Abstract
Emergence and re-emergence of pathogens bearing the risk of becoming a pandemic threat are on the rise. Increased travel and trade, growing population density, changes in urbanization, and climate have a critical impact on infectious disease spread. Currently, the world is confronted with the emergence of a novel coronavirus SARS-CoV-2, responsible for yet more than 800 000 deaths globally. Outbreaks caused by viruses, such as SARS-CoV-2, HIV, Ebola, influenza, and Zika, have increased over the past decade, underlining the need for a rapid development of diagnostics and vaccines. Hence, the rational identification of biomarkers for diagnostic measures on the one hand, and antigenic targets for vaccine development on the other, are of utmost importance. Peptide microarrays can display large numbers of putative target proteins translated into overlapping linear (and cyclic) peptides for a multiplexed, high-throughput antibody analysis. This enabled for example the identification of discriminant/diagnostic epitopes in Zika or influenza and mapping epitope evolution in natural infections versus vaccinations. In this review, we highlight synthesis platforms that facilitate fast and flexible generation of high-density peptide microarrays. We further outline the multifaceted applications of these peptide array platforms for the development of serological tests and vaccines to quickly encounter pandemic threats.
Keywords: infectious diseases, epitope mapping, microarrays, array synthesis technologies
1. Introduction
In a pandemic situation, the outbreak of an infectious disease has spread globally with a major impact on morbidity and mortality. Besides a severe negative impact on health, pandemics are accompanied by an enormous economic loss, as well as social and political implications. Pandemics have threatened humankind for centuries, such as the Bubonic plague (Black Death) pandemic (14th century), the fifth cholera pandemic (19th century), as well as the influenza and human immunodeficiency virus (HIV) pandemics in the 20th and 21st century.1 Most pandemics found their origin from zoonotic transmissions from domesticated animals (e.g., avian influenza) or wildlife (e.g., Ebola). Since late 2019, humanity is faced with the currently ongoing Coronavirus disease (COVID)-19 pandemic responsible for more than 23 million infections and over 800 000 deaths worldwide (as of 24th of August 2020).2,3
The definition of a pandemic is still the subject of debate because of its multifactorial and multidisciplinary extent. A recent definition by the dictionary of epidemiology states that a pandemic is “an epidemic occurring over a very wide area, crossing international boundaries, and usually affecting a large number of people. Only some pandemics cause severe disease in some individuals or at a population level.4 In contrast, an epidemic just differs in size of the area where a new or re-emerging pathogen causes disease, while an outbreak is even more localized.4,5 Factors that define the severity of a pandemic, such as spread and transmissibility, case/fatality rate, immunity of a population, time span of the asymptomatic phase (facilitating undetected distribution), a challenging clinical picture (difficult to be differentiated or diagnosed), and economic impact, are not included in such definitions.6,7
The dramatic Ebola virus (EBOV) outbreak in West Africa in 2013–2016 has shown the need for a preparedness strategy against pathogens with epidemic potential. In the aftermath of the outbreak, the World Health Organization (WHO) initiated a blueprint for research and development to accelerate diagnostics, therapeutics, and vaccines.8 This initiative is a response to the experience with past epidemics, highlighting the need to improve emergency preparedness.
Thus, the rapid development of diagnostic measures and intervention strategies is of utmost importance to combat the emergence of (novel) pathogens, causing life-threatening diseases. Understanding the host immune defense mechanisms and identifying the pathogens Achilles heel can guide the design of drugs and vaccines. Humoral responses are known to play a vital role in clearing many infections.9 Moreover, pathogen-specific antibody responses are often used as a basis for serological diagnostics.10 Hence, the in-depth analysis of the underlying antigens of (protective) immune responses eliciting (1) neutralizing antibodies, (2) (early) antibodies that can be used for diagnostics, and (3) antibodies that can be applied for epidemiological or immune monitoring studies is necessary to control pandemic infections.
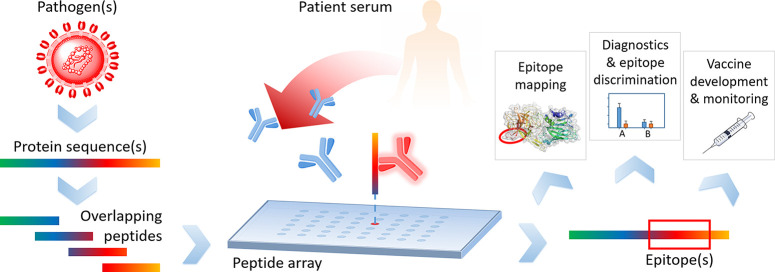
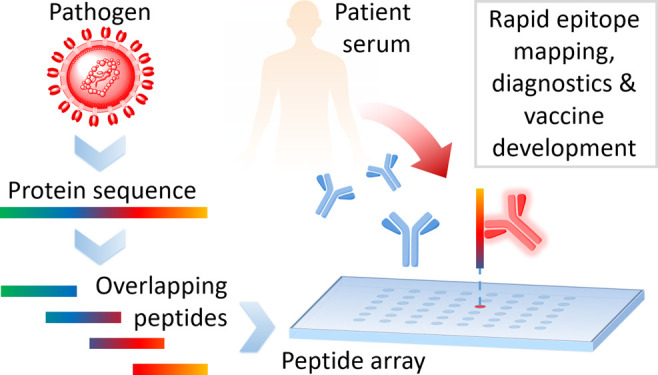
Peptide microarrays11 are an ideal tool to decipher epitope-specific humoral immune responses toward the (full) proteome of an emerging pathogen (Figure 1). They enable the analysis of tens of thousands of peptides simultaneously in a fast and cost-effective way for applications, such as epitope mapping, diagnostics, epitope discrimination, vaccine development, and vaccine monitoring. Other technologies, enabling insights into antibody responses on epitope level encompass phage display12−18 (or related display technologies), bead-based multiplex systems,19−22 and peptide-based enzyme-linked immunosorbent assays (ELISA).23−26 A difference in the mentioned technologies is certainly the number of parameters (here peptides), which can be measured simultaneously per sample. ELISA, with the lowest number of possible parameters per sample (1 peptide per sample), is followed by bead-based multiplex systems (up to ∼500 peptides per sample),20 peptide microarrays (typically 500–50 000 peptides per sample), and phage display (library size up to 109–1010).12 Unique for the latter one is the generation of phage particles, expressing an unrivaled diversity of peptides. However, it apparently also has some inherent bias, since it is a biological workflow, prone to for example unspecific binding (e.g., the VirScan approach14 did not identify a highly common polio virus epitope). The prerequisite of a presynthesis of peptides in larger scale is common for ELISA and bead-based multiplex systems. For peptide microarrays, pre- or in situ synthesis of peptides is possible, depending on the technology used for microarray production, as described in the next section.
Figure 1.
Typical workflow of a peptide microarray experiment. The pathogens of interest are selected; their protein sequences are cut into overlapping peptides, and these peptides are then synthesized on peptide microarrays. Patient samples are incubated on the arrays and serum antibodies bind to distinct epitopes. This information is the basis for many different applications.
In this Review, we discuss the currently available peptide microarray synthesis technologies, highlighting those techniques with the capacity to rapidly generate thousands of peptides. Then, we give an overview on their applications in the field of epidemic/pandemic infectious diseases, where parts or the full proteome of an emerging or mutated pathogen were screened, to quickly respond to a pandemic threat.
2. Peptide Microarrays and Rapid Production Technologies
Since the advent of parallel peptide synthesis in the 1980s, peptide and array production methods have become much more refined and highly automated. Today, peptide arrays are versatile, widespread, and easily accessible tools for research. They are used for antibody profiling, for the mapping of epitopes, to study ligand–receptor interactions, or to determine substrate specificities of enzymes.11,27 Analogous to the enzyme-linked immunosorbent assay (ELISA), they offer the parallel analysis of thousands of peptides, resulting immunogenicity maps of proteins or full proteomes in single amino acid resolution.
In 1992, Ronald Frank invented the SPOT synthesis,28 which revolutionized the automated production of peptides and peptide arrays: Metal pins or pipettes spot dissolved amino acid building blocks onto a cellulose surface in order to parallelize Merrifield’s Nobel Prize awarded solid-phase peptide synthesis (Figure 2).29 Together with advanced activation and protecting group strategies, peptides are elongated separately on each spot. The standard protocol for a synthesis cycle consists of (1) printing of building blocks, (2) coupling reaction, (3) washing, and (4) deprotection. Nowadays, chain lengths of more than 25 amino acids can be reached. For decades, the SPOT method represented the gold standard in peptide and peptide array synthesis. Yet, the method has one decisive drawback: it only achieves a spot density of ∼25 spots per cm2, which translates into a limited parallelized production output and expensive arrays with rather high chemical costs. To manufacture peptide arrays with higher density, the presynthesized peptides are solubilized and afterward respotted in higher density onto glass substrates. This procedure is widespread and readily available in many different facets. It is commonly used to produce many array replicates, but it is still relatively slow and costly. An interesting concept to miniaturize this approach is to adapt principles from atomic force microscopy to deliver minute amounts of liquids selectively on a surface for synthesis.30 This approach can enable arrays with very high spot densities, although an automated production setup is still lacking.
Figure 2.
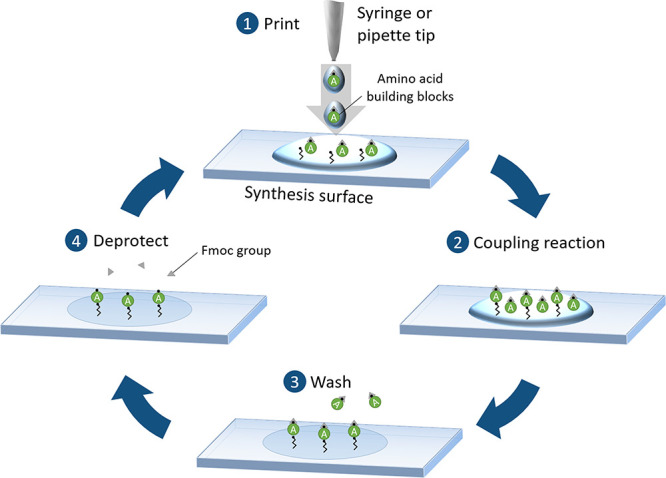
SPOT synthesis technology. (1) Syringe or pipet tip is used to dispense solvents with solubilized amino acid building blocks onto a surface. (2) Coupling reaction proceeds directly upon contact with the surface-bound free reactive groups. (3) Excess and nonreacted building blocks are removed by washing and (4) the fluorenylmethyloxycarbonyl (Fmoc) protecting group is removed in a subsequent chemical washing step. SPOT synthesis is the gold standard in the field, offering reliable access to peptides, but with a limited number of peptides.
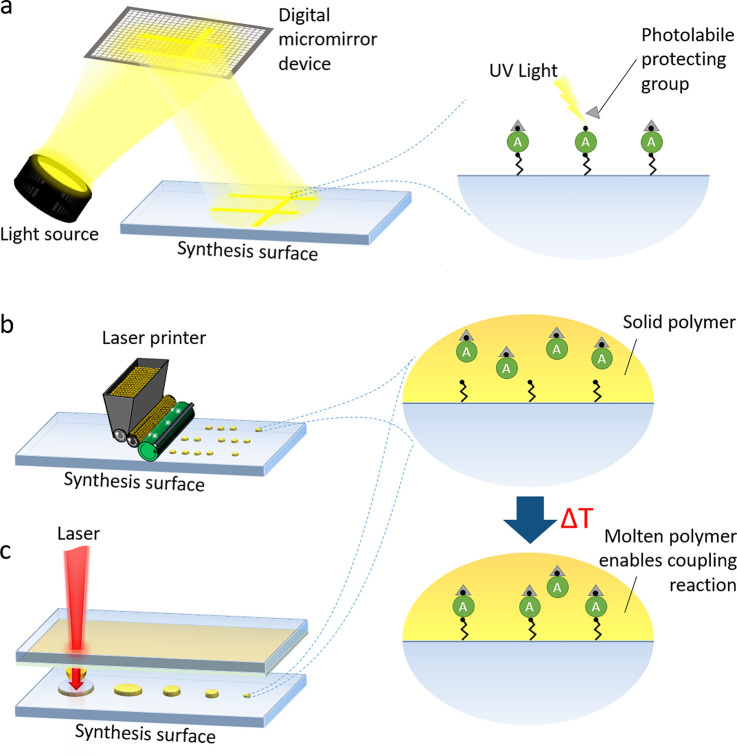
To immediately react to an emerging pathogen, rapid production of individual arrays with thousands of sequences is needed. Here, in situ synthesis technologies have favorable lead times and much higher parallelization capabilities. They allow for an on-demand combinatorial preparation of minimal amounts of peptides directly on a microarray surface. Although these peptides are of somewhat lower quality (i.e., unpurified, including truncated sequences), they are attractive for serum profiling and antibody screenings, since antibodies can still find their binding counterpart within the crude peptide mixture on the surface. In principle, all of these in situ techniques are directly or indirectly based on selective light irradiation.
A prominent in situ synthesis approach is based on the photolithographic principle (Figure 3a), which is lent from semiconductor chip fabrication. Almost 30 years ago, Fodor et al.31 laid the foundation and showed that amino acids, carrying a photolabile protecting group, can be selectively deprotected by light, using a lithographic mask. Meanwhile, projection technology has significantly progressed due to advanced microelectromechanical systems engineering, and nowadays, digital micromirror devices have replaced laborious mask production. While Fodor et al. successfully switched their focus to lithographic synthesis of DNA arrays, others followed the path of lithographically synthesizing peptide microarrays. Several sophisticated variants of this approach have emerged.32−35 However, the main disadvantages regarding deprotection efficiency (photocleavage efficiencies is often <80%), throughput (synthesis setup is occupied for the synthesis of one complete array), contamination issues (stray light or diffusion of photo acids), and the requirement of sophisticated equipment have apparently limited general market access.
Figure 3.
Selected high-throughput technologies for the synthesis of peptide microarrays. Lithographic methods (a) use a digital micromirror device to cleave photolabile protecting groups from amino acid building blocks via illumination, offering much higher spot densities. Solid material-based synthesis methods, such as the laser printer technology (b) or the combinatorial laser-induced forward transfer approach (c), offer a highly parallelized peptide array synthesis. Both rely on the deposition of at room temperature solid polymer, which embeds the amino acid building blocks. Only after several minutes of heating in an oven, the coupling reaction begins. Since this allows for the separation of patterning and coupling steps, these approaches can yield shorter process times for a rapid production of arrays.
Other approaches leverage solid polymers, which serve as at room temperature solid solvents for peptide synthesis. These solid material-based peptide synthesis technologies facilitate a highly flexible and affordable production of high-density peptide microarrays. In contrast to other approaches with dissolved building blocks, a solid polymer embedding the amino acid building blocks is delivered as spots to a surface (Figure 3b and c). The solid solvent has two major advantages: It does not spread on the synthesis surface, facilitating high spot densities, and it does not evaporate, allowing for the parallelization of chemical processing. As a key difference to the SPOT synthesis, after deposition of the solid solvent (i.e., polymer), a several minutes long heating step induces melting and the amino acids couple within the polymer spots to the surface. A modified laser printer (Figure 3b) with 20 amino acid printing units is one of the technologies for the deposition of amino acid toner particles onto a functionalized glass surface.36 Variants of this technology employed a semiconductor chip and laser fusing to selectively deposit polymer toner particles.37,38 More recently, the combinatorial laser-induced forward transfer technology (Figure 3c) was developed,39,40 which can be used for an automated peptide array synthesis. Furthermore, a low-budget variant is available for research laboratories.41
While many sophisticated peptide synthesis technologies have been invented, only the SPOT synthesis with its many variants and the laser printer approach are currently broadly commercially available. Technologies, which enable the rapid synthesis of thousands of different biomolecules, can play a crucial part in future diagnostics and vaccine development. Yet, there are still challenges for the production of peptide microarrays to be resolved. Usually, only linear peptides are produced but also cyclic constrained peptides are possible. These can mimic looped epitope structures, enabling the detection of (continuous) conformational epitopes. However, discontinuous conformational epitopes are still difficult to construct. Other, more cumbersome synthesis routes have to be followed to enable such arrays.42,43 Another challenge concerns posttranslationally modified peptides (phosphorylation, glycosylation, hydroxamic acid-modification,44 etc.), which are currently difficult and expensive to be synthesized in situ in the array format.
In the following sections, we give an overview on the applications of peptide microarrays for the analysis of pathogens with epidemic or pandemic potential. Not all arrays were produced with the aforementioned rapid production technologies (lithography, laser printing), since many pathogens have been circulating for longer whiles. Yet, the current SARS-CoV-2 outbreak highlights the need for technologies, which enable a rapid response to tackle research questions arising from endemic or pandemic threats.
3. Application of Peptide Microarrays for Pandemic Diseases
Peptide microarrays have been applied for many different infectious diseases. Table 1 gives a literature overview of peptide array applications for SARS-CoV-2, Ebola, influenza, flaviviruses, HIV, and Chikungunya virus, spanning from infection- to vaccine-induced screenings.
Table 1. Overview on the Diverse Applications of Peptide Microarrays in the Field of Pandemic Pathogensa.
| pathogen | keywords with respect to peptide microarray applicationsb | peptide array content | studied immune response | sample origin | ref |
|---|---|---|---|---|---|
| SARS-CoV-2 | proteome-wide profiling of antibody responses on epitope level | proteome as overlapping peptidesc | infection-induced | human | (45, 46) |
| epitope mapping | spike protein as overlapping peptides | infection-induced | human | (47) | |
| peptide-PNA library of Spike protein | infection-induced | human | (48) | ||
| Ebola | epitope mapping | surface glycoprotein as overlapping peptides | infection- and vaccination-induced | human | (49) |
| influenza | proteome-wide profiling of antibody responses on epitope level | proteome of influenza A virus (H1N1) and HA proteins of other Influenza A subtypes as overlapping peptides | infection- and vaccination-induced | human | (50) |
| immunosignatures to predict vaccine efficacy | random peptide array | vaccination-induced | mouse | (51) | |
| HA peptides | vaccination-induced | human | (52) | ||
| comparative study on different vaccine approaches | 4 HA proteins as overlapping peptides | vaccination-induced | mouse | (53) | |
| peptide-based influenza inhibitors | 1 peptide in 152 mutant variants (site directed substitution of amino acids) | N/D | labeled virus strain | (54) | |
| antibody characterization, epitope mapping | nonstructural protein 1 as overlapping peptides | N/D | mAb | (55) | |
| flaviviruses | antibody profiling, discriminative epitopes, differential diagnostics | proteomes of different arboviruses | infection-induced | monkey | (56) |
| peptides derived from different arboviruses | infection-induced | human | (57) | ||
| proteome of TBEV as overlapping peptides | infection- and vaccination-induced | human | (58) | ||
| ZIKV proteome as overlapping peptides | infection-induced | human | (59) | ||
| proteomes of different arboviruses as overlapping peptides | infection- and vaccination induced | human | (60) | ||
| epitope mapping, molecular basis on differential antibody binding | ZIKV nonstructural protein 1 sequences as overlapping peptides | infection-induced | human | (61) | |
| immune-diagnostics, linear epitopes vs discontinuous epitopes | ZIKV nonstructural protein 1-derived peptides | infection-induced | human | (43) | |
| antibody characterization, epitope mapping | DENV2 nonstructural protein 4B as overlapping peptides | N/D | mAb | (62) | |
| ZIKV envelope protein-derived overlapping peptides | N/D | purified Ab (rabbit) | (63) | ||
| epitope mapping, potential correlates of protection | DENV nonstructural protein 1-derived overlapping peptides | infection (human)- and vaccination (mouse)-induced | mouse, human | (64) | |
| immunosignature analyses | non-natural randomized peptides | N/D | purified Ab (mouse) | (65) | |
| epitope mapping | ZIKV envelope protein-derived peptides | vaccination-induced | monkey | (66) | |
| HIV | specificities of bNAbs | HIV envelope proteins as overlapping peptides (different subtypes) | infection-induced | human | (67) |
| humoral immune response post vaccination (clinical) | HIV envelope proteins as overlapping peptides (different subtypes) | vaccination-induced | human | (68−70) | |
| HIV envelope proteins as overlapping peptides (different subtypes) | vaccination- and infection induced | human | (71) | ||
| GB virus C/HIV-1 coinfections, diagnostics | GBV-C E2 protein-derived peptides | infection-induced | human | (72) | |
| humoral immune response post vaccination (preclinical) | HIV envelope protein-derived overlapping peptides | vaccination-induced | rabbit | (73) | |
| HIV envelope protein-derived overlapping peptides (different subtypes) | vaccination-induced | monkey | (74−76) | ||
| library of peptides derived from HIV-1 proteome covering the global sequence diversity | vaccination-induced | guinea pig | (77,78) | ||
| epitope diversity | library of peptides derived from HIV-1 proteome covering the global sequence diversity | infection- and vaccination-induced | human, monkey, guinea pig | (79) | |
| microarray-based diagnostics, antibody monitoring | HIV-1 clade C peptides and proteins covering the proteome | infection-induced | human | (80) | |
| antibody response, analytical treatment interruption | library of peptides derived from HIV-1 proteome covering the global sequence diversity | infected/vaccinated/ART | human | (81) | |
| bNAbs, CDR-H3, epitope recognition | HIV-1 envelope protein as overlapping peptides | vaccination-induced | mouse | (82) | |
| CHIKV | proteome-wide profiling of antibody responses on epitope level | proteome as overlapping peptides | infection- and vaccination-induced | human | (83) |
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; refs, references; mAb, monoclonal antibodies; PNA, peptide nucleic acid; HA, hemagglutinin; TBEV, tick-borne encephalitis virus; bNAbs, broadly neutralizing antibodies; ART, antiretroviral therapy; HIV, Human immunodeficiency virus; CDR-H3, heavy chain complementarity determining region 3; CHIKV, Chikungunya virus.
See original articles for more detailed information.
Different peptide to peptide overlaps.
3.1. Coronaviruses
Coronaviruses (CoV) belong to the subfamily Coronavirinae, in the family Coronaviridae of the order Nidovirales. CoVs can infect humans and may cause respiratory, enteric, and central nervous system diseases. Prior to 2002, four human CoVs were identified (HCoV-229E, HCoV-OC43, HCoV-NL63, HKU1) that showed only mild disease progression in humans.84 Thus, CoVs were considered to be not highly pathogenic, until the emergence of the severe acute respiratory syndrome (SARS)-CoV in 2002/2003.85−87 Ten years later, the Middle East respiratory syndrome (MERS)-CoV was identified 2012 in Saudi Arabia, which causes severe disease in humans and had a fatality rate of 34.4%.88 The virus is generally transferred to humans from infected dromedary camels. Now, seven years later, the world is facing another CoV outbreak with SARS-CoV-2, causing the Coronavirus disease (COVID)-19.89 It is assumed that SARS-CoV-2 already began to spread in December 2019. In March 2020, the WHO declared this outbreak as a pandemic. With more than 800 000 deaths (as of 24 August 2020), this outbreak is the most dramatic one in the history of CoVs.
All three viruses, SARS-CoV, MERS-CoV, and SARS-CoV-2, are highly pathogenic and, therefore, are listed in the WHO Blueprint as priority pathogens,8 warranting epidemic preparedness strategies.
In contrast to SARS-CoV, SARS-CoV-2 is easily transmitted, generally through respiratory droplets and aerosols. Tracking the virus is a challenge due to the generally mild symptoms, lack of testing, and the high infectivity of the virus. Although SARS-CoV-2 has only been identified around January 2020, there is a large magnitude of peer-reviewed (>7000 COVID-19-related publications)90 and non-peer-reviewed (>1900 COVID-19-related preprints)90 articles, describing many aspects of the virus and its course of disease. However, because of the short period of time since the outbreak, there is still scarce knowledge on SARS-CoV-2 and COVID-19. Reverse transcription polymerase chain reaction (RT-PCR) diagnostics and ELISA tests have their limitations, since PCR can only detect an acute infection with sufficient viral loads, while ELISA tests have some sensitivity issues, as well as false positive results.91 Thus, many questions regarding immune responses to SARS-CoV-2 are still open and need to be addressed in the near future.
While acute infections are identified by PCR, antibody responses to SARS-CoV-2 are generally measured by ELISA against the spike glycoprotein (S GP), and more specifically against the S1 subunit. Serological assays are crucial for patient contact tracing, identifying asymptomatic spreads, as well as seroconversion in populations. Amanat et al.92 developed an ELISA with the full-length S GP ectodomain (AA1–1213) and the receptor-binding domain (AA319–541) with the signal peptide (AA1–14) using expression in mammalian cells or in insect cells. Testing patient sera from different time points after onset of symptoms revealed that the full-length S GP has improved performance in the detection of seroconversion. Here, peptide microarrays can give a more detailed picture, enabling the differential binding studies toward the complete sequence. This can yield important biomarkers (some may directly correlate with protection or disease progression), as well as information on domain specificity and, together with structural protein data, epitope accessibility.
In a preprint, Wang and colleagues45 identified IgG and IgM targeted epitopes from ten COVID-19 patient sera with whole proteome peptide microarrays. They found one epitope of a potential neutralizing antibody in the receptor-binding domain (RBD, AA456–460) of the spike protein. Furthermore, in another preprint, Dahlke et al.46 reported the kinetics of the development of SARS-CoV-2-specific IgA, IgG, and IgM antibody responses within patients in relation to clinical features using whole proteome peptide microarrays. The promptness of antibody development in patients may be related to the severity of symptoms and progression of the disease. Furthermore, results showed an early IgA response within a few days after onset of symptoms, which may be picked up earlier than standard ELISA testing. In both studies, overlapping peptide sequences of the spike glycoprotein were identified in the RBD (AA446–463) and the fusion peptide (AA806–831). The use of peptides with more sequence overlap (15-mer peptides with 13 amino acids overlap) enables a more precise detection of linear epitopes. Others reported in a preprint47 that they have identified three linear epitopes using 12-mer peptides with an overlap of 6 amino acids with 55 convalescent sera, which were used for the enrichment of spike protein neutralizing antibodies. Finally, the Winssinger lab reported in a recent preprint study48 on a variant peptide array approach. They used 12-mer peptide–peptide nucleic acid (PNA) conjugates with an overlap of 6 amino acids of the spike ectodomain, coupled to a DNA microarray via hybridization. These investigations revealed epitopes in the spike glycoprotein regions between AA553–684 and AA764–829. For the development of immunity, neutralizing antibodies against the spike glycoprotein will be of vast impact.
The SARS-CoV-2 research field is progressing quickly and more samples are currently analyzed with microarrays and should be publicly available soon. Screening longitudinal samples from patients will allow to follow the individual epitope development (epitope spread or reduction) over the course of the disease and afterward, as well as to analyze long-term epitope stability many months after the infection.
3.2. Ebola Virus Disease
The Ebola virus disease (EVD) is one of the most threatening illnesses due to high fatality rates. The 2013–2016 outbreak of the Ebola virus (EBOV) in West Africa was the largest since its discovery in 1976 and resulted in 11 308 deaths.93 This outbreak affected for the first time more than one country, with a high number of cases in Guinea, Liberia, and Sierra Leone. The world was ill-prepared to limit the spread of the virus, highlighting the need to improve emergency response.
The disease can be diagnosed by RT-PCR or immunoassays, such as enzyme-linked immunosorbent assay (ELISA) or simple lateral flow immunoassays for antigen detection.
However, the need for high biosafety standards results in limited access to human samples. Becquart et al.94 used overlapping 15-mer peptides of the surface glycoprotein (GP), nucleoprotein (NP), and matrix viral proteins VP40 and VP35 for an ELISA study of pooled sera, collected from EVD survivors 7 days after the end of symptoms and 7–12 years postinfection in comparison with seropositive asymptomatic individuals. They found early humoral responses mostly against GP peptides.
Neutralizing antibodies against the EBOV GP can prevent infections, demonstrating a straightforward way for an efficient vaccination strategy or antibody-based treatment. Saphire et al.95 analyzed a global collection of monoclonal antibodies against EBOV GP regarding neutralization, effector function, and binding site. To identify the binding epitopes, ELISA assays, together with time-consuming studies with electron microscopy and alanine scanning mutagenesis of the EBOV GP were performed.
Several vaccine candidates were tested during the outbreak, but so far, only the recombinant vesicular stomatitis virus (VSV)-based vector carrying the EBOV GP (rVSV-ZEBOV) became a licensed EBOV vaccine. To draw a clear picture of the antibody response to rVSV-ZEBOV in patients, Ehrhardt et al.96 investigated the B cell and humoral immune response 18.5 to 26 months after single-dose vaccination in comparison to seven EVD survivors. The vaccination elicits potent neutralizing antibodies with a broad spectrum of targeted epitopes. Additionally, the distribution of targeted epitopes coincides with antibodies identified by others97,98 and those from EVD survivors.
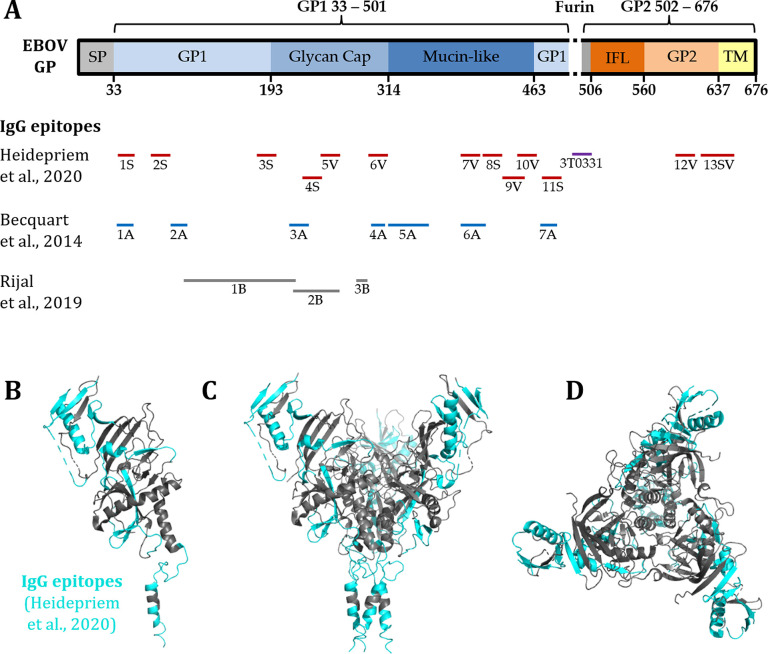
Showing a simple and rapid approach with high-density peptide arrays, Heidepriem and Krähling et al.49 identified epitopes of EBOV S GP targeted by the antibodies of vaccinees with rVSV-ZEBOV and an EVD survivor (Figure 4). By mapping the whole S GP as overlapping peptide fragments in single amino acid resolution, new epitopes were recognized, and overlaps with epitopes from the literature were confirmed. In addition, a highly selective binding epitope of one neutralizing monoclonal antibody elicited from vaccination with rVSV-ZEBOV could be verified.
Figure 4.
Immunogenic epitopes of the EBOV GP (676 AA), identified with peptide microarrays (figure derived from Heidepriem et al.;49 reprinted with permission). (A) Comparison of the immunogenic IgG epitopes from Heidepriem et al. (S, survivor; V, vaccinee) in the EBOV GP with the published epitopes from other human response studies (Becquart et al., Rijal et al.).94,99 (B–D) 3D view of the EBOV GP trimer structure with the in Heidepriem et al. identified IgG epitopes highlighted in cyan.
3.3. Influenza Virus
Influenza pandemics have threatened the humanity in the past century, with the Spanish flu (1918) being the most fatal, responsible for at least 50 million deaths globally, followed by the Asian flu (1957), Hong Kong flu (1968), and the most recent Swine flu in 2009.1,100 Moreover, several outbreaks of avian flu are of major concern.100 The vast potential of influenza to mutate and spread as well as a wide range of hosts makes the emergence of novel flu pandemics inevitable and unpredictable. Despite influenza vaccines being available, the high rate of mutation requires a constant redesign.101,102 The versatile application of peptide microarrays in unraveling epitope-specific antibody responses, drug and vaccine development approaches for influenza are highlighted by several studies.50−55
A peptide microarray, covering the entire proteome of the influenza A virus, responsible for the pandemic swine flu in 2009 (H1N1) and hemagglutinin (HA) proteins from 12 other influenza A subtypes were used to investigate the epitope landscape of natural pandemic infections and vaccinees receiving an H1N1 vaccine.50 The study showed that natural flu infection leads to a different epitope recognition pattern than flu vaccination and identified one differential peptide specific in vaccinated subjects, which was suggested to serve as correlate of protection in the context of pandemic flu.50 With the aim being to predict vaccine performance, we applied peptide microarrays in conjunction with a mouse flu model system to define immunosignatures by profiling the antibody repertoire.51 A further study evaluated the efficacy of certain flu vaccines in the mouse model53 and determined the vaccine-induced immune responses. Among others, linear antibody epitope analyses were performed, comparing the antibody reaction pattern elicited by the used vaccine approaches.
Peptide microarrays are ideally suited for epitope mapping of monoclonal and serum antibodies. For example, a peptide microarray was used to map the epitope of a monoclonal antibody generated against the NS1 (nonstructural) protein of avian influenza virus.55 As monoclonal antibodies are important in the field of diagnostics and treatment, characterizing the epitope (and eventually cross-reactivity) is essential for further applications. The authors reported a 4-mer epitope on the avian influenza virus NS1 protein that is recognized by a monoclonal antibody.
Beside vaccines, drugs are essential anti-infective measures. In a study addressing the development of peptide-based influenza inhibitors, which may interfere with host cell entry, peptide microarray-based site directed substitution of amino acids assisted in the identification of HA binding peptides with higher binding affinity.54 The peptides displayed a broader specificity, binding on HA of human and avian pathogenic influenza strains.
Although not addressing pandemic influenza, Price and colleagues utilized the antibody reactivity profile obtained by peptide microarray immunostaining to predict the outcome of vaccination for seasonal influenza.52 Epitope recognition was associated with effective or ineffective responses to influenza vaccination.
3.4. Flaviviruses
Many flaviviruses pathogenic to humans are arthropod-borne viruses, transmitted by either mosquitoes or ticks, belonging to the family of Flaviviridae. Among other viruses, the genus includes Dengue (DENV), Zika (ZIKV), Yellow Fever (YFV), West Nile virus (WNV), Japanese encephalitis (JEV), or tick-borne encephalitis virus (TBEV).103 Clinically, most of these viruses are associated with an influenza-like febrile illness syndrome, developing into more severe disease in a subset of patients. DENV is estimated to affect almost half of the world’s population with around 400 million cases annually,104 while ZIKV and YFV recently caused large epidemics (ZIKV) or local outbreaks (YFV) in Latin America and Africa, respectively.105,106
The challenge for the development of vaccines and diagnostics against flaviviruses is the considerably high proteome homology between some of its members, causing cross-reactive immune responses. Hence, results of serological diagnostic tests may be false positive due to previous vaccination or natural infection with another flavivirus.107 In addition, cross-reactive antibodies can be subneutralizing and enhance viral uptake as well as replication in monocytes exacerbating disease outcome, known as antibody-dependent enhancement (ADE).108,109 Currently, vaccines are licensed for YFV, JEV, TBEV, and DENV. Postlicensure follow-up studies of the DENV vaccine DENVaxia showed higher risk of hospitalization in vaccinated individuals that were serologically naïve prior to vaccinations,110 highlighting the potential of immunological enhancement as pathomechanism.
Thus, vaccine approaches and diagnostics need to find a balance between highly conserved immunogenic epitopes, neutralizing cross-viral and highly variable epitopes, identifying only specific viruses of the genus. Peptide microarrays covering whole proteomes as linear overlapping peptides represent a promising tool for the investigation of potential epitopes for diagnostic or prognostic intervention.
Indeed, several studies using peptide microarrays focused on the identification of discriminating epitopes between certain flaviviruses.56−60 For example, Viedma and colleagues57 used a peptide microarray presenting 866 peptides of DENV serotypes 1–4, ZIKV, WNV, as well as CHIKV, a member of the family Togaviridae. By combination of experimental and computational work, they could identify peptides bound by virus specific antibodies, hence, having possible diagnostic potential. Another study discovered 13 ZIKV specific epitopes by using serum samples from ZIKV, DENV, WNV, YFV, and TBEV infected individuals on a ZIKV peptide microarray.59 An outstanding example was shown by Mishra et al.,60 where a ZIKV specific NS2B epitope was identified, which was translated into a highly selective and specific ELISA test. Therefore, they tested serum samples of flavivirus vaccinated or naturally infected individuals on peptide microarrays derived from different flavivirus proteomes.
While the microarray surface (e.g., functionalization and protein resistance) plays a major role in antibody binding, the impact of linear vs conformational peptides, as well as discontinuous epitopes, is often difficult to study, due to technological barriers. The study by Freire and co-workers61 presented insights into the molecular basis on how monoclonal antibodies differentially recognize ZIKV NS1 and DENVs NS1 proteins. By using bioinformatic predictions and modeling in conjunction with peptide microarray data, two conformational and one linear epitope within in the NS1 protein of ZIKV were described. Comparing these epitopes to their homologous regions in NS1 from DENV2 revealed differences in the epitope flanking regions, responsible for a difference in the electrostatic surface potential and, hence, differential antibody discrimination.61 A further study by Sola et al.43 focused on a comparison of linear epitopes (i.e., single peptides) versus discontinuous epitopes (i.e., multiple linear peptides) of ZIKV. The authors used a peptide microarray with single linear peptides of ZIKV NS1 as well as combinations of peptides mimicking the proximity in the native protein and, hence, discontinuous epitopes. Results showed that single linear peptides did not lead to antibody recognition, while combination of peptides showed an antibody response.
Since antibodies are the key to diagnostics and treatments against these viruses, mapping their precise epitopes is essential. Applying peptide microarrays, Xie et al.62 found two monoclonal antibodies, recognizing one highly conserved as well as one highly variable peptide sequence within the DENV NS4B protein. In contrast, another study investigated polyclonal antibodies, recognizing three domains of the Env protein of DENV serotypes 1–3, but not WNV, highlighting that antibodies can be used as possible distinguishable diagnostic tools.63
As the magnitude, specificity, and breadth of antibody response induced by vaccines is crucial for protection, investigation of these parameters is of crucial importance. Therefore, high-density peptide microarrays provide an opportunity to give rapid insights into the immune response.64−66 The cited studies used different adjuvants, as well as vaccine platforms, and could show by peptide microarrays that magnitude and antibody patterns differ between approaches dependent on adjuvant and platform of vaccination.
3.5. Human Immunodeficiency Virus (HIV)
First cases of HIV were reported in West Africa in 1981. However, the HIV epidemic increased to pandemic dimensions by the late 20th century.1,5 According to the WHO, approximately 75 million individuals have become infected since its first report. Extraordinary progress in the development of diagnostics111,112 and antiretroviral therapy has been made, however an efficacious vaccine is still elusive.113,114 In the field of HIV research, peptide microarrays are widely used to profile antibody responses in the context of vaccination, infection, and coinfection.67−82
Broadly neutralizing antibodies (bNAbs) are considered to be crucial for an effective treatment and prevention of infections with highly variable pathogens such as HIV.115,116 With the use of peptide microarrays, Tomaras and colleagues studied the specificities of existing bNAbs and the identification of potentially new targets of bNAbs in samples of HIV-1 infected individuals. Overall, distinct bNAb specificities were identified in the plasma of each donor, which differ in the contribution to neutralization.67 A deeper understanding of bNAbs provided a study by Wang and co-workers.82 Specific features of bNAbs are an often uncommon heavy chain complementarity determining region 3 (CDR-H 3s) and a high level of somatic hypermutations.117 Peptide microarrays have been applied to study the effect of the CDR-H3 amino acid content on the patterns of epitope recognition of HIV-1 envelope protein.82
Several studies utilized peptide microarrays to monitor vaccine-induced antibody responses in preclinical73−79 and clinical settings.68−71,79,81 For example, a global HIV-1 peptide microarray was used to evaluate the epitope diversity of antibody responses in HIV-1 infected and HIV-1 vaccinated humans, respectively. Furthermore, antibody responses in HIV-1 vaccinated rhesus monkeys and guinea pigs were profiled, which represent two commonly used animal models in HIV vaccine research.79 Differences in recognized epitope variants were uncovered, corroborating the application of peptide microarrays for a better understanding of HIV- and vaccine-induced humoral immune responses.
3.6. Chikungunya Virus (CHIKV)
Although not yet declared as a pandemic agent, the WHO assigned CHIKV infections as “epidemic and pandemic-prone diseases”.118 As an emerging mosquito-borne virus, CHIKV has spread effectively since its first discovery in Tanzania in 1952.119 Cases of CHIKV have been reported in Africa, Asia, Europe, the Americas and Oceania/pacific islands.120 So far, neither a licensed specific antiviral treatment nor a licensed vaccine is available.119 Since humoral responses are key against CHIKV and were known to correlate with protection,121 gaining in-depth knowledge of the targets will pave the way for vaccine development.
Recently, a whole proteome CHIKV peptide microarrays allowed the analysis of epitope-specific antibody responses in CHIKV-infected individuals in a longitudinal setting as well as in vaccinees.83 The study uncovered epitopes in the E2-B domain and the flanking acid-sensitive regions (ASRs) as the main antigenic targets in natural infection. In vaccinees, however, the overall antibody reactivity was lower, detecting epitope-specific responses only in one flanking acid-sensitive region as major target. Hence, the data may assist in future vaccine design.
3.7. Other Infectious Diseases
Peptide microarrays are also widely used in other infectious diseases, which are not necessarily of pandemic magnitude, but also cause large burdens on public health. To give several examples of the extensive list, peptide arrays are employed to decipher epitope-specific antibody responses, to evaluate or map vaccine-induced immune responses, or to identify novel targets for vaccine development and diagnostics in (1) malaria,122−133 (2) Lyme disease,134−137 (3) schistosomiasis,138 (4) Chagas disease,139,140 (5) toxoplasmosis,141−143 (6) Crimean-Congo hemorrhagic fever,144,145 (7) tick-borne diseases,137 and (8) tick-borne encephalitis.58
3.8. Random Peptide Array Approach for Disease Research
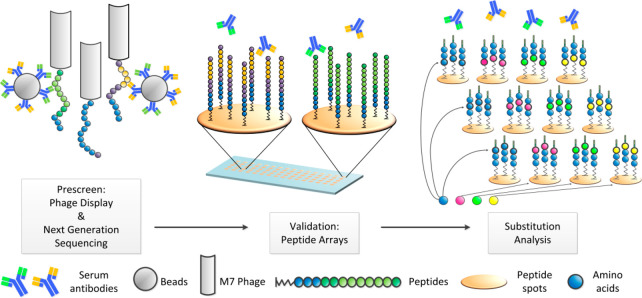
All aforementioned approaches have one prerequisite in common: knowledge about the cause of the disease. This makes the design of the peptide array content straightforward, by concentrating on the proteome of a pathogen. However, many enigmatic diseases lack in any indication of origin. To investigate, whether the antibody repertoire contains hints to the disease etiology, for example an unknown pathogen or an autoimmune reaction, peptide microarrays can be employed. In contrast to the previous approaches, the complete humoral antibody response has to be scrutinized without a priori knowledge of a pathogen to decipher possible relations to causative agents. Since proteomic assays can display only a limited number of proteins/peptides, a random content approach is currently the only way to tackle this problem. In a proof of principle study,146 a human serum was first prescreened in a random peptide phage display approach (Figure 5). Since this (biological) display technology usually shows bias due to detection of unspecific binders or missing out low affinity binders, all positive peptide binders were subsequently validated on peptide arrays. Finally, by employing the aforementioned rapid peptide microarrays synthesis technologies, in-depth substitution analyses can be performed. Similar to the alanine-scan approach (one amino acid in a peptide is stepwise replaced by alanine), these analyses allow for the identification of exact binding motifs of antibodies by stepwise substituting one amino acid in a given peptide by all other 19 amino acids. The result is an antibody “binding fingerprint”, listing all amino acids that are indispensable or irrelevant in a given epitope for antibody binding. Now, protein databases can be queried for this “binding fingerprint”, to link putatively causing agents or pathogens. With this approach, Weber et al. found a fingerprint, which was likely elicited by the polio vaccine.146
Figure 5.
In an initial prescreen, up to 109 random peptides displayed on phage were screened for their binding to serum antibodies, immobilized on beads. Next, the identified epitope peptides were validated with solid material-based peptide microarray technology. Finally, the validated epitopes were fine mapped by comprehensive substitution analysis. The resulting “binding fingerprints” enable the identification of those proteins that match the antibody specificity, and, eventually, the correlation to disease causing agents. Reprinted with permission from ref (146). Copyright 2017 Elsevier B.V.
Another interesting concept by Johnston et al. is the immunosignaturing approach.147 It relies on microarrays with a defined set of some 10 000 random peptides with a very high peptide density, that is, a small interpeptide–peptide distance in a spot of the chip. Here, the high density of the peptides (peptide–peptide spacing of <3 nm) can increase the general affinity of serum antibodies to the peptides more than 1000-fold.148 Thus, each serum of a patient results in a unique binding pattern with hundreds to thousands of strongly bound peptides on these immunosignaturing arrays. Because of the high density of the peptides, they seem to form artificial epitopes on the array surface, which do not (necessarily) correlate with any proteome sequence. With this approach, the binding patterns of healthy donors were compared to patient serum samples. Although the peptides are not (necessarily) related to a pathogen sequence, applying different statistical analyses, these patterns (a limited set of peptide binders) may be correlated to distinct diseases and even disease stages. Not only diseases, but also different vaccines were investigated, revealing that immunosignaturing is a promising tool to predict vaccine efficacy. Examples of immunosignaturing are the diagnosis of infections,147 the monitoring of vaccine responses,149 and diagnosing cancer.150 In similar approaches, fixed-complexity random-sequence peptide arrays were used for epitope identification and characterization of monoclonal antibodies.135,151 For myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS),152,153 which is a disease with unknown etiology, two studies resulted in two sets of 25 and 256 peptides, which could discriminate the samples. Because of the underlying technology of random artificial epitopes, no relation to an infectious agent could be drawn. However, little overlap between the two sets was found. This might be caused by differences in patient samples or may be attributed the immunosignaturing technology: its dependency on exact peptide–peptide distances, forming artificial high-affinity epitopes. This requires highly precise and reproducible control of surface functionalization, which might be challenging.
In conclusion, the number of studies utilizing random peptide libraries emphasizes their large potential for diagnostics and therapeutics.
4. Conclusion
Peptide microarrays have become a versatile, widespread, and easily accessible tool to examine epitope-specific antibody responses in many disease areas. This Review highlighted the diverse applications of peptide microarrays in the field of infectious diseases with the focus on pandemic pathogens. As experienced with the current COVID-19 pandemic, prompt strategies are needed for (sero-)diagnostics and vaccine development. In this context, the identification of immunodominant (protective) or discriminating epitopes is crucial—an investigation where peptide microarrays are an ideal methodology.
Recent peptide microarray technologies allow for a highly flexible and timely production, which enables the immediate analysis of humoral responses against tens of thousands of peptides in parallel. Moreover, it combines multiple advantages in a single experiment, such as (i) reduced laboratory efforts due to parallel screenings, (ii) minimal sample volumes, (iii) flexibility to be used with diverse samples, and (iv) simultaneous detection of different classes of antibodies (e.g., IgG, IgM, and IgA). For in-depth analysis of essential amino acids within the epitope sequences, amino acid substitution analysis can be performed, screening many similar peptide variants of a peptide binder. This can help to elucidate the crucial amino acids for binding or to identify potential cross-reactivities. Such amino acid- or epitope-specific antibodies may also be involved in antibody-dependent enhancement.
In conclusion, peptide arrays are a useful research tool to analyze antibody responses and to deduce important epitopes for the development of serological assays and vaccine design. Thus, rapid and high-throughput array synthesis technologies can significantly accelerate research during pandemic outbreaks.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors gratefully acknowledge the financial support of the German Federal Ministry of Education and Research [BMBF, Grant 13XP5050A], the Max Planck Society, and the Fraunhofer-Max Planck cooperation project [Glyco3Display].
The authors declare the following competing financial interest(s): K.H. and L.K.W. are employees of PEPperPRINT GmbH, Germany, producing and selling peptide microarrays with a laser printer. F.F.L. is named on a patent for the production of microarrays via laser transfer. The funders had no role in the design, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish.
References
- Madhav N.; Oppenheim B.; Gallivan M.; Mulembakani P.; Rubin E.; Wolfe N.. Pandemics: Risks, Impacts, and Mitigation. In Disease Control Priorities: Improving Health and Reducing Poverty; Jamison D. T., Gelband H., Horton S., Jha P., Laxminarayan R., Mock C. N., Nugent R., Eds.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, 2017. DOI: 10.1596/978-1-4648-0527-1_ch17. [DOI] [PubMed] [Google Scholar]
- Johns Hopkins University . https://coronavirus.jhu.edu/map.html (accessed April 9, 2020).
- Dong E.; Du H.; Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20 (5), 533–534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta M. S.; Greenland S.; Herna′n M.; Silva I. d. S.; Last J. M.; International Epidemiological Association . A Dictionary of Epidemiology, 6th ed.; Oxford University Press: Oxford, 2014; p xxxii, 343 pages. [Google Scholar]
- Grennan D. What Is a Pandemic?. JAMA 2019, 321 (9), 910. 10.1001/jama.2019.0700. [DOI] [PubMed] [Google Scholar]
- Doshi P. The elusive definition of pandemic influenza. Bull. World Health Organ 2011, 89 (7), 532–8. 10.2471/BLT.11.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D. J. Pandemic influenza and its definitional implications. Bull. World Health Organ 2011, 89 (7), 539. 10.2471/BLT.11.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . https://www.who.int/blueprint (accessed June 29, 2020).
- Lu L. L.; Suscovich T. J.; Fortune S. M.; Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018, 18 (1), 46–61. 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohst C.; Saschenbrecker S.; Stiba K.; Steinhagen K.; Probst C.; Radzimski C.; Lattwein E.; Komorowski L.; Stocker W.; Schlumberger W. Reliable Serological Testing for the Diagnosis of Emerging Infectious Diseases. Adv. Exp. Med. Biol. 2018, 1062, 19–43. 10.1007/978-981-10-8727-1_3. [DOI] [PubMed] [Google Scholar]
- Szymczak L. C.; Kuo H. Y.; Mrksich M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90 (1), 266–282. 10.1021/acs.analchem.7b04380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. F.; Yu M. Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr. Drug Targets 2004, 5 (1), 1–15. 10.2174/1389450043490668. [DOI] [PubMed] [Google Scholar]
- Aghebati-Maleki L.; Bakhshinejad B.; Baradaran B.; Motallebnezhad M.; Aghebati-Maleki A.; Nickho H.; Yousefi M.; Majidi J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23 (1), 66. 10.1186/s12929-016-0285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. J.; Kula T.; Xu Q.; Li M. Z.; Vernon S. D.; Ndung’u T.; Ruxrungtham K.; Sanchez J.; Brander C.; Chung R. T.; O’Connor K. C.; Walker B.; Larman H. B.; Elledge S. J. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348 (6239), aaa0698. 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y.; Cai J.; Zhang C.; Xing X.; Qin E.; He J.; Mao P.; Cheng J.; Liu K.; Xu D.; Song H. Mimotopes selected with neutralizing antibodies against multiple subtypes of influenza A. Virol. J. 2011, 8, 542. 10.1186/1743-422X-8-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S.; Fuentes S.; Coyle E. M.; Ravichandran S.; Davey R. T. Jr.; Beigel J. H. Human antibody repertoire after VSV-Ebola vaccination identifies novel targets and virus-neutralizing IgM antibodies. Nat. Med. 2016, 22 (12), 1439–1447. 10.1038/nm.4201. [DOI] [PubMed] [Google Scholar]
- Ravichandran S.; Hahn M.; Belaunzaran-Zamudio P. F.; Ramos-Castaneda J.; Najera-Cancino G.; Caballero-Sosa S.; Navarro-Fuentes K. R.; Ruiz-Palacios G.; Golding H.; Beigel J. H.; Khurana S. Differential human antibody repertoires following Zika infection and the implications for serodiagnostics and disease outcome. Nat. Commun. 2019, 10 (1), 1943. 10.1038/s41467-019-09914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N.; Dimitrova M.; Carter D. M.; Crevar C. J.; Ross T. M.; Golding H.; Khurana S. Influenza virus H1N1pdm09 infections in the young and old: evidence of greater antibody diversity and affinity for the hemagglutinin globular head domain (HA1 Domain) in the elderly than in young adults and children. J. Virol 2012, 86 (10), 5515–22. 10.1128/JVI.07085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin E. D. The use of multiplexing technology in the immunodiagnosis of infectious agents. J. Immunoassay Immunochem. 2019, 40 (1), 109–122. 10.1080/15321819.2018.1563551. [DOI] [PubMed] [Google Scholar]
- Liechti T.; Kadelka C.; Ebner H.; Friedrich N.; Kouyos R. D.; Gunthard H. F.; Trkola A. Development of a high-throughput bead based assay system to measure HIV-1 specific immune signatures in clinical samples. J. Immunol. Methods 2018, 454, 48–58. 10.1016/j.jim.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Drummond J. E.; Shaw E. E.; Antonello J. M.; Green T.; Page G. J.; Motley C. O.; Wilson K. A.; Finnefrock A. C.; Liang X.; Casimiro D. R. Design and optimization of a multiplex anti-influenza peptide immunoassay. J. Immunol. Methods 2008, 334 (1–2), 11–20. 10.1016/j.jim.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Yufenyuy E. L.; Parekh B. S. Development of a Multiplex Assay for Concurrent Diagnoses and Detection of HIV-1, HIV-2, and Recent HIV-1 Infection in a Single Test. AIDS Res. Hum. Retroviruses 2018, 34 (12), 1017–1027. 10.1089/aid.2017.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Wan Z.; Li T.; Xie Q.; Sun H.; Chen H.; Liang G.; Shao H.; Qin A.; Ye J. A novel linear epitope crossing Group 1 and Group 2 influenza A viruses located in the helix A of HA2 derived from H7N9. Vet. Microbiol. 2019, 228, 39–44. 10.1016/j.vetmic.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Liu L.; Wang P.; Nair M. S.; Yu J.; Rapp M.; Wang Q.; Luo Y.; Chan J. F.; Sahi V.; Figueroa A.; Guo X. V.; Cerutti G.; Bimela J.; Gorman J.; Zhou T.; Chen Z.; Yuen K. Y.; Kwong P. D.; Sodroski J. G.; Yin M. T.; Sheng Z.; Huang Y.; Shapiro L.; Ho D. D. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584 (7821), 450–456. 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Changula K.; Yoshida R.; Noyori O.; Marzi A.; Miyamoto H.; Ishijima M.; Yokoyama A.; Kajihara M.; Feldmann H.; Mweene A. S.; Takada A. Mapping of conserved and species-specific antibody epitopes on the Ebola virus nucleoprotein. Virus Res. 2013, 176 (1–2), 83–90. 10.1016/j.virusres.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. H.; Chou F. P.; Wang Y. K.; Huang S. C.; Cheng C. H.; Wu T. K. Characterization and epitope mapping of Dengue virus type 1 specific monoclonal antibodies. Virol. J. 2017, 14 (1), 189. 10.1186/s12985-017-0856-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz C.; Levy-Beladev L.; Rotem-Bamberger S.; Rito T.; Rudiger S. G. D.; Friedler A. Studying protein-protein interactions using peptide arrays. Chem. Soc. Rev. 2011, 40 (5), 2131–2145. 10.1039/c0cs00029a. [DOI] [PubMed] [Google Scholar]
- Frank R. Spot-Synthesis - an Easy Technique for the Positionally Addressable, Parallel Chemical Synthesis on a Membrane Support. Tetrahedron 1992, 48 (42), 9217–9232. 10.1016/S0040-4020(01)85612-X. [DOI] [Google Scholar]
- Merrifield R. B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85 (14), 2149–2154. 10.1021/ja00897a025. [DOI] [Google Scholar]
- Atwater J.; Mattes D. S.; Streit B.; von Bojnicic-Kninski C.; Loeffler F. F.; Breitling F.; Fuchs H.; Hirtz M. Combinatorial Synthesis of Macromolecular Arrays by Microchannel Cantilever Spotting (microCS). Adv. Mater. 2018, 30 (31), e1801632. 10.1002/adma.201801632. [DOI] [PubMed] [Google Scholar]
- Fodor S. P. A.; Read J. L.; Pirrung M. C.; Stryer L.; Lu A. T.; Solas D. Light-Directed, Spatially Addressable Parallel Chemical Synthesis. Science 1991, 251 (4995), 767–773. 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Pellois J. P.; Zhou X. C.; Srivannavit O.; Zhou T. C.; Gulari E.; Gao X. L. Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol. 2002, 20 (9), 922–926. 10.1038/nbt723. [DOI] [PubMed] [Google Scholar]
- Buus S.; Rockberg J.; Forsstrom B.; Nilsson P.; Uhlen M.; Schafer-Nielsen C. High-resolution Mapping of Linear Antibody Epitopes Using Ultrahigh-density Peptide Microarrays. Mol. Cell. Proteomics 2012, 11 (12), 1790–1800. 10.1074/mcp.M112.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. V.; Tangsombatvisit S.; Xu G. Y.; Yu J. T.; Levy D.; Baechler E. C.; Gozani O.; Varma M.; Utz P. J.; Liu C. L. On silico peptide microarrays for high-resolution mapping of antibody epitopes and diverse protein-protein interactions. Nat. Med. 2012, 18 (9), 1434. 10.1038/nm.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legutki J. B.; Zhao Z. G.; Greving M.; Woodbury N.; Johnston S. A.; Stafford P. Scalable high-density peptide arrays for comprehensive health monitoring. Nat. Commun. 2014, 5, 4785. 10.1038/ncomms5785. [DOI] [PubMed] [Google Scholar]
- Stadler V.; Felgenhauer T.; Beyer M.; Fernandez S.; Leibe K.; Guttler S.; Groning M.; Konig K.; Torralba G.; Hausmann M.; Lindenstruth V.; Nesterov A.; Block I.; Pipkorn R.; Poustka A.; Bischoff F. R.; Breitling F. Combinatorial synthesis of peptide arrays with a laser printer. Angew. Chem., Int. Ed. 2008, 47 (37), 7132–5. 10.1002/anie.200801616. [DOI] [PubMed] [Google Scholar]
- Beyer M.; Nesterov A.; Block I.; Konig K.; Felgenhauer T.; Fernandez S.; Leibe K.; Torralba G.; Hausmann M.; Trunk U.; Lindenstruth V.; Bischoff F. R.; Stadler V.; Breitling F. Combinatorial synthesis of peptide arrays onto a microchip. Science 2007, 318 (5858), 1888. 10.1126/science.1149751. [DOI] [PubMed] [Google Scholar]
- Maerkle F.; Loeffler F. F.; Schillo S.; Foertsch T.; Muenster B.; Striffler J.; Schirwitz C.; Bischoff F. R.; Breitling F.; Nesterov-Mueller A. High-density peptide arrays with combinatorial laser fusing. Adv. Mater. 2014, 26 (22), 3730–4. 10.1002/adma.201305759. [DOI] [PubMed] [Google Scholar]
- Loeffler F. F.; Foertsch T. C.; Popov R.; Mattes D. S.; Schlageter M.; Sedlmayr M.; Ridder B.; Dang F. X.; von Bojnicic-Kninski C.; Weber L. K.; Fischer A.; Greifenstein J.; Bykovskaya V.; Buliev I.; Bischoff F. R.; Hahn L.; Meier M. A.; Brase S.; Powell A. K.; Balaban T. S.; Breitling F.; Nesterov-Mueller A. High-flexibility combinatorial peptide synthesis with laser-based transfer of monomers in solid matrix material. Nat. Commun. 2016, 7, 11844. 10.1038/ncomms11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris G.; Klinkusch A.; Heidepriem J.; Tsouka A.; Zhang J.; Mende M.; Mattes D. S.; Mager D.; Riegler H.; Eickelmann S.; Loeffler F. F. Laser-induced forward transfer of soft material nanolayers with millisecond pulses shows contact-based material deposition. Appl. Surf. Sci. 2020, 508, 144973. 10.1016/j.apsusc.2019.144973. [DOI] [Google Scholar]
- Eickelmann S.; Tsouka A.; Heidepriem J.; Paris G.; Zhang J. F.; Molinari V.; Mende M.; Loeffler F. F. A Low-Cost Laser-Based Nano-3D Polymer Printer for Rapid Surface Patterning and Chemical Synthesis of Peptide and Glycan Microarrays. Adv. Mater. Technol-Us 2019, 4 (11), 1900503. 10.1002/admt.201900503. [DOI] [Google Scholar]
- Mattes D. S.; Rentschler S.; Foertsch T. C.; Münch S. W.; Loeffler F. F.; Nesterov-Mueller A.; Bräse S.; Breitling F. A Trifunctional Linker for Purified 3D Assembled Peptide Structure Arrays. Small Methods 2018, 2 (2), 1700205. 10.1002/smtd.201700205. [DOI] [Google Scholar]
- Sola L.; Gagni P.; D’Annessa I.; Capelli R.; Bertino C.; Romanato A.; Damin F.; Bergamaschi G.; Marchisio E.; Cuzzocrea A.; Bombaci M.; Grifantini R.; Chiari M.; Colombo G.; Gori A.; Cretich M. Enhancing Antibody Serodiagnosis Using a Controlled Peptide Coimmobilization Strategy. ACS Infect. Dis. 2018, 4 (6), 998–1006. 10.1021/acsinfecdis.8b00014. [DOI] [PubMed] [Google Scholar]
- Moreno-Yruela C.; Vrsanova A.-E.; Maric H. M.; Olsen C. A.. Hydroxamic Acid-Modified Peptide Microarrays for Profiling Isozyme-Selective Interactions and Inhibition of Histone Deacetylases. chemRxiv, 2020. https://chemrxiv.org/articles/Hydroxamic_Acid-Modified_Peptide_Microarrays_for_Profiling_Isozyme-Selective_Interactions_and_Inhibition_of_Histone_Deacetylases/11513163/1. [DOI] [PMC free article] [PubMed]
- Wang H.; Hou X.; Wu X.; Liang T.; Zhang X.; Wang D.; Teng F.; Dai J.; Duan H.; Guo S.; Li Y.; Yu X.. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. bioRxiv, 2020. https://www.biorxiv.org/content/10.1101/2020.03.26.994756v3. [DOI] [PMC free article] [PubMed]
- Dahlke C.; Heidepriem J.; Kobbe R.; Santer R.; Koch T.; Fathi A.; Ly M. L.; Schmiedel S.; Seeberger P. H.; Addo M. M.; Loeffler F. F.. Distinct early IgA profile may determine severity of COVID-19 symptoms: an immunological case series. medRxiv, 2020. https://www.medrxiv.org/content/10.1101/2020.04.14.20059733v1.
- Li Y.; Lai D.-y.; Zhang H.-n.; Jiang H.-w.; Tian X.-l.; Ma M.-l.; Qi H.; Meng Q.-f.; Guo S.-j.; Wu Y.-l.; Wang W.; Yang X.; Shi D.-w.; Dai J.-b.; Ying T.-l.; Zhou J.; Tao S.-c.. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. medRxiv, 2020. https://www.medrxiv.org/content/10.1101/2020.06.07.20125096v1. [DOI] [PMC free article] [PubMed]
- Farrera L.; Daguer J.-P.; Barluenga S.; Cohen P. R.; Pagano S.; Yerly S.; Kaiser L.; Vuilleumier N.; Winssinger N.. Identification of immunodominant linear epitopes from SARS-CoV-2 patient plasma. medRxiv, 2020. https://www.medrxiv.org/content/10.1101/2020.06.15.20131391v1. [DOI] [PMC free article] [PubMed]
- Heidepriem J.; Krahling V.; Dahlke C.; Wolf T.; Klein F.; Addo M. M.; Becker S.; Loeffler F. F. Epitopes of Naturally Acquired and Vaccine-Induced Anti-Ebola Virus Glycoprotein Antibodies in Single Amino Acid Resolution. Biotechnol. J. 2020, 15, 2000069. 10.1002/biot.202000069. [DOI] [PubMed] [Google Scholar]
- Ambati A.; Valentini D.; Montomoli E.; Lapini G.; Biuso F.; Wenschuh H.; Magalhaes I.; Maeurer M. H1N1 viral proteome peptide microarray predicts individuals at risk for H1N1 infection and segregates infection versus Pandemrix((R)) vaccination. Immunology 2015, 145 (3), 357–66. 10.1111/imm.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legutki J. B.; Johnston S. A. Immunosignatures can predict vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (46), 18614–9. 10.1073/pnas.1309390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. V.; Jarrell J. A.; Furman D.; Kattah N. H.; Newell E.; Dekker C. L.; Davis M. M.; Utz P. J. Characterization of influenza vaccine immunogenicity using influenza antigen microarrays. PLoS One 2013, 8 (5), e64555. 10.1371/journal.pone.0064555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A.; Bullard B. L.; Weaver E. A. Efficacy of an Adenoviral Vectored Multivalent Centralized Influenza Vaccine. Sci. Rep. 2017, 7 (1), 14912. 10.1038/s41598-017-14891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak H.; Lauster D.; Kar P.; Di Lella S.; Volkmer R.; Knecht V.; Herrmann A.; Ehrentreich-Forster E.; Bier F. F.; Stocklein W. F. Anti-Hemagglutinin Antibody Derived Lead Peptides for Inhibitors of Influenza Virus Binding. PLoS One 2016, 11 (7), e0159074. 10.1371/journal.pone.0159074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.; Wang X.; Wen X.; Bao H.; Shi L.; Tao Q.; Jiang Y.; Zeng X.; Xu X.; Tian G.; Zheng S.; Chen H. Identification of a Highly Conserved Epitope on Avian Influenza Virus Non-Structural Protein 1 Using a Peptide Microarray. PLoS One 2016, 11 (3), e0149868. 10.1371/journal.pone.0149868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron A. S.; Mohr E. L.; Baker D.; Haj A. K.; Buechler C. R.; Bailey A.; Dudley D. M.; Newman C. M.; Mohns M. S.; Koenig M.; Breitbach M. E.; Rasheed M.; Stewart L. M.; Eickhoff J.; Pinapati R. S.; Beckman E.; Li H.; Patel J.; Tan J. C.; O’Connor D. H. Antibody responses to Zika virus proteins in pregnant and non-pregnant macaques. PLoS Neglected Trop. Dis. 2018, 12 (11), e0006903. 10.1371/journal.pntd.0006903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Viedma M. D. P.; Kose N.; Parham L.; Balmaseda A.; Kuan G.; Lorenzana I.; Harris E.; Crowe J. E. Jr.; Pickett B. E. Peptide arrays incubated with three collections of human sera from patients infected with mosquito-borne viruses. F1000Research 2019, 8, 1875. 10.12688/f1000research.20981.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen S.; Hepojoki J.; Vene S.; Vaheri A.; Vapalahti O. Identification of linear human B-cell epitopes of tick-borne encephalitis virus. Virol. J. 2014, 11, 115. 10.1186/1743-422X-11-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.; Hotop S. K.; Faye O.; Ndiaye O.; Bohlken-Fascher S.; Pessoa R.; Hufert F.; Stahl-Hennig C.; Frank R.; Czerny C. P.; Schmidt-Chanasit J.; Sanabani S. S.; Sall A. A.; Niedrig M.; Bronstrup M.; Fritz H. J.; Abd El Wahed A. Diagnosing Zika virus infection against a background of other flaviviruses: Studies in high resolution serological analysis. Sci. Rep. 2019, 9 (1), 3648. 10.1038/s41598-019-40224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N.; Caciula A.; Price A.; Thakkar R.; Ng J.; Chauhan L. V.; Jain K.; Che X.; Espinosa D. A.; Montoya Cruz M.; Balmaseda A.; Sullivan E. H.; Patel J. J.; Jarman R. G.; Rakeman J. L.; Egan C. T.; Reusken C.; Koopmans M. P. G.; Harris E.; Tokarz R.; Briese T.; Lipkin W. I. Diagnosis of Zika Virus Infection by Peptide Array and Enzyme-Linked Immunosorbent Assay. mBio 2018, 9 (2), e00095-18. 10.1128/mBio.00095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire M.; Pol-Fachin L.; Coêlho D. F.; Viana I. F. T.; Magalhães T.; Cordeiro M. T.; Fischer N.; Loeffler F. F.; Jaenisch T.; Franca R. F.; Marques E. T. A.; Lins R. D. Mapping Putative B-Cell Zika Virus NS1 Epitopes Provides Molecular Basis for Anti-NS1 Antibody Discrimination between Zika and Dengue Viruses. ACS Omega 2017, 2 (7), 3913–3920. 10.1021/acsomega.7b00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Zou J.; Wang Q. Y.; Noble C. G.; Lescar J.; Shi P. Y. Generation and characterization of mouse monoclonal antibodies against NS4B protein of dengue virus. Virology 2014, 450–451, 250–7. 10.1016/j.virol.2013.12.025. [DOI] [PubMed] [Google Scholar]
- Akhras S.; Herrlein M. L.; Elgner F.; Holzhauser T.; Hildt E. ZIKV Envelope Domain-Specific Antibodies: Production, Purification and Characterization. Viruses 2019, 11 (8), 748. 10.3390/v11080748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz T.; Beatty P. R.; MacMillen Z.; Killingbeck S. S.; Wang C.; Harris E. Antibody Epitopes Identified in Critical Regions of Dengue Virus Nonstructural 1 Protein in Mouse Vaccination and Natural Human Infections. J. Immunol. 2017, 198 (10), 4025–4035. 10.4049/jimmunol.1700029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda D.; Batista M. T.; Pereira L. R.; de Jesus Cintra M.; Amorim J. H.; Mathias-Santos C.; Pereira S. A.; Boscardin S. B.; Silva S. D. R.; Faquim-Mauro E. L.; Silveira V. B.; Oliveira D. B. L.; Johnston S. A.; Ferreira L. C. S.; Rodrigues J. F. Adjuvant-Mediated Epitope Specificity and Enhanced Neutralizing Activity of Antibodies Targeting Dengue Virus Envelope Protein. Front. Immunol. 2017, 8, 1175. 10.3389/fimmu.2017.01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbink P.; Larocca R. A.; De La Barrera R. A.; Bricault C. A.; Moseley E. T.; Boyd M.; Kirilova M.; Li Z.; Ng’ang’a D.; Nanayakkara O.; Nityanandam R.; Mercado N. B.; Borducchi E. N.; Agarwal A.; Brinkman A. L.; Cabral C.; Chandrashekar A.; Giglio P. B.; Jetton D.; Jimenez J.; Lee B. C.; Mojta S.; Molloy K.; Shetty M.; Neubauer G. H.; Stephenson K. E.; Peron J. P.; Zanotto P. M.; Misamore J.; Finneyfrock B.; Lewis M. G.; Alter G.; Modjarrad K.; Jarman R. G.; Eckels K. H.; Michael N. L.; Thomas S. J.; Barouch D. H. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science 2016, 353 (6304), 1129–32. 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras G. D.; Binley J. M.; Gray E. S.; Crooks E. T.; Osawa K.; Moore P. L.; Tumba N.; Tong T.; Shen X.; Yates N. L.; Decker J.; Wibmer C. K.; Gao F.; Alam S. M.; Easterbrook P.; Abdool Karim S.; Kamanga G.; Crump J. A.; Cohen M.; Shaw G. M.; Mascola J. R.; Haynes B. F.; Montefiori D. C.; Morris L. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol 2011, 85 (21), 11502–19. 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasavvas N.; Billings E.; Rao M.; Williams C.; Zolla-Pazner S.; Bailer R. T.; Koup R. A.; Madnote S.; Arworn D.; Shen X.; Tomaras G. D.; Currier J. R.; Jiang M.; Magaret C.; Andrews C.; Gottardo R.; Gilbert P.; Cardozo T. J.; Rerks-Ngarm S.; Nitayaphan S.; Pitisuttithum P.; Kaewkungwal J.; Paris R.; Greene K.; Gao H.; Gurunathan S.; Tartaglia J.; Sinangil F.; Korber B. T.; Montefiori D. C.; Mascola J. R.; Robb M. L.; Haynes B. F.; Ngauy V.; Michael N. L.; Kim J. H.; de Souza M. S.; Collaboration M. T. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res. Hum. Retroviruses 2012, 28 (11), 1444–57. 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D. H.; Liu J.; Peter L.; Abbink P.; Iampietro M. J.; Cheung A.; Alter G.; Chung A.; Dugast A. S.; Frahm N.; McElrath M. J.; Wenschuh H.; Reimer U.; Seaman M. S.; Pau M. G.; Weijtens M.; Goudsmit J.; Walsh S. R.; Dolin R.; Baden L. R. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001). J. Infect. Dis. 2013, 207 (2), 248–56. 10.1093/infdis/jis671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadai Y.; Held K.; Joseph S.; Ahmed M. I. M.; Hoffmann V. S.; Peterhoff D.; Missanga M.; Bauer A.; Joachim A.; Reimer U.; Zerweck J.; McCormack S.; Cope A. V.; Tatoud R.; Shattock R. J.; Robb M. L.; Sandstroem E. G.; Hoelscher M.; Maboko L.; Bakari M.; Kroidl A.; Wagner R.; Weber J.; Pollakis G.; Geldmacher C. Envelope-Specific Recognition Patterns of HIV Vaccine-Induced IgG Antibodies Are Linked to Immunogen Structure and Sequence. Front. Immunol. 2019, 10, 717. 10.3389/fimmu.2019.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo R.; Bailer R. T.; Korber B. T.; Gnanakaran S.; Phillips J.; Shen X.; Tomaras G. D.; Turk E.; Imholte G.; Eckler L.; Wenschuh H.; Zerweck J.; Greene K.; Gao H.; Berman P. W.; Francis D.; Sinangil F.; Lee C.; Nitayaphan S.; Rerks-Ngarm S.; Kaewkungwal J.; Pitisuttithum P.; Tartaglia J.; Robb M. L.; Michael N. L.; Kim J. H.; Zolla-Pazner S.; Haynes B. F.; Mascola J. R.; Self S.; Gilbert P.; Montefiori D. C. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One 2013, 8 (9), e75665. 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L.; Bleda M. J.; Gomara M. J.; Haro I. Design and application of GB virus C (GBV-C) peptide microarrays for diagnosis of GBV-C/HIV-1 co-infection. Anal. Bioanal. Chem. 2013, 405 (12), 3973–82. 10.1007/s00216-012-6585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J. F.; Morin T. J.; Yu B.; Tatsuno G. P.; O’Rourke S. M.; Theolis R. Jr.; Mesa K. A.; Berman P. W. HIV-1 envelope proteins and V1/V2 domain scaffolds with mannose-5 to improve the magnitude and quality of protective antibody responses to HIV-1. J. Biol. Chem. 2014, 289 (30), 20526–42. 10.1074/jbc.M114.554089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Stephenson K. E.; Kang Z. H.; Lavine C. L.; Seaman M. S.; Barouch D. H. Common features of mucosal and peripheral antibody responses elicited by candidate HIV-1 vaccines in rhesus monkeys. J. Virol. 2014, 88 (22), 13510–5. 10.1128/JVI.02095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. S.; Pollara J.; Kunz E. L.; Jeffries T. L. Jr.; Duffy R.; Beck C.; Stamper L.; Wang M.; Shen X.; Pickup D. J.; Staats H. F.; Hudgens M. G.; Kepler T. B.; Montefiori D. C.; Moody M. A.; Tomaras G. D.; Liao H. X.; Haynes B. F.; Ferrari G.; Fouda G. G. A.; Permar S. R. Combined HIV-1 Envelope Systemic and Mucosal Immunization of Lactating Rhesus Monkeys Induces a Robust Immunoglobulin A Isotype B Cell Response in Breast Milk. J. Virol. 2016, 90 (10), 4951–4965. 10.1128/JVI.00335-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G.; Shen X.; Zurawski S.; Tomaras G. D.; Montefiori D. C.; Roederer M.; Ferrari G.; Lacabaratz C.; Klucar P.; Wang Z.; Foulds K. E.; Kao S. F.; Yu X.; Sato A.; Yates N. L.; LaBranche C.; Stanfield-Oakley S.; Kibler K.; Jacobs B.; Salazar A.; Self S.; Fulp W.; Gottardo R.; Galmin L.; Weiss D.; Cristillo A.; Pantaleo G.; Levy Y. Superiority in Rhesus Macaques of Targeting HIV-1 Env gp140 to CD40 versus LOX-1 in Combination with Replication-Competent NYVAC-KC for Induction of Env-Specific Antibody and T Cell Responses. J. Virol. 2017, 91 (9), e01596-16. 10.1128/JVI.01596-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricault C. A.; Kovacs J. M.; Badamchi-Zadeh A.; McKee K.; Shields J. L.; Gunn B. M.; Neubauer G. H.; Ghantous F.; Jennings J.; Gillis L.; Perry J.; Nkolola J. P.; Alter G.; Chen B.; Stephenson K. E.; Doria-Rose N.; Mascola J. R.; Seaman M. S.; Barouch D. H. Neutralizing Antibody Responses following Long-Term Vaccination with HIV-1 Env gp140 in Guinea Pigs. J. Virol. 2018, 92 (13), e00369-18. 10.1128/JVI.00369-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricault C. A.; Yusim K.; Seaman M. S.; Yoon H.; Theiler J.; Giorgi E. E.; Wagh K.; Theiler M.; Hraber P.; Macke J. P.; Kreider E. F.; Learn G. H.; Hahn B. H.; Scheid J. F.; Kovacs J. M.; Shields J. L.; Lavine C. L.; Ghantous F.; Rist M.; Bayne M. G.; Neubauer G. H.; McMahan K.; Peng H.; Cheneau C.; Jones J. J.; Zeng J.; Ochsenbauer C.; Nkolola J. P.; Stephenson K. E.; Chen B.; Gnanakaran S.; Bonsignori M.; Williams L. D.; Haynes B. F.; Doria-Rose N.; Mascola J. R.; Montefiori D. C.; Barouch D. H.; Korber B. HIV-1 Neutralizing Antibody Signatures and Application to Epitope-Targeted Vaccine Design. Cell Host Microbe 2019, 25 (1), 59–72. 10.1016/j.chom.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson K. E.; Neubauer G. H.; Reimer U.; Pawlowski N.; Knaute T.; Zerweck J.; Korber B. T.; Barouch D. H. Quantification of the epitope diversity of HIV-1-specific binding antibodies by peptide microarrays for global HIV-1 vaccine development. J. Immunol. Methods 2015, 416, 105–23. 10.1016/j.jim.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallerano D.; Wollmann E.; Lupinek C.; Schlederer T.; Ebner D.; Harwanegg C.; Niespodziana K.; Schmetterer K.; Pickl W.; Puchhammer-Stockl E.; Sibanda E.; Valenta R. HIV microarray for the mapping and characterization of HIV-specific antibody responses. Lab Chip 2015, 15 (6), 1574–89. 10.1039/C4LC01510J. [DOI] [PubMed] [Google Scholar]
- Stephenson K. E.; Neubauer G. H.; Bricault C. A.; Shields J.; Bayne M.; Reimer U.; Pawlowski N.; Knaute T.; Zerweck J.; Seaman M. S.; Rosenberg E. S.; Barouch D. H. Antibody Responses After Analytic Treatment Interruption in Human Immunodeficiency Virus-1-Infected Individuals on Early Initiated Antiretroviral Therapy. Open Forum Infect Dis 2016, 3 (2), ofw100. 10.1093/ofid/ofw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Kapoor P.; Parks R.; Silva-Sanchez A.; Alam S. M.; Verkoczy L.; Liao H. X.; Zhuang Y.; Burrows P.; Levinson M.; Elgavish A.; Cui X.; Haynes B. F.; Schroeder H. Jr. HIV-1 gp140 epitope recognition is influenced by immunoglobulin DH gene segment sequence. Immunogenetics 2016, 68 (2), 145–55. 10.1007/s00251-015-0890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henss L.; Yue C.; Von Rhein C.; Tschismarov R.; Lewis-Ximenez L. L.; Dolle A.; Baylis S. A.; Schnierle B. S. Analysis of Humoral Immune Responses in Chikungunya Virus (CHIKV)-Infected Patients and Individuals Vaccinated With a Candidate CHIKV Vaccine. J. Infect. Dis. 2020, 221 (10), 1713–1723. 10.1093/infdis/jiz658. [DOI] [PubMed] [Google Scholar]
- Liu D. X.; Liang J. Q.; Fung T. S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Reference Module in Life Sciences 2020, 10.1016/B978-0-12-809633-8.21501-X. [DOI] [Google Scholar]
- Peiris J. S.; Lai S. T.; Poon L. L.; Guan Y.; Yam L. Y.; Lim W.; Nicholls J.; Yee W. K.; Yan W. W.; Cheung M. T.; Cheng V. C.; Chan K. H.; Tsang D. N.; Yung R. W.; Ng T. K.; Yuen K. Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361 (9366), 1319–25. 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T. G.; Erdman D.; Goldsmith C. S.; Zaki S. R.; Peret T.; Emery S.; Tong S.; Urbani C.; Comer J. A.; Lim W.; Rollin P. E.; Dowell S. F.; Ling A. E.; Humphrey C. D.; Shieh W. J.; Guarner J.; Paddock C. D.; Rota P.; Fields B.; DeRisi J.; Yang J. Y.; Cox N.; Hughes J. M.; LeDuc J. W.; Bellini W. J.; Anderson L. J.; SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348 (20), 1953–66. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Drosten C.; Gunther S.; Preiser W.; van der Werf S.; Brodt H. R.; Becker S.; Rabenau H.; Panning M.; Kolesnikova L.; Fouchier R. A.; Berger A.; Burguiere A. M.; Cinatl J.; Eickmann M.; Escriou N.; Grywna K.; Kramme S.; Manuguerra J. C.; Muller S.; Rickerts V.; Sturmer M.; Vieth S.; Klenk H. D.; Osterhaus A. D.; Schmitz H.; Doerr H. W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348 (20), 1967–76. 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Zaki A. M.; van Boheemen S.; Bestebroer T. M.; Osterhaus A. D.; Fouchier R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367 (19), 1814–20. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H. R.; Zhu Y.; Li B.; Huang C. L.; Chen H. D.; Chen J.; Luo Y.; Guo H.; Jiang R. D.; Liu M. Q.; Chen Y.; Shen X. R.; Wang X.; Zheng X. S.; Zhao K.; Chen Q. J.; Deng F.; Liu L. L.; Yan B.; Zhan F. X.; Wang Y. Y.; Xiao G. F.; Shi Z. L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579 (7798), 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- All that’s fit to preprint. Nat. Biotechnol. 2020, 38 ( (5), ), 507. 10.1038/s41587-020-0536-x [DOI] [PubMed] [Google Scholar]
- Lou B.; Li T. D.; Zheng S. F.; Su Y. Y.; Li Z. Y.; Liu W.; Yu F.; Ge S. X.; Zou Q. D.; Yuan Q.; Lin S.; Hong C. M.; Yao X. Y.; Zhang X. J.; Wu D. H.; Zhou G. L.; Hou W. H.; Li T. T.; Zhang Y. L.; Zhang S. Y.; Fan J.; Zhang J.; Xia N. S.; Chen Y. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur. Respir. J. 2020, 56 (2), 2000763. 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F.; Stadlbauer D.; Strohmeier S.; Nguyen T. H. O.; Chromikova V.; McMahon M.; Jiang K.; Arunkumar G. A.; Jurczyszak D.; Polanco J.; Bermudez-Gonzalez M.; Kleiner G.; Aydillo T.; Miorin L.; Fierer D. S.; Lugo L. A.; Kojic E. M.; Stoever J.; Liu S. T. H.; Cunningham-Rundles C.; Felgner P. L.; Moran T.; Garcia-Sastre A.; Caplivski D.; Cheng A. C.; Kedzierska K.; Vapalahti O.; Hepojoki J. M.; Simon V.; Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26 (7), 1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . https://apps.who.int/ebola/current-situation/ebola-situation-report-30-march-2016 (accessed June 29, 2020).
- Becquart P.; Mahlakoiv T.; Nkoghe D.; Leroy E. M. Identification of Continuous Human B-Cell Epitopes in the VP35, VP40, Nucleoprotein and Glycoprotein of Ebola Virus. PLoS One 2014, 9 (6), e96360. 10.1371/journal.pone.0096360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire E. O.; Schendel S. L.; Gunn B. M.; Milligan J. C.; Alter G. Antibody-mediated protection against Ebola virus. Nat. Immunol. 2018, 19 (11), 1169–1178. 10.1038/s41590-018-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt S. A.; Zehner M.; Krahling V.; Cohen-Dvashi H.; Kreer C.; Elad N.; Gruell H.; Ercanoglu M. S.; Schommers P.; Gieselmann L.; Eggeling R.; Dahlke C.; Wolf T.; Pfeifer N.; Addo M. M.; Diskin R.; Becker S.; Klein F. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 2019, 25 (10), 1589–1600. 10.1038/s41591-019-0602-4. [DOI] [PubMed] [Google Scholar]
- Bornholdt Z. A.; Turner H. L.; Murin C. D.; Li W.; Sok D.; Souders C. A.; Piper A. E.; Goff A.; Shamblin J. D.; Wollen S. E.; Sprague T. R.; Fusco M. L.; Pommert K. B.; Cavacini L. A.; Smith H. L.; Klempner M.; Reimann K. A.; Krauland E.; Gerngross T. U.; Wittrup K. D.; Saphire E. O.; Burton D. R.; Glass P. J.; Ward A. B.; Walker L. M. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 2016, 351 (6277), 1078–83. 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak A. I.; Shen X.; Murin C. D.; Turner H. L.; David J. A.; Fusco M. L.; Lampley R.; Kose N.; Ilinykh P. A.; Kuzmina N.; Branchizio A.; King H.; Brown L.; Bryan C.; Davidson E.; Doranz B. J.; Slaughter J. C.; Sapparapu G.; Klages C.; Ksiazek T. G.; Saphire E. O.; Ward A. B.; Bukreyev A.; Crowe J. E. Jr. Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell 2016, 164 (3), 392–405. 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal P.; Elias S. C.; Machado S. R.; Xiao J.; Schimanski L.; O’Dowd V.; Baker T.; Barry E.; Mendelsohn S. C.; Cherry C. J.; Jin J.; Labbé G. M.; Donnellan F. R.; Rampling T.; Dowall S.; Rayner E.; Findlay-Wilson S.; Carroll M.; Guo J.; Xu X. N.; Huang K. A.; Takada A.; Burgess G.; McMillan D.; Popplewell A.; Lightwood D. J.; Draper S. J.; Townsend A. R. Therapeutic Monoclonal Antibodies for Ebola Virus Infection Derived from Vaccinated Humans. Cell Rep. 2019, 27 (1), 172–186. 10.1016/j.celrep.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Kain T.; Fowler R. Preparing intensive care for the next pandemic influenza. Crit Care 2019, 23 (1), 337. 10.1186/s13054-019-2616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning J.; Thwaites R. S.; Openshaw P. J. M. Seasonal and pandemic influenza: 100 years of progress, still much to learn. Mucosal Immunol. 2020, 13 (4), 566–573. 10.1038/s41385-020-0287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. J.; Crank M. C.; Shiver J.; Graham B. S.; Mascola J. R.; Nabel G. J. Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discovery 2020, 19 (4), 239–252. 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T. C.; Diamond M. S. The continued threat of emerging flaviviruses. Nat. Microbiol 2020, 5 (6), 796–812. 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S.; Gething P. W.; Brady O. J.; Messina J. P.; Farlow A. W.; Moyes C. L.; Drake J. M.; Brownstein J. S.; Hoen A. G.; Sankoh O.; Myers M. F.; George D. B.; Jaenisch T.; Wint G. R.; Simmons C. P.; Scott T. W.; Farrar J. J.; Hay S. I. The global distribution and burden of dengue. Nature 2013, 496 (7446), 504–7. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Gonzalez F. J.; Peres C. A.; Steiner Sao Bernardo C.; Hunter P. R.; Lake I. R. After the epidemic: Zika virus projections for Latin America and the Caribbean. PLoS Neglected Trop. Dis. 2017, 11 (11), e0006007. 10.1371/journal.pntd.0006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M. U. G.; Faria N. R.; Reiner R. C. Jr.; Golding N.; Nikolay B.; Stasse S.; Johansson M. A.; Salje H.; Faye O.; Wint G. R. W.; Niedrig M.; Shearer F. M.; Hill S. C.; Thompson R. N.; Bisanzio D.; Taveira N.; Nax H. H.; Pradelski B. S. R.; Nsoesie E. O.; Murphy N. R.; Bogoch I. I.; Khan K.; Brownstein J. S.; Tatem A. J.; de Oliveira T.; Smith D. L.; Sall A. A.; Pybus O. G.; Hay S. I.; Cauchemez S. Spread of yellow fever virus outbreak in Angola and the Democratic Republic of the Congo 2015–16: a modelling study. Lancet Infect. Dis. 2017, 17 (3), 330–338. 10.1016/S1473-3099(16)30513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwinn R.; Doerr H. W.; Emmerich P.; Schmitz H.; Preiser W. Cross-reactivity in flavivirus serology: new implications of an old finding?. Med. Microbiol. Immunol. 2002, 190 (4), 199–202. 10.1007/s00430-001-0107-9. [DOI] [PubMed] [Google Scholar]
- Katzelnick L. C.; Gresh L.; Halloran M. E.; Mercado J. C.; Kuan G.; Gordon A.; Balmaseda A.; Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017, 358 (6365), 929–932. 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S. B.; Nimmannitya S.; Cohen S. N. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J. Biol. Med. 1970, 42 (5), 311–28. [PMC free article] [PubMed] [Google Scholar]
- Halstead S. B. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine 2017, 35 (47), 6355–6358. 10.1016/j.vaccine.2017.09.089. [DOI] [PubMed] [Google Scholar]
- Parekh B. S.; Ou C.-Y.; Fonjungo P. N.; Kalou M. B.; Rottinghaus E.; Puren A.; Alexander H.; Hurlston Cox M.; Nkengasong J. N. Diagnosis of Human Immunodeficiency Virus Infection. Clin. Microbiol. Rev. 2019, 32 (1), e00064-18. 10.1128/CMR.00064-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.; Tang Y. W. Myths in the laboratory diagnosis of HIV infection. Emerging Microbes Infect. 2019, 8 (1), 1240–1242. 10.1080/22221751.2019.1656550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; McKay P. F.; Mann J. F. S. Advances in HIV-1 Vaccine Development. Viruses 2018, 10 (4), 167. 10.3390/v10040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D. R. Advancing an HIV vaccine; advancing vaccinology. Nat. Rev. Immunol. 2019, 19 (2), 77–78. 10.1038/s41577-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.; Qureshi H.; Deshpande S.; Bhattacharya J. Broadly neutralizing antibodies in HIV-1 treatment and prevention. Ther Adv. Vaccines Immunother 2018, 6 (4), 61–68. 10.1177/2515135518800689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaraman H.; Schanz M.; Trkola A. Broadly neutralizing antibodies: What is needed to move from a rare event in HIV-1 infection to vaccine efficacy?. Retrovirology 2018, 15 (1), 52. 10.1186/s12977-018-0433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D. R.; Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu. Rev. Immunol. 2016, 34, 635–59. 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . http://www.emro.who.int/pandemic-epidemic-diseases/chikungunya/index.html (accessed June 20, 2020).
- Silva J. V. J. Jr.; Ludwig-Begall L. F.; Oliveira-Filho E. F.; Oliveira R. A. S.; Durães-Carvalho R.; Lopes T. R. R.; Silva D. E. A.; Gil L. A scoping review of Chikungunya virus infection: epidemiology, clinical characteristics, viral co-circulation complications, and control. Acta Trop. 2018, 188, 213–224. 10.1016/j.actatropica.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . https://www.cdc.gov/chikungunya/geo/index.html (accessed June 29, 2020).
- Milligan G. N.; Schnierle B. S.; McAuley A. J.; Beasley D. W. C. Defining a correlate of protection for chikungunya virus vaccines. Vaccine 2019, 37 (50), 7427–7436. 10.1016/j.vaccine.2018.10.033. [DOI] [PubMed] [Google Scholar]
- Bailey J. A.; Berry A. A.; Travassos M. A.; Ouattara A.; Boudova S.; Dotsey E. Y.; Pike A.; Jacob C. G.; Adams M.; Tan J. C.; Bannen R. M.; Patel J. J.; Pablo J.; Nakajima R.; Jasinskas A.; Dutta S.; Takala-Harrison S.; Lyke K. E.; Laurens M. B.; Niangaly A.; Coulibaly D.; Kouriba B.; Doumbo O. K.; Thera M. A.; Felgner P. L.; Plowe C. V. Microarray analyses reveal strain-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 variants following natural infection and vaccination. Sci. Rep. 2020, 10 (1), 3952. 10.1038/s41598-020-60551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou N.; Jiang N.; Ma Y.; Zou Y.; Piao X.; Liu S.; Chen Q. Low-Complexity Repetitive Epitopes of Plasmodium falciparum Are Decoys for Humoural Immune Responses. Front. Immunol. 2020, 11, 610. 10.3389/fimmu.2020.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj D. K.; Das Mohapatra A.; Jnawali A.; Zuromski J.; Jha A.; Cham-Kpu G.; Sherman B.; Rudlaff R. M.; Nixon C. E.; Hilton N.; Oleinikov A. V.; Chesnokov O.; Merritt J.; Pond-Tor S.; Burns L.; Jolly G.; Ben Mamoun C.; Kabyemela E.; Muehlenbachs A.; Lambert L.; Orr-Gonzalez S.; Gnadig N. F.; Fidock D. A.; Park S.; Dvorin J. D.; Pardi N.; Weissman D.; Mui B. L.; Tam Y. K.; Friedman J. F.; Fried M.; Duffy P. E.; Kurtis J. D. Anti-PfGARP activates programmed cell death of parasites and reduces severe malaria. Nature 2020, 582 (7810), 104–108. 10.1038/s41586-020-2220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A. E.; Berry A. A.; Bailey J. A.; Pike A.; Dara A.; Agrawal S.; Stucke E. M.; Ouattara A.; Coulibaly D.; Lyke K. E.; Laurens M. B.; Adams M.; Takala-Harrison S.; Pablo J.; Jasinskas A.; Nakajima R.; Niangaly A.; Kouriba B.; Kone A. K.; Rowe J. A.; Doumbo O. K.; Thera M. A.; Patel J. J.; Tan J. C.; Felgner P. L.; Plowe C. V.; Travassos M. A. Antibodies to Peptides in Semiconserved Domains of RIFINs and STEVORs Correlate with Malaria Exposure. mSphere 2019, 4 (2), e00097-19. 10.1128/mSphere.00097-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank A.; Furle K.; Jaschke A.; Mikus G.; Lehmann M.; Husing J.; Heiss K.; Giese T.; Carter D.; Bohnlein E.; Lanzer M.; Haefeli W. E.; Bujard H. Immunization with full-length Plasmodium falciparum merozoite surface protein 1 is safe and elicits functional cytophilic antibodies in a randomized first-in-human trial. NPJ. Vaccines 2020, 5, 10. 10.1038/s41541-020-0160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. W.; Jongwutiwes S.; Putaporntip C.; Jackson A. P. Clinical expression and antigenic profiles of a Plasmodium vivax vaccine candidate: merozoite surface protein 7 (PvMSP-7). Malar. J. 2019, 18 (1), 197. 10.1186/s12936-019-2826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch T.; Heiss K.; Fischer N.; Geiger C.; Bischoff F. R.; Moldenhauer G.; Rychlewski L.; Sie A.; Coulibaly B.; Seeberger P. H.; Wyrwicz L. S.; Breitling F.; Loeffler F. F. High-density Peptide Arrays Help to Identify Linear Immunogenic B-cell Epitopes in Individuals Naturally Exposed to Malaria Infection. Mol. Cell. Proteomics 2019, 18 (4), 642–656. 10.1074/mcp.RA118.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.; Sack B. K.; Oyen D.; Zenklusen I.; Piccoli L.; Barbieri S.; Foglierini M.; Fregni C. S.; Marcandalli J.; Jongo S.; Abdulla S.; Perez L.; Corradin G.; Varani L.; Sallusto F.; Sim B. K. L.; Hoffman S. L.; Kappe S. H. I.; Daubenberger C.; Wilson I. A.; Lanzavecchia A. A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nat. Med. 2018, 24 (4), 401–407. 10.1038/nm.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon C. E.; Park S.; Pond-Tor S.; Raj D.; Lambert L. E.; Orr-Gonzalez S.; Barnafo E. K.; Rausch K. M.; Friedman J. F.; Fried M.; Duffy P. E.; Kurtis J. D. Identification of Protective B-Cell Epitopes within the Novel Malaria Vaccine Candidate Plasmodium falciparum Schizont Egress Antigen 1. Clin. Vaccine Immunol. 2017, 24 (7), e00068-17. 10.1128/CVI.00068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler F. F.; Pfeil J.; Heiss K. High-Density Peptide Arrays for Malaria Vaccine Development. Methods Mol. Biol. 2016, 1403, 569–82. 10.1007/978-1-4939-3387-7_32. [DOI] [PubMed] [Google Scholar]
- Angeletti D.; Albrecht L.; Wahlgren M.; Moll K. Analysis of antibody induction upon immunization with distinct NTS-DBL1alpha-domains of PfEMP1 from rosetting Plasmodium falciparum parasites. Malar. J. 2013, 12, 32. 10.1186/1475-2875-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S.; Moll K.; Angeletti D.; Albrecht L.; Kursula I.; Jiang N.; Sun X.; Berzins K.; Wahlgren M.; Chen Q. Characterization of the Duffy-Binding-Like Domain of Plasmodium falciparum Blood-Stage Antigen 332. Malar. Res. Treat. 2011, 2011, 671439. 10.4061/2011/671439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A.; Latov N.; Wormser G. P.; Marques A. R.; Alaedini A. Epitope mapping of antibodies to VlsE protein of Borrelia burgdorferi in post-Lyme disease syndrome. Clin. Immunol. 2011, 141 (1), 103–10. 10.1016/j.clim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer J.; Johnston S. A.; Stafford P. Epitope identification from fixed-complexity random-sequence peptide microarrays. Mol. Cell. Proteomics 2015, 14 (1), 136–47. 10.1074/mcp.M114.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L. K.; Isse A.; Rentschler S.; Kneusel R. E.; Palermo A.; Hubbuch J.; Nesterov-Mueller A.; Breitling F.; Loeffler F. F. Antibody fingerprints in lyme disease deciphered with high density peptide arrays. Engineering in Life Sciences 2017, 17 (10), 1078–1087. 10.1002/elsc.201700062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R.; Mishra N.; Tagliafierro T.; Sameroff S.; Caciula A.; Chauhan L.; Patel J.; Sullivan E.; Gucwa A.; Fallon B.; Golightly M.; Molins C.; Schriefer M.; Marques A.; Briese T.; Lipkin W. I. A multiplex serologic platform for diagnosis of tick-borne diseases. Sci. Rep. 2018, 8 (1), 3158. 10.1038/s41598-018-21349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. H.; Vance G. M.; Cartwright J.; Cao J. P.; Wilson R. A.; Castro-Borges W. Mapping the epitopes of Schistosoma japonicum esophageal gland proteins for incorporation into vaccine constructs. PLoS One 2020, 15 (2), e0229542. 10.1371/journal.pone.0229542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante I. M.; La Spina P. E.; Carmona S. J.; Agüero F.; Buscaglia C. A. High-resolution profiling of linear B-cell epitopes from mucin-associated surface proteins (MASPs) of Trypanosoma cruzi during human infections. PLoS Neglected Trop. Dis. 2017, 11 (9), e0005986. 10.1371/journal.pntd.0005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona S. J.; Nielsen M.; Schafer-Nielsen C.; Mucci J.; Altcheh J.; Balouz V.; Tekiel V.; Frasch A. C.; Campetella O.; Buscaglia C. A.; Agüero F. Towards High-throughput Immunomics for Infectious Diseases: Use of Next-generation Peptide Microarrays for Rapid Discovery and Mapping of Antigenic Determinants. Mol. Cell. Proteomics 2015, 14 (7), 1871–84. 10.1074/mcp.M114.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimov P.; Zerweck J.; Maksimov A.; Hotop A.; Gross U.; Pleyer U.; Spekker K.; Daubener W.; Werdermann S.; Niederstrasser O.; Petri E.; Mertens M.; Ulrich R. G.; Conraths F. J.; Schares G. Peptide microarray analysis of in silico-predicted epitopes for serological diagnosis of Toxoplasma gondii infection in humans. Clin. Vaccine Immunol. 2012, 19 (6), 865–74. 10.1128/CVI.00119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimov P.; Zerweck J.; Maksimov A.; Hotop A.; Gross U.; Spekker K.; Daubener W.; Werdermann S.; Niederstrasser O.; Petri E.; Mertens M.; Ulrich R. G.; Conraths F. J.; Schares G. Analysis of clonal type-specific antibody reactions in Toxoplasma gondii seropositive humans from Germany by peptide-microarray. PLoS One 2012, 7 (3), e34212. 10.1371/journal.pone.0034212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz-Solis D.; Cordeiro C.; Young L. H.; Darde M. L.; Commodaro A. G.; Grigg M. E.; Saeij J. P. J. Serotyping of Toxoplasma gondii Infection Using Peptide Membrane Arrays. Front. Cell. Infect. Microbiol. 2019, 9, 408. 10.3389/fcimb.2019.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzen A.; Risinger C.; Korukluoglu G.; Christova I.; Corli Hitzeroth A.; Viljoen N.; Burt F. J.; Mirazimi A.; Blixt O. Epitope-mapping of the glycoprotein from Crimean-Congo hemorrhagic fever virus using a microarray approach. PLoS Neglected Trop. Dis. 2018, 12 (7), e0006598. 10.1371/journal.pntd.0006598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M.; Guerrero L. I. W.; Albarino C. G.; Bergeron E.; Nichol S. T.; Spiropoulou C. F. Identification of broadly neutralizing monoclonal antibodies against Crimean-Congo hemorrhagic fever virus. Antiviral Res. 2017, 146, 112–120. 10.1016/j.antiviral.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L. K.; Palermo A.; Kugler J.; Armant O.; Isse A.; Rentschler S.; Jaenisch T.; Hubbuch J.; Dubel S.; Nesterov-Mueller A.; Breitling F.; Loeffler F. F. Single amino acid fingerprinting of the human antibody repertoire with high density peptide arrays. J. Immunol. Methods 2017, 443, 45–54. 10.1016/j.jim.2017.01.012. [DOI] [PubMed] [Google Scholar]
- Navalkar K. A.; Johnston S. A.; Woodbury N.; Galgiani J. N.; Magee D. M.; Chicacz Z.; Stafford P. Application of immunosignatures for diagnosis of valley fever. Clin. Vaccine Immunol. 2014, 21 (8), 1169–77. 10.1128/CVI.00228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford P.; Halperin R.; Legutki J. B.; Magee D. M.; Galgiani J.; Johnston S. A. Physical characterization of the ″immunosignaturing effect″. Mol. Cell. Proteomics 2012, 11 (4), M111.011593. 10.1074/mcp.M111.011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legutki J. B.; Magee D. M.; Stafford P.; Johnston S. A. A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine 2010, 28 (28), 4529–37. 10.1016/j.vaccine.2010.04.061. [DOI] [PubMed] [Google Scholar]
- Stafford P.; Cichacz Z.; Woodbury N. W.; Johnston S. A. Immunosignature system for diagnosis of cancer. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (30), E3072–80. 10.1073/pnas.1409432111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford P.; Johnston S. A.; Kantarci O. H.; Zare-Shahabadi A.; Warrington A.; Rodriguez M. Antibody characterization using immunosignatures. PLoS One 2020, 15 (3), e0229080. 10.1371/journal.pone.0229080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.; Stafford P.; Schlauch K. A.; Tillett R. R.; Gollery M.; Johnston S. A.; Khaiboullina S. F.; De Meirleir K. L.; Rawat S.; Mijatovic T.; Subramanian K.; Palotas A.; Lombardi V. C. Humoral Immunity Profiling of Subjects with Myalgic Encephalomyelitis Using a Random Peptide Microarray Differentiates Cases from Controls with High Specificity and Sensitivity. Mol. Neurobiol. 2018, 55 (1), 633–641. 10.1007/s12035-016-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther O. P.; Gardy J. L.; Stafford P.; Fluge O.; Mella O.; Tang P.; Miller R. R.; Parker S. M.; Johnston S. A.; Patrick D. M. Immunosignature Analysis of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). Mol. Neurobiol. 2019, 56 (6), 4249–4257. 10.1007/s12035-018-1354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]