Abstract
Small RNAs are important regulators of gene expression and are involved in human development and disease. Next generation sequencing (NGS) allows for scalable, genome-wide studies of small RNA; however, current methods are challenged by low sensitivity and high bias, limiting their ability to capture an accurate representation of the cellular small RNA population. Several studies have shown that this bias primarily arises during the ligation of single-strand adapters during library preparation, and that this ligation bias is magnified by 2′-O-methyl modifications (2′OMe) on the 3′ terminal nucleotide. In this study, we developed a novel library preparation process using randomized splint ligation with a cleavable adapter, a design which resolves previous challenges associated with this ligation strategy. We show that a randomized splint ligation based workflow can reduce bias and increase the sensitivity of small RNA sequencing for a wide variety of small RNAs, including microRNA (miRNA) and tRNA fragments as well as 2′OMe modified RNA, including Piwi-interacting RNA and plant miRNA. Finally, we demonstrate that this workflow detects more differentially expressed miRNA between tumorous and matched normal tissues. Overall, this library preparation process allows for highly accurate small RNA sequencing and will enable studies of 2′OMe modified RNA with new levels of detail.
INTRODUCTION
Small RNAs (sRNAs) are a diverse class of RNA that have a fundamental role in transcriptional and post-transcriptional gene regulation. Members of this category range in size from approximately 18–33 nucleotides and include microRNA (miRNA), small interfering RNA (siRNA), PIWI-interacting RNA (piRNA) and tRNA derived fragments (tRFs). Typically, sRNAs associate with members of the Argonaut protein family to form ribonucleoprotein complexes and act as guides for targeted RNA silencing through complementary base-pairing (1). sRNA based RNA silencing regulates a wide variety of biological processes including development, maintenance and determination of cell fate, fine tuning of gene expression, silencing of transposons and antiviral defenses (1,2). Furthermore, aberrant expression of sRNAs are involved in many human diseases. miRNAs in particular are often aberrantly expressed in tumor cells and are useful biomarkers for both diagnosis and prognosis in a variety of cancer types (3). tRNA fragments are a newly discovered and important class of sRNAs. tRFs are organized into two main categories: longer tRNA-halves and shorter tRNA fragments. Longer 3′ and 5′ tRNA-halves have a role in regulating protein synthesis and their biogenesis is triggered by cellular stress such as infection, oxidative or nutritional stress (2). Less is known about shorter 3′-tRFs and 5′-tRFs, however it has been shown that they can be loaded onto Argonaute proteins and guide mRNA silencing on a variety of targets using mechanisms similar to miRNA induced silencing (4,5).
Several methods are available to quantify sRNAs, including hybridization-based techniques, qPCR and next generation sequencing (NGS). NGS is a particularly attractive method that allows for low cost, genome-wide quantification of sRNA. Furthermore, it is the only technique that can identify novel sRNAs of unknown sequence, distinguish closely related sRNAs, and identify post-transcriptionally modified sequences (6). Typical sRNA library preparation workflows involve adding adapters using sequential single-stranded ligations, followed by reverse transcription and PCR. Several studies have identified the ligation steps as the main source of bias in the library preparation process (7–11). The ligation efficiency and ligation bias of the single-stranded ligations depends on the sequence of the target and the adapter, therefore different adapter sequences can cause profound changes in library content (8,12,13). Cofolding structure between the target sRNAs and the adapters has been identified as a key determinant of ligation efficiency (9,11). Some RNAs form favorable cofold structures with the adapters that allow for ligation at a much higher rate than targets that don’t base-pair with the adapter, or form cofold structures that are unfavorable to ligation, leading to biased representation in the library.
Several methods have been employed to ameliorate this bias. PEG, an intramolecular crowding agent, improves ligation efficiency and reduces bias, a finding that has been incorporated into several commercially available kits (14,15). Furthermore, it was found that adding randomized bases into the adapter helps to increase the diversity of favorable cofold structures and reduces bias (8,11,16). These findings have been commercialized in the NEXTflex kit (Perkin Elmer), which includes adapters that have 4 bp degenerate sequences incorporated at the ligation junctions. Furthermore, several kits have been designed to reduce or eliminate the ligation steps involved in the process. The SMARTer small RNA kit (Takara) uses poly-adenylation followed by reverse transcription and template switching to make ligation-free libraries. While this technique does eliminate ligation associated biases, template switching on uncapped sRNAs itself has some sequence bias (17). Furthermore, for reasons that are not well understood, template switching has a very low detection sensitivity for miRNAs compared to ligation-based approaches when performed on total RNA, due to a large amplification of background RNA such as rRNA (18–20). Finally, because template switching approaches involve non-templated tailing at both ends of the target molecule, precise determination of the original 3′ and 5′ ends is not possible making it more difficult to confidently identify miRNAs from the same family or post-transcriptionally modified miRNAs (21).
Some classes of sRNAs contain a 2′-O-methylation (2′OMe) modification on the ribose moiety of the 3′ terminal nucleotide. This modification stabilizes the sRNA and is present in endogenous siRNAs, miRNAs in plants and piRNAs in animals (1). The 2′OMe modification severely impacts ligation efficiency to ssDNA adapters, as well as the efficiency of the 3′ polyadenylation or polyuridylation required for template-switching approaches (22). Combined with structural and sequence biases, this modification can make sequencing and discovery of 2′OMe modified RNA difficult and bias sequencing libraries against modified sRNA (18).
Randomized splint ligation is a technique in which a double-stranded adapter with a short single-stranded degenerate extension is used to anneal to unknown target nucleic acids. After hybridization to the degenerate portion of the adapter, ligation can occur. This method has been shown to be effective in ssDNA library preparations (23–25) and to reduce bias compared to the ligation of ssDNA adapters (25). Randomized splint ligation has been also used for sRNA library preparations; however, it has not gained widespread adoption in the field presumably due to comparably lower accuracy and sensitivity (6). In this study, we have overcome these challenges through a novel adapter design and an optimized workflow that significantly reduces bias and increases yields, accuracy and sensitivity of sRNA sequencing. We show that our method significantly outperforms the leading sRNA sequencing methods on a variety of sRNA classes including examples of human miRNA and tRFs as well as 2′OMe modified small RNA such as human piRNA and plant miRNAs.
MATERIALS AND METHODS
RNA samples and oligonucleotides
The miRXplore synthetic RNA mix was obtained from Miltenyi Biotec Inc., (Auburn, CA, USA). All total RNA samples were obtained from BioChain Inc., (Newark, CA, USA). Total RNA was extracted using guanidine thiocyanate techniques, treated with DNase I and verified as DNA free using PCR by BioChain. RNA integrity and purity were checked using gel electrophoresis and NanoDrop. A subset of samples were analyzed by Agilent bio-chip and all had RIN values >6. All oligonucleotides were synthesized by Integrated DNA Technologies Inc. (Coralville, IA, USA). See Supplementary Table S2 for oligonucleotide sequences used in this study.
Preparation of randomized splint adapters
The components of the 3′ and 5′ adapters were resuspended in annealing buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 0.1 mM EDTA). The adapter strand of the 3′ adapter was preadenylated using the 5′ DNA adenylation kit (NEB E2610) and purified using the Monarch DNA cleanup kit (NEB T1030). The splint strands of each adapter (oligos 5 and 7, Supplementary Table S2) were diluted to a concentration of 20 μM and mixed with the corresponding adapter strands (oligos 4 and 6, Supplementary Table S2) at a 10 μM concentration in annealing buffer. The 3′ adapter was annealed in a thermocycler by heating to 95°C for 2 min, followed by 70 cycles with the temperature decreasing 1°C per minute for each cycle. The 5′ adapter was annealed by heating to 82°C for 2 min followed by a 0.1°C/s cooling ramp to 4°C. Annealed adapters were stored at −20°C and thawed before use.
Capillary electrophoresis measurement of ligation efficiency
To test the ligation efficiency of sequential 3′ and 5′ ligations, we used a library of oligos containing 20 degenerate ribonucleotides, with an internal fluorescent FAM label in the center (Oligo 1, Supplementary Table S2). The oligo library was heat denatured and treated with T4 Polynucleotide Kinase (NEB M0201) to add a 5′ phosphate and purified using the Monarch RNA cleanup kit (NEB T2030). For testing the effect of the 2′OMe modification on ligation efficiency, we used libraries containing 21 degenerate oligoribonucleotides with 5′ FAM fluorescent labels. Two versions were synthesized, one with the terminal nucleotide including the 2′OMe modification (Oligo 3, Supplementary Table S2) while the other contained a normal ribonucleotide at the terminus (Oligo 2, Supplementary Table S2). All experiments were carried out in quadruplicate along with ligase-free and adapter-free negative controls. Randomized splint reactions were carried out as follows: 1 pmol of FAM labeled oligo library was diluted to a final volume of 5 μl in nuclease free water, heated to 70°C for 2 min and then cooled on ice. The following components were then added: 1× final concentration of T4 RNA ligase buffer (NEB M0204), 20% final concentration of PEG (NEB M0204), 0.05% final concentration of Tween 20 (VWR Radnor, PA), 10 pmol annealed 3′ adapter (Substrate to adapter ratio of 1:10; 10 pmol top adapter strand, 20 pmol bottom splint strand), 200 units of T4 RNA Ligase 2, truncated KQ (NEB M0373) and nuclease free water to a final volume of 20 μl. These reactions were incubated in a thermocycler at 25°C for 1 h. The ligase was heat inactivated by heating the reaction to 75°C and cooled to 4°C. 4 μl samples of each reaction were then diluted to a concentration of 20 nM and capillary electrophoresis was performed on a 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Reactions containing the internal FAM substrate continued on to the sequential 5′ ligation. First, the volume removed for sampling from the reaction was replaced with 4 μl of 1× reaction buffer. The 5′ ligation was then performed by adding ATP to a final concentration of 1 mM, 20 pmol of the 5′ adapter (Substrate to adapter ratio of 1:20; 20 pmol adapter top strand, 40 pmol splint bottom strand) and 20 units of T4 RNA ligase 2 (NEB M0239) in a final volume of 29 μl. These reactions were incubated at 37°C for 1 h, the ligase was heat inactivated at 75°C for 5 min and the samples were diluted to 20 nM and run on capillary electrophoresis as described above. For comparison to the standard single-stranded ligations, the NEBNext sRNA kit was used with some modifications to the procedure to standardize the adapter concentrations across both methods: 1 pmol of the FAM labeled oligo libraries was combined with the 10 pmol of the 3′ SR Adaptor for Illumina in a final volume of 7 μl (Substrate to adapter ratio of 1:10). The mixture was heated to 70°C for 2 min and cooled to 4°C. The 3′ Ligation Reaction Buffer and enzyme mix were added as described in the kit and the mixture was incubated at 25°C for 1 h. The ligase was heat inactivated at 75°C for 5 min and 4 μl samples of each reaction were then diluted to a concentration of 20 nM and capillary electrophoresis was performed as described above. Reactions containing the internal FAM substrate continued on to the sequential 5′ ligation. First the volume removed for sampling from the reaction was replaced with 4 μl of 1× reaction buffer. The 5′ ligation was then performed by adding 20 pmol of heat-denatured 5′ SR adapter (Substrate to adapter ratio of 1:20), 1 μl of the 5′ ligation buffer, 2.5 μl of the 5′ ligation enzyme mix and water to a final volume of 30 μl. The reaction was incubated at 25°C for 1 h, the ligase was heat inactivated at 75°C for 5 min and the samples were diluted to 20 nM and run on capillary electrophoresis as described above.
2′OMe spike-in
Spike-in oligos were designed by Dard-Dascot et al. (18). Briefly, 12 oligos were synthesized in six pairs (Oligos 8–19, Supplementary Table S2). These oligos do not map to any known miRNA sequences and each pair has the same sequence except for a single nucleotide polymorphism (SNP) that is used to uniquely identify the oligo. The SNP was designed in such a way as to not affect the predicted secondary structure of the oligos (18). Finally, one member of each pair carries the 2′OMe modification in the 3′ terminal nucleotide. Oligos were resuspended at a concentration of 1.5 μM in TE buffer. Oligo concentrations were measured using the Qubit miRNA Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) and 10 ng of each oligo was added to the spike-in mix. TE buffer was added to create a final mix with all 12 oligos and a final concentration of 1 ng/μl. 1 ng of the spike-in mix was spiked into a background of 500 ng total human brain or testes RNA and libraries were constructed as described below.
Library construction
50 fmol of synthetic miRXplore RNA, or 500 ng of total RNA was used as input to the libraries. Libraries were constructed with NEBNext (New England Biolabs Inc., Ipswich, MA), NEXTflex (PerkinElmer Inc., Waltham, MA) and TruSeq (Illumina Inc., San Diego, CA, USA) kits following the manufacturer's directions. For experiments testing a range of RNA inputs, between 1 and 1000 ng was used as input and the adapter concentrations were adjusted as noted in Supplementary Table S1. Based on initial testing PCR cycles were adjusted so that all libraries would be amplified to approximately the same concentration, which generally entailed amplifying the libraries made with the randomized splint method 2–3 cycles less than the other methods, see Supplementary Table S1. Randomized splint ligation libraries were constructed using the following method. Total RNA samples were diluted to a volume of 5 μl in nuclease free water and heated to 70°C for 2 min and then cooled on ice. The following components were then added: 1× final concentration of T4 RNA ligase buffer (NEB M0204), 20% final concentration of PEG (NEB M0204), 0.05% final concentration of Tween 20 (VWR Radnor, PA), 2.5 pmol annealed 3′ adapter (2.5 pmol top strand, 5 pmol bottom strand), 200 units of T4 RNA Ligase 2, truncated KQ (NEB M0373) and nuclease free water to a final volume of 20 μl. These reactions were incubated in a thermocycler at 25°C for 1 h. Following ligation 2.5 units of lambda exonuclease (NEB M0262) and 25 units of 5′ deadenylase (NEB M0331) were added and the reactions were incubated for 15 min at 30°C, 15 min at 37°C and 5 min at 75°C. Five units of Uracil-DNA Glycosylase (NEB M0280) and 20 units of Endonuclease IV (M0304) were added and reactions were incubated for an additional hour at 37°C. The 5′ ligation was then performed by adding ATP to a final concentration of 1 mM, 5 pmol of the 5′ adapter (5 pmol top strand, 10 pmol bottom strand) and 20 units of T4 RNA ligase 2 (NEB M0239). The reaction was incubated at 37°C for 1 h. Reverse transcription was performed by adding 50 mM final concentration of Tris–HCl buffer (pH 7.5), 75 mM final concentration of potassium chloride, 10 mM final concentration of DTT, 500 μM final concentration of each DNTP, 20 units of Murine RNase inhibitor (M0314), 200 units of Protoscript II reverse transcriptase (NEB M0368) and nuclease free water to bring the final volume to 50 μl. This reaction was then incubated for 1 h at 42°C. First strand cDNA products were purified using 70 μl NEBNext sample purification beads (NEB E7767) and 70 μl of 100% Isopropanol. Reactions were washed and eluted in 10 μl of nuclease free water according to the manufacturer's directions. PCR amplification of the library was performed using NEBNext High-Fidelity 2X PCR Master Mix (NEB M0541) and 25 pmol each of the forward and reverse primers. PCR was performed with the following program: An initial denaturation of 98°C for 30 s followed by a varying number of cycles depending on input (see Supplementary Table S1) of: 98°C for 10 s, 62°C for 30 s and 72°C for 30 s. Followed by a final elongation step of 72°C for 5 min. Libraries were size selected using the NEBNext sample purification beads (NEB E7767) and using the small RNA library size selection protocol from the NEBNext small RNA library kit (NEB E7330). Purified libraries were assayed on the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) to assess purity and concentration before being pooled and sequenced using 50 cycles of single-end Illumina sequencing. Representative bioanalyzer traces are shown in Supplementary Figure S1.
qPCR
cDNA was synthesized using human brain RNA and the miRCURY LNA RT Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. miRNAs were amplified in quadruplicate using the primers from the miRCURY LNA miRNA miRNome PCR Panels I and II (Qiagen, Hilden Germany) and Luna Universal qPCR mastermix (NEB M3003). Assays were performed on a CFX-384 real time PCR detection system (Bio-Rad, Hercules, CA, USA).
Bioinformatic analysis
MiRXplore analysis: Raw reads were trimmed using cutadapt (26). Low quality reads and reads <15 bp were removed. Additionally, the eight degenerate bases incorporated into the NEXTflex adapters were removed. Reads were then subsampled to a total 1 million reads per replicate using the reformat function in BBtools (https://sourceforge.net/projects/bbmap/). Reads were mapped to the miRXplore reference sequences using bowtie (27). Possible alignments were considered with up to 1 mismatch in the first 10 bp seed region of the read. Reads with >100 possible alignments were considered unmapped and only the single best alignment was reported for each read. Sequence counts were generated from mapped reads using the idxstats function in samtools. For each library, we generated an expected read count by dividing the total number of mapped reads by 962, since the 962 miRNAs in the mix are expected to be equimolar. We then divided the raw read counts by the expected read count to normalize them. Analysis was then performed on the normalized read counts.
Analysis of human brain RNA: Brain samples were trimmed using the cutadapt method described above. Reads were mapped to the 2′OMe spike-in mix allowing no mismatches using BBMap (https://sourceforge.net/projects/bbmap/) and coverage statistics were generated after mapping using samtools. Spike-in read counts were divided by the total number of reads mapping to the spike-in mix and multiplied by 1 × 106 to obtain the counts in reads per million (rpm). They were then normalized by dividing by the expected value of 8333 rpm, given that there were 12 equimolar oligos in the mixture. Remaining reads that did not map to the spike-in set were subsampled to a total 2 million reads per replicate using the reformat function in BBtools (https://sourceforge.net/projects/bbmap/). Reads were mapped to the human genome (build GrCH38) using bowtie (27) with the settings described above. HTSeq (28) was used to generate counts of miRNAs from the mapped reads using the genomic coordinates from miRbase (29). For analysis of sensitivity we repeatedly randomly subsampled the trimmed reads, increasing the number of reads sampled by 5000 in each sample until we reached 2 × 106 reads. Each of these subsamples was then subjected to the mapping pipeline described above. Trimmed and subsampled reads were also mapped to tRFs using MINTmap and coverage statistics were generated during mapping, considering only unambiguous tRFs (30).
piRNA analysis: Libraries were made from 500 ng human testes total RNA. Technical replicates were pooled to create a large set of reads for each method. Raw reads were trimmed using cutadapt (26). Low quality reads, reads mapping to the spike-in mix and reads <15 bp were removed. Reads were then subsampled to a total of 16.5 million reads per method using BBtools (https://sourceforge.net/projects/bbmap/). These datasets were further analyzed using the PILFER pipeline to characterize the piRNA (31). Briefly, reads were mapped to the human genome (build hg19) and piRNAbank (32) using bowtie. Reads between the sizes of 26 and 33 nucleotides mapping to piRNAbank were considered to be canonical piRNAs. Additionally, reads were considered to be putative piRNA if they met the following criteria: (i) mapped to the genome but were not in piRNAbank, (ii) between the sizes of 26 and 33nts and (iii) did not match any other non-coding RNA annotations in ensemble. Finally, piRNA clusters were identified in each dataset using the PILFER algorithm. The sets of canonical and putative piRNA were then filtered to contain unique species and the number of piRNAs were counted using custom R scripts. Plots of sequence content and 5′ uridine bias of the unique piRNA species were created using ggseqlogo (33). To plot genome coverage, we divided the raw read counts by the number of times they mapped to the genome to normalize the values of multimapping reads. We then used the bedmap function from BEDOPS (34) to divide the genome into 100kb non-overlapping windows and calculate the sum of the normalized read counts within each window. Additionally, we downloaded the RepeatMasker bed file from the UCSC genome browser (35) and used bedmap to calculate the percentage of each 100 kb window covered by repetitive elements such as retrotransposons. A circos plot of the results was created using the R-package BioCircos (36,37).
Arabidopsis: Reads were trimmed with cutadapt and mapped to the Arabidopsis thaliana genome (build TAIR10) using bowtie (27) with the settings described above. HTSeq (28) was used to generate counts of miRNAs from the mapped reads using the genomic coordinates from miRbase (29).
Cancer samples: Each sample was sequenced with two technical replicates per sample, per technique. Reads were trimmed, mapped and counted using the workflow described above for human brain samples. Differential expression was performed on the read counts. Because we did not have biological replicates, we used a non-parametric method which uses the technical replicates to model noise distribution as implemented in the R package NOIseq (38).
qPCR: qPCR data were analyzed using the CFX Manager software (Version 3.1, Bio-Rad, Hercules, CA, USA). An inter-plate calibrator (IPC) target provided by the manufacturer was used to correct the quantification cycle values (Cqs) for any batch effects. To generate the correction-factor, the Cq of each IPC was subtracted from the mean IPC. The correction factor for each replicate was then subtracted from the raw Cq values. Targets were included in the analysis if they met the following criteria: (i) Were detected in the qPCR assay with an average Cq of <37, (ii) had a coefficient of variation of <5% among the four qPCR replicates and (iii) were detected by at least one of the sequencing methods with an average of at least 10 reads per million.
Statistics
All analyses were conducted in R (R Core Team, version 3.6.3. https://www.R-project.org/). Statistical comparisons were done using ANOVA followed by post-hoc testing using the estimated marginal means (R package, version 1.4.5. https://CRAN.R-project.org/package=emmeans). P-values were corrected for multiple comparisons using a Tukey correction. Detection and expression patterns were plotted using pheatmap (R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap) and ggVennDiagram (R package version 0.3. https://CRAN.R-project.org/package=ggVennDiagram).
RESULTS
Randomized splint ligation is more efficient than single-stranded ligation
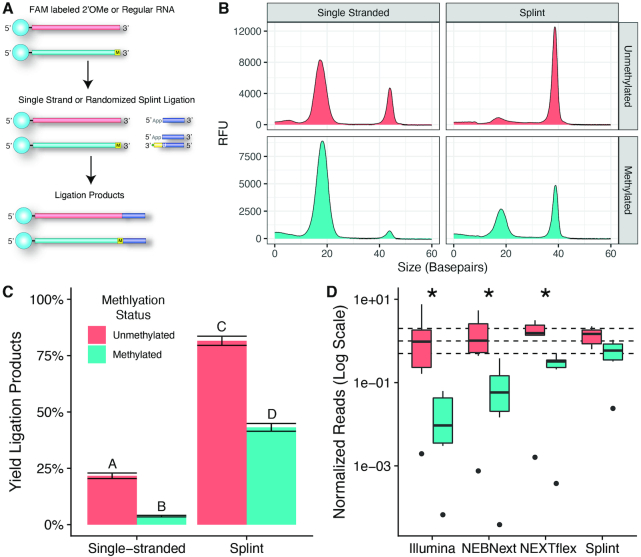
We compared the ligation efficiency of randomized splint ligation to the ligation efficiency of single-stranded ligation for small RNA (sRNA). We used a FAM labeled fully degenerate RNA oligo as a test substrate. The adapters used in this experiment had the same sequence but differed on the presence or absence of the splint strand. The substrate was first ligated at the 3′ end to an adapter and then ligated at the 5′ to another adapter, as diagramed in Figure 1A. Capillary electrophoresis (CE) was used to measure the percentage of the substrate that was converted to the ligation products by integrating the area under the peaks, which we define as ligation efficiency (Figure 1B). In the first ligation, the randomized splint ligation had a significantly higher ligation efficiency, converting 3-fold more of the RNA oligo substrate to ligation product (Figure 1C, P < 1 × 10−4). In the subsequent 5′ ligation, the randomized splint ligation produced >6-fold more of the full ligation product (Figure 1C, P < 1 × 10−4). Furthermore, randomized splint ligation resulted in much less side product formation during the 5′ adapter ligation than the single-stranded ligation method resulting in higher overall efficiency (Figure 1B).
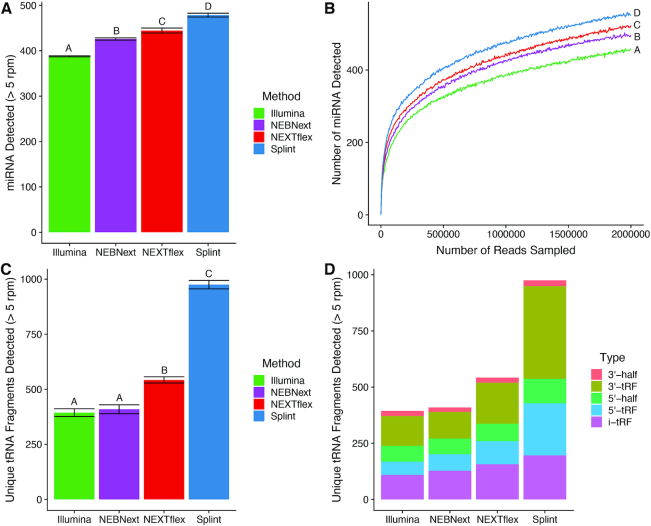
Figure 1.
Capillary electrophoresis measurements of ligation efficiency. (A) Schematic of the CE assay. A FAM labeled degenerate RNA oligo was used as a substrate for sequential 3′ and 5′ ligations using either the single-stranded or randomized splint methods. (B) Example CE traces showing single-stranded (blue) and randomized splint (red) reactions. The major products and side products are labeled with letters according to the key below the traces. (C) Quantification of the main ligation products as a percentage of the total amount of FAM labeled oligo. Experiments were performed in quadruplicate.
Design of novel randomized splint ligation based sRNA library preparation
Based on our findings that randomized splint ligation exhibits higher ligation efficiency for sRNA than the widely used ssDNA adapter ligation, we developed a novel library preparation workflow (Figure 2). The workflow involves sequential randomized splint ligations to attach adapters to each side of the sRNA, followed by PCR to enrich adapter-containing molecules for sequencing. We have several novel design features of the 3′ adapter that serve to increase efficiency and reduce adapter dimer formation. We included an inverted deoxythymidine blocking nucleotide on the 3′ end of the degenerate portion of the bottom strand of the adapter, which increases efficiency of ligation to the targets by blocking adapter to adapter ligations (Supplementary Figure S2A). Secondly, we included a deoxyuridine nucleotide before the degenerate portion of the adapter. This allowed us to cleave off the degenerate nucleotides and use the fixed portion of the splint strand as a primer in the reverse transcription, ensuring that the degenerate nucleotides are not incorporated into the cDNA product. Cleaving off the degenerate portion of the adapter before the 5′ ligation also strongly reduced the amount of adapter dimer formation, which is a major challenge in small RNA sequencing (Supplementary Figure S2B). Finally, we used a preadenylated top strand adapter to reduce the formation of side products and sequential ligations to reduce the formation of adapter dimers. We then compared our optimized workflow with several leading sRNA library preparation methods.
Figure 2.
Schematic of randomized splint ligation library preparation. First the preadenylated 3′ adapter is ligated on using randomized splint ligation. Following adapter ligation, the excess adapter is depleted using 5′ deadenylase and lambda exonuclease, and the degenerate portion of the adapter is cleaved off by excising the deoxyuracil using USER. Next the 5′ adapter is ligated on using randomized splint ligation and cDNA is synthesized using the remaining portion of the 3′ adapter splint strand as a primer for the reverse transcription. Finally, library molecules containing both adapters are enriched and extended using PCR.
Randomized splint ligation based method reduces bias and increases sensitivity
To evaluate the sequencing bias of our library preparation workflow, we used a synthetic reference RNA called miRXplore. This RNA is an equimolar mix of 962 synthetic miRNA sequences from several species including human, mouse and rat and is commonly used to benchmark the bias of sRNA workflows. We prepared libraries from 50 fmol of the miRXplore RNA using Illumina's TruSeq, New England Biolab's NEBNext, Perkin Elmer's NEXTflex and our randomized splint ligation library prep workflow. Reads were mapped, counted and normalized such that each miRNA was expected to have a normalized read count of 1. Values < 1 represent miRNA that are underrepresented in the library compared to their expected values, while values > 1 are overrepresented. We found very high technical reproducibility for all the methods tested (Spearman ρ > 0.99, Supplementary Figure S3). Normalized read values are plotted in log scale (Figure 3A). We found that our randomized splint ligation based method could achieve similar yields to other methods with 2–5 cycles less PCR, indicating higher efficiency of miRNA capture (Supplementary Table S1). We found a large bias mainly towards under-representation in TruSeq library preparations (>50 000-fold between the highest and lowest represented sequences). This is improved using the NEBNext kit and further improved using the NEXTflex kit. However, even in libraries produced with the NEXTflex kit, <40% of sequences are represented in the sequenced library within 2-fold of their expected values, and only 66.5% within 4-fold. Using our randomized splint method, we were able to capture a significantly higher percentage of targets (>75%) within 2-fold of their expected values and the representation of all targets was improved, with 96.3% of targets within 4-fold of their expected value. Comparing the number of targets within 2-fold of their expected values we found that the randomized splint method was significantly higher than the 3 other methods (Figure 3B, Tukey corrected P < 1 × 10−4 in all three comparisons). Furthermore, the randomized splint method was the only one that captured all 962 targets in the library. To investigate potential sequence bias caused by the randomized splint method, we focused on the first six bases (5′ end of miRNA) and last six bases (3′ end of miRNA), the regions of the miRNAs that would hybridize to the adapters. First, we investigated the GC bias in these regions, under the assumption that high GC sequences might hybridize to the adapter more efficiently and therefore be overrepresented in the library. We grouped the data in three categories, either >2-fold (overrepresented), within 2-fold (correctly represented) or <2-fold (underrepresented). In fact, there is no pronounced enrichment for high GC sequences using the randomized splint method (Supplementary Figure S4). Furthermore, we analyzed the sequence motifs for these regions in each category using sequence logos (39). We did not see any strong motifs, aside from a slight overrepresentation of purines in the 5′ end among the overrepresented sequences in the randomized splint method, which may be due to the fact that randomized splint method has much less miRNAs in the overrepresented category compared to the other three methods. (Supplementary Figure S5). Overall, the splint method has much lower bias than the other methods.
Figure 3.
Comparing bias of leading library prep methods with randomized splint ligation. (A) Normalized read counts of libraries prepared from an equimolar mix of 962 synthetic miRNA are plotted in logarithmic scale. Each miRNA was expected to have a normalized read value of 1 (central dashed line). The upper and lower dashed lines correspond to the interval of 2-fold over- or underrepresented. The total number of miRNA detected with each method is listed below the X-axis label. (B) Percentage of miRNAs falling within 2-fold of their expected values shown as the average of two technical replicates per method. Error bars represent the standard deviation. Letter codes indicate groups that are significantly different from each other with Tukey corrected P < 0.001. (C) Quantification of human brain miRNAs using qPCR compared to each sequencing method. qPCR values are represented as the average Cq of 4 technical replicates while sequencing values are represented as the average number of reads per million for 3–4 technical replicates per method, both are plotted on a logarithmic scale. Linear models were used to plot the line of best fit for each correlation and the R2 value of the correlation is printed in the lower left-hand corner of each plot.
To confirm our results in libraries prepared from biological samples, we made libraries with 500 ng of total human brain RNA as an input using each of the four library preparation methods. We evaluated the bias by comparing miRNA sequencing read counts to qPCR measurements from the same sample on 379 miRNA targets. The results were plotted on a log scale with the average qPCR threshold cycle (Cq) values on the y-axis and the average number of reads per million for each method on the x-axis (Figure 3C). We found that the randomized splint method had a higher correlation to the qPCR data than any other method. To test if the randomized splint method was significantly better correlated to the qPCR data than the other methods, we used Hittner, May and Silver's Z-statistic for comparing overlapping dependent correlations (40). We found that the randomized splint method was significantly better correlated to the qPCR data than any other library preparation method (Figure 3C, Corrected P < 1 × 10−2 in all comparisons).
Further analyses of human brain miRNA showed that the randomized splint method detected significantly more miRNA sequences than any other method (Figure 4A, corrected P < 10−3 in all comparisons) and the most unique detections (Supplementary Figure S6A). Furthermore, the randomized splint method had the highest detection sensitivity at all subsampled read depths (Figure 4B). Overall expression patterns were similar across all four methods, with NEBNext and NEXTflex being the most similar to each other (Supplementary Figure S6B). We were also able to use the method to detect miRNA with a variety of input RNA amounts ranging from 1ug to 1ng of total RNA. Correlations between technical replicates were highly repeatable within an input level as well across input levels (Supplementary Figure S7, Spearman ρ values > 0.75).
Figure 4.
sRNAs detected in human brain. (A) The number of miRNAs from human brain detected at a threshold of >5 reads per million (rpm), represented as an average of 3–4 technical replicates per method. Error bars represent the standard deviation of the mean and letters represent groups that are significantly different from each other with a Tukey corrected P-value < 0.001. (B) Number of miRNAs detected with a read depth >5 rpm at various subsampled read depths. Datasets were randomly sampled with the number of reads increasing by 5000 in each sample. Values represent the mean number detected in four technical replicates individually sampled at each read depth. Letters represent groups that are significantly different from each other with a Tukey corrected P-value < 0.001 at the 2 million read sampling endpoint. (C) Number of unique and unambiguously identifiable tRNA fragments detected with each method at a threshold of >5 rpm. Values represent the average ± standard deviation of 3–4 technical replicates per method. Letters represent groups that are significantly different from each other with a Tukey corrected P-value < 0.0001. (D) Composition of tRNA fragment categories identified by each method represented as an average of 3–4 technical replicates. Categories were defined as follows: 3′ and 5′ tRNA-haves are fragments that terminate or begin at the known angiogenin cleavage sites and contain the rest of the 5′ or 3′ end of the tRNA. 5′ tRNA fragments (5′-tRFs) begin a the 5′ end of the tRNA but end either before or after the angiogenin cleavage sites. 3′ tRNA fragments (3′-tRFs) begin either before or after the angiogenin cleavage sites and end within the CCA tail of the 3′ terminus of the tRNA. Internal tRNA fragments (i-tRF) are fragments that begin after the 5′ terminus and end before the CCA tail of the 3′ end of the tRNA.
In addition, the randomized splint method detected more unique tRFs than the other methods (Figure 4C, corrected P < 10−4 in all comparisons and Supplementary Figure S6C). The overall profile of the tRFs detected using the randomized splint method was notably different from the other methods tested, mainly due to the large number (44% of all detected species) that were only detected by the splint method (Supplementary Figure S6D). The randomized splint method detected a greater number of tRF species in all categories but in particular had a much richer sampling of shorter 5′ and 3′-tRFs (Figure 4D).
Randomized splint ligation based method improves detection of 2′-O-methyl modified RNA
The 2′OMe modification at the 3′ end of sRNA has posed challenges for library preparation. We compared the ligation efficiency between single-stranded ligation and randomized splint ligation methods using a 5′-FAM labeled degenerate RNA oligos which either contained a 2′OMe modified ribonucleotide at the 3′ terminus or a normal ribonucleotide (Figure 5A). We found that the 2′OMe modification caused a decrease in ligation efficiency compared to the unmodified substrate in both the single-stranded and randomized splint ligations (Figure 5B). However, randomized splint ligation was much more efficient on both substrates (Figure 5C, corrected P < 10−4 in all comparisons). Furthermore, we found that splint ligation significantly decreased the impact of the 2′OMe modification (1.9-fold less efficient compared to 5.7-fold less efficient with single-stranded ligation; interaction P-value = 9.88 × 10−9).
Figure 5.
Ligation and sequencing of 2′OMe modified RNA. (A) Schematic of the CE assay. A FAM labelled degenerate RNA oligo containing either a normal ribonucleotide or a 2′OMe modified at the 3′ terminus was used as a substrate for ligation reactions using single-stranded or randomized splint methods. (B) Example CE traces. Shorter unligated substrate runs faster at around 18 bp, while the longer ligation products run slower at >35 bp. (C) Percentage of the FAM labeled oligo that was converted to the ligation product. Values represent the mean of four replicates ± the standard deviation and letters represent groups that are significantly different from each other with a Tukey corrected P-value < 0.0001. (D) Normalized number of 2′OMe spike-in reads plotted on a logarithmic scale. Each miRNA was expected to have a normalized read value of 1 (central dashed line). The upper and lower dashed lines correspond to the interval of 2-fold over- or under-represented. Asterisks denote groups where the normalized read counts of the unmethylated oligos was significantly different from the methylated oligos at a Tukey corrected P-value of < 0.05.
Next we compared the ability of different library preparation methods to capture 2′OMe modified RNA in a background of total RNA. We spiked a mixture of 2′OMe modified synthetic oligos and their unmodified counterparts into total human brain RNA. The 2′OMe spike-in mix contained six pairs of oligos; oligos in each pair were of the same sequence except for a non-structural single nucleotide polymorphism to allow for mapping. Oligos in each pair contained one with a 2′OMe modified 3′ terminal nucleotide and the other with a normal nucleotide. We found that the methods varied with respect to their ability to detect the 2′OMe modified RNA (Figure 5D). TruSeq had the lowest detection, followed by NEBNext and NEXTflex, while the randomized splint method had the best detection. Comparing unmethylated and methylated oligos within each method, we found significantly reduced sensitivity for 2′OMe in all methods except the randomized splint method (corrected P-values < 0.05 in all methods except randomized splint).
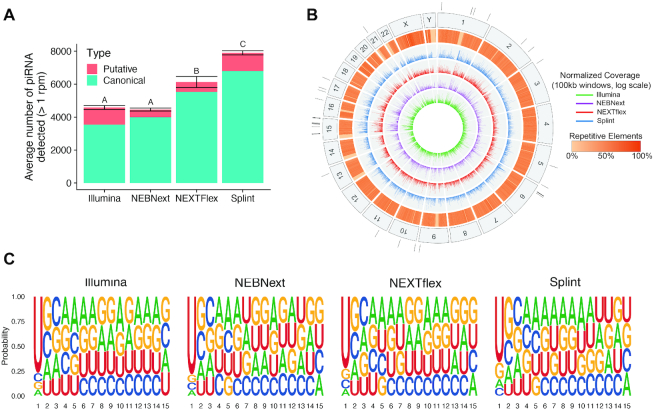
Furthermore, we compared the sensitivity of different library preparation methods to detect naturally occurring sRNAs containing the 2′OMe modification. First, we sequenced human testes sRNAs and mapped the piRNAs. We found that the randomized splint method detected the highest number of the unique species of piRNA, followed by NEXTflex while NEBNext and TruSeq were not significantly different from each other in detection (Figure 6A and Supplementary Figure S6E). We found that the piRNA mapped to repeat containing regions of the genome and exhibited similar genomic clustering patterns across methods (Figure 6B), while expression levels of individual piRNAs varied among methods (Supplementary Figure S6F). Furthermore, we found a prominent 5′ U bias in the sequences identified with each method, which is a feature of piRNA related to their biogenesis (Figure 6C). We also sequenced total RNA from a plant, Arabidopsis thaliana. Most plant miRNAs naturally contain the 2′OMe modification. We found that the randomized splint method detected significantly more miRNAs compared to the NEBNext method (Supplementary Figure S8, P < 1 × 10−3, other methods were not tested).
Figure 6.
piRNAs detected in human testes. (A) Number of piRNA detected at an RPM > 1, by each method in human testes. Values represent the average ± standard deviation of 2–4 technical replicates per method. Letters represent groups that are significantly different from each other with a Tukey corrected P-value < 0.001. (B) Circos plot representing piRNA mapping on the human genome. The outermost track represents the human chromosome numbers with black tick marks showing the locations of piRNA clusters identified in at least three of the methods. The inner five tracks were broken up into 100 kb non-overlapping windows. The second outermost track represents the percentage of each window containing repetitive elements such as retrotransposons. The 4 innermost tracks represent the number of piRNA mapping within each window, normalized by the number of times they mapped to the genome and plotted on a log scale. Each method is represented by its own track with the Illumina as the innermost track, followed by NEBNext, NEXTflex and randomized splint. (C) SeqLogo plots showing the sequence motifs for the piRNA species detected by each method.
Randomized splint ligation based method detects more differentially expressed miRNA
Finally, we tested our method in miRNA differential expression. We obtained archived total RNA samples from tumors and adjacent normal tissue from the same donor. Libraries were constructed using four matched pairs from stomach, lung, kidney and breast tissue using the randomized splint and NEBNext methods. In all cases we found that the randomized splint method detected more miRNA than the NEBNext method (Table 1). Furthermore, the randomized splint method found more differentially expressed miRNA in most cases, except for the breast tissue samples where we detected the same number of differential expressed genes using both methods.
Table 1.
Differential expression of miRNAs. miRNAs were compared between tumor and adjacent normal tissue in four human donors. Each sample was assayed using the randomized splint workflow and the NEBNext workflow in duplicate
| Tissue | Biochain Lot number | Donor age | Donor sex | Technique | Tissue type | Average number of miRNA detected (rpm > 5) | Number differentially expressed (probability > 0.9) | Shared differential expression |
|---|---|---|---|---|---|---|---|---|
| Stomach | A612105 | 51 | M | NEBNext | Normal | 302.0 | 45 | 38 |
| Tumor | 333.0 | |||||||
| Randomized Splint | Normal | 441.0 | 57 | |||||
| Tumor | 479.0 | |||||||
| Lung | B501175 | 67 | M | NEBNext | Normal | 315.0 | 22 | 18 |
| Tumor | 328.0 | |||||||
| Randomized Splint | Normal | 404.5 | 73 | |||||
| Tumor | 476.5 | |||||||
| Kidney | A610274 | 2 | M | NEBNext | Normal | 349.0 | 72 | 53 |
| Tumor | 349.5 | |||||||
| Randomized Splint | Normal | 504.0 | 86 | |||||
| Tumor | 496.0 | |||||||
| Breast | B610021 | 56 | F | NEBNext | Normal | 330.5 | 29 | 15 |
| Tumor | 332.5 | |||||||
| Randomized Splint | Normal | 447.5 | 29 | |||||
| Tumor | 464.0 |
DISCUSSION
sRNAs are important regulators of gene expression which present unique challenges in quantification due to their size. Low bias and highly sensitive methods are necessary to provide accurate and complete information in small RNA sequencing. In this study we designed a novel randomized splint ligation-based workflow which improves many long-standing challenges in the field. Randomized splint ligation is a highly efficient and low bias process, requiring only transient hybridization to the degenerate extension, as opposed to more complex cofolding interactions necessary for single-stranded ligation reactions. High ligation efficiency is important to increase yield and allow for low input samples. With our optimized workflow we found that randomized splint ligation could be used with inputs as low as 1 ng of total RNA and generally required fewer PCR cycles to achieve the same yield as the other commercially available methods and does not require gel-purification.
We further showed that our randomized splint ligation workflow has lower bias and higher sensitivity compared to the commercially available kits. In our tests using 962 equimolar synthetic miRNAs, the randomized splint method detected by far the most targets within 2-fold of their expected value, indicating that it has minimal sequence bias. Further, it was the only method tested to detect all sequences in the synthetic library. The bias was much lower than any other commercially available methods, even compared with the NEXTflex kit which also makes use of degenerate nucleotides and is frequently cited as the best low-bias method currently available (16,18). Furthermore, in our tests comparing each method to qPCR, randomized splint ligation detected the most miRNAs that were also detected by qPCR and had the best correlation to the qPCR data. In our testing on human brain RNA, we found that randomized splint ligation detected more miRNA than any other method.
Bias in miRNA detection is important for several reasons. During miRNA maturation, the double stranded precursor miRNA is cleaved, and one strand is loaded on to the Argonaut, while the passenger strand is degraded. For annotation in databases, the proposed functional strand is mainly determined by the relative sequencing depth of the two species (41). Accurate quantification of the relative abundance of each is critical to make that determination and biased methods may not detect it correctly, leading to incorrect annotations. Furthermore, it is possible for the passenger strand to become functional in a poorly understood process known as ‘arm-switching’ (21). A low-bias sequencing method will enable comprehensive genome-wide studies of arm switching and corrections of functional annotations in existing miRNA databases.
The most common use-case for sRNA sequencing is to look for differential expression between samples. We examined the performance of our method in this type of experiment using archived matched tumor samples. Because we have minimal information on the patients and we do not have biological replicates of the tumors, we can’t evaluate the biological significance of our results. However, we do have technical replicates and can compare between different techniques done on the same samples. We found that the randomized splint method detected more miRNA than the NEBNext method and importantly more differentially expressed miRNA between the matched tumor and normal tissue. Although many of the differentially expressed genes were found by both methods, having a low-bias method permits accurate quantitation of the expression levels. This is important for ranking possible therapeutic targets and avoids losing targets that are not captured correctly by biased methods.
We found that our method detected a much wider diversity of tRNA fragments than any other method. In particular, we detected a much larger diversity of short tRFs, falling into the category of 5′-tRFs and 3′-tRFs. While the functional relevance of these fragments is not well understood, our method will enable investigations into their targets and role in cellular physiology and disease that may not be possible with other methods that don’t detect them well. In all methods, we detected very few tRNA half species. This is likely because all of the surveyed methods require a free 3′ hydroxyl group and a 5′ phosphate group in the targets in order for ligation to occur. tRNA halves are mainly generated during stress induced cleavage by angiogenin. Angiogenin cleavage results in a 2–3′ cyclic phosphate on the 3′ end of the 5′ cleavage product and a 5′ hydroxyl on the 3′ cleavage product, thus we would not detect them unless these products were repaired by cellular kinases or phosphatases prior to library construction (42). Future studies could employ an end repair strategy using T4 polynucleotide kinase or a similar enzyme to better capture these species.
Finally, we have shown that our method is particularly well suited to studies of 3′ terminal 2′OMe modified sRNA, such as plant miRNA and piRNA. The randomized splint ligation significantly relieves the impairment in ligation efficiency caused by the modification, perhaps by forming a more favorable structure for the enzyme to complete the phosphodiester bond formation. Munafó and Robb (22) showed that increasing incubation time and enzyme concentration can improve the ligation efficiency in single-stranded ligations, it is possible that the same strategies could be used with our workflow to optimize the recovery of this class of sRNAs even more. Our technique will significantly aide in discovery and quantification of plant miRNAs which are significantly underrepresented in miRbase, probably due to structural and modification based biases that lead to large numbers of miRNA not being represented in sequencing datasets (43). Furthermore, our method will be useful for the study of piRNA. As we demonstrated in this study, we were able to identify a higher diversity of piRNA than any other method. The piRNA we identified showed the characteristic clustering patterns and 5′ uridine bias related to their biogenesis and feed-forward amplification mechanism (31,44).
In conclusion, we have developed a novel method that significantly improves the accuracy and sensitivity of sRNA library preparations. Our method is particularly well suited for low-input samples and 2′OMe modified sRNAs which have been challenging to study in the past and will therefore provide new insights into the biology of sRNAs.
DATA AVAILABILITY
Sequencing data have been uploaded to the NCBI sequencing reads archive (SRA) and are available under project accession: PRJNA603337. qPCR data are available as a supplementary data file.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank their NEB colleagues William Jack, Tom Evans, Eileen Dimalanta, Brittany Sexton and Jennifer Ong for their helpful discussions and critical feedback on the manuscript. We also thank Laurie Mazzola and Danielle Fuchs for performing capillary electrophoresis. We are grateful to Donald Comb and Rich Roberts for their research support.
Contributor Information
Sean Maguire, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Gregory J S Lohman, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Shengxi Guan, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs, Inc. Funding for open access charge: New England Biolabs, Inc.
Conflict of interest statement. Authors are employees of New England Biolabs, Inc. New England Biolabs is a manufacturer and vendor of molecular biology reagents, including several enzymes and buffers used in this study. This affiliation does not affect the authors’ impartiality, adherence to journal standards and policies, or availability of data. New England Biolabs has filed a patent application based on the inventions in this paper. Subjects of this paper may be potential products of New England Biolabs.
REFERENCES
- 1. Ghildiyal M., Zamore P.D.. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009; 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keam S., Hutvagner G.. tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life. 2015; 5:1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bottani M., Banfi G., Lombardi G.. Circulating miRNAs as diagnostic and prognostic biomarkers in common solid Tumors: Focus on lung, breast, prostate cancers, and osteosarcoma. J. Clin. Med. 2019; 8:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kumar P., Anaya J., Mudunuri S.B., Dutta A.. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014; 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shigematsu M., Kirino Y.. tRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul. Syst. Biol. 2015; 9:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mestdagh P., Hartmann N., Baeriswyl L., Andreasen D., Bernard N., Chen C., Cheo D., D’Andrade P., DeMayo M., Dennis L. et al.. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods. 2014; 11:809–815. [DOI] [PubMed] [Google Scholar]
- 7. Hafner M., Renwick N., Brown M., Mihailović A., Holoch D., Lin C., Pena J.T.G., Nusbaum J.D., Morozov P., Ludwig J. et al.. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011; 17:1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jayaprakash A.D., Jabado O., Brown B.D., Sachidanandam R.. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011; 39:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhuang F., Fuchs R.T., Sun Z., Zheng Y., Robb G.B.. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012; 40:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raabe C.A., Tang T.-H., Brosius J., Rozhdestvensky T.S.. Biases in small RNA deep sequencing data. Nucleic Acids Res. 2014; 42:1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fuchs R.T., Sun Z., Zhuang F., Robb G.B.. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015; 10:e0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baran-Gale J., Kurtz C.L., Erdos M.R., Sison C., Young A., Fannin E.E., Chines P.S., Sethupathy P.. Addressing bias in small RNA library preparation for Sequencing: a new protocol recovers MicroRNAs that evade capture by current methods. Front. Genet. 2015; 6:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McLaughlin L.W., Romaniuk E., Romaniuk P.J., Neilson T.. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur. J. Biochem. 1982; 125:639–643. [DOI] [PubMed] [Google Scholar]
- 14. Song Y., Liu K.J., Wang T.-H.. Elimination of ligation dependent artifacts in T4 RNA ligase to achieve high efficiency and low bias MicroRNA capture. PLoS One. 2014; 9:e94619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Lee J.E., Riemondy K., Anderson E.M., Yi R.. High-efficiency RNA cloning enables accurate quantification of miRNA expression by deep sequencing. Genome Biol. 2013; 14:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorefan K., Pais H., Hall A.E., Kozomara A., Griffiths-Jones S., Moulton V., Dalmay T.. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012; 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wulf M.G., Maguire S., Humbert P., Dai N., Bei Y., Nichols N.M., Corrêa I.R., Guan S.. Non-templated addition and template switching by MMLV-based reverse transcriptases co-occur and compete with each other. J. Biol. Chem. 2019; 294:18220–18231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dard-Dascot C., Naquin D., d’Aubenton-Carafa Y., Alix K., Thermes C., van Dijk E.. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics. 2018; 19:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heinicke F., Zhong X., Zucknick M., Breidenbach J., Sundaram A.Y.M., Flåm S.T., Leithaug M., Dalland M., Farmer A., Henderson J.M. et al.. Systematic assessment of commercially available low-input miRNA library preparation kits. RNA Biology. 2020; 17:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wright C., Rajpurohit A., Burke E.E., Williams C., Collado-Torres L., Kimos M., Brandon N.J., Cross A.J., Jaffe A.E., Weinberger D.R. et al.. Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genomics. 2019; 20:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim H., Kim J., Kim K., Chang H., You K., Kim V.N.. Bias-minimized quantification of microRNA reveals widespread alternative processing and 3′ end modification. Nucleic Acids Res. 2019; 47:2630–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Munafó D.B., Robb G.B.. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010; 16:2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raine A., Manlig E., Wahlberg P., Syvänen A.C., Nordlund J.. SPlinted ligation adapter tagging (SPLAT), a novel library preparation method for whole genome bisulphite sequencing. Nucleic Acids Res. 2017; 45:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu J., Dai W., Wu L., Wang J.. SALP, a new single-stranded DNA library preparation method especially useful for the high-throughput characterization of chromatin openness states. BMC Genomics. 2018; 19:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gansauge M.T., Gerber T., Glocke I., Korlević P., Lippik L., Nagel S., Riehl L.M., Schmidt A., Meyer M.. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. 2017; 45:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 27. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loher P., Telonis A.G., Rigoutsos I.. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 2017; 7:41184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ray R., Pandey P.. piRNA analysis framework from small RNA-Seq data by a novel cluster prediction tool - PILFER. Genomics. 2018; 110:355–365. [DOI] [PubMed] [Google Scholar]
- 32. Lakshmi S.S., Agrawal S.. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008; 36:D173–D177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wagih O. ggseqlogo: A ‘ggplot2’ extension for drawing publication-ready sequence logos. Bioinformatics. 2017; 33:3645–3647. [DOI] [PubMed] [Google Scholar]
- 34. Neph S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K., Rynes E., Maurano M.T., Vierstra J., Thomas S. et al.. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012; 28:1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krzywinski M., Schein J., Birol İ., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cui Y., Chen X., Luo H., Fan Z., Luo J., He S., Yue H., Zhang P., Chen R.. BioCircos.js: an interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics. 2016; 32:1740–1742. [DOI] [PubMed] [Google Scholar]
- 38. Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A.. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011; 21:2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneider T.D., Stephens R.M.. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990; 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hittner J.B., May K., Silver N.C.. A Monte Carlo evaluation of tests for comparing dependent correlations. J. Gen. Psychol. 2003; 130:149–168. [DOI] [PubMed] [Google Scholar]
- 41. Axtell M.J. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013; 19:740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lyons S.M., Fay M.M., Akiyama Y., Anderson P.J., Ivanov P.. RNA biology of angiogenin: current state and perspectives. RNA Biology. 2017; 14:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Axtell M.J., Meyers B.C.. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell. 2018; 30:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stein C.B., Genzor P., Mitra S., Elchert A.R., Ipsaro J.J., Benner L., Sobti S., Su Y., Hammell M., Joshua-Tor L. et al.. Decoding the 5′ nucleotide bias of PIWI-interacting RNAs. Nat. Commun. 2019; 10:828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been uploaded to the NCBI sequencing reads archive (SRA) and are available under project accession: PRJNA603337. qPCR data are available as a supplementary data file.