Abstract
It has been recently shown that many proteins are lacking from reference databases used in mass spectrometry analysis, due to their translation templated on alternative open reading frames. This questions our current understanding of gene annotation and drastically expands the theoretical proteome complexity. The functions of these alternative proteins (AltProts) still remain largely unknown. We have developed a large-scale and unsupervised approach based on cross-linking mass spectrometry (XL-MS) followed by shotgun proteomics to gather information on the functional role of AltProts by mapping them back into known signalling pathways through the identification of their reference protein (RefProt) interactors. We have identified and profiled AltProts in a cancer cell reprogramming system: NCH82 human glioma cells after 0, 16, 24 and 48 h Forskolin stimulation. Forskolin is a protein kinase A activator inducing cell differentiation and epithelial–mesenchymal transition. Our data show that AltMAP2, AltTRNAU1AP and AltEPHA5 interactions with tropomyosin 4 are downregulated under Forskolin treatment. In a wider perspective, Gene Ontology and pathway enrichment analysis (STRING) revealed that RefProts associated with AltProts are enriched in cellular mobility and transfer RNA regulation. This study strongly suggests novel roles of AltProts in multiple essential cellular functions and supports the importance of considering them in future biological studies.
INTRODUCTION
It is accepted that the eukaryotic mature messenger ribonucleic acids (mRNAs) are monocistronic, leading to the translation of a single protein product. According to the rules described by Kozak (1), the coding DNA sequence (CDS) region is the longest nucleotide sequence flanked on each side by a START and a STOP codon. The CDS defines the reference open reading frame (RefORF) that is translated into a protein. Nevertheless, the proteome has been shown to be more complex than initially expected. Indeed, >10% of proteomic data remain unmatched by interrogation of reference protein (RefProt) databases; however, the quality of the tandem mass spectrometry (MS/MS) data is sufficient (number of characteristic fragment ions) to lead to protein identification. These unmatched data were used to demonstrate that proteins encoded by alternative ORFs (AltORFs) can be translated from the mRNA, in addition to the predicted protein encoded by the RefORF (2). These AltORFs are translated to so-called alternative proteins (AltProts) that altogether are considered as a hidden or ghost proteome. AltProts can be issued from different parts of mRNAs, including 5′UTR, 3′UTR, overlapping regions between 5′UTR and CDS or CDS and 3′UTR, and +2 and +3 frameshifts in the CDS. Therefore, they are separated into three groups: AltORF5′UTR, AltORFCDS and AltORF3′UTR. As supported by different studies and given the more permissive AltORF annotation rules, it is expected that AltProts outnumber RefProts in terms of diversity (2,3). Moreover, high-throughput genome and transcriptome sequencing have led to validate an important number of small ORFs (sORFs) containing <100 codons that were previously arbitrary considered as non-coding. The translation of these sORFs into peptides or small proteins (microproteins, SEPs) was demonstrated by different strategies such as ribosome profiling (4,5) and peptidomics combined to massively parallel RNA-seq (6). Since protein databases used in large-scale proteomic approaches [e.g. UniProt (7) or NCBI (8)] are built on genomic annotations, AltProts have been missing and hence could not be identified through MS-based proteomics. Considering these accumulating evidences, the genome annotation and protein translation dogma had to be reconsidered, and the development of new databases including the AltProts was then a mandatory step to their identification in large-scale proteomic studies. Over the past years, several initiatives have led to predict and annotate AltProt sequences. Recently, the OpenProt database has been publicly released and can be used to identify AltProts in large-scale proteomic datasets (9). In silico studies using these AltProt databases show that the average AltProt size, and thus molecular weight, is lower than their reference counterpart with ∼57 versus ∼344 amino acid length for the AltProts and the RefProts, respectively (2,10). Before the advent of these new databases, a few AltProts have been nonetheless reported; however, they have been considered as an epiphenomenon despite showing central biological functions (11). The implementation of AltProt database interrogation in large-scale proteomic experiments has opened the way to their massive identification moving forward with the characterization of their physiological and physiopathological role [e.g. ovarian cancer (12), brain physiome (13) and viral infection (14)]. Not only AltProts are believed to have a central role in signalling pathways, but it was also demonstrated that some of them (e.g. AltMRVI1) were showing higher expression than the RefProts though with shorter half-lives (10).
As of today, numerous AltProts have been identified but very few studies have been focusing on their functions, which hence remain largely unknown. Recently, it has been described that the ‘Humanin’ AltProt could be involved in Asian population longevity (15), while other AltProts would play a role in the metabolism regulation (16). The acquired data and knowledge on AltProts support their involvement in the regulation of protein expression, though this assumption must be confirmed. However, investigating AltProt functions remains challenging. Since there is no commercial antibody raised against AltProts available yet, most conventional strategies such as co-immunoprecipitation cannot be used. Moreover, a candidate-by-candidate targeted strategy is very time consuming and cannot depict the AltProts’ global organization at a cellular scale. In that sense, large-scale and unsupervised approaches should be preferred. In particular, using an unsupervised approach to identify the AltProt interaction partners and assigning them in known protein networks and signalling pathways is one approach to get insights into their functions.
Over the past decade, various methods have been developed to detect protein–protein interactions (PPIs). MS-based proteomic strategies are particularly well suited for measuring PPIs at a larger scale from a limited amount of complex mixture (17). This includes affinity purification (18) and tandem affinity purification (19,20), proximity labelling (21) such as APEX (22) and BioID (23–25), viral particle sorting approach (Virotrap) (26) and cross-linking mass spectrometry (XL-MS) (27,28). XL-MS is based on the formation of covalent bonds using a chemical linker of a defined length to freeze the interaction between partners. In the XL-MS strategy, the sample is first submitted to cross-linking, which is followed by enzymatic digestion and subsequent liquid chromatography–mass spectrometry (LC–MS) analysis. The MS/MS data gathered during the LC–MS run are then used to identify the interacting peptides and deduce the PPIs. Both intra- and interprotein cross-links are observed depending on the protein conformation, its interaction and the length of the cross-linker. Very advantageously, XL-MS not only provides knowledge on the interaction partners but can also be used to predict the structure of the protein or the protein complex, most often in combination with other modalities such as molecular modelling, X-ray crystallography, nuclear magnetic resonance spectroscopy and cryo-electron microscopy (cryo-EM). The latter provide higher structural resolution than XL-MS but the analyses are performed from large amounts of highly purified proteins or complexes not in their native environment and only representing a static snapshot of a protein without consideration for the cell dynamic. Therefore, despite lower structural resolution, XL-MS appears as a straightforward strategy with a wide range of applications to provide structural information on protein complexes as they exist within a biological system. For a decade, XL-MS development has faced several challenges. The first caveat sits in the difficulty of interpreting MS/MS data generated by two cross-linked peptides. This can be overcome by using CID cleavable cross-linkers such as the commercialized disuccinimidyl sulfoxide (DSSO) (29) and disuccinimidyl dibutyric urea (DSBU) (30). Another issue derives from the low abundance of cross-linked versus non-cross-linked peptides that limits their detection during the LC–MS analysis. To get over this hurdle, various methodologies have been proposed to enrich cross-linked peptides, including size exclusion chromatography, ion exchange using either cartridges or columns (31) and affinity purification for tagged cross-linkers (32–34). At last, data analysis is far more complex in XL-MS experiments than in conventional shotgun proteomics and requires dedicated software and platforms (35–40). So far, finding PPIs from all identified proteins versus the entire protein database is still not possible due to limited computational power in conventional labs. However, searching for candidates against the database is feasible. Another important aspect to be careful about the data analysis is the possible misassignments (41). Some of the most advanced tools for XL-MS data analysis such as XlinkX computing node (42), which can be included within the usual proteomic identification pipeline of Proteome Discoverer 2.2 (PD2.2), are designed to avoid misassignments and include a score of false discovery rate (FDR) for the cross-link identification. With all this progress, robust XL-MS workflows have emerged and are routinely in use by different groups (43–46). Very interestingly, it has been shown that XL-MS could be performed in cellulo if picking up a cross-linker diffusing through cell or cell compartment membranes (47,48). Because XL-MS is truly unsupervised, unlike other MS-based strategies that at least require the knowledge of a bait candidate, it is a well-suited strategy in investigating the role of AltProts. We recently re-analysed HeLa cell data (49) to demonstrate the importance of AltProts in multiple cellular compartments and processes. This analysis has confirmed the potential of XL-MS strategies to get new insights into AltProt functions (49).
In this work, we applied XL-MS to study the role of AltProts during cancer cell reprogramming. To address this question, we stimulated glioma cells with Forskolin that activates the enzyme adenylate cyclase and increases the intracellular level of cyclic AMP (cAMP). Among the cAMP effectors, the role of protein kinase A (PKA) in the regulation of multiple cellular processes has been highlighted in several studies. Thus, PKA and downstream regulators are involved in promoting cell differentiation by the activation of epithelial–mesenchymal transition or conversely the mesenchymal–epithelial transition (50,51). In glioblastoma tumours, the maintenance of the cancer cell differentiation status is dependent on cAMP/PKA signalling (51,52). Combined shotgun proteomics and XL-MS were used to identify AltProts, their interaction partners and their signalling pathways. The XL-MS workflow was optimized from NCH82 human grade IV glioma cells to enable XL-MS to be performed on nuclear and cytoplasmic fractions. The resulting network was then enriched based on the interactions already described in the literature. By comparing these networks and their combinations, functions can then be suggested for the identified AltProts as a result of the biological process in which they are involved. Here, we demonstrate for the first time the AltProt regulation in response to PKA-induced phenotypic modifications.
MATERIALS AND METHODS
Chemicals and materials
DSSO, Dulbecco’s modified Eagle’s medium (DMEM), foetal bovine serum (FBS), l-glutamine, penicillin, streptomycin and phosphate-buffered saline (PBS) were obtained from Thermo Fisher Scientific (Les Ulis, France). Formic acid (FA), HPLC grade water, trifluoroacetic acid (TFA), acetonitrile (ACN), methanol, ethanol, acetone and trichloroacetic acid were all purchased from Biosolve (Dieuze, France). dl-Dithiothreitol (DTT), iodoacetamide (IAA), chloroform, dimethyl sulfoxide (DMSO), ammonium bicarbonate (AB), 4-(2-hydroxyethyl)piperazine-1-ethane sulfonic acid, N-(2-hydroxyethyl)piperazine-N-(2-ethane sulfonic acid) (HEPES), sodium chloride (NaCl) and magnesium chloride (MgCl2) were obtained from Sigma-Aldrich. Tris was purchased from Bio-Rad (Steenvoorde, France). Extraction Illustra triplePrep Kit was from GE Healthcare. LysC/trypsin was obtained from Promega (Charbonnières-les-Bains, France). Amicon centrifugal filters and C18 ZipTip pipette tips were from Merck Millipore (Merck KGaA, Darmstadt, Germany).
Cell culture
NCH82 and U87-MG human glioma cell lines were grown as monolayers up to 80–90% confluency in complete media [high-glucose DMEM supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM l-glutamine (Sigma-Aldrich)], before harvesting and passing in a new flask. The cells were kept at 37°C in humidified air containing 5% CO2. NCH82 cells <500 000 for the time course and over 1 million for the total analysis (1 × 106 per T75 flask) were stimulated in DMEM complete media for 16, 24 or 48 h supplemented with 50 μM Forskolin (BIOTREND Chemicals AG), a cell-permeable activator of adenylyl cyclase that leads to an increase in intracellular concentration of cAMP and, consequently, to PKA stimulation. Forskolin was prepared as 50 mM stock solution in DMSO.
Shotgun proteomics
For whole cell analysis, protein extraction was carried out using the Illustra triplePrep Kit (GE Healthcare 28-94259-44) to separate DNA, RNA and proteins. The isolated protein fraction was kept for large-scale shotgun proteomics. The protein extraction, reduction/alkylation and enzymatic digestion were performed using the FASP method (53). Briefly, the sample was in 30 μl of 8 M urea in 0.1 M Tris/HCl, pH 8.5 (UA buffer), and an equivalent volume of 100 mM in UA DTT. The sample was then incubated for 40 min at 56°C. Total proteins were loaded onto 10 kDa Amicon filters, supplemented with 200 μl of UA buffer and centrifuged for 15 min at 14 000 × g. Then, 100 μl of 0.05 M IAA in AU was added and incubated for 20 min in the dark before centrifugation for 15 min at 14 000 × g. Finally, a 0.05 M ammonium bicarbonate solution in water (AB) was added and centrifuged for 15 min at 14 000 × g twice. For the digestion, 50 μl LysC/trypsin at 20 μg/ml in AB buffer was added and incubated at 37°C overnight. The digested peptides were then recovered after centrifugation for 15 min at 14 000 × g after transferring the filter into new tubes, reconstitution in 50 μl of AB buffer, followed by a second centrifugation step for 15 min at 14 000 × g. The eluted peptides were then acidified with 10 μl of 0.1% TFA and vacuum dried.
Cellular cross-linking
The separation between cytoplasm and nuclei was carried out according to the method described by Liu et al. (48). Briefly, the pellet of 2 million cells was recovered in 100 μl of stabilizing buffer (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.4% NP-40, pH 7.8) containing protease inhibitors (AEBSF 2 mM, phosphoramidon 1 μM, bestatin 130 μM, 14 μM E-64, 1 μM leupeptin, 0.2 μM aprotinin, 10 μM pepstatin A), incubated on ice for 10 min and centrifuged at 3200 × g for 10 min. The supernatant was discarded, and the pellet was resuspended in 100 μl of buffer (20 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT, pH 7.8) containing the protease inhibitor. The cells were lysed by three cycles of sonication of 30 s each at 50% of the maximum power, on the ice. The extract was then centrifuged for 20 min at 13 800 × g to remove cell debris. 500 mM stock solution of DSSO cross-linker was prepared in DMSO. The cross-linking reaction was performed on the nuclear fraction at a concentration of 1 mM DSSO cross-linker by addition of 2 μl of the DSSO stock solution in 100 μl of sample (∼100-fold excess of cross-linker). The reaction was then carried out at room temperature under gentle stirring and stopped after 1 h by adding 2 μl of 500 mM Tris pH 8.5 solution and gentle stirring for 20 min for quenching the N-hydroxysulfosuccinimide function of the cross-linker. After reaction stopping, the sample was vacuum dried. Cross-linked proteins were suspended in 30 μl of 8 M urea, loaded onto 30 kDa Amicon filters and processed according to the FASP protocol described in the previous section.
LC–MS/MS analysis
The samples were reconstituted in 20 μl of 0.1% TFA aqueous solution and desalted using a C18 ZipTip (Millipore, Saint-Quentin-en-Yvelines, France). They were eluted from the ZipTip using 20 μl of ACN/0.1% aqueous TFA (80:20, v/v) and then vacuum dried. For LC–MS analysis, samples were reconstituted in 0.1% FA in water/ACN (98:2, v/v), and separated by reverse-phase LC using a nanoAcquity UPLC equipped with a C18 pre-column (180 μm ID × 20 mm length, 5 μm PD, Waters) and a Peptide BEA C18 column (25 cm length, 75 μm ID, 1.7 μm PD, Waters). Separation was performed using a linear gradient starting at 95% solvent A (0.1% FA in water) and 5% solvent B (0.1% FA in ACN) up to 70% solvent A and 30% solvent B for 120 min at 300 nl/min. The LC system was in line with a Thermo Scientific Q-Exactive mass spectrometer set to select up to the 10 most intense precursors in data-dependent acquisition mode for fragmentation, with a voltage of 2.8 kV. The survey scans were set to a resolving power of 70 000 at full width at half-maximum (m/z 400), in positive mode and using an AGC target of 3E+6. For the shotgun proteomics, the instrument was set to perform MS/MS only from >+2 and <+8 charge state but for XL-MS experiments where larger peptides are measured only >+3 charge state ions were selected, excluding unassigned load states, +1, +2 and >+8.
Shotgun proteomic data analysis
Raw data obtained from the nLC–MS/MS run were treated using MaxQuant v1.6.1.0 using the LFQ (label-free quantification) annotation of the protein identified. UniProtKB database for reviewed human proteins of April 2018 containing 20 303 protein sequences was used. Statistical analyses were carried out using Perseus software after filtering for ‘reverse’ and ‘contaminant’ proteins. For the comparison between control and Forskolin-treated groups, t-test was performed with a permutation-based FDR of 0.05, and P-values <0.05 were considered to be statistically significant. A heat map of differentially expressed proteins across the two different groups was generated. Gene Ontology (GO) analysis was performed using ClueGO (54) on Cytoscape v3.7.1 (55). For AltProt identification, a database (named HaltORF) corresponding to the annotation of long non-coding RNAs (ncRNAs), ncRNAs and mRNA uncoding regions was used. The protein sequences are predicted using the Homo sapiens genome database (release hg38, assembly: GCF_000001405.26). To ensure that unbiased AltProt identification is obtained, the human AltProt database HaltORF (reference name ‘HS_GRCh38_altorf_20170421’, which contains 182 709 entries) was combined with the UniProtKB (7) database that encompasses the RefProts (April 2018 release, 20 303 protein sequences) in a single database for a total of 203 012 entries. Since MaxQuant was described as not suitable for such a large database, the identification was performed using PD2.2. Additional online databases such as ‘Ensembl’ (https://www.ensembl.org) and ‘RefSeq’ (https://www.ncbi.nlm.nih.gov/refseq) were also used to trace back the origin of the identified AltProts after HaltORF data interrogation.
Cross-link data analysis
Data were analysed using PD2.2 implemented with the XlinkX node (48). Interrogation of data was performed according to the following workflow: first spectra were selected and DSSO was defined as cross-linker (characteristic mass 158.003765 Da). Then, the workflow was divided into two paths: (i) The first was dedicated to the cross-link identifications using the XlinkX with the following search parameters: precursor mass tolerance: 10 ppm, FTMS fragment: 20 ppm, ITMS fragment: 0.5 Da, and searching a compiled database comprising both HaltORF and UniProtKB. The validation was performed using percolator with an FDR set to 0.01. (ii) The second path was dedicated to the shotgun protein identification using SequestHT and considering the following parameters: trypsin as an enzyme, two missed cleavages, methionine oxidation as variable modification, DSSO hydrolysed and carbamidomethylation of cysteines as static modification, precursor mass tolerance: 10 ppm and fragment mass tolerance: 0.6 Da. The validation was performed using Percolator with an FDR set to 0.01. A consensus workflow was then applied for the statistical arrangement. A de-isotope and a TopX filter were used to determine the m/z error with a selectivity of ∼10% FDR. The PPIs identified (Supplementary Table S1) were manually checked (Supplementary Figure S1, Supplementary Table S2) and then displayed in Cytoscape 3.7.1 (55). The accession number of each protein showing an interaction was extracted with its XlinkScore and transformed into a text file to get the visualization of the partners. The network was obtained according to the XlinkScore by plotting the distance between the nodes and the recurrence numbers of a given interaction.
Modelling and prediction of interactions
Structure modelling of AltProts and RefProts was performed with the I-TASSER software (56). For both RefProts and AltProts, the most stable models (C-score between −5 and +2) were retained. The prediction of PPIs was performed with the ClusPro software (57). The RefProt was identified as a receiver and the AltProt as a ligand. The interaction model was carried out by docking the ligand onto the receiver without cross-linker size restriction. ClusPro then generates multiple interaction models ranked in the order of stability. The selected models are still part of the top 5 ‘balanced’ models taking into account the best compromise of stability. The selected interactions were then illustrated with Chimera (58) to measure the distance between the atoms observed during XL-MS analysis. The model is split between the ligand and the receptor to form two independent chains; the lysines found to be involved in interactions on PD2.2 are assigned as reference points to measure the distance in the model and have it compared to the length of the cross-linker.
Immunofluorescence
The three AltProts (AltEPHA5, AltMAP2 and AltTRNAU1AP) identified as interactors of TPM4 were amplified from NCH82 cDNA and cloned in fusion with a GFP tag and TPM4 with a 3xFLAG tag. These constructs were then co-transfected in NCH82 cells, each AltProt being co-transfected with TPM4 independently of the other AltProts. Staining was realized as in (59). Briefly, transiently transfected cells were grown on coverslips, and rinsed with PBS before fixation with 4% paraformaldehyde for 15 min at room temperature. After fixation, they were washed three times with PBS 0.1% Triton X-100. Cells were then blocked in PBS 0.01% Triton and 5% bovine serum albumin for 30 min at 4°C. Transfected cells were then incubated overnight at 4°C with monoclonal anti-FLAG M2 mouse antibody (dilution 1:500, Sigma-Aldrich). After three washes for 10 min with PBS 0.01% Triton X-100, cells were incubated for 1 h at 37°C with the Alexa Fluor 647-conjugated secondary antibody donkey anti-mouse IgG (H + L) (1:2000, Thermo Fisher Scientific). Cells were rinsed with PBS and counterstained with phalloidin tetramethylrhodamine B for 30 min at 4°C (5 μg/ml, sc 301530, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Cells nuclei were stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific) in PBS for 5 min. Finally, after a last PBS wash, cells were mounted on a slide using Dako Fluorescent Mounting Medium (Agilent Dako, Santa Clara, CA, USA). Control experiments were performed following the same immunostaining protocol without the primary antibody incubation. Acquisition was performed on a Zeiss LSM700 confocal microscope connected to a Zeiss Axiovert 200 M equipped with an EC Plan-Neofluar 40×/1.30 numerical aperture and an oil immersion objective (Carl Zeiss AG, Oberkochen, Germany). The image acquisition characteristics (pinhole aperture, laser intensity, scan speed) were the same throughout the experiments to ensure comparability of the results. Processing of the images was performed using ImageJ software and applied to the entire images including the controls.
RESULTS
Protein regulation under cell reprogramming by Forskolin
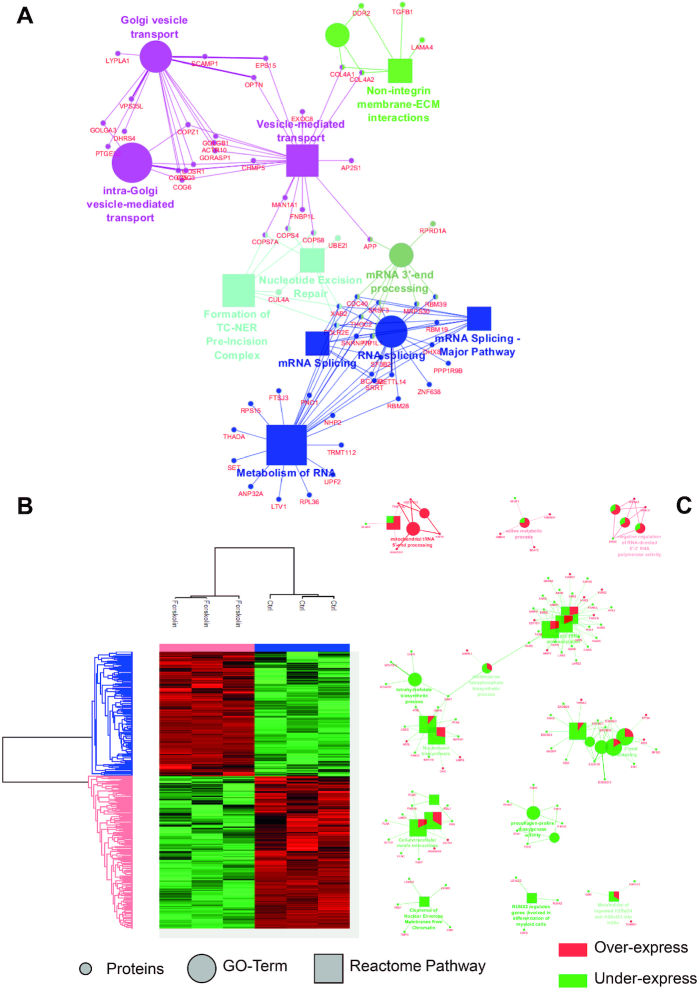
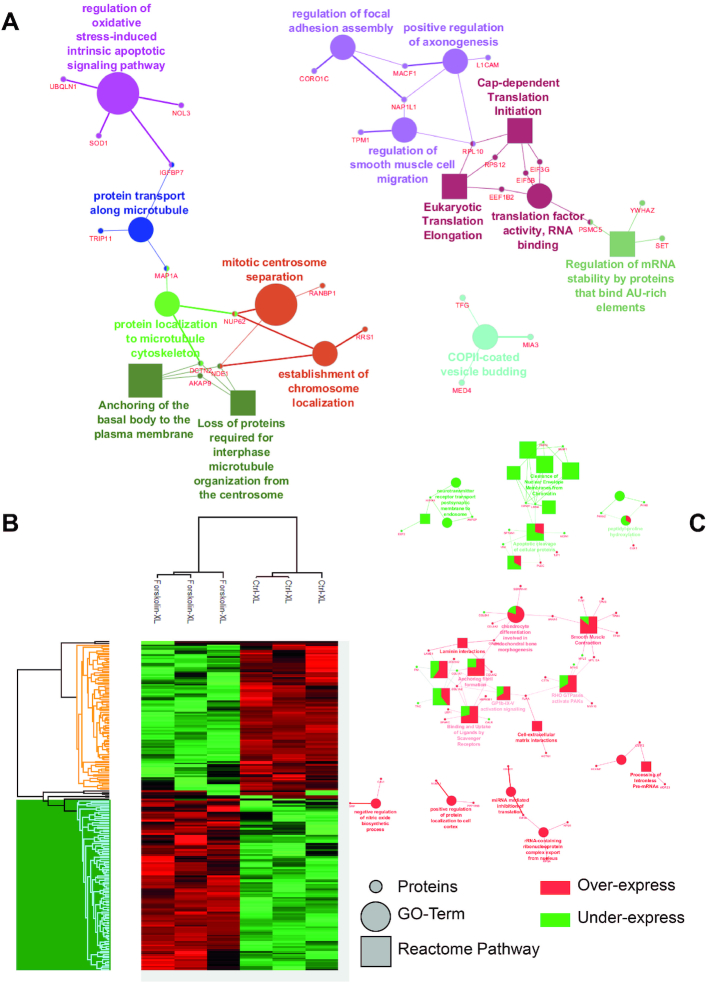
To assess the PPI changes following PKA-induced cell reprogramming, we performed large-scale protein identification with LFQ of NCH82 human glioma cells treated with Forskolin for 48 h (51) (Figure 1). A total of 3363 proteins were identified, among which 41 are exclusive to the non-treated cells and 201 to the Forskolin condition (Supplementary Table S3). Of these 201 proteins, 148 originate from cell organelles according to the extraction protocol for nuclei and organelles, and some are known to be involved in cell reprogramming. For example, transforming growth factor β1 (TGF-β1), the death-inducer obliterator 1 (DIDO1), the mitogen-activated protein kinase 4 (MAP4K4) and the protein Hook homolog 3 (HOOK3) are known to be involved in regulating self-renewal of embryonic stem cells, which is a way of research of cancer cell reprogramming and therapy (60,61). Cytoscape combined with the ClueGO application was then used to retrieve the cellular processes associated with these specific proteins (Figure 1A). Two main processes were found to be associated with Forskolin treatment: one related to the mRNA splicing and metabolism and another to the intracellular trafficking, including Golgi vesicle transport. Other enrichments such as non-integrin membrane–extracellular matrix interaction and nucleotide excision repair were identified. A t-test (P < 0.05 threshold) was then applied to generate the heat map representing the over- and underexpressed proteins between the two conditions. The 1797 proteins found to be significantly over- and underexpressed are distributed in two main clusters: one (991 proteins) associated with untreated cells and another (806 proteins) with Forskolin treatment (Figure 1B). Overexpressed proteins under Forskolin are involved in mitochondrial transfer RNA (tRNA) processing, valine metabolic process and the negative regulation of the 5′–3′ RNA-directed polymerase activity. In contrast, underexpressed proteins are related to nucleotide biosynthesis, rRNA 3′-end processing and tRNA aminoacetylation. PKA activation by Forskolin triggers a complex network of cellular pathways involving numerous nuclear factors (62). To better elucidate the molecular events occurring upon Forskolin treatment, we then enriched nuclei from untreated and treated cells and analysed their protein content by shotgun proteomics (Figure 2). From the nuclear fraction, 936 proteins were identified with 69 exclusives to Forskolin condition and 11 to the control. Cellular processes enriched in Forskolin-treated samples are divided into four main pathways, including (i) translation, (ii) axogenesis and neuritogenesis, (iii) centrosome and (iv) oxidative stress linked to apoptotic pathways. The heat map representing the hierarchical clustering of the differentially expressed proteins revealed a clear segregation between the two conditions (Figure 2B). A set of signalling events associated with Forskolin treatment is observed. For instance, proteins involved in the regulation of alternative splicing, the translation and the modulation of the translation by miRNA were identified. On the contrary, networks related to neurotransmitter receptor transport from postsynaptic membrane to endosome and to nuclear envelope membrane clearance were downregulated after treatment (Figure 2C).
Figure 1.
Identified proteins and their related signalling pathways from NCH82 glioma total cells with and without Forskolin stimulation. (A) GO terms and signalling pathways associated with the proteins identified as unique to the Forskolin-treated cells. (B) Heat map representation generated after raw nLC–MS/MS data interrogated by MaxQuant with LFQ and further processed in Perseus using a t-test showing the proteins over- and underexpressed under Forskolin treatment. (C) GO terms and signalling pathways associated with the over- and underexpressed proteins (Cytoscape; ClueGo app). In Cytoscape, the proteins overexpressed under Forskolin stimulation are represented in red and the underexpressed ones in green.
Figure 2.
Identified proteins and their related signalling pathways from NCH82 glioma cell nuclei fraction with and without Forskolin. (A) GO terms and signalling pathways associated with the proteins identified as unique to the Forskolin-treated cells. (B) Heat map representation generated after raw nLC–MS/MS data interrogated by MaxQuant with LFQ and further processed in Perseus using a t-test showing the proteins over- and underexpressed upon Forskolin treatment. (C) GO terms and signalling pathways associated with the over- and underexpressed proteins obtained by Cytoscape with the application ClueGo. In Cytoscape, the proteins overexpressed under Forskolin stimulation are represented in red and the underexpressed ones in green.
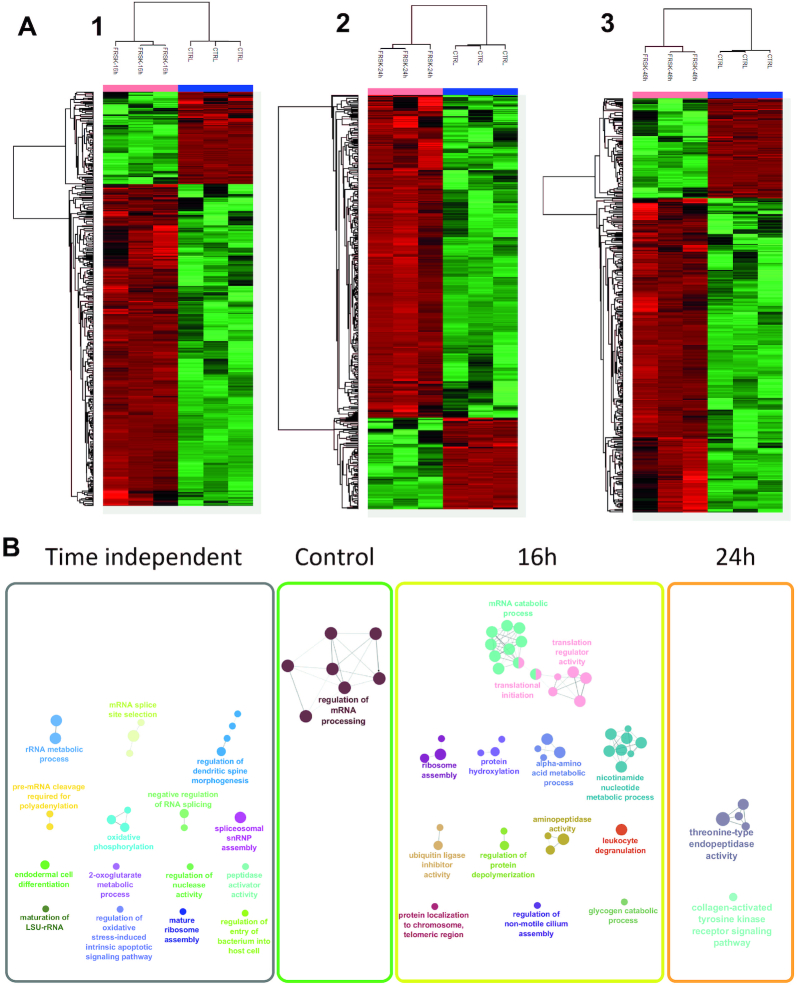
To get a better understanding of the signalling events’ dynamics associated with Forskolin reprogramming, a time-course study at 16, 24 and 48 h of treatment was performed using again the enriched nuclear fraction from untreated and treated cells (Figure 3). Each treatment dataset point was compared to untreated cells to identify specific changes in the protein regulation. Notably, specific clusters of up- and downregulated proteins were identified after analysis (Figure 3A), suggesting that significant changes occur at the nuclear level after treatment. Enrichment within each cluster of the time-course experiment reveals that specific molecular events undergo temporal changes while others are found to be time independent (Figure 3B). In detail, the GO term analysis of the identified proteins shows significantly altered signalling pathways after 16 h of Forskolin treatment (Figure 3B). In line with the role of PKA in transcriptional and translational regulation, proteins related to translation initiation and regulation, ribosome assembly and nucleic acid metabolism are identified. Moreover, pathways involving protein degradation through ubiquitin tagging, regulation of protein depolymerization and enzymatic activity such as aminopeptidase are also observed. At 24 h after PKA activation, only two pathways are modulated: the threonine-type endopeptidase activity and the collagen-activated tyrosine kinase receptor signalling, while none are enriched after 48 h (Figure 3B). The other identified pathways do not show a specific temporal regulation (Supplementary Figure S2). Taken together, this reveals the dynamic modulation of the cellular processes under the PKA activation.
Figure 3.
Identified over- and underexpressed proteins and their related signalling pathways from NCH82 glioma cell nucleus fraction following the Forskolin treatment time course (16, 24 and 48 h) compared to the control condition. (A) Heat maps of the LFQ variation for 16 h (1), 24 h (2) and 48 h (3) treatment with Forskolin, each compared to the control condition, generated after raw nLC–MS/MS data interrogated by MaxQuant with LFQ and further processed in Perseus using a t-test showing the proteins over- and underexpressed. (B) GO terms associated with overexpressed proteins (in A) under Forskolin treatment time course. The untreated condition presents the mRNA processing regulation underexpression that was observed for the corresponding stimulation times. After 16 h Forskolin, an overexpression of proteins in specific pathways such as the initiation of the translation and the nucleotide metabolic process appears. At 24 h, two specific enrichments were found related to collagen formation and cytoskeleton. At 48 h, no specific enrichment was observed. All the other cellular processes are shared between all Forskolin treatment times (time independent).
Time-course identification of PPIs
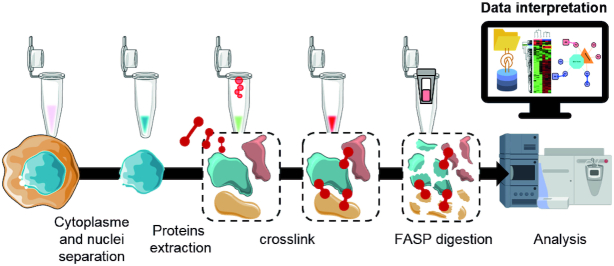
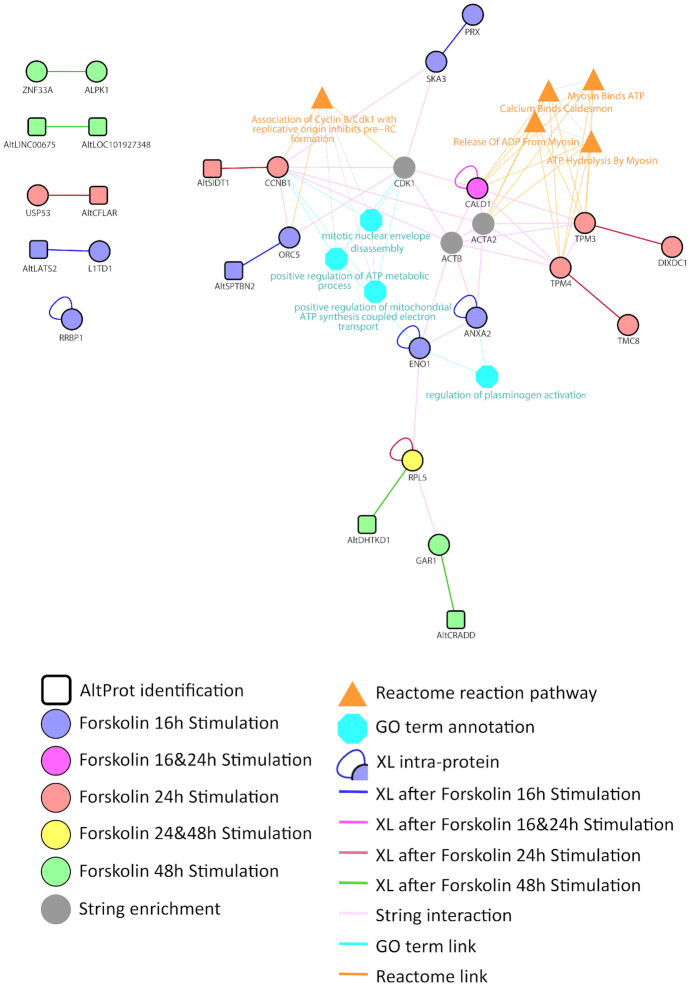
To validate our comparative enrichment analysis, we searched for PPI modulation following Forskolin treatment by large-scale XL-MS strategy. Nuclear fractions were submitted to XL-MS using the membrane-permeable DSSO cross-linker, and the samples were processed using the FASP method (Figure 4). In this time-course analysis, a total of 20 cross-link spectra matches were detected after XL-MS, representing 16 interactions between proteins. The interaction networks known from the literature for these enriched proteins and plotted using STRING (63) interrogation are presented in Figure 5. Cytoscape and ClueGo were applied to correlate the resulting identifications to known processes using Reactome and GO term databases. Most proteins identified at 16 h belong to the ATP synthesis pathway, including the electron transport signalling. According to Reactome, at 24 h, identified proteins are related to the hydrolysis of ATP by myosin pathway. CALD1, which is identified by XL-MS with an internal cross-link, appears to be at the interface of changes in the signalling pathways activated at 16 and 24 h. Based on literature prediction, as obtained by STRING enrichment, CDK1, ACTA2 and ACTB represent a possible link with the proteins identified by XL-MS. The modulation of these signalling pathways clearly highlights the role of PKA activation in the regulation of ATP and its link to myosin and caldesmon at 24 h. Hydrolysis of ATP by myosin is known to stimulate elongation of actin filaments. This effect is particularly well described in neurons upon filopodia elongation (64). RPL5, which was identified at 24 and 48 h, is a common actor of the transition occurring between these two time points. Interestingly, at 48 h, RPL5 is found to be in direct interaction with an AltProt. Other AltProts are also identified at the different time points and a precise temporal modulation was observed for these proteins. Specific AltProts were detected at 48 h (AltDHTKD1, AltCRADD, AltLNC00675 and AltLOC101927348) compared to 16 h (AltSPTBN2, AltLATS2) and 24 h (AltSIDT1, AltCFLAR). Overall, this analysis provides important insights into the timing and composition of PPI networks modulated after PKA stimulation and reveals that AltProts might have a role in their regulation.
Figure 4.
Schematic representation of the XL-MS workflow used in the study. After removing cell cytoplasm by cell lysis using appropriated buffers and centrifugation, the cell nuclei proteins are extracted and cross-linked. Cross-linked proteins are then digested, and peptides are analysed by LC–MS/MS. Recorded data are processed to get the LFQ variations of the cross-linked proteins using either UniProt or a combination of UniProt and AltProt databases. Networks of proteins and their associated pathways are then built in Cytoscape and associated with GO terms with ClueGo. The measured interactions are as well enriched from the bibliographic data by STRING.
Figure 5.
Network of proteins and their associated GO terms generated from the XL-MS data and enriched from STRING and known pathways for the NCH82 cell nucleus fraction upon time-course treatment by Forskolin for 16 h (purple), 24 h (red) and 48 h (green). The proteins are depicted in the colour according to the time point at which they were observed. Data were interrogated using a combined database between RefProts and AltProts. Circles correspond to the identified RefProts and squares to the AltProts. The networks were enriched through addition of STRING network to the identified RefProts using ClueGO application on Cytoscape.
PPI network at 48 h Forskolin treatment
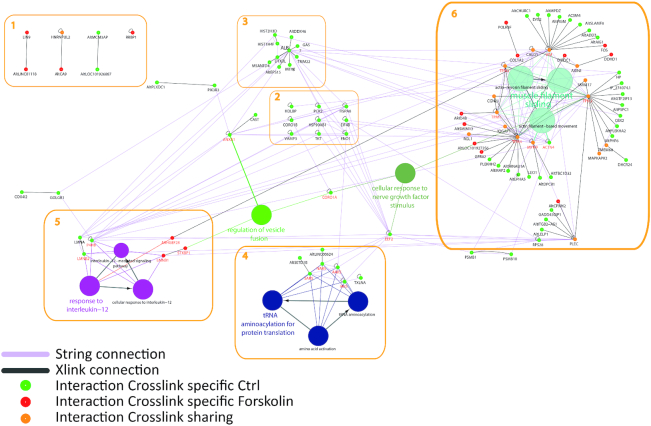
Since Forskolin is known to induce a complete reprogramming after 48 h treatment (51), we dedicated a particular attention to the events occurring after 24 h, expecting to catch intermediate rewiring of PPIs. XL-MS analysis was performed from the nuclear fraction of NCH82 glioma cells leading to the identification of a total of 219 cross-links, including 138 specifics to untreated samples and 81 to Forskolin-treated samples (Figure 6). Five functional categories were identified from the cross-linked peptides, plus a few outsider proteins that were not assigned to known networks. The main network (numbered 6 in Figure 6) is related to cell mobility and cytoskeleton reorganization (Figure 7, boxes A and D). The second and third networks (4,5) are related to tRNA amino acylation for protein translation and response to interleukin-12 (IL-12; Figure 7, box C). tRNA amino acylation for protein translation was also found enriched in untreated cells. Four proteins were found to be directly related to this signalling pathway: SARS, NARS, AARS and IARS. These four proteins are involved in mRNA translation, the attachment of amino acids (aa) to tRNAs and aminoacyl-tRNA synthetases (aaRS), which are essential to ensure proper translation. This group comprises as well two AltProts interacting with these aaRS, respectively, AltSETD1B and AltLINC00624. If AltLINC00624 is derived from a non-coding RNA, AltSETD1B is from the mRNA encoding the SETD1B histone-lysine N-methyltransferase. Based on the observation of the cross-link interaction between these two AltProts and the aaRS (Supplementary Figure S1), one may assume that these two AltProts have a central role in this signalling pathway. AltProts have been described as being involved in regulating other proteins translation, here indirectly by participating in aa-tRNA assembly (Figure 7, box B). Network 5 show proteins that switch from the control (LMNA, LMNB2 and P4HB) to the Forskolin condition (LMNB1, STXBP1, ARHGEF28). ARHGEF28 is directly connected to vimentin, which is involved in the production of microtubules and therefore cell mobility and cytoskeletal organization. STXBP1 is also linked to the regulation of vesicle fusion. The scheme is augmented by CORO1As and EEF2 proteins that connect the different networks together as shown by STRING analysis (63) though only intramolecular was observed for these two proteins. CORO1A links the GO terms of IL-12 regulation, vesicular fusion and cytoskeletal structuring. EEF2 is known to be involved in the regulation of tRNA, but it is also described in cell mobility and cytoskeletal regulation. Network 3 is centred on ALB. By contrast, proteins in network 2 were only found with intra-cross-links and they are known to be connected according to the literature. Among the proteins not known to be involved in specific networks, not surprisingly four are AltProts. One of them interacts with LIN9, which was shown to be involved in embryonic stem cell reprogramming and to exert an antitumour effect (65,66).
Figure 6.
Network of proteins assembled on Cytoscape and their associated GO terms generated from the XL-MS data and enriched from STRING and known pathways for the NCH82 cell nucleus fraction for 48 h treatment by Forskolin by comparison to the control condition (untreated). Proteins were identified by interrogating a RefProt and AltProt merged database. The networks were enriched by addition of STRING network to the identified RefProts using ClueGO application on Cytoscape. Green circles represent proteins only found in the control, red circles represent proteins specific to 48 h Forskolin samples and yellow circles represent proteins found in both untreated and 48 h Forskolin samples. The dark grey lines represent the measured intermolecular cross-links and the small black circles the intramolecular ones. The pink lines correspond to the known connections retrieved in STRING. The global network was subdivided into six groups. In some of these groups, various AltProts were identified. The attribution of GO terms to the RefProts provides a link between the networks involving AltProts to those involving the RefProts, and brings an initial information on the function of these AltProts (see Supplementary Table S3 for more information).
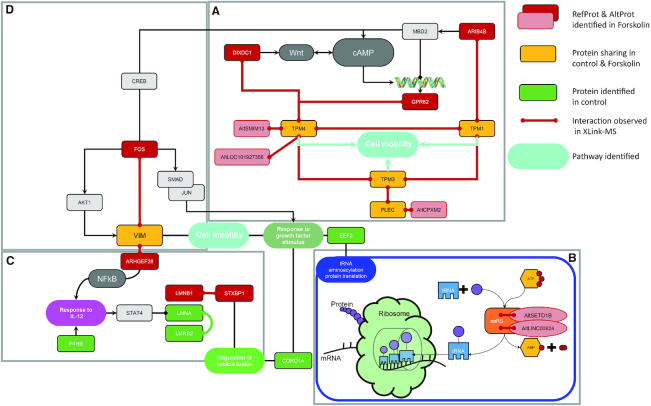
Figure 7.
Schematic representation of over-regulated pathway after 48 h Forskolin stimulation. The AltProts have been positioned within this pathway. The Forskolin stimulates the cAMP pathway that involves different factors such as ARIB4B, DIXDC1 and GPR82 as previously described. The cell mobility pathway seems tightly connected to the cAMP one. The tRNA modulation and the regulation of proteins synthesis are also downstream pathways of cAMP. AltProts are associated with different levels, both in the cAMP pathway and in the tRNA modulation and protein synthesis with the involvement of AltSETD1B and AltLINC00624. The RefProt SETD18 associated with AltSETD1B was described to be involved in the epigenetic control of chromatin structure and gene expression.
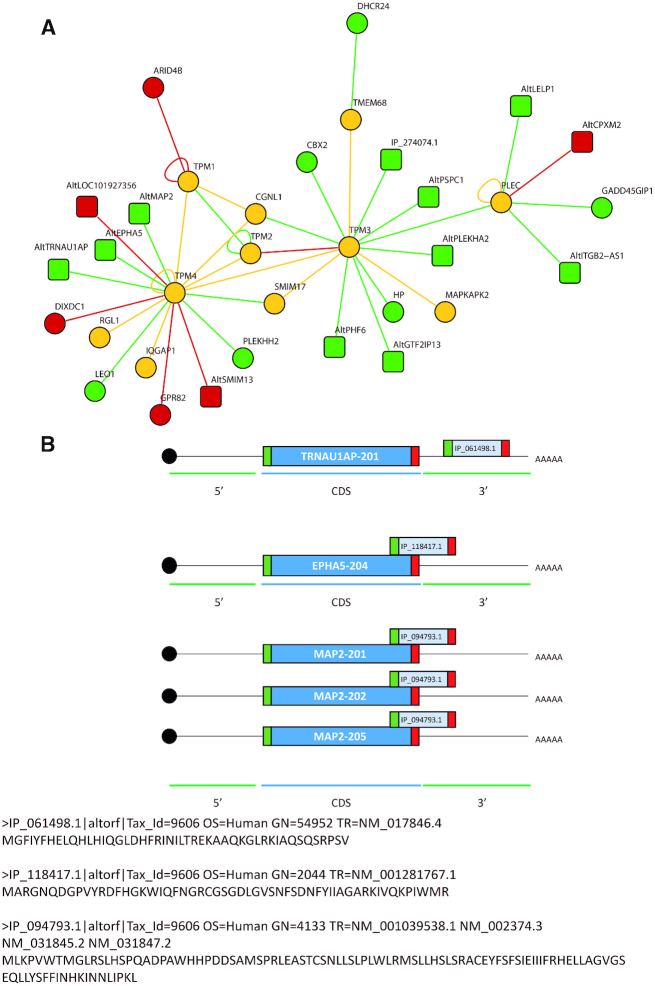
The main network (network 6) is centred on the formation of microtubules and in particular the formation of actin filaments. This network presents 15 common proteins (yellow) to controls and Forskolin stimulation. The main hub of this network is made up of TPM4, TPM3, TPM2, TPM1 and CGNL1. These interactions observed by XL-MS enable to identify under both conditions IQGAP1 and RGL1 connected to TPM4, as well as SMIM17, TMEM68 and MAPKAPK2 connected to TPM3. Condition-specific interactions are also observed with 10 AltProts and 6 RefProts for the control condition and 3 AltProts plus 3 RefProts under Forskolin stimulation (Table 1).
Table 1.
List of the AltProts identified to be in direct interaction with the TPM RefProt family members after 48 h of Forskolin treatment in NCH82 human glioma cells
| Control | Forskolin | ||
|---|---|---|---|
| AltMAP2 | LEO1 | AltLOC101927356 | ARID4B |
| AltEPHA5 | PLEKHH2 | AltSMIM13 | DIXDC1 |
| AltTRNAU1AP | HP | AltCPXM2 | GPR82 |
| AltPHF6 | CBX2 | ||
| AltGTF2IP13 | DHCR24 | ||
| AltPLEKHA2 | GADD45GIP1 | ||
| AltPSPC1 | |||
| IP_274074.1 | |||
| AltLELP1 | |||
| AltITGB2-AS1 | |||
The treated cells’ nuclear fraction is collected and submitted to XL-MS after protein extraction using DSSO as cross-linker. The cross-links are identified from the shotgun proteomic analysis using the XlinkX node in addition to PD2.2.
TPM network reveals hidden AltProt interactions
In network 6 (Figure 6), TPMs are the major protein family for which PPIs were detected by XL-MS. Interestingly, TPM1, TPM2, TPM3 and TPM4 are in direct interaction regardless of the Forskolin treatment. However, some of the PPI partners are specific to one of the two conditions. In particular, in the control condition, TPM4 is interacting with AltProts such as AltTRAU1AP, AltMAP2 and AltEPHA5. The other members TPM3 and PLEC also interact with AltProts (Table 2). Two of these identified AltProts were previously included in transcript databases but were removed (XR_428143.1 and XM_006723305.1). Eight other identified AltProts were issued from an overlapping between CDS and 3′UTR (CDS–3′UTR) (2 of them), 3′UTR (1), 5′UTR (CDS–5′UTR) (1), +2 on the CDS (3 of them) and an ncRNA (1) (Figure 8A). For the three AltProts interacting with TPM4, AltTRNAU1AP is translated from the 3′UTR part of the transcript 201 of the TRNAU1AP gene. AltEPHA5 is coming from an overlap between the CDS domain and the 3′UTR of the transcript EPHA5-204 also coding for an EPHA5 RefProt. Finally, the sequence coding for AltMAP2 can be issued from the transcripts 201, 202 and 205 of MAP2 as overlapping the CDS and the 3′UTR (Figure 8B).
Table 2.
Sequence, gene entry and name, transcript number and origin of the AltProts identified to be in direct interaction with the TPM RefProt family members after 48 h of Forskolin treatment in NCH82 cells
| AltProt | Sequence | Gene | Gene name | Transcript number | Origin |
|---|---|---|---|---|---|
| TPM4 | |||||
| IP_061498.1 | MGFIYFHELQHLHIQGLDHFRINILTREKAAQKGLRKIAQSQSRPSV | 54952 | TRNAU1AP | NM_017846.4 | 3′UTR |
| IP_118417.1 | MARGNQDGPVYRDFHGKWIQFNGRCGSGDLGVSNFSDNFYIIAGARKIVQKPIWMR | 2044 | EPHA5 | NM_001281767.1 | CDS-3′UTR |
| IP_094793.1 | MLKPVWTMGLRSLHSPQADPAWHHPDDSAMSPRLEASTCSNLLSLPLWLRMSLLHSLSRACEYFSFSIEIIIFRHELLAGVGSEQLLYSFFINHKINNLIPKL | 4133 | MAP2 | NM_001039538.1 | CDS-3′UTR |
| TPM3 | |||||
| IP_218514.1 | MHLDGRLLMKWKSSSVSRLIETSEKPKRNWRQKWKQLGMNTN | 55269 | PSPC1 | NM_001042414.2 | CDS+2 |
| IP_159909.1 | MLVKNRKLCLAWPRREDPSGQEQPSNMAQPHLCSPTSDPQLLEPKGSRLQ | 101930567 | Removed | XR_428143.1 | – |
| IP_274074.1 | MGIHVVFIKTLMYTRKFIMRSSINVRNTEGPLKELEKLLHFKEFMMVRNTLNAHSVGNPLECMHNLLDIRKSILMRKLTNVWNVARTSDFIHSLPNIREFILVRNPTNVCTVRRFLELVHSSLNIREFTLVRNLMHVRNVGRLLEYVENLLVIREFILENTVDGFNR | 57711 | Removed | XM_006723305.1 | – |
| IP_163557.1 | MRNVINPEKRAEGRERRRGHWGRPERGEREARRVWQADPEIPGARRTRRPEGRPRPM | 59339 | PLEKHA2 | NM_021623.1 | 5′UTR (NCBI) |
| IP_303632.1 | MIKLKYERNLHKEFTWSIAENTRKLHITPKQLI | 84295 | PHF6 | NM_032335.3 | CDS+2 |
| PLEC | |||||
| IP_070624.1 | MIKVNQMTPRLSPRTAIPSVNKSVSPNASPAV | 149018 | LELP1 | NM_001010857.2 | CDS+2 |
| IP_291098.1 | MERRNQPRRIKRWEQARKPGLSLENNEEGQKENTILLHPLMT | 100505746 | ITGB2-AS1 | NR_038311.1 | ncRNA |
Figure 8.
(A) Focus on the TPM protein network and the different PPIs found by the XL-MS experiment from the NCH82 glioma cell nucleus fraction upon 48 h stimulation with Forskolin versus untreated cells. In green are represented the proteins only found in the untreated condition, in red the overexpressed ones after 48 h Forskolin treatment and in yellow those observed in both conditions. The circles are the RefProts and the squares the AltProts. Interaction maps are generated from XL-MS experiments performed in triplicates (set of files N0, N1, N2, N3). The triplicates were grouped together to identify the significant interactions. (B) Representation of the AltORF encoding for the AltProts interacting with TPM4 location on the mRNA. AltTRNAU1AP is templated on the 3′UTR of the transcript 201 coding for TRNAU1AP. AltEPHA5 coding sequence is located on the overlapping region between the CDS and the 3′UTR region of the transcript 204 coding for EPHA5. AltMAP2 is encoded on the overlapping region between the CDS and the 3′UTR for the three transcripts 201, 202 and 203 coding for MAP2.
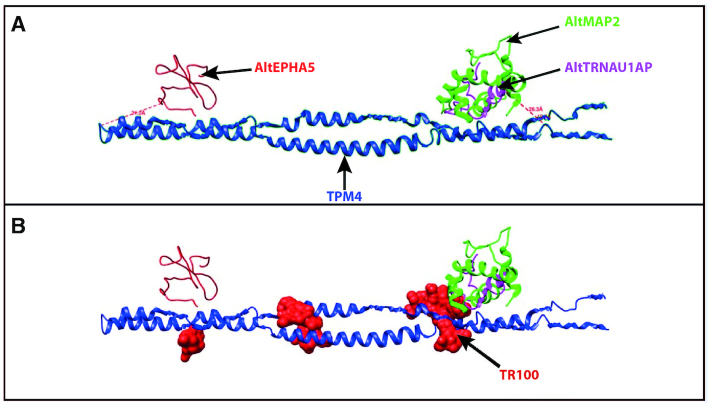
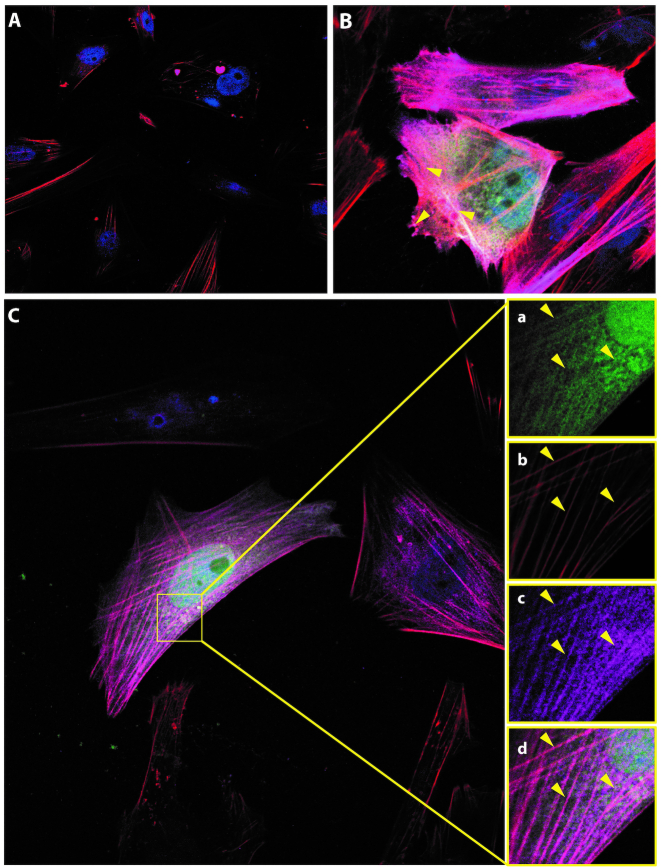
To model the interaction observed by XL-MS between TPM4 and the three AltProts, a 3D model was created. First, the protein structures were predicted in I-TASSER and only the best conformations for each of them, according to the C-score, were kept (Supplementary Figure 3). Then, ClusPro was used to predict the possible interaction between these partners. To avoid any bias in the prediction, the cross-linker length was not considered. Docking analysis was realized between TPM4 and the three AltProts. The best interaction model was visualized in Chimera (Figure 9A) and the distances between the amino acids found to be cross-linked were then measured. These distances are in good agreement with what is expected for the DSSO cross-linker (26 Å theoretical length) (29). Indeed, the distance between TPM4 and the three AltProts was found to be in the 10–30 Å range. Therefore, this prediction supports the interaction between TPM4 and the three AltProts AltMAP2, AltTRNAU1AP and AltEPHA5 (Figure 9A), with AltEPHA5 (measured cross-link distance 21 Å) interacting with the middle of the TPM4 sequence, while AltMAP2 (cross-link distance 18.7 Å) and AltTRNAU1AP (cross-link distance 26.3 Å) would be located at the TPM4 ends. The cross-link distances retrieved from the modelization comfort the interaction found by XL-MS in the absence of other data for these proteins, such as cryo-EM (67). Finally, to confirm the PPIs discovered by XL-MS and sustained by the in silico binding predictions, an immunofluorescence-based colocalization assay was performed using confocal microscopy on NCH82 cells co-transfected with GFP-tagged AltEPHA5, AltMAP2 and AltTRNAU1AP and 3xFLAG-tagged TPM4. Five triple-staining combinations were tested: TPM4 isoforms 1 and 2, in order to see whether both tagged TPM4 isoforms were still able to associate with phalloidin-stained actin cytoskeleton, and each GFP-tagged AltProt to test their colocalization with TPM4 and actin cytoskeleton. These experiments show that both isoforms 1 and 2 of TPM4 are located on the cytoskeleton (Figure 10B). Moreover, in line with the XL-MS analysis and the 3D model prediction, GFP-tagged AltEPHA5, AltMAP2 (Supplementary Figure S4) and AltTRNAU1AP collocate with TPM4 and actin (Figure 10). As shown by its IF profile, GFP-AltTRNAU1AP (Figure 10C, a) follows the signal of the cytoskeleton (Figure 10C, b) as well as the one of 3XFlag-TPM4 (Figure 10C, c), which is well observed when the images are merged (Figure 10C, d).
Figure 9.
(A) ClusPro in silico docking representation of the interaction between TPM4 and the three AltProts that were detected as TMP4 interactors by XL-MS. The folded model is the one that best allows these AltProts to be positioned. AltEPHA5 is predicted to bind the central region of TPM4, while AltMAP2 and AltTRNAU1AP are likely to interact at the TPM4 ends. The interaction models are generated without considering the cross-linker length. Then, the cross-linking distances obtained from the model are compared to the distances expected for the DSSO cross-linker to confirm the probability of interaction. AltEPHA5 cross-link distance = 21 Å; AltTRNAU1AP cross-link distance = 26.3 Å; AltMAP2 cross-link distance = 18.7 Å. (B) Prediction of the binding sites of the TPM inhibitor. Performed docking using Chimera predicts that TR100 could interact at the same location as the two AltProts AltTRNAU1AP and AltMAP2. TR100 could also bind at a site close to the TPM4–AltEPHA5 interaction site.
Figure 10.
Immunofluorescence microscopy of NCH82 cells co-transfected with GFP-tagged (green channel) and 3xFLAG-tagged (mouse anti-FLAG M2 with secondary anti-mouse Alexa 647) proteins, as indicated in each thumbnail. (A) Control with empty vector GFP-only and 3xFLAG-only, without protein. (B) Colocalization of TPM4 isoforms 1 (green) and 2 (magenta) on the actin cytoskeleton (phalloidin red) allowed us to confirm proper tagged TPM4 addressing, for both isoforms. (C) Representative localization pattern of GFP-tagged AltProts and 3xFLAG-TPM4. Here, AltTRNAU1AP (green, a) is collocated (d), with a clear enrichment on the actin filaments (b), also stained by 3xFLAG-TPM4 (c). This colocalization (d) strongly supports the XL-MS results and the predictive interaction 3D model between AltTRNAU1AP and TPM4.
DISCUSSION
Translation initiation from an AltORF, different from the RefORF of a given mRNA and encoding a different protein in terms of primary structure, has been demonstrated as a genetic optimization strategy in viruses and bacteria four decades ago (68). In human cells, AltProt identification was occasionally reported then (69), but this mechanism has long been considered as anecdotal in eukaryotes. Nonetheless, based on the tremendous increase of large-scale proteomic and transcriptomic studies over the last decade, we (2,12,13) and other teams (6,70) were able to demonstrate the presence of AltORF-encoded proteins. The discovery of this ‘ghost’ proteome inherently raised many questions about the potential function exerted by these small proteins and how to analyse and predict their role through large-scale and unsupervised analysis. Thus, our objective was to investigate their function by correlating these proteins to GO terms through the identity of their RefProt interactors. This was achieved by large-scale XL-MS study using NCH82 human glioma cells under reprogramming by Forskolin as a physiopathological model. First, with large-scale proteomics from whole cell extracts, the modulation of proteins known to be involved in cell reprogramming was confirmed. Especially embryonic stem cell proteins like TGF-β1 or DIDO1, MAP4K4 and HOOK3 were found to be modulated under Forskolin treatment. Further, we validated by XL-MS combined with in silico analyses, on the nucleus-enriched fraction, the hypothesis that Forskolin induces cell growth arrest and neural differentiation targeting the cAMP/CREB signalling pathway. Indeed, we highlighted the modulation of the ATP pathway at 16 h post-Forskolin treatment. STRING enrichment adds CDK1, ACTA2 and ACTB proteins to the identified proteins and connects ATP synthesis (16–24 h) and ATP hydrolysis by myosin pathways. Hydrolysis of ATP by myosin is a mechanism already described in neurons during the development of cellular protrusions including filopodia (71,72). Such a morphological modification under Forskolin treatment has also been reported in U87 glioma cells (51). Forskolin stimulates the expression of cAMP-related protein CREB and pCREB as well as apoptosis-related proteins. Thus, Forskolin would inhibit the proliferation as well as the invasion of U87 and promote their apoptosis. This matches our findings that highlight the prominent role of signalling pathways controlling cytoskeletal organization and remodelling. At 48 h Forskolin stimulation, the main observed network is enriched for proteins related to cell mobility and cytoskeleton reorganization, but is also linked to the cAMP pathway. For example, DIXDC1 protein found in this network has been shown to be involved in Wnt5 signalling pathway, which is known to modulate the increase of Ca2+ under Forskolin treatment and to stimulate the cAMP pathway. ARID4B is known to interact with MBD2, a transcription regulator under the influence of cAMP signalling pathway. The GPR82 orphan receptor also appears to be under the regulation of factors involved in the cAMP pathway. The other identified networks are related to tRNA amino acylation for protein translation and response to IL-12. All these networks are linked together through STXBP1, CORO1As and EEF2. CORO1A links the GO terms of IL-12 regulation, vesicular fusion and cytoskeletal structuring. EEF2 is known to be involved in the regulation of tRNA as in cell mobility and cytoskeletal regulation. Nevertheless, the main network is centred on the formation of microtubules and actin filaments, which is related to tumour phenotype switching in neuronal profile as illustrated by the axon guidance proteins such as protein canopy homolog 2 (CNPY2), debrin, plectin and synaptopodin.
We also identified several AltProts in these networks (81) that are grouped in the same networks and pathways as the RefProts, leading to the assumption that they share related functions. For example, AltProts AltSMIM13, AltLOC101927356 and AltCPXM2 are specific to Forskolin stimulation, thus potentially acting as downstream mediators. TPM4 is known to play a role in tumour migration. Several AltProts were found in connection with TPM4 such as AltSMIM13 and AltLOC101927356, which are directly connected to TPM4, while AltCPXM2 shows a specific interaction with PLEC known to play a role in cellular mobility. Other identified AltProts are shown to be involved in protein synthesis regulation. For example, AltSETD1B and AltLINC00624 are connected to the tRNA aminoacylation for protein translation correlating with the protein networks identified in the whole proteome analysis. These results suggest a functional correlation between RefProts and AltProts in modulating PKA signalling in glioblastoma cells. AltProts AltSETD1B and AltLINC00624 may play a role in the regulation of protein expression, corroborating the hypothesis that multiple layers of regulation are involved in the cellular reprogramming (73). In this context, the role of AltProts as possible drivers of this complexity is still unexplored, thus offering novel opportunities to identify therapeutic targets and biomarkers. The XL-MS data therefore show consistency with enriched signalling pathways identified in whole cell proteome analysis. Two major pathways are identified: (i) the protein translation, by the interaction with the aaRS proteins involved in proteins biosynthesis, and (ii) the cell mobility, which is significantly modulated after Forskolin stimulation. These two pathways are in agreement with the previous prediction of several AltProt functions (74).
In a nutshell, XL-MS-based identification of different PPI networks enables to connect AltProts to specific GO terms. Specific AltProts were indeed demonstrated to be involved in cytoskeletal and intracellular transport, as supported by in silico 3D docking and IF colocalization. Despite the fact that phenotypic tests could not be conducted in this study, the confirmed colocalization of these proteins under recombinant tagged overexpression is a first cross-validation to demonstrate that they are indeed expressed and interact in cells. TPM4 association with two AltProts highlights the role of these AltProts in the cellular response to external stress. Interestingly, modelling of the binding sites of the TPM inhibitor, TR100 (2-cyano-3-[1-[3-(dimethylamino)propyl]-2-methyl-1H-indol-3-yl]-N-octyl-2-propenamide), predicts that TR100 would interact at the same location as the two AltProts AltTRNAU1AP and AltMAP2. TR100 could also bind close to AltEPHA5 (Figure 9B). In this context, no cross-link with these three AltProts is observed under TR100 treatment, which suggests a competitive binding between TR100 and these AltProts, reinforcing the hypothesis of their interaction with TPM4. Taken together, the implementation of a non-targeted strategy by XL-MS has also enabled replacing multiple AltProts within the cellular interactome landscape. Moreover, this study has brought strong evidence that AltProts have an important functional role in the Forskolin-induced PKA activation process. This provides a rationale for further investigating the role of ghost proteins in the cell reprogramming regulation. We also linked numerous AltProts to RefProts in specific functional networks and showed that they were both regulated upon Forskolin-induced phenotypic changes, suggesting their cooperation in these specific functions. This will allow to investigate in more detail the precise functional characterization of these novel AltProts. We strongly believe that this work will open new avenues in cell biology studies and will shed light on how important it is to consider AltORFs and AltProts as functional entities in future studies.
DATA AVAILABILITY
Proteomic datasets including MaxQuant files and annotated MS/MS datasets were uploaded to the ProteomeXchange Consortium via the PRIDE database, and then assigned the dataset identifier PXD014642.
Supplementary Material
ACKNOWLEDGEMENTS
Author contribution: Conceptualization, I.F., J.F. and M.S.; methodology, I.F., J.F., E.C, E.M.N.L, M.D., T.C. and M.S.; software, T.C.; validation, I.F., J.F., T.C. and M.S.; formal analysis, T.C.; investigation, I.F., J.F., T.C. and M.S.; resources, I.F., M.M. and M.S.; data curation, T.C.; writing—original draft, T.C., I.F. and M.S; writing—review and editing, E.C., E.M.N.L, D.V., I.F. and M.S.; supervision, I.F., J.F. and M.S.; project administration, I.F. and M.S.; funding acquisition, I.F. and M.S.
Contributor Information
Tristan Cardon, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France.
Julien Franck, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France.
Etienne Coyaud, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France.
Estelle M N Laurent, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France.
Marina Damato, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France; Department of Biological and Environmental Sciences and Technologies, University of Salento, 73100 Lecce, Italy.
Michele Maffia, Department of Biological and Environmental Sciences and Technologies, University of Salento, 73100 Lecce, Italy.
Daniele Vergara, Department of Biological and Environmental Sciences and Technologies, University of Salento, 73100 Lecce, Italy.
Isabelle Fournier, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France; Institut Universitaire de France (IUF),75005 Paris, France.
Michel Salzet, Univ. Lille, Inserm, CHU Lille, U1192—Protéomique Réponse Inflammatoire Spectrométrie de Masse (PRISM), F-59000 Lille, France; Institut Universitaire de France (IUF),75005 Paris, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministère de l'Enseignement Supérieur, de la Recherche et de l'Innovation (MESRI); Institut National de la Santé et de la Recherche Médicale (Inserm); University of Lille.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999; 234:187–208. [DOI] [PubMed] [Google Scholar]
- 2. Vanderperre B., Lucier J.-F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.-M., Roucou X. et al.. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013; 8:e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mouilleron H., Delcourt V., Roucou X.. Death of a dogma: eukaryotic mRNAs can code for more than one protein. Nucleic Acids Res. 2016; 44:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A.S., Brocard M., Couso J.-P.. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife. 2014; 3:e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R. et al.. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunet M.A., Brunelle M., Lucier J.F., Delcourt V., Levesque M., Grenier F., Samandi S., Leblanc S., Aguilar J.D., Dufour P. et al.. OpenProt: a more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Res. 2019; 47:D403–D410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delcourt V., Staskevicius A., Salzet M., Fournier I., Roucou X.. Small proteins encoded by unannotated ORFs are rising stars of the proteome, confirming shortcomings in genome annotations and current vision of an mRNA. Proteomics. 2018; 18:1700058. [DOI] [PubMed] [Google Scholar]
- 11. Hashimoto Y., Niikura T., Tajima H., Yasukawa T., Sudo H., Ito Y., Kita Y., Kawasumi M., Kouyama K., Doyu M. et al.. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc. Natl Acad. Sci. U.S.A. 2001; 98:6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delcourt V., Franck J., Leblanc E., Narducci F., Robin Y.M., Gimeno J.P., Quanico J., Wisztorski M., Kobeissy F., Jacques J.F. et al.. Combined mass spectrometry imaging and top-down microproteomics reveals evidence of a hidden proteome in ovarian cancer. EBioMedicine. 2017; 21:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delcourt V., Franck J., Quanico J., Gimeno J.P., Wisztorski M., Raffo-Romero A., Kobeissy F., Roucou X., Salzet M., Fournier I.. Spatially-resolved top-down proteomics bridged to MALDI MS imaging reveals the molecular physiome of brain regions. Mol. Cell. Proteomics. 2018; 17:357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Razooky B., Obermayer B., O’May J., Tarakhovsky A.. Viral infection identifies micropeptides differentially regulated in smORF-containing lncRNAs. Genes. 2017; 8:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuku N., Pareja-Galeano H., Zempo H., Alis R., Arai Y., Lucia A., Hirose N.. The mitochondrial-derived peptide MOTS-c: a player in exceptional longevity?. Aging Cell. 2015; 14:921–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Couso J.-P., Patraquim P.. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017; 18:575–589. [DOI] [PubMed] [Google Scholar]
- 17. Bensimon A., Heck A.J.R., Aebersold R.. Mass spectrometry–based proteomics and network biology. Annu. Rev. Biochem. 2012; 81:379–405. [DOI] [PubMed] [Google Scholar]
- 18. Dunham W.H., Mullin M., Gingras A.C.. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012; 12:1576–1590. [DOI] [PubMed] [Google Scholar]
- 19. Gavin A.C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M. et al.. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002; 415:141–147. [DOI] [PubMed] [Google Scholar]
- 20. Maeda K., Poletto M., Chiapparino A., Gavin A.C.. A generic protocol for the purification and characterization of water-soluble complexes of affinity-tagged proteins and lipids. Nat. Protoc. 2014; 9:2256–2266. [DOI] [PubMed] [Google Scholar]
- 21. Li P., Li J., Wang L., Di L.J.. Proximity labeling of interacting proteins: application of BioID as a discovery tool. Proteomics. 2017; 17:1700002. [DOI] [PubMed] [Google Scholar]
- 22. Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., Ting A.Y.. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2014; 12:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li P., Meng Y., Wang L., Di L.J.. BioID: a proximity-dependent labeling approach in proteomics study. Methods Mol. Biol. 2019; 1871:143–151. [DOI] [PubMed] [Google Scholar]
- 25. Roux K.J., Kim D.I., Burke B., May D.G.. BioID: a screen for protein–protein interactions. Curr. Protoc. Protein Sci. 2018; 91:19.23.1–19.23.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eyckerman S., Titeca K., Van Quickelberghe E., Cloots E., Verhee A., Samyn N., De Ceuninck L., Timmerman E., De Sutter D., Lievens S. et al.. Trapping mammalian protein complexes in viral particles. Nat. Commun. 2016; 7:11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu C., Huang L.. Mass spectrometry: an emerging technology for interactomics and structural biology. Anal. Chem. 2018; 90:144–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chavez J.D., Bruce J.E.. Chemical cross-linking with mass spectrometry: a tool for systems structural biology. Curr. Opin. Chem. Biol. 2019; 48:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kao A., Chiu C., Vellucci D., Yang Y., Patel V.R., Guan S., Randall A., Baldi P., Rychnovsky S.D., Huang L.. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics. 2011; 10:M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mü M.Q., Dreiocker F., Ihling C.H., Schä M., Sinz A., Müller M.Q., Dreiocker F., Ihling C.H., Schäfer M., Sinz A.. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 2010; 82:6958–6968. [DOI] [PubMed] [Google Scholar]
- 31. Fritzsche R., Ihling C.H., Götze M., Sinz A.. Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun. Mass Spectrom. 2012; 26:653–658. [DOI] [PubMed] [Google Scholar]
- 32. Rey M., Dupré M., Lopez-Neira I., Duchateau M., Chamot-Rooke J.. EXL-MS: an enhanced cross-linking mass spectrometry workflow to study protein complexes. Anal. Chem. 2018; 90:10707–10714. [DOI] [PubMed] [Google Scholar]
- 33. Nury C., Redeker V., Dautrey S., Romieu A., Van Der Rest G., Renard P.Y., Melki R., Chamot-Rooke J.. A novel bio-orthogonal cross-linker for improved protein/protein interaction analysis. Anal. Chem. 2015; 87:1853–1860. [DOI] [PubMed] [Google Scholar]
- 34. Burke A.M., Kandur W., Novitsky E.J., Kaake R.M., Yu C., Kao A., Vellucci D., Huang L., Rychnovsky S.D.. Synthesis of two new enrichable and MS-cleavable cross-linkers to define protein–protein interactions by mass spectrometry. Org. Biomol. Chem. 2015; 13:5030–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riffle M., Jaschob D., Zelter A., Davis T.N.. ProXL (protein cross-linking database): a platform for analysis, visualization, and sharing of protein cross-linking mass spectrometry data. J. Proteome Res. 2016; 15:2863–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Götze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., Fritzsche R., Kühn U., Sinz A.. StavroX: a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 2012; 23:76–87. [DOI] [PubMed] [Google Scholar]
- 37. Müller F., Fischer L., Chen Z.A., Auchynnikava T., Rappsilber J.. On the reproducibility of label-free quantitative cross-linking/mass spectrometry. J. Am. Soc. Mass Spectrom. 2018; 29:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leitner A., Walzthoeni T., Aebersold R.. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC–MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc. 2014; 9:120–137. [DOI] [PubMed] [Google Scholar]
- 39. Walzthoeni T., Joachimiak L.A., Rosenberger G., Röst H.L., Malmström L., Leitner A., Frydman J., Aebersold R.. XTract: software for characterizing conformational changes of protein complexes by quantitative cross-linking mass spectrometry. Nat. Methods. 2015; 12:1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Combe C.W., Fischer L., Rappsilber J.. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteomics. 2015; 14:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iacobucci C., Sinz A.. To be or not to be? Five guidelines to avoid misassignments in cross-linking/mass spectrometry. Anal. Chem. 2017; 89:7832–7835. [DOI] [PubMed] [Google Scholar]
- 42. Du X., Chowdhury S.M., Manes N.P., Wu S., Mayer M.U., Adkins J.N., Anderson G.A., Smith R.D.. Xlink-Identifier: an automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. J. Proteome Res. 2011; 10:923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leitner A., Faini M., Stengel F., Aebersold R.. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 2016; 41:20–32. [DOI] [PubMed] [Google Scholar]
- 44. Klykov O., Steigenberger B., Pektaş S., Fasci D., Heck A.J.R., Scheltema R.A.. Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 2018; 13:2964–2990. [DOI] [PubMed] [Google Scholar]
- 45. Sinz A. The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks. Expert Rev. Proteomics. 2014; 11:733–743. [DOI] [PubMed] [Google Scholar]
- 46. Chen Z.A., Rappsilber J.. Quantitative cross-linking/mass spectrometry to elucidate structural changes in proteins and their complexes. Nat. Protoc. 2019; 14:171–201. [DOI] [PubMed] [Google Scholar]
- 47. Kaake R.M., Wang X., Burke A., Yu C., Kandur W., Yang Y., Novtisky E.J., Second T., Duan J., Kao A. et al.. A new in vivo cross-linking mass spectrometry platform to define protein–protein interactions in living cells. Mol. Cell. Proteomics. 2014; 13:3533–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu F., Rijkers D.T.S.S., Post H., Heck A.J.R.R.. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods. 2015; 12:1179–1184. [DOI] [PubMed] [Google Scholar]
- 49. Cardon T., Salzet M., Franck J., Fournier I.. Nuclei of HeLa cells interactomes unravel a network of ghost proteins involved in proteins translation. Biochim. Biophys. Acta: Gen. Subj. 2019; 1863:1458–1470. [DOI] [PubMed] [Google Scholar]
- 50. Pattabiraman D.R., Bierie B., Kober K.I., Thiru P., Krall J.A., Zill C., Reinhardt F., Tam W.L., Weinberg R.A.. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016; 351:aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xing F., Luan Y., Cai J., Wu S., Mai J., Gu J., Zhang H., Li K., Lin Y., Xiao X. et al.. The anti-Warburg effect elicited by the cAMP-PGC1α pathway drives differentiation of glioblastoma cells into astrocytes. Cell Rep. 2017; 18:468–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang H., Sun T., Hu J., Zhang R., Rao Y., Wang S., Chen R., McLendon R.E., Friedman A.H., Keir S.T. et al.. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J. Clin. Invest. 2014; 124:4489–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiśniewski J.R., Zougman A., Nagaraj N., Mann M.. Universal sample preparation method for proteome analysis. Nat. Methods. 2009; 6:359–362. [DOI] [PubMed] [Google Scholar]
- 54. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J.. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roy A., Kucukural A., Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010; 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J.. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004; 32:W96–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 59. Raffo-Romero A., Arab T., Al-Amri I., Le Marrec-Croq F., Van Camp C., Lemaire Q., Salzet M., Vizioli J., Sautiere P.-E., Lefebvre C.. Medicinal leech CNS as a model for exosome studies in the crosstalk between microglia and neurons. Int. J. Mol. Sci. 2018; 19:4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Y., Kim H., Liang J., Lu W., Ouyang B., Liu D., Songyang Z.. The death-inducer obliterator 1 (Dido1) gene regulates embryonic stem cell self-renewal. J. Biol. Chem. 2014; 289:4778–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saito S., Lin Y.C., Nakamura Y., Eckner R., Wuputra K., Kuo K.K., Lin C.S., Yokoyama K.K.. Potential application of cell reprogramming techniques for cancer research. Cell. Mol. Life Sci. 2019; 76:45–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shabb J.B. Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 2001; 101:2381–2411. [DOI] [PubMed] [Google Scholar]
- 63. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. et al.. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mitchison T., Kirschner M.. Cytoskeletal dynamics and nerve growth. Neuron. 1988; 1:761–772. [DOI] [PubMed] [Google Scholar]
- 65. Boheler K.R. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J. Cell. Physiol. 2009; 221:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gagrica S., Hauser S., Kolfschoten I., Osterloh L., Agami R., Gaubatz S.. Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J. 2004; 23:4627–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu F., Heck A.J.. Interrogating the architecture of protein assemblies and protein interaction networks by cross-linking mass spectrometry. Curr. Opin. Struct. Biol. 2015; 35:100–108. [DOI] [PubMed] [Google Scholar]
- 68. Normark S., Bergstrom S., Edlund T., Grundstrom T., Jaurin B., Lindberg F.P., Olsson O.. Overlapping genes. Annu. Rev. Genet. 2003; 17:499–525. [DOI] [PubMed] [Google Scholar]
- 69. Wang R.F., Parkhurst M.R., Kawakami Y., Robbins P.F., Rosenberg S.A.. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J. Exp. Med. 1996; 183:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappé J., Gevaert K., Van Damme P.. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics. 2013; 12:1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang F.S., Wolenski J.S., Cheney R.E., Mooseker M.S., Jay D.G.. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 1996; 273:660–663. [DOI] [PubMed] [Google Scholar]
- 72. Berg J.S., Cheney R.E.. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002; 4:246–250. [DOI] [PubMed] [Google Scholar]
- 73. Simeone P., Trerotola M., Franck J., Cardon T., Marchisio M., Fournier I., Salzet M., Maffia M., Vergara D.. The multiverse nature of epithelial to mesenchymal transition. Semin. Cancer Biol. 2018; 58:1–10. [DOI] [PubMed] [Google Scholar]
- 74. Samandi S., Roy A. V, Delcourt V., Lucier J.F., Gagnon J., Beaudoin M.C., Vanderperre B., Breton M.A., Motard J., Jacques J.F. et al.. Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. Elife. 2017; 6:e27860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic datasets including MaxQuant files and annotated MS/MS datasets were uploaded to the ProteomeXchange Consortium via the PRIDE database, and then assigned the dataset identifier PXD014642.