Abstract
Glycosaminoglycan (GAG)-protein interactions mediate critical physiological and pathological processes, such as neuronal plasticity, development, and viral invasion. However, mapping GAG-protein interaction networks is challenging as these interactions often require specific GAG sulfation patterns and involve trans-membrane receptors or extracellular matrix-associated proteins. Here, we report the first GAG polysaccharide-based photoaffinity probes for the system-wide identification of GAG-binding proteins in living cells. A general platform for the modular, efficient assembly of various chondroitin sulfate (CS)-based photoaffinity probes was developed. Systematic evaluations led to benzophenone-containing probes that efficiently and selectively captured known CS-E-binding proteins in vitro and in cells. Importantly, the probes also enabled the identification of >50 new proteins from living neurons that interact with the neuroplasticity-relevant CS-E sulfation motif. Several candidates were independently validated and included membrane receptors important for axon guidance, innate immunity, synapse development, and synaptic plasticity. Overall, our studies provide a powerful approach for mapping GAG-protein interaction networks, revealing new potential functions for these polysaccharides and linking them to diseases such as Alzheimer’s and autism.
Graphical Abstract

Glycosaminoglycans (GAGs) are a family of structurally complex, linear polysaccharides that are covalently attached to proteoglycans at the cell surface and in the extracellular matrix (ECM).1–3 Chondroitin sulfate (CS) and heparan sulfate (HS), the most prevalent GAGs, are composed of repeating disaccharide units of uronic acid and hexosamine sugars. Tightly-regulated epimerization, along with N- or O-sulfation of these units, gives rise to highly diverse structural motifs.1 These discrete sulfation motifs serve as recognition elements for a range of proteins, including growth factors, chemokines, axon guidance molecules, and cell-surface receptors.1–8 GAG-protein interactions depend on the physiological context and regulate processes such as inflammation,4,9 development,10–12 neuroplasticity,7,13–16 and cell–ECM communication.17 A notable example is the ability of a specific CS sulfation motif, CS-E (Figure 1A), to inhibit axon regeneration after central nervous system injury through its interactions with the cell-surface receptors protein tyrosine phosphatase-sigma (PTPσ)7,15,16 and Nogo (NogoR).18 Mapping GAG-protein interaction networks in their native cellular contexts is therefore key to understanding molecular mechanisms underlying critical biological processes. However, while more than 100 HS GAG-binding proteins (GAG-BPs) have been described,19,20 the specific sulfation motifs mediating those interactions are mostly unknown, and general approaches towards the systems-level identification of GAG-BPs have been lacking. Furthermore, relatively few chondroitin, dermatan, and keratan sulfate GAG-BPs have been reported, suggesting that the vast majority of the GAG-protein interactome remains undiscovered.
Figure 1.

(A) CS sulfation motifs used in this study. (B) Design of CS photoaffinity probes. n = 9–34.
Chemical tools are needed to identify sulfation pattern-specific and context-dependent GAG-protein interactions. The discovery of GAG-BPs presents unique challenges due to the large structural diversity of GAGs, as well as the weak, graded affinities and multivalency of many GAG-protein interactions.21,22 Affinity chromatography approaches rely on non-covalent interactions and require cell lysis,19,20,23,24 which disrupts the native cellular environment and can alter the activity, structure, and binding interactions of GAG-BPs. Such methods thus fail to provide important context-dependent information and are poorly suited to capturing plasma membrane and extracellular proteins. Furthermore, GAGs often bind to shallow cavities rather than deep pockets in proteins1,2,16 and their interactions depend highly on physiological salt concentrations.25 As a result, weaker, but equally important, interactions can be lost during the washing procedures, limiting protein identification to the strongest binders. Here, we report a general approach for mapping GAG-protein interaction networks in living cells. We developed a modular strategy to synthesize the first GAG polysaccharide-based photoaffinity probes. Our studies provide a ‘global snapshot’ of the diverse network of GAG-protein interactions mediating the communication between neurons and the ECM, and reveal new potential functions for these polysaccharides.
As a starting point, we chose to identify CS-E binding proteins to further our understanding of neural plasticity and regeneration. A panel of photoaffinity probes was efficiently constructed by coupling natural CS-E-enriched polysaccharides to trifunctional photo-crosslinking reagents (TPRs, 1–4; Figure 1B and Scheme S1). These reagents contained three key elements: 1) an amine functionality for coupling to GAG chains via carboxylic acids on D-glucuronic acid (GlcA); 2) a photo-crosslinking group (PC) to covalently crosslink interacting proteins; and 3) an alkyne functionality for attaching a biotin handle for affinity enrichment. Alternatively, a biotin moiety was incorporated by functionalizing the peptide N-terminus at the reducing end of the GAG chain. We envisaged that these TPRs could be appended to any GAG oligo- or polysaccharide. Here, we chose polysaccharides to favor higher affinity, multivalent binding of proteins.
A series of CS polysaccharide probes enriched in the CS-E or CS-C motifs were designed and synthesized to examine the effects of different photo-crosslinking labels, linkage types, and linker lengths on crosslinking. Diazirine (DA)- or benzophenone (BP)-containing probes 5–10 were prepared by coupling TPRs 1–3 to end-biotinylated CS polysaccharides via isourea or amide linkages (Figure 2). The average photo-crosslinker to CS polysaccharide ratios (PC:CS) were determined using a carbazole assay26 and UV-Vis spectroscopy (Figure S1). The coupling reactions conditions were optimized to obtain reproducible PC:CS ratios and enable direct comparisons (Figure S1 and Table S1). Importantly, probes 5–9 retained the ability to bind a known CS-E-binding protein, PTPσ, and CS-E-specific monoclonal antibody (mAb),15,27 as determined by enzyme-linked immunosorbent assays (ELISAs; Figures 3A–B and S2A). As expected, CS-C-based probe 10 showed 100-fold weaker binding to the proteins. These results suggested that the addition of PC moieties did not significantly alter the affinity or specificity of CS polysaccharides for proteins.
Figure 2.
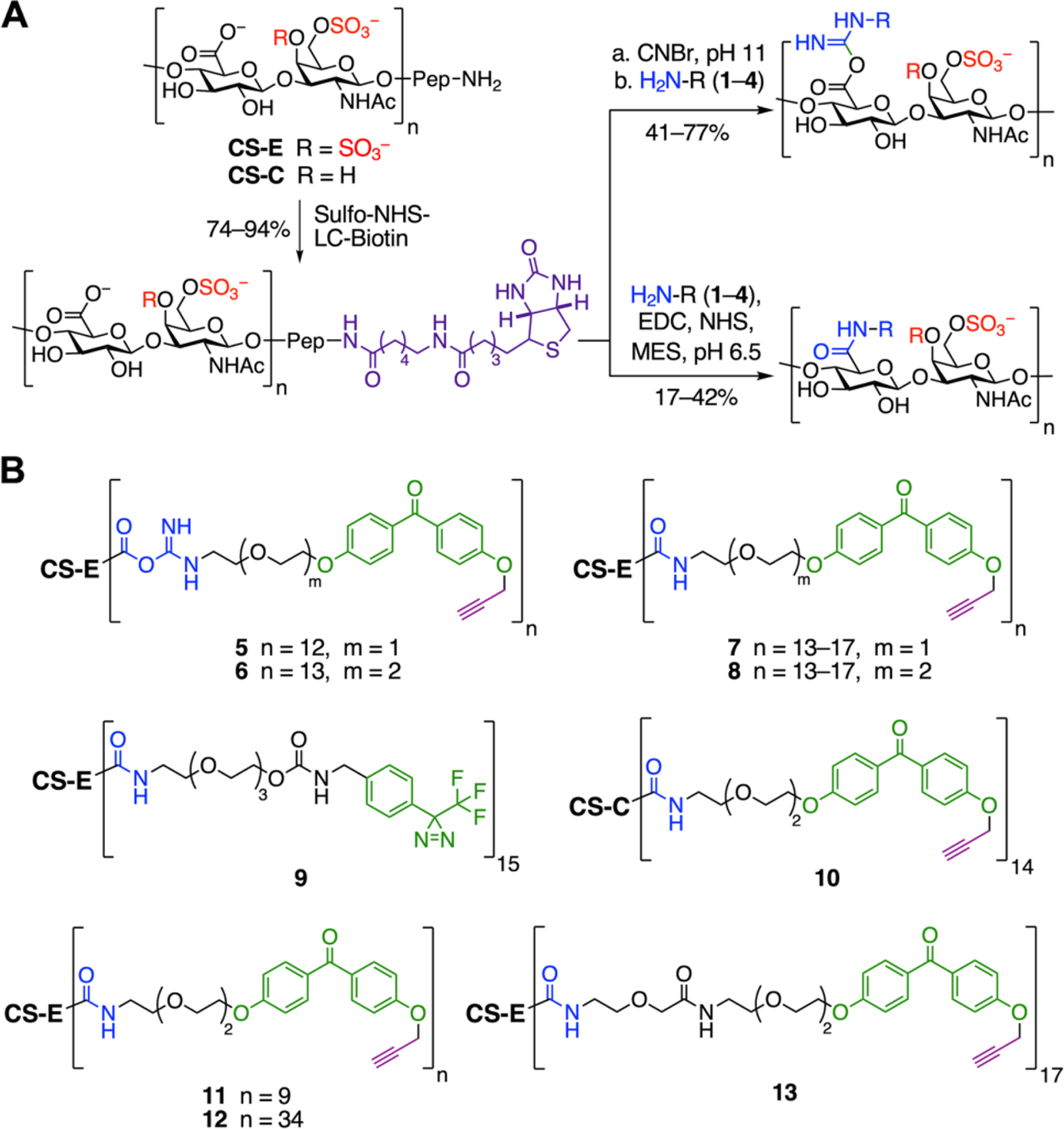
(A) Synthesis of GAG-based photoaffinity probes. (B) Structures of probes 5–13. The average n (PC:CS ratio) was calculated as described in the Supporting Information. Pep = peptide.
Figure 3.
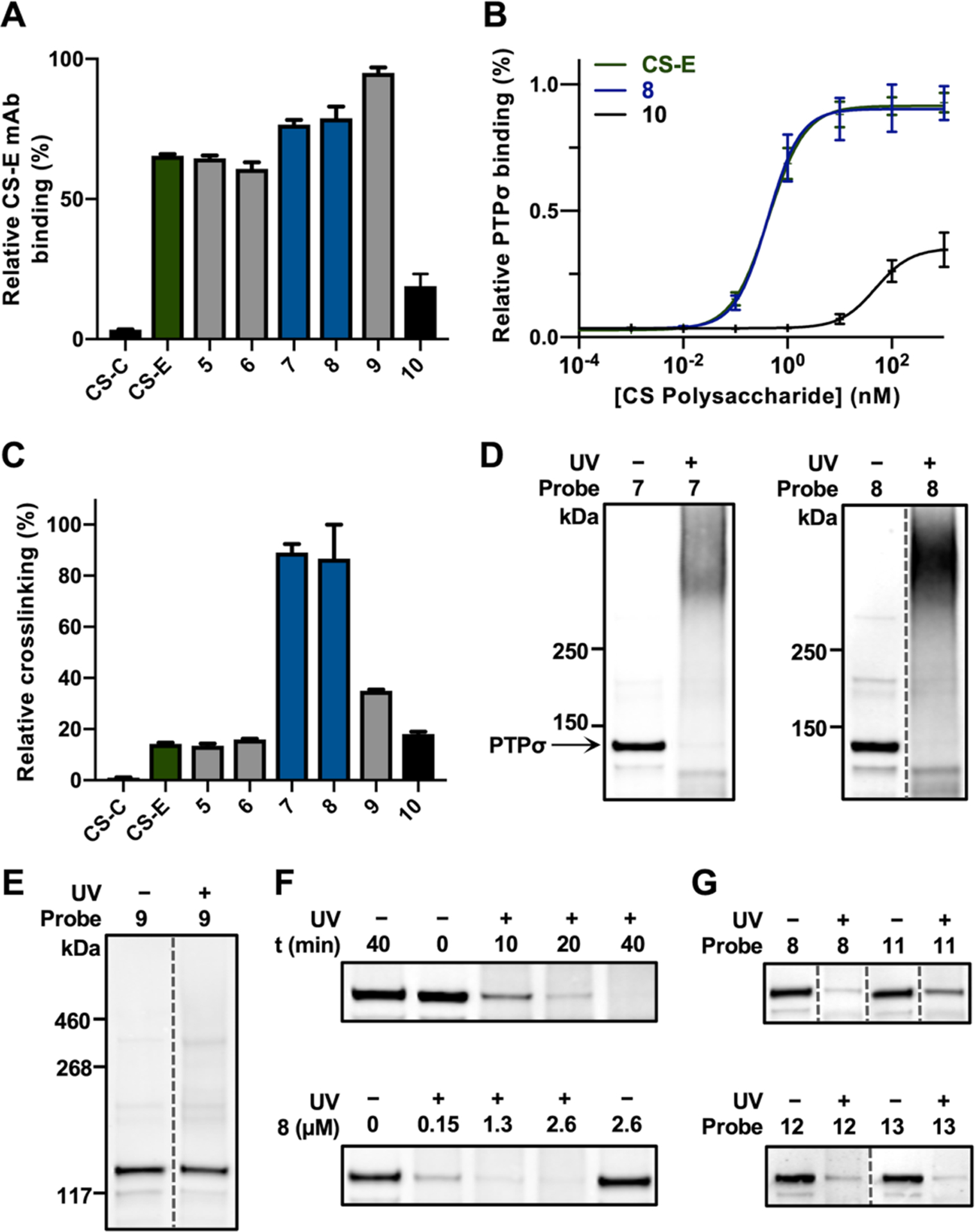
(A) Probes 5–9 retain binding to a CS-E mAb (n = 4). (B) The PTPσ ectodomain fused to an immunoglobulin Fc domain (PTPσ-Fc) binds with similar affinities to 8 and end-biotinylated CS-E, but not to CS-C-based probe 10 (n = 2). (C) Probes 7 and 8 label the CS-E mAb most efficiently, as determined by photo-ELISA (n = 2). (D) Labeling of PTPσ-Fc with 7 and 8, as detected by immunoblotting. (E) Probe 9 does not label PTPσ-Fc. (F) Time- and concentration-dependent labeling of PTPσ-Fc with 7 (top) and 8 (bottom). (G) Effects of the PC:CS ratio and linker length on photo-crosslinking efficiency. Data represent the mean ± SEM. Dotted lines indicate non-adjacent lanes within the same blot where irrelevant lanes were cropped out for clarity.
Next, we compared the photo-crosslinking efficiencies of probes 5–10 using a novel photo-ELISA assay (Figure S2B). The probes were adhered to streptavidin-coated plates, and the CS-E mAb was added. After irradiation and stringent washes with 1% SDS to remove non-covalent interactions, the covalently-crosslinked CS-E mAb was detected using an anti-mouse IgG streptavidin-horseradish peroxidase (HRP) conjugate. We found that the amide-linked probes 7 and 8 labeled the CS-E mAb more efficiently than the equivalent isourea-linked probes 5 and 6 (Figure 3C). Moreover, photo-crosslinking was greater for the amide-linked BP-containing probes 7 and 8 compared to amide-linked DA probe 9, presumably due to the longer-lived triplet diradical formed upon BP pho-toreduction. Increasing the linker length between the photo-crosslinking group and CS (7 versus 8) had no significant effect on crosslinking efficiency. Furthermore, negligible photo-crosslinking of CS-C-based probe 10 to the CS-E mAb was observed by photo-ELISA, indicating that photo-crosslinking was binding dependent and thus specific.
To confirm these results, probes 7– 9 were incubated with PTPσ, irradiated, and resolved by SDS-PAGE. Complete disappearance of the PTPσ band and formation of higher molecular weight PTPσ adducts were observed for BP probes 7 and 8 (Figure 3D), but not for DA probe 9 (Figure 3E), supporting the results obtained by photo-ELISA. Formation of these crosslinked adducts depended on the probe concentration and irradiation time (Figure 3F). To determine the optimal PC:CS ratio and linker length, we synthesized probes 11 (PC:CS 9:1) and 12 (PC:CS 34:1) (Figure 2B). Covalent labeling of PTPσ with 11 was less efficient compared to 8 and 12 (Figure 3G), suggesting that a PC:CS ratio higher than 9:1 was required for optimal photo-crosslinking. As extending the linker on 8 did not improve the PTPσ labeling efficiency (13, Figures 2B and 3G), probe 8 was chosen for additional studies.
We performed competition experiments to assess the selectivity of 8. Photo-crosslinking to PTPσ was significantly reduced in the presence of excess polysaccharides enriched in the CS-E motif, but not the CS-A or CS-C motif (Figure 4A). Consistent with the photo-ELISA results (Figure 3C), the corresponding CS-C probe 10 failed to label PTPσ, demonstrating the ability of 8 to capture GAG-BPs that recognize specific sulfation motifs. We next confirmed that photo-crosslinking had occurred via the interaction of 8 with a defined binding site on PTPσ. CS-E recognizes a conserved, positively-charged surface within the first immunoglobulin (Ig)-like domain of PTPσ.7,16 Accordingly, mutation of lysine residues K67–71 in this region significantly reduced the photo-cross-linking of PTPσ to 8 (Figure S2C).
Figure 4.
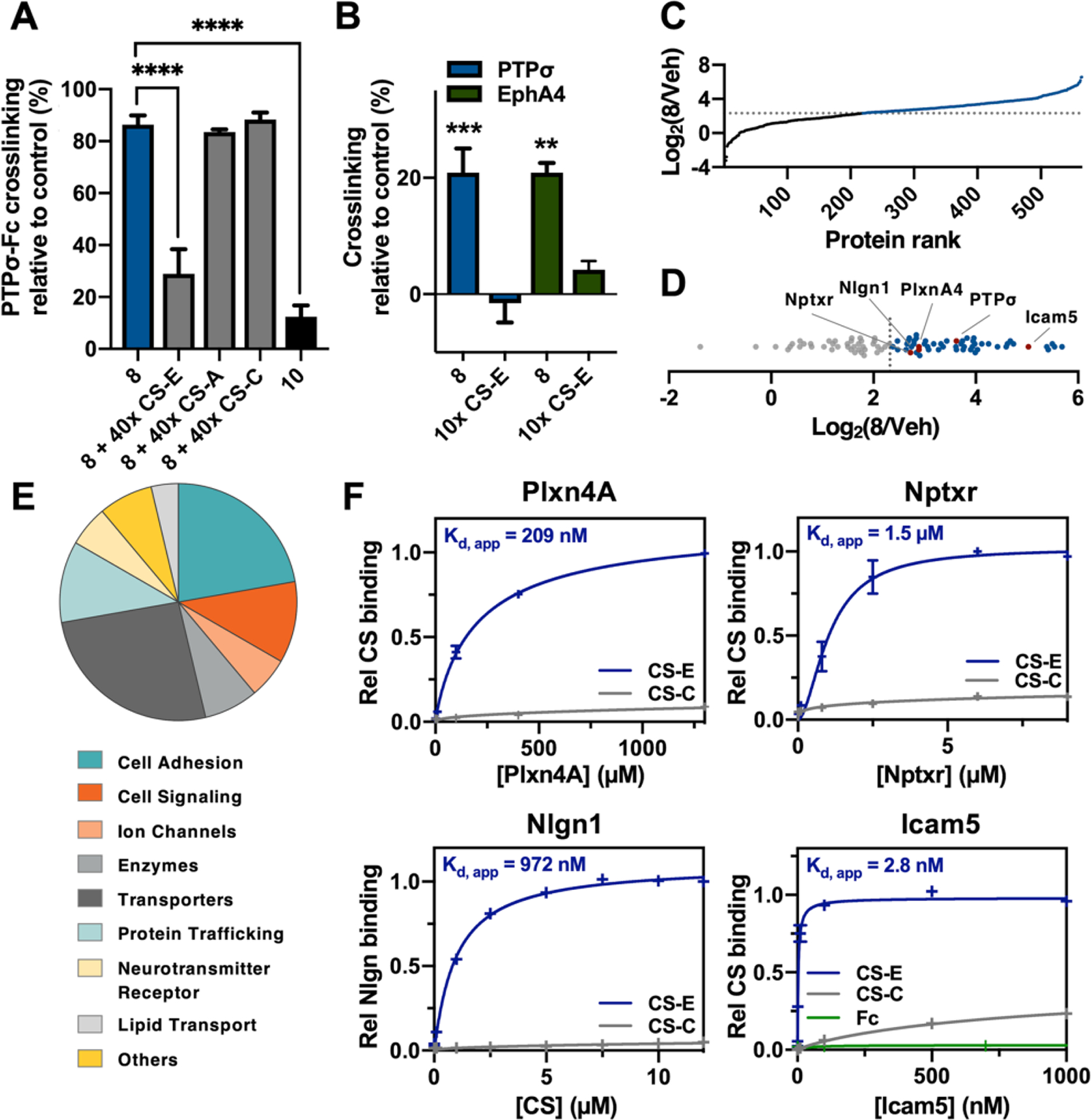
(A) Competition experiments show that 8 selectively photo-crosslinks PTPσ-Fc in vitro (n = 3). (B) Selective photo-crosslinking of endogenous PTPσ and EphA4 with 8 in cortical neurons (n = 4). (C) TMT ratios of 8-treated versus vehicle (Veh)-treated neurons showing proteins enriched by ⩾5-fold (blue). (D) TMT ratio plot of the 54 membrane-associated and ECM proteins (blue). (E) Functions of the identified CS-E interactors. (F) Validation by ELISA of new CS-E-binding proteins (n = 2). Data represent the mean ± SEM. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01.
Next, we evaluated whether photo-crosslinked proteins could be efficiently pulled down via the end-terminal biotin handle. PTPσ was incubated with 8 and irradiated prior to pull down using streptavidin-coated beads. After washing, the PTPσ-Fc–8 adduct was eluted, resolved by SDS-PAGE, and detected by immunoblotting. Unfortunately, low pull-down efficiencies were observed (Figure S3). We reasoned that the poor capture efficiency might be due to interference from the long, anionic CS chains. Thus, we trimmed the polysaccharides using chondroitinase ABC (ChABC) after the irradiation step and appended biotin moieties using copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) chemistry. This time, PTPσ-Fc was efficiently pulled down (Figure S3).
Having demonstrated the selectivity and efficiency of the probe in vitro, we investigated whether 8 could crosslink endogenous proteins in cells. The probe was incubated with cortical neurons, and after irradiation, photo-crosslinked proteins were detected by immunoblotting. Notably, robust labeling of the CS-E-binding proteins PTPσ7,15 and ephrin type-A receptor 4 (EphA4)28 was observed (Figure 4B). This labeling was competitively inhibited by the addition of excess CS-E-enriched polysaccharides, confirming the specificity of these proteins for the CS-E motif. Together, our results demonstrate that probe 8 selectively photo-crosslinks CS-E-binding proteins in living cells.
Finally, we investigated whether novel CS-E interacting proteins could be discovered using probe 8. Cortical neurons were incubated with 8 or vehicle control, then irradiated and lysed (Figure S4). The samples were treated with ChABC, and biotin moieties were appended via Cu-AAC chemistry. After streptavidin pull down, the captured proteins were eluted, resolved by SDS-PAGE, and subjected to in-gel proteolytic digestion. The resulting peptides were quantified by isotopic labeling with tandem mass tags (TMTs),29 fractionated, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
In total, we identified 54 ECM and membrane-associated CS-E-binding proteins that showed ⩾5-fold enrichment in the probe versus the control samples in two independent experiments (Figures 4C–D and S5; Table S2). Interestingly, six of the proteins possess Ig-like folds,7,16 suggesting a potential CS-E recognition motif. Moreover, three previously reported CS-binding proteins were identified, including PTPσ,7 neural cell adhesion molecule 2 (NCAM2),30,31 and apolipoprotein E (ApoE).32 The novel CS-E interactors included known regulators of axon guidance, neurite outgrowth, and synaptic plasticity, as well as proteins involved in cell adhesion and membrane trafficking (Figure 4E and Table S2). For instance, we identified plexin-A4, a receptor for class 3 semaphorins (Sema3As) that plays important roles in axon guidance and development.33 Interestingly, CS-E is a known binder of Sema3A,34 but its interaction with plexin-A4 had not been described. Our findings suggest a new potential role for CS-E in the regulation of Sema3A/plexin-A4 complexes and semaphorin signaling. Intercellular adhesion molecule 5 (Icam5) and neuroligin-1 (Nlgn1) are cell adhesion molecules linked to neuropsychiatric disorders, including autism, and they play important roles in synapse development and function.35–37 The neuronal pentraxin receptor (Nptxr) is involved in synapse organization and is a potential biomarker of Alzheimer’s disease progression.38,39 Plexin-A4, Nlgn1, Icam5, and Nptxr were independently validated by ELISA, and all showed robust and specific binding to CS-E-, but not CS-C-enriched, polysaccharides (Figure 4F). Together, these results demonstrate the ability of our GAG photo-crosslinking probes to selectively capture and identify novel GAG-BPs.
In summary, we have developed a modular, versatile, and efficient strategy for the generation of GAG-based photo-crosslinking probes. Notably, this approach can be extended to map sulfation-specific GAG-BPs for other GAG classes, including heparan and dermatan sulfate. These probes directly address the shortcomings of traditional methods for GAG-BP identification and enable the discovery of context-dependent interactions involving cell-surface and ECM-associated proteins. Given the important regulatory roles of GAGs, mapping GAG-protein interactions in different cellular contexts will provide a deeper understanding of critical physiological and pathological processes. Ultimately, we envision that these discoveries will contribute to the development of new approaches to tackling diseases with unmet medical needs, including cancer, neurodegenerative, neuropsychiatric, and autoimmune disorders.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Michael Sweredoski, Dr. Annie Moradian, and Dr. Brett Lomenick from the Protein Exploration Laboratory (PEL) at Caltech. This work was supported by the NIH (R01 GM093627).
Footnotes
The authors declare no competing financial interest.
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
Experimental procedures and characterization data, Scheme S1, Figures S1–S5, Tables S1–S2 (PDF).
REFERENCES
- (1).Capila I; Linhardt RJ Heparin-Protein Interactions. Angew. Chem., Int. Ed 2002, 41, 391–412. [DOI] [PubMed] [Google Scholar]
- (2).Xu D; Esko JD Demystifying Heparan Sulfate-Protein Interactions. Ann. Rev. Biochem 2014, 83, 129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Karamanos NK; Piperigkou Z; Theocharis AD; Watanabe H; Franchi M; Baud S; Brézillon S; Götte M; Passi A; Vigetti D; Ricard-Blum S; Sanderson RD; Neill T; Iozzo RV Proteoglycan Chemical Diversity Drives Multifunctional Cell Regulation and Therapeutics. Chem. Rev 2018, 118, 9152–9232. [DOI] [PubMed] [Google Scholar]
- (4).Handel TM; Johnson Z; Crown SE; Lau EK; Sweeney M; Proudfoot AE Regulation of Protein Function by Glycosaminoglycans - As Exemplified by Chemokines. Ann. Rev. Biochem 2005, 74, 385–410. [DOI] [PubMed] [Google Scholar]
- (5).Gama CI; Tully SE; Sotogaku N; Clark PM; Rawat M; Vaidehi N; Goddard WA; Nishi A; Hsieh-Wilson LC Sulfation Patterns of Glycosaminoglycans Encode Molecular Recognition and Activity. Nat. Chem. Biol 2006, 2, 467–473. [DOI] [PubMed] [Google Scholar]
- (6).Da Costa DS; Reis RL; Pashkuleva I Sulfation of Glycosaminoglycans and Its Implications in Human Health and Disorders. Annu. Rev. Biomed. Eng 2017, 19, 1–26. [DOI] [PubMed] [Google Scholar]
- (7).Shen Y; Tenney AP; Busch SA; Horn KP; Cuascut FX; Liu K; He Z; Silver J; Flanagan JG PTPσ Is a Receptor for Chondroitin Sulfate Proteoglycan, an Inhibitor of Neural Regeneration. Science 2009, 326, 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Petitou M; van Boeckel CAA A Synthetic Antithrombin III Binding Pentasaccharide Is Now a Drug! What Comes Next? Angew. Chem., Int. Ed 2004, 43, 3118–3133. [DOI] [PubMed] [Google Scholar]
- (9).Taylor KR; Gallo RL Glycosaminoglycans and Their Proteoglycans: Host-Associated Molecular Patterns for Initiation and Modulation of Inflammation. FASEB J. 2006, 20, 9–22. [DOI] [PubMed] [Google Scholar]
- (10).Carulli D; Laabs T; Geller HM; Fawcett JW Chondroitin Sulfate Proteoglycans in Neural Development and Regeneration. Curr. Opin. Neurobiol 2005, 15, 116–120. [DOI] [PubMed] [Google Scholar]
- (11).Schwartz NB; Domowicz MS Proteoglycans in Brain Development and Pathogenesis. FEBS Lett. 2018, 592, 3791–3805. [DOI] [PubMed] [Google Scholar]
- (12).Häcker U; Nybakken K; Perrimon N Heparan Sulphate Proteoglycans: the Sweet Side of Development. Nat. Rev. Mol. Cell Biol 2005, 6, 530–541. [DOI] [PubMed] [Google Scholar]
- (13).Miller GM; Hsieh-Wilson LC Sugar-Dependent Modulation of Neuronal Development, Regeneration, and Plasticity by Chondroitin Sulfate Proteoglycans. Exp. Neurol 2015, 274, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Vo T; Carulli D; Ehlert EME; Kwok JCF; Dick G; Mecollari V; Moloney EB; Neufeld G; De Winter F; Fawcett JW; Verhaagen J The Chemorepulsive Axon Guidance Protein semaphoring 3A Is a Constituent of Perineuronal Nets in the Adult Rodent Brain. Mol. Cell. Neurosci 2013, 56, 186–200. [DOI] [PubMed] [Google Scholar]
- (15).Brown JM; Xia J; Zhuang B; Cho K-S; Rogers CJ; Gama CI; Rawat M; Tully SE; Uetani N; Mason DE; Tremblay ML; Peters EC; Habuchi O; Chen DF; Hsieh-Wilson LC A Sulfated Carbohydrate Epitope Inhibits Axon Regeneration After Injury. Proc. Nat. Acad. Sci. U. S. A 2012, 109, 4768–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Griffith AR; Rogers CJ; Miller GM; Abrol R; Hsieh-Wilson LC; Goddard WA Predicting Glycosaminoglycan Surface Protein Interactions and Implications for Studying Axonal Growth. Proc. Nat. Acad. Sci. U. S. A 2017, 114, 13697–13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Linhardt RJ; Toida T Role of Glycosaminoglycans in Cellular Communication. Acc. Chem. Res 2004, 37, 431–438. [DOI] [PubMed] [Google Scholar]
- (18).Dickendesher TL; Baldwin KT; Mironova YA; Koriyama Y; Raiker SJ; Askew KL; Wood A; Geoffroy CG; Zheng B; Liepmann CD; Katagiri Y; Benowitz LI; Geller HM; Giger RJ NgR1 and NgR3 Are Receptors for Chondroitin Sulfate Proteoglycans. Nat. Neurosci 2012, 15, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Esko JD; Prestegard JH; Linhardt RJ Proteins That Bind Sulfated Glycosaminoglycans In Essentials of Glycobiology, edition;Varki A; Cummings RD; Esko JD; Stanley P; Hart GW; Aebi M; Darvill AG; Kinoshita T; Packer NH; Prestegard JH; Schnaar RL; Seeberger PH Eds.; Cold Spring Harbor: NY, 2017. [Google Scholar]
- (20).Ori A; Wilkinson MC; Fernig DG A Systems Biology Approach for the Investigation of the Heparin/Heparan Sulfate Interactome. J. Biol. Chem 2011, 286, 19892–19904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kreuger J; Salmivirta M; Sturiale L; Giménez-Gallego G; Lindahl U Sequence Analysis of Heparan Sulfate Epitopes with Graded Affinities for Fibroblast Growth Factors 1 and 2. J. Biol. Chem 2001, 276, 30744–30752. [DOI] [PubMed] [Google Scholar]
- (22).Collins BE; Paulson JC Cell Surface Biology Mediated by Low Affinity Multivalent Protein-Glycan Interactions. Curr. Opin. Chem. Biol 2004, 8, 617–625. [DOI] [PubMed] [Google Scholar]
- (23).Gesslbauer B; Derler R; Handwerker C; Seles E; Kungl AJ Exploring the Glycosaminoglycan-Protein Interaction Network by Glycan-Mediated Pull-Down Proteomics. Electrophoresis 2016, 37, 1437–1447. [DOI] [PubMed] [Google Scholar]
- (24).Zhang Y; Jiang N; Lu H; Hou N; Piao X; Cai P; Yin J; Wahlgren M; Chen Q Proteomic Analysis of Plasmodium Falciparum Schizonts Reveals Heparin-Binding Merozoite Proteins. J. Proteome Res 2013, 12, 2185–2193. [DOI] [PubMed] [Google Scholar]
- (25).Zhao J; Liu X; Kao C; Zhang E; Li Q; Zhang F; Linhardt RJ Kinetic and Structural Studies of Interactions Between Glycosaminoglycans and Langerin. Biochemistry 2016, 55, 4552–4559. [DOI] [PubMed] [Google Scholar]
- (26).Taylor KA; Buchanan-Smith JG A Colorimetric Method for the Quantitation of Uronic Acids and a Specific Assay for Galacturonic Acid. Anal. Biochem 1992, 201, 190–196. [DOI] [PubMed] [Google Scholar]
- (27).Tully SE; Rawat M; Hsieh-Wilson LC Discovery of a TNF-a Antagonist Using Chondroitin Sulfate Microarrays. J. Am. Chem. Soc 2006, 128, 7740–7741. [DOI] [PubMed] [Google Scholar]
- (28).Miller GM; Griffin ME; Brown JM; Rogers CJ; Goddard WA; Zhuang B; Hsieh-Wilson LC Unpublished Work. [Google Scholar]
- (29).Thompson A; Schäfer J; Kuhn K; Kienle S; Schwarz J; Schmidt G; Neumann T; Johnstone R; Mohammed AKA; Hamon C Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Anal. Chem 2003, 75, 1895–1904. [DOI] [PubMed] [Google Scholar]
- (30).Friedlander DR; Milev P; Karthikeyan L; Margolis RK; Margolis RU; Grumet M The Neuronal Chondroitin Sulfate Proteoglycan Neurocan Binds to the Neural Cell Adhesion Molecules Ng-CAM/L1/NILE and N-CAM, and Inhibits Neuronal Adhesion and Neurite Outgrowth. J. Cell Biol 1994, 125, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Djerbal L; Lortat-Jacob H; Kwok JCF Chondroitin Sulfates and Their Binding Molecules in the Central Nervous System. Glycoconjugate J. 2017, 34, 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Burgess JW; Liang P; Vaidyanath CA; Marcel YL ApoE of the HepG2 Cell Surface Includes a Major Pool Associated with Chondroitin Sulfate Proteoglycans. Biochemistry 1998, 38, 524–531. [DOI] [PubMed] [Google Scholar]
- (33).Pasterkamp RJ Getting Neural Circuits Into Shape with Semaphorins. Nat. Rev. Neurosci 2012, 13, 605–618. [DOI] [PubMed] [Google Scholar]
- (34).Dick G; Tan CL; Alves JN; Ehlert EME; Miller GM; Hsieh-Wilson LC; Sugahara K; Oosterhof A; van Kuppevelt TH; Verhaagen J; Fawcett JW; Kwok JCF Semaphorin 3A Binds to the Perineuronal Nets via Chondroitin Sulfate Type E Motifs in Rodent Brains. J. Biol. Chem 2013, 288, 27384–27395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jeong J; Pandey S; Li Y; Badger JD; Lu W; Roche KW PSD-95 Binding Dynamically Regulates Nlgn1 Trafficking and Function. Proc. Nat. Acad. Sci. U. S. A 2019, 116, 12035–12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pei Y-P; Wang Y-Y; Liu D; Lei H-Y; Yang Z-H; Zhang Z-W; Han M; Cheng K; Chen Y-S; Li J-Q; Cheng G-R; Xu L; Wu Q-M; McClintock SM; Yang Y; Zhang Y; Zeng Y ICAM5 as a Novel Target for Treating Cognitive Impairment in Fragile X Syndrome. J. Neurosci 2020, 40, 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Glessner JT; Wang K; Cai G; Korvatska O; Kim CE; Wood S; Zhang H; Estes A; Brune CW; Bradfield JP; Imielinski M; Frackelton EC; Reichert J; Crawford EL; Munson J; Sleiman PMA; Chiavacci R; Annaiah K; Thomas K; Hou C; Glaberson W; Flory J; Otieno F; Garris M; Soorya L; Klei L; Piven J; Meyer KJ; Anagnostou E; Sakurai T; Game RM; Rudd DS; Zurawiecki D; McDougle CJ; Davis LK; Miller J; Posey DJ; Michaels S; Kolevzon A; Silverman JM; Bernier R; Levy SE; Schultz RT; Dawson G, Owley T; McMahon WM; Wassink TH; Sweeney JA; Nurnberger JI Jr; Coon H; Sutcliffe JS; Minshew NJ; Grant SFA; Bucan M; Cook EH Jr; Buxbaum JD; Devlin B; Schellenberg GD; Hakonarson H Autism Genome-Wide Copy Number Variation Reveals Ubiquitin and Neuronal Genes. Nature 2009, 459, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bryant L; Tsolaki M; Soosaipillai A; Brown M; Zilakaki M; Tagaraki F; Fotiou D; Koutsouraki E; Grosi E; Prassas I; Diamandis EP Liquid Biopsy of Cerebrospinal Fluid Identifies Neuronal Pentraxin Receptor (Nptxr) as a Biomarker of Progression of Alzheimer’s Disease. Clin. Chem. Lab. Med 2019, 57, 1875–1881. [DOI] [PubMed] [Google Scholar]
- (39).Lee S-J; Wei M; Zhang C; Maxeiner S; Pak C; Botelho SC; Trotter J; Sterky FH; Südhof TC Presynaptic Neuronal Pentraxin Receptor Organizes Excitatory and Inhibitory Synapses. J. Neurosci 2017, 37, 1062–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


