagent (Invitrogen, USA) and reverse transcription was performed using the PrimeScript™II 1st Strand cDNA synthesis kit (Takara Bio, Inc., Tokyo, Japan) following manufacturer’s instructions. The quantitative real-time PCR was conducted using the SYBR Green® Realtime PCR Master Mix assay kit (Toyobo, Osaka, Japan) and analyzed by the Applied Biosystems ViiA™ 7 Real-time PCR system (Applied Bio-systems, Foster City, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene. The forward primer 5′-CTGGCCGAAACTGAGTATGT-3′ and reverse primer 5′-GGACACTGCCACTTCCTTTA-3′ were used for MGMT, while the forward primer 5′-CTGGCCGAAACTGAGTATGT-3′ and reverse primer 5′-GGACACTGCCACTTCCTTTA-3′ were used for GAPDH. ΔCt and 2−ΔΔCt were calculated to represent the mRNA expression levels in PBLCs and HBE cells, respectively.
Immunoblotting analysis
To determine H3K36me3 levels in HBE cells, 30 μg proteins per sample was separated by 8–16% SDS–polyacrylamide gel electrophoresis and then transferred onto nitrocellulose membranes by electroblotting. The membranes were blocked in 5% milk in PBST for 2 h at room temperature and probed with antibodies against H3K36me3 (1:3000 dilution; Abnova, Taiwan) or H3 (1:2000 dilution; Abcam, Cambridge, MA, USA) overnight at 4°C. Then, membranes were incubated with secondary antibody (1:2000 dilutions, Santa Cruz, CA, USA) for 1 h at room temperature. After washing with PBST, the membranes were developed using an enhanced chemiluminescence substrate (Santa Cruz, CA, USA) and exposed to X-ray film for visualization of the antigenic bands.
Analysis of PAHs internal exposure
Urinary 1-OHP was measured using high-pressure liquid chromatography as described previously [25]. The lowest detection limit was 0.14 μg l−1 urine, with signal-to-noise ratio ~3. Measurements below the limit of detection were replaced with limits of detection (LOD) .
.
Analysis of DNA damage
Comet assay was performed as previously reported [14, 26]. Briefly, 5 μl EDTA–blood was mixed well with 200 μl of 0.8% low-melting point agarose, then 30 μl mixture was evenly layered onto a CometAssay® HT 20-well slide (Trevigen). After lysing for 3 h, the samples were subjected to electrophoresis (25 V, 300 mA), neutralization, dehydration, and staining. About 100 cells from each sample were randomly examined under a fluorescent microscope (Nikon Eclipse Ti-E, Tokyo, Japan) and scored by Comet Assay Software Project-1.2.2 (University of Wroclaw, Poland). The differences in Olive tail moment (OTM) and tail DNA percentage (% Tail DNA) were measured to indicate the degree of DNA damage.
Mediation analysis
In this study, we hypothesize that the associations of urinary 1-OHP and epigenetic alterations might be partially mediated by DNA damage. Thus, mediation analysis was performed as previously reported [27]. Exposure level of PAHs, extent of DNA damage, and alterations of epigenetic modifications are significantly correlated pairwise, which meets the criteria for mediation analysis. Mediation analysis was conducted with DNA damage as mediator, adjusting for age, body mass index, smoking status, alcohol abuse, grilled food consumption, lymphocyte percentage, and white blood cell count. All analyses were performed using R software (version 3.1.2, mediation package).
Statistics analyses
Urinary 1-OHP, histone modification, and DNA damage indexes were adjusted by natural logarithmic transformation to fit normal distribution. The LINE-1 and MGMT methylation data were logit transformed before statistical analysis. Median and interquartile ranges (IQR) were calculated for data of urinary 1-OHP, OTM, % Tail DNA, LINE-1 and MGMT methylation, and H3K36me3. Mann–Whitney U test was used to analyze DNA methylation between different groups. General linear models were employed to test the associations of epigenetic alterations with categorized urinary 1-OHP in all subjects. The associations between different epigenetic modifications, as well as epigenetic modifications and DNA damage were also examined using general linear models. Since the coke oven workers consumed significantly higher amounts of grilled food than the controls and grilled food consumption is well-known to produces PAH–DNA damage, we conducted three separate general linear models to adjust for potential confounding factors. Data in model 1 showed crude results. Data in model 2 were adjusted for grilled food consumption. Data in model 3 were additionally adjusted for age, BMI, smoking status, alcohol use, lymphocyte percentage, and WBC count. Spearman correlation test was conducted to analyze the correlation between MGMT expression with MGMT methylation and H3K36me3 modification. Statistical analyses were performed using SPSS software (SPSS 22.0, Chicago, USA) and all test results were considered to be of statistically significance at two-sided level of P < 0.05.
Results
General characteristics of study subjects
About 173 male coke oven workers and 94 male control workers were included in this study. As shown in Table 1, there were no differences between two groups of workers in terms of age, BMI, smoking status, alcohol use, WBC count, and lymphocyte percentage. However, the proportion of grilled food consumption more than once a week was significantly higher in the coke oven workers (P < 0.001). Urinary 1-OHP level in coke oven workers (median at 63.49, IQR: 19.28–160.54) was ~10-fold higher than that in the control workers (median at 6.69, IQR: 4.66–9.09). Meanwhile, the degree of DNA damage indicated by levels of OTM and % Tail DNA (Comet assay) was 5.9- and 8.2-fold greater in PAH-exposed workers than the controls (both P < 0.001) (Table 1). The mRNA level of MGMT in PAH-exposed workers was increased by 6.6% than that in control group (P < 0.001) (Table 1). Taken together, the data show that the coke oven workers, exposed to higher levels of PAHs than the controls, also had higher levels of DNA damage and significantly increased MGMT expression.
Table 1.
Demographic and exposure characteristics of two groups of workers
| Variables | Control group (n = 94) | PAH-exposed group (n = 173) | P |
|---|---|---|---|
| Age (years, mean ± SD) | 45.55 ± 8.74 | 44.12 ± 5.68 | 0.152a |
| BMI (kg m−2, mean ± SD) | 25.43 ± 3.80 | 24.59 ± 3.18 | 0.055a |
| Current smokers [yes (%)] | 62 (66.0%) | 120 (69.4%) | 0.568b |
| Alcohol user [yes (%)] | 62 (66.0%) | 112 (64.7%) | 0.842b |
| Grilled food consumption ≥ weekly [yes (%)] | 5 (5.3%) | 49 (18.4%) | <0.001b |
| Urinary 1-OHP [μg g−1 creatinine, median (IQR)] | 6.69 (4.66–9.09) | 63.49 (19.28–160.54) | <0.001c |
| OTM [median (IQR)] | 3.66 (0.94–5.63) | 21.50 (12.18–31.61) | <0.001c |
| % Tail DNA [median (IQR)] | 4.81 (2.61–6.18) | 39.64 (27.64–50.51) | <0.001c |
| WBC (109 l−1, mean ± SD) | 6.35 ± 1.35 | 6.42 ± 1.78 | 0.529a |
| Lymphocyte % (mean ± SD) | 31.27 ± 6.48 | 32.84 ± 7.43 | 0.085a |
| MGMT mRNA (∆Ct, mean ± SD) | 6.41 ± 0.62 | 5.99 ± 0.66 | <0.001c |
SD, standard deviations; BMI, body mass index; 1-OHP, 1-hydroxypyrene; IQR, interquartile range; OTM, Olive Tail Moment; WBC, white blood cell.
aTwo-sided two-sample t test between control and coke oven workers.
bTwo-tailed chi-square test between control and coke oven workers.
cTwo-sided two-sample t test for natural logarithmic-transformed data between control and coke oven workers.
Hypomethylation of LINE-1 and MGMT and enhanced H3K36me3 modification in PBCLs of PAH-exposed group
The DNA methylation levels of LINE-1 and MGMT were measured by pyrosequencing and H3K36me3 modification were analyzed by ELISA in PBLCs of all workers. As a result, the average methylation rate of LINE-1 and MGMT decreased by 3.9 and 40.8%, respectively in PAH-exposed group compared to that in control group (both P < 0.001) (Table 2). In contrast, the H3K36me3 modification in PAH-exposed group was 1.7 times higher than that of control group (P < 0.001) (Table 2). In addition, we showed that an increasing urinary 1-OHP was associated with hypomethylation of LINE-1 and MGMT and enhanced H3K36me3 modification before or after adjustment of confounder factors in all subjects (all P < 0.001) (Table 3). These findings indicate that PAH exposure is correlated with specific epigenetic alteration in a dose-dependent manner.
Table 2.
Methylation of LINE-1 and MGMT and H3K36me3 modification in two groups of workers
| Epigenetic modifications | Control group | PAH-exposed group | P b | ||
|---|---|---|---|---|---|
| N a | Median (IQR) | N a | Median (IQR) | ||
| LINE-1 (% 5mC) | 94 | 83.66 (82.07–85.84) | 169 | 80.42 (78.90–82.84) | <0.001 |
| MGMT (% 5mC) | 92 | 5.51 (3.61–7.21) | 168 | 3.26 (2.78–4.30) | <0.001 |
| H3K36me3 (relative %) | 94 | 7.35 (4.00–11.15) | 173 | 12.58 (8.36–18.99) | <0.001 |
ELISA, sandwich enzyme-linked immunosorbent assay; 5mC, 5-methylcytosine; IQR, interquartile range.
a N varies obtained based on the success of the pyrosequencing analysis and ELISA.
bMann–Whitney U test.
Table 3.
DNA methylation (LINE-1 and MGMT) and histone modification (H3K36me3) of all subjects stratified by quartiles of urinary 1-OHP concentration
| Epigenetic modifications | Urinary 1-OHP concentration (μg g−1 creatinine) | P-trenda | P-trendb | P-trendc | |||
|---|---|---|---|---|---|---|---|
| Q1 (n = 66) | Q2 (n = 68) | Q3 (n = 66) | Q4 (n = 67) | ||||
| LINE-1 (% 5mC) | |||||||
| Mean (95% CI) | 83.65 (82.97, 84.33) | 82.80 (82.24, 83.35) | 81.74 (81.13, 82.35) | 81.72 (81.09, 82.34) | <0.001 | <0.001 | <0.001 |
| Median (IQR) | 84.06 (81.69–85.81) | 82.61 (81.07–84.49) | 81.39 (79.97–83.00) | 81.59 (80.46–83.19) | |||
| MGMT (% 5mC) | |||||||
| Mean (95% CI) | 6.14 (5.06, 7.22) | 4.55 (3.82, 5.27) | 3.85 (3.49, 4.22) | 3.69 (3.25, 4.13) | <0.001 | <0.001 | <0.001 |
| Median (IQR) | 5.11 (3.63–7.13) | 3.48 (2.17–5.09) | 3.42 (2.83–4.60) | 3.19 (2.71–4.21) | |||
| H3K36me3 (relative %) | |||||||
| Mean (95% CI) | 9.32 (7.96, 10.68) | 12.36 (8.28, 16.44) | 17.68 (13.58, 21.78) | 14.99 (11.96, 16.02) | <0.001 | <0.001 | <0.001 |
| Median (IQR) | 8.44 (4.11–12.94) | 8.49 (5.05–13.40) | 13.15 (8.44–19.97) | 11.93 (7.99–18.77) | |||
1-OHP, 1-hydroxypyrene; 5mC, 5-methylcytosine; IQR: interquartile ranges; 95% CI: 95% confidence intervals.
aCrude.
bAdjusted for grilled food consumption.
cAdjusted for grilled food consumption, age, BMI, smoking status, alcohol use, lymphocyte percentage, and white blood cell count. Values of histone modifications was natural logarithmic transformed, and DNA methylation levels were logit transformed.
Different epigenetic modes might be collaborating in response to DNA damage at a whole genome level
To clarify the associations between the epigenetic alterations and the degree of DNA damage, we conducted general linear regression analyses between the analyzed parameters of the Comet assay (% Tail DNA and OTM) and epigenetic modifications (methylation of LINE-1 and MGMT, and H3K36me3 modification). Results showed that the extent of DNA damage was negatively associated with methylation levels of LINE-1 (β% Tail DNA = −0.333, βOTM = −0.272, both P < 0.001) and MGMT (β% Tail DNA = −0.329, βOTM = −0.321, both P < 0.001) even after confounder adjustments. In contrast, it was positively associated with H3K36me3 modification after confounder adjustments (β% Tail DNA = 0.337, βOTM = 0.277, both P < 0.001) (Table 4). Notably, the level of H3K36me3 modification was negatively correlated with DNA methylation of LINE-1 even after confounder adjustments (β = −0.198, P = 0.002) (Table 5), indicating a synergistic effect between histone modification and DNA methylation at whole genome level.
Table 4.
Associations between epigenetic modifications and extent of DNA damage
| Epigenetic modifications | % Tail DNA | OTM | ||
|---|---|---|---|---|
| β (95% CI) | P | β (95% CI) | P | |
| LINE-1 (% 5mC) | ||||
| Model 1a | −0.333 (−8.258, −3.639) | <0.001 | −0.270 (−8.190, −2.871) | <0.001 |
| Model 2b | −0.326 (−8.113, −3.582) | <0.001 | −0.265 (−8.084, −2.773) | <0.001 |
| Model 3c | −0.333 (−8.255, −3.677) | <0.001 | −0.272 (−8.247, −2.889) | <0.001 |
| MGMT (% 5mC) | ||||
| Model 1a | −0.320 (−2.858,-1.215) | <0.001 | −0.310 (−3.181, −1.309) | <0.001 |
| Model 2b | −0.316 (−2.824, −1.202) | <0.001 | −0.308 (−3.156, −1.295) | <0.001 |
| Model 3c | −0.329 (−2.917, −1.274) | <0.001 | −0.321 (−3.264, −1.382) | <0.001 |
| H3K36me3 (relative %) | ||||
| Model 1a | 0.350 (0.441, 0.936) | <0.001 | 0.287 (0.355, 0.933) | <0.001 |
| Model 2b | 0.336 (0.414, 0.909) | <0.001 | 0.277 (0.332, 0.912) | <0.001 |
| Model 3c | 0.337 (0.414, 0.915) | <0.001 | 0.277 (0.328, 0.914) | <0.001 |
OTM, Olive Tail Moment; 95% CI: 95% confidence intervals; 5mC, 5-methylcytosine.
aCrude.
bAdjusted for grilled food consumption.
cAdjusted for grilled food consumption, age, BMI, smoking status, alcohol use, lymphocyte percentage, and white blood cell count. Values of DNA damage and histone modification were natural logarithmic transformed and DNA methylation levels were logit transformed.
Table 5.
Associations between histone modification (H3K36me3) and DNA methylation (LINE-1 and MGMT)
| H3K36me3 (relative %) | LINE-1 (% 5mC) | MGMT (% 5mC) | ||
|---|---|---|---|---|
| β (95% CI) | P | β (95% CI) | P | |
| Model 1a | −0.202(−0.036, −0.009) | 0.001 | −0.009 (−0.042, 0.036) | 0.844 |
| Model 2b | −0.198 (−0.036, −0.009) | 0.001 | −0.007 (−0.042, 0.037) | 0.909 |
| Model 3c | −0.198 (−0.036, −0.009) | 0.002 | −0.003 (−0.040, 0.039) | 0.967 |
95% CI: 95% confidence intervals; 5mC, 5-methylcytosine.
aCrude.
bAdjusted for grilled food consumption.
cAdjusted for grilled food consumption, age, BMI, smoking status, alcohol use, lymphocyte percentage, and white blood cell count. The variables of DNA methylation were logit transformed, while H3K36me3 was natural logarithmic transformed to improve model fit.
To statistically assess how much PAH exposure-induced epigenetic modification is accounted for by DNA damage, we performed mediation analyses. In line with our hypothesis, DNA damage mediated 19.2–33.7% of the negative association between urinary 1-OHP and % 5mC in LINE-1 sequence, 25.7–30.5% of the negative association between urinary 1-OHP and % 5mC in MGMT promoter, and 34.2–55.9% of the positive association between urinary 1-OHP and H3K36me3, respectively (Table 6). These results revealed an association between PAHs-induced epigenetic modifications and DNA damage.
Table 6.
The mediation proportion by DNA damage in associations of urinary 1-OHP and epigenetic alterations
| DNA damage | Epigenetic modifications | Mediation proportion (%) [mean (95% CI)] | P |
|---|---|---|---|
| % Tail DNA | LINE-1 (% 5mC) | 33.7 (9.4, 88.0) | <0.01 |
| OTM | LINE-1 (% 5mC) | 19.2 (0.0, 59.0) | <0.05 |
| % Tail DNA | MGMT (% 5mC) | 30.5 (13.0, 57.0) | <0.001 |
| OTM | MGMT (% 5mC) | 25.7 (10.6, 49.0) | <0.001 |
| % Tail DNA | H3K36me3 (relative %) | 55.9 (26.2, 131.0) | <0.001 |
| OTM | H3K36me3 (relative %) | 34.2 (13.4, 76.0) | <0.01 |
Note: The variables of DNA methylation were logit transformed, and the other variables were natural logarithmic transformed. OTM, Olive Tail Moment; 1-OHP, 1-hydroxypyrene; 95% CI: 95% confidence intervals; 5mC, 5-methylcytosine.
H3K36me3 modification is involved in regulation of MGMT expression
Next, we explore whether different epigenetic alterations cooperate in regulation of the expression of DNA repair gene, MGMT in response to PAH exposure. Although it is well recognized that DNA methylation of gene promoter leads to suppression of gene expression, we failed to show MGMT methylation inversely associated with mRNA level of MGMT in all subjects. On the contrary, we found that hypomethylation of MGMT was positively associated with the expression of MGMT (r = 0.202, P = 0.003). Meanwhile, we showed a positive correlation between H3K36me3 modification and MGMT expression (r = 0.253, P < 0.001) in PBLCs of all subjects. Previously, we revealed that enhanced H3K36me3 modification participated in upregulation of MGMT expression in HBE cells following PAH exposure [14]. Moreover, by bioinformatics analysis (https://www.ncbi.nlm.nih.gov/genome/gdv/browser/geo/), we found that the CHIP-seq results of human lung tissue, liver cells, kidney cells, and peripheral blood mononuclear cells etc., indicated physical binding of H3K36me3 and gene body of MGMT (Supplementary Fig. 1). To test whether MGMT methylation leads to suppression of MGMT expression, we treated HBE cells with COEs at doses of 1, 5, and 10 μg ml−1 for 4 weeks. As a result, we found that treatment of COEs led to suppression of MGMT expression and decreased H3K36me3 modification at concentrations of 5 and 10 μg ml−1. However, no significant change was observed in MGMT methylation (Fig. 1). The in vitro results reinforced the notion that expression of MGMT might be primarily regulated by H3K36me3 modifications following exposure of PAHs.
Figure 1.
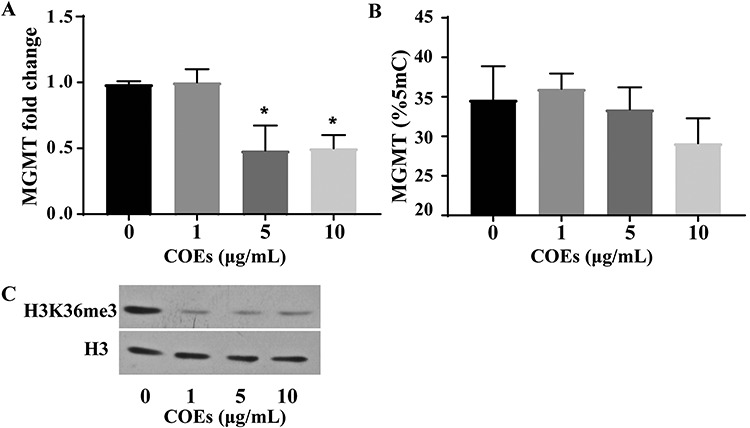
MGMT expression and epigenetic modification changes after COEs treatment. The HBE cells were exposed to COEs weekly at doses of 1, 5, 10 μg/mL for 4 weeks, DMSO was used as vehicle control. MGMT expression levels were determined by qRT-PCR (A). The methylation of MGMT (B) and H3K36me3 modification (C) were detected by pyrosequencing and immunoblotting analysis, respectively. COEs, coke oven emissions; DMSO, Dimethyl sulfoxide; qRT-PCR, quantitative real-time PCR. *P < 0.05 compared with the DMSO group.
Discussion
In this study, we sought to investigate the epigenetic alterations in response to PAH exposure and crosstalk between different epigenetic modifications in coke oven workers exposed to high level of PAHs. We revealed that PAHs-induced DNA damage may partially contribute to epigenetic alterations, including hypomethylation of LINE-1 and MGMT and enhanced H3K36me3 modifications. Importantly, we demonstrate that DNA methylation and histone modification might collaborate in response to DNA damage at whole genome level. Moreover, results from population-based and in vitro human cell-based studies and bioinformatic analysis suggest that H3K36me3 modification is involved in regulation of MGMT expression. This study shed light on the associations between PAH exposure, DNA damage and epigenetic alterations.
In this study, we revealed enhanced H3K36me3 modifications in PBLCs of coke oven workers and these changes were positively correlated with the level of PAH exposure and degree of DNA damage. These findings suggested that epigenetic alterations might play an important role in mediating cellular response to environmental stimuli. LINE-1, a widely spread transposable element (TE), is mostly repressed by DNA methylation and its’ methylation level is related to genomic instability [28, 29]. It has been reported that PAH-induced DNA damages led to hypomethylation of LINE-1, which is in agreement with a notion that DNA damage leads to genomic instability [13]. H3K36me3 modification plays pivotal role in regulation of DNA repair and maintenance of genomic stability in response to DNA damage [30, 31]. At a genome-wide scale, these epigenetic modifications might keep in a state of coordination to maintain genomic stability.
The epigenome changes can response to environmental pollutants, and the interplay between different epigenetic modification may occur in the course of cellular damage and repair [32–35]. Recent study reports that active LINE-1 plays a critical role in establishment of the repressive H3K9me3 modification in germ cell, providing evidence that a crosstalk may take place between LINE-1 methylation and histone modifications [36]. Hypomethylation of LINE-1 tends to activate LINE-1, which leads to increased genomic instability. In this study, mediation analysis suggested that PAHs-induced DNA damage may contribute to LINE-1 hypomethylation and enhanced H3K36me3 modification. Besides, we also revealed that decreased genome methylation and enhanced H3K36me3 modifications may collaborate in the regulation of network, which might facilitate DNA repair in PBLCs of PAH-exposed workers. Taken together, these findings indicate that different epigenetic marks might collaborate in DNA damage response at a genome-wide level.
MGMT is one of important enzymes involved in DNA damage repair. It has been reported that the expression of MGMT is regulated by histone modification and DNA methylation. Previous studies have shown that MGMT expression could be regulated by DNA methylation and the demethylation in MGMT promoter leads to an activation of gene expression [37, 38]. MGMT gene expression changed following chemical exposure through alteration of DNA methylation status [12, 13, 39]. A number of studies have revealed that MGMT could be involved in regulation of DNA damage response upon PAH exposure [12–14, 40]. However, the correlation between hypomethylation of MGMT and transcriptional activation of MGMT expression was absent in PBLCs of coke oven workers. Unexpectedly, this study revealed that DNA methylation status of MGMT was positively correlated with mRNA level of MGMT. We speculate the main probable reasons are as follows. In this study, methylation of 9 CpG sites after the transcription start site (sequence range: 14–63 bp) [12] was determined by pyrosequencing assay. Pyrosequencing can detect DNA methylation level in a high throughput manner, but the length of target sequence is very limited, which might lead to decreased efficiency [41, 42]. Another possible reason might be that the population size is not big enough, and in future study more subjects should be enrolled to clarify that point. However, the main reason we suspect is that histone modification might be more essential in regulation of MGMT expression upon PAH exposure-induced DNA damage. Recent studies reveal that H3K36me3 can participate in transcriptional activation of DNA repair genes mainly through regulation of chromatin accessibility [30, 43, 44]. Bioinformatic analysis also indicated that H3K36me3 can physically bind with MGMT gene body. The in vitro cell-based analysis provided additional results that supported these speculations. These findings indicate that DNA methylation is not a unique mechanism in regulation of MGMT expression, which is consistent to a previous study [45]. In conclusion, H3K36me3 modification might be involved in regulation of MGMT expression in response to PAH exposure.
Conclusions
Epigenetic regulation is an important and complex mechanism to regulate the toxic effects of xenobiotics, including PAHs. Crosstalk may take place between different epigenetic modes upon chemical exposure. Yet, the interactions among different epigenetic modes are underestimated in previous research on PAHs. The findings of our study shed light on the associations between histone modification and DNA methylation in response to DNA damage following PAH exposure, which help to elucidate the epigenetic mechanism underlying PAH exposure-induced DNA damage.
Supplementary Material
Funding
This work was supported by the Major Research Plan Integration Project of National Natural Science Foundation of China (91943301), National Natural Science Foundation of China (81872656, 81602882, 81402715), Guangdong Provincial Natural Science Foundation Team Project (2018B030312005).
Contributor Information
Xiumei Xing, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Zhini He, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China; Food Safety and Health Research Center, School of Public Health, Southern Medical University, 1023 South Shatai Road, Guangzhou 510515, China.
Ziwei Wang, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Ziying Mo, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Liping Chen, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Boyi Yang, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Zhengbao Zhang, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Shen Chen, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Lizhu Ye, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Rui Zhang, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Yuxin Zheng, Department of Toxicology, School of Public Health, Qingdao University 38 Dengzhou Road, Qingdao 266021, China.
Wen Chen, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Daochuan Li, Department of Toxicology, Guangdong Provincial Key Laboratory of Food, Nutrition and Health, School of Public Health, Sun Yat-sen University, 74 Zhongshan Road 2, Guangzhou 510080, China.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1. Berger SL, Kouzarides T, Shiekhattar R et al. An operational definition of epigenetics. Genes Dev 2009;23:781–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kundaje A, Meuleman W, Ernst J et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LeBaron MJ, Rasoulpour RJ, Klapacz J et al. Epigenetics and chemical safety assessment. Mutat Res 2010;705:83–95. [DOI] [PubMed] [Google Scholar]
- 4. Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics 2011;6:791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burris HH, Baccarelli AA. Environmental epigenetics: from novelty to scientific discipline. J Appl Toxicol 2014;34:113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perera BPU, Faulk C, Svoboda LK et al. The role of environmental exposures and the epigenome in health and disease. Environ Mol Mutagen 2020;61:176–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J 2009;50:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagamori I, Kobayashi H, Nishimura T et al. Relationship between PIWIL4-mediated H3K4me2 Demethylation and piRNA-dependent DNA methylation. Cell Rep 2018;25:350–6. [DOI] [PubMed] [Google Scholar]
- 9. Chung FF, Herceg Z. The promises and challenges of Toxico-Epigenomics: environmental chemicals and their impacts on the Epigenome. Environ Health Perspect 2020;128:15001–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J, Xing X, Zhang X et al. Enhanced H3K4me3 modifications are involved in the transactivation of DNA damage responsive genes in workers exposed to low-level benzene. Environ Pollut 2018;234:127–35. [DOI] [PubMed] [Google Scholar]
- 11. He Z, Zhang R, Chen S et al. FLT1 hypermethylation is involved in polycyclic aromatic hydrocarbons-induced cell transformation. Environ Pollut 2019;252:607–15. [DOI] [PubMed] [Google Scholar]
- 12. Duan H, He Z, Ma J et al. Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon. Arch Toxicol 2013;87:2013–22. [DOI] [PubMed] [Google Scholar]
- 13. Zhang X, Li J, He Z et al. Associations between DNA methylation in DNA damage response-related genes and cytokinesis-block micronucleus cytome index in diesel engine exhaust-exposed workers. Arch Toxicol 2016;90:1997–2008. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Z, Chen L, Xing X et al. Specific histone modifications were associated with the PAH-induced DNA damage response in coke oven workers. Toxicol Res (Camb) 2016;5:1193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaina B, Christmann M, Naumann S et al. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6:1079–99. [DOI] [PubMed] [Google Scholar]
- 16. Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol 2005;83:429–37. [DOI] [PubMed] [Google Scholar]
- 17. Cabrini G, Fabbri E, Lo Nigro C et al. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (review). Int J Oncol 2015;47:417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakagawachi T, Soejima H, Urano T et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene 2003;22:8835–44. [DOI] [PubMed] [Google Scholar]
- 19. Meng C, Zhu X, Peng G et al. Role of histone modifications and DNA methylation in the regulation of O6-methylguanine-DNA methyltransferase gene expression in human stomach cancer cells. Cancer Invest 2010;28:331–9. [DOI] [PubMed] [Google Scholar]
- 20. Chase KA, Rosen C, Rubin LH et al. Evidence of a sex-dependent restrictive epigenome in schizophrenia. J Psychiatr Res 2015;65:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cantone L, Nordio F, Hou L et al. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ Health Perspect 2011;119:964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bollati V, Baccarelli A, Hou L et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 2007;67:876–80. [DOI] [PubMed] [Google Scholar]
- 23. Zhang W, Zhang J, Yan W et al. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer 2013;119:814–24. [DOI] [PubMed] [Google Scholar]
- 24. Zhai Q, Duan H, Wang Y et al. Genetic damage induced by organic extract of coke oven emissions on human bronchial epithelial cells. Toxicol In Vitro 2012;26:752–8. [DOI] [PubMed] [Google Scholar]
- 25. Jongeneelen FJ, Bos RP, Anzion RB et al. Biological monitoring of polycyclic aromatic hydrocarbons. Metabolites in urine. Scand J Work Environ Health 1986;12:137–43. [DOI] [PubMed] [Google Scholar]
- 26. Kumaravel TS, Jha AN. Reliable comet assay measurements for detecting DNA damage induced by ionising radiation and chemicals. Mutat Res 2006;605:7–16. [DOI] [PubMed] [Google Scholar]
- 27. Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol 1986;51:1173–82. [DOI] [PubMed] [Google Scholar]
- 28. Shin YJ, Kim Y, Wen X et al. Prognostic implications and interaction of L1 methylation and p53 expression statuses in advanced gastric cancer. Clin Epigenetics 2019;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu N, Lee CH, Swigut T et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 2018;553:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Z, Zhang Y, Jia J et al. H3K36me3, message from chromatin to DNA damage repair. Cell Biosci 2020;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Misri S, Pandita S, Kumar R et al. Telomeres, histone code, and DNA damage response. Cytogenet Genome Res 2008;122:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ooi SK, Qiu C, Bernstein E et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007;448:714–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295–304. [DOI] [PubMed] [Google Scholar]
- 34. Brinkman AB, Gu H, Bartels SJ et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 2012;22:1128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo X, Wang L, Li J et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 2015;517:640–4. [DOI] [PubMed] [Google Scholar]
- 36. Pezic D, Manakov SA, Sachidanandam R et al. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 2014;28:1410–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- 38. Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: in the right place at the right time. Science 2018;361:1336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ji W, Yang L, Yu L et al. Epigenetic silencing of O6-methylguanine DNA methyltransferase gene in NiS-transformed cells. Carcinogenesis 2008;29:1267–75. [DOI] [PubMed] [Google Scholar]
- 40. Caiment F, Gaj S, Claessen S et al. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res 2015;43:2525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reed K, Poulin ML, Yan L et al. Comparison of bisulfite sequencing PCR with pyrosequencing for measuring differences in DNA methylation. Anal Biochem 2010;397:96–106. [DOI] [PubMed] [Google Scholar]
- 42. Wojdacz TK, Møller TH, Thestrup BB et al. Limitations and advantages of MS-HRM and bisulfite sequencing for single locus methylation studies. Expert Rev Mol Diagn 2010;10:575–80. [DOI] [PubMed] [Google Scholar]
- 43. Li F, Mao G, Tong D et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013;153:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfister SX, Ahrabi S, Zalmas LP et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014;7:2006–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kreth S, Thon N, Eigenbrod S et al. O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One 2011;6:e17156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


