Abstract
Propionic acidemia (PA) is an autosomal recessive metabolic liver disease caused by defects in propionyl CoA carboxylase. Propionyl CoA carboxylase is a dodecameric enzyme consisting of multiple copies of alpha and beta subunits encoded by the PCCA and PCCB genes. Mutations in either PCCA or PCCB can cause the disease. PA is categorized as a rare disease and accessing patients' cells to study the disease has been challenging. Here we describe the generation of two isogenic induced pluripotent stem cell (iPSC) lines in which exon 2 of the PCCB gene was mutated using CRISPR Cas9 gene editing. The PCCB−/− iPSCs express characteristic pluripotency proteins, are competent to differentiate into cell lineages from each of the three embryonic germ layers and display a normal karyotype.
Resource Table:
| Unique stem cell lines identifier | MUSCSDi001-A-1 |
| MUSCSDi001-A-2 | |
| Alternative names of stem cell lines | PCCB Δ14/Δ29 (MUSCSDi001-A-1) |
| PCCB Δ19/Δ481 (MUSCSDi001-A-2) | |
| Institution | Medical University of South Carolina |
| Contact information of distributor | Stephen Duncan, Duncanst@musc.edu |
| Type of cell lines | iPSCs that have been gene edited using CRISPR/Cas9 |
| Origin | Human |
| Cell Source | Original cell type induced: Human foreskin fibroblast. |
| Clonality | Clonal |
| Method of reprogramming | Transgene free plasmid transfection |
| Multiline rationale | Isogenic clones |
| Gene modification | YES |
| Type of modification | Indel mutation (deletion) |
| Associated disease | Autosomal recessive propionic acidemia (PA) |
| Gene/locus | Exon2 of PCCB 3q22.3 |
| Method of modification | CRISPR Cas9 |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 2016–2019 |
| Cell line repository/bank | The lines had been registered with hpscreg.eu with these unique identifiers, MUSCSDi001-A-1, MUSCSDi001-A-2 |
| Ethical approval | Medical University of South Carolina Stem Cell Research Oversight Committee (SCRO) protocol 8. |
1. Resource utility
The differentiation of PCCB deficient iPSC lines (Table 1) toward affected cell types, such as hepatocytes, will provide a cell culture model to study the mechanisms underlying PA and a platform for the discovery of potential therapeutics.
Table 1.
Summary of lines.
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| MUSCSDi001-A-1 | PCCB Δ14/Δ29 | Male | Newborn | N/A | Allele 1: g.5578_5592del Allele 2: g.5563_5592del |
Autosomal recessive propionic acidemia |
| MUSCSDi001-A-2 | PCCBΔ19/Δ481 | Male | Newborn | N/A | Allele 1: g.5582_5601del Allele 2: g.5586_6067del |
Autosomal recessive propionic acidemia |
2. Resource details
Propionic acidemia (PA, OMIM #606054) is a rare inborn metabolic disease caused by loss of function mutations in the PCCA or PCCB genes that encode the alpha and beta subunits of the propionyl CoA carboxylase enzyme. In the mitochondria, Propionyl CoA carboxylase is responsible for conversion of propionyl CoA to methylmalonyl CoA during branched chain amino acid metabolism. Loss of enzyme activity leads to accumulation of toxic products in the mitochondria. Intermediate metabolites including propionic acid, methylcitrate and propionyl carnitine can be detected in the urine and blood of afflicted patients (Chapman et al., 2016). Here, we describe the generation of iPSCs in which the PCCB gene was disrupted using CRISPR Cas9 mediated gene editing. The parental iPSCs (iPSC-K3) were derived previously from human foreskin fibroblasts (ATCC CRL2097) using reprogramming factors encoding plasmids (Si-Tayeb et al., 2010). Human iPSCs-derived hepatocyte like cells have been used previously to study inborn errors in hepatic metabolism and as a platform for drug discovery (Pournasr and Duncan, 2017). We propose that iPSCs deficient in propionyl CoA carboxylase, will therefore be useful for the study of PA (Table 2).
Table 2.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Data not shown |
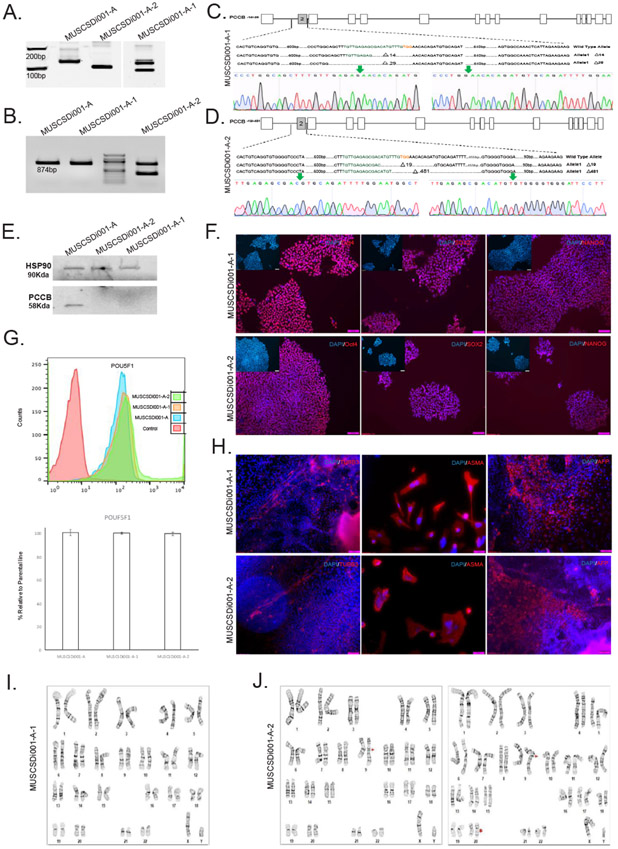
| Phenotype | Qualitative analysis by Immunocytochemistry | Assess staining/expression of pluripotency markers: Pou5f1, Nanog, Sox2 | Fig. 1 panel E |
| Quantitative analysis Flow cytometry | Assess % of positive cells for cell surface marker Pou5f1 more that 85% | Fig. 1 panel F | |
| Genotype | Karyotype (G-banding) and resolution | MUSCSDi001-A-1: 46XY, Resolution 450–525 and MUSCSDi001-A-2: 46XY, add(9)(q21.2)[ll]/46,XY, add(9)(q21.2),dyp(20) (q11.2q11.2)[9] | Fig. 1 panel H, I |
| Identity | STR analysis | 27 allelic polymorphisms across the 15 STR loci analyzed | Submitted in archive with journal |
| Mutation analysis (IF APPLICABLE) | Sequencing | Compound heterozygous frameshift deletions in both cell lines | Fig. 1 panel A, B, and C.I |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by PCR-ELISA. Negative | Supplementary File-1 |
| Differentiation potential | Embryoid body formation | Mesoderm: smooth muscle actin | Fig. 1 panel GI |
| Ectoderm: β-III-tubulin | |||
| Endoderm α-fetoprotein | |||
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
A guide RNA was designed to target PCCB exon 2 (TGTTGAGAGCGACATGTTTG) and was cloned into a plasmid containing Cas9 from S. pyogenes in-frame with a T2A-Puromycin selectable marker (pSpCAs9(BB)-2A-Puro V2.0, PX459) as described elsewhere (Ran et al., 2013). Parental K3 iPSCs (Si-Tayeb et al., 2010) were electroporated with pSpCas9(BB)-2A-Puro-sgPCCB, followed by 48 h of selection in Puro containing medium. After 7–10 days individual colonies were manually collected. Half of each colony was used for genomic DNA extraction while the remainder was plated on a matrigel-coated plate. INDELs were identified by electrophoretic separation of PCR amplicons on acrylamide gels (Fig. 1A). Two clones harbouring compound heterozygous deletions were selected for further analyses. Nucleotide sequencing of the of PCR products (Fig. 1B) using primers described in Table 3 revealed that the MUSCSDi001-A-1 carried a 14 bp deletion (g.5578_5592del, p.S82Rfs*92) on one allele and a 29 bp deletion (g.5563_5592, p.S78Tfs*87) on the other allele. MUSCSDi001-A-2 carried a 19 bp deletion (g.5582_5601del, p.M84Vfs*142) on one allele and a 481 bp deletion (g.5586_6067del, p.V86Sfs*142) on the other allele. (Fig. 1C, D). All these frameshift mutations were predicted to lead to the loss of propionyl CoA carboxylase, which was subsequently demonstrated by western blot analysis in iPSCs (Fig. 1E). The PCCB knockout iPSCs had a characteristic pluripotent morphology with a small cytoplasmic morphology, epithelial characteristic, and growth in colonies. Both iPSC lines expressed the pluripotency proteins Pou5f1 (also called Oct4), Nanog, and Sox2 (Fig. 1F). FACS analysis of Pou5f1 showed comparable levels to the parental cell line (Fig. 1G). Quantification of SOX2 immunostaining also showed more than %95 positive cells in both cell lines (Supplementary Fig. 1). Moreover, the cells were able to spontaneously differentiate as embryoid bodies into derivatives of all three germ layers (Fig. 1H). G-band chromosome analyses revealed that MUSCSDi001-A-1 had a normal karyotype (Fig. 1I). However, in iPSC MUSCSDi001-A-2 11 of 20 cells contained an unbalanced structural aberration in the long (q) arm of chromosome 9 (image on left Fig. 1J). The additional genetic material that had translocated to chromosome 9q, could not be characterized by G-banding. The cells (9 of 20 cells; image on right Fig. 1J) also contain an interstitial duplication in the long (q) arm of chromosome 20. There is a known recurrent acquired duplication at this location in human pluripotent stem cell cultures. Finally, both iPSC lines were confirmed to originate from K3 iPSCs using Short Tandem Repeat (STR) analysis and were confirmed to be free mycoplasma.
Fig 1.
Table 3.
Reagents details.
| Antibodies used for immunocytochemistry/flow-citometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Rabbit anti-NANOG | 1:200 | Cell Signaling Technology Cat# 4903, RRID:AB_10559205 |
| Rabbit anti-Sox2 | 1:100 | Abcam Cat# ab15830, RRID:AB_443255 | |
| Rabbit anti-Pou5f1 | 1:100 | Santa Cruz Biotechnology Cat# sc-9081, RRID:AB_2167703 | |
| Differentiation Markers | Mouse Anti-AFP | 1:100 | Sigma-Aldrich Cat# A8452, RRID:AB_258392 |
| Rabbit Anti- αSMA | 1:100 | Abcam Cat# ab124964, RRID:AB_11129103 | |
| Mouse Anti- TUBB3 | 1:100 | BioLegend Cat# 801202, RRID:AB_10063408 | |
| Secondary antibodies | Alexa flour 594 donkey anti rabbit | 1:1000 | Thermo Fisher Scientific Cat# A-21207, RRID:AB_141637 |
| Alexa flour 488 donkey anti rabbit | 1:1000 | Thermo Fisher Scientific Cat# A-21206, RRID:AB_2535792 | |
| Alexa flour 594 goat anti mouse | 1:1000 | Thermo Fisher Scientific Cat# A-21203, RRID:AB_2535789 | |
| Thermo Fisher Scientific Cat# A-21203, RRID:AB_2535789 | |||
| Primers | |||
| Target | Forward/Reverse primer (5′-3′) | ||
| Genotyping | PCCB | Fwd-120:GGAAAGCTAACAGCCAG/Rev120:CTTATTCTTATCAGCAGC | |
| Fwd131:TTCCATTGTAGGGAAAGC/Rev131:CTTATTCTTATCAGCAGC | |||
| Targeted mutation analysis/sequencing | PCCB | Fwd-XbaI:TTTTCTAGAGGAAAGCTAACAGCCAG/Rev-EcoRI:AACGAATTCCTTATTCTTATCAGCAGC | |
| Fwd-XbaI:TTTTCTAGATTCCATTGTAGGGAAAGA//Rev-EcoRI:AACGAATTCCTTATTCTTATCAGCAGC | |||
| Fwd615: CTTCCACAGTTGGGTGGCT /Rev615: AAGGCAGGGTACGGTCGCT | |||
| Fwd1230: CACTGTCAGGTGTGGGGTCCCT/Rev1230: ATGAGTTTGGCCACTATGCCTCA | |||
In summary, we have generated two iPSCs with deletions in the PCCB gene using CRISPR Cas9-mediated gene editing. Both lines displayed characteristics of iPSCs including the capacity to differentiate into all three germ layers. Although one of the lines had a normal chromosomal content and will be useful to examine the mechanistic basis of PA, the other line had genetic abnormalities that, if used, must be considered when interpreting any results.
3. Materials and methods
3.1. Cell culture
The iPSCs were cultured on an eCadherin-coated plate in mTeSR1 medium (Nagaoka et al., 2010) containing 40 ng/ml zebrafish fibroblast growth factor (zbFGF) in a humidified chamber at 37 °C, 4% O2, and 5% CO2. Cells were split one to ten approximately 4–6 days after reaching optimal confluency using Versene solution (Termofisher/Gibco, #15040-066). Cell lines were confirmed free of mycoplasma contamination using the Mycoplasma Detection PCR ELISA Kit (Roche, Germany, #11663925910).
3.2. CRISPR Cas9-mediated PCCB mutation
For PCCB mutation, 10 × 107 iPSCs were electroporated with 60 μg pSpCas9(BB)-2A-puro-sgPCCB plasmid using a BTX electroporator. pSpCas9(BB)-2A-Puro (PX459) V2.0 was a gift from Feng Zhang (Addgene plasmid # 62,988; http://n2t.net/addgene:62988; RRID:Addgene_62988). Soon after electroporation, cells were plated on Matrigel-coated plates in mTeSR1 supplemented with 10 μM Y-27632 (StemRD, CA, #146986-50-7). Twenty-four hours later, medium was supplemented with 1 μg/ml puromycin and cells cultured for 48 h. The medium was then replaced with mTeSR1 and culture continued until individual colonies could be collected. Colonies were treated with 10 μM Y-27632 for 2 h before individual clones were collected by scratching. Around half of each colony was expanded in a single well from a 24-well Matrigel coated plate. The remainder of the colony provided genomic DNA for PCR analyses.
3.3. Genotyping and sequencing
Genomic DNA was extracted from iPSC clones using QuickExtract™ DNA extraction solution (Epicenter, WI, #QE09050). The targeted region of PCCB gene was amplified using Herculase Fusion Polymerase (Agilent, CA, #600675) with the primers listed in Table 3. PCR amplicons were run on Novex 4%–20% TBE gels (ThermoFisher/Invitrogen, CA, #EC6225BOX) to identify INDELs (Fig. 1A,B). Deletions were confirmed within amplicons by DNA sequencing and loss of protein was confirmed by western blot. Nucleotide sequencing was performed by Retrogen Inc. The results were aligned using SnapGene and CLC sequence viewer software (Fig. 1C, D).
3.4. Western blot
Whole cell lysates were collected from the iPSCs using RIPA buffer with protease inhibitor cocktail (ThermoFisher Scientific, NY, #78443). 20-30ug total protein was separated by SDS-PAGE using Any kD Mini-protean TGX stain-freeTM precast gels (BioRad, CA,#4568123), and transferred to PVDF membrane using the Trans-Blot TurboTM Transfer System (BioRad, CA, #1704155). Membranes were incubated overnight with antibodies listed at Table 3 at 4 °C. HRP-conjugated secondary antibodies were used at a dilution of 1:5000. Protein levels were quantified using BioRad stain-free Imaging System using Image Lab software from BioRad. Antibodies used are listed in Table 3.
3.5. Immunocytochemistry for pluripotency and differentiation markers
Cultured cells were fixed with 4% PFA for 20 min and made permeable using 0.4% Triton X-100 in PBS for 20 min. Cells were treated with 3% BSA in PBS for 1 h at room temperature followed by overnight incubation with the primary antibody at 4 °C. Cells were rinsed with PBS 3 X 5 min and incubated with secondary antibody for 1 h at room temperature. Control and experimental wells were processed identically.
Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich). Images were taken using IX51 Olympus fluorescence microscope and merged and quantified using Image J software. Antibodies used are listed in Table 3.
3.6. Embryoid body (EB) formation
The iPSCs were detached from eCadherin matrix using Versene solution (Life technologies, #15040–066) and transferred into suspension culture plates in complete mTESRl medium containing 10 μM Y-27632. Forty-eight hours after plating, Y-27632 was removed and spheres were cultured for another 8 days as suspension cultures in DMEM/Ham’s F12 containing 20% knock-out serum replacement, NEAA and Pen/Strep without bFGF. Thereafter, EBs were then transferred onto a gelatin or matrigel-coated plate and maintained in the same medium for another 8 days.
4. Karyotyping and cell authentication
G-banded karyotyping and Short Tandem Repeat Analyses were performed by WiCell .
Supplementary Material
Acknowledgement
Work was supported by funds from the National Institutes of Health (DK102716, DK119728, DK123704, GM130457, and CA138313).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2020.101953.
References
- Chapman KA, Collado MS, Figler RA, Hoang SA, Armstrong AJ, Cui W, Purdy M, Simmers MB, Yazigi NA, Summar ML, Wamhoff BR, Dash A, 2016. Recapitulation of metabolic defects in a model of propionic acidemia using patient-derived primary hepatocytes. Mol. Genet. Metab 117 (3), 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka M, Si-Tayeb K, Akaike T, Duncan SA, 2010. Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev. Biol 10, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournasr B, Duncan SA, 2017. Modeling Inborn Errors of Hepatic Metabolism Using Induced Pluripotent Stem Cells. Arterioscler. Thromb. Vase. Biol 37 (11), 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8 (11), 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW, Duncan SA, 2010. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev. Biol 10, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.