Abstract
Marine flavobacteria possess dedicated Polysaccharide Utilization Loci (PULs) enabling efficient degradation of a variety of algal polysaccharides. The expression of these PULs is tightly controlled by the presence of the substrate, yet details on the regulatory mechanisms are still lacking. The marine flavobacterium Zobellia galactanivorans DsijT digests many algal polysaccharides, including alginate from brown algae. Its complex Alginate Utilization System (AUS) comprises a PUL and several other loci. Here, we showed that the expression of the AUS is strongly and rapidly (<30 min) induced upon addition of alginate, leading to biphasic substrate utilization. Polymeric alginate is first degraded into smaller oligosaccharides that accumulate in the extracellular medium before being assimilated. We found that AusR, a GntR family protein encoded within the PUL, regulates alginate catabolism by repressing the transcription of most AUS genes. Based on our genetic, genomic, transcriptomic and biochemical results, we propose the first model of regulation for a PUL in marine bacteria. AusR binds to promoters of AUS genes via single, double or triple copies of operator. Upon addition of alginate, secreted enzymes expressed at a basal level catalyze the initial breakdown of the polymer. Metabolic intermediates produced during degradation act as effectors of AusR and inhibit the formation of AusR/DNA complexes, thus lifting transcriptional repression.
INTRODUCTION
Polysaccharides account for up to 50% of the dry weight of marine micro- and macroalgae (1–3). Consequently, polysaccharides abound in marine organic matter, comprising up to 50 and 25% of high molecular weight compounds in surface and deep waters, respectively (4). Heterotrophic microorganisms make this pool of organic matter accessible to higher trophic levels via the microbial loop and therefore control the fluxes of carbon and energy between compartments of the ecosystem (5). Furthermore, they are a rich source of enzymatic tools for algal biomass valorization. Marine Flavobacteriia (phylum Bacteroidetes) are recognized as key players for the degradation of polysaccharides (6–8). This specialization relies on dedicated genomic regions called Polysaccharide Utilization Loci (PULs) that encode a suite of proteins necessary for the assimilation of a specific polysaccharide (9). This includes (i) carbohydrate-active enzymes for the breakdown of the substrate, (ii) modification enzymes catalyzing the removal of substituents (e.g. acetylases, sulfatases), (iii) membrane proteins favoring binding of the substrate to cell surface, (iv) transporters for oligosaccharides and (v) transcriptional regulators that control the PUL expression depending on substrate availability. This organization was first described in the gut symbiont Bacteroides thetaiotaomicron, with the characterization of the Starch Utilization System (Sus) that became the paradigm for PUL systems (10,11). Numerous PULs have subsequently been described in intestinal Bacteroidetes. They target food-derived terrestrial plant polysaccharides such as cellulose, xyloglucans, rhamnogalacturonans and fructans as well as host-derived polysaccharides (12–17). More recently, the biological and ecological importance of PULs from marine flavobacteria has been increasingly recognized. Several PULs targeting marine polysaccharides have been thoroughly characterized, including PULs targeting alginate and laminarin from brown algae (18,19), agars and carrageenans from red algae (20–22) and ulvans from green algae (23).
Expression of all genes from a PUL is co-regulated and tightly controlled by the presence of the substrate. This is achieved by transcriptional regulators encoded within or in the vicinity of the PUL (9). Predicted PULs encompass a large diversity of regulators from different families, including hybrid two-component systems (HTCS), extra-cytoplasmic functioning (ECF) family of σ/anti-σ factors and LacI, CRP, AraC (non-HTCS), SARP-OmpR and GntR-like proteins (24). However, the regulatory mechanisms have only been characterized in a few PULs from intestinal Bacteroidetes. The first identified PUL regulator was SusR from the Sus system in B. thetaiotaomicron. It encodes an inner membrane-spanning receptor that binds starch-derived oligosaccharides and controls the expression of other sus genes (25). Subsequent studies highlighted different regulatory strategies depending on the PUL. In B. thetaiotaomicron, the expression of the fructan PUL is controlled by a fructose-binding HTCS (15). Arabinan utilization is regulated by a HTCS transcriptional activator that responds to arabinan-derived oligosaccharides and a NrtR-family transcriptional repressor AraR that responds to arabinose (26,27), while utilization of host mucosal glycans is controlled by ECF-σ transcription factors (16). Furthermore, integration of several regulatory pathways allows a fine-tuned interpretation of complex glycan signals, leading to substrate prioritization and coordinated regulation of other cellular responses (28–30). By contrast, PUL regulatory pathways are unknown in marine Bacteroidetes. Previous studies have shown that the expression of marine PULs is induced by their respective substrates, both in cultured isolates (18–19,22,31–32) and in field experiments (33–35). This underlines the importance of hitherto underexplored regulatory mechanisms that allow cells to adapt their metabolism to the complex, rapidly changing glycan landscape in marine environments.
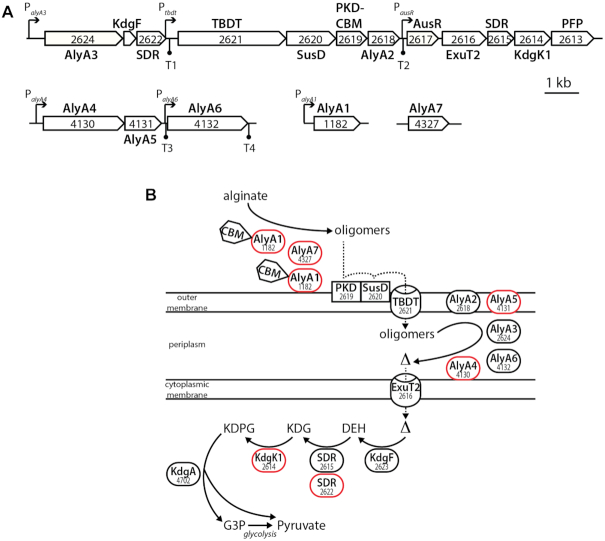
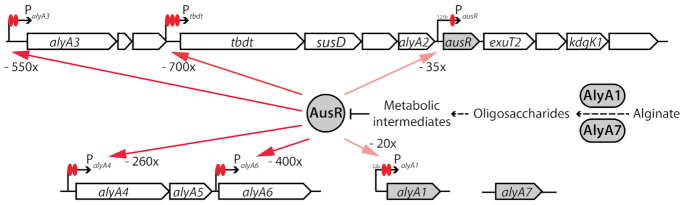
Zobellia galactanivorans DsijT is a marine heterotrophic flavobacterium that has become a model organism to study PUL-mediated polysaccharide degradation, complementing the knowledge primarily based on gut Bacteroidetes. Originally isolated from red seaweed in Roscoff, Brittany, France (36), it degrades a large set of complex polysaccharides such as agars and carrageenans from red algae, as well as alginate, laminarin and fucoidans from brown algae (37,38). Z. galactanivorans DsijT features several adaptive traits for interaction with macroalgae, including formation of biofilms, consumption of algal exudates, resistance to algal defense as well as 50 predicted PULs (38). Several of these PULs have been biochemically characterized (18,20,22). One of the best-studied PUL in Z. galactanivorans targets alginate, a major anionic cell wall polysaccharide from brown algae made of β-D-mannuronate and α-L-guluronate. In Z. galactanivorans, the Alginate Utilization System (AUS) is encoded by two loci transcribed as polycistronic mRNAs and two genes isolated in the genome (Figure 1A) (18). The first and larger locus (zgal_2624-2613) is centered on genes encoding a TonB-dependent transporter (TBDT) and a SusD-like protein, which are considered hallmarks of archetypal PULs (39). It notably encodes an uncharacterized transcriptional regulator AusR of the GntR family (zgal_2617). Members of this family are widespread regulators that undergo conformational changes upon binding of an effector molecule, leading to a change in DNA-binding properties and repression or activation of transcription (40). The current model of alginate degradation by Z. galactanivorans relies on biochemical characterizations and bioinformatic annotations (18,41) (Figure 1B). Alginate is initially depolymerized by the extracellular alginate lyases AlyA1 (polysaccharide lyase family PL7, with appended carbohydrate binding module of family CBM32) and AlyA7 (PL14) that release oligosaccharides with 4,5-unsaturation at the non-reducing end. Two surface-exposed proteins (ZGAL_2619 containing PKD and CBM domains, and the SusD-like protein ZGAL_2620) likely recruit the polysaccharide and/or oligosaccharides to the cell surface to facilitate the production of shorter oligomers by cell-bound alginate lyases. The TBDT ZGAL_2621 could import unsaturated oligosaccharides into the periplasm, where they are further degraded into unsaturated mono-uronic acid (Δ) by the alginate lyases AlyA2 (PL7), AlyA3 (PL17), AlyA4 (PL6), AlyA5 (PL7) and AlyA6 (PL6). Monomers Δ may enter the cytoplasm via the ExuT2 transporter (Major Facilitator Superfamily), where they are further converted into 2-keto-3-deoxy-6-phosphogluconate (KDPG) through the consecutive action of the ring-opening enzyme KdgF (42), two 2-dehydro-3-deoxy-D-gluconate 6-dehydrogenases (ZGAL_2615 and ZGAL_2622) and the 2-dehydro-3-deoxy-glucono-kinase KdgK1 (18). KDPG is eventually assimilated into the central metabolism through the Entner-Doudoroff pathway. We previously showed that the expression of Z. galactanivorans AUS is tightly controlled by the presence of alginate in the medium (18,32), but a mechanistic understanding of this regulation is still lacking.
Figure 1.
The AUS in Zobellia galactanivorans DsiJT. Gene numbers are indicated together with the names of the encoded proteins. (A) Genetic organization of the AUS. Potential promoter and Rho-independent terminator regions detected in a previous study (18) are indicated by arrows and circles, respectively. (B) Schematic representation of alginate metabolism based on gene annotation and functional studies. Enzymes (ovals), substrate-binding proteins (rectangles) and membrane transporters (barrels) are depicted. The activity of enzymes depicted in red has been experimentally validated in previous studies. CBM: carbohydrate-binding module; TBDT: TonB-dependent transporter; Δ: unsaturated mono-uronic acid; DEH: 4-deoxy-L-erythro-5-hexoseulose uronic acid; KDG: 2-keto-3-deoxy-gluconate; KDPG: 2-keto-3-deoxy-6-phosphogluconate; G3P: glyceraldehyde-3-phosphate.
In the present study, we investigated the fine-scale kinetics of alginate degradation and AUS expression by Z. galactanivorans DsijT, to better characterize the regulation processes. We further showed that the PUL-encoded AusR regulator represses AUS expression in the absence of alginate. This report constitutes the first characterization of the regulatory mechanism for a PUL in marine bacteria.
MATERIALS AND METHODS
Chemicals
Unless otherwise stated, all chemicals were from Sigma-Aldrich. Two sources of alginate were used for growth experiments, provided by Algaia (Satialgine S1600NS) or Danisco (Grindsted FD176). Estimation of molecular weight using Multi-Angle Laser Light Scattering (MALLS) showed Mw = 3.9 105 and 2.0 105, respectively. To prepare low molecular weight oligoalginates, Algaia alginate (4.5 g.l−1) was digested by the recombinant alginate lyase AlyA1 from Z. galactanivorans DsijT (41) and further ultrafiltered on a 10-kDa membrane. To obtain defined alginate trisaccharide degradation products, fractions of oligoguluronates (G blocks), oligomannuronates (M blocks) and mixed MG blocks were prepared by acid hydrolysis according to Haug et al. (43). G and MG blocks were degraded by the guluronate-specific alginate lyase AlyA1, and products were purified by size exclusion chromatography as described previously (41) to obtain ΔGG and ΔMG oligosaccharides, respectively (Δ: 4-deoxy-L-erythro-hex-4-enepyranosyluronate). M blocks were degraded by the mannuronate-specific alginate lyase Aly from Pseudomonas alginovora X017, and products were purified by size exclusion chromatography as described previously (44) to obtain ΔMM oligosaccharides. D-mannuronic acid sodium salt and L-guluronic acid sodium salt were from Carbosynth (Compton, UK).
Bacterial strains, plasmids and primers
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Escherichia coli strains were routinely grown in Luria-Bertani (LB) medium at 37°C, 180 rpm. Zobellia galactanivorans strains were routinely grown at 20°C in Zobell medium 2216E [per liter: 5 g tryptone, 1 g yeast extract, 800 ml filtered seawater, 200 ml distilled water (45)]. If needed, antibiotics were used at the following concentrations: ampicillin: 100 μg.ml−1; erythromycin: 50 μg.ml−1. A calibration curve was obtained to convert Z. galactanivorans OD600 measurements to bacterial biomass dry weight (Supplementary Figure S1), by drying 14 ml of bacterial suspension with known OD600 for 4 days at 60°C and weighing the dry cell residue on a digital scale (precision 10−4 g). Primers used in this study are listed in Supplementary Table S2.
Construction of ausR deletion mutant
A Z. galactanivorans deletion mutant for ausR was generated using the previously described method (46). A 2081-bp fragment spanning the first 30 bp of ausR and 2051 bp of upstream sequences was amplified from genomic DNA using primers OFT0015 and OFT0017. The fragment was digested using BamHI and PstI and ligated into pYT313 that had been digested with the same enzymes, to generate pFT7. A 2037-bp fragment spanning the final 42 bp of ausR and 1995 bp of downstream sequence was amplified using primers OFT0016 and OFT0018. The fragment was cloned into PstI and SphI sites of pFT7 to generate the ausR deletion construct pFT8, which was introduced into the wild-type Z. galactanivorans DsijT by conjugation from an E. coli S17-1 strain. Conjugants with plasmids integrated in the genome were isolated on Cytophaga-agar containing 50 μg.ml−1 erythromycin. Single erythromycin-resistant colonies were grown overnight in Cytophaga medium in the absence of antibiotics at 30°C. The cells in which a second recombination event resulted in loss of the plasmid were selected on Cytophaga-agar containing 5% sucrose. Isolated colonies were checked for erythromycin sensitivity. ausR deletion was confirmed by polymerase chain reaction (PCR) and sequencing on isolated colonies using primers OFT0019 and OFT0020.
Culture conditions for physiology experiments
The Z. galactanivorans strains were cultivated in liquid Zobell medium 2216E or marine mineral medium (MMM) supplemented with different substrates (4 g.l−1) as the sole carbon source (37). Pre-cultures in Zobell medium were rinsed with two volumes of sterile saline solution prior to inoculation. Cell density was monitored by measuring turbidity at 600 nm using a Spark Tecan spectrophotometer. Aliquots for RNA extraction were mixed with 0.5 vol. of ice-cold killing buffer (20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 20 mM NaN3) and centrifuged at 4200 rpm at 4°C for 3 min. Pellets were immediately frozen in liquid nitrogen and stored at −80°C until further processing. To monitor alginate degradation products, aliquots were centrifuged for 5 min at 13 000 rpm and supernatants were frozen at −20°C until analysis.
To test growth on jellified alginate, plates were prepared by adding 15 mM CaCO3 and 20 mM glucono-D-lactone to a 2% alginate FD176 solution in MMM (47). Plates were inoculated with 2 μl of cell suspensions (OD600 normalized to 1.7) deposited on the center and incubated at 25°C.
RNA extraction
Total RNA was extracted using phenol–chloroform extraction as in (22), resuspended in 30 μl of RNase-free water, treated with Turbo DNAse (Thermofisher) and purified using RNeasy Mini Kit (QIAGEN). Genomic DNA elimination was checked by PCR amplification on RNA samples using the OFT0007 and OFT0008 primers. RNA was quantified using a Qubit fluorometer, its purity from proteins (A260/A280) and residual chemical contaminants (A260/A230) was checked on a Nanodrop and its integrity was checked by gel electrophoresis or with Bioanalyzer.
Gene expression analysis
Gene expression was analyzed using RT-qPCR. cDNAs were synthesized with the ImProm-II Reverse kit Transcription System (Promega) using random hexamers, diluted to 0.5 ng.μl−1 eq. RNA and stored at −20°C. Quantitative PCR was performed following conditions described in (37), in 5 μl reactions containing 1× LightCycler 480 Sybr Green I Master (Roche), 400 nM of each primer and 2.1 μl of diluted cDNA sample. The expression of genes from the alginolytic system was quantified using 17 validated primer pairs (Supplementary Table S2) and normalized by the geometric mean of the reference genes gmkA, glyA and icdA (37). MIQE information related to RT-qPCR is gathered in Supplementary Table S3. Additional whole-genome transcriptomic results obtained on Z. galactanivorans DsijT were recovered from previously published, publicly available datasets (NCBI GEO accessions GSE99940 and GSE101142) (22,32).
Northern Blot analysis
One μg of total RNA was separated by electrophoresis for 4 h at 40 V on 1% agarose gel containing 2% formaldehyde. As a control of sample quantity and integrity, duplicate samples were run on the same gel and stained with ethidium bromide. Nucleic acids were transferred by capillarity to a nylon membrane (Hybond N+, Amersham) and cross-linked with UV (1 min, 125 mJ). A DIG-labeled ausR-specific DNA probe was constructed by PCR (PCR DIG Probe synthesis kit, Roche Applied Science), using 0.2 μM of primers zg_2617_fwN and zg_2617_rvN (Supplementary Table S2) and 10 ng of Z. galactanivorans genomic DNA. RNA was hybridized with 2 μl of probe (final concentration 1 ng.μl−1) for 16 h at 50°C and detected using anti-DIG/alkaline phosphatase antibody and CDP-Star (Roche Applied Science). The membrane was exposed for 30 min to a detection film (Kodak). Approximate sizes of the hybridizing mRNA were estimated using a 0.2–10 kb RNA ladder (RNA marker R7020, Sigma).
Monitoring of alginate degradation products
Sugar reducing ends were quantified with the ferricyanide assay (48) in a microtiter plate reader (TECAN Spark 10M) on 10 μl of sample using a glucose calibration curve (0.25–1.5 mM). Soluble unsaturated uronic acids were quantified by measuring A235 on samples diluted 1:4 with distilled water in a microtiter plate, using the molar extinction coefficient ϵ = 8500 l.mol−1.cm−1 (49). Size distribution profiles of products were determined by MALLS. Samples (0.1 ml, filtered at 0.45 μm) were separated on Shodex OH pak SB-805, 804 and 803-HQ columns connected in series (mobile phase LiNO3 0.1 M, flow rate 0.5 ml.min−1) and analyzed using a DAWN® HELEOS® II MALLS detector (Wyatt Technology). Generated data were analyzed with Astra software. Oligosaccharides were further analyzed with HPAEC coupled with pulsed amperometry, by injecting 20 μl of filtered sample (30 kDa cut-off) onto a Carbopac PA200 column (Thermofisher), with 100 mM NaOH as eluent (1 ml.min−1) and a gradient of sodium acetate up to 700 mM. Oligosaccharides produced by the Z. galactanivorans alginate lyase AlyA1 (41) were used as size standards.
Cloning, expression and purification of recombinant AusR
The ausR gene (zgal_2617) was amplified by PCR on Z. galactanivorans genomic DNA using the primers zg2617_fwC and zg2617_rvC. The PCR product (726 bp) was digested with BamHI and MfeI and cloned into the expression vector pFO4, followed by transformation into E. coli NEB5α. All cloning steps were performed as described in (50). After confirmation of successful cloning, plasmids were transformed into the E. coli BL21(DE3) expression strain. For protein production, 250 ml of ZYP autoinducible medium with ampicillin (51) were inoculated with 25 μl of overnight pre-culture in LB medium and incubated at 20°C, 150 rpm for 3 days. Cells were centrifuged for 20 min at 4°C, 5000 rpm and resuspended in 4 ml of buffer A (25 mM Tris–HCl, 400 mM NaCl, 100 mM imidazole, pH 7.5) containing 5U.μl−1 DNAse, 1 mg.ml−1 lysozyme and an anti-protease cocktail (Complete EDTA-free, Roche). Samples were lysed by sonication (GE-50 ultrasonic processor, 20 kHz, amplitude 55%, 3 × 10 s on ice) and centrifuged for 30 min at 4°C, 20 000 g. The supernatant was filtered at 0.45 μm and loaded on a 5 ml HisTrap FF column (GE Healthcare) charged with NiSO4 and equilibrated with buffer A. Proteins were eluted with a linear gradient between buffer A and buffer B (25 mM Tris–HCl, 400 mM NaCl, 1 M imidazole, pH 7.5) in 60 ml at 5 ml.min−1. Fractions showing the presence of the recombinant protein on SDS-PAGE (12% acrylamide) were pooled. Imidazole was removed using a PD-10 Sephadex G-25M desalting column (GE Healthcare), proteins were eluted in buffer C (25 mM Tris–HCl pH 7.5, 400 mM NaCl) and concentrated by centrifugation on Amicon Ultra-0.5 10 kDa (Millipore). Protein concentration was determined by Bradford assay using BSA as a standard.
Electrophoretic Mobility Shift Assay (EMSA)
Cy-5 labeled DNA fragments covering the PalyA3 (201 bp) and PausR (211 bp) promoters were amplified by PCR from Z. galactanivorans DsijT genomic DNA with the Q5 High-Fidelity DNA polymerase (New England Biolabs) using primer pairs listed in Supplementary Table S2. PCR reactions were purified using QIAquick PCR Purification kit (QIAGEN). Sixty fmol of Cy-5 labeled probe were incubated for 30 min at 20°C with increasing concentrations of purified AusR in 10 μl of binding buffer (20 mM Tris-HCl pH 7.5, 2 mM DTT, 5% glycerol, 2 mM MgCl2, 14 μg.ml−1 polyd(I-C), 200 μg.ml−1 BSA). Samples were analyzed by electrophoresis on 5% native polyacrylamide gels in 0.5X Tris-Glycine buffer for 50 min at 170 V and 4°C. Gels were scanned on a Typhoon Imager (Amersham) with automatic PMT settings and images were analyzed using ImageQuant TL 1D v8.1. The fraction of free DNA was fitted to the Hill equation using non-linear least square model in R v3.5.0, as follows:
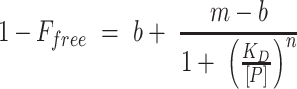 |
where Ffree is the fraction of free DNA, [P] is the protein concentration in binding reaction, KD is the apparent dissociation constant, n is the Hill coefficient, and m and b represent the fraction of bound DNA at the upper and lower asymptotes of the titration (52). To test the potential effectors of AusR-DNA interaction, the protein was incubated with 100 nmol D-glucose, potassium gluconate, alginate trisaccharides, D-mannuronate or L-guluronate for 15 min at 20°C, before addition of the DNA fragments (60 fmol) and further incubation for 30 min at 20°C.
Bioinformatic analysis
The AusR protein sequence was analyzed using InterProScan (53), ProtParam (https://web.expasy.org/protparam/) and BLAST against the UniprotKB/Swiss-Prot and PDB databases. Cellular localization was predicted using PSORTb v3.0 (54). Ungapped motifs were searched upstream all AUS genes from Z. galactanivorans DsijT and 12 related flavobacterial species (Supplementary Table S4), using MEME v5.1.0 (55), with a motif site distribution of zero or one site per sequence and a motif width >10 and <50. The detected motif was further searched in the entire Z. galactanivorans DsijT genome using FIMO (56). The RNAfold webserver was used to predict the optimal secondary structure of the 230-bp intergenic region zgal_2617-2616, using minimum free energy prediction (57).
RESULTS
Alginate degradation follows a biphasic kinetics
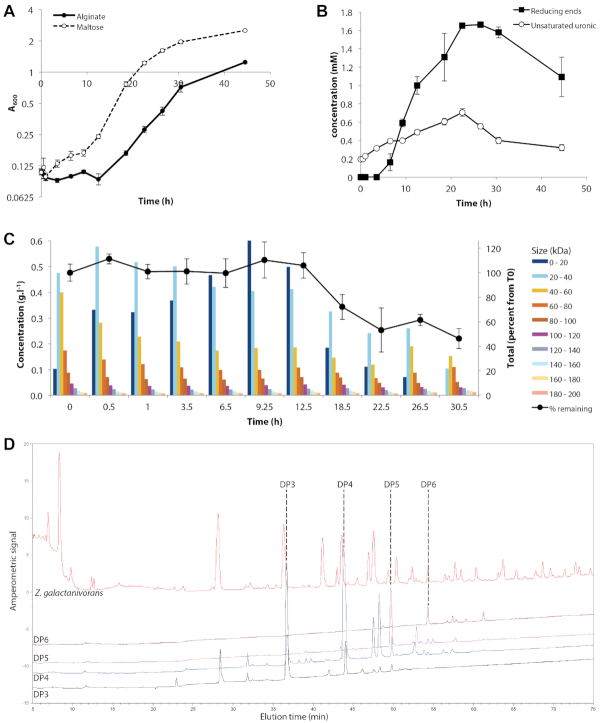
We followed the growth of Z. galactanivorans DsijT in minimum medium supplemented with either alginate or maltose as sole carbon and energy source (Figure 2). A longer lag-phase was observed with alginate, compared to maltose (ca. 12.5 and 1 h, respectively) (Figure 2A). Cell growth rate was 20% lower with alginate compared to maltose (0.113 ± 0.002 and 0.141 ± 0.003 h−1, respectively). The final cell density was twice as high with maltose compared to alginate and corresponded to 2.068 and 1.039 grams of dry bacterial biomass per liter for maltose and alginate-supplemented cultures, respectively (conversion factor 0.9555, see Supplementary Figure S1). Assuming a carbon / dry weight ratio of 0.5 in bacterial biomass (58) and knowing maltose and sodium alginate carbon content of 42% and 33.3%, respectively, we estimated that 62% and 40% of carbon from maltose and alginate were used for biomass production, respectively.
Figure 2.
Monitoring the growth of Zobellia galactanivorans on alginate. (A) Growth at 20°C, 180 rpm in minimum medium supplemented with either maltose or alginate FD176 as a sole carbon source (mean ± s.e.m., n = 4). (B, C, D) Analysis of soluble degradation products measured in spent medium of Z. galactanivorans grown in MMM supplemented with alginate FD176. (B) Concentration of soluble sugar reducing ends (expressed as glucose equivalent) and unsaturated uronic residues (mean ± s.e.m., n = 4). (C) MALLS analysis of degradation products. Bars denote the mean concentration (left axis) of products from different size fractions (from 0 to 200 kDa). Black circles denote the remaining percentage of total products detected compared to T0 (right axis, mean ± s.e.m., n = 4). (D) Representative HPAEC analysis of degradation products after 30.5 hours of cultivation, as compared to standards enriched in alginate tri-, tetra, penta- and hexa-saccharides.
Degradation products were monitored in alginate-fed cultures using biochemical assays (Figure 2B) and MALLS analysis (Figure 2C). The concentration of alginate oligosaccharides in spent medium was assessed by measuring both sugar reducing ends and unsaturated uronic residues specific to alginate degradation by lyases (Figure 2B). Oligosaccharides first accumulated in the medium, suggesting that their production exceeded consumption by the cells, followed by a steady decrease. They reached a maximum concentration after 22.5 h, in the middle of the exponential growth phase. MALLS analysis confirmed this biphasic assimilation pattern (Figure 2C). The total concentration of molecules detected by MALLS in spent medium remained stable during the lag-phase (until 12.5 h of cultivation) before decreasing during the exponential phase. MALLS also revealed changes in the alginate size profile along the degradation kinetics. The initial alginate substrate was mostly in the range 20–80 kDa (ca. 100–400 degree of polymerization, DP) with estimated average Mw = 58 kDa. Although the total concentration remained stable, changes in alginate size profile were detected as early as 0.5 h of cultivation. There was a gradual degradation of molecules >40 kDa into smaller products. This led first to an accumulation of products with intermediate size (20–40 kDa, peaking at 0.5h), followed by accumulation of smaller products (<20 kDa, peaking at 9.25 h) that were ultimately assimilated. Half of the initial substrate was consumed after 30.5 h. Molecules > 80 kDa appeared refractory to degradation, as their concentration only decreased from 0.2 to 0.16 g.L−1. Anion exchange chromatography showed that dimer and trimer oligoalginates were the main products accumulating in spent medium at the end of the cultivation, with additional contributions of tetra- and pentamers (Figure 2D).
A tight control allows a surge of AUS expression upon addition of alginate
We compared the expression of 17 genes from the AUS along the growth curve on alginate and maltose using RT-qPCR (Figure 3). In the control maltose-supplemented medium, the expression of most genes from locus 1 (zgal_2624-2613) and 2 (zgal_4130-4132) showed less than 4-fold variations compared to T0 until 30.5 hours. A transient increase was detected during the first hours, potentially due to cell manipulation during the change of medium or fast entry into exponential phase. After 44.5 hours with maltose, the more robust increase in expression levels could be due to cells scouting for other carbon sources in substrate-depleted medium.
Figure 3.
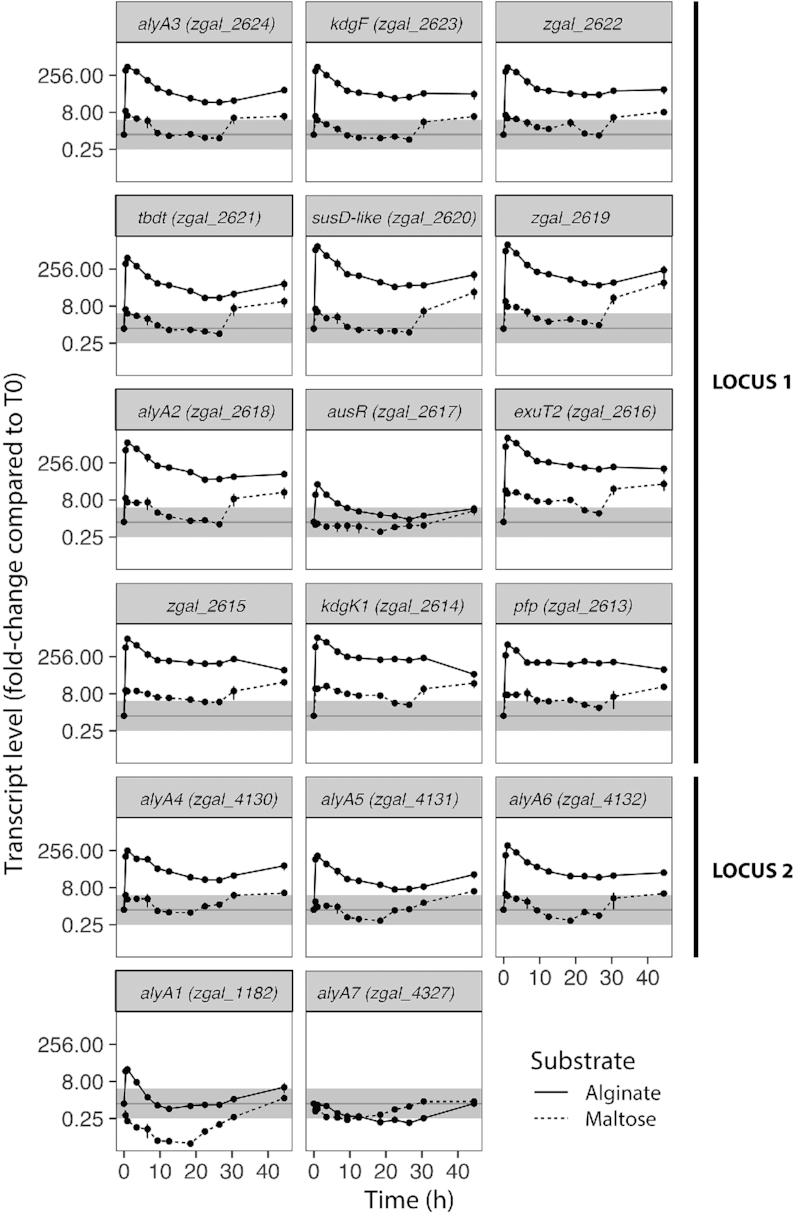
Expression of genes from the AUS after addition of maltose (dotted lines) or alginate FD176 (plain lines). Values are expressed as fold change compared to pre-cultures in Zobell medium used to prepare the inoculum (i.e. T0). Differential expression compared to T0 was considered significant for fold-change below 0.25 or above 4 (outside the gray zone). Values are mean ± s.e.m. (n = 4).
When cells were transferred from Zobell medium to alginate-supplemented medium, the transcription of all genes from locus 1 and 2 was first strongly and rapidly induced within 1 hour followed by a slow decrease, resulting in a surge in mRNA levels (Figure 3). This surge coincided with a 3-fold increase in the concentration of oligosaccharides < 20 kDa in the first 30 minutes (Figure 2C). Genes from both loci followed the same expression pattern, suggesting they are part of the same regulon. Genes downstream the promoters Ptbdt (zgal_2621-2618) and Paly6 (zgal_4132) were more expressed than the upstream part of their respective locus. This is likely a cumulative effect of both the activation of these promoters and of read-through expression from the upstream promoters PalyA3 and PalyA4, respectively. The gene ausR was the least induced gene of locus 1 (fold-change only 32). Altogether, this indicates that transcription can start from several promoters. This was confirmed by Northern blot analysis of an independent experiment (Supplementary Figure S2A). A probe targeting the ausR gene revealed two RNA species (770 and 950 bases) in the absence of alginate. Since ausR is a 729 bp ORF, the two products might correspond to the gene transcribed alone with different initiation or termination sites. The Northern blot profile changed for alginate-grown cells. The transcription of the 950 bases ausR transcript was favored in this culture condition. Moreover, new RNA species were detected. The larger transcript (9.5 kb) could correspond to a transcription initiated from Ptbdt and covering the region zgal_2621 to zgal_2616. The products of 5 kb and 2.6 kb could correspond to initiations from PausR but with different termination sites, or to differential mRNA decay (see Discussion). This is in line with the observation that the genes zgal_2616-2613 showed a much higher induction in the presence of alginate compared to ausR (Figure 3).
The genes alyA1 and alyA7 followed different expression patterns from those in loci 1 and 2 (Figure 3). The expression of alyA1 increased transiently (0.5–3.5 h, 32-fold) before returning to its initial level after 9 hours. By contrast, it was strongly down-regulated in the presence of maltose. The expression of alyA7 decreased both with alginate and maltose and was lowest in exponential phase, before returning to its initial level in stationary phase.
Role of AusR as regulator of the alginate utilization system
To explain the inducible expression of the AUS, we hypothesized that the PUL-encoded AusR protein is an essential regulator of alginate metabolism in Z. galactanivorans. ausR encodes a 27.6 kDa cytoplasmic protein comprising 242 amino-acids. InterProScan analysis predicted that AusR belongs to the GntR family of transcriptional regulators (59), featuring a N-terminal Helix-Turn-Helix DNA-binding domain (residues 12–80, IPR000524) and a C-terminal effector-binding domain of the FadR subfamily (residues 85–231, IPR008920). AusR best BLAST hit against the characterized protein database UniprotKB/Swiss-prot was LutR from Bacillus subtilis strain 168 (accession O07007, 31.6% sequence identity, e-value 2.10−23) that negatively regulates L-lactate utilization. The closest structurally characterized homolog to Z. galactanivorans AusR was a GntR-like regulator from Streptococcus agalactiae (PDB accession 6AZ6, 30.6% sequence identity, e-value 1.10−13) encoded in a genomic cluster dedicated to the degradation of glucuronated substrates that notably comprises genes for 2-keto-3-deoxy-gluconate kinase, β-D-glucuronidase and uronate isomerase (60).
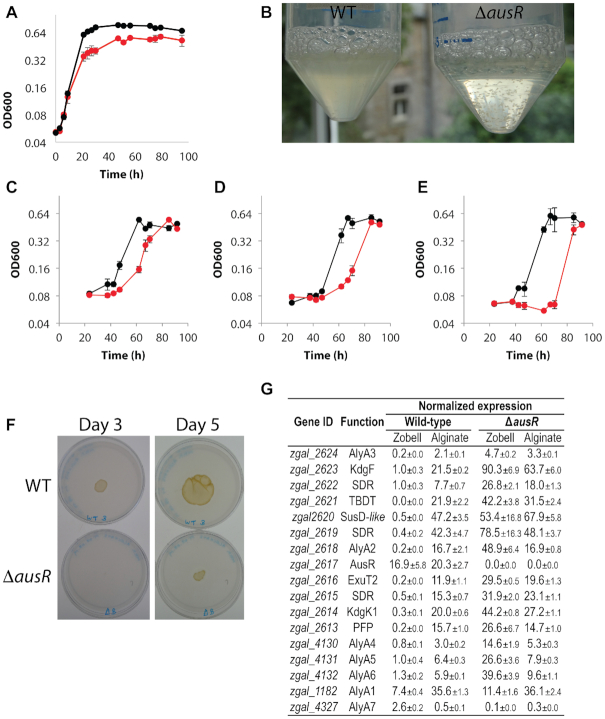
To assess the role of AusR as a regulator of alginate metabolism in Z. galactanivorans DsijT, we compared the growth of a ΔausR strain to the wild type (WT) strain. No growth delay or difference in growth rate could be detected in the absence of alginate in Zobell rich medium (Figure 4A). The formation of cell aggregates by Z. galactanivorans (ΔausR) in the stationary phase (Figure 4B) prevented accurate OD600 measurements in Zobell medium and explains the apparent lower final cell density. Growth was further tested in minimum medium containing alginate of different sizes as sole carbon source, ie. polymeric alginate of average Mw 3.9 105 (hereafter S1600NS, Figure 4C) or 2.0 105 (hereafter FD176, Figure 4D) and oligomers < 10 kDa (Figure 4E). In all three cases, growth of Z. galactanivorans (ΔausR) was delayed compared to WT. The length of the Z. galactanivorans (ΔausR) lag-phase was inversely proportional to the size of the alginate substrate (S1600NS, 42–47 h; FD176, 47–62 h; oligomers, 70–85 h), whereas it was below 40 h in all three conditions for WT. The deletion did not significantly affect doubling times measured on S1600NS alginate (WT, G = 6.9±0.9 h; ΔausR, G = 6.1 ±1.2 h; Student t-test t = -0.86, P = 0.45), FD176 alginate (WT, G = 7.0±0.7 h; ΔausR, G = 8.6 ±0.3 h; t = 3.32, P = 0.06) or oligomers (WT, G = 6.9±0.8 h; ΔausR, G = 5.6 ±1.2 h; t = -1.57, P = 0.21). Final cell density was similar in all conditions. No cell aggregates were formed in the stationary phase in alginate medium. The growth delay was confirmed on solid medium with alginate as sole carbon source (Figure 4F). The WT strain formed a liquefied patch within 3 days, while only faint growth and no liquefaction was detectable for the ΔausR strain. After 5 days, Z. galactanivorans (ΔausR) cells covered a surface 85% smaller than WT.
Figure 4.
Phenotype analysis of the Zobellia galactanivorans (ΔausR) deletion mutant. (A) Growth of WT (black) and mutant (red) strains in liquid Zobell medium at 25°C (mean ± s.e.m., n = 3). (B) Photograph of cultures in Zobell medium 9 h post-inoculation. (C, D, E) Growth of WT (black) and mutant (red) strains at 25°C in MMM supplemented with (C) alginate S1600NS (Mw = 3.9 105), (D) alginate FD176 (Mw = 2.0 105) or (E) oligoalginates < 10 kDa (mean ± s.e.m., n = 3). (F) Growth of equal amount of WT or ΔausR strain spotted on jellified alginate. Photographs show representative results of independent triplicate experiments. (G) Effect of ausR deletion on the expression of the AUS for cells in steady state in Zobell medium or alginate-supplemented minimum medium. Transcript levels were normalized against the geometric mean of glyA, icdA and gmkA. Values are mean ± s.e.m. of triplicate measurements.
We compared the expression of AUS genes in WT and ΔausR strains for cells in steady-state in Zobell medium or in alginate-complemented minimum medium (Figure 4G). For the WT strain in Zobell medium, alyA1 and alyA7 were the most expressed of the seven alginate lyase-encoding genes. Furthermore, expression was low for all genes of loci 1 and 2 except ausR, suggesting this gene could have a higher basal expression than the rest of the locus. To confirm these results, we compared the expression of ausR to that of upstream and downstream genes in Z. galactanivorans WT cells grown with 8 different mono- and polysaccharides as sole carbon sources (Supplementary Figure S3). In all cases except with alginate, ausR was more expressed than neighboring genes within the locus.
In Zobell medium, expression values were higher in Z. galactanivorans (ΔausR) compared to WT for all genes of loci 1 and 2 and for alyA1 (Figure 4G). This indicates that ausR represses the expression of these genes in the absence of alginate. Their expression was comparable between WT and ΔausR cells grown on alginate, showing that AusR was inactive in this condition. The deletion-induced derepression effect observed in Zobell was highest for genes within locus 1 (maximum 245-fold for alyA2) and only minor for alyA1 (1.5-fold). By contrast, alyA7 expression was 26-fold lower in Z. galactanivorans (ΔausR) compared to WT in Zobell medium, suggesting a different regulation mechanism. In Z. galactanivorans (ΔausR), the presence of alginate did not increase the expression of genes compared to the derepressed state in Zobell medium except for alyA1 and alyA7 that showed 3-fold higher expression with alginate compared to Zobell medium.
AusR binds to promoters within the AUS genomic regions
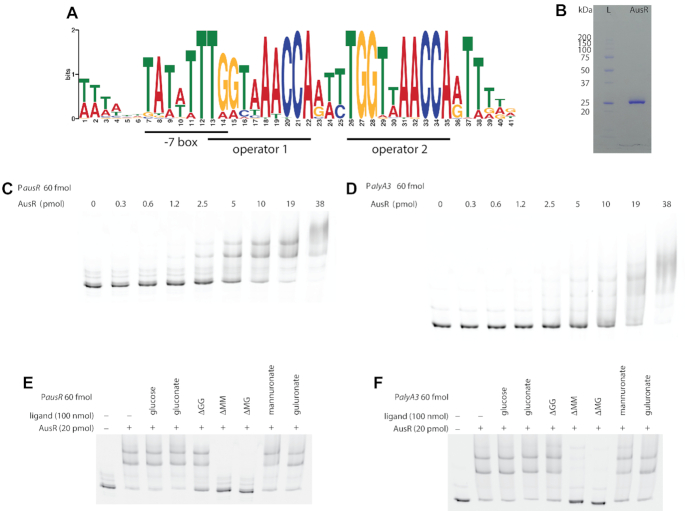
To precisely determine the AusR-mediated regulation mechanism, we searched for conserved DNA motifs upstream all genes of the AUS from Z. galactanivorans DsijT and 12 additional marine flavobacteria with public genomes (Supplementary Table S4). The MEME software highlighted the presence of a strong motif (Figure 5A, e-value 6.10−478). The detected DNA motif included the -7 box [TAnnTTTG] and sites that match the consensus operator sequence [NyGTNxACNy] found in the GntR family (61) and more specifically the consensus [TKGTN2–3ACMA] of the FadR subfamily to which AusR belongs (62). Pairs of this putative AusR operator motif were detected upstream alyA3, alyA4, alyA6 and alyA1 (Supplementary Figure S4A, D, E, F) and were separated by three bases. A third operator sequence was present in the promoter Ptbdt upstream zgal_2621 (Supplementary Figure S4B). A single canonical operator was present upstream ausR (Supplementary Figure S4C). Sequence alignment of the conserved DNA motifs showed that the palindromic structure of a second operator in PausR was lost by mutation of two guanines into adenines (Supplementary Figure S4H). No motif could be detected upstream alyA7. In promoters PalyA3, Ptbdt, PalyA4 and PalyA6, the putative operators overlapped the predicted -7 box. Binding of AusR to these operators would thus strongly hinder the recognition of the promoters by the RNA polymerase to initiate transcription. Larger distances between the -7 box and the operator were found for PalyA1 (32 bases) and PausR (125 bases). FIMO was used to scan the entire Z. galactanivorans DsijT genome for additional occurrences of the conserved motif. A single instance was found in the promoter of the gene zgal_2637 (Supplementary Figure S4G), suggesting it is part of the AusR regulon. This genes encodes the predicted gluconokinase GntK1. Z. galactanivorans possesses a second paralogous gluconokinase gene gntK2 (zgal_4757, 33% sequence identity) that lacks the operator sequence in its promoter. Transcriptomic analysis using public DNA microarray data (32) showed that the expression of gntK1, but not of gntK2, was induced two-fold in the presence of alginate compared to glucose (Supplementary Figure S5).
Figure 5.
AusR binds to promoters containing the predicted operator sequence. (A) Sequence logo of the conserved DNA motif detected upstream genes from the AUS. Individual sequences are shown in Supplementary Figure S4. (B) SDS-PAGE analysis of the purified recombinant His-tagged AusR protein. Lane L, molecular weight ladder. (C, D) Electrophoretic mobility shift assays of DNA fragments covering the promoters PausR (C) and PalyA3 (D) with increasing amount of purified His-tagged AusR (from 0 to 38 pmol). (E, F) Alginate trisaccharides prevent AusR binding to its promoters. EMSAs were performed with 20 pmol of purified His-tagged AusR and 60 fmol of Cy5-labeled PausR (E) or PalyA3 (F), in the absence of any ligand or with 100 nmol D-glucose, D-gluconate, ΔGG, ΔMM, ΔMG, D-mannuronate or L-guluronate. The first lane without AusR shows the mobility of free DNA. Δ: 4-deoxy-L-erythro-hex-4-enepyranosyluronate; M: D-mannuronate; G: L-guluronate.
We further tested whether AusR interacts directly with promoters containing the predicted operator sequence. The ausR gene was cloned into the pFO4 expression vector (N-terminal His-Tag fusion) and the recombinant protein was overexpressed in E. coli BL21(DE3) and purified to homogeneity by nickel ion affinity chromatography. The purified protein had an estimated mass of ca. 27 kDa on SDS-PAGE (Figure 5B), in line with the 28.4 kDa mass calculated based on His-tagged AusR sequence. Electrophoretic mobility shift assays (EMSAs) were conducted using purified His-tagged AusR and increasing concentrations of Cy5-labeled DNA covering fragments of PausR (Figure 5C) and PalyA3 (Figure 5D). In both cases, AusR bound the promoters in a concentration-dependent manner. Two PausR-AusR complexes were detected: a first complex was detected with 1.2 pmol AusR, while the second, more delayed complex, appeared only with AusR concentrations above 2.5 pmol. Similarly, three PalyA3-AusR complexes were detected. This could indicate cooperative interaction of DNA fragments with one or multiple monomers of AusR. Apparent dissociation constants KD estimated using gel densitometry analysis showed a higher in vitro affinity of AusR for PausR (KD = 587 nM) than for PalyA3 (KD = 1570 nM). Estimates of Hill coefficient were > 1 for both PausR and PalyA3 (n = 2.7 and 1.7, respectively), corroborating cooperative binding of AusR to these promoters. The relatively high molar excess of AusR needed to observe the first complex might be due to the presence of the His-tag on the purified protein used for EMSA, which could affect binding. To confirm that the direct interaction of AusR to the promoters was dependent on the predicted operator sequence, we performed EMSAs using DNA probes covering or excluding the predicted AusR operator of PausR (Supplementary Figure S6). Shorter versions excluding the operator showed only a very weak binding, with >95% of probes remaining as free DNA when incubated with 25 pmol purified AusR.
Identification of AusR effectors
We screened potential effector molecules that could mediate the interaction of AusR with PausR and PalyA3 (Figure 5E and F). Glucose, gluconate, mannuronate and guluronate had no effect on the binding of AusR to the promoters. By contrast, the alginate trisaccharides ΔMM and ΔMG strongly inhibited the interaction with both promoters. A slight effect could also be detected for ΔGG with PausR, where a higher proportion of probe remained as free DNA compared to the control without ligand. Therefore, alginate oligosaccharides are effectors of AusR.
DISCUSSION
Based on these results, we proposed a model for the control of alginate utilization by the AusR transcriptional regulator in Z. galactanivorans DsijT (Figure 6). AusR belongs to the GntR family, FadR subfamily of regulators and strongly represses the expression of most AUS genes by binding to specific operators in their promoters. Repression by AusR is lower for alyA1 and its own gene ausR. The expression of alyA7 appears to be controlled by a yet-unknown mechanism. The lower alyA7 expression in Z. galactanivorans (ΔausR) (Figure 4G) suggests an indirect control by AusR, although further research is needed to elucidate the signaling pathway. Wild-type Z. galactanivorans therefore produces the AusR regulator and the alginate lyases AlyA1 and AlyA7 at a basal level in the absence of alginate. Upon addition of alginate, the extracellular secreted enzymes AlyA1 and AlyA7 catalyze the initial degradation of the polymer. Metabolic intermediates (unsaturated uronic oligosaccharides) act as effectors of AusR and prevent binding to its operators, hence relaxing the repression. This triggers a rapid increase in the expression of the whole AUS. The activities of the AUS-encoded enzymes would subsequently reduce the levels of the AusR effectors, potentially allowing a negative feedback mechanism. Such control of mRNA surges by metabolic intermediates was previously shown to operate in the degradation of chondroitin sulfate and heparin in B. thetaiotaomicron (63). The repression of alyA7 expression in alginate-containing medium (lowest expression in the exponential phase, Figure 3) could reflect its main role as a ‘sentry’ enzyme releasing the first oligosaccharides upon addition of alginate, whereas it would be less efficient afterwards when the AUS is expressed and most of the substrate is oligomeric. Our results also show an uncoupling of the expression of ausR and the downstream genes zgal_2616-2613 (Figure 3), although they are currently believed to be transcribed as polycistronic mRNA from the promoter PausR (Figure 1a). We propose three non-exclusive explanations: (1) the presence of an additional promoter; (2) differential decay mediated by ribonuclease(s) and (3) conditional termination of transcription in the intergenic region ausR-exuT2. First, the presence of another transcription starting site upstream exuT2 is unlikely, since we did not find the consensus -7 box [TAnnTTTG] that is conserved in Flavobacterium and Bacteroides species (64). Furthermore, no AusR binding site was predicted in the intergenic region ausR-exuT2, although one would be necessary to mediate transcriptional repression of the zgal_2616-2613 mRNA in the absence of alginate. Second, the downstream part of the zgal_2617-2613 mRNA might be degraded by 3′-exonuclease(s) in the absence of alginate. RNAfold analysis predicted a secondary structure within the 230-bp ausR-exuT2 intergenic region (Supplementary Figure S2B), which might protect ausR from this degradation. This type of regulation has already been documented in other bacteria (65,66). Third, the same or another secondary structure may conditionally block mRNA elongation after the ausR gene, in a manner similar to riboswitches that fold into termination or anti-termination RNA structures (67). Whether it involves differential decay and/or conditional termination, the regulation would require an ON/OFF switch depending on physiological conditions, signaling alginate availability. Folding of the predicted mRNA secondary structure may depend on direct binding of a signaling molecule (eg. alginate metabolic intermediate), or on interaction with a regulatory RNA-binding protein. Finally, it remains to be elucidated whether alginate utilization is additionally controlled by catabolic repression and/or substrate prioritization, as other PULs from human gut bacteria (27–29). Previous studies showed that the gammaproteobacterium Alteromonas macleodii 83-1, which possesses a AUS distantly related to Z. galactanivorans (including a predicted FadR subfamily regulator), prioritizes laminarin over alginate through an unknown mechanism (68).
Figure 6.
Proposed model for the regulation of alginate utilization by AusR in Zobellia galactanivorans DsijT. The repression of the AUS promoters by AusR that occurs in the absence of alginate is represented by red arrows. The level of repression estimated from qPCR results (Figure 3) is indicated. The number of AusR operators in each promoter is depicted by red ellipses, together with their distance to the -7 box (no indicated distance means that operators overlap the -7 box). Genes and proteins depicted in grey are those with no or lower direct repression by AusR, allowing basal expression in the absence of alginate.
The Z. galactanivorans (ΔausR) mutant strain had two visible phenotypes (Figure 4). First, the mutant strain showed a longer lag phase than the wild type when transferred from Zobell medium to alginate-containing medium. This was unexpected since most genes from the AUS were strongly de-repressed in the absence of alginate in Z. galactanivorans (ΔausR). However, these genes encode cell-bound proteins that process small alginate degradation products rather than polymeric alginate. By contrast, alyA7 was only minimally expressed in Z. galactanivorans (ΔausR). The lack of secreted AlyA7 would slow down the initial breakdown of alginate polymers into short oligosaccharides processable by the rest of the AUS, leading to a longer lag phase. While the other secreted alginate lyase AlyA1 was still expressed in Z. galactanivorans (ΔausR), it might not compensate for the 26-fold lower expression of AlyA7. AlyA1 is an endolytic alginate lyase specific for guluronate motifs (41). Although the exact specificity of AlyA7 is currently unknown, other members of the PL14 family are endolytic mannuronate lyases (69–71). AlyA1 and AlyA7 might therefore have complementary activities to breakdown polymeric alginate.
The second phenotype of Z. galactanivorans (ΔausR) was the formation of aggregates in rich medium (Figure 4). This auto-aggregation might be due to a change in self-recognizing surface structures, such as capsular exopolysaccharides (72). Indeed, a Bacteroides fragilis strain defective for a capsule biosynthesis locus auto-aggregated during liquid growth, likely because surface adhesive molecules usually shielded by the capsule became exposed in the mutant (73). Although the presence of a capsule has never been assessed in Z. galactanivorans, it features potential capsule biosynthesis loci (i.e. zgal_1742-1751, zgal_3751-3762, zgal_3780-3787) encoding the putative capsule synthesis proteins CapC, CapD, CapJ and CapM together with O-acetylases, O antigen biosynthesis proteins and glycosyltransferases (38). Furthermore, previous results showed that B. thetaiotaomicron co-regulates the degradation of host glycans with capsular polysaccharide synthesis, in an unknown signaling pathway that depends on Sus-like outer membrane proteins (30). A similar coordinated regulation of the alginate PUL and capsule biosynthesis loci might exist in Z. galactanivorans and have been disrupted in the ΔausR mutant. Such a concerted expression might protect cells against immune defense reactions by secreting/modifying their capsule while degrading algal host cell walls.
The AusR operator sequence matched the FadR-subfamily consensus [TKGTN2–3ACMA] that was previously inferred mostly from Proteobacteria, Actinobacteria and Firmicutes species (62), confirming that DNA recognition via the HTH domain is widely conserved across the phylogenetic tree. In Z. galactanivorans, the presence of multiple operators in immediate proximity to the -7 box (PalyA3, Ptbdt, PalyA4 and PalyA6) was associated with a strong transcriptional repression by AusR (Figure 6). By contrast, a longer distance to the -7 box (Paly1, PausR) and the presence of a single operator (PausR) were linked to a lower repression effect. Such a modular organization of operators resembles that found in the ara and gan gene clusters of Bacillus subtilis (74,75), which encode catabolic pathways for arabinose and galactan, respectively. In both systems, two consecutive operators control the expression of genes encoding metabolic enzymes, leading to a maximal repression effect. The regulator genes themselves only have one copy of the operator sequence that allows a moderate self-negative feedback regulation. Dual operators may be involved in cooperative binding of regulators, alternative oligomerization, recruitment of additional subunits and DNA bending ensuring a more flexible and precise control of gene expression (62,76–77). In line with this hypothesis, EMSA analysis (Figure 5) evidenced cooperative binding and an additional protein/DNA complex when AusR was incubated with PalyA3 (two operators) compared to PausR (one operator). The present study indicates that the AusR regulon comprises several distant loci in Z. galactanivorans. It is worth noting that alyA1 features a putative AusR operator site, although this gene was recently acquired by horizontal transfer from Actinobacteria (46). This suggests alyA1 was rapidly recruited into the regulon via the gain of AusR binding sites within its promoter. The detection of a pair of AusR operators in the promoter of the alginate-induced gntK1 gene suggests it also belongs to the AusR regulon. Future genome-wide mapping of AusR binding sites using DNA affinity purification sequencing (DAP-seq) or chromatin immunoprecipitation sequencing (ChIP-seq) will help define the exact recognized DNA motif and potentially expand the list of AusR-regulated targets within Z. galactanivorans genome.
AusR utilizes short oligosaccharides released by alginate lyases as its effectors (Figure 5E and F). This is reminiscent of previous studies in the soil bacteria Sphingomonas sp. strain A1. This strain possesses an alginate-degrading system different from most heterotrophic bacteria, whereby cells form a mouth-like pit that directly imports the polysaccharide into the cytoplasm before degradation (78). This system is controlled by the LacI family transcription repressor AlgO. Interaction of the C-terminal effector-binding domain of AlgO with the unsaturated alginate oligosaccharides ΔGGG, ΔGGGG, ΔMMM and ΔMMMM induces a conformational change in the N-terminal HTH domain and inhibits the formation of the AlgO/DNA complex (79). In Z. galactanivorans, it was intriguing that unsaturated trisaccharides can act as effectors of the cytoplasmic regulator AusR. Indeed, the current degradation model predicts that unsaturated monosaccharides (Δ) are imported into the cytoplasm via the transporter ExuT2 from the Major Facilitator Superfamily (MFS), while longer oligosaccharides would be confined to the periplasm (Figure 1). The closest characterized homolog of ExuT2 from Z. galactanivorans is the hexuronate transporter ExuT from E. coli K-12 (Uniprot accession P0AA78, 31% sequence identity), which imports glucuronate, galacturonate and glucose into the cytoplasm (80,81). The apparent in vitro interaction of trisaccharides ΔMM and ΔMG with AusR might reflect specific recognition of the Δ motif, an hypothesis supported by the lack of interaction with the saturated monosaccharides D-mannuronate and L-guluronate (Figure 5E and F). The instability of the Δ monomer, which spontaneously converts to 4-deoxy-L-erythro-5-hexoseulose uronic acid within minutes (49), makes it technically difficult to test its interaction with AusR in vitro. Alternatively, Z. galactanivorans ExuT2 might import unsaturated trisaccharides into the cytoplasm, where they could interact with AusR. Other members of the MFS superfamily can transport oligosaccharides such as maltotriose (82) and cellodextrins (83).
To our knowledge, AusR is the first characterized PUL-encoded regulator in marine bacteria. Members of the FadR subfamily control the metabolism of various oxidized substrates, including fatty acids, amino acids and central metabolites such as aspartate, pyruvate, glycolate, galactonate, lactate, malonate and gluconate (61). The present study extends the range of targets of the FadR subfamily to polysaccharide catabolism and constitutes the first characterization of a FadR-like regulator within the Bacteroidetes phylum. AUS featuring ausR orthologs, similar genetic organizations and high sequence similarities were previously detected in Flavobacteriia, Bacteroidia, Cytophagia, Alphaproteobacteria and Gammaproteobacteria (18). Here, we show that the DNA motif recognized by AusR in Z. galactanivorans DsijT is conserved in closely related alginate-degrading flavobacteria (Supplementary Table S4). Within the PULDB database that lists PULs in Bacteroidetes species (84), 69 out of 70 predicted alginolytic PULs (encoding both PL17 and PL7) possessed a ausR homolog (March 2020). Therefore, our results illuminate a AusR-dependent regulatory mechanism for alginate degradation globally conserved in marine bacteria, as well as in gut microbes that laterally acquired the AUS (18,85). This fine control may participate to the adaptation of bacteria to complex, highly variable ecological niches, as proposed for other GntR-like regulators (40). During the degradation of alginate by Z. galactanivorans, oligosaccharides first accumulate in the medium before being assimilated by cells (Figure 2). From an ecological perspective, these soluble degradation products released in the environment might act as public goods (86) and fuel scavenger bacteria less equipped for polymeric alginate degradation. This corroborates recent findings that few specialized bacteria can act as pioneer degraders of complex marine polysaccharides (87–89), including Z. galactanivorans (46). This behavior is particularly relevant for the efficient utilization of substrates immobilized within brown algal cell walls. Furthermore, the elucidation of the regulatory controls governing alginate degradation might inspire ongoing efforts to bioengineer microbial platforms for brown algal biomass valorization (90–92).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Anne-Catherine Dock-Brégeon for invaluable advice on EMSA protocols and to Murielle Jam, Dr. Sabine Joncourt and Dr. Lionel Cladière for their help with protein purification.
Author contributions: Author contributions following the CRediT taxonomy (https://casrai.org/credit/) are as follows: Conceptualization: FT; Formal analysis: MD, AD, TR, FT; Funding acquisition: BS, FT; Investigation: MD, AD, DJ, FT; Project administration: FT; Supervision: GM, BS, FT; Visualization: MD, AD, FT; Writing – original draft: FT; Writing – review & editing: MD, AD, DJ, TR, GM, BS, FT.
Notes
Present address: Anissa Dieudonné, Aix Marseille Univ, CEA, CNRS, BIAM, UMR 7265, Saint Paul-Lez-Durance, France.
Contributor Information
Magda Dudek, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Anissa Dieudonné, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Diane Jouanneau, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Tatiana Rochat, Université Paris-Saclay, INRAE, UVSQ, VIM, 78350, Jouy-en-Josas, France.
Gurvan Michel, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Benoit Sarels, Sorbonne Université, CNRS, Laboratoire Jacques-Louis Lions, Université de Paris, 75252 Paris, France.
François Thomas, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by an internal grant from the LBI2M department, the RegAl project (Brittany regional council grant SAD2016-9613), the French ANR investment expenditure program IDEALG (grant agreement ANR-10-BTBR-04) and the French ANR project ALGAVOR (grant agreement ANR-18-CE02-0001-01).
Conflict of interest statement. None declared.
REFERENCES
- 1. Kloareg B., Quatrano R.S.. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. An Annu. Rev. 1988; 26:259–315. [Google Scholar]
- 2. Myklestad S.M. Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci. Total Environ. 1995; 165:155–164. [Google Scholar]
- 3. Biddanda B., Benner R.. Carbon, nitrogen, and carbohydrate fluxes during the production of particulate and dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 1997; 42:506–518. [Google Scholar]
- 4. Benner R., Pakulski J.D., Mccarthy M., Hedges J.I., Hatcher P.G., Pakulski J.D., Mccarthy M., Hedges J.I., Hatcher P.G.. Bulk chemical characteristics of dissolved organic matter in the ocean. Science. 1992; 255:1561–1564. [DOI] [PubMed] [Google Scholar]
- 5. Azam F., Malfatti F.. Microbial structuring of marine ecosystems. Nat Rev Micro. 2007; 5:782–791. [DOI] [PubMed] [Google Scholar]
- 6. Kirchman D.L. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 2002; 39:91–100. [DOI] [PubMed] [Google Scholar]
- 7. Thomas F., Hehemann J.-H., Rebuffet E., Czjzek M., Michel G.. Environmental and gut Bacteroidetes: the food connection. Front. Microbiol. 2011; 2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchan A., LeCleir G.R., Gulvik C.A., González J.M.. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014; 12:686–698. [DOI] [PubMed] [Google Scholar]
- 9. Grondin J.M., Tamura K., Déjean G., Abbott D.W., Brumer H.. Polysaccharide Utilization Loci: Fuelling microbial communities. J. Bacteriol. 2017; 199:e00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson K.L., Salyers A.A.. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 1989; 171:3192–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson K.L., Salyers A.A.. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 1989; 171:3199–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martens E.C., Lowe E.C., Chiang H., Pudlo N.A., Wu M., McNulty N.P., Abbott D.W., Henrissat B., Gilbert H.J., Bolam D.N. et al.. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011; 9:e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naas A.E., Mackenzie A.K., Mravec J., Schückel J., Willats W.G.T., Eijsink V.G.H., Pope P.B.. Do Rumen Bacteroidetes Utilize an Alternative Mechanism for cellulose degradation. MBio. 2014; 5:e01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsbrink J., Thompson A.J., Lundqvist M., Gardner J.G., Davies G.J., Brumer H.. A complex gene locus enables xyloglucan utilization in the model saprophyte Cellvibrio japonicus. Mol. Microbiol. 2014; 94:418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sonnenburg E.D., Zheng H., Joglekar P., Higginbottom S.K., Firbank S.J., Bolam D.N., Sonnenburg J.L.. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010; 141:1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martens E.C., Chiang H.C., Gordon J.I.. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008; 4:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao Y., Rocha E.R., Smith C.J.. Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc. Natl. Acad. Sci. USA. 2014; 111:12901–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas F., Barbeyron T., Tonon T., Génicot S., Czjzek M., Michel G.. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ. Microbiol. 2012; 14:2379–2394. [DOI] [PubMed] [Google Scholar]
- 19. Kabisch A., Otto A., König S., Becher D., Albrecht D., Schüler M., Teeling H., Amann R.I., Schweder T.. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J. 2014; 8:1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hehemann J.-H., Correc G., Thomas F., Bernard T., Barbeyron T., Jam M., Helbert W., Michel G., Czjzek M.. Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J. Biol. Chem. 2012; 287:30571–30584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hehemann J.-H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G.. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010; 464:908–912. [DOI] [PubMed] [Google Scholar]
- 22. Ficko-Blean E., Préchoux A., Thomas F., Rochat T., Larocque R., Zhu Y., Stam M., Génicot S., Jam M., Calteau A. et al.. Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria. Nat. Commun. 2017; 8:1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reisky L., Préchoux A., Zühlke M.-K., Bäumgen M., Robb C.S., Gerlach N., Roret T., Stanetty C., Larocque R., Michel G. et al.. A complex enzyme cascade degrades the polysaccharide ulvan from green algae. Nat. Chem. Biol. 2019; 15:803–812. [DOI] [PubMed] [Google Scholar]
- 24. Terrapon N., Lombard V., Gilbert H.J., Henrissat B.. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. 2015; 31:647–655. [DOI] [PubMed] [Google Scholar]
- 25. D’Elia J.N., Salyers A.A.. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 1996; 178:7180–7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang C., Tesar C., Li X., Kim Y., Rodionov D.A., Joachimiak A.. A novel transcriptional regulator of L-arabinose utilization in human gut bacteria. Nucleic Acids Res. 2015; 43:10546–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwalm N.D., Townsend G.E., Groisman E.A.. Multiple signals govern utilization of a polysaccharide in the gut bacterium Bacteroides thetaiotaomicron. MBio. 2016; 7:e01342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lynch J.B., Sonnenburg J.L.. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol. Microbiol. 2012; 85:478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tuncil Y.E., Xiao Y., Porter N.T., Reuhs B.L., Martens E.C., Hamaker B.R.. Reciprocal prioritization to dietary glycans by gut bacteria in a competitive coexistence. MBio. 2017; 8:e01068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martens E.C., Roth R., Heuser J.E., Gordon J.I.. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J. Biol. Chem. 2009; 284:18445–18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xing P., Hahnke R.L., Unfried F., Markert S., Huang S., Barbeyron T., Harder J., Becher D., Schweder T., Glöckner F.O. et al.. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 2015; 9:1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomas F., Bordron P., Eveillard D., Michel G.. Gene expression analysis of Zobellia galactanivorans during the degradation of algal polysaccharides reveals both substrate-specific and shared transcriptome-wide responses. Front. Microbiol. 2017; 8:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Unfried F., Becker S., Robb C.S., Hehemann J.-H., Markert S., Heiden S.E., Hinzke T., Becher D., Reintjes G., Krüger K. et al.. Adaptive mechanisms that provide competitive advantages to marine Bacteroidetes during microalgal blooms. ISME J. 2018; 12:2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krüger K., Chafee M., Ben Francis T., Glavina del Rio T., Becher D., Schweder T., Amann R.I., Teeling H.. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019; 13:2800–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kappelmann L., Krüger K., Hehemann J.H., Harder J., Markert S., Unfried F., Becher D., Shapiro N., Schweder T., Amann R.I. et al.. Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 2019; 13:76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barbeyron T., L’Haridon S., Corre E., Kloareg B., Potin P.. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2001; 51:985–997. [DOI] [PubMed] [Google Scholar]
- 37. Thomas F., Barbeyron T., Michel G.. Evaluation of reference genes for real-time quantitative PCR in the marine flavobacterium Zobellia galactanivorans. J. Microbiol. Methods. 2011; 84:61–66. [DOI] [PubMed] [Google Scholar]
- 38. Barbeyron T., Thomas F., Barbe V., Teeling H., Schenowitz C., Dossat C., Goesmann A., Leblanc C., Oliver Glöckner F., Czjzek M. et al.. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: Example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ. Microbiol. 2016; 18:4610–4627. [DOI] [PubMed] [Google Scholar]
- 39. Martens E.C., Koropatkin N.M., Smith T.J., Gordon J.I.. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 2009; 284:24673–24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoskisson P.A., Rigali S.. Chapter 1 Variation in Form and Function. The Helix-Turn-Helix Regulators of the GntR Superfamily. Adv. Appl. Microbiol. 2009; 69:1–22. [DOI] [PubMed] [Google Scholar]
- 41. Thomas F., Lundqvist L.C.E., Jam M., Jeudy A., Barbeyron T., Sandström C., Michel G., Czjzek M.. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 2013; 288:23021–23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hobbs J.K., Lee S.M., Robb M., Hof F., Barr C., Abe K.T., Hehemann J.-H., McLean R., Abbott D.W., Boraston A.B.. KdgF, the missing link in the microbial metabolism of uronate sugars from pectin and alginate. Proc. Natl. Acad. Sci. 2016; 113:6188–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haug A., Larsen B., Smidsrød O.. Uronic acid sequence in alginate from different sources. Carbohydr. Res. 1974; 32:217–225. [Google Scholar]
- 44. Lundqvist L.C.E., Jam M., Barbeyron T., Czjzek M., Sandström C.. Substrate specificity of the recombinant alginate lyase from the marine bacteria Pseudomonas alginovora. Carbohydr. Res. 2012; 352:44–50. [DOI] [PubMed] [Google Scholar]
- 45. Zobell C. Studies on marine bacteria I The cultural requirements of heterotrophic aerobes. J. Mar. Res. 1941; 4:42–75. [Google Scholar]
- 46. Zhu Y., Thomas F., Larocque R., Li N., Duffieux D., Cladière L., Souchaud F., Michel G., McBride M.J.. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ. Microbiol. 2017; 19:2164–2181. [DOI] [PubMed] [Google Scholar]
- 47. Draget K.I., Ostgaard K., Smidsrød O.. Alginate-based solid media for plant tissue culture. Appl. Microbiol. Biotechnol. 1989; 31:79–83. [Google Scholar]
- 48. Kidby D.K., Davidson D.J.. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal. Biochem. 1973; 55:321–325. [DOI] [PubMed] [Google Scholar]
- 49. Preiss J., Ashwell G.. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 1962; 237:309–316. [PubMed] [Google Scholar]
- 50. Groisillier A., Herve C., Jeudy A., Rebuffet E., Pluchon P., Chevolot Y., Flament D., Geslin C., Morgado I., Power D. et al.. MARINE-EXPRESS: taking advantage of high throughput cloning and expression strategies for the post-genomic analysis of marine organisms. Microb. Cell Fact. 2010; 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005; 41:207–234. [DOI] [PubMed] [Google Scholar]
- 52. Ryder S.P., Recht M.I., Williamson J.R.. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008; 488:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. et al.. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu N.Y., Wagner J.R., Laird M.R., Melli G., Rey S., Lo R., Dao P., Cenk Sahinalp S., Ester M., Foster L.J. et al.. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010; 26:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grant C.E., Bailey T.L., Noble W.S.. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L.. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Watson S.W., Novitsky T.J., Quinby H.L., Valois F.W.. Determination of bacterial number and biomass in the marine environment. Appl. Environ. Microbiol. 1977; 33:940–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haydon D., Guest J.. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 1991; 79:291–296. [DOI] [PubMed] [Google Scholar]
- 60. Little M.S., Pellock S.J., Walton W.G., Tripathy A., Redinbo M.R.. Structural basis for the regulation of β-glucuronidase expression by human gut Enterobacteriaceae. Proc. Natl. Acad. Sci. USA. 2017; 115:E152–E161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rigali S., Derouaux A., Giannotta F., Dusart J.. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 2002; 277:12507–12515. [DOI] [PubMed] [Google Scholar]
- 62. Suvorova I.A., Korostelev Y.D., Gelfand M.S.. GntR Family of Bacterial Transcription Factors and Their DNA Binding Motifs: Structure, Positioning and Co-Evolution. PLoS One. 2015; 10:e0132618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raghavan V., Lowe E.C., Townsend G.E., Bolam D.N., Groisman E.A.. Tuning transcription of nutrient utilization genes to catabolic rate promotes growth in a gut bacterium. Mol. Microbiol. 2014; 93:1010–1025. [DOI] [PubMed] [Google Scholar]
- 64. Chen S., Bagdasarian M., Kaufman M.G., Bates A.K., Walker E.D.. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J. Bacteriol. 2007; 189:5108–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rochat T., Bouloc P., Repoila F.. Gene expression control by selective RNA processing and stabilization in bacteria. FEMS Microbiol. Lett. 2013; 344:104–113. [DOI] [PubMed] [Google Scholar]
- 66. Trinquier A., Durand S., Braun F., Condon C.. Regulation of RNA processing and degradation in bacteria. Biochim. Biophys. Acta - Gene Regul. Mech. 2020; 1863:194505. [DOI] [PubMed] [Google Scholar]
- 67. Pavlova N., Kaloudas D., Penchovsky R.. Riboswitch distribution, structure, and function in bacteria. Gene. 2019; 708:38–48. [DOI] [PubMed] [Google Scholar]
- 68. Koch H., Dürwald A., Schweder T., Noriega-Ortega B., Vidal-Melgosa S., Hehemann J.H., Dittmar T., Freese H.M., Becher D., Simon M. et al.. Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 2019; 13:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shimizu E., Ojima T., Nishita K.. cDNA cloning of an alginate lyase from abalone, Haliotis discus hannai. Carbohydr. Res. 2003; 338:2841–2852. [DOI] [PubMed] [Google Scholar]
- 70. Rahman M.M., Wang L., Inoue A., Ojima T.. cDNA cloning and bacterial expression of a PL-14 alginate lyase from a herbivorous marine snail Littorina brevicula. Carbohydr. Res. 2012; 360:69–77. [DOI] [PubMed] [Google Scholar]
- 71. Rahman M.M., Inoue A., Tanaka H., Ojima T.. cDNA cloning of an alginate lyase from a marine gastropod Aplysia kurodai and assessment of catalytically important residues of this enzyme. Biochimie. 2011; 93:1720–1730. [DOI] [PubMed] [Google Scholar]
- 72. Trunk T., Khalil H.S., Leo J.C.. Bacterial autoaggregation. AIMS Microbiol. 2018; 4:140–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu C.H., Lee S.M., VanLare J.M., Kasper D.L., Mazmanian S.K.. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. USA. 2008; 105:3951–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mota L.J., Tavares P., Sá-Noguelra I.. Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 1999; 33:476–489. [DOI] [PubMed] [Google Scholar]
- 75. Habib C., Yu Y., Gozzi K., Ching C., Shemesh M., Chai Y.. Characterization of the regulation of a plant polysaccharide utilization operon and its role in biofilm formation in Bacillus subtilis. PLoS One. 2017; 12:e0179761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Müller-Hill B. The function of auxiliary operators. Mol. Microbiol. 1998; 29:13–18. [DOI] [PubMed] [Google Scholar]
- 77. Mota L.J., Sarmento L.M., Isabel D.S.-N.. Control of the Arabinose Regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity and DNA looping. J. Bacteriol. 2001; 183:4190–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hashimoto W., Kawai S., Murata K.. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng. Bugs. 2010; 1:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hayashi C., Takase R., Momma K., Maruyama Y., Murata K., Hashimoto W.. Alginate-dependent gene expression mechanism in Sphingomonas sp. Strain A1. J. Bacteriol. 2014; 196:2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nemoz G., Robert Baudouy J., Stoeber F.. Physiological and genetic regulation of the aldohexuronate transport system in Escherichia coli. J. Bacteriol. 1976; 127:706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim H.J., Jeong H., Lee S.J.. Short-Term adaptation modulates anaerobic metabolic flux to succinate by activating ExuT, a Novel D-Glucose transporter in Escherichia coli. Front. Microbiol. 2020; 11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Magalhães F., Vidgren V., Ruohonen L., Gibson B.. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016; 16:fow053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Galazka J.M., Tian C., Beeson W.T., Martinez B., Glass N.L., Cate J.H.D.. Cellodextrin transport in yeast for improved biofuel production. Science. 2010; 330:84–86. [DOI] [PubMed] [Google Scholar]
- 84. Terrapon N., Lombard V., Drula É., Lapébie P., Al-Masaudi S., Gilbert H.J., Henrissat B.. PULDB: The expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018; 46:D677–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mathieu S., Touvrey-Loiodice M., Poulet L., Drouillard S., Vincentelli R., Henrissat B., Skjak-Bræk G., Helbert W.. Ancient acquisition of ‘alginate utilization loci’ by human gut microbiota. Sci. Rep. 2018; 8:8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cavaliere M., Feng S., Soyer O.S., Jiménez J.I.. Cooperation in microbial communities and their biotechnological applications. Environ. Microbiol. 2017; 19:2949–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hehemann J.-H., Arevalo P., Datta M.S., Yu X., Corzett C.H., Henschel A., Preheim S.P., Timberlake S., Alm E.J., Polz M.F.. Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat. Commun. 2016; 7:12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Enke T.N., Datta M.S., Schwartzman J., Cermak N., Schmitz D., Barrere J., Pascual-García A., Cordero O.X.. Modular assembly of Polysaccharide-Degrading marine microbial communities. Curr. Biol. 2019; 29:528–1535. [DOI] [PubMed] [Google Scholar]
- 89. Ebrahimi A., Schwartzman J., Cordero O.X.. Cooperation and spatial self-organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proc. Natl. Acad. Sci. 2019; 116:23309–23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wargacki A.J., Leonard E., Win M.N., Regitsky D.D., Santos C.N.S., Kim P.B., Cooper S.R., Raisner R.M., Herman A., Sivitz A.B. et al.. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012; 335:308–313. [DOI] [PubMed] [Google Scholar]
- 91. Enquist-Newman M., Faust A.M.E., Bravo D.D., Santos C.N.S., Raisner R.M., Hanel A., Sarvabhowman P., Le C., Regitsky D.D., Cooper S.R. et al.. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature. 2014; 505:239–243. [DOI] [PubMed] [Google Scholar]
- 92. Poblete-Castro I., Hoffmann S.L., Becker J., Wittmann C.. Cascaded valorization of seaweed using microbial cell factories. Curr. Opin. Biotechnol. 2020; 65:102–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.