Abstract
With the central role of nucleic acids there is a need for development of fluorophores that facilitate the visualization of processes involving nucleic acids without perturbing their natural properties and behaviour. Here, we incorporate a new analogue of adenine, 2CNqA, into both DNA and RNA, and evaluate its nucleobase-mimicking and internal fluorophore capacities. We find that 2CNqA displays excellent photophysical properties in both nucleic acids, is highly specific for thymine/uracil, and maintains and slightly stabilises the canonical conformations of DNA and RNA duplexes. Moreover, the 2CNqA fluorophore has a quantum yield in single-stranded and duplex DNA ranging from 10% to 44% and 22% to 32%, respectively, and a slightly lower one (average 12%) inside duplex RNA. In combination with a comparatively strong molar absorptivity for this class of compounds, the resulting brightness of 2CNqA inside double-stranded DNA is the highest reported for a fluorescent base analogue. The high, relatively sequence-independent quantum yield in duplexes makes 2CNqA promising as a nucleic acid label and as an interbase Förster resonance energy transfer (FRET) donor. Finally, we report its excellent spectral overlap with the interbase FRET acceptors qAnitro and tCnitro, and demonstrate that these FRET pairs enable conformation studies of DNA and RNA.
INTRODUCTION
While DNA stores the genetic information for a cell to produce proteins, RNA is actively involved in the (bio)chemistry and regulation of gene expression. In addition to early established mRNA, tRNA and rRNA, RNAs like microRNA (miRNA), small interfering RNA (siRNA), small nuclear RNA (snRNA), small nucleolar (snoRNA), piwi-interacting RNA (piRNA), long non-coding RNA (lncRNA), ribozymes and riboswitches have more recently emerged as vital players in the regulation of protein synthesis. These fundamental molecules of life have spurred an increasing need to develop tools and techniques for their study in multiple contexts, such as transcription, RNA transport, the ribosomal machinery and mRNA translation, mRNA degradation, and several complex, RNA-mediated mechanisms of gene silencing and epigenetic modification (1–3). Molecular biology and biochemistry assays, supported by radioactive labelling and studies employing external fluorophores (fluorophores attached covalently to the 5′ or 3′ end of oligonucleotides; e.g. Cy-, Alexa- and ATTO-dyes), were and still are vital techniques to provide insights about nucleic acid function and related processes (4–6). DNA is relatively static and structurally less versatile than RNA. Hence, techniques like NMR, X-ray crystallography and Cryo-EM have successfully provided significant structure data for DNA (7,8). This is also true for certain, structurally less dynamic RNAs (9–14). However, obtaining structure and dynamics data (including information on supertertiary structure, conformational ensembles and spatiotemporal dynamics) for various forms of the structurally more pliable RNAs calls for novel and/or improved techniques that can complement existing ones.
Förster resonance energy transfer (FRET) is a fluorescence-based technique that can provide both structural and dynamics information, and in this respect represents a complementary method to NMR and X-ray crystallography to study DNA and, especially, RNA. Such detailed structural and dynamics information has, for example, been obtained using single-molecule FRET combined with comprehensive analysis and modelling (15). Unfortunately, the number of intrinsically fluorescent entities in DNA and RNA that could be employed in sensitive spectroscopy and microscopy techniques is scarce. One exception is the mRNA cap - a naturally occurring N7-modified guanosine that is present on the mRNA 5′ end. As a fluorophore, it has been successfully applied in spectroscopic studies of mRNA-related processes, such as protein synthesis (16) and mRNA turnover (17), as well as in the characterisation of mRNA-binding proteins (18–20). To overcome the shortage of intrinsic and minimally perturbing fluorophores in nucleic acids, we are continuously expanding our library of fluorescent base analogues (FBAs) (21–25). This class of fluorophores (24) represents a versatile alternative to the commonly used external fluorophores; they have the advantage of being (i) non-bulky base-complementary mimics of natural nucleobases suitable for incorporation into DNA/RNA, preserving their canonical conformation and (ii) reporter fluorophores that can be located close to or inside a ligand (e.g. a small molecule, a protein, another nucleic acid or a drug) binding site. A remaining challenge with the utilization of fluorescent base analogues is their lower brightness (ϵΦF; molar absorptivity multiplied by the fluorescence quantum yield) compared with conventional external fluorophores such as those mentioned above. However, recent studies show that the brightness gap between internal and external fluorophores is decreasing (26–28).
In particular, we demonstrated that the brightness of our parent adenine analogue, quadracyclic adenine (qA) (29) can be significantly improved by replacing its outer ring with a pyridine or a naphthalene, resulting in fluorophores of a brightness that is excellent for a fluorescent base analogue in aqueous solution, qAN1 (ϵΦF = 1700 M−1 cm−1) and pA (ϵΦF = 10100 M−1 cm−1), respectively (28,30). Unfortunately, when incorporated into DNA, the fluorescence quantum yields and lifetimes of the two adenine derivatives decrease and are sensitive to the neighbouring bases (28,30). The brightness of pA inside DNA is still respectable (on average 1400 M−1 cm−1) and close to the highest average brightness inside DNA (2000 M−1 cm−1, as reported for tCO) (31), but compared with commonly used external fluorophores, which in general have brightness values well above 104 M−1 cm−1, there is still room for improvement. In our recent study on outer ring substituents of qA, the 2CNqA nucleobase (Figure 1), bearing a cyano (CN) group in the 2-position of the outer ring, was found to possess a high and stable quantum yield in a variety of solvents (32). Therefore, in our search for an adenine analogue which, like tCO (31), would exhibit a high and stable quantum yield regardless of sequence context, rendering a high brightness and therefore applicability as a nucleic acid label and as an interbase FRET donor (30,33,34), we decided to investigate 2CNqA in nucleic acids.
Figure 1.

Structure of the studied 2CNqA, qAnitro and tCnitro nucleosides; R1 is H for DNA and OH for RNA.
Herein, we characterise the photophysical properties of 2CNqA in both DNA and RNA and show that it can be successfully used as a donor in FRET pairs with qAnitro (30) and tCnitro (35) as acceptor nucleobases (Figure 1). We investigate the properties of 2CNqA in both B-form DNA and A-form RNA, and its selectivity in base-pairing. We find that our novel 2CNqA analogue is a highly promising nucleic acid fluorophore for future utilisation in biophysical, biochemical and molecular biology contexts and that it displays the highest average brightness inside DNA ever reported for a fluorescent base analogue.
MATERIALS AND METHODS
Synthesis of the 2CNqA deoxyribonucleoside and ribonucleoside
The method developed for the preparation of the previously published qA derivatives, qAN1, qAnitro and pA, was adopted for the synthesis of the 2CNqA-containing deoxyribonucleoside (28,30). The DMTr-protected phosphoramidite of 2CNqA (5) was synthesized over 13 steps with an overall yield of 25% from commercially available 6-chloro-7-iodo-7-deazapurine (1) (Scheme 1; for details, see Supporting information (SI)).
Scheme 1.
Synthesis of the 2CNqA-DNA (5) and 2CNqA-RNA (13) phosphoramidite building blocks: (a) NaH, MeCN, 0°C, 30 min; (b) 1.2 eq 3, 0°C, 5 min, then RT, 30 min, 72% over two steps; (c) Bis(trimethylsilyl)acetamide, TMSOTf, MeCN, 80°C, 1 h, 60%; (d) 2 mol% Pd(PPh3)4, HBPin, Et3N, THF, 80°C, 36 h, 76%; (e) Boc2O, NaHMDS, THF, −78°C, 1 h, 81%; (f) 1 eq 10, 5 mol% PdCl2(PPh3)2, K2CO3, DME, 80°C, 55 h, 83%; (g) DBU, DABCO, DMF, 70°C, 12 h; (h) NaOMe, MeOH, RT, 1 h, 46% over two steps; (i) DMTrCl, Pyridine, RT, 3 h, 80%; (j) TBDMSCl, AgNO3, THF, Pyridine, RT, 7 h, 67%; (k) CEPCl, DIPEA, THF, RT, 20 h, 96%.
The 2CNqA ribonucleoside was initially thought to be accessible through the N-glycosylation of compound 2 with a protected ribose sugar. However, this strategy turned out to be incompatible with the majority of commercially available protected ribose derivatives. Instead, N-glycosylation was performed as the first step, as in the original qA synthesis protocol (29,36). Silyl–Hilbert–Johnson glycosylation of deazapurine 1 with protected ribose 6 furnished compound 7 in moderate yield (37). Following our previously reported protocol, Miyaura-borylation of 7 provided pinacol ester 8 in a good yield (30,38). 3-Amino-4-iodobenzonitrile (9) was reacted with di-tert-butyl dicarbonate to yield 10, which was used in a Suzuki-Miyaura cross-coupling with 4 to produce compound 11. Intramolecular cyclisation was achieved using DABCO and DBU to furnish compound 12 after a global deprotection using sodium methoxide (29). The final phosphoramidite building block for solid-phase synthesis was prepared using standard protection and phosphorylation chemistry to yield compound 13 over a total of eight steps and an overall yield of 9% from 1.
Synthesis of modified and unmodified DNA and RNA oligonucleotides
The sequences of 2CNqA-containing DNA and RNA oligonucleotides used in this study are presented in Tables 1–3. See the SI for synthesis and characterisation details. Complementary oligonucleotides to these sequences (unmodified and modified, which either contain qAnitro (DNA) or tCnitro (RNA)), as well as the mismatched oligonucleotides of CT, GA and TA DNA sequences (Table 1) with thymine in position 5, substituted by A, C or G, were either synthesised (see the SI for details) or purchased from ATDBio Ltd.
Table 1.
Sequences of 10 nt 2CNqA-ssDNAs and melting temperature (Tm) of their duplexes and their unmodified duplex counterpartsa
| Sequence namea | 5′-3′ Sequence (X = 2CNqA) | 2CNqA-dsDNA Tm [°C] | Unmodified dsDNA Tm [°C] | ΔTm [°C] |
|---|---|---|---|---|
| AA | CGC AAX ATC G | 42.4 ± 0.3 | 43.3 | −0.9 |
| AC | CGC AAX CTC G | 48.8 ± 0.3 | 47.0 | 1.8 |
| AG | CGC AAX GTC G | 45.5 ± 0.2 | 45.8 | −0.3 |
| AT | CGC AAX TTC G | 46.1 ± 0.1 | 43.5 | 2.6 |
| CA | CGC ACX ATC G | 52.1 ± 0.2 | 46.9 | 5.2 |
| CC | CGC ACX CTC G | 58.1 ± 0.3 | 50.5 | 7.6 |
| CG | CGC ACX GTC G | 55.8 ± 0.1 | 49.5 | 6.3 |
| CT | CGC ACX TTC G | 56.0 ± 0.3 | 47.0 | 9.0 |
| GA | CGC AGX ATC G | 45.0 ± 0.1 | 45.3 | −0.3 |
| GC | CGC AGX CTC G | 51.6 ± 0.3 | 49.0 | 2.6 |
| GG | CGC AGX GTC G | 49.1b | 48.0 | 1.1 |
| GT | CGC AGX TTC G | 48.7b | 45.6 | 3.1 |
| TA | CGC ATX ATC G | 44.4 ± 0.3 | 41.1 | 3.3 |
| TC | CGC ATX CTC G | 49.5 ± 0.3 | 43.5 | 6.0 |
| TG | CGC ATX GTC G | 47.4b | 43.5 | 3.9 |
| TT | CGC ATX TTC G | 47.4 ± 0.4 | 40.5 | 6.9 |
aSequence names indicate bases surrounding 2CNqA on the 5′ and 3′ side. Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5.
b Measured only once.
Table 3.
Sequences of 25 nt 2CNqA-ssRNAs and complementary tCnitro-ssRNAs used in the RNA FRET study
| Sequence namea | 5′-3′ Sequence (X = 2CNqA, Y = tCnitro) |
|---|---|
| RNA-D0 | CGA CAA AAU CAA AAU GCG UGA UUG G |
| RNA-D6 | CGA CAX AAU CAA AAU GCG UGA UUG G |
| RNA-D7 | CGA CAA XAU CAA AAU GCG UGA UUG G |
| RNA-D12 | CGA CAA AAU CAX AAU GCG UGA UUG G |
| RNA-D13 | CGA CAA AAU CAA XAU GCG UGA UUG G |
| RNA-A0 | CCA AUC ACG CAU UUU GAU UUU GUC G |
| RNA-A6 | CCA AUY ACG CAU UUU GAU UUU GUC G |
| RNA-A8 | CCA AUC AYG CAU UUU GAU UUU GUC G |
| RNA-A10 | CCA AUC ACG YAU UUU GAU UUU GUC G |
aSequence names indicate position of the donor (D) or the acceptor (A) in the oligonucleotide counted from its 5′ end, whereas RNA-D0 and RNA-A0 denote unmodified reference sequences.
General information on photophysical measurements
All photophysical measurements were carried out in 0.3 or 0.4 cm pathlength quartz cuvettes (Hellma). The concentration of both single- and double-stranded DNA/RNA oligonucleotides was 4.0 μM, unless stated otherwise.
The molar absorptivities of DNA oligonucleotides were approximated by the linear combination of molar absorptivities of individual nucleotides and multiplied by 0.9 to account for the base-stacking interactions. The following molar absorptivities at 260 nm were used: ϵA = 15300 M−1 cm−1, ϵC = 7400 M−1 cm−1, ϵG = 11800 M−1 cm−1, ϵT = 9300 M−1 cm−1 (39), ϵ2CNqA = 14600 M−1 cm−1 (32) and ϵqAnitro = 12900 M−1 cm−1 (30). For RNA oligonucleotides, molar absorptivities at 260 nm were calculated using the Integrated DNA Technologies (IDT) OligoAnalyzer (http://eu.idtdna.com), with the modified base replaced by its unmodified counterpart, and correcting for the difference in molar absorptivity at 260 nm between unmodified base and 2CNqA (ϵ2CNqA = 14600 M−1 cm−1) or tCnitro (ϵtCnitro = 9700 M−1 cm−1) (33). Using the Beer−Lambert law, the molar absorptivity of the 2CNqA ribonucleoside at 260 nm and the lowest-energy absorption maximum (λAbs,Max) = 356 nm was determined by measuring the absorption of its aqueous solution (MQ water, Millipore) at three different concentrations. Experiments on DNA were performed in 13 mM sodium phosphate buffer, containing 100 mM NaCl, pH 7.50 ± 0.05 and on RNA in 10 mM sodium phosphate buffer, containing 100 mM NaCl and 1.0 mM EDTA, pH 7.40 ± 0.05. All measurements were conducted at room temperature (RT, ca 22°C), unless stated otherwise. To form duplexes, a 2CNqA-modified strand was mixed with 15% excess of its complementary strand. Prior to measurements, DNA duplex samples were heated to 95°C for 10 min, incubated at 95°C for 5 min and cooled to 5°C at a rate of 0.13°C/min. To hybridize RNA duplexes, the samples were heated to 85°C over 15 min, incubated at 85°C for 15 min and cooled to 5°C at a rate of 1°C/min. In the FRET and quantum yield measurements, a 30% excess of acceptor strand was used to ensure complete hybridisation, i.e. that every donor strand molecule is bound to an acceptor strand. Data were obtained from two or more measurements and are presented as the arithmetic mean ± standard error of the mean (SEM), unless stated otherwise.
Absorption and absorption-based nucleic acid melting curves
Absorption spectra were recorded from 230 to 600 nm on Varian Cary spectrophotometer (model 4000 or 5000) using a spectral bandwidth (SBW) of 1.0 nm. The data interval was 0.5 nm, and the signal averaging time was 0.1 s. The absorption (UV) DNA and RNA melting curves were measured using the same instrument equipped with a multicell temperature block by monitoring the absorption at 260 nm versus temperature. Both the heating and the cooling curves over the range 20–90°C were measured at 1.0°C intervals. The temperature ramp was 1.0°C/min. The duplex concentration was 2.0 μM in all melting experiments. The melting temperature (Tm) was determined as the maximum of the first derivative of the melting curve after FFT filter smoothing.
Circular dichroism (CD)
CD spectra were recorded from 200 to 450 nm for the 10 bp 2CNqA-DNAs (Table 1) and from 200 to 500 nm for the 25 bp 2CNqA-RNAs (Table 3) on a Chirascan CD spectrometer (Applied Photophysics), using an SBW of 1.0 nm. Three subsequent scans were collected with 1.0 nm intervals and 0.5 s integration time and then averaged and background subtracted. Samples were measured at a concentration of 2.0 μM.
Steady-state fluorescence
Steady-state fluorescence measurements were carried out using a Spex Fluorolog 3 spectrofluorometer (JY Horiba). Emission spectra were recorded from 365 to 650 nm, with the excitation at 355 nm for DNAs and at 354 nm for RNAs. The slit widths for excitation and emission were set at 1.6 nm. The data interval was 1.0 nm and the integration time was 0.1 s. The FRET efficiency in steady-state experiments (EFRET,SS) was determined from the equation:
 |
where FDA is the integrated fluorescence emission intensity of the donor in the presence of the acceptor and FD is the integrated fluorescence emission intensity of the donor alone.
Time-resolved fluorescence
Time-resolved fluorescence decays were measured using a time-correlated single-photon counting (TCSPC) fluorescence lifetime spectrometer (LifeSpec II, Edinburgh Analytical Instruments), equipped with a 377 nm pulsed diode laser (PicoQuant model LDH-P-C-375) as the excitation source (FWHM pulse width was 1.0 nm and 70 ps with respect to wavelength and time, respectively), and a R3809U-50 microchannel plate photomultiplier (Hamamatsu). The emission monochromator was set to 446 nm. The data were collected using a pulse frequency of 5.0 MHz and 200 ns time range until 10 000 counts were recorded in the top channel out of 2048 channels used. The instrument response function (IRF) was measured at the same settings of the apparatus. Average fluorescence lifetimes (<τ>) were determined by fitting bi- or triexponential functions with reconvolution to the decay curves and calculating the amplitude-weighted averages:
 |
where τi is the fluorescence lifetime of the ith component and αi is the amplitude of the ith component. The fitting was performed using FluoFit Pro v.4 software (PicoQuant). The FRET efficiency in the time-resolved experiments (EFRET,TR) was determined from the equation:
 |
where <τDA> is the average fluorescence lifetime of the donor in the presence of the acceptor and <τD> is the average fluorescence lifetime of the donor in the absence of the acceptor.
The final experimental FRET efficiency (EFRET,Exp) was calculated as the weighted average of EFRET,SS and EFRET,TR, with the weights given by the reciprocal squares of the corresponding SEMs. The error in EFRET,Exp is the higher of the internal and external weighted variances (40; NIST/SEMATECH e-Handbook of Statistical Methods, http://www.itl.nist.gov/div898/handbook/).
The radiative decay rate constant, kR is given by:
 |
where ΦF is the fluorescence quantum yield and the non-radiative decay rate constant, kNR is given by:
 |
For details regarding the determination of the fluorescence quantum yield (ΦF), the spectral overlap integral (JDA) and theoretical FRET efficiency (EFRET,The), see the Supplementary methods.
Photodegradation quantum yield determination
The photodegradation quantum yield of the 2CNqA donor in the absence of the acceptor (ΦPd,D) for the single- and double-stranded DNAs and RNAs (containing 2CNqA flanked by two adenines) was determined by measuring the rate of fluorescence decay (2CNqA bleaching) and comparing it to the ring closing reaction of DAE6 (a diarylethene photoswitch employed as an actinometric reference; Supplementary Figure S9) in 1,4-dioxane (ΦPd,R = 0.51 (41)) using the same irradiation conditions and sample volume. DAE6 can be isomerized photonically from its non-fluorescent open-ring isomer to the fluorescent closed-ring isomer using irradiation in the UV region. The ring closing reaction of DAE6 and the 2CNqA bleaching were both performed in the Spex Fluorolog 3 spectrofluorometer (JY Horiba) using λExc = 365 nm (excitation slit at 7.0 nm) while observing the emission (emission slit at 0.5 nm) at 445 nm for 2CNqA and 580 nm for DAE6. The excitation slit was selected to let through a considerable amount of excitation light in order to capture the photodegradation event on the hour time-scale. Continuous irradiation and sample stirring were applied and the post-irradiation isomeric state of DAE6 was confirmed by absorption spectroscopy (Supplementary Figure S9). To determine ΦPd,D, a mono-exponential function was fitted to the fluorescence intensity decay of the sample (and fluorescence intensity growth of the reference), yielding the time constant, τ:
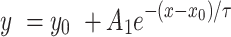 |
ΦPd,D was then obtained from the equation:
 |
where ϵ(λExc) is the molar absorptivity at the excitation wavelength, and subscripts S and R refer to the sample and reference, respectively.
Orientation of the donor and acceptor transition dipoles
Time-dependent density functional theory (TDDFT) methodology was used as described previously (32) to computationally predict the orientation of the lowest energy transition dipole moment of 2CNqA and qAnitro. These values and the experimentally determined orientation for the transition dipole moment of the tCnitro acceptor were used together with nucleic acid B- and A-form, respectively, to calculate the phase angle.
RESULTS AND DISCUSSION
Design of the 2CNqA-containing DNA and RNA oligonucleotides
To investigate the photophysical and physicochemical properties of 2CNqA as an emissive adenosine mimic inside nucleic acids, 16 DNA sequences were synthesised, bearing 2CNqA at the same position within a 10-mer oligonucleotide, but flanked with all possible combinations of neighbouring canonical bases (Table 1). To evaluate the potential of 2CNqA as a FRET donor, two previously developed acceptor entities were used, qAnitro (30) and tCnitro (33). One of the FRET pairs, 2CNqA-qAnitro, was incorporated into DNA oligonucleotides (33 nt; Table 2) and the second one, 2CNqA-tCnitro into RNA oligonucleotides (25 nt; Table 3), to examine this new FRET donor in both types of nucleic acids. The RNA sequences of the FRET investigation were also used in a less comprehensive study (compared to the one of DNA described above) of the structure, base analogue and photophysical properties of 2CNqA inside RNA.
Table 2.
Sequences of 33 nt 2CNqA-ssDNAs and complementary qAnitro-ssDNAs used in the DNA FRET study
| Sequence namea | 5′-3′ Sequence (X = 2CNqA, Y = qAnitro) |
|---|---|
| DNA-D0 | CGA TCA AAA AAA ATT ACG ATT ATA AGG AGG AGG |
| DNA-D7 | CGA TCA XAA AAA ATT ACG ATT ATA AGG AGG AGG |
| DNA-D9 | CGA TCA AAX AAA ATT ACG ATT ATA AGG AGG AGG |
| DNA-D11 | CGA TCA AAA AXA ATT ACG ATT ATA AGG AGG AGG |
| DNA-A0 | CCT CCT CCT TAT AAT CGT AAT TTT TTT TGA TCG |
| DNA-A13 | CCT CCT CCT TAT YAT CGT AAT TTT TTT TGA TCG |
| DNA-A14 | CCT CCT CCT TAT AYT CGT AAT TTT TTT TGA TCG |
| DNA-A19 | CCT CCT CCT TAT AAT CGT YAT TTT TTT TGA TCG |
| DNA-A20 | CCT CCT CCT TAT AAT CGT AYT TTT TTT TGA TCG |
aSequence names indicate position of the donor (D) or the acceptor (A) in the oligonucleotide counted from its 5′ end, whereas DNA-D0 and DNA-A0 denote unmodified reference sequences.
Conformation, stability and base-pairing selectivity of 2CNqA-containing DNA- and RNA-duplexes
To study the impact of 2CNqA on the conformation of nucleic acids, CD spectra were recorded for the 2CNqA-dsDNAs (Figure 2, left; Supplementary Figures S1 and S2) and 2CNqA-dsRNAs (Figure 2, right) in comparison to their unmodified counterparts. As for unmodified DNA duplexes, the CD spectra of the 2CNqA-containing DNA duplexes are characterised by a positive band at about 260–280 nm and a negative band around 245 nm (Figure 2, left; Supplementary Figures S1 and S2). This indicates that 2CNqA-dsDNAs adopt the canonical double helix conformation, B-DNA. The deviations from the CD of unmodified duplexes, which are general to all duplexes investigated (Supplementary Figure S2), can be ascribed to differences in molar absorptivity of the adenine that is exchanged with a 2CNqA in the modified duplex. As our previously studied fluorescent adenine analogues (qA (29), qAN1 (30) or pA (28)), no CD band outside the nucleic acid wavelength range and corresponding to the long-wavelength absorption band of 2CNqA can be observed (28–31).
Figure 2.
Left: Representative circular dichroism spectra of a 10 bp 2CNqA-dsDNA (AA; black line) and its unmodified counterpart (blue line). Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5 at RT. Right: Circular dichroism spectra of 25 bp dsRNAs used in the FRET study: 2CNqA-dsRNA (RNA-D13:RNA-A0; red line) and the corresponding donor-acceptor duplex, 2CNqA-tCnitro-dsRNA (RNA-D13:RNA-A10; blue line), acceptor-only duplex, tCnitro-dsRNA (RNA-D0:RNA-A10; navy line) and unmodified dsRNA (RNA-D0:RNA-A0; black line). Measured in 10 mM sodium phosphate buffer, 100 mM NaCl, 1.0 mM EDTA, pH 7.4 at RT.
The UV-melting experiments also clearly suggest that the B-form secondary structure of dsDNA is preserved upon 2CNqA-incorporation. The melting curves of the 16 DNA duplexes containing the 2CNqA have overall the same shape as the curves of their unmodified counterparts (data not shown). The melting temperature (Tm) of unmodified and corresponding 2CNqA-dsDNAs are summarised in Table 1. On average, 2CNqA incorporation increases the stability of DNA duplexes by 3.6°C and ranges from a minor destabilising effect (less than -1.0°C) to a stabilising effect of up to 9.0°C. It is noteworthy that DNA duplexes containing 2CNqA demonstrate higher average thermal stability than the ones previously reported for FBAs, pA (ΔTm = 1.1°C (28)), qAN1 (ΔTm = 2.9°C (30)) and qA (ΔTm = 3.0°C (29)). In a similar manner to qAN1, a higher stabilization impact on dsDNAs is observed when 2CNqA has pyrimidines as nearest neighbours. For AA, AG and GA as nearest neighbours, a small decrease in Tm is observed compared to unmodified duplexes: −0.9°C, −0.3°C and −0.3°C, respectively. An analogous decrease in melting temperature has previously been observed for qA with AA neighbours (29), for qAN1 with AA and GA neighbours (30) and for pA with AA, AG and GA neighbours (28).
Importantly, we also find 2CNqA to be highly specific for base-pairing with thymine (T) in DNA duplexes (Figure 3 and Supplementary Table S1). If the thymine of the complementary strand opposite the 2CNqA is replaced by adenine (A), cytosine (C) or guanine (G), the Tm of the duplex significantly decreases (by −10.5 to −19.6°C; Figure 3 and Supplementary Table S1). Regardless of the nearest neighbours to 2CNqA, pyrimidines (CT), purines (GA) or a combination of the two (TA), the highest destabilization effect is observed for the 2CNqA-A mismatch (−15.2 to −19.6°C).
Figure 3.

The difference in melting temperature (ΔTm) between three 10 bp 2CNqA-dsDNA (CT, GA and TA) containing the complementary 2CNqA-T pair and the corresponding duplexes containing the 2CNqA-A, 2CNqA-C or 2CNqA-G mismatches. Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5.
We next investigated the behaviour of 2CNqA as an adenine analogue in RNA duplexes. As can be seen in Figure 2, right, the unmodified and modified (2CNqA, tCnitro or both) RNA duplexes display a positive band near 265 nm and a negative band at 210 nm. Such spectral features in the CD of nucleic acids correspond to the canonical A-form RNA. As for DNA, the long-wavelength absorption band of 2CNqA in RNA is not observed in the CD (Figure 2, right, RNA-D13:RNA-A0). The same is true for the FRET acceptor, tCnitro (Figure 2, right, RNA-D0:RNA-A10). The shape of the UV-melting curves of the 2CNqA-containing and doubly modified RNA duplexes corresponds well to the profile of their unmodified counterpart, RNA-D0:RNA-A0 (Supplementary Figure S3), although slightly shifted towards higher temperatures (Table 4). This gives further support that the A-form RNA is formed when an adenine is replaced by 2CNqA and a cytosine is replaced by tCnitro. The Tm of the RNA-D13:RNA-A0 2CNqA-modified duplex is 1.5°C higher than that of the unmodified RNA-D0:RNA-A0, indicating that 2CNqA incorporation into RNA duplex in place of an adenine slightly increases RNA stability and that 2CNqA serves well as an adenine analogue also inside RNA. The small effect of 2CNqA positioned between AA neighbours on Tm is in line with the small effect found on Tm for 2CNqA between AA neighbours in DNA duplexes (Table 1).
Table 4.
The melting temperature (Tm) of a 25 bp unmodified sequence (RNA-D0:RNA-A0) and the corresponding 2CNqA-dsRNA (RNA-D13:RNA-A0), tCnitro-dsRNA (RNA-D0:RNA-A10) and doubly-labelled dsRNA (RNA-D13:RNA-A10)a
| dsRNA | T m [°C] | ΔTm [°C] |
|---|---|---|
| RNA-D0:RNA-A0 | 67.0 ± 0.1 | – |
| RNA-D13:RNA-A10 | 68.7 ± 0.2 | 1.7 |
| RNA-D13:RNA-A0 | 68.5 ± 0.2 | 1.5 |
| RNA-D0:RNA-A10 | 68.2 ± 0.3 | 1.2 |
aMeasured in 13 mM sodium phosphate buffer, 100 mM NaCl, 1.0 mM EDTA, pH 7.4.
In summary, based on the results of the conformation, stability and base-pairing study of 2CNqA in DNA and RNA, we conclude that 2CNqA is an excellent mimic of natural adenine in both nucleic acids. It preserves the specificity of adenine for thymine, and slightly stabilizes the canonical B-form helix structure of DNA. The effect of a 2CNqA modification in RNA is similar to that in DNA: the canonical A-form conformation of RNA is being adopted and a slight increase in duplex stability is observed.
Photophysical properties of 2CNqA in single- and double-stranded DNA
2CNqA has a high fluorescence quantum yield as a monomer in aqueous solution (42%) as well as a high brightness value (ϵΦF = 4500 M−1 cm−1) (32). Here, we investigate its fluorophore properties inside DNA (Table 1) and will return in the RNA FRET study below to a smaller photophysical characterisation of 2CNqA in RNA employing the RNA FRET sequences (Table 3). To understand how various neighbouring bases inside DNA influence the photophysical properties of 2CNqA, we investigated all 16 possible base surroundings (see Table 1 for sequences and sequence names). A detailed summary of the photophysical properties of 2CNqA incorporated into the 16 ssDNA sequences as well as their duplex counterparts can be found in Table 5.
Table 5.
Photophysical properties of the sixteen 10 nt 2CNqA-ssDNAs and -dsDNAsa
| ssDNA | dsDNA | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| λ Abs | λ Em | ϵ | ΦF | ϵΦF | <τ> | k R | k NR | λ Abs | λ Em | ϵ | ΦF | ϵΦF | <τ> | k R | k NR | |
| NN | [nm] | [nm] | [M−1 cm−1] | [%] | [M−1 cm−1] | [ns] | [107 s−1] | [108 s−1] | [nm] | [nm] | [M−1 cm−1] | [%] | [M−1 cm−1] | [ns] | [107 s−1] | [108 s−1] |
| AA | 357 | 452 | 10300 | 42 | 4300 | 11 | 3.8 | 0.53 | 355 | 453 | 9800 | 31 | 3000 | 9.5 | 3.3 | 0.73 |
| AC | 358 | 453 | 10400 | 35 | 3600 | 9.4 | 3.7 | 0.69 | 355 | 451 | 10100 | 29 | 2900 | 9.2 | 3.2 | 0.77 |
| AG | 358 | 453 | 10200 | 35 | 3500 | 8.6 | 4.0 | 0.76 | 356 | 454 | 9700 | 32 | 3100 | 9.7 | 3.3 | 0.70 |
| AT | 357 | 453 | 10600 | 18 | 1900 | 4.7 | 3.8 | 1.7 | 355 | 449 | 10000 | 32 | 3200 | 9.6 | 3.4 | 0.71 |
| CA | 358 | 457 | 9700 | 33 | 3200 | 8.6 | 3.9 | 0.78 | 356 | 458 | 8800 | 26 | 2300 | 8.5 | 3.0 | 0.87 |
| CC | 358 | 454 | 9500 | 27 | 2600 | 7.1 | 3.8 | 1.0 | 356 | 455 | 8700 | 28 | 2400 | 9.1 | 3.0 | 0.79 |
| CG | 359 | 453 | 9400 | 25 | 2400 | 6.2 | 4.1 | 1.2 | 356 | 459 | 8500 | 27 | 2300 | 9.2 | 3.0 | 0.79 |
| CT | 357 | 456 | 9700 | 15 | 1500 | 4.0 | 3.8 | 2.1 | 357 | 451 | 9000 | 31 | 2700 | 9.7 | 3.2 | 0.72 |
| GA | 358 | 453 | 9700 | 40 | 3900 | 11 | 3.8 | 0.56 | 355 | 455 | 9400 | 26 | 2400 | 8.5 | 3.0 | 0.87 |
| GC | 359 | 454 | 10100 | 28 | 2800 | 7.4 | 3.8 | 0.97 | 355 | 451 | 9500 | 25 | 2400 | 8.3 | 3.0 | 0.90 |
| GG | 358 | 453 | 9500 | 40 | 3800 | 9.9 | 4.0 | 0.61 | 357 | 456 | 9000 | 29 | 2700 | 9.2 | 3.2 | 0.77 |
| GT | 357 | 454 | 10000 | 17 | 1700 | 4.2 | 4.0 | 2.0 | 356 | 448 | 9700 | 29 | 2800 | 9.4 | 3.1 | 0.76 |
| TA | 358 | 456 | 9800 | 15 | 1400 | 3.7 | 4.0 | 2.3 | 356 | 456 | 9400 | 22 | 2100 | 7.4 | 3.0 | 1.0 |
| TC | 359 | 455 | 9600 | 13 | 1300 | 3.4 | 3.9 | 2.5 | 357 | 456 | 8800 | 26 | 2300 | 8.8 | 3.0 | 0.83 |
| TG | 360 | 453 | 9600 | 14 | 1300 | 3.3 | 4.2 | 2.7 | 358 | 459 | 8200 | 27 | 2200 | 8.9 | 3.0 | 0.82 |
| TT | 358 | 456 | 10000 | 10 | 1000 | 2.8 | 3.6 | 3.2 | 357 | 449 | 9200 | 29 | 2700 | 9.3 | 3.1 | 0.76 |
a λ Abs: absorption maximum, λEm: emission maximum, ϵ: molar absorptivity, ΦF: fluorescence quantum yield, ϵΦF: brightness, <τ>: average fluorescence lifetime, kR: radiative decay rate constant, kNR: non-radiative decay rate constant; Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5 at RT. The ΦF was determined at λExc = 355 nm with quinine sulfate in 0.5 M H2SO4 (ΦF,R = 0.55) as the reference. The <τ> was determined at λExc = 377 nm and λEm = 446 nm. The reported ϵ, ΦF and <τ> values are the arithmetic mean of two or more measurements. Their standard errors of the mean (SEM) can be found in Supplementary Table S2. The reported kR and kNR values were calculated as described in Materials and Methods.
The 2CNqA-DNA monomer has its lowest-energy absorption (λAbs,Max) and emission maxima (λEm,Max) at 356 and 480 nm, respectively (32). The corresponding absorption maxima of 2CNqA in ssDNA and dsDNA are found at virtually the same positions as for the monomer and are centered at approximately 358 and 356 nm, respectively, across the different combinations of neighbouring bases (Figure 4, left, Table 5 and Supplementary Figures S4 and S5). Since the natural nucleobases do not absorb above 300 nm (42), this important feature of 2CNqA allows for its selective excitation inside DNA.
Figure 4.
Absorption (left) and emission (right; λExc = 355 nm) spectra of a representative 10 nt 2CNqA-ssDNA with adenines as neighbours (AA) of 2CNqA (blue) and its duplex counterpart (black). Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5 at RT.
The emission peak of 2CNqA in DNA is blue-shifted compared to the monomer and positioned between 452 nm and 457 nm in ssDNA and 448 nm and 459 nm in dsDNA (Figure 4, right, Table 5 and Supplementary Figures S6 and S7). The observed blue-shift of 2CNqA inside DNA is most likely an effect of the less polar environment, which leads to a reduced solvent relaxation - an effect commonly observed for fluorescent base analogues inside nucleic acids (4).
When incorporated into ssDNA, 2CNqA displays a high fluorescence quantum yield, ΦF, ranging from 10% for double-T neighbours to 44% for double-A neighbours (Table 5). The lowest ΦF values were obtained for sequences containing thymine as the nearest neighbour (TT, TC, TG, TA, CT, GT and AT; 10–18%), whereas the highest ones were found for sequences containing two purines as nearest neighbours (AG, GG, GA and AA; 36–44%). These latter, high quantum yields of 2CNqA in ssDNA are comparable to the quantum yield of 2CNqA alone (42% in water (32)), suggesting a minimal quenching in these sequences relative to aqueous surroundings. Similar quenching properties (strong for neighbouring thymine and minimal for neighbouring purines) have been observed for our recently reported quadracyclic adenine analogue, qAN1 (30).
Remarkably, the quantum yield of 2CNqA shows significantly less variation between different sequences when incorporated into dsDNA (ΦF ranges from 22 to 32%; Table 5). Such low dependence of sequence surroundings on the quantum yield of a fluorescent base analogue has previously only been reported for the tricyclic cytosine analogues tC (43) and tCO (31). As was observed for qAN1 (30), the quenching effect of neighbouring thymine is decreased when going from single- to double-stranded DNA. Moreover, the difference in quenching between purines and pyrimidines is not as pronounced for dsDNAs as for ssDNAs (Table 5). On average, the quantum yield of 2CNqA in dsDNA is 28.1%, which is comparable to that in single strands (26.1%) and 1.5-fold lower compared to the 2CNqA monomer (42% (32)). The reported quantum yields inside DNA are high in comparison to those of most reported fluorescent base analogues and together with molar absorptivities of, on average, 9900 M−1 cm−1 in ssDNA and 9200 M−1 cm−1 in dsDNA (Table 5), the average brightness of 2CNqA reaches 2500 M−1 cm−1 and 2600 M−1 cm−1 for single- and double-stranded DNA, respectively. These values are the highest single-photon excitation brightness values reported so far for a fluorescent base analogue in nucleic acid systems (for a comparative table, see the report by Bood et al. (28)); e.g. average brightness in dsDNA is 510 M−1 cm−1 for qAN1, 760 M−1 cm−1 for tC, 1400 M−1 cm−1 for pA, 1700 M−1 cm−1 for 6-MI and 2000 M−1 cm−1 for tCO). Moreover, unlike previously reported A analogues such as our bright qAN1 and pA, 2CNqA has a high quantum yield and is bright regardless of sequence context in duplex DNA. These features are important for future applications of 2CNqA as an internal nucleic label and as a donor in interbase FRET (vide infra).
The average fluorescence lifetime, <τ> of 2CNqA, like the fluorescence quantum yield, is comparably insensitive to the sequence surroundings in double strands. As for most fluorescent base analogues inside nucleic acids, more than one lifetime is needed to fit the decay data properly. For 2CNqA two or three lifetimes are required to get an accurate fit to the experimental data. The average lifetimes are fairly long compared with many fluorophores of this class (21,22) inside DNA duplexes and vary between 7.4 ns and 10.4 ns (Table 5 and Supplementary Figure S8), which is comparable to the lifetime of the 2CNqA monomer in aqueous solution (8.1 ns (32)). In contrast, the fluorescence lifetime of 2CNqA in single-stranded DNA depends more on the sequence and is in the range of 2.8 ns (TT) to 10.9 ns (AA) (Table 5 and Supplementary Figure S8). The fact that the lifetimes vary more in single strands than in duplexes reflects well the corresponding effect found for the fluorescence quantum yields (vide supra). Moreover, a considerable consistency between the variations in lifetime and fluorescence quantum yield of single-stranded DNA sequences can be observed (Table 5): those with a thymine neighbour, having the lowest ΦF values, also have the shortest lifetimes (2.8–4.7 ns), whereas those where 2CNqA is embedded between two purines have the longest lifetimes (8.6–10.9 ns), accompanied by the highest ΦF values. The average lifetime of all 16 single strands is 6.6 ns, which is slightly lower than the corresponding value for duplexes (9.0 ns). The longer lifetimes of 2CNqA in ss- and dsDNA compared to previously reported fluorescence base analogue probes with high emission inside nucleic acids (31,43) could be useful for biomolecule size determination experiments using fluorescence anisotropy of slightly larger size ranges than what is presently possible.
Finally, the calculated radiative decay rate constant, kR of 2CNqA is between 3.6 and 4.3 × 107 s−1 for ssDNA sequences and between 3.0 and 3.4 × 107 s−1 for dsDNA sequences (Table 5). These low variations of kR in different environments are also reflected in a fairly stable low-energy absorption band molar absorptivities of 2CNqA inside single- and double-stranded DNA (Table 5). The non-radiative decay rate constant, kNR ranges between 0.52 and 3.2 × 108 s−1 for the 2CNqA in ssDNAs and between 0.70 and 1.0 × 108 s−1 in dsDNAs. These values can be compared with the corresponding values for the 2CNqA monomer that are 5.2 × 107 s−1 and 0.7 × 108 s−1 for kR and kNR, respectively. As can be expected from trends in quantum yields, the non-radiative rate constant varies significantly more in the single-stranded case compared with the duplex case, where the quantum yield is fairly stable throughout the sequences.
Our study of the fluorescence properties of 2CNqA also comprises an evaluation of its photodegradation quantum yield (ΦPd,D) in DNA and RNA (Supplementary Figure S10). In single-stranded DNA (ΦPd,D = 1.4 ± 0.05%) 2CNqA is more photostable than in single-stranded RNA (ΦPd,D = 2.61 ± 0.10%). However, when comparing the corresponding duplexes, the RNA (ΦPd,D = 0.17 ± 0.02%) is slightly more resistant to photodegradation than the DNA (ΦPd,D = 0.25 ± 0.01%). The ΦPd,D values also highlight that, regardless of the nucleic acid type, photobleaching is significantly less prominent for the duplexes (5–15 times lower quantum yield) compared to the corresponding single-strands. It should be noted that the excitation light intensity used to promote the photodegradation of the samples in this work was deliberately excessive, to accommodate the quantification of the photodegradation quantum yields on a reasonable time-scale (a measurement not longer than a few hours). In fact, as a comment on practical usability, a trivial adjustment of excitation and emission slits, as well as integration time in a fluorimeter setup allows for the consecutive acquisition of hundreds of spectra without notable photodegradation.
FRET between 2CNqA and qAnitro in dsDNA
We have previously reported on interbase FRET pairs consisting of a cytosine donor and acceptor (33) as well as adenine donors and an acceptor (28,30). Here, we investigate a novel adenine-adenine FRET pair, 2CNqA-qAnitro, in DNA, and also a new interbase FRET pair between analogues of two different nucleobases, 2CNqA-tCnitro, inside RNA.
For FRET to occur, the donor and acceptor need a significant spectral overlap. It can be seen in Figure 5, left, that there is a large spectral overlap between the 2CNqA emission and the qAnitro absorption. The value of the spectral overlap integral, JDA,Exp = 1.6 × 1014 M−1 cm−1 nm4 (Table 6) derived from the 2CNqA-dsDNA emission and the qAnitro-dsDNA absorption spectra (Figure 5, left) suggests that the two bases constitute a useful FRET pair for studying DNA. The spectral overlap is larger for this FRET pair than for all DNA interbase FRET pairs that have been reported so far (28,30,33,34), suggesting that longer distances in duplex DNA can be investigated with this new FRET pair. To experimentally examine the FRET efficiency, EFRET,Exp, for the 2CNqA-qAnitro pair in DNA, three ssDNA donor sequences (33 nt) with 2CNqA in position 7, 9 and 11, and with adenines as nearest neighbours (DNA-D7, DNA-D9 and DNA-D11; Table 2), and four complementary ssDNA acceptor sequences containing qAnitro in position 13, 14, 19 and 20 (DNA-A13, DNA-A14, DNA-A19 and DNA-A20; Table 2) were synthesised. By annealing all possible combinations of these single strands, twelve duplexes were obtained, covering the separations between the donor and the acceptor from 2–13 base pairs. The DNA-D7:DNA-A0, DNA-D9:DNA-A0 and DNA-D11:DNA-A0 duplexes, containing the complementary unmodified acceptor-less strand (DNA-A0; Table 2), were used as references (no-FRET samples). The fluorescence quantum yield, ΦF,D, of 2CNqA at the three donor positions is 0.23 on average. This is lower than what was found for the dsDNA AA (0.31; Table 5) and indicates that other factors such as small local structural changes around the donor not resulting from the immediate neighbours could affect the quantum yield. The FRET efficiency between 2CNqA and qAnitro in dsDNA was determined as a function of donor–acceptor separation (in bp) employing two independent techniques: steady-state fluorescence measurement of the decrease in the 2CNqA donor emission and time-resolved fluorescence measurement of the shortening of the 2CNqA donor lifetime. The final experimental FRET efficiency, EFRET,Exp, (Figure 6, left, and Supplementary Table S3) was obtained as a weighted average of the steady-state (EFRET,SS; Supplementary Table S3) and time-resolved (EFRET,TR; Supplementary Table S3) FRET efficiencies. As can be seen in Figure 6, left, the EFRET,Exp of 2CNqA-qAnitro in dsDNA is high at short donor–acceptor distances (2–4 bp), but it sharply decreases at about 5 bp separation, and then oscillates between higher and lower values at longer distances. Such a behaviour is typical for rigidly stacked interbase FRET-pairs, has been seen in previous investigations (28,30,33,34) and is due to that both the distance and the orientation between the two base analogues change with the increasing number of separating bases. The FRET efficiencies measured at longer separations are higher than reported for previous interbase FRET pairs (28,30,33,34). This is an expected effect due to the larger overlap integral for our new FRET pair and the high fluorescence quantum yield of the donor.
Figure 5.
Left: The spectral overlap between 2CNqA emission (black line; λExc = 355 nm) and qAnitro absorption (red line) in dsDNA. Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5 at RT. Right: The spectral overlap between 2CNqA emission (black line; λExc = 354 nm) and tCnitro absorption (red line) in dsRNA. Measured in 10 mM sodium phosphate buffer, 100 mM NaCl, 1.0 mM EDTA, pH 7.4 at RT. For both DNA and RNA, the spectra are normalised at their long-wavelength maxima.
Table 6.
The spectral overlap integral of the 2CNqA-qAnitro FRET pair in dsDNA and of the 2CNqA-tCnitro FRET pair in dsRNA from experiment (JDA,Exp) and theory (JDA,The)
| J DA,Exp | J DA,The | |||
|---|---|---|---|---|
| Donor | Acceptor | Oligonucleotide | [M−1 cm−1 nm4] | [M−1 cm−1 nm4] |
| 2CNqA | qAnitro | dsDNA | 1.6 × 1014 | 2.8 × 1014 |
| tCnitro | dsRNA | 1.9 × 1014 | 2.3 × 1014 |
Figure 6.
Left: Experimental FRET efficiency (EFRET,Exp; dots) as a function of the number of base-pairs separating the 2CNqA donor and qAnitro acceptor in dsDNA compared with the theoretical FRET efficiency (EFRET,The; line). Measured in 13 mM sodium phosphate buffer, 100 mM NaCl, pH 7.5 at RT. Right: Experimental FRET efficiency (EFRET,Exp; dots) as a function of the number of base-pairs separating the 2CNqA donor and tCnitro acceptor in dsRNA compared with the theoretical FRET efficiency (EFRET,The; line). Measured in 10 mM sodium phosphate buffer, 100 mM NaCl, 1.0 mM EDTA, pH 7.4 at RT. For both DNA and RNA, the experimental values are weighted averages of FRET efficiencies determined in steady-state (EFRET,SS) and time-resolved (EFRET,TR) fluorescence experiments. The error bars represent the weighted variance of EFRET,Exp. The EFRET,The depicted as a line shows the predicted values also at unnatural non-integer separations.
An in-house designed MATLAB script was developed and used to fit a theoretical curve, EFRET,The (bp) (see SI for details; Supplementary Figure S11), to the experimentally determined EFRET,Exp values (Figure 6, left). The optimal fit was obtained for the spectral overlap integral, JDA,The of 2.8 × 1014 M−1 cm−1 nm4 and for a phase angle (i.e. the angle between the donor and acceptor transition dipole moments at 0 bp separation) equal to 46°. Using time-dependent density functional theory (TDDFT) to calculate the orientation of the transition dipole moment of 2CNqA and applying that in pair with the previously calculated orientation of the transition dipole moment of qAnitro (30) both being present in a B-DNA double helix, resulted in a phase angle of 36°. The agreement between computationally and experimentally determined values for the spectral overlap integral and the phase angle is good and similar to previous reports (28,30,33). Overall, the fit between theory (EFRET,The) and experimentally determined FRET efficiencies is excellent, except for the very short distances (Figure 6, left; base separations 2–4). A similar trend has been observed before (28,30,34) and is most likely an effect of FRET theory being increasingly insufficient to describe the complete energy transfer processes at these short distances; other short-range effects like Dexter energy transfer will have an increased impact as the separation between the donor and acceptor shortens. Overall, we find that our new interbase FRET pair, 2CNqA-qAnitro, is a useful tool for DNA studies. With the stable and high quantum yield of 2CNqA in various DNA sequence surroundings and the large spectral overlap with its FRET acceptor, enabling long-distance interbase FRET measurements, 2CNqA-qAnitro is the most powerful interbase FRET pair reported for DNA so far.
FRET between 2CNqA and tCnitro in dsRNA
Next, the photophysical properties of 2CNqA for the development of a new interbase FRET system in RNA were investigated. There is a significant spectral overlap between the 2CNqA donor and the non-fluorescent acceptor used in this RNA study, tCnitro (Figure 5, right). The JDA,Exp between them in dsRNA equals to 1.9 × 1014 M−1 cm−1 nm4 (Table 6), which is slightly higher than for the DNA system presented above.
To study the FRET efficiency at different separations of the 2CNqA and tCnitro inside RNA, four 2CNqA-containing donor sequences (RNA-D6, RNA-D7, RNA-D12 and RNA-D13 with adenines as nearest neighbours for all donor strands; Table 3) with three complementary tCnitro-containing acceptor sequences (RNA-A6, RNA-A8 and RNA-A10; Table 3) were synthesised and hybridized, resulting in twelve duplexes with 2–13 base pairs separating the donor and acceptor. As no-FRET reference duplexes, RNA-D6:RNA-A0, RNA-D7:RNA-A0, RNA-D12:RNA-A0 and RNA-D13:RNA-A0, each containing the complementary acceptor-less strand (RNA-A0; Table 3) were used. The average fluorescence quantum yield of the no-FRET RNA duplexes is 0.12 which is lower than the corresponding value of the DNA FRET study above (0.23) and indicates a lower overall quantum yield of 2CNqA in RNA than in DNA systems, possibly as an effect of their different secondary structures. The donor–acceptor FRET efficiencies in dsRNA as a result of various base separations are presented in Figure 6, right, and Supplementary Table S4. The EFRET,Exp is close to the maximum value (1.0) for duplexes in which the acceptor is in the close proximity to the donor (2–4 bp), but at larger separations it significantly changes due to an increase in distance and also varies in a periodical fashion due to the change in orientation factor, similar to the DNA system. These data suggest that both the 2CNqA and tCnitro bases are firmly stacked inside the RNA. Moreover, and as in the case reported for DNA above, this suggests the great applicability of 2CNqA in RNA interbase FRET experiments.
Using FRETmatrix (44), the expected FRET efficiency between 2CNqA and tCnitro at different separations can be calculated (line in Figure 6, right). The software uses a static A-form RNA model and the photophysical properties of the probes as an input. The best overlap between the calculations and the experimental data was obtained for the spectral overlap integral, JDA,The, of 2.3 × 1014 M−1 cm−1 nm4 and an angle between the 2CNqA transition dipole and the X-axis (tD) set to 45.5°. From TDDFT calculations this angle is predicted to be 27.5°. Hence, the agreement between JDA from spectra and from the FRET data is good. Additionally, the TDDFT-predicted orientation of the 2CNqA transition dipole moment fits satisfactorily with what is suggested by the FRET measurements. Compared with in DNA, the theoretical FRET values fit better to experimental values at shorter distances in RNA (Figure 6, right). Overall, our new interbase FRET pair, 2CNqA-tCnitro, performs excellently in the RNA system studied and preserves the A-form secondary structure. Given the quantum yield of 2CNqA in RNA and the large spectral overlap with its FRET acceptor, the 2CNqA-tCnitro interbase FRET pair constitutes a valuable complement to the tCO-tCnitro FRET pair (45) – the only FRET pair studied in RNA to date, which is limited to cytosine substitutions, has a lower spectral overlap but a higher donor quantum yield than the FRET pair we report herein.
CONCLUSIONS
We have here performed a thorough photophysical and biophysical study of a new fluorescent adenine analogue, 2CNqA, inside DNA and RNA. Using circular dichroism, duplex melting and mismatch investigations, we show that 2CNqA is an excellent analogue of adenine in both DNA and RNA contexts. In the photophysical characterisation, we demonstrate that 2CNqA retains a high quantum yield inside nucleic acids. Unlike our previously reported bright adenine analogues, qAN1 and pA, the quantum yield of 2CNqA in duplex DNA is high irrespective of sequence context, and we find 2CNqA to be the brightest single-photon excitation fluorescent base analogue inside DNA reported until now. These powerful spectroscopic properties can be combined with our previously reported interbase FRET acceptors, qAnitro and tCnitro, to constitute excellent FRET pairs in DNA and RNA, respectively. Thanks to a large spectral overlap and high donor quantum yield, 2CNqA and qAnitro constitute the most powerful FRET pair reported for DNA so far. We envision that our new 2CNqA adenine analogue, due to its significant brightness, is a promising alternative to external fluorophores as an internal label for nucleic acids. Finally, we anticipate its use in a significant number of applications as an interbase FRET donor in pair with our acceptors reported herein; for example, we are currently utilizing it in assays reporting on interactions between drug molecules and pre-miRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
A.W.d.N. had the main responsibility for writing the manuscript, measured, assembled all the measured data and analysed it. A.F.F. and M.B. wrote and performed the synthesis part of the manuscript and measured and analysed (A.F.F.). J.N. measured and analysed and was part of writing the photodegradation part. M.S.W. was main responsible for the FRET fitting analysis part including writing of SI. S.S., P.P. and V.S.R. made measurements. A.H.E.-S. helped with synthesis and purification of oligonucleotides and supervised A.F.F. and M.B. in this process. A.D. and M.G. supervised the synthesis part and contributed in the development of synthetic routes. T.B. supervised A.F.F. and M.B. during work in his lab. L.M.W. conceived and supervised major parts of the project and revised drafts of manuscript texts. Everyone except S.S. contributed with feedback to the manuscript. We also wish to thank Mr. Tristan Giraud for contributions to a few of the measurements in the study, Assistant Professor Martin Rahm (Chalmers University of Technology, Department of Chemistry and Chemical Engineering) for help with the TDDFT calculations and Professor Andrzej Majhofer (University of Warsaw, Faculty of Physics) for an insightful discussion and advice on weighted average.
Contributor Information
Anna Wypijewska del Nogal, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
Anders F Füchtbauer, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden; Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg SE-412 96, Sweden.
Mattias Bood, Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg SE-412 96, Sweden; Medicinal Chemistry, Research and EarlyDevelopment, Cardiovascular, Renal and Metabolism (CVRM), BioPharmaceuticals R&D, AstraZeneca, Gothenburg, Pepparedsleden 1, Mölndal, SE-431 83, Sweden.
Jesper R Nilsson, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
Moa S Wranne, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
Sangamesh Sarangamath, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
Pauline Pfeiffer, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
Vinoth Sundar Rajan, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
Afaf H El-Sagheer, Chemistry Branch, Faculty of Petroleum and Mining Engineering, Suez University, Suez 43721, Egypt.
Anders Dahlén, Discovery Sciences, BioPharmaceuticals R&D, AstraZeneca, Gothenburg, Pepparedsleden 1, Mölndal, SE-431 83, Sweden.
Tom Brown, Chemistry Research Laboratory, Department of Chemistry, University of Oxford, Oxford OX1 3TA, UK.
Morten Grøtli, Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg SE-412 96, Sweden.
L Marcus Wilhelmsson, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Actions Individual Fellowship [H2020-MSCA-IF-2016, grant agreement no. 753595 to A.W.d.N.]; Swedish Foundation for Strategic Research [SSF, grant no. IS14-0041 and IRC15-0065 to L.M.W., ID14-0036 to M.G.]; Swedish Research Council [VR, grant no. 2017-03707 to L.M.W.]. Funding for open access charge: VR (The Swedish Research Council).
Conflict of interest statement. None declared.
REFERENCES
- 1. Shyu A.B., Wilkinson M.F., van Hoof A.. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008; 27:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kazimierczyk M., Kasprowicz M.K., Kasprzyk M.E., Wrzesinski J.. Human long noncoding RNA Interactome: detection, characterization and function. Int. J. Mol. Sci. 2020; 21:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mongelli A., Atlante S., Bachetti T., Martelli F., Farsetti A., Gaetano C.. Epigenetic signaling and RNA regulation in cardiovascular diseases. Int. J. Mol. Sci. 2020; 21:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lakowicz J.R. Principles of Fluorescence Spectroscopy. 2006; 3rd ednNY: Springer. [Google Scholar]
- 5. Zearfoss N.R., Ryder S.P.. End-labeling oligonucleotides with chemical tags after synthesis. Methods Mol. Biol. 2012; 941:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larson J.D., Rodgers M.L., Hoskins A.A.. Visualizing cellular machines with colocalization single molecule microscopy. Chem. Soc. Rev. 2014; 43:1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mirkin S.M. Discovery of alternative DNA structures: a heroic decade (1979-1989). Front. Biosci. 2008; 13:1064–1071. [DOI] [PubMed] [Google Scholar]
- 8. Watson J.D., Crick F.H.C.. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. [DOI] [PubMed] [Google Scholar]
- 9. Kim S.H., Suddath F.L., Quigley G.J., McPherson A., Sussman J.L., Wang A.H., Seeman N.C., Rich A.. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974; 185:435–440. [DOI] [PubMed] [Google Scholar]
- 10. Robertus J.D., Ladner J.E., Finch J.T., Rhodes D., Brown R.S., Clark B.F., Klug A.. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974; 250:546–551. [DOI] [PubMed] [Google Scholar]
- 11. Yan C., Hang J., Wan R., Huang M., Wong C.C., Shi Y.. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015; 349:1182–1191. [DOI] [PubMed] [Google Scholar]
- 12. Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A.. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000; 289:905–920. [DOI] [PubMed] [Google Scholar]
- 13. Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F. et al.. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000; 102:615–623. [DOI] [PubMed] [Google Scholar]
- 14. Wimberly B.T., Brodersen D.E., Clemons W.M. Jr., Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V.. Structure of the 30S ribosomal subunit. Nature. 2000; 407:327–339. [DOI] [PubMed] [Google Scholar]
- 15. Peulen T.O., Opanasyuk O., Seidel C.A.M.. Combining graphical and analytical methods with molecular simulations to analyze time-resolved FRET measurements of labeled macromolecules accurately. J. Phys. Chem. B. 2017; 121:8211–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kowalska J., Wypijewska del Nogal A., Darzynkiewicz Z.M., Buck J., Nicola C., Kuhn A.N., Lukaszewicz M., Zuberek J., Strenkowska M., Ziemniak M. et al.. Synthesis, properties, and biological activity of boranophosphate analogs of the mRNA cap: versatile tools for manipulation of therapeutically relevant cap-dependent processes. Nucleic Acids Res. 2014; 42:10245–10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wypijewska A., Bojarska E., Lukaszewicz M., Stepinski J., Jemielity J., Davis R.E., Darzynkiewicz E.. 7-Methylguanosine diphosphate (m7GDP) is not hydrolyzed but strongly bound by decapping scavenger (DcpS) enzymes and potently inhibits their activity. Biochemistry. 2012; 51:8003–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wypijewska A., Bojarska E., Stepinski J., Jankowska-Anyszka M., Jemielity J., Davis R.E., Darzynkiewicz E.. Structural requirements for Caenorhabditis elegans DcpS substrates based on fluorescence and HPLC enzyme kinetic studies. FEBS J. 2010; 277:3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wypijewska del Nogal A., Surleac M., Kowalska J., Lukaszewicz M., Jemielity J., Bisaillon M., Darzynkiewicz E., Milac A., Bojarska E.. Analysis of decapping scavenger (DcpS)-cap complex using modified cap analogs reveals molecular determinants for efficient cap binding. FEBS J. 2013; 280:6508-6527. [DOI] [PubMed] [Google Scholar]
- 20. Milac A.L., Bojarska E., Wypijewska del Nogal A.. Decapping scavenger (DcpS) enzyme: advances in its structure, activity and roles in the cap-dependent mRNA metabolism. Biochim. Biophys. Acta. 2014; 1839:452–462. [DOI] [PubMed] [Google Scholar]
- 21. Wilhelmsson L.M. Fluorescent nucleic acid base analogues. Q. Rev. Biophys. 2010; 43:159–183. [DOI] [PubMed] [Google Scholar]
- 22. Sinkeldam R.W., Greco N.J., Tor Y.. Fluorescent analogs of biomolecular building blocks: Design, properties, and applications. Chem. Rev. 2010; 110:2579–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dodd D.W., Hudson R.H.E.. Intrinsically fluorescent base-discriminating nucleoside analogs. Mini-Rev. Org. Chem. 2009; 6:378–391. [Google Scholar]
- 24. Xu W., Chan K.M., Kool E.T.. Fluorescent nucleobases as tools for studying DNA and RNA. Nat. Chem. 2017; 9:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matarazzo A., Hudson R.H.E.. Fluorescent adenosine analogs: a comprehensive survey. Tetrahedron. 2015; 71:1627–1657. [Google Scholar]
- 26. Fisher R.S., Nobis D., Füchtbauer A.F., Bood M., Grøtli M., Wilhelmsson L.M., Jones A.C., Magennis S.W.. Pulse-shaped two-photon excitation of a fluorescent base analogue approaches single-molecule sensitivity. Phys. Chem. Chem. Phys. 2018; 20:28487–28498. [DOI] [PubMed] [Google Scholar]
- 27. Nobis D., Fisher R.S., Simmermacher M., Hopkins P.A., Tor Y., Jones A.C., Magennis S.W.. Single-molecule detection of a fluorescent nucleobase analogue via multiphoton excitation. J. Phys. Chem. Lett. 2019; 10:5008–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bood M., Füchtbauer A.F., Wranne M.S., Ro J.J., Sarangamath S., El-Sagheer A.H., Rupert D.L.M., Fisher R.S., Magennis S.W., Jones A.C. et al.. Pentacyclic adenine: a versatile and exceptionally bright fluorescent DNA base analogue. Chem. Sci. 2018; 9:3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dierckx A., Miannay F.A., Gaied N., Preus S., Björck M., Brown T., Wilhelmsson L.M.. Quadracyclic adenine: a non-perturbing fluorescent adenine analogue. Chem. Eur. J. 2012; 18:5987–5997. [DOI] [PubMed] [Google Scholar]
- 30. Wranne M.S., Füchtbauer A.F., Dumat B., Bood M., El-Sagheer A.H., Brown T., Gradén H., Grøtli M., Wilhelmsson L.M.. Toward complete sequence flexibility of nucleic acid base analogue FRET. J. Am. Chem. Soc. 2017; 139:9271–9280. [DOI] [PubMed] [Google Scholar]
- 31. Sandin P., Börjesson K., Li H., Mårtensson J., Brown T., Wilhelmsson L.M., Albinsson B.. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008; 36:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foller Larsen A., Dumat B., Wranne M.S., Lawson C.P., Preus S., Bood M., Gradén H., Wilhelmsson M.L., Grøtli M.. Development of bright fluorescent quadracyclic adenine analogues: TDDFT-calculation supported rational design. Sci. Rep. 2015; 5:12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Börjesson K., Preus S., El-Sagheer A.H., Brown T., Albinsson B., Wilhelmsson L.M.. Nucleic acid base analog FRET-pair facilitating detailed structural measurements in nucleic acid containing systems. J. Am. Chem. Soc. 2009; 131:4288–4293. [DOI] [PubMed] [Google Scholar]
- 34. Han J.H., Yamamoto S., Park S., Sugiyama H.. Development of a Vivid FRET system based on a highly emissive dG-dC analogue pair. Chem. Eur. J. 2017; 23:7607–7613. [DOI] [PubMed] [Google Scholar]
- 35. Preus S., Kilså K., Wilhelmsson L.M., Albinsson B.. Photophysical and structural properties of the fluorescent nucleobase analogues of the tricyclic cytosine (tC) family. Phys. Chem. Chem. Phys. 2010; 12:8881–8892. [DOI] [PubMed] [Google Scholar]
- 36. Buhr C.A., Matteucci M.D., Froehler B.C.. Synthesis of a tetracyclic 2′-deoxyadenosine analog. Tetrahedron Lett. 1999; 40:8969–8970. [Google Scholar]
- 37. Niedballa U., Verbrüggen H.. Synthesis of nucleosides. 17. A general synthesis of N-glycosides. 6. On the mechanism of the stannic chloride catalyzed silyl Hilbert-Johnson reaction. J. Org. Chem. 1976; 41:2084–2086. [DOI] [PubMed] [Google Scholar]
- 38. Dumat B., Bood M., Wranne M.S., Lawson C.P., Larsen A.F., Preus S., Streling J., Gradén H., Wellner E., Grøtli M. et al.. Second-generation fluorescent quadracyclic adenine analogues: environment-responsive probes with enhanced brightness. Chem. Eur. J. 2015; 21:4039–4048. [DOI] [PubMed] [Google Scholar]
- 39. Dawson R.M.C. Data for Biochemical Research. 1986; Oxford: Clarendon Press. [Google Scholar]
- 40. Taylor J.R. An Introduction to Error Analysis. 1992; Oxford: Oxford University Press. [Google Scholar]
- 41. Uno K., Niikura H., Morimoto M., Ishibashi Y., Miyasaka H., Irie M.. In situ preparation of highly fluorescent dyes upon photoirradiation. J. Am. Chem. Soc. 2011; 133:13558–13564. [DOI] [PubMed] [Google Scholar]
- 42. Onidas D., Markovitsi D., Marguet S., Sharonov A., Gustavsson T.. Fluorescence properties of DNA nucleosides and nucleotides: a refined steady-state and femtosecond investigation. J. Phys. Chem. B. 2002; 106:11367–11374. [Google Scholar]
- 43. Sandin P., Wilhelmsson L.M., Lincoln P., Powers V.E.C., Brown T., Albinsson B.. Fluorescent properties of DNA base analogue tC upon incorporation into DNA - negligible influence of neighbouring bases on fluorescence quantum yield. Nucleic Acids Res. 2005; 33:5019–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Preus S., Kilså K., Miannay F.A., Albinsson B., Wilhelmsson L.M.. FRETmatrix: a general methodology for the simulation and analysis of FRET in nucleic acids. Nucleic Acids Res. 2013; 41:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Füchtbauer A.F., Wranne M.S., Bood M., Weis E., Pfeiffer P., Nilsson J.R., Dahlén A., Grøtli M., Wilhelmsson L.M.. Interbase FRET in RNA: from A to Z. Nucleic Acids Res. 2019; 47:9990–9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







