Abstract
Chitinolyticbacter meiyuanensis SYBC-H1, a bacterium capable of hydrolyzing chitin and shrimp shell to N-acetyl glucosamine (GlcNAc) as the only product, was isolated previously. Here, the hydrolysis mechanism of this novel strain toward chitin was investigated. Sequencing and analysis of the complete genome of SYBC-H1 showed that it encodes 32 putatively chitinolytic enzymes including 30 chitinases affiliated with the glycoside hydrolase (GH) families 18 (26) and 19 (4), one GH family 20 β-N-acetylglucosaminidase (NAGase), and one Auxiliary Activities (AA) family 10 lytic polysaccharide monooxygenase (LPMO). However, only eight GH18 chitinases, one AA10 LPMO, and one GH20 NAGase were detected in the culture broth of the strain, according to peptide mass fingerprinting (PMF). Of these, genes encoding chitinolytic enzymes including five GH18 chitinases (Cm711, Cm3636, Cm3638, Cm3639, and Cm3769) and one GH20 NAGase (Cm3245) were successfully expressed in active form in Escherichia coli. The hydrolysis of chitinous substrates showed that Cm711, Cm3636, Cm3638, and Cm3769 were endo-chitinases and Cm3639 was exo-chitinase. Moreover, Cm3639 and Cm3769 can convert the GlcNAc dimer and colloidal chitin (CC) into GlcNAc, which showed that they also possess NAGase activity. In addition, NAGase Cm3245 possesses a very high exo-acting activity of hydrolyzing GlcNAc dimer. These results suggest that chitinases and NAGase from SYBC-H1 both play important roles in conversion of N-acetyl chitooligosaccharides to GlcNAc, resulting in the accumulation of the final product GlcNAc. To our knowledge, this is the first report of the complete genome sequence and chitinolytic enzyme genes discovery of this strain.
Keywords: Chitinolyticbacter meiyuanensis SYBC-H1, complete genome sequencing, chitinolytic enzymes, chitin, N-acetyl glucosamine
Introduction
Chitin, a β-1,4-linked N-acetyl glucosamine (GlcNAc) polymer, is the second most abundant polysaccharide in nature with production of 100 billon tons per year (Kaur and Dhillon, 2015). GlcNAc prepared from chitin exhibits specific bioactivities that have been used widely in various fields such as dietary supplements, cosmetic ingredients, and osteoarthritis therapeutics (Chen et al., 2010; Liu et al., 2013). Thus, the conversion of abundant chitin biomass to valuable GlcNAc is both economically and ecologically significant. At present, GlcNAc production from chitin on an industrial scale relies mainly on the hydrolysis of strong acids, which lead to low yield and acidic wastes during the synthesis procedure (Zhang et al., 2016b). Thus, enzymatic production of GlcNAc from chitin using chitinolytic enzymes is a better approach because it is generally environmentally friendly, produces high yields, and generates GlcNAc with high bioactivities (Zhang et al., 2018a).
The degradation of chitin to GlcNAc requires cooperation of chitinolytic enzymes such as chitinase [mainly belonging to glycoside hydrolase (GH) families 18 and 19; hydrolyzes chitin to N-acetyl chitooligosaccharides (N-acetyl COSs)]; β-N-acetylglucosaminidase (GH family 20; hydrolyzes N-acetyl COSs to GlcNAc); and lytic polysaccharide monooxygenase (LPMO) [Auxiliary Activity (AA) families 10 and 11; cleavage of chitin chains with oxidation to enhance the hydrolysis of chitin] (Zhang et al., 2018b). Many microbes have been identified as inexpensive sources of chitinolytic enzymes. However, most reported crude chitinolytic enzymes always produce a mixture of GlcNAc and N-acetyl COSs during enzymatic hydrolysis of chitin. For example, the crude enzymes derived from Trichoderma sp. and Bacillus sp. produce GlcNAc of 36 mg and (GlcNAc)2 of 28 mg from 200 mg of chitin (Kudan et al., 2010). The crude enzymes from Bacillus licheniformis SK-1 chitinase completely hydrolyzed chitin within 6 days, resulting in 75% GlcNAc and 20% (GlcNAc)2 (Pichyangkura et al., 2002). To date, there are few reports of crude chitinolytic enzymes capable of hydrolyzing chitin to produce GlcNAc as the only product (Sashiwa et al., 2002).
A Gram-negative bacterium Chitinolyticbacter meiyuanensis SYBC-H1 belonging to the family Neisseriaceae was isolated previously (Hao et al., 2011). Crude chitinolytic enzymes secreted by SYBC-H1 efficient degrade chitin and shrimp shells to GlcNAc without N-acetyl COSs (Zhang et al., 2016b; Wei et al., 2017). However, the specific mechanism is still unknown. In this study, the complete genome of SYBC-H1 was sequenced and genes encoding potential chitinolytic enzymes were analyzed. Furthermore, the key chitinolytic enzymes in culture broth of SYBC-H1 were determined based on the results of peptide mass fingerprinting (PMF). The genes encoding key chitinolytic enzymes were heterologous expressed in Escherichia coli. In addition, the activity and hydrolysis pattern of these recombinant chitinolytic enzymes were also investigated. This study will help us understand the mechanism by which chitinolytic enzymes from SYBC-H1 convert chitin to GlcNAc as the sole product and advance potential biotechnological applications of SYBC-H1.
Materials and Methods
Strains, Plasmids, and Chemicals
Chitinolyticbacter meiyuanensis SYBC-H1 [CGMCC 3438 T, ATCC BAA-2140(T)] was previously isolated from a soil sample at Meiyuan Park (33°56′N, 120°18′E), Wuxi city, China (Hao et al., 2011). E. coli DH5α and E. coli BL21(DE3) (Novagen Co., Shanghai, China) were, respectively, used in sub-cloning and expression of chitinolytic enzyme genes from C. meiyuanensis SYBC-H1. E. coli strains were cultivated in Luria-Bertani (LB) broth or on agar plates containing 50 μg/mL kanamycin. The pMD19-T and pET-28a(+) plasmids were, respectively, used as the subclone and expression vector of the chitinolytic enzyme genes.
Chitin, GlcNAc, p-nitrophenyl N-acetyl glucosaminide (pNP-GlcNAc), p-nitrophenyl N,N′-diacetyl chitobiose [pNP-(GlcNAc)2], and p-nitrophenyl N,N′,N″-triacetyl chitotriose [pNP-(GlcNAc)3] were purchased from Sigma–Aldrich Co. Ltd. (Shanghai, China). The N-acetyl chitooligosaccharides [(GlcNAc)2-(GlcNAc)6] were acquired from Qingdao BZ Biotechnology Co., Ltd. (Qingdao, China). Peptone and yeast extract were purchased from the Oxoid Co., Ltd. (Beijing, China). The molecular reagents were purchased from Takara Bio Inc. (Dalian, China). Colloidal chitin (CC) was prepared as described in a previous study (Gao et al., 2015). Other chemicals used in this study were purchased from local suppliers at analytical grade or higher purity.
Complete Genome Sequencing and Assembly
SYBC-H1 was cultured according to our previous studies (Zhang et al., 2016a, 2018b). The genomic DNA was extracted using the TIANamp Bacteria DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China). The full genome was sequenced by Personal Biotechnology Co., Ltd. (Shanghai, China). A whole genome shotgun strategy was used in this study. The libraries of different genome inserts (0.2-67 kb) were constructed and sequenced using the third generation single-molecule sequencing technology on the PacBio sequencer. The data obtained by PacBio were assembled using HGAP4 (Chin et al., 2016) and CANU (V1.6) (Koren et al., 2017) software to obtain scaffold sequences. Contigs obtained by splicing the second-generation and third-generation sequencing data were collinearly analyzed using MUMmer (v3) (Delcher et al., 1999) software, the assembly results were confirmed again and the contigs were determined. The positional relationship between the contigs was filled with gaps. Finally, the results were corrected using pilon (version 1.22) (Walker et al., 2014) and then spliced to obtain the complete sequence.
Genome Annotation and Analysis
Sequence alignment of protein-coding genes was performed using DIAMOND (v0.9.10.111) software (Buchfink et al., 2015) based on the National Center for Biotechnology Information (NCBI1) nr database. Swiss-Prot2, Kyoto Encyclopedia of Genes and Genomes (KEGG3), and Clusters of Orthologous Groups (COG4) databases were used for functional annotation. Pairwise BLAST-based Average Nucleotide Identity (ANI) were obtained using JSpecies5 and digital DNA–DNA Hybridization (dDDH) values were calculated with the Genome-to-Genome Distance Calculator 99 (GGDC 2.1), using formula 26. The KO and Pathway annotations of the protein-coding genes were mainly performed using KEGG’s automated annotation system (KAAS Ver. 2.1) (Moriya et al., 2007). The promoter was analyzed using Promoter 2.0 Prediction Server7. Nucleotide and amino acid sequences were analyzed using Snap GeneTM 1.1.3 software8 and the ExPASy ProtParam tool9. The Carbohydrate-Active enZyme (CAZy) enzyme genes present in the genomic sequence were predicted using hmmscan (3.1b2, February 2015) (Yoon, 2009) software. Enzyme locations were analyzed using the SignalP 5.0 server10 and Gneg-mPLoc server v.2.011. The conserved domains and classifications were identified using SMART database12. The DNA and protein sequence alignments were performed via the NCBI server with the programs BLASTN and BLASTP13, respectively.
Identification of Chitinolytic Enzymes in Fermentation Broth of C. meiyuanensis SYBC-H1
The production of chitinolytic enzymes of the strain was conducted according to our previous study (Zhang et al., 2018b). The supernatant from the culture was used for PMF analysis with an electrospray ionization quadrupole time-of-flight mass spectrometer (ESI-Q-TOF MS/MS) technique (PROTTECH, Inc., Suzhou, China). The detected peptide masses were then compared to theoretical mass values in the Mascot website databases14 to reveal the amino acid sequences of the peptide fragments. The peptide sequence was then aligned with the genome of C. meiyuanensis SYBC-H1 (GenBank accession number, CP041335) to find the corresponding proteins and their coding genes.
Cloning and Expression of Genes Encoding Chitinolytic Enzymes in Culture Broth
The genomic DNA of SYBC-H1 was used as the template for PCR amplification. Primers with suitable restriction sites designed to amplify the genes are listed in Table 1. The primers were used to amplify DNA fragments corresponding to the whole ORF without any putative signal peptide. The amplified PCR products were purified by gel electrophoresis, digested with restriction enzymes, and then ligated into pMD19-T vector and sequenced by Invitrogen Corporation (Shanghai, China). The positive recombinant plasmids were digested with corresponding restriction enzymes, and these genes were inserted into the pET-28a(+) vector expression plasmid with a C-terminal His6 tag. The recombinant plasmids were introduced into E. coli BL21 (DE3) and then cultured in LB medium (containing 50 μg/mL kanamycin) at 37°C in a shaker with a rotation speed of 200 r/min. When the cell density (OD600) was 0.6, isopropyl β-D-thiogalactoside was added at a final concentration of 1 mM for protein induction, and was further grown at 25°C for 12 h.
TABLE 1.
Oligonucleotide primers used for PCR in this study.
| Primers | Oligo-nucleotides (5′-3′) | Restriction sites |
| ORF711-F | GAATTCCATATGCGTTCCCG GCCTGGCAG | NdeI |
| ORF711-R | TCCGCTCGAGCTGCAGGGTA GCGGCCG | XhoI |
| ORF3245-F | GAATTCCATATGATGAGCCG TCCCGCCGGATC | NdeI |
| ORF3245-R | TCCGCTCGAGGGCGCCCACC TGCACCG-3′ | XhoI |
| ORF3636-F | GAATTCCATATGGCCCATGC CGCACCGGCATG | NdeI |
| ORF3636-R | CCCAAGCTTGTACTTCAACG CGCTGCAG | HindIII |
| ORF3638-F | GAATTCCATATGGCTGAATG GGCGGAAGGCAAC | NdeI |
| ORF3638-R | TCCGCTCGAGGTGTGATGTC CGGCGTACGC | XhoI |
| ORF3639-F | GAATTCCATATGCGTACCCG GTATGGCAGG | NdeI |
| ORF3639-R | TCCGCTCGAGCATGGGCAAC ATGAACAAG | XhoI |
| ORF3769-F | GAATTCCATATACCGAGATC GCCCCGTACTTC | NdeI |
| ORF3769-R | TCCGCTCGAGGGTGCTGGGC AACGGCCGC | XhoI |
The underlined sequences are restriction enzyme cutting sites.
The cells were harvested by centrifugation at 6000 g and 4°C for 5 min, after which the cell precipitation was re-suspended with His-tag binding buffer (20 mM Tris-HCl, 500 mM NaCl, 50 mM imidazole [pH 7.0]) and lyzed by JY92-IIN ultrasonication (Ningbo Xinzhi biotechnology, Ltd., Ningbo, China). Cell debris was removed by centrifugation at 8000 g for 5 min and the supernatant was purified using a fast protein liquid chromatography (FPLC) system (GE AKTA Pure 150; General Electric Co., Boston, MA, United States) with a Ni-nitrilotriacetic acid affinity chromatography column (His Trap FF 5 mL). The target protein was eluted with elution buffer (20 mM Tris-HCl, 500 mM NaCl, 250 mM imidazole [pH 7.0]). The eluted fractions were passed through 10 kDa ultrafiltration tubes (Millipore, United States) to remove the imidazole with sodium phosphate (pH 7.0) and concentrate the recombinant enzymes.
The purified enzymes were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 3% stacking gel and a 10% separating gel, according to the method described by Laemmli (1970). The protein marker (Takara Biotechnology Co., Ltd., Nanjing, China) containing 180-, 140-, 100-, 75-, 60-, 45-, 30-, and 25-kDa proteins was used as the molecular mass standard. Protein concentrations were determined using the Bradford method with bovine serum albumin as the standard (Bradford, 1976).
Determination of Enzyme Activities
The assay of enzyme activities used CC and chromogenic substrates. For CC, a mixture containing 0.5 mL CC of 1% (w/v), 1.4 mL 50 mM sodium phosphate buffer (pH 7.0), and 100 μg enzyme, was incubated for 30 min at 37°C. After incubating at 37°C for 30 min, the reaction was terminated by boiling for 5 min. Reducing sugars released during the reaction were quantified using GlcNAc standards at concentrations ranging from 0.25–1.25 μmol/mL, according to the 3,5-dinitrosalicylic acid method. For (GlcNAc)2, the standard reaction mixture (1 mL) contained 50 mM sodium phosphate buffer (pH 7.0), 10 g/L (GlcNAc)2, and 5 μg enzyme. After incubation at 37°C for 30 min, the reaction was terminated by boiling for 5 min. The GlcNAc released was measured by liquid chromatography (LC). One unit (1 U) of chitinase activity was defined as the amount of enzyme required to produce 1 μmol GlcNAc per minute at 37°C.
For chromogenic substrates, pNP-GlcNAc, pNP-(GlcNAc)2, and pNP-(GlcNAc)3 were used as substrates, the standard reaction mixture (1 mL) contained 50 mM sodium phosphate buffer (pH 7.0), 0.5 mM chromogenic substrate, and 5 μg enzyme. The mixture was incubated at 37°C for 30 min, and then the reaction was terminated by adding 1 mL of 0.2 M NaOH. The amount of pNP released during the reaction was measured by reading the optical density at 405 nm. One unit of chitinase activity was defined as the amount of enzyme required to release 1 μmol pNP from the substrate per minute at 37°C.
The Hydrolysis Pattern of Chitinolytic Enzymes Toward CC
Reaction mixtures (1 mL) containing purified chitinolytic enzymes (50 μg) and 0.5 mL CC at a final concentration of 10 g/L were incubated in 0.4 mL of 50 mM sodium phosphate buffer (pH 7.0) at 37°C for 12 h. In each case, the supernatant after hydrolysis was centrifuged at 12,000 g for 5 min. Reaction products were analyzed using an Agilent 1260 series LC system (Agilent Technologies, Santa Clara, CA, United States) according to our previous study (Zhang et al., 2018b).
Nucleotide Sequence Accession Number
The complete genome sequence of C. meiyuanensis SYBC-H1 was deposited in GenBank under the accession number CP041335.
Results and Discussion
General Features and Gene Annotation of the Complete Genome of C. meiyuanensis SYBC-H1
To explore the genetic basis for the ability of chitin degradation, the complete genome of strain C. meiyuanensis SYBC-H1 was sequenced. The genome shows the highest similarity to the reported genome of Chitiniphilus shinanonensis strain SAY3 (95.85%) (Sato et al., 2009), followed by Chitinibacter tainanensis DSM15459 (94.58%) (Chern et al., 2004). ANI and dDDH values were used to accurately measure nucleotide-level genomic similarity between SYBC-H1 and the two strains. The results showed that the ANI and dDDH values between SYBC-H1 and SAY3 genomes were 78.73 and 22.30%, respectively. The ANI and dDDH values between the SYBC-H1 and DSM15459 genomes were 71.09 and 19.20%, respectively. These ANI and dDHH results are far lower than the species cut-off thresholds of 96 and 70% for ANI and dDDH, respectively, suggesting that SYBC-H1 is not the same species as SAY3 or DSM15459.
The characteristics of the complete genome of SYBC-H1 are shown in Table 2. The final assembly of the SYBC-H1 genome was 4.3 Mb in size with a high GC content of 63.52%, 57 tRNA genes, 12 rRNA genes, and three other ncRNA genes (Figure 1 and Table 2). These data show some differences from SAY3 and DSM15459 genomes.
TABLE 2.
Genome features of C. meiyuanensis SYBC-H1.
| Features | SYBC-H1 | SAY3 | DSM15459 |
| Genome size (Mb) | 4.33 | 4.15 | 3.42 |
| GC content (%) | 63.5 | 66.4 | 56.1 |
| proteins | 4,018 | 3,658 | 3,141 |
| ORF total length (bp) | 3,815,517 | – | – |
| ORF average length (bp) | 928.8 | – | – |
| Longest ORF length (bp) | 37,698 | – | – |
| ORF density | 0.949 genes per kb | – | – |
| tRNAs | 56 | 54 | 54 |
| rRNAs (5S, 16S, 23S) | 12 | 6 | 13 |
| ncRNAs | 3 | 4 | 4 |
| GenBank accession number | CP041335 | ARGN00000000 | AUCN00000000 |
FIGURE 1.
Classical circular genome maps of the C. meiyuanensis SYBC-H1. From the inner circle to the outermost, each circle represents (1) genome size (0.5 Mb/scale), (2) GC skew, (3) GC content reverse CDS, (4) COG groups of each CDS, (5) CDS, (6) rRNA/tRNA.
A total of 4108 ORFs, which account for 88.20% of the total nucleotides, were obtained in the SYBC-H1 genome. Among these genes, 3734 (90.9%), 3436 (83.64%), 1932 (47.03%), 2691 (65.51%), and 2838 (69.08%) coding genes were functionally annotated using NR (NCBI), COG, KEGG, Swiss-Prot, and Gene Ontology (GO) databases, respectively (Table 3).
TABLE 3.
The functional annotation of protein coding gene.
| Annotation in database | No. of genes | Proportion of annotation (%) |
| NR | 3,734 | 90.90 |
| COG | 3,436 | 83.64 |
| KEGG | 1,932 | 47.03 |
| Swiss-Prot | 2,691 | 65.51 |
| GO | 2,838 | 69.08 |
Prediction and Analysis of Genes Encoding Chitinolytic Enzymes in the SYBC-H1 Genome
Bacteria utilize chitin as a carbon source by degrading it into GlcNAc, a process that requires synergy of several chitinolytic enzymes such as chitinase and NAGase (Bhattacharya et al., 2007). Analysis of the SYBC-H1 genome showed that it encodes 32 enzymes that could potentially be involved in chitin utilization, including 26 GH18 chitinases, four GH19 chitinases, one GH20 β-N-acetyl glucosaminidase (NAGase), and one AA10 LPMO (Figure 2), which is more than most bacteria such as SAY3 (12 GH18 chitinases, one GH19 chitinase, and one GH20 NAGase) (Huang et al., 2012a), Streptomyces coelicolor A3 (11 GH18 chitinases, two GH19 chitinases, and five exo-NAGases) (Bentley et al., 2002), Paenibacillus xylanilyticus W4 (six GH18 chitinases) (Liao et al., 2019). The 32 chitinolytic enzymes, except for Cm3769 with two GH18 catalytic domains, have only one catalytic domain. Chitin-binding domain (ChBD) can efficiently anchor the domains on the surface of chitin and help catalytic domains to digest insoluble chitin efficiently (Boraston et al., 2004). Except for GH 18 chitinases (Cm2058, Cm2059, and Cm2362) and GH20 NAGase (Cm3245), most of them contain one or two ChBDs, which is similar to most reported chitinolytic enzymes (Huang et al., 2012a; Yang et al., 2016; Lee et al., 2018; Tanaka et al., 2018).
FIGURE 2.
Domain organization of chitinolytic enzymes. Protein domains were identified using SMART databases. aa, amino acid; SP, signal peptide; GH18, Glycoside Hydrolase family 18; GH19, Glycoside Hydrolase family 19; GH20, Glycoside Hydrolase family 20; ChBD, chitin-binding domain; AA10, Auxiliary Activity family 10; Black line, no function area.
SAY3, the most similar bacterium to SYBC-H1, encodes 15 chitinolytic enzymes (ChiA–O), consisting of 12 GH18 chitinases, one GH19 chitinase, one GH20 family NAGase, and a protein with only two ChBDs and no catalytic domains (ChiF). Seven genes encoding GH18 ChiB–H form a cluster in the SAY3 genome. Of these, five genes encoding GH18 ChiC–G are likely to constitute an operon, as well as the genes encoding GH18 ChiL and ChiM (Huang et al., 2012a,b). A similar result was found in the SYBC-H1 genome, with seven genes encoding GH18 chitinases Cm3633-3639 likely to form a cluster, as well as genes encoding GH18 chitinases Cm2058-2059 and genes encoding chitinases GH18 Cm2362-2363. Analysis of the operon identified short distances in ORF3635-3638 (74–177 bp) and ORF2058-2059 (174 bp). Moreover, the promoter-like sequence was also identified upstream of the ORF3638 and ORF2058. The results showed that ORF3635-3638 likely form an operon as well as ORF2058-2059.
Identification of Chitinolytic Enzymes in Culture Broth of C. meiyuanensis SYBC-H1
Although we identified 32 genes encoding chitinolytic enzymes in the SYBC-H1 genome, the expression patterns of these genes in medium containing chitin is not known. Thus, the chitinolytic enzymes in culture broth of SYBC-H1 were identified by combining PMF and genomic data. As shown in Table 4, 10 chitinolytic enzymes containing eight GH18 chitinases (Cm711, Cm3635, Cm3636, Cm3637, Cm3638, Cm3639, Cm3653, and Cm3769), one AA10 LPMO (Cm3403), and one GH20 NAGase (Cm3245) were determined in culture broth. Among them, the relative contents of Cm711 (10.2%), Cm3245 (5.2%), Cm3636 (12.4%), Cm3638 (8.9%), Cm3639 (19.3%), and Cm3769 (11.3%) in broth were high, which showed that they play a key role in degrading chitin to GlcNAc. BLAST analyses revealed that these 10 chitinolytic enzymes showed significant similarity (60.54–99.70%) to various bacterial chitinolytic enzymes. However, their enzymatic characteristics have not been clarified.
TABLE 4.
Chitinolytic enzymes in culture broth of C. meiyuanensis SYBC-H1.
| Chitinolytic enzymes | Relative content | Probability | Family | aMolecular Weight (kDa) | Signal peptide | The closest relative sequence (similarity) | Accession number |
| Cm711 | 10.2% | 99.0% | GH18 | 96.8 | Yes | Chitinilyticum litopenaei chitinase (81.06%) | WP_028454508 |
| Cm3245 | 5.2% | 99.0% | GH20 | 91.7 | No | Chitiniphilus shinanonensis NAGase (81.68%) | WP_018749679 |
| Cm3403 | 3.6% | 99.0% | AA10 | 53.7 | Yes | Iodobacter sp. BJB302 chitinase (60.54%) | WP_099398589 |
| Cm3635 | 1.3% | 99.0% | GH18 | 81.1 | Yes | Chitinibacter sp. ZOR0017 chitinase (74.24%) | WP_047395740 |
| Cm3636 | 12.4% | 99.0% | GH18 | 55.9 | Yes | Andreprevotia lacus DSM23236 chitinase (62.95%) | SMC28638 |
| Cm3637 | 3.9% | 99.0% | GH18 | 84.9 | Yes | Jeongeupia sp. USM3 chitinase (77.83%) | WP_070528194 |
| Cm3638 | 8.9% | 99.4% | GH18 | 63.7 | Yes | Chitiniphilus eburneus chitinase (78.41%) | WP_136773598 |
| Cm3639 | 19.3% | 99.0% | GH18 | 67.7 | Yes | Staphylococcus sp. J2 chitinase (99.70%) | AGC59908 |
| Cm3653 | 0.2% | 88.3% | GH18 | 68.8 | Yes | Chitiniphilus eburneus chitinase (78.62%) | WP_136774653 |
| Cm3769 | 11.3% | 99.0% | GH18 | 100.7 | Yes | Chitiniphilus eburneus chitinase (74.52%) | WP_136773598 |
aMolecular weight without signal peptide.
Bacteria need to degrade chitin to GlcNAc before they can use it. However, the substrate chitin has a large molecular weight and cannot enter cells. Thus, bacterial chitinolytic enzymes must be secreted extracellularly to utilize chitin. Prediction of signal peptides showed that eight GH18 chitinases and one AA10 LPMO possess a putative N-terminal signal peptide, which showed that the locations of these enzymes were extracellular. To date, most reported NAGases (hydrolyze the N-acetyl COSs to GlcNAc) are secretory proteins (Konno et al., 2012; Zhou et al., 2017; Visnapuu et al., 2020). However, there was no putative signal peptide in the sequence of GH20 NAGase Cm3245. On the contrary, the prediction using Gneg-mPLoc 2.0 showed that the location of Cm3245 was in the periplasm. In addition, Cm3245 was detected in the fermentation broth of SYBC-H1. This phenomenon suggests that Cm3245 is a secretory protein.
Our previous study showed that chitinolytic enzymes purified from the culture broth of SYBC-H1 based on the affinity adsorption between ChBD and substrate chitin can also hydrolyze chitin to GlcNAc as the sole product (Zhang et al., 2016b). However, NAGase Cm3245 cannot be purified in this process, because it does not have ChBD. Therefore, we infer that other detected chitinolytic enzymes (besides Cm3245) may also possess NAGase activity.
Cloning and Expression of Genes Encoding Key Chitinolytic Enzymes
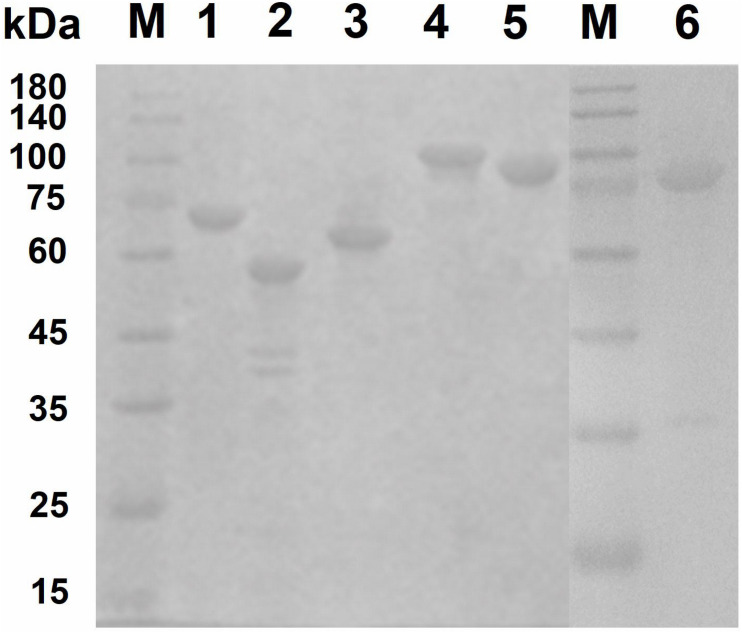
To investigate the key enzymes (Cm711, Cm3245, Cm3636, Cm3638, Cm3639, and Cm3769) noted above, the genes encoding these enzymes were cloned without signal peptide and expressed in E. coli BL21(DE3). The recombinant proteins with a C-terminal His6 tag were successfully expressed in a soluble form and were purified by Ni-NTA affinity chromatography (Figure 3). The molecular weights of recombinant Cm711 (96.2 kDa), Cm3245 (92.1 kDa), Cm3636 (56.3 kDa), Cm3638 (63.4 kDa), Cm3639 (67.3 kDa), and Cm3769 (100.1 kDa) were consistent with those calculated from the amino acid sequence. The purified recombinant proteins were used in the characterization of chitin degrading activity.
FIGURE 3.
SDS–PAGE analysis of purified recombinant proteins. The purified recombinant proteins (20 μg) were analyzed by SDS-PAGE using a 3% stacking gel and a 10% separating gel with addition of SDS. Lane M, molecular weight markers; lane 1, Cm3639; lane 2, Cm3636; lane 3, Cm3638; lane 4, Cm3769; lane 5, Cm711; lane 6, Cm3245.
Properties of Key Recombinant Chitinolytic Enzymes
The activities of six purified recombinant proteins were examined using CC and chromogenic substrates, pNP-(GlcNAc)1–3. As shown in Table 5, the recombinant GH18 chitinases (Cm711, Cm3636, Cm3638, and Cm3769) exhibited hydrolysis activity toward CC, as well as pNP-(GlcNAc)2 and pNP-(GlcNAc)3, which showed that they were endo-chitinases. Interestingly, Cm3639 and Cm3769 showed some hydrolysis activity toward pNP-GlcNAc and (GlcNAc)2, which revealed that they possess NAGase activity, which is similar to chitinase PbChi74 from Paenibacillus barengoltzii (Fu et al., 2014). In our previous study, we confirmed that Cm3639 (CmChi1) was a multifunctional chitinase with mainly exo-acting activity and trace endo-acting and NAGase activities, which can hydrolyze CC to only GlcNAc over a long reaction time (Zhang et al., 2018b). However, the hydrolysis products of other enzymes toward CC were unknown. Thus, the hydrolysis pattern toward CC was investigated in this study.
TABLE 5.
Enzyme activities of recombinant key chitinolytic enzymes toward chitinous substrates.
| Enzymes |
Enzyme activity (U/mg of protein) |
||||
| CC | (GlcNAc)2 | pNP-GlcNAc | pNP-(GlcNAc)2 | pNP-(GlcNAc)3 | |
| Cm711 | 3.6 ± 0.2 | nda | nd | 2.3 ± 0.1 | 1.7 ± 0.1 |
| Cm3245 | 0.02 ± 0.001 | 4,233 ± 210 | 3,902.9 ± 130 | 2,140 ± 160 | 1,082 ± 62 |
| Cm3636 | 2.1 ± 0.1 | nd | nd | 6.2 ± 0.3 | 0.7 ± 0.02 |
| Cm3638 | 4.6 ± 0.2 | nd | nd | 8.5 ± 0.7 | 1.6 ± 0.1 |
| Cm3639 | 15.3 ± 0.3 | 0.5 ± 0.02 | 0.2 ± 0.01 | 27.3 ± 0.6 | nd |
| Cm3769 | 4.1 ± 0.4 | 13.5 ± 0.9 | 10.3 ± 0.6 | 13.3 ± 1.2 | 3.2 ± 0.2 |
aActivity was not detected. Reaction mixture (1 mL) containing enzyme and 10 g/L CC, (GlcNAc)2, or 0.5 mM pNP-(GlcNAc)1–3 were incubated at 37°C and pH 7.0.
Cm711 and Cm3636 digested the CC into a mixture of GlcNAc and (GlcNAc)2 (Figure 4). Cm3638 mainly produces (GlcNAc)2–4 from CC. These results showed that Cm711, Cm3636, and Cm3638 possess typical characteristics of endo-type chitinases, consistent with results above. Interestingly, Cm3769 completely converted CC into GlcNAc as the only product from beginning to end. The result is different from CmChi1 and PbChi74, which yield mainly (GlcNAc)2 at the initial hydrolysis stage and (GlcNAc)2 needed a longer reaction time to achieve full conversion into GlcNAc. The phenomena can be explained that NAGase activity (13.5 U/mg) of Cm3769 is higher than its endo activity (5.1 U/mg), causing the produced N-acetyl COSs to be immediately degraded to GlcNAc. The result accounts for our previous study, which showed that chitinases from SYBC-H1 without NAGase degraded chitin to GlcNAc as the sole product (4). In addition, the NAGase Cm3245 possesses very high activity (1082-4233 U/mg) for (GlcNAc)2 and pNP-(GlcNAc)1–3, which showed that it represents the typical exo NAGase activity. Moreover, Cm3245 showed some activity (0.02 U/mg) toward CC, making it unique among NAGases. Characterization of this enzyme is ongoing.
FIGURE 4.
HPLC analysis of the hydrolysate of CC generated by purified recombinant chitinolytic enzymes (A) Cm711, (B) Cm3636, (C) Cm3638, and (D) Cm3769. A total of 10 g/L CC was incubated with enzymes (5 μg) in a 2 mL volume at pH 7 and 37°C for different time intervals. 1-6: Standards of GlcNAc, (GlcNAc)2, (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6.
In conclusion, NAGase activity of chitinolytic enzymes from SYBC-H1 is much higher than its chitinase activity, which leads to the accumulation of GlcNAc.
Analysis of Genes Encoding GlcNAc Metabolic Pathway in the SYBC-H1 Genome
In nature, bacteria use chitin by degrading insoluble chitin into GlcNAc for their own growth (Meibom et al., 2004). Thus, the genes involved in GlcNAc metabolism in the SYBC-H1 genome were also analyzed. As shown in Figure 5, GlcNAc metabolic essential genes encoding N-acetylglucosamine kinase (Cm281, catalyzes GlcNAc to GlcNAc-6-phosphate), N-acetylglucosamine-6-phosphate deacetylase (Cm3796, catalyzes GlcNAc-6-phosphate to glucosamine 6-phosphate), and glucosamine-6-phosphate deaminase (Cm2530, catalyzes glucosamine 6-phosphate to fructose-6-phosphate, that enters central glycolytic pathway for cell growth) were both found (Beier and Bertilsson, 2013).
FIGURE 5.
Proposed pathway of chitin degradation in C. meiyuanensis SYBC-H1. GH18, Glycoside Hydrolase family 18 chitinase; GH20, Glycoside Hydrolase family 20 β-N-acetylglucosaminidase; AA10, Auxiliary Activity family 10 lytic polysaccharide monooxygenase; ABC, ABC-sugar transporter; kinase, N-acetylglucosamine kinase; deacetylase, N-acetylglucosamine-6-phosphate deacetylase; deaminase, glucosamine-6-phosphate deaminase.
Data Availability Statement
The datasets generated for this study can be found in the NCBI database, under BioProject PRJNA576511 and genome assemblies GCA_011149675.1, GCA_009184605.1, GCA_009183735.1, GCA_009183705.1 and GCA_009183685.1.
Author Contributions
AZ and ZH conceived and designed the research. AZ, XM, YW, and NZ performed the experiments. AZ, XM, and GW analyzed the data. AZ and KC wrote the manuscript. All authors commented on the manuscript and approved the content.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Key Research and Development Program (2016YFA0204300), the National Natural Science Foundation of China (31700092), the National Nature Science Foundation for Young Scientists of China (Grant Nos. 21908101 and 21576134), and China Postdoctoral Science Foundation (2018M642237 and 2020T130295).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.572053/full#supplementary-material
References
- Beier S., Bertilsson S. (2013). Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front. Microbiol. 4:149. 10.3389/fmicb.2013.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley S. D., Chater K. F., Cerdeno-Tarraga A. M., Challis G. L., Thomson N. R., James K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417 141–147. 10.1038/417141a [DOI] [PubMed] [Google Scholar]
- Bhattacharya D., Nagpure A., Gupta R. K. (2007). Bacterial chitinases: Properties and potential. Crit. Rev. Biotechnol. 27 21–28. 10.1080/07388550601168223 [DOI] [PubMed] [Google Scholar]
- Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382 769–781. 10.1042/bj20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Chen J.-K., Shen C.-R., Liu C.-L. (2010). N-Acetylglucosamine: production and applications. Mar. Drugs 8 2493–2516. 10.3390/md8092493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern L. L., Stackebrandt E., Lee S. F., Lee F. L., Chen J. K., Fu H. M. (2004). Chitinibacter tainanensis gen. nov., sp nov., a chitin-degrading aerobe from soil in Taiwan. Int. J. Syst. Evol. Microbiol. 54 1387–1391. 10.1099/ijs.0.02834-0 [DOI] [PubMed] [Google Scholar]
- Chin C.-S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., Clum A., et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13:1050. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Kasif S., Fleischmann R. D., Peterson J., White O., Salzberg S. L. (1999). Alignment of whole genomes. Nucleic Acids Res. 27 2369–2376. 10.1093/nar/27.11.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Yan Q., Yang S., Yang X., Guo Y., Jiang Z. (2014). An acidic, thermostable exochitinase with beta-N-acetylglucosaminidase activity from Paenibacillus barengoltzii converting chitin to N-acetyl glucosamine. Biotechnol. Biof. 7:174. 10.1186/s13068-014-0174-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Zhang A., Chen K., Hao Z., Tong J., Ouyang P. (2015). Characterization of extracellular chitinase from Chitinibacter sp GC72 and its application in GlcNAc production from crayfish shell enzymatic degradation. Biochem. Eng. J. 97 59–64. 10.1016/j.bej.2015.02.010 [DOI] [Google Scholar]
- Hao Z. K., Cai Y. J., Liao X. R., Liang X. H., Liu J. Y., Fang Z. Y., et al. (2011). Chitinolyticbacter meiyuanensis SYBC-H1(T), Gen. Nov., sp Nov., a Chitin-Degrading Bacterium Isolated From Soil. Curr. Microbiol. 62 1732–1738. 10.1007/s00284-011-9921-5 [DOI] [PubMed] [Google Scholar]
- Huang L., Garbulewska E., Sato K., Kato Y., Nogawa M., Taguchi G., et al. (2012a). Isolation of genes coding for chitin-degrading enzymes in the novel chitinolytic bacterium, Chitiniphilus shinanonensis, and characterization of a gene coding for a family 19 chitinase. J. Biosci. Bioeng. 113 293–299. 10.1016/j.jbiosc.2011.10.018 [DOI] [PubMed] [Google Scholar]
- Huang L., Shizume A., Nogawa M., Taguchi G., Shimosaka M. (2012b). Heterologous expression and functional characterization of a novel chitinase from the chitinolytic bacterium Chitiniphilus shinanonensis. Biosci. Biotechnol. Biochem. 76 517–522. 10.1271/bbb.110822 [DOI] [PubMed] [Google Scholar]
- Kaur S., Dhillon G. S. (2015). Recent trends in biological extraction of chitin from marine shell wastes: a review. Crit. Rev. Biotechnol. 35 44–61. 10.3109/07388551.2013.798256 [DOI] [PubMed] [Google Scholar]
- Konno N., Takahashi H., Nakajima M., Takeda T., Sakamoto Y. (2012). Characterization of beta-N-acetylhexosaminidase (LeHex20A), a member of glycoside hydrolase family 20, from Lentinula edodes (shiitake mushroom). Amb. Express 2:29. 10.1186/2191-0855-2-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., Phillippy A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudan S., Eksittikul T., Pichyangkura R., Park R. D. (2010). Preparation of N-acetyl-D-glucosamine and N,N ’-diacetylchitobiose by enzymatic hydrolysis of chitin with crude chitinases. J. Biotechnol. 150 S89–S89. 10.1016/j.jbiotec.2010.08.229 [DOI] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lee H. J., Lee Y. S., Choi Y. L. (2018). Cloning, purification, and characterization of an organic solvent-tolerant chitinase, MtCh509, from Microbulbifer thermotolerans DAU221. Biotechnol. Biof. 11:14. 10.1186/s13068-018-1299-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Liu P., Liao W., Miao L. (2019). Complete Genome of the Chitin-Degrading Bacterium, Paenibacillus xylanilyticus W4. Genome Biolo. Evol. 11 3252–3255. 10.1093/gbe/evz241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu Y., Shin H.-D., Chen R., Li J., Du G., et al. (2013). Microbial production of glucosamine and N-acetylglucosamine: advances and perspectives. Appl. Microbiol. Biotechnol. 97 6149–6158. 10.1007/s00253-013-4995-6 [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Li X. B. B., Nielsen A. T., Wu C. Y., Roseman S., Schoolnik G. K. (2004). The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101 2524–2529. 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35 W182–W185. 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichyangkura R., Kudan S., Kuttiyawong K., Sukwattanasinitt M., Aiba S. (2002). Quantitative production of 2-acetamido-2-deoxy-D-glucose from crystalline chitin by bacterial chitinase. Carbohydrate Res. 337 557–559. 10.1016/s0008-6215(02)00007-1 [DOI] [PubMed] [Google Scholar]
- Sashiwa H., Fujishima S., Yamano N., Kawasaki N., Nakayama A., Muraki E., et al. (2002). Production of N-acetyl-D-glucosamine from alpha-chitin by crude enzymes from Aeromonas hydrophila H-2330. Carbohydr. Res. 337 761–763. 10.1016/s0008-6215(02)00034-4 [DOI] [PubMed] [Google Scholar]
- Sato K., Kato Y., Taguchi G., Nogawa M., Yokota A., Shimosaka M. (2009). Chitiniphilus shinanonensis gen. nov., sp nov., a novel chitin-degrading bacterium belonging to Betaproteobacteria. J. Gen. Appl. Microbiol. 55 147–153. 10.2323/jgam.55.147 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Akutsu H., Yabuta I., Hara M., Sugimoto H., Ikegami T., et al. (2018). A novel chitin-binding mode of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12 revealed by solid-state NMR. FEBS Lett. 592 3173–3182. 10.1002/1873-3468.13226 [DOI] [PubMed] [Google Scholar]
- Visnapuu T., Teze D., Kjeldsen C., Lie A., Duus J. O., Andre-Miral C., et al. (2020). Identification and Characterization of a beta-N-Acetylhexosaminidase with a Biosynthetic Activity from the Marine Bacterium Paraglaciecola hydrolytica S66(T). Int. J. Mol. Sci. 21:22. 10.3390/ijms21020417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Zhang A., Chen K., Ouyang P. (2017). Enzymatic production of N-acetyl-D-glucosamine from crayfish shell wastes pretreated via high pressure homogenization. Carbohydr. Polym. 171 236–241. 10.1016/j.carbpol.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Yang S., Fu X., Yan Q., Guo Y., Liu Z., Jiang Z. (2016). Cloning, expression, purification and application of a novel chitinase from a thermophilic marine bacterium Paenibacillus barengoltzii. Food Chem. 192 1041–1048. 10.1016/j.foodchem.2015.07.092 [DOI] [PubMed] [Google Scholar]
- Yoon B.-J. (2009). Hidden markov models and their applications in biological sequence analysis. Curr. Genom. 10 402–415. 10.2174/138920209789177575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Gao C., Chen K., Wei C., Ouyang P. (2016a). Enhanced chitinase production by Chitinolyticbacter meiyuanensis SYBC-H1 using staged pH control. J. Gen. Appl. Microbiol. 62 126–131. 10.2323/jgam.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Zhang A., Gao C., Wang J., Chen K. Q., Ouyang P. K. (2016b). An efficient enzymatic production of N-acetyl-D-glucosamine from crude chitin powders. Green Chem. 18 2147–2154. 10.1039/c5gc02242h [DOI] [Google Scholar]
- Zhang A., Wei G. G., Mo X. F., Zhou N., Chen K. Q., Ouyang P. K. (2018a). Enzymatic hydrolysis of chitin pretreated by bacterial fermentation to obtain pure N-acetyl-D-glucosamine. Green Chem. 20 2320–2327. 10.1039/c8gc00265g [DOI] [Google Scholar]
- Zhang A., He Y. M., Wei G. G., Zhou J., Dong W. L., Chen K. Q., et al. (2018b). Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-D-glucosamine production. Biotechnol. Biof. 11:14. 10.1186/s13068-018-1169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Song Z., Zhang R., Chen C., Wu Q., Li J., et al. (2017). A Shinella beta-N-acetylglucosaminidase of glycoside hydrolase family 20 displays novel biochemical and molecular characteristics. Extremophiles 21 699–709. 10.1007/s00792-017-0935-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI database, under BioProject PRJNA576511 and genome assemblies GCA_011149675.1, GCA_009184605.1, GCA_009183735.1, GCA_009183705.1 and GCA_009183685.1.