Abstract
Inhibition of the kinase ATR, a central regulator of the DNA damage response, eliminates subsets of cancer cells in certain tumors. As previously shown, this is at least partly attributable to synthetic lethal interactions between ATR and POLD1, the catalytic subunit of the polymerase δ. Various POLD1 variants have been found in colorectal cancer, but their significance as therapeutic targets for ATR pathway inhibition remains unknown. Using CRISPR/Cas9 in the colorectal cancer cell line DLD-1, which harbors four POLD1 variants, we established heterozygous POLD1-knockout clones with exclusive expression of distinct variants to determine the functional relevance of these variants individually by assessing their impact on ATR pathway activation, DNA replication, and cellular sensitivity to inhibition of ATR or its effector kinase CHK1. Of the four variants analyzed, only POLD1R689W affected POLD1 function, as demonstrated by compensatory ATR pathway activation and impaired DNA replication. Upon treatment with ATR or CHK1 inhibitors, POLD1R689W strongly decreased cell survival in vitro, which was attributable at least partly to S phase impairment and apoptosis. Similarly, treatment with the ATR inhibitor AZD6738 inhibited growth of murine xenograft tumors, harboring the POLD1R689W variant, in vivo. Our POLD1-knockout model thus complements algorithm-based models to predict the pathogenicity of tumor-specific variants of unknown significance and illustrates a novel and potentially clinically relevant therapeutic approach using ATR/CHK1 inhibitors in POLD1-deficient tumors.
Subject terms: Cancer, Molecular biology, Biomarkers, Diseases, Medical research, Molecular medicine, Oncology
Introduction
The relationship between two genes, in which single mutations alone are not lethal, but in combination are incompatible with cell survival, is defined as synthetic lethality1,2. As tumors frequently harbor defects in genes involved in DNA repair3 and those defects are often compensated by other DNA repair genes3,4, synthetic lethality represents a novel approach for the individualized genotype-based treatment of tumors by pharmacologically targeting the compensatory partner of defective DNA repair genes. One prominent example of such an approach is the treatment of patients harboring BRCA1/2-deficient cancers with PARP inhibitors5.
ATR is a phosphoinositide 3-kinase-related kinase and acts as central regulator of the replication checkpoint during DNA damage response6. Activated by the accumulation of single-stranded DNA at sites of replication stress or DNA damage, ATR initiates replication fork stabilization, cell cycle arrest and DNA repair via homologous recombination (HR)6,7. Inhibitors of ATR are currently investigated in clinical trials8,9 and appear to enable the effective elimination of certain subsets of cancer cells. Despite the identification of a variety of potential biomarkers (comprehensively reviewed by Lecona & Fernandez-Capetillo7), the specific determinants of this therapeutic response remain incompletely defined, particularly in regard to synthetically lethal relationships between ATR and certain DNA repair genes.
We and others previously identified POLD1 as acting synthetically lethal with ATR10,11. POLD1 is the catalytic subunit of the polymerase (Pol) δ which is responsible for the elongation of the lagging strand during DNA replication. Related essential components of DNA replication are the Polα-primase complex and Polε12,13. POLD1 variants have been recently identified in colorectal and other cancers14–16. The significance of these variants as potential biomarkers for the targeted treatment of tumors with ATR inhibitors, however, remains enigmatic.
Therefore, we applied CRISPR/Cas9 to establish a POLD1-knockout (KO) model in the human colorectal cancer (CRC) cell line DLD-1, which harbors four heterozygous POLD1 variants17, yielding cell clones with exclusive expression of distinct POLD1 variants. On the one hand, this model facilitated the determination of the functional impact of specific POLD1 variants and could thus functionally complement existing prediction models for the pathogenicity of variants of unknown significance (VUS). On the other hand, it allowed defining the impact of different POLD1 variants on the sensitivity to inhibitors of ATR or other components of the ATR pathway in vitro and in vivo.
Results
CRISPR/Cas9-mediated generation of a POLD1-knockout model
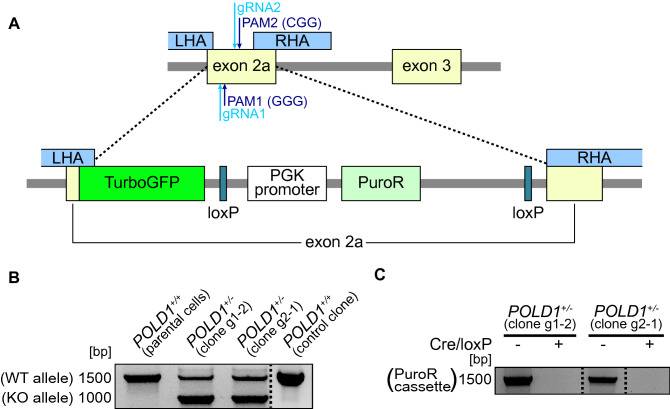
To establish a POLD1-KO in the DLD-1 cell line, we used the CRISPS/Cas9 technique to integrate a repair template into exon 2a of the POLD1 locus via HR (Fig. 1A), causing allele inactivation. Multiple single-cell colonies were genotyped and two distinct heterozygous POLD1-KO clones obtained (termed g1-2 and g2-1). Clones derived from scrambled guide RNA (gRNA)-transfected cells served as POLD1+/+ control (ctrl) (Fig. 1B). Immunoblotting of g1-2 and g2-1 revealed no changes in POLD1 protein expression as compared to POLD1+/+ parental cells and the ctrl clone (data not shown). To enable the consecutive generation of homozygous POLD1-KO clones, we excised the floxed puromycin resistance cassette (Fig. 1A) in both g1-2 and g2-1 cell clones using Cre recombinase (Fig. 1C). Upon targeting of the second allele, however, no homozygous POLD1-KO clones were obtained despite multiple targeting rounds.
Figure 1.
CRISPR/Cas9-mediated generation of a POLD1-knockout model. (A) Schematic overview of the targeting procedure, displaying CRISPR/Cas9-mediated integration of the repair template in exon 2a of the POLD1 locus. (B) PCR detecting the POLD1-KO and –WT alleles as well as (C) the Cre/loxP-mediated excision of the puromycin resistance cassette. Dashed lines indicate cropping, and the original gels are displayed in Figure S1.
Identification of the allele-specific localization of previously reported POLD1 variants in DLD-1
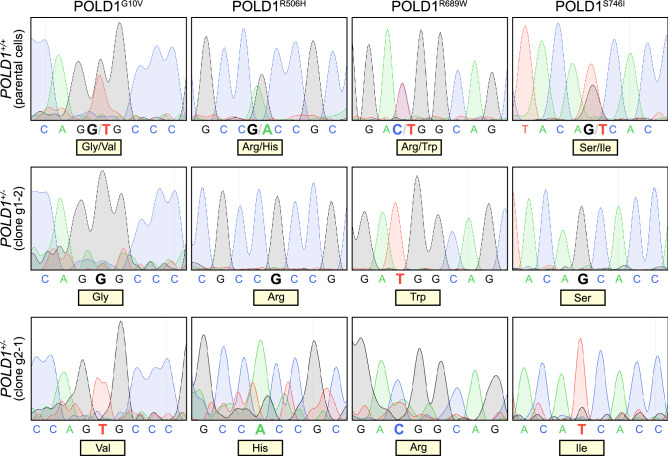
Four heterozygous POLD1 variants have been reported in DLD-1 cells, namely G10V, R506H, R689W and S746I17. These were confirmed in our DLD-1 POLD1+/+ cells by genome sequencing (Fig. 2, upper panel). However, the allele specific localization of these variants remained unknown. Using mRNA-sequencing, we demonstrated that the intact POLD1 allele of the heterozygous cell clone g1-2 exhibits only the R689W variant (termed POLD1R689W/-, Fig. 2, middle panel), while the intact POLD1 allele of the heterozygous cell clone g2-1 exhibits the other three variants G10V, R506H and S746I (POLD1G10V,R506H,S746I/-, according to the data obtained later termed here POLD1+/- for convenience, Fig. 2, lower panel).
Figure 2.
Identification of the allele-specific localization of previously reported POLD1 variants in DLD-1. Sequencing of POLD1 variants in POLD1+/+ parental cells (upper panel) as well as the POLD1+/- g1-2 (middle panel) and g2-1 (lower panel) cell clones.
Functional characterization of the pre-existing POLD1 variants in DLD-1
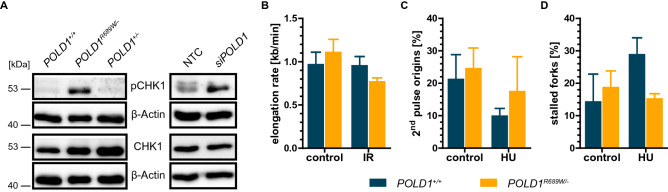
As defects in DNA replication lead to the activation of DNA repair pathways4, the functional inactivation of POLD1 is expected to cause compensatory activation of the ATR pathway10. We therefore assessed protein and phosphorylation levels of CHK1, the major effector kinase of ATR, to determine the functional significance of the pre-existing POLD1 variants in DLD-1. We observed a strong constitutive CHK1 phosphorylation exclusively in POLD1R689W/- but not in POLD1+/- and POLD1+/+ cells (Fig. 3A, left panel), an effect qualitatively comparable to the effects of small interfering RNA (siRNA)-mediated subtotal POLD1-depletion (Fig. 3A, right panel). Therefore, only the POLD1R689W variant had a measurable impact on POLD1 function in regard to compensatory ATR/CHK1 pathway activation, while the other variants POLD1G10V, POLD1R506H and POLD1S746I caused no discernible effect. As POLD1 encodes the catalytic subunit of Polδ, we next investigated the impact of this POLD1R689W variant on DNA replication upon treatment with ionizing radiation (IR) or hydroxyurea (HU), using DNA fiber assays. We found that the constitutive overall elongation rate was slightly higher in POLD1R689W/- than in POLD1+/+ cells. Upon IR-treatment, however, the elongation rate decreased in POLD1R689W/- cells, but remained unchanged in POLD1+/+ cells (Fig. 3B). Upon HU-treatment, 2nd pulse origins were decreased to a higher extent in POLD1+/+ than in POLD1R689W/- cells (Fig. 3C). Consistently, stalled replication forks were increased only in POLD1+/+ but not in POLD1R689W/- cells (Fig. 3D). Although, these data did not reach statistical significance, there appeared to be a clear trend suggesting that the POLD1R689W variant functionally impaired the stability of DNA fibers, as shown by the decreased elongation rate upon IR, while having no effect on the ongoing replication process despite replicational stress, as demonstrated by virtually unchanged second pulse origins and stalled replication forks specifically in POLD1R689W/- cells.
Figure 3.
Functional characterization of the pre-existing POLD1 variants in DLD-1. (A) Representative results of constitutive protein levels (n = 3) in POLD1+/+, POLD1R689W/- and POLD1+/- cells as well as protein levels 120 h after siPOLD1 transfection in POLD1+/+ cells by immunoblotting. β-Actin served as loading control and original immunoblots are displayed in Figure S2. (B) Quantification of the elongation rate, (C) 2nd pulse origins and (D) stalled replication forks upon treatment of POLD1+/+ and POLD1R689W/- cells with IR or HU via DNA fiber assay. Error bars represent mean ± SEM of three independent experiments with at least 100 DNA fibers analyzed in each experiment. Using a two-tailed, unpaired Student’s t test, statistical significance could not be reached.
Mechanistic characterization of different POLD1 variants in DLD-1
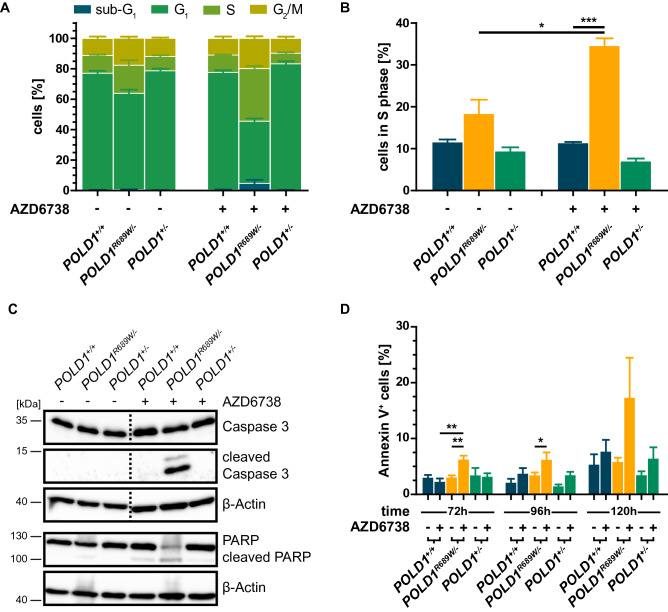
To analyze the molecular mechanism underlying R689W-mediated impairment of POLD1 function, we compared the cell cycle profiles of POLD1+/+, POLD1R689W/- and POLD1+/- cells upon treatment with the ATR inhibitor AZD6738 (Fig. 4A). Constitutively, POLD1R689W/- cells displayed a slight increase of the S phase fraction compared to POLD1+/+ and POLD1+/- cells (Fig. 4A + B). Treatment with the ATR inhibitor AZD6738 resulted in an additional, significant increase of the S phase fraction exclusively in POLD1R689W/- cells. Furthermore, we found an increase of the sub-G1 fraction exclusively in POLD1R689W/- cells upon AZD6738 treatment, indicating that not only cell cycle perturbations but also apoptosis appeared to contribute to the POLD1R689W-dependent effects of ATR inhibition on cell viability. Consistently, we observed cleavage of caspase 3, the main effector protease of apoptosis, and its substrate PARP nearly exclusively in POLD1R689W/- cells upon treatment with AZD6738 (Fig. 4C). To quantify the extent of apoptosis, we next applied annexin V-staining at 72 h, 96 h and 120 h after treatment with AZD6738 in POLD1+/+, POLD1R689W/- and POLD1+/- cells. We observed a significant increase of annexin V+ cells in AZD6738-treated POLD1R689W/- 72 h and 96 h post-treatment as compared to AZD6738-treated POLD1+/+ or untreated POLD1R689W/- cells, respectively, an effect which numerically increased strongly further (approximately threefold) but did not reach statistical significance at 120 h post-treatment (Fig. 4D). Thus, the detrimental effects of POLD1R689W observed upon treatment with the ATR inhibitor AZD6738 are mechanistically at least partially attributable to an S phase impairment and apoptosis.
Figure 4.
Mechanistic characterization of the pre-existing POLD1 variants in DLD-1. (A) Cell cycle profile (n = 3) of DLD-1 POLD1+/+, POLD1R689W/-, and POLD1+/- cells 96 h after treatment with 1 µM AZD6738 as assessed by flow cytometry. (B) S phase fraction of the cell cycle profile from (A) in detail. (C) Representative results of protein levels (n = 5) in POLD1+/+, POLD1R689W/-, and POLD1+/- cells by immunoblotting 72 h after treatment with 1 µM AZD6738. β-Actin served as loading control. Dashed lines indicate cropping, and the original immunoblots are displayed in Figure S3. (D) Quantification (n = 5) of annexin V+ apoptotic cells (including early apoptotic PI-/annexin V+ cells and late apoptotic PI+/annexin V+ cells) in POLD1+/+, POLD1R689W/-, and POLD1+/- cells at 72 h, 96 h and 120 h after treatment with 1 µM AZD6738 as assessed by flow cytometry. Statistically significant outliers were identified by Grubbs’ test and excluded. Error bars represent mean ± SEM, and asterisks (* p < 0.05, ** p < 0.01, *** p < 0.001) mark statistical significance using a one-tailed (D) or two-tailed (B), unpaired Student’s t test.
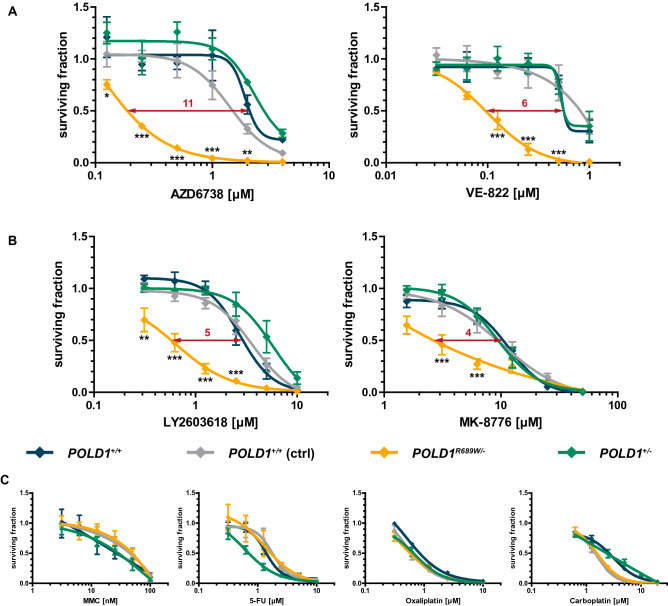
POLD1R689W-mediated sensitization of DLD-1 cells to ATR and CHK1 inhibitors in vitro
Having established a functional role of the POLD1R689W variant and its mechanistic impact on treatment with the ATR inhibitor AZD6738, we next tested, whether these effects were generalizable to other inhibitors targeting either ATR or its main effector kinase CHK1. Compared to parental and ctrl POLD1+/+ cells, only POLD1R689W/- but not POLD1+/- cells displayed an increased sensitivity not only to AZD6738, but also to another ATR inhibitor VE-8228,9 with IC50 ratios of 11 and 6, respectively (Fig. 5A). Similarly, only POLD1R689W/- but not POLD1+/- cells displayed an increased sensitivity to the CHK1 inhibitors LY2603618 and MK-877618–20 with IC50 ratios of 5 and 4, respectively (Fig. 5B). To exclude an unspecific drug hypersensitivity phenotype, the cells were additionally treated with common chemotherapeutics including mitomycin C (MMC), 5-fluorouracil (5-FU), carboplatin and oxaliplatin, none of which caused POLD1R689W-dependent inhibition of proliferation (Fig. 5C). Thus, only POLD1R689W but not POLD1G10V, POLD1R506H or POLD1S746I sensitizes DLD-1 cells specifically to ATR and CHK1 inhibitors, but not to common chemotherapeutics.
Figure 5.
POLD1R689W-mediated sensitization of DLD-1 cells to ATR and CHK1 inhibitors in vitro. (A) Effects of ATR inhibitors (n = 3), (B) CHK1 inhibitors (n = 4), and (C) common chemotherapeutics (n = 3 for MMC and carboplatin; n = 4 for 5-FU and oxaliplatin) on the proliferation of DLD-1 POLD1R689W/- cells as compared to POLD1+/- and POLD1+/+ parental and ctrl cells, measured 120 h after drug treatment. Data are presented as mean ± SEM and each data point reflects triplicate wells of multiple independently performed experiments. Asterisks (* p < 0.05, ** p < 0.01, *** p < 0.001) mark statistical significance using a two-way ANOVA with Bonferroni post-test.
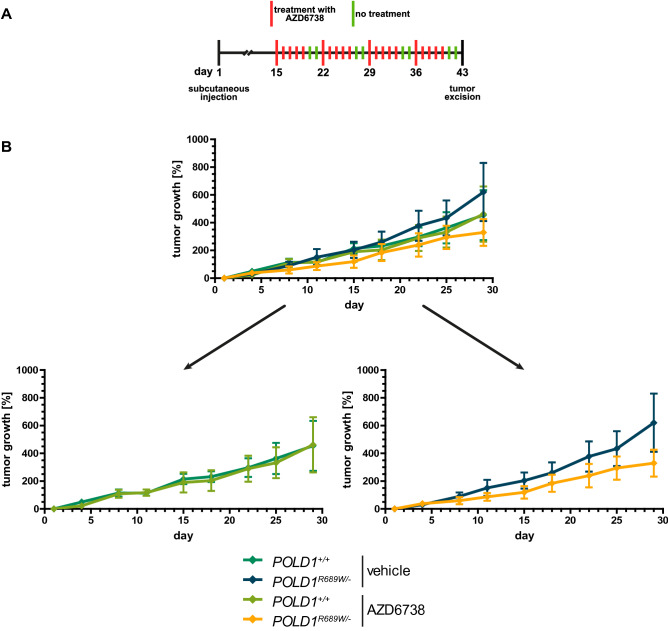
POLD1R689W-mediated sensitization of DLD-1 cells to the ATR inhibitor AZD6738 in vivo
To extend our in vitro data we next used a murine xenograft tumor model to assess the sensitivity of POLD1+/+ and POLD1R689W/- cells to the ATR inhibitor AZD6738 in vivo, as this inhibitor showed the strongest proliferation inhibitory effects in the in vitro experiments (Fig. 5A + B). Female nude mice were treated two weeks after subcutaneous injection of DLD-1 POLD1+/+ or POLD1R689W/- cells with the ATR inhibitor AZD6738 or vehicle once daily for five consecutive days, followed by two days without treatment, over a time period of four weeks (Fig. 6A). When comparing vehicle- and AZD6738-treated POLD1+/+ tumors, we observed virtually no discernible differences in tumor growth, with both quintupling their original tumor size during treatment (Fig. 6B, upper and lower left panel). In contrast, while the overall strongest tumor growth (> 600%) was observed in vehicle-treated POLD1R689W/- tumors, AZD6738-treated POLD1R689W/- tumors showed the slowest tumor growth (Fig. 6B, upper and lower right panel), although these data did not reach statistical significance. Nevertheless, these data at least support the hypothesis that AZD6738 decreases tumor growth specifically of POLD1R689W/- tumors in vivo. Taken together with our in vitro data, these results illustrate the general suitability of a cellular POLD1-KO model system to define POLD1 VUS as either pathogenic or non-pathogenic, as has similarly been shown for BRCA2 VUS21.
Figure 6.
POLD1R689W-mediated sensitization of DLD-1 cells to the ATR inhibitor AZD6738 in vivo. (A) Schematic representation of the experimental procedure. For clarity, the datasets illustrating vehicle- and AZD6738-treated POLD1+/+ tumors (B, lower left panel) as well as vehicle- and AZD6738-treated POLD1R689W/- tumors (B, lower right panel) are displayed separately. Error bars represent mean ± SEM of eight (vehicle-treated POLD1R689W/- tumors) or nine mice (vehicle-treated POLD1+/+ tumors, AZD6738-treated POLD1+/+ tumors, and AZD6738-treated POLD1R689W/- tumors), respectively.
Discussion
Synthetic lethality offers new approaches for a targeted and individualized tumor therapy with reduced side effects1,2. By screening an siRNA library, we previously identified POLD1, coding for the catalytic subunit of Polδ, to act synthetically lethal with ATR, a DNA damage sensing kinase10. Recently, variants in POLD1 have been identified in colorectal and other cancers14–16. Therefore, we established a POLD1-KO model system to determine the pathogenicity of individual POLD1 variants and their impact on the sensitivity to inhibitors of the ATR pathway in vitro and in vivo.
DLD-1, a human CRC cell line, harbors the four heterozygous POLD1 variants G10V, R506H, R689W and S746I17. We applied the CRISPR/Cas9 technique in order to separately disrupt each POLD1 allele followed by genome/mRNA sequencing to identify the allele-specific localization of the above variants, yielding KO clones expressing either POLD1G10V/R506H/S746I (termed POLD1+/-) or POLD1R689W (termed POLD1R689W/-), respectively. This approach provides a well-suited model system for the functional characterization of POLD1 VUS. Despite multiple targeting rounds, we were not able to establish a homozygous POLD1-/- clone, indicating POLD1 to be an essential gene in cancer cells. This hypothesis is further supported by a study of Uchimura et al.22, demonstrating in a murine KO model that POLD1 is essential during embryonic development.
In our system, only POLD1R689W but not POLD1G10V/R506H/S746I displayed functional significance with regard to compensatory activation of the ATR pathway, as shown by strongly increased CHK1 phosphorylation as a surrogate marker for DNA replication stress, exclusively in POLD1R689W/- cells. Transferred to a clinical setting, CHK1 phosphorylation could thus provide a predictive biomarker for increased sensitivity to ATR inhibitors7. Consistently, the S. cerevisiae analog of POLD1R689W (POLD1R696W) causes catastrophic genomic instability and a mutator phenotype in yeast23–25.
To assess the impact of POLD1R689W during DNA replication more accurately, we also applied DNA fiber assays. We observed a constitutively increased elongation rate in POLD1R689W/- cells, which could be attributable to a reduction of nucleotide selectivity caused by POLD1R689W and its mediated increase of nucleotide levels as has been demonstrated previously in yeast and also other human cancer cell lines23–25. In contrast, IR-treatment led to a decreased elongation rate in POLD1R689W/- cells. This could be explained by an increased mutational burden caused by additional induction of extrinsic DNA damage in cells that already harbor a mutator phenotype – predominantly GC→TA transversions and GC→AT transitions—due to POLD1R689W24,25, ultimately leading to irreversible instability of DNA fibers. Unexpectedly, POLD1R689W did not have an impact on the ongoing replication process, even upon treatment with HU, which inhibits the ribonucleotide reductase26 resulting in decreased levels of nucleotides and eventually a blocked replication process. This could be explained by the previously described POLD1R689W-induced increase of nucleotide levels24, consecutively antagonizing the effects of HU.
Investigating the role of cell cycle impairment or apoptosis as potential molecular mechanisms for the increased sensitivity of POLD1R689W/- cells to ATR/CHK1 inhibitors, we observed a constitutive slight increase of the S phase fraction in POLD1R689W/- cells, which significantly increased upon AZD6738 treatment. Perhaps even more importantly, AZD6738-treatment also induced apoptosis in POLD1R689W/- cells as indicated by the increased sub-G1 fraction in the cell cycle experiments, and consecutively verified by detection of cleaved caspase 3 and its substrate PARP, as well as by quantification of annexin V+ apoptotic cells. Interestingly, the observed S phase impairment along with caspase 3-mediated apoptosis in POLD1-deficient colorectal cancer cells upon AZD6738 treatment has similarly been demonstrated in ATM-dysfunctional gastric cancer cells upon AZD6738 treatment, supporting the known synthetically lethal relationship between ATR and ATM27.
We next assessed the impact of POLD1R689W on the cellular sensitivity to ATR pathway-inhibiting chemical agents in vitro, using clinically relevant inhibitors of ATR8,9 and CHK118–20. In fact, both ATR and CHK1 inhibitors decreased proliferation specifically of POLD1R689W/- but not POLD1+/- cells. Importantly, no significant proliferation differences were observable between these cells upon treatment with common chemotherapeutics, excluding a general drug hypersensitivity phenotype of POLD1R689W/- cells. As an extension to our in vitro data, we further tested the effects of the ATR inhibitor AZD6738 on POLD1R689W/- cells in a murine xenograft tumor model in vivo. Although the tumor growth rates did not differ in a statistically significant manner overall among the four groups (vehicle- versus AZD6738-treatment of POLD1+/+ versus POLD1R689W/- tumors), a discernible inhibition of tumor growth upon AZD6738 treatment was only observable in POLD1R689W/- tumors, while POLD1+/+ tumors displayed virtually identical tumor growth rates when compared to vehicle-treated animals. While statistically, these results do not allow conclusions beyond the recognition of a trend, the reasons for the weaker effects of our in vivo as compared to our in vitro data might simply be attributable to typical problems inherent to animal models such as route of administration, dosing or certain artifacts, and merit further studies in modified in vivo models. In addition, future studies should address the impact of other ATR and CHK1 inhibitors in vivo—including VE-822, LY2603618, and MY-8776—to allow a more accurate estimation of the potential clinical relevance of our data, and as a prerequisite prior to the establishment of clinical trials.
In addition, we compared our functional data defining the significance of the investigated POLD1 variants with predictive data from various pathogenicity tools, including PON-P2, a machine learning-based classified, which groups variants into pathogenic, neutral, or unknown classes, based on random forest probability score28,29; PolyPhen-2, calculating the naïve Bayes posterior probability of a variant’s impact on protein function to be possibly/probably damaging or benign (non-damaging)30,31; PROVEAN, which uses an alignment-based score to discriminate between deleterious and neutral effects on protein function32,33; and MutationAssessor, calculating a functional impact score by using evolutionary conservation patterns34,35. According to these prediction tools, POLD1G10V and POLD1S746I variants have no or only low functional impact (Table 1), which is in concordance with our data on these variants. In contrast, POLD1R506H and POLD1R689W are both predicted by PON-P2 and PROVEAN to be pathogenic, while they are differently classified by PolyPhen-2 and MutationAssessor regarding their damaging potential and functional impact score, respectively, with POLD1R689W achieving a stronger functional impairment. However, despite the predicted functional significance and pathogenicity of POLD1R506H, we demonstrated that in our model only POLD1R689W had a functional impact in regard to CHK1 phosphorylation, DNA replication, cell cycle impairment, apoptosis induction, and cellular sensitivity to inhibitors of the ATR pathway. Besides the obvious interpretation, i.e. the false classification of these VUS by either the prediction tools or our assays, these inconsistencies could also be explained by the fact that we did not analyze the POLD1R506H variant alone, but only in combination with the variants POLD1G10V and POLD1S746I, which could cause antagonizing or protective effects in terms of “revertant” variants, a phenomenon previously described for BRCA2 in a similar clinical context36,37. This hypothesis should be tested in future studies through the individual introduction of single POLD1 variants (including the POLD1R506H variant), and consecutive characterization of their functional relevance in colorectal cancer cells. Nevertheless, the combination of algorithm-based, theoretical prediction tools to identify pathogenic variants along with functional testing in suitable KO/Knock-in models likely represents the most accurate approach currently available to estimate pathogenicity and define clinical-therapeutic exploitability of specific VUS in tumors.
Table 1.
Pathogenicity for POLD1 variants according to different prediction tools with investigated variants are in bold.
| Prediction tool | |||||
|---|---|---|---|---|---|
| Variant | References | PON-P2 | PolyPhen-2 | PROVEAN | Mutation assessor |
|
p.G10V c.29G > T |
17 | Unknown (0.398) | Benign (0.041) | Neutral (− 1.407) | Neutral (0.55) |
|
p.V295M c.883G > A |
16 | Unknown (0.703) | Benign (0.410) | Neutral (0.412) | Neutral (0.09) |
|
p.D316H c.946G > C |
16 | Pathogenic (0.941) | Probably damaging (1.000) | Deleterious (− 6.857) | High (3.69) |
|
p.D316G c.947A > G |
16 | Pathogenic (0.919) | Probably damaging (1.000) | Deleterious (− 6.857) | High (3.69) |
|
p.P327L c.981C > G |
14 | Pathogenic (0.947) | Probably damaging (0.999) | Deleterious (− 9.824) | High (3.74) |
|
p.S370R c.1110C > G |
14 | Unknown (0.560) | Benign (0.407) | Deleterious (− 4.259) | Medium (3.115) |
|
p.R409W c.1225C > T |
16 | Pathogenic (0.963) | Probably damaging (1.000) | Deleterious (− 7.859) | High (3.74) |
|
p.G426S c.1276G > A |
14 | Unknown (0.237) | Benign (0.042) | Neutral (− 0.579) | Neutral (− 0.485) |
|
p.L474P c.1421T > C |
15,16 | Pathogenic (0.937) | Probably damaging (1.000) | Deleterious (− 6.735) | High (3.74) |
|
p.S478N c.1433G > A |
14–16 | Pathogenic (0.759) | Probably damaging (0.998) | Deleterious (− 2.820) | High (4.215) |
|
p.R506H c.1517G > A |
17 | Pathogenic (0.955) | Possibly damaging (0.755) | Deleterious (− 4.922) | Medium (2.86) |
|
p.R521Q c.1562G > A |
16 | Pathogenic (0.957) | Possibly damaging (0.816) | Deleterious (− 3.464) | Medium (3.255) |
|
p.R689W c.2065C > T |
17 | Pathogenic (0.973) | Probably damaging (0.987) | Deleterious (− 7.978) | High (4.56) |
|
p.S746I c.2237G > T |
17 | Unknown (0.475) | Benign (0.143) | Deleterious (− 3.178) | Medium (2.92) |
Multiple heterozygous germline variants in POLD1 have been identified in various cancer types, including colon, endometrium, breast, or brain tumors14–16. Most of those variants that are predicted to be functionally significant (Table 1) are located in the proofreading exonuclease domain of POLD1, presumably impairing the repair capacity of Polδ, and thereby conferring a mutator phenotype24. In contrast, the role of variants in the polymerase active domain of POLD1 is less well understood. In addition to POLD1R689W24,25 only one further variant within the polymerase active domain, namely POLD1L604K of murine Polδ, has been described to be potentially pathogenic38. Yet, there is growing evidence that base selectivity defects in tumors—as induced by POLD1R689W—might result in dramatic consequences for genome stability24. Our data now add another example that variants not only within the proofreading exonuclease domain but also in the polymerase active domain of POLD1, e.g. R689W, are functionally relevant and could thus confer a pathogenic phenotype.
In summary, we used CRISPR/Cas9 to establish a POLD1-KO model for the functional characterization of POLD1 VUS. This model could complement and improve the accuracy of algorithm-based theoretical prediction models of POLD1 VUS pathogenicity. In this model, we demonstrated and mechanistically characterized the impact of the POLD1R689W variant on the cellular sensitivity to ATR and CHK1 inhibitors in vitro, confirming the previously described synthetically lethal relationship between ATR and POLD110. In addition, we demonstrated in a murine in vivo model that treatment with the ATR inhibitor AZD6738 caused a discernible inhibition of tumor growth rate specifically in tumors harboring the POLD1R689W variant. Taken together, our study defines the POLD1R689W variant as target for ATR pathway inhibition and thereby illustrates the emerging role of tumor-specific alterations in polymerases39–43 along with the need of their comprehensive identification and functional characterization in suitable model systems to aid the development of novel and rational approaches towards improved tumor therapy44.
Methods
Cell lines and culture conditions
The human CRC cell line DLD-1 was purchased from the Leibniz Institut DSMZ (Braunschweig, Germany). All cell lines and clones were maintained in Roswell Park Memorial Institute (RPMI 1640) medium supplemented with 10% fetal bovine serum (FBS) and incubated at 37 °C and 5% CO2.
Drugs
AZD6738 and VE-822 were purchased from MedKoo Biosciences (Morrisville, NC, USA), MK-8776 and LY2603618 from Selleckchem (Munich, Germany), and MMC and 5-FU from Sigma-Aldrich (Hamburg, Germany). Oxaliplatin and carboplatin were kindly donated from the cytostatic drug department of the University Hospital Marburg. AZD6738, used in the xenograft in vivo assay, was purchased from AdooQ Bioscience (Irvine, CA, USA).
Transfection
Reverse transfection was used for the transfection experiments. siRNA targeting POLD1 (CGGGACCAGGGAGAATTAATA) (Qiagen, Hilden, Germany) at a final concentration of 10 nM or 1 μg plasmid DNA, respectively, was incubated with HiPerFect from Qiagen in RPMI 1640 medium free of FBS for 20 min at room temperature and then added to freshly seeded cells.
POLD1 knockout with CRISPR/Cas9
The CRISPR POLD1 Knockout Kit was purchased from OriGene (Rockville, MD, USA), containing two pCas9 plasmids (targeted sequence of gRNA1 TGCCCCCAAAGCGGGCCCGT and gRNA2 GGGATGATGATGATGCACCT, respectively), one pCas9 plasmid with a scrambled sequence as ctrl and a repair template with a left homology arm (LHA) and right homology arm (RHA) of POLD1 together with a GFP-Puromycin functional cassette (Figure S4). DLD-1 cells were reverse co-transfected using 1 μg of the repair template together with 1 μg of one of the three pCas9-Guide vectors. 72 h later the transfection complex containing medium was replaced with puromycin containing medium for selection of stably transfected cells. Selected cells were allowed to grow in single-cell colonies and genotyped, using the primers #1–#4 (Table S1). Heterozygous clones show two PCR products, one representing the KO allele (approx. 1000 bp) and the other one representing the intact wild type (WT) allele (1656 bp). To engineer homozygous KO clones, heterozygous clones were first transfected with a Cre vector to remove the puromycin resistance gene. Single cell colonies with removed puromycin resistance gene were verified via PCR using the primers #5 and #6. Single-cell colonies with no puromycin resistance gene were then repeatedly transfected as described, applying three independent targeting rounds with at least 80 genotyped single-cell colonies during each targeting round.
PCR and sequencing
DNA and RNA were isolated using the Mini Kits QIAamp and RNeasy from QIAGEN, respectively. RNA to complementary DNA transcription was performed using the Omniscript RT Kit from Qiagen. For the PCR reactions Promega’s GoTaq G2 DNA Polymerase was used (Madison, WI, USA). Primers are listed in Table S1 and PCR conditions are available on request. Sequencing was performed by GATC Biotech AG (Constance, Germany), samples were prepared as asked by the provider. Primers used for sequencing are listed in Table S2.
Immunoblotting
Immunoblotting was performed as described previously39. In short, cells were lysed and protein extracts were boiled and loaded on 10% polyacrylamide gels. After electrophoretic separation, the proteins were transferred to PVDF membranes, which were blocked with 5% milk powder in Tris-buffered saline + 0.1% Tween 20 (TBS-T) for 1 h. Incubation of primary antibody in TBS-T was performed at 4 °C overnight. Membranes were then washed and stained with secondary antibody. Chemiluminescence was elicited using Western Lightning Ultra from PerkinElmer (Waltham, MA, USA) or Clarity Western ECL Substrate from Bio-Rad Laboratories (Hercules, CA, USA), respectively, according to the manufacturers’ instructions. The following primary antibodies were used: anti-caspase 3, anti-cleaved caspase 3 (Asp175), anti-PARP, and anti-pCHK1(Ser345) (133D3) from Cell Signaling (Cambridge, UK), anti-CHK1 (G-4) and anti-POLD1 (A9) from Santa Cruz Biotechnology (Dallas, TX, USA), and peroxidase-conjugated anti-β-Actin (AC-15) from Sigma-Aldrich (Hamburg, Germany). HRP-conjugated anti-rabbit, anti-goat and anti-mouse antibodies from Santa Cruz Biotechnology were used as secondary antibodies.
DNA fiber assay
Exponentially growing cells were pulse labeled with 25 μM chlorodeoxyuridine (CldU) followed by 250 μM iododeoxyuridine (IdU), both from Sigma-Aldrich (Hamburg, Germany), for 30 min each. For analysis of fork stability cells were treated with 2 mM HU for 4 h or irradiated with 6 Gy between both labels. Labeled cells were harvested and DNA fiber spreads prepared and stained as described45–47. Fibers were examined using an Axioplan 2 fluorescence microscope from Carl Zeiss (Jena, Germany). CldU and IdU tracks were measured using ImageJ and micrometer values were converted into kilobases. At least 100 forks were analyzed. Different classes of labeled tracks were classified: red-green (ongoing replication), red (stalled forks) and green (2nd pulse origin). Labeled tracks were counted using ImageJ and at least three independent experiments were performed.
Analysis of cell cycle and apoptosis
Cells were seeded and allowed to adhere overnight before treated with 1 µM AZD6738. After 96 h of treatment, cells were collected, washed, and stained with propidium iodide (PI) (0.1% sodium citrate, 0.1% Triton X-100, and 50 µg/ml PI) as described previously48. For analysis of apoptosis, cells were collected at various time points after treatment with 1 µM AZD6738, washed with cold Hank’s Balanced Salt Solution, and stained for 20 min in the dark at room temperature with 25 µl/ml of a FITC-conjugated annexin V-antibody from Biolegend (San Diego, CA, USA). Directly before flow cytometric measurement, 10 µg/ml PI was added to the staining solution. Cell cycle distribution and apoptotic cells were quantified by using the BD FACSCanto II from BD Biosciences (San Jose, CA) and the FlowJo v10 software from FlowJo, LLC (Ashland, OR). At least 20,000 gated events per sample were analyzed.
Cell proliferation assays
Cell proliferation assays were performed over a broad range of concentrations covering 100% to 0% cell survival. 1500–1800 cells of the DLD-1 POLD1+/+ parental and ctrl clones as well as of the POLD1R689W/- and POLD1+/- clones were plated in 96 well plates and allowed to adhere overnight, before being treated with various drugs at multiple concentrations for 120 h. Following incubation, the cells were washed, lysed in 100 μl H2O and 0.2% SYBRGreen (Lonza, Cologne, Germany) was added. Fluorescence was measured using a Victor3 V plate reader (PerkinElmer, Waltham, MA, USA) and growth inhibition was calculated as compared to the untreated control samples.
Xenograft model
Xenograft tumors were induced in 6 week old female nude NMRI-Foxn1nu/nu mice from Charles River Laboratories (Wilmington, MA, USA) by subcutaneously injecting 106 POLD1+/+ or POLD1R689W/- DLD-1 cells, respectively, suspended in phosphate buffered saline. Two weeks after injection, mice were randomly divided into two groups for each cell clone and the ATR inhibitor AZD6738 at a dose of 50 mg/kg or the vehicle solution alone were administered by gavage once daily for five consecutive days followed by two days of no administration. This application schedule was conducted for four weeks (Fig. 6A). Solid tumors were measured twice a week with calipers and the calculated tumor volume () on day one of the application schedule was set as reference to calculate the percentage change of tumor volume. Additionally, mice were weighed before every inhibitor/vehicle application. AZD6738 was diluted in 100% dimethyl sulfoxide (DMSO) and the vehicle solutions consisted of 10% DMSO (with or without AZD6738), 40% 1,3-propanediol and 50% H2O.
All animal experiments were performed according to the guidelines of the German law for animal life protection and approved by the commission for animal protection of the veterinary office of the regional commission of Gießen with the file numbers G44/2017 and G89/2018.
Statistical analysis
All statistical analyses were performed using Prism 5 from GraphPad Software Inc. (La Jolla, CA, USA). Surviving fractions of the proliferation assays were calculated by curve fitting with nonlinear regression. Using Grubbs’ test statistically significant outliers were determined and not included. Data are presented as mean ± SEM, and a one-tailed or two-tailed, unpaired Student’s t-test, or a two-way ANOVA with Bonferroni post-test were used for statistical interpretation. P-values of p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***) were considered statistically significant.
Supplementary information
Acknowledgements
We thank Bettina Geisel for expert technical assistance as well as Madeleine Thamm and Julia Kirch for supporting the in vivo experiments. This work was funded in part by the Deutsche Forschungsgemeinschaft (DFG, German Resarch Foundation) – Project No 391498046 (Clinical Research Unit 325, subprojects A1 and A2).
Author contributions
A.J. designed, performed and analyzed the experiments, drafted the manuscript and wrote the animal test request. M.T. helped writing the animal test request and performing the animal experiments. C.S. helped establishing heterozygous knockout cell clones. V.L., H.S. and B.L.-B. performed some experiments. A.Z. and K.B. performed the DNA fiber assays. C.B., T.M.G. and M.B. provided important intellectual content. E.G. designed, analyzed and supervised the study, and wrote the final manuscript. These data are part of A.J.’s doctoral thesis. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-76033-1.
references
- 1.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 2.Nijman SMB. Synthetic lethality: General principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 4.Hartman JL., IV Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 5.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minchom, A., Aversa, C. & Lopez, J. Dancing with the DNA damage response: Next-generation anti-cancer therapeutic strategies. Ther. Adv. Med. Oncol.10 (2018). [DOI] [PMC free article] [PubMed]
- 7.Lecona E, Fernandez-Capetillo O. Targeting ATR in cancer. Nat. Rev. Cancer. 2018;18:586–595. doi: 10.1038/s41568-018-0034-3. [DOI] [PubMed] [Google Scholar]
- 8.Pihlak R, Valle JW, McNamara MG. Germline mutations in pancreatic cancer and potential new therapeutic options. Oncotarget. 2017;8:73240–73257. doi: 10.18632/oncotarget.17291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnitz LM, Zou L. Molecular pathways: Targeting ATR in cancer therapy. Clin. Cancer Res. 2015;21:4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hocke S, et al. A synthetic lethal screen identifies ATR-inhibition as a novel therapeutic approach for POLD1-deficient cancers. Oncotarget. 2016;7:7080–7095. doi: 10.18632/oncotarget.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohni KN, et al. A synthetic lethal screen identifies DNA repair pathways that sensitize cancer cells to combined ATR inhibition and cisplatin treatments. PLoS ONE. 2015;10:e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garg P, Burgers PMJ. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas E, Golemis EA, Arora S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene. 2016;590:128–141. doi: 10.1016/j.gene.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valle L, et al. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum. Mol. Genet. 2014;23:3506–3512. doi: 10.1093/hmg/ddu058. [DOI] [PubMed] [Google Scholar]
- 16.Bellido F, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: Review of reported cases and recommendations for genetic testing and surveillance. Genet. Med. 2016;18:325–332. doi: 10.1038/gim.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flohr T, et al. Detection of mutations in the DNA polymerase delta gene of human sporadic colorectal cancers and colon cancer cell lines. Int. J. Cancer. 1999;80:919–929. doi: 10.1002/(SICI)1097-0215(19990315)80:6<919::AID-IJC19>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Scagliotti G, et al. Phase II evaluation of LY2603618, a first-generation CHK1 inhibitor, in combination with pemetrexed in patients with advanced or metastatic non-small cell lung cancer. Invest. New Drugs. 2016;34:625–635. doi: 10.1007/s10637-016-0368-1. [DOI] [PubMed] [Google Scholar]
- 19.Daud AI, et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J. Clin. Oncol. 2015;33:1060–1066. doi: 10.1200/JCO.2014.57.5027. [DOI] [PubMed] [Google Scholar]
- 20.Webster JA, et al. Randomized phase II trial of cytosine arabinoside with and without the CHK1 inhibitor MK-8776 in relapsed and refractory acute myeloid leukemia. Leuk. Res. 2017;61:108–116. doi: 10.1016/j.leukres.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hucl T, et al. A syngeneic variance library for functional annotation of human variation: Application to BRCA2. Cancer Res. 2008;68:5023–5030. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchimura A, Hidaka Y, Hirabayashi T, Hirabayashi M, Yagi T. DNA polymerase δ is required for early mammalian embryogenesis. PLoS ONE. 2009;4:e4184. doi: 10.1371/journal.pone.0004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase variant modeled in yeast causes a catastrophic increase in genomic instability. Proc. Natl. Acad. Sci. 2010;107:157–162. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertz TM, Sharma S, Chabes A, Shcherbakova PV. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. 2015;112:E2467–E2476. doi: 10.1073/pnas.1422934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mertz TM, Baranovskiy AG, Wang J, Tahirov TH, Shcherbakova PV. Nucleotide selectivity defect and mutator phenotype conferred by a colon cancer-associated DNA polymerase δ mutation in human cells. Oncogene. 2017;36:4427–4433. doi: 10.1038/onc.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarbro JW. Mechanism of action of hydroxyurea. Semin. Oncol. 1992;19:1–10. [PubMed] [Google Scholar]
- 27.Min A, et al. AZD6738, a novel oral inhibitor of ATR, induces synthetic lethality with ATM deficiency in gastric cancer cells. Mol. Cancer Ther. 2017;16:566–577. doi: 10.1158/1535-7163.MCT-16-0378. [DOI] [PubMed] [Google Scholar]
- 28.Niroula A, Urolagin S, Vihinen M. PON-P2: Prediction method for fast and reliable identification of harmful variants. PLoS ONE. 2015;10:e0117380. doi: 10.1371/journal.pone.0117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.PON-P2. https://structure.bmc.lu.se/PON-P2/. Accessed: 27 Nov 2018.
- 30.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.PolyPhen-2. https://genetics.bwh.harvard.edu/pph2/. Accessed: 27 Nov 2018.
- 32.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.PROVEAN Protein. https://provean.jcvi.org/seq_submit.php. Accessed: 27 Nov 2018.
- 34.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118–e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MutationAssessor. https://mutationassessor.org/r3/. Accessed: 27th Nov 2018.
- 36.Barber LJ, et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J. Pathol. 2013;229:422–429. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- 37.Edwards SL, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 38.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase δ increases genomic instability and accelerates tumorigenesis. Mol. Cell. Biol. 2007;27:7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Job A, et al. Inactivation of PRIM1 function sensitizes cancer cells to ATR and CHK1 inhibitors. Neoplasia. 2018;20:1135–1143. doi: 10.1016/j.neo.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spier I, et al. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int. J. Cancer. 2015;137:320–331. doi: 10.1002/ijc.29396. [DOI] [PubMed] [Google Scholar]
- 41.Elsayed FA, et al. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur. J. Hum. Genet. 2015;23:1080–1084. doi: 10.1038/ejhg.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silvestri R, Landi S. DNA polymerases in the risk and prognosis of colorectal and pancreatic cancers. Mutagenesis. 2019 doi: 10.1093/mutage/gez031. [DOI] [PubMed] [Google Scholar]
- 43.Rogers RF, et al. CHK1 inhibition is synthetically lethal with loss of B-family DNA polymerase function in human lung and colorectal cancer cells. Cancer Res. 2020;80:1735–1747. doi: 10.1158/0008-5472.CAN-19-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bøe CA, Håland TW, Boye E, Syljuåsen RG, Grallert B. A novel role for ATR/Rad3 in G1 phase. Sci. Rep. 2018;8:6880. doi: 10.1038/s41598-018-25238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parplys AC, Petermann E, Petersen C, Dikomey E, Borgmann K. DNA damage by X-rays and their impact on replication processes. Radiother. Oncol. 2012;102:466–471. doi: 10.1016/j.radonc.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Nikkilä J, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat. Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wurster S, et al. PARP1 inhibition radiosensitizes HNSCC cells deficient in homologous recombination by disabling the DNA replication fork elongation response. Oncotarget. 2016;7:9732–9741. doi: 10.18632/oncotarget.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.