Abstract
The Metabolomics Quality Assurance and Quality Control Consortium (mQACC) evolved from the recognized need for a community-wide consensus on improving and systematizing quality assurance (QA) and quality control (QC) practices for untargeted metabolomics. As an initial step, members of the consortium and several non-members who used liquid chromatography-mass spectrometry (LC-MS) untargeted metabolomics were asked to voluntarily participate in a collaborative research project and took part by providing the QA and QC practices utilized in their laboratories, via a six-page questionnaire composed of over 120 questions and comment fields. All contributors to this project are authors. Responses were then analyzed to identify common and divergent QA and QC practices among the contributing laboratories. For QA, many laboratories reported documenting maintenance, calibration and tuning (82%); having established data storage and archival processes (71%); depositing data in public repositories (55%); having standard operating procedures (SOPs) in place for all laboratory processes (68%) and training staff on laboratory processes (55%). For QC, universal practices included using system suitability procedures (100%) and using a robust system of identification (Metabolomics Standards Initiative level 1 identification standards) for at least some of the detected compounds. Most laboratories used QC samples (>86%); used internal standards (91%); used a designated analytical acquisition template with randomized experimental samples (91%); and manually reviewed peak integration following data acquisition (86%). A minority of laboratories included technical replicates of experimental samples in their workflows (36%). Due to the recruitment method for participants and its voluntary nature, although the 23 contributors were researchers with diverse and international backgrounds from academia, industry and government, most being current members of mQACC, they are not necessarily representative of the worldwide pool of practitioners. The findings presented here, in addition to other data gathering efforts within mQACC, will be used to guide discussions for recommendations of best practices within the community and to establish internationally agreed upon reporting standards.
Introduction
Untargeted metabolomics detects and identifies as many small molecules (typically compounds < 2000 Da) present in specimens as is technically possible for a given platform, usually in the range of hundreds to low thousands. Often referred to as a discovery tool, untargeted metabolomics provides an open evaluation of detectable small molecules and can identify novel signatures of exposures or disease, in animals, plants and microbes and provide a route to understand biological processes at a mechanistic level. Unlike measures generated from genomics, small molecules represent the products of endogenous metabolism, in addition to products of environmental exposure (including lifestyle, diet, and other environmental exposures), individual-specific metabolism driven by underlying genetics, gut microbial influences, and the presence or absence of disease or pathological processes. Data derived from untargeted metabolomics have been used to interrogate the mechanisms underlying exposure-disease relationships and treatment or intervention effects as well as to understand the metabolic phenotype at the human population level (Dunn et al., 2015, Ilhan et al., 2019, Gafson et al., 2019, Chen et al., 2019, Bouhifd et al., 2013, Ramirez et al., 2013, Crestani et al., 2019, Hollister et al., 2019, Hu et al., 2019, Lains et al., 2019, Rangel-Huerta et al., 2019, Yu et al., 2019, Cao et al., 2019, Shi et al., 2019, Kelly et al., 2019, Blacher et al., 2019, Wilmanski et al., 2019, Tang et al., 2019, Plaza-Diaz et al., 2019, de Groot et al., 2019, Wittemans et al., 2019, Tziotzios et al., 2019, Burrage et al., 2019, McCullough et al., 2019, Olson et al., 2018, Zambrana et al., 2019, Sato et al., 2019, Rebholz et al., 2019, Isganaitis et al., 2019, Gangler et al., 2019, Cirulli et al., 2019, Shin et al., 2014).
Due to the enormous potential of untargeted metabolomics to enrich scientific research, investigators increasingly include these data in their research. This has led to a rise in the number of laboratories and core facilities with untargeted metabolomics capabilities. However, the lack of clear standard operating procedures (SOPs), or recommendations regarding untargeted metabolomics quality management, including quality assurance (QA) and quality control (QC), has called into question the quality and integrity of metabolomics data (Bouhifd et al., 2015). Given the current mandates for submission of data into public repositories according to the Findable, Accessible, Interoperable and Reusable (FAIR) principles for the benefit of the wider community (Wilkinson et al., 2016), the lack of clear quality management practice guidelines makes discerning valid and reliable data very difficult, if not impossible. The concern within the untargeted metabolomics community, and the agencies that fund them, is that data collected under poor quality management could generate biased results, waste valuable resources throughout the scientific process, halt progress, or even harm the field’s reputation for high quality science. This would be a great disadvantage considering the potential of metabolomics to substantially contribute to the advancement of scientific discovery.
Multiple organizations have set forth formal definitions for the terms Quality Management System (QMS), Quality Assurance (QA) and Quality Control (QC) including ISO (2015) and Eurachem (2016). For the purposes of this document, mQACC used the following definitions which built on these formal definitions:
Quality Management System (QMS) is a management system to direct and control an organization with regard to quality where the organization can be small (e.g. an academic research group or start-up company) or large (e.g. a government organization or large pharmaceutical company).
Quality Assurance (QA) is part of a quality management system that is focused on providing confidence that quality requirements will be fulfilled and includes practices focused on providing confidence that quality requirements will be fulfilled consistently over time. QA processes are performed independent of data acquisition processes and include, but are not limited to, training, calibrating instrument, writing SOPs and performing audits.
Quality Control (QC) is part of a quality management system that is focused on fulfilling quality requirements. Practices are focused on demonstrating that quality requirement(s) have been fulfilled. In general terms, most QC processes are performed during or after data acquisition in each study. Examples of QC processes include, but are not limited to, analysis of QC samples, analysis of sample blanks, and reporting of study-specific quality metrics.
Several initiatives have begun to address the lack of recommendations regarding untargeted metabolomics quality management including the Metabolomics Standards Initiative in Toxicology (MERIT), through the European Centre for Ecotoxicology and Toxicology of Chemicals (ECETOC); and the Metabolomics Standards Initiative (MSI) through the international Metabolomics Society. These groups and others have convened to define and highlight quality management processes (Viant et al., 2019, Kirwan et al., 2018, Sumner et al., 2007, Dunn et al., 2017, Playdon et al., 2019, Dudzik et al., 2018, Bouhifd et al., 2015). Another such initiative was the “Think Tank on Quality Assurance and Quality Control for Untargeted Metabolomics Studies” sponsored by the US National Institutes of Health (NIH), where the inaugural meeting convened at the NIH in 2017 (Beger et al., 2019). Metabolomics practitioners from government, academic and industrial laboratories from around the globe were invited to discuss QA and QC practices in untargeted metabolomics. As such, participants represented a diversity of untargeted metabolomics research applications and regulated environments as well as repository providers. One of the outcomes of this Think Tank was the formation of the Metabolomics Quality Assurance and Quality Control Consortium (mQACC). The mission of mQACC is to communicate and promote the development, dissemination and harmonization of best QA and QC practices in untargeted metabolomics. The objectives of mQACC that support the consortium’s mission are to: 1) identify, catalog, harmonize and disseminate QA/QC best practices for untargeted metabolomics; 2) establish mechanisms to enable the metabolomics community to adopt QA/QC best practices; 3) promote and support systematic training in QA/QC best practices for the metabolomics community; and 4) to encourage the prioritization and development of reference materials applicable to metabolomics research.
An initial step to achieve these objectives was to summarize the current quality management practices, both QA and QC, of laboratories that were willing and able to share them. The goals of this manuscript were to: (1) highlight common QA/QC practices as a framework for minimum requirements, (2) highlight the differences between laboratories in QA/QC practices and (3) report and discuss practices that are not currently routine in any lab. The differences found can indicate specific potential challenging areas within the community for establishing guidelines and can also serve to highlight areas that warrant further discussion and development. Since QA/QC practices were recently detailed elsewhere (Broadhurst et al., 2018, Viant et al., 2019, Dudzik et al., 2018), we focus here on identifying common practices and differences in QA/QC workflows across 23 laboratories and provide references for detailed descriptions of the practices themselves.
Methods
Members of the mQACC consortium and several non-member contributors who utilized liquid chromatography-mass spectrometry (LC-MS) untargeted metabolomics were asked to voluntarily participate in a collaborative research project and took part by providing the QA and QC practices utilized in their laboratories, via a six-page questionnaire composed of over 120 questions and open comment fields. All individuals who contributed their laboratory’s protocols are listed as authors. Representatives from participating laboratories filled out a questionnaire (Supplementary Material 1) and/or submitted their relevant QA/QC standard operating procedures (SOPs). One laboratory submitted solely its QA/QC SOP, which was then used to fill out the questionnaire as fully as possible by the primary author. The scope of the questionnaire was limited to LC-MS untargeted metabolomics practitioners to ensure a manageable questionnaire length for participants. The questionnaire posed a series of yes/no questions on the use of specific QA/QC procedures, followed by more granular questions and open comment fields regarding the frequency and purpose of those practices for those who answered affirmatively. These data were used to determine the proportion of laboratories that used the various QA/QC practices, to quantify QC sample frequency, and to summarize how the data from QC samples was utilized by the laboratory.
If a question was unanswered or the answer provided insufficient detail, it was considered “Not Specified” and was excluded from both the numerator and denominator in the calculation of percent use. We present the proportion of “Yes” responses out of the number of total responses to the question, in addition to the count. Contributors did not always answer all questions, and no criteria was used to exclude a participant based on a minimum percentage of responses. This led to instances of seemingly incongruent data where the denominators of different questions could be different. The difference in the denominator was solely a result of the number of laboratories choosing to or not to respond to each question. In specific cases, the number of non-respondents was highlighted. Instances where respondents noted that they were “In process” of development of specific QA or QC practices were counted as a “no” responses.
Results
Twenty-three (23) laboratories chose to participate in the questionnaire. Seventy percent of respondents (16/23) were from academic institutions, 17% (4/23) were from government institutions and 13% (3/23) were from industry. Sixteen contributors represented institutions located in North America, six were from Europe, two were from Australasia, and one was from South America. The questionnaire data transposed into an excel file and percent usage rates of all QA and QC practices can be found in Supplementary Material 2 and 3, respectively. Seventeen (17) of the contributing laboratories had a response rate of over 95% of the questions, and twenty (20) of the laboratories answered over 90% of the questions. There was one laboratory whose response rate was significantly lower than the other contributors having only answered 46% of the questions (Supplementary Material 6). We could not assess the reasons why contributors did not answer any particular question, but it does appear to be more related to the individual filling out the questionnaire, rather than the question itself as the response rate per QA/QC topic area was well conserved across the questionnaire (Supplementary Material 7).
1.0. General Quality Assurance (QA) Practices
Quality assurance is the part of a quality management system that ensures that a laboratory is be able to meet its quality objectives. Many QA activities are, therefore, related to assessing the workflow to identify areas where errors may occur and making sure processes are in place and adhered to that track and monitor the processes that are independent of data collection (Kauffmann et al., 2017). Various QA procedures were queried and organized into general categories as follows.
1.1. Review/Audits
Some QA activities involve reviewing processes and workflows with a focus on finding and mitigating potential sources of error. This can be done through a variety of processes including formal regulatory accreditation where external organizations such as the International Organization for Standardization (ISO) or the College of American Pathologists (CAP) that provide established metrics for assessing a method and auditing the organization for compliance as part of accreditation. Only 10% (2/20) of respondents reported that their laboratories were fully regulated or were maintaining accreditation from an external entity, which included one industrial and one academic laboratory. One additional laboratory was in the process of accreditation, and another two laboratories were part of larger organizations that abided by set regulations (e.g. operating at Good Laboratory Practices (GLP) standards or Good Clinical Practices (GCP) standards), which were not directly incorporated by the laboratories. About a third (29%: 6/21) of responding laboratories utilized an independent QA group within their organization, and 27% (6/22) of responding laboratories reported regular laboratory audits by either internal or external reviewers.
Multiple rounds of data quality review and approval, ideally performed by staff not directly involved in the data acquisition, is generally considered a QA practice because reviewer(s) assess whether the initial data review and approval processes were performed according to established quality management system specifications. Half (50%: 11/22) of responding laboratories reported having a formalized review process with multiple levels of approval, with review processes under development in two additional laboratories.
1.2. Traceability and Trackability
Part of a quality system is the traceability/trackability of processes that track how a sample is handled or stored and whether there are any changes to or deviations from SOPs. Quality assurance establishes processes needed to easily track and periodically review these procedures for potential improvement. A third (32%: 7/22) of responding laboratories reported using sample tracking, management and chain of custody software (e.g. LIMS system). Half of respondents (50%: 11/22) reported having established a formal document control system with version control and approval processes (under development in one laboratory). Half (50%: 11/22) of responding laboratories had formal documentation control processes for tracking at least some laboratory phenomenon, such as deviations from SOPs, change control documentation (formal tracking, traceability and approval for any changes to methodology), incidents and corrective and preventative action plans (CAPAs: a formal process to track errors in process and put in place and report). Many (82%: 18/22) responding laboratories utilized logbooks or files to document activities such as maintenance, calibration and tuning, while a third (32%: 7/22) also reported tracking/logging new mobile phase production lots/batches, new column installation and column lots/batch. Seventy-one percent (15/21) of the responding laboratories reported having an established data storage and archival process (under development in one laboratory). Over half (55%: 12/22) of responding laboratories reported uploading data into a public repository with three noting that this was only done for specific projects/studies or when required by the publication journal or project funding agency. This process was under development in two additional laboratories. Two thirds (68%: 15/22) of responding laboratories reported having SOPs in place for all laboratory processes, with SOPs under development for two additional laboratories (e.g. SOPs for sample handling, storage and shipping).
1.3. Equipment Testing and Monitoring
As previously stated, QA activities ensure that a laboratory will be able to meet its quality metrics, though quality metrics can differ between laboratories. It is, therefore, important to maintain established processes that demonstrate the functionality of the laboratory and equipment to perform the methods independent of actual sample analyses. Sixty-two percent (13/21) of responding laboratories reported having established systems to set and monitor laboratory atmospheric conditions with six also reporting having established an automated alarm notification system that sent out alerts (e.g. emails, texts) to user-specified personnel if the laboratory conditions deviated outside of user specified set bounds. The majority (91%: 20/22) of responding laboratories reported having established systems to set and monitor cold storage temperature units with 16 laboratories also using an automated alarm notification system that sent out alerts (e.g. emails, texts) to user-specified personnel if the cold storage units deviated from set parameters. Most (81%: 17/21) respondents reported having established new equipment and instrument qualifications to ensure instrument functionality. In addition to installation and operational qualifications, 12 of these laboratories also established performance qualifications that had to be met. All responding laboratories (22/22) reported having established preventative maintenance (PM), calibration and tuning schedules for their LC and MS systems. Eighty-six percent (19/22) of responding laboratories had established calibration and/or maintenance schedules for some secondary equipment including pipettes and analytical weighing balances and 18% (4/22) of the responding laboratories reported comprehensive scheduled primary and secondary equipment maintenance and calibration processes which further included centrifuges, drying apparatus, homogenization equipment, and water supply.
1.4. Staff Training and Proficiency Testing
In addition to processes that demonstrate the ability of equipment to meet quality metrics, laboratory staff can also be assessed for their ability to meet quality metrics through training and proficiency testing. Over half (55%: 12/22) of responding laboratories reported having established formal or semi-formal training processes for staff and 27% (6/22) reported also conducting proficiency testing, albeit with varying degrees of formality. Three laboratories conducted specific and routine proficiency testing of staff annually.
2.0. General Quality Control (QC) Practices
2.1. Use of a System Suitability Procedure
System suitability procedures determine whether instrumentation and methods are performing to a defined, appropriate and acceptable level before acquisition of project samples (Viant et al., 2019, Broadhurst et al., 2018). All respondents (23/23) reported using a system suitability procedure to establish instrument and method readiness. Over 90% of responding laboratories reported that system readiness was defined by the correct retention time (RT) of standards within acceptance criteria (22/22), correct mass accuracy of defined compounds within acceptance criteria (23/23) and sensitivity within acceptance criteria (22/23) (Figure 1A). Sixty-four percent of responding laboratories (14/22) also reviewed chromatographic peak width as part of determining instrument readiness. Over 70% of laboratories also required instrument stability over several injections (17/23) and reviewed peaks observed in blank samples (18/23). The specific acceptance criteria that each laboratory used was not evaluated in this questionnaire.
Figure 1.
System suitability user metrics. A. Parameters assessed to determine instrument readiness. B. Percentage of different sample types used for system suitability testing.
Different types of samples were used to determine instrument readiness. Forty-three percent (9/21) of responding laboratories reported assessing system suitability with sole use of a solution of authentic chemical standards of varying complexity, which ranged from a small number of chemical standards to a mixture of over 40 chemical standards (Figure 1B); 10% (2/21) of responding laboratories reported the sole use of a biological sample spiked with chemical standards; 48% (10/21) of responding laboratories reported using multiple sample types for system suitability assessment including various combinations of a solution of chemical standards (10/10), a biological sample spiked with chemical standards (9/10) and a non-spiked biological sample (2/10).
2.2. Use of Quality Control Samples
2.2.1. Blanks
2.2.1.1. Process/Extraction Blanks
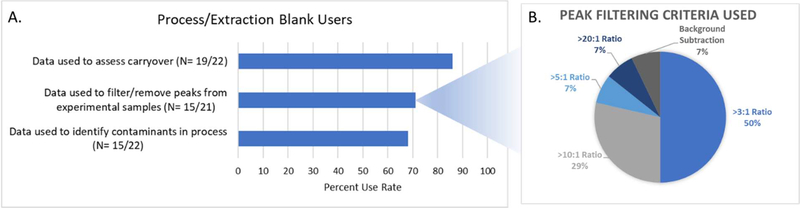
A process or extraction blank (Broadhurst et al., 2018, Viant et al., 2019) is defined as a blank sample (for example, an aliquot of water) passed through the analytical process including the extraction and instrument analysis followed by comparison to a biological sample’s data. Most (86%: 19/22) of the responding laboratories reported analyzing process/extraction blanks with every study. With regards to frequency of use, 58% (11/19) of responding laboratories analyzed these blanks at multiple points throughout a batch, specifically one or two blanks at the beginning and end of each batch, and additional blanks analyzed intermittently placed throughout the batch at approximately every 10 to 20 injections. The remaining 42% (8/19) of responding laboratories analyzed one or two process/extraction blanks per batch. Questions were also asked, in the questionnaire, to elucidate the use of the blank data. The majority of responding laboratories (86%: 19/22) reported using the process/extraction blanks data to assess carryover in the methods, particularly during method development (Figure 2A). Two thirds (66%: 15/22) of responding laboratories reported using process/extraction blanks data to identify contaminants in the processes arising from storage and handling. Most (71%: 15/21) responding laboratories used the process/extraction blank data to filter or remove peaks detected in the experimental samples, of which one laboratory used background subtraction methodology and the remaining laboratories filtered peaks based on blank levels by using a defined “must exceed” ratio of the experimental peak intensity to blank peak intensity. Of those laboratories that chose to set a defined “must exceed” ratio, seven reported using a minimum 3:1 experimental-to-blank ratio for inclusion of peaks; four laboratories used a 10:1 ratio; one laboratory used a 5:1 ratio and one laboratory used a conservative 20:1 ratio (Figure 2B).
Figure 2.
Process/extraction blank user metrics. A. Uses of the data; B. Peak filtering criteria when removing peaks from experimental samples.
2.2.1.2. System Suitability Sample/Solvent Blank with Standards
Most (70%: 16/23) laboratories reported injecting a system suitability sample (Broadhurst et al., 2018) (a mixture of standards in a solvent blank) at defined intervals throughout a batch while the frequency of use mirrored that of the process/extraction blanks, where slightly over half of the laboratories analyzed these blanks multiple times throughout a batch. The reported use of this blank was mostly to isolate and assess instrument performance independent of the sample extraction process.
2.2.1.3. Solvent Blank (No Internal Standards)
Over a third (39%: 9/23) of laboratories reported analyzing a pure solvent blank (Broadhurst et al., 2018) (a solvent that has not passed through the sample preparation process applied to biological samples and which contains no internal standards) injected periodically throughout a batch, with eight of these nine laboratories utilizing this blank in addition to the process/extraction blank. This solvent blank was reported as being used to assess carryover, including the carryover of the internal standards that have generally been spiked into all other samples. Solvent blanks were also used to assess contamination coming solely from the solvent system.
2.2.2. Pooled-QC/Intra-Study QC Samples
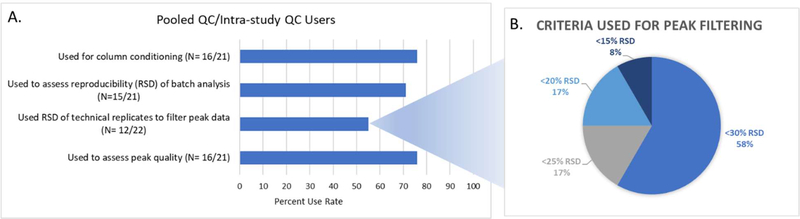
A pooled QC or intra-study QC sample is a technical replicate of a sample that has the same matrix as the sample type in the study (Sangster et al., 2006). Pooled/intra-study QC samples are created by pooling an aliquot of all or a subset of the experimental biological samples in the study (Broadhurst et al., 2018, Viant et al., 2019, Dunn et al., 2012, Gika et al., 2007, Bijlsma et al., 2006, Gika et al., 2008). Almost all (96%: 22/23) laboratories reported using pooled QC/intra-study QC samples. The frequency was consistent with most responding laboratories running one to three injections at the beginning of a batch (excluding column conditioning injections), one to two at the end, and intermittently throughout the batch, generally every 10 to 20 injections. Approximately three-quarters (76%: 16/21) of responding laboratories used these samples for column conditioning before batch analysis, 71% (15/21) used them to assess relative standard deviation (RSD) for a study and 76% (16/21) reported using the pooled/intra-study QC samples to assess peak quality (Figure 3A). About half (55%: 12/22) of responding laboratories reported using these data to filter/remove peaks based on the relative standard deviation (RSD) of response and detection rate criteria. Specifically, in seven laboratories, the RSD of the experimental peak in the technical replicates were required to be <30%; two laboratories used <25%; two laboratories used <20% and one laboratory reported using a <15% RSD criteria (Figure 3B). Detection rate filter criteria (Smilde et al., 2005) ranged from a required detection rate of greater than 50%, to a required 100% in all technical replicates for inclusion in the subsequent data analyses.
Figure 3.
Pooled QC/intra-study QC metrics. A. Pooled/intra-study QC data uses. B. RSD criteria of technical replicate peaks when removing peaks from experimental samples.
2.2.3. Long-Term Reference (LTR)/Intra-Laboratory QC Samples
A long-term reference (LTR) or intra-laboratory QC sample is a large pool of samples that is analyzed consistently and repeatedly over time within a laboratory, though typically at a lower frequency than for pooled-QC/intra-study QC samples. To support this QC sample type, a large quantity of this sample is required, and the sample needs to be stable over the time-period of use (Viant et al., 2019, Broadhurst et al., 2018). Half (52%: 11/21) of responding laboratories reported using a long-term reference QC sample (with three additional laboratories in the development stage). Eight of these laboratories used their own pool of sample(s) for this purpose, one laboratory did not specify the LTR source and the rest used the National Institute of Standards and Technologies (NIST) SRM 1950 (Simon-Manso et al., 2013). Some laboratories noted that LTR samples were not applicable to their laboratory since they did not analyze samples from longitudinal or large studies that would require assessment of long-term instrument stability. The laboratories that utilized LTR samples analyzed them multiple times per batch, generally following the same trend as blanks with one to two at the beginning of the batch, one to two at the end of the batch and additional samples interspersed throughout the batch. One laboratory reported that if an LTR sample was being used for bridging long-term studies, it was incorporated approximately every 10 samples. Reasons for incorporating LTR samples were to: 1) assess the stability of laboratory processes over time and/or 2) bridge/normalize batch-to-batch data over time (Zelena et al., 2009, Dunn et al., 2011).
2.2.4. Standard Reference Material (SRM)/Inter-Laboratory QC Samples
A standard reference material (SRM)/inter-laboratory QC sample is an externally tested, validated and maintained sample that is available for purchase by the metabolomics community (Broadhurst et al., 2018, Viant et al., 2019). SRMs can range in compound complexity but within the context of untargeted metabolomics, it is generally a biological sample of appropriate biological origin (human, plant, fish, etc.) and matrix (plasma, urine, etc.). A third (33%: 7/21) of responding laboratories reported the use of an SRM, specifically commercially available SRM 1950 (NIST Metabolites in Frozen Human Plasma (Simon-Manso et al., 2013)), and two laboratories also used several different SRMs (all NIST standards). Two laboratories noted that SRM samples were used as their LTR QC samples while the other laboratories reported the use of an SRM only in the context of cross-platform or site performance comparisons.
2.3. General Practices
2.3.1. Use of Internal Standards
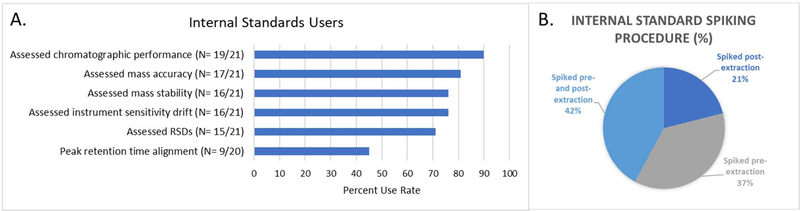
Internal standards are defined here as isotopically labelled or otherwise not endogenously present chemicals which are intentionally spiked into some or all samples in a process and are used to help assess data quality. Most (91%: 21/23) laboratories reported using internal standards in their laboratories. Two laboratories used them in some but not all of their methods. Of the laboratories/methods that used them, internal standards were consistently spiked into all samples. Eight of the laboratories spiked different standards at two different points in the process (pre-extraction and post-extraction); seven laboratories spiked all standards pre-extraction; four laboratories spiked all standards post-extraction; and two laboratories did not respond to their spiking procedures (Figure 4B). Among laboratories spiking standards at multiple times in the extraction process, the rationale was to separately assess quality throughout the entire process, including sample extraction and the quality of the instrumental analysis and ionization matrix effects. The number of standards utilized per method varied greatly across the laboratories from one to 800 standards, with one to fifteen standards being the most common. Internal standards were specified to be chosen to cover both the analytical method mass range and chromatographic time, in addition to not interfering with other compounds present in the sample. A list of specific standards used by each laboratory was not requested, therefore, information on their origin and/or if these standards were isotopically labelled were not available.
Figure 4.
Internal standards user metrics. A. Parameters assessed and data usage from internal chemical standards; B. Percentage of different internal standard spiking procedures.
More than 80% of responding laboratories used the spiked internal standards to assess the chromatographic performance (19/21) and mass accuracy (17/21) of the data. Over 70% of responding laboratories reported using the standards to assess RSDs (15/21), to monitor for instrument sensitivity drift (16/21) and mass stability (16/21) across the data. Just under half (45%: 9/20) used their standards for peak retention time alignment post-acquisition (Figure 4A).
2.3.2. Use of Technical Replicates of Experimental Samples
Technical replicates are repeat analyses of the same biological sample and should not be confused with biological replicates which are repeat biological samples of a specific study design, i.e. six mice in a control group are considered biological replicates. Technical replicates can be applied in two primary ways. One is to perform two or more injections from the same post-extraction sample tube (“injection replicates”) and another is by re-extracting the same source of biological sample multiple times and analyzing each sample once and independently (“extraction replicates”). Over a third (36%: 8/22) of responding laboratories reported the use of technical replicates on at least a subset of experimental samples in addition to QC samples. An additional six laboratories used technical replicates on experimental samples “sometimes” or in relation to specific methods or when the number of biological replicates was low. Five laboratories reported that their experimental technical replicates were injection replicates (i.e. injections from the same post-extraction tube), three laboratories used extraction replicates (i.e. analysis of the same source sample extracted multiple times) and other laboratories used a combination of both replicate types depending on the method or study. The investigators reported using the technical replicate data to monitor reproducibility while other investigators used them for batch normalization to correct for instrument drift within a batch or to merge/integrate data from different batches/studies analyzed at different times. When technical replicates of experimental samples were used, most investigators reported that data from the replicates were averaged.
2.3.3. Run Order Randomization and Balancing
Run (or analysis) order is defined as the order in which the experimental and QC samples are analyzed in a batch (Broadhurst et al., 2018). Run order randomization is the process by which experimental samples are randomly distributed within a batch to ensure that important metadata elements are not inadvertently accumulated during any phase of the batch, such as running more control samples at the beginning of the batch than at the end. Balancing is the process by which the experimental samples are intentionally distributed based on metadata to ensure that important metadata elements are not inadvertently accumulated across multiple batches, such as running significantly more females or control samples in one batch compared to other batches. Balancing is more critical for multiday analyses than for single day analyses if the run order is randomized. Paired case/control sets can be randomized across batches to address this issue. Most (91%: 20/22) responding laboratories reported randomizing experimental samples within a defined template of QC samples and one additional laboratory randomizing all samples (experimental and QC). Eighty-one percent (17/21) of responding laboratories reported balancing their study samples. Laboratories that did not balance their study samples did not provide a specific reason, however, they all reported randomization of experimental samples.
2.4. Post Analysis Quality Management
2.4.1. Quality of Compound Identification
Compound identification is the process by which an LC-MS peak is assigned a chemical identity. The act of compound identification is not a QC practice, however, the verification of the correctness of the identification is considered to be a QC practice. The rigor with which a compound is identified has been classified into distinct levels by the Metabolomics Society in order to promote a clear understanding of the confidence of the reported identification (Sumner et al., 2007). MSI level 1 is the highest confidence and most rigorous identification and is defined as “a minimum of two independent and orthogonal data relative to an authentic compound analyzed under identical experimental conditions”. For LC-MS applications, retention time and MS/MS data are utilized for identification. Analysis of authentic chemical standards in the research laboratory is essential for the retention time data because different laboratories utilize different LC assays (column type, solvents, gradient elution, temperature, stationary phase), thus retention time data collected in one laboratory may not be transferable to another laboratory. MS/MS data can be collected in different laboratories using the same analytical conditions and so data collected in one laboratory are transferable to other laboratories. All laboratories (22/22) reported establishing MSI level 1 (Sumner et al., 2007) confidence identifications on at least some of the detected compounds (Supplementary Material 4). Thirteen respondents stated that they reported the MSI confidence levels for all reported/published compounds, and three additional laboratories were in the development process. Eighty percent (16/20) of responding laboratories established their own in-house library in order to have MSI level 1 confidence identifications. In all cases, the compound retention time and mass were stored and in all except one laboratory, the fragmentation spectra were also stored within the compound libraries. These libraries ranged in size from 40 to 4,800 compounds (median = 600; mean=884). The majority (91%: 20/22) of responding laboratories also used a public database in their compound identification process. Close to two thirds (65%: 11/17) of responding laboratories reported using workflows that identified statistically significant compounds only. It should be noted that there are four MSI levels for reporting metabolite identifications and that these levels are currently being reviewed by the metabolomics community (personal communication, Warwick Dunn, April 2020).
2.4.2. Data Quality Review
Many (86%: 19/22) laboratories reported the manual review of peak integrations. Four laboratories reviewed only a subset of peak integrations which includes: named compounds only, compounds with poor integration track records or those compounds highlighted by an automated software system that flagged peaks of concern. Greater than 75% of responding laboratories assessed resolution of chromatographic peaks (16/18), consistency of integration throughout a study (14/18) and signal-to-noise ratio (19/19) (Supplementary Material 5). Two laboratories also used positive correlation analysis either across methods or across metabolite groups to assess quality of peak integration.
Approximately half (11/21) of the responding laboratories manually reviewed of peak alignment across samples in a study. Two additional laboratories used an automated process to check alignment quality. Several laboratories used retention time window criteria to assess and monitor alignment quality; for example, no greater than 25% of peaks must be outside a 0.1 min window, or all aligned peaks must be within 0.2 min. Several laboratories also used retention time deviation plots to help identify problem alignments. Most (85%: 17/20) laboratories used principal component analysis (PCA) score plots to look for clustering of replicate samples and to identify potential QC and/or biological outliers during the quality review process. Several groups also used additional minimum required detection rates within the experimental samples based on group/meta-data criteria; for example, for a peak to be included in a study, it must be found in at least 80% of the treatment samples.
Discussion
We designed and administered a questionnaire to assess quality assurance (QA) and quality control (QC) management practices of a cross section of international LC-MS untargeted metabolomics research groups (including members of the Metabolomics QA and QC Consortium (mQACC)) drawing from academic, industry and government facilities (Figure 5). The over-arching aim of the project was to establish a consensus of QA/QC best practice within this specific group of experts such that it could be formally documented and disseminated to the broader metabolomics community. We also sought to identify differences in QA/QC practices across laboratories to prompt discussion within the metabolomics community on areas requiring development, promotion or training. These data are intended to inform best practices and reporting standards within the consortium and serve as a basis for the expansion of content through further community engagement efforts. Future planned activities include conducting similar studies to gather information on QA/QC practices for nuclear magnetic resonance (NMR) spectroscopy and gas chromatography-mass spectrometry (GC-MS) based metabolomics methods, lipidomics methods and to determine specific pass/fail metrics and criteria used in quality processes. It is important to note that the goal here was not to define which criteria should or should not be used by the community but rather to define acceptable practices to use and report such data and processes.
Figure 5.
Summary of example QMS practices. Note that the practices highlighted in this figure as QA or QC will likely change as the community continues to discuss these topics and reach a consensus.
This initiative builds upon the continued efforts of the Metabolomics Society (Sumner et al., 2007, Dunn et al., 2017, Kirwan et al., 2018), MERIT (Viant et al., 2019), the Johns Hopkins Center for Alternatives to Animal Testing (CAAT) (Bouhifd et al., 2015) and independent researchers (Broadhurst et al., 2018) to encourage the acceptance and use of quality management practices. As such, this publication bridges the various differences in terminology among the current publications when referring to the same quality practices.
We found that some practices were commonly adopted among laboratories (>80% usage rate)(Figure 6). These included system suitability assessments, sample run-order randomization and balancing, the use of authentic chemical standards, and the use of various quality control samples such as pooled-QC/intra-study QC samples and blanks. In addition, a high degree of concordance occurred among post-analytical quality control practices, such as compound identification practices, assessment of integration accuracy and the use of PCA for quality assessments of sample clustering. This speaks to the success of the previous efforts to bring awareness of these practices to the community, and also to the value of the practices to the users (and the metabolomics field) to monitor and assess quality.
Figure 6.
Common (>80% usage rate) QA and QC practices
For laboratories adopting a specific QC practice, the use of practice-specific procedures was comprehensive (Figures 1 to 4, and Supplementary Material 4 and 5). For example, over 70% of respondents assessed the same factors in system suitability practices, namely: sensitivity, mass accuracy, retention time, blank levels and instrument stability (Figure 1). Likewise, greater than 70% of laboratories used internal standards to assess chromatographic performance, mass stability, mass accuracy, sensitivity/response stability and sample-to-sample reproducibility (Figure 4); and used pooled-QC/intra-study QC samples to assess process reproducibility, peak quality and to condition columns.
We evaluated post-analysis quality practices such as establishing MSI level 1 identifications (Sumner et al., 2007) to ensure high confidence compound identification, manual review and confirmation of peak integrations and alignment, and the use of PCA as a quality assessment tool. Accurate compound identification is fundamental to the process of deriving correct biological insight. The rigor with which a compound is identified has been classified into distinct levels by the Metabolomics Society in order to promote a clear understanding of the confidence of the reported identification (Sumner et al., 2007). MSI level 1 is the highest confidence and most rigorous identification and is defined as “a minimum of two independent and orthogonal data relative to an authentic compound analyzed under identical experimental conditions”. Lower confidence identifications rely on matching experimental data to public or commercial spectral libraries that were not collected under identical experimental conditions. These data showed that the metabolomics field relies upon public and/or commercially available databases for tentative annotations, with over 90% of practitioners utilizing them. However, all laboratories also reported establishing MSI level 1 confidence identifications on at least a subset of metabolites (Supplementary Material 4). This was predominantly done by establishing in-house authentic standard libraries which ranged in size from 40 compounds to over 4,800, with a median of 600 compounds and a mean of 884 compounds. Establishing comprehensive in-house authentic standard libraries to support MSI level 1 confidence identification requires a large investment in resources. Therefore, some laboratories reported only doing so for a subset of reported molecules, presumably focusing on the statistically relevant compounds in their study. An additional community effort is underway through the Metabolomics Society’s Metabolite Identification Task Group to further “build consensus on metabolite identification reporting standards” and “educate the community on best practices” (www.metabolomicssociety.org/board/scientific-task-groups/metabolite-identification-task-group).
Other post-analysis quality practices were well conserved with over 80% of practitioners reviewing peak integrations (Supplementary Material 5). and using PCA to assess clustering of replicate injections and identification of outliers. Half of the laboratories also reported a manual review of data alignment using various metrics. As many of the processes involved in untargeted metabolomics rely on automated processes such as peak integration, alignment and compound identification; data quality is directly impacted by the algorithms at the core of the processes, which do not always perform as desired.
Many of the differences in quality control practices were notably related to practices that were either inappropriate or unnecessary for some workflows. For example, laboratories that rarely analyzed data over long periods of time had no need for a long-term reference standard/intra-laboratory QC sample. SRMs/inter-laboratory QC samples were only utilized when a comparison of performances across sites or laboratories was required, further emphasizing the need for future best practice guidelines to consider the context of their use and analysis of the data.
Other differences were relatively minor such as whether blanks included internal standards or were solvent blanks with no spiked standards or were differences related to the point at which the internal standards were added in the sample preparation process (Figure 4b). Ultimately, these differences may or may not need to be defined in best practices. Rather, it is more useful to recommend the inclusion of blanks and internal standards with explanations for their use, but leave these finer details to the discretion of laboratory practitioners to decide what is most appropriate for their laboratory and/or a given study.
Differences were seen in the frequency of injections utilized by practitioners and the specific acceptance criteria. For example, for respondents using a ratio of peak levels detected in an experimental sample to peak levels detected in a process blank to remove or filter low quality peaks, the specific acceptance criteria ranged from a greater than 3-to-1 ratio to a greater than 20-to-1 ratio for acceptance (Figure 2B). A similar diversity of acceptance criteria was apparent for the process of filtering/removing peaks based on the reproducibility of aligned peaks in the technical replicates of a pooled QC/intra-study QC sample; acceptance criteria ranged from filtering the data based on an RSD of technical replicates (with cut-off values ranging from 15% to 30%) to no filtering criteria based on RSDs at all (Figure 3B). These differences in acceptance criteria could contribute to inter-laboratory differences, where some labs could remove/disqualify certain compounds as a result of more strict acceptance criteria. Laboratories using more strict criteria usually did so for specific methods or studies only. Reasons for choosing the method or study-specific criteria were not probed, but a better understanding of what drives the choice for stricter quality criteria is warranted.
There appeared to be limited consensus on most quality assurance practices among the responders. Few laboratories incorporated various QA practices into their workflow outside of instrument care, maintenance and sample cold storage. The definition of QA was another area of debate among authors and the wider mQACC community. The definition as settled upon after debate was that QA practices provide confidence that quality requirements will be fulfilled consistently over time and that these practices are performed independent of data acquisition processes. Many QA processes are focused on traceability, establishing defined processes with documentation, and optimizing workflows through reviews/audits. Informally, part of quality assurance is analogous to having a “second pair of eyes” which critically reviewing the entire process to identify areas in need of improvement and to ensure traceability. Examples of quality assurance practices include requiring: secondary signatures in laboratory notebooks, peer-review of scientific manuscripts, secondary review of data, establishing systems for sample and data tracking, regular auditing of all processes, and establishing processes and documentation that show alternations to or deviations from SOPs. Untargeted metabolomics laboratory practitioners may not be exposed to quality assurance processes since these are typically only required in regulated environments. However, their added value is important and is mandated if the data generated is destined for regulatory agencies or involves the study of humans. The area of QA within the untargeted metabolomics community is certainly wide open for discussion regarding which of the QA activities are of high enough value for potential incorporation into practice recommendations (Manghani, 2011, Dudzik et al., 2018).
Strengths and Limitations of the Study
Strengths of this study include participation from over 20 laboratories from across the globe with inclusion of academic, industry and government laboratories. Also, the success of the last two mQACC workshops at the annual Metabolomics Society meeting indicate there is a strong demand for a forum to discuss these topics, which the current analysis aimed to facilitate. Paradoxically, this strength is also a weakness, as the results presented are likely biased toward greater incorporation of quality assurance and quality control practices than actually occurs in the wider metabolomics community; participants willing to fill out a questionnaire on QA/QC are more likely to be concerned with QA/QC practices. The data presented here reflects the overall experience and commitment to QA and QC by the questionnaire participants. We also acknowledge that many metabolomics practitioners fit their QA/QC practices to a specific purpose. For example, some laboratories did not use specific quality control practices because the practices were, justifiably, not relevant to their workflow. Consequently, any “good practices” recommendations and defined reporting standards must be flexible enough to permit these natural and expected differences among users with appropriate knowledge transfer. It will be important to expand our questionnaire beyond mQACC to gain a more representative snapshot of current practices in the field. In summary, there was a high level of consensus among untargeted metabolomics users regarding QA and QC practices, however, there were also differences, which warrants open discussion and establishment of best minimal practices by the community.
Conclusion
There was a surprisingly high level of consensus regarding best practices across the international laboratories sampled for this study. These results provide a useful blueprint for QA and QC assessment when developing new assays within a laboratory as well as a teaching resource for analytical chemistry/biochemistry. We are optimistic that through mQACC the metabolomics community at large will be able to develop a common set of best minimum practices and reporting standards to ensure that metabolomics research is robust and repeatable.
Supplementary Material
Acknowledgments
Disclaimer
The opinions expressed in this publication are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency, the U.S. Food and Drug Administration or the National Institute of Standards and Technology.
Footnotes
Conflict of Interest Declaration
The authors have no conflict of interest to declare.
Compliance with Ethical Standards
This article does not contain any studies with human and/or animal participants performed by any of the authors.
Data Availability Statement
The questionnaire data reported in this paper has been organized and blinded into an excel spreadsheet and are available in Supplementary Material 2.
References
- 2015. ISO 9000:2015: Quality Management Systems - Fundamentals and Vocabulary, Switzerland, International Organization for Standardization. [Google Scholar]
- 2016. Eurachem/CITAC Guide: Guide to Quality in Analytical Chemistry: An Aid to Accreditation. In: BARWICK V. (ed.) 3rd ed. [Google Scholar]
- Beger RD, Dunn WB, Bandukwala A, Bethan B, Broadhurst D, Clish CB, Dasari S, Derr L, Evans A, Fischer S, Flynn T, Hartung T, Herrington D, Higashi R, Hsu PC, Jones C, Kachman M, Karuso H, Kruppa G, Lippa K, Maruvada P, Mosley J, Ntai I, O’Donovan C, Playdon M, Raftery D, Shaughnessy D, Souza A, Spaeder T, Spalholz B, Tayyari F, Ubhi B, Verma M, Walk T, Wilson I, Witkin K, Bearden DW & Zanetti KA 2019. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics, 15, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B & Smilde AK 2006. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem, 78, 567–74. [DOI] [PubMed] [Google Scholar]
- Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, Zabari M, Brik RB, Kviatcovsky D, Zmora N, Cohen Y, Bar N, Levi I, Amar N, Mehlman T, Brandis A, Biton I, Kuperman Y, Tsoory M, Alfahel L, Harmelin A, Schwartz M, Israelson A, Arike L, Johansson MEV, Hansson GC, Gotkine M, Segal E & Elinav E 2019. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature, 572, 474–480. [DOI] [PubMed] [Google Scholar]
- Bouhifd M, Beger R, Flynn T, Guo L, Harris G, Hogberg H, Kaddurah-Daouk R, Kamp H, Kleensang A, Maertens A, Odwin-DaCosta S, Pamies D, Robertson D, Smirnova L, Sun J, Zhao L & Hartung T 2015. Quality assurance of metabolomics. ALTEX, 32, 319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhifd M, Hartung T, Hogberg HT, Kleensang A & Zhao L 2013. Review: toxicometabolomics. J Appl Toxicol, 33, 1365–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst D, Goodacre R, Reinke SN, Kuligowski J, Wilson ID, Lewis MR & Dunn WB 2018. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics, 14, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage LC, Thistlethwaite L, Stroup BM, Sun Q, Miller MJ, Nagamani SCS, Craigen W, Scaglia F, Sutton VR, Graham B, Kennedy AD, Members of the U, Milosavljevic A, Lee BH & Elsea SH 2019. Untargeted metabolomic profiling reveals multiple pathway perturbations and new clinical biomarkers in urea cycle disorders. Genet Med, 21, 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Miller MS, Lubet RA, Grubbs CJ & Beger RD 2019. Pharmacometabolomic Pathway Response of Effective Anticancer Agents on Different Diets in Rats with Induced Mammary Tumors. Metabolites, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, He FJ, Dong Y, Huang Y, Harshfield GA & Zhu H 2019. Sodium Reduction, Metabolomic Profiling, and Cardiovascular Disease Risk in Untreated Black Hypertensives. Hypertension, 74, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Guo L, Leon Swisher C, Shah N, Huang L, Napier LA, Kirkness EF, Spector TD, Caskey CT, Thorens B, Venter JC & Telenti A 2019. Profound Perturbation of the Metabolome in Obesity Is Associated with Health Risk. Cell Metab, 29, 488–500 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crestani E, Harb H, Charbonnier LM, Leirer J, Motsinger-Reif A, Rachid R, Phipatanakul W, Kaddurah-Daouk R & Chatila TA 2019. Untargeted Metabolomic Profiling Identifies Diseasespecific Signatures in Food Allergy and Asthma. J Allergy Clin Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Khan MT, Herrema H, Ackermans M, Serlie MJM, de Brauw M, Levels JHM, Sales A, Gerdes VE, Stahlman M, Schimmel AWM, Dallinga-Thie G, Bergman JJ, Holleman F, Hoekstra JBL, Groen A, Backhed F & Nieuwdorp M 2019. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudzik D, Barbas-Bernardos C, Garcia A & Barbas C 2018. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. J Pharm Biomed Anal, 147, 149–173. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R & Human Serum Metabolome C 2011. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc, 6, 1060–83. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Edison A, Guillou C, Viant MR, Bearden DW & Beger R 2017. Quality assurance and quality control processes: summary of a metabolomics community questionnaire. Metabolomics, 13. [Google Scholar]
- Dunn WB, Lin W, Broadhurst D, Begley P, Brown M, Zelena E, Vaughan AA, Halsall A, Harding N, Knowles JD, Francis-McIntyre S, Tseng A, Ellis DI, O’Hagan S, Aarons G, Benjamin B, Chew-Graham S, Moseley C, Potter P, Winder CL, Potts C, Thornton P, McWhirter C, Zubair M, Pan M, Burns A, Cruickshank JK, Jayson GC, Purandare N, Wu FC, Finn JD, Haselden JN, Nicholls AW, Wilson ID, Goodacre R & Kell DB 2015. Molecular phenotyping of a UK population: defining the human serum metabolome. Metabolomics, 11, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WB, Wilson ID, Nicholls AW & Broadhurst D 2012. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis, 4, 2249–64. [DOI] [PubMed] [Google Scholar]
- Gafson AR, Savva C, Thorne T, David M, Gomez-Romero M, Lewis MR, Nicholas R, Heslegrave A, Zetterberg H & Matthews PM 2019. Breaking the cycle: Reversal of flux in the tricarboxylic acid cycle by dimethyl fumarate. Neurol Neuroimmunol Neuroinflamm, 6, e562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangler S, Waldenberger M, Artati A, Adamski J, van Bolhuis JN, Sorgjerd EP, van Vliet-Ostaptchouk J & Makris KC 2019. Exposure to disinfection byproducts and risk of type 2 diabetes: a nested case-control study in the HUNT and Lifelines cohorts. Metabolomics, 15, 60. [DOI] [PubMed] [Google Scholar]
- Gika HG, Macpherson E, Theodoridis GA & Wilson ID 2008. Evaluation of the repeatability of ultra-performance liquid chromatography-TOF-MS for global metabolic profiling of human urine samples. J Chromatogr B Analyt Technol Biomed Life Sci, 871, 299–305. [DOI] [PubMed] [Google Scholar]
- Gika HG, Theodoridis GA, Wingate JE & Wilson ID 2007. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J Proteome Res, 6, 3291–303. [DOI] [PubMed] [Google Scholar]
- Hollister EB, Oezguen N, Chumpitazi BP, Luna RA, Weidler EM, Rubio-Gonzales M, Dahdouli M, Cope JL, Mistretta TA, Raza S, Metcalf GA, Muzny DM, Gibbs RA, Petrosino JF, Heitkemper M, Savidge TC, Shulman RJ & Versalovic J 2019. Leveraging Human Microbiome Features to Diagnose and Stratify Children with Irritable Bowel Syndrome. J Mol Diagn, 21, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JR, Grams ME, Coresh J, Hwang S, Kovesdy CP, Guallar E, Rhee EP & Shafi T 2019. Serum Metabolites and Cardiac Death in Patients on Hemodialysis. Clin J Am Soc Nephrol, 14, 747–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilhan ZE, Laniewski P, Thomas N, Roe DJ, Chase DM & Herbst-Kralovetz MM 2019. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine, 44, 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isganaitis E, Venditti S, Matthews TJ, Lerin C, Demerath EW & Fields DA 2019. Maternal obesity and the human milk metabolome: associations with infant body composition and postnatal weight gain. Am J Clin Nutr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann HM, Kamp H, Fuchs R, Chorley BN, Deferme L, Ebbels T, Hackermuller J, Perdichizzi S, Poole A, Sauer UG, Tollefsen KE, Tralau T, Yauk C & van Ravenzwaay B 2017. Framework for the quality assurance of ‘omics technologies considering GLP requirements. Regul Toxicol Pharmacol, 91 Suppl 1, S27–S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RS, Boulin A, Laranjo N, Lee-Sarwar K, Chu SH, Yadama AP, Carey V, Litonjua AA, Lasky-Su J & Weiss ST 2019. Metabolomics and Communication Skills Development in Children; Evidence from the Ages and Stages Questionnaire. Metabolites, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan JA, Brennan L, Broadhurst D, Fiehn O, Cascante M, Dunn WB, Schmidt MA & Velagapudi V 2018. Preanalytical Processing and Biobanking Procedures of Biological Samples for Metabolomics Research: A White Paper, Community Perspective (for “Precision Medicine and Pharmacometabolomics Task Group”-The Metabolomics Society Initiative). Clin Chem, 64, 1158–1182. [DOI] [PubMed] [Google Scholar]
- Lains I, Chung W, Kelly RS, Gil J, Marques M, Barreto P, Murta JN, Kim IK, Vavvas DG, Miller JB, Silva R, Lasky-Su J, Liang L, Miller JW & Husain D 2019. Human Plasma Metabolomics in Age-Related Macular Degeneration: Meta-Analysis of Two Cohorts. Metabolites, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghani K 2011. Quality assurance: Importance of systems and standard operating procedures. Perspect Clin Res, 2, 34–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough ML, Maliniak ML, Stevens VL, Carter BD, Hodge RA & Wang Y 2019. Metabolomic markers of healthy dietary patterns in US postmenopausal women. Am J Clin Nutr, 109, 1439–1451. [DOI] [PubMed] [Google Scholar]
- Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ & Hsiao EY 2018. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell, 174, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Playdon MC, Joshi AD, Tabung FK, Cheng S, Henglin M, Kim A, Lin T, van Roekel EH, Huang J, Krumsiek J, Wang Y, Mathe E, Temprosa M, Moore S, Chawes B, Eliassen AH, Gsur A, Gunter MJ, Harada S, Langenberg C, Oresic M, Perng W, Seow WJ & Zeleznik OA 2019. Metabolomics Analytics Workflow for Epidemiological Research: Perspectives from the Consortium of Metabolomics Studies (COMETS). Metabolites, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza-Diaz J, Alvarez-Mercado AI, Ruiz-Marin CM, Reina-Perez I, Perez-Alonso AJ, Sanchez-Andujar MB, Torne P, Gallart-Aragon T, Sanchez-Barron MT, Reyes Lartategui S, Garcia F, Chueca N, Moreno-Delgado A, Torres-Martinez K, Saez-Lara MJ, Robles-Sanchez C, Fernandez MF & Fontana L 2019. Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC Cancer, 19, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez T, Daneshian M, Kamp H, Bois FY, Clench MR, Coen M, Donley B, Fischer SM, Ekman DR, Fabian E, Guillou C, Heuer J, Hogberg HT, Jungnickel H, Keun HC, Krennrich G, Krupp E, Luch A, Noor F, Peter E, Riefke B, Seymour M, Skinner N, Smirnova L, Verheij E, Wagner S, Hartung T, van Ravenzwaay B & Leist M 2013. Metabolomics in toxicology and preclinical research. ALTEX, 30, 209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Huerta OD, Gomez-Fernandez A, de la Torre-Aguilar MJ, Gil A, Perez-Navero JL, Flores-Rojas K, Martin-Borreguero P & Gil-Campos M 2019. Metabolic profiling in children with autism spectrum disorder with and without mental regression: preliminary results from a cross-sectional case-control study. Metabolomics, 15, 99. [DOI] [PubMed] [Google Scholar]
- Rebholz CM, Surapaneni A, Levey AS, Sarnak MJ, Inker LA, Appel LJ, Coresh J & Grams ME 2019. The Serum Metabolome Identifies Biomarkers of Dietary Acid Load in 2 Studies of Adults with Chronic Kidney Disease. J Nutr, 149, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T, Major H, Plumb R, Wilson AJ & Wilson ID 2006. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst, 131, 1075–1078. [DOI] [PubMed] [Google Scholar]
- Sato S, Basse AL, Schonke M, Chen S, Samad M, Altintas A, Laker RC, Dalbram E, Barres R, Baldi P, Treebak JT, Zierath JR & Sassone-Corsi P 2019. Time of Exercise Specifies the Impact on Muscle Metabolic Pathways and Systemic Energy Homeostasis. Cell Metab, 30, 92–110 e4. [DOI] [PubMed] [Google Scholar]
- Shi M, Bazzano LA, He J, Gu X, Li C, Li S, Yaffe K, Kinchen JM, Stuchlik P, Mi X, Nierenberg JL, Razavi AC & Kelly TN 2019. Novel serum metabolites associate with cognition phenotypes among Bogalusa Heart Study participants. Aging (Albany NY), 11, 5124–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E, Multiple Tissue Human Expression Resource, C., Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmuller G, Spector TD & Soranzo N 2014. An atlas of genetic influences on human blood metabolites. Nat Genet, 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Manso Y, Lowenthal MS, Kilpatrick LE, Sampson ML, Telu KH, Rudnick PA, Mallard WG, Bearden DW, Schock TB, Tchekhovskoi DV, Blonder N, Yan X, Liang Y, Zheng Y, Wallace WE, Neta P, Phinney KW, Remaley AT & Stein SE 2013. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal Chem, 85, 11725–31. [DOI] [PubMed] [Google Scholar]
- Smilde AK, van der Werf MJ, Bijlsma S, van der Werff-van der Vat BJ & Jellema RH 2005. Fusion of mass spectrometry-based metabolomics data. Anal Chem, 77, 6729–36. [DOI] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriot P, Nicholls AW, Reily MD, Thaden JJ & Viant MR 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Putluri V, Ambati CR, Dorsey TH, Putluri N & Ambs S 2019. Liver- and Microbiome-derived Bile Acids Accumulate in Human Breast Tumors and Inhibit Growth and Improve Patient Survival. Clin Cancer Res, 25, 5972–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tziotzios C, Petridis C, Dand N, Ainali C, Saklatvala JR, Pullabhatla V, Onoufriadis A, Pramanik R, Baudry D, Lee SH, Wood K, Liu L, Seegobin S, Michelotti GA, Lwin SM, Christou EAA, Curtis CJ, de Rinaldis E, Saxena A, Holmes S, Harries M, Palamaras I, Cunningham F, Parkins G, Kaur M, Farrant P, McDonagh A, Messenger A, Jones J, Jolliffe V, Ali I, Ardern-Jones M, Mitchell C, Burrows N, Atkar R, Banfield C, Alexandroff A, Champagne C, Cooper HL, Vano-Galvan S, Molina-Ruiz AM, Perez NO, Patel GK, Macbeth A, Page M, Bryden A, Mowbray M, Wahie S, Armstrong K, Cooke N, Goodfield M, Man I, de Berker D, Dunnill G, Takwale A, Rao A, Siah TW, Sinclair R, Wade MS, Dlova NC, Setterfield J, Lewis F, Bhargava K, Kirkpatrick N, Estivill X, Stefanato CM, Flohr C, Spector T, Watt FM, Smith CH, Barker JN, Fenton DA, Simpson MA & McGrath JA 2019. Genome-wide association study in frontal fibrosing alopecia identifies four susceptibility loci including HLA-B*07:02. Nat Commun, 10, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant MR, Ebbels TMD, Beger RD, Ekman DR, Epps DJT, Kamp H, Leonards PEG, Loizou GD, MacRae JI, van Ravenzwaay B, Rocca-Serra P, Salek RM, Walk T & Weber RJM 2019. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat Commun, 10, 3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, t Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J & Mons B 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data, 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, Omenn GS, Hood L, Gibbons SM & Price ND 2019. Blood metabolome predicts gut microbiome alpha-diversity in humans. Nat Biotechnol, 37, 1217–1228. [DOI] [PubMed] [Google Scholar]
- Wittemans LBL, Lotta LA, Oliver-Williams C, Stewart ID, Surendran P, Karthikeyan S, Day FR, Koulman A, Imamura F, Zeng L, Erdmann J, Schunkert H, Khaw KT, Griffin JL, Forouhi NG, Scott RA, Wood AM, Burgess S, Howson JMM, Danesh J, Wareham NJ, Butterworth AS & Langenberg C 2019. Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat Commun, 10, 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Flexeder C, McGarrah RW 3rd, Wyss A, Morrison AC, North KE, Boerwinkle E, Kastenmuller G, Gieger C, Suhre K, Karrasch S, Peters A, Wagner GR, Michelotti GA, Mohney RP, Schulz H & London SJ 2019. Metabolomics Identifies Novel Blood Biomarkers of Pulmonary Function and COPD in the General Population. Metabolites, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrana LE, McKeen S, Ibrahim H, Zarei I, Borresen EC, Doumbia L, Bore A, Cissoko A, Douyon S, Kone K, Perez J, Perez C, Hess A, Abdo Z, Sangare L, Maiga A, Becker-Dreps S, Yuan L, Koita O, Vilchez S & Ryan EP 2019. Rice bran supplementation modulates growth, microbiota and metabolome in weaning infants: a clinical trial in Nicaragua and Mali. Sci Rep, 9, 13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelena E, Dunn WB, Broadhurst D, Francis-McIntyre S, Carroll KM, Begley P, O’Hagan S, Knowles JD, Halsall A, Consortium H, Wilson ID & Kell DB 2009. Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Anal Chem, 81, 1357–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.