Abstract
The tumor suppressor BRCA2 plays a key role in initiating homologous recombination by facilitating RAD51 filament formation on single-stranded DNA. The small acidic protein DSS1 is a crucial partner to BRCA2 in this process. In vitro and in cells (1,2), BRCA2 associates into oligomeric complexes besides also existing as monomers. A dimeric structure was further characterized by electron microscopic analysis (3), but the functional significance of the different BRCA2 assemblies remains to be determined. Here, we used biochemistry and electron microscopic imaging to demonstrate that the multimerization of BRCA2 is counteracted by DSS1 and ssDNA. When validating the findings, we identified three self-interacting regions and two types of self-association, the N-to-C terminal and the N-to-N terminal interactions. The N-to-C terminal self-interaction of BRCA2 is sensitive to DSS1 and ssDNA. The N-to-N terminal self-interaction is modulated by ssDNA. Our results define a novel role of DSS1 to regulate BRCA2 in an RPA-independent fashion. Since DSS1 is required for BRCA2 function in recombination, we speculate that the monomeric and oligomeric forms of BRCA2 might be active for different cellular events in recombinational DNA repair and replication fork stabilization.
INTRODUCTION
Carriers of germline mutations in the breast cancer susceptibility gene BRCA2 are predisposed to breast cancer with high penetrance, ovarian and other cancers with lower penetrance (4,5). Extensive studies have revealed that BRCA2 is a critical factor in homologous recombination (HR), a high-fidelity DNA repair pathway for double-strand breaks (DSBs) and interstrand crosslinks (ICLs) with additional functions in DNA replication fork support (6,7). BRCA2 is required to orchestrate the formation of RAD51 filaments on single-stranded DNA (ssDNA), the catalytic scaffold of homology search and DNA strand invasion during HR. Molecularly, BRCA2 facilitates overcoming the inhibition from RPA, the eukaryotic ssDNA-binding protein (SSB), to form RAD51-ssDNA filaments, an essential activity directly demonstrated with purified full-length BRCA2 protein in vitro (8–10).
BRCA2 contains multiple domains including eight BRC repeats, a C-terminal DNA binding domain (C-termDBD) that binds ssDNA and double-stranded DNA (dsDNA), a separate ssDNA-binding site in the N-terminal region, and a C-terminal domain containing two nuclear localization signals (NLS) and an additional RAD51 interaction site (6,11–13). The sequence and spacing of the BRC repeats are conserved in mammals, and they bind RAD51 with different affinities by mimicking the structure of the individual RAD51 subunit at its polymerization interface (13–16). The C-termDBD of BRCA2 contains an α-helical domain and three oligonucleotide/oligosaccharide-binding (OB) folds, OB1, OB2 and OB3 (12). The structure of a small N-terminal region defining the PALB2 interaction has also been determined (17). Nevertheless, the structure and function of large regions of BRCA2 remain to be determined. A first low-resolution electron microscopic structure of full-length BRCA2 presents a dimeric BRCA2 configuration in a kidney bean shape (3). Upon binding to RAD51, this BRCA2 dimer extends into a heart shape and interacts with 4–5 RAD51 molecules symmetrically through each subunit (3). BRCA2 associated with the ends of RAD51–ssDNA filaments was suggested to promote the unidirectional growth of RAD51 on ssDNA (3). It remains a conceptual issue of how a symmetric BRCA2 dimer with RAD51 on both sides would support the nucleation of RAD51 on ssDNA with unidirectional growth, as discussed in (3). The analysis could not conclude whether only one subunit of the dimer is active or whether the BRCA2 in the filament is monomeric instead. Thus, more investigations of the full-length BRCA2 protein are needed to address this issue.
Among the BRCA2 interaction partners, DSS1 stands out since its depletion phenocopies a BRCA2 defect in mammals and fungi (18,19). DSS1 is a highly acidic protein with 70 amino acids. Mutational studies suggest that the BRCA2-DSS1 interaction is essential in vivo. Point mutations in the DSS1 binding site abolish the BRCA2 function (20,21). Moreover, a cancer-associated mutant of BRCA2, D2723H, fails to interact with DSS1 and is mislocalized to the cytoplasm due to an aberrantly exposed nuclear export signal (22). Several models have been proposed to account for how DSS1 modulates the function of BRCA2. These include improving protein stability, inhibiting DNA binding, controlling the monomer-dimer transition, and modulating the BRCA2–RAD51 interaction (18,20,23–27). Recently, it was shown that DSS1 displaces RPA from ssDNA acting as a DNA mimic, independent of BRCA2 (28). Human BRCA2 does not interact with RPA directly (8), and it is possible that DSS1 bound to BRCA2 has a different, BRCA2-dependent, role in HR. The versatility of DSS1 is reflected in its involvement in other biological processes. It is a component of the TREX-2 transcription export complex with Sac3 and Thp1, as well as of a transcriptional regulatory complex with Csn12 and Thp3 (29–32). DSS1 also functions in other functional contexts, including the 26S proteasome, where it forms a complex with Rpn3 and Rpn7 (33,34). Overall, the prevailing evidence suggests DSS1 modulates protein–protein and protein–DNA interactions, and therefore modulates the composition or architecture of multi-domain proteins and multi-protein complexes.
In this study, we show that negatively stained human full-length BRCA2 particles mainly exist as a mixture of monomers, dimers and multimers, consistent with previously published in vitro and in vivo observations (1–3). DSS1 or ssDNA alone could disrupt BRCA2 multimers. Simultaneous binding of DSS1 and ssDNA stabilizes the monomeric state of BRCA2 and improves its structural homogeneity. Structural analysis and comparisons between the dimeric and two independently derived monomeric forms suggest a self-interaction between the N- and C-terminus of BRCA2, which was validated by biochemical investigations of a series of BRCA2 mutants. This N-to-C terminal interaction is modulated by DSS1 and ssDNA, whereas a weaker N-to-N terminal interaction is modulated by ssDNA. Our results show how self-interactions influence the architecture of BRCA2 and how DSS1 disrupts multimerization to potentially regulate BRCA2 function in DNA repair.
MATERIALS AND METHODS
Establishment of full-length human BRCA2 expressing stable cell lines
The tagged full-length human BRCA2 expression construct, 2× MBP-BRCA2-FLAG-His10, was codon optimized to improve protein expression and cloned into phCMV1 (a gift from Dr. Kowalczykowski) and pcDNA5/FRT/TO vectors (a gift from Dr. Zhong). The 2× MBP-BRCA2-FLAG-His10 construct contains an 8 Glycine linker engineered between the first and second MBP, and a four Glycine linker engineered between the BRCA2 ORF and the FLAG tag. Additionally, a PreScission Protease site was engineered between the second MBP sequence and the start codon of the BRCA2 ORF. The BRCA2 ORF was custom synthesized with codon optimized sequence for human expression (GeneScript). All cloning steps and final constructs were verified by DNA sequencing. Thereafter, the two constructs expressing engineered BRCA2 will be referred as phCMV1-BRCA2 and pcDNA5-BRCA2 for simplicity. The phCMV1-BRCA2 expression plasmid was stably transfected into the BRCA2 mutant cells V-C8 (gift from Dr. Kowalczykowski) using Lipofectamine 2000 (Invitrogen), and cells were selected with Ham's F-10 media containing 15% FBS and 1mg/ml G418 (Sigma). G418-resistant colonies were further cultured and tested with Ham's F-10 media containing 15% FBS and 0.5 mg/ml G418. Cells were collected, and the expression of recombinant BRCA2 in established stable cell lines was confirmed by immunoprecipitation/blots. Cell lysates were immunoprecipitated with anti-BRCA2 antibody (Ab-1, EMD) first, and blotted with either anti-MBP (NEB) or another anti-BRCA2 antibody (H-300, Santa Cruz Biotechnology). A confirmed cell line stably expressing BRCA2 was used for the complementation assay of mitomycin C sensitivity of BRCA2-deficient cells to validate the function of dually tagged BRCA2 protein in vivo. Using the same method, the pcDNA5-BRCA2 purification plasmid was stably co-transfected with pOG44 into HEK293 Flp-In host cell line (gift from Dr. Zhong), followed by selection using 75 μg/ml Hygromycin B (EMD). Hygromycin B-resistant colonies were further cultured with DMEM media containing 10% FBS and 50 μg/ml Hygromycin B. Stable cell lines were induced with 10 μg/ml tetracycline (Sigma) and harvested after 48 h. Cell lysates were screened by immunoblots (anti-BRCA2 antibody H-300, Santa Cruz Biotechnology), and cell lines with the highest BRCA2 expressing level were used for BRCA2 purification.
Cell culture
Chinese hamster cells V79 (BRCA2 wild type cells) and V-C8 (brca2−/− cells) (both gifts from Dr. Kowalczykowski) were grown in Ham's F-10 media (Invitrogen) with 15% FBS (Sigma). V-C8 cells stably expressing BRCA2 were grown in Ham's F-10 media with 15% FBS and 0.5 mg/ml G418 (Sigma). BRCA2 purification cells were grown in DMEM (Corning Cellgro) with 10% Tet-free serum (Denville Scientific) and 50 μg/ml Hygromycin B (EMD). Cells were cultured at 37°C with humidity and 5% CO2.
Clonogenic survival assay
A V-C8 clone stably expressing recombinant BRCA2 (Clone #14) was used in the mitomycin C (MMC, Sigma) clonogenic survival assay. BRCA2 wild type V79 cells, BRCA2 mutant V-C8 cells, and V-C8 cells transfected with control phCMV1 or phCMV1-BRCA2 vectors were each plated in triplicate in 6-well plates. Different numbers of cells were plated for each different cell line to obtain reliable colony numbers after MMC treatment. With 0, 0.1, 0.25, 0.5, 0.75 μM MMC, 200, 300, 800, 3000 and 5000 cells were plated for V-C8 cells, respectively. For V-C8 cells transfected with phCMV1 empty vector, 300, 600, 2000, 10 000 and 30 000 cells were plated, respectively. For V-C8 cells transfected with phCMV1-BRCA2, 300, 300, 400, 500 and 500 cells were plated respectively. For V79 cells, 150 cells were plated for each MMC concentration. After overnight incubation, cells were treated with different final concentrations of MMC for 1 h. After treatment, MMC was immediately removed and cells were washed with PBS. Cells were cultured in Ham's F-10 with 15% FBS for 8–11 days to form colonies. Colonies were washed with PBS and stained with 0.5% crystal violet (Sigma) for 0.5–1 h. Colonies on each plate were counted and the surviving fraction was determined for each drug treatment.
Purification of full-length human BRCA2
BRCA2 was purified from selected stable cell lines according to published protocols with slight modifications (8,9). BRCA2 cells (180 15-cm plates, ∼4.5 l) were lysed with 150 ml lysis buffer containing 50 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA, 1 mM TCEP, 3 mM MgCl2, 1 mM ATP, 1 mM PMSF, 1% Igepal CA-630 and Protease Inhibitor Cocktail (Roche). The cell suspension was rotated for 1 h at 4°C and then harvested at 35 000 rpm for 45 min. The cleared supernatant was incubated with 1 ml of amylose resin (New England Biolabs) per 50 ml of cell lysate. After incubating overnight at 4°C, the amylose resin was harvested at 1500 rpm for 5 min to remove the supernatant and then poured into a disposable plastic column (Pierce) before extensive washing with buffer A containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 1 mM EDTA, 1 mM TCEP, 0.2 mM PMSF, 10% glycerol and 0.05% Igepal CA-630. BRCA2 was eluted from the amylose column with 10 mM maltose in buffer EA containing 50 mM HEPES (pH 7.5), 250 mM NaCl, 0.5 mM EDTA, 0.25 mM TCEP, 0.2 mM PMSF, 10% glycerol and 0.05% Igepal CA-630. Fractions were analyzed by 4–15% gradient SDS-PAGE and pooled fractions were incubated with anti-FLAG M2 affinity gel (Sigma) for 1 h at 4°C. The anti-FLAG affinity gel was poured into a disposable plastic column (Pierce) before extensive washing with buffer F containing 50 mM HEPES (pH 7.5), 250 mM NaCl, 0.2 mM EDTA, 0.25 mM TCEP, 10% glycerol, 0.05% Igepal CA-630). BRCA2 was eluted from the FLAG resin with 100 μg/ml FLAG peptide (Sigma) in buffer F. Full-length human BRCA2 protein containing the 2× MBP and FLAG-His10 tags was verified by 4–15% gradient SDS-PAGE and Denville Blue (Denville Scientific) staining.
Plasmid construction of truncated BRCA2 fragments
All truncated BRCA2 fragments were PCR amplified from the phCMV1-BRCA2 expression plasmid and subcloned into pFastBac1, pET28b+ or pET15b vectors. BRCA2CT (aa 2300–3418) and BRCA2NT (aa 1–714) fragments were cloned into pFastBac1 and fused with two MBP tags at the amino terminus and a FLAG-His10 tag at the carboxyl terminus, similar to the full-length BRCA2 expression construct. Thereafter, they will be referred as pFB1-MBP-BRCA2CT and pFB1-MBP-BRCA2NT for simplicity. Another tagged version of BRCA2NT fragment was generated similarly into pFastBac1 vector with a GST tag inserted and separated by a TEV cleavage site at the amino terminus, and a HA-His10 tag at the carboxyl terminus, referred as pFB1-GST-BRCA2NT. For truncations of the BRCA2NT fragment, BRCA2N3 (aa 1–106), BRCA2N4 (aa 107–211), and BRCA2N5 (aa 212–320) were cloned into pFastBac1 vector with a GST tag inserted and separated by TEV cleavage site at the amino terminus, and two HA tags and six Histidines at the carboxyl terminus, referred as pFB1-GST-BRCA2N3, N4 and N5, respectively. Another set of BRCA2NT truncations, including BRCA2N7 (aa 250–370), BRCA2N8 (aa 371–480) and BRCA2N9 (aa 481–714) was integrated into pET28b+ vector and fused with GST and Histidine tags, but no HA tag. They are referred as pET28b+-GST-BRCA2N7, N8 and N9, respectively. The OB2 fragment (aa 2780–3080) was cloned into pET15b vector and fused with two MBP tags at the amino terminus, and 10 histidines at the carboxyl terminus, referred as pET15b-MBP-OB2. All constructs were sequence verified by DNA sequencing.
MBP-truncated BRCA2 fragment purification
2× MBP-BRCA2CT-FLAG-His10 (thereafter referred as MBP-BRCA2CT) and 2× MBP-BRCA2OB2-His10 (referred as MBP-OB2) were purified by the same procedure. Proteins from baculovirus-infected insect cells obtained from 3-liter cultures were lysed in 200 ml Buffer A (50 mM Tris–Cl (pH 8.0), 1 mM EDTA, 10% glycerol) containing 250 mM NaCl, 0.5% NP-40, 1 mM TCEP, 1 mM PMSF, 2 μM Leupeptin, 2 μM Pepstatin A and 1 mM Benzamidin. The cell suspension was rotated for 1 h at 4°C. After high-speed centrifugation, the cleared lysates were incubated with 1 ml of amylose resin (New England Biolabs) per 40 ml of cell lysate for 4 h at 4 C°, and then packed into a disposable plastic column (Pierce). The beads were extensively washed with Buffer A containing 1.5 M NaCl, 0.05% NP-40, 1 mM TCEP, 0.2 mM PMSF, 2 μM Leupeptin, 1 μM Pepstatin A and 0.1 mM Benzamidin. The bound proteins were eluted with 20 mM Maltose in Buffer A containing 50 mM NaCl, 0.05% NP-40, 1 mM TCEP, 0.2 mM PMSF, 2 μM Leupeptin, 1 μM Pepstatin A and 0.1 mM Benzamidin. Fractions were analyzed by 4–20% gradient SDS-PAGE (BioRads) and pooled fractions were passed through a MonoQ column. The bound proteins were eluted by a 50–600 mM NaCl gradient in buffer A containing 1 mM TCEP. MBP-BRCA2CT containing fractions were identified by 4–20% gradient SDS-PAGE with SYPRO Orange (ThermoFisher Scientific) staining and immunoblotting with anti-MBP antibody (New England Biolabs), anti-BRCA2 H-300 (Santa Cruz Biotechs), anti-FLAG (Sigma). The quality of MBP-BRCA2OB2 fractions were evaluated by 4–20% gradient SDS-PAGE with SYPRO Orange (ThermoFisher Scientific) staining. 2× MBP-BRCA2NT-FLAG-His10 (thereafter referred as MBP-BRCA2NT) was purified similarly as MBP-BRCA2CT, but using Buffer M (50 mM HEPES pH 7.8, 1 mM EDTA, 10% glycerol, 1 mM ATP and 3 mM MgCl2) as a base buffer instead of Buffer A. Eluted fractions from MonoQ column were verified by 4–20% gradient SDS-PAGE with SYPRO Orange (ThermoFisher Scientific) staining.
GST-truncated BRCA2 fragment purification
Truncated BRCA2 N-terminal fragments were purified from either 500-ml cultures of insect cells for GST-BRCA2N3, N4, and N5 or 10 liters of Escherichia coli Arctic (DE21) carrying pET28b+-GST-BRCA2N7, N8, or N9 after induction with 1 mM IPTG (USBiological) for GST-BRCA2N7, N8 and N9. Cells were harvested and lysed in 200 ml Buffer A containing 250 mM NaCl, 0.5% NP-40, 1 mM TCEP, 1 mM PMSF, 2 μM Leupeptin, 2 μM Pepstatin A and 1 mM Benzamidin. The cell suspension was rotated for 1 h at 4 °C. After high-speed centrifugation, the cleared lysates were incubated with 5 ml of Pierce™ Glutathione Agarose resin (ThermoFisher Scientific) for overnight at 4°C and then packed into a disposable plastic column (Pierce). The beads were extensively washed with Buffer A containing 1.5 M NaCl, 0.05% NP-40, 1 mM TCEP, 0.2 mM PMSF, 2 μM Leupeptin, 1 μM Pepstatin A and 0.1 mM Benzamidin. The bound proteins were eluted with 30 mM Glutathione in PBS (137 mM NaCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 and 2.7 mM KCl) containing 1 mM TCEP. Fractions were analyzed by 4–20% gradient SDS-PAGE and pooled fractions were batch bound to 6 ml HisPur™ Ni-NTA Superflow Agarose resin (ThermoFisher Scientific) for 4 h at 4°C, and then packed into a disposable plastic column. The beads were extensively wash with PBS containing 1 M NaCl and 10 mM Imidazole. The bound proteins were eluted with 500 mM Imidazole in Buffer A containing 50 mM NaCl, 5 mM EDTA and 1 mM DTT. Fractions were analyzed by 4–20% gradient SDS-PAGE and pooled fractions were passed through a MonoQ column. The bound proteins were eluted by a 50–800 mM NaCl gradient in buffer A containing 5 mM EDTA and 1 mM DTT. Fractions were analyzed by 4–20% gradient SDS-PAGE with SYPRO Orange (ThermoFisher Scientific) staining. GST-BRCA2NT-HA-His10 (thereafter referred as GST-BRCA2NT) was purified similarly as GST-BRCA2N3, but using Buffer M (50 mM HEPES pH 7.8, 1 mM EDTA, 10% glycerol, 1 mM ATP and 3 mM MgCl2) as a base buffer instead of Buffer A and the Ni-NTA column was skipped. Eluted fractions from MonoQ column were analyzed by 4–20% gradient SDS-PAGE with SYPRO Orange (ThermoFisher Scientific) staining.
Human DSS1 protein
Human DSS1 protein was expressed in E. coli and purified without modification as described (9).
Crosslinking of BRCA2 complexes
About 2.5–5 μg of BRCA2 (0.5 nM) alone, with either 28.7 μM in nucleotides/0.287 μM molecule of 5′-biotinylated oligo (Supplementary Table S1), 0.64 μM of DSS1, or both were incubated in buffer R containing 20 mM HEPES (pH 7.5), 1 mM DTT, and 100 mM NaCl, at at 37°C for 30 min. The different BRCA2 species was crosslinked with 0.1% glutaraldehyde for an hour before EM imaging. We collected about 150 images each for BRCA2, BRCA2–DSS1 and BRCA2–ssDNA, and 1359 images for BRCA2 + DSS1 + ssDNA, from different squares and areas of the grids with identical imaging settings. For analysis of the largest diameters, 600–900 particles were measured manually for each condition.
Negative stain electron microscopy
5 μl of BRCA2 or BRCA2 complexes was applied to glow-discharged copper grids with a layer of continuous carbon and incubated for 2 min. Grids were washed briefly with 5 mM Mg(OAc)2 for three times to remove buffer, sucrose, and glycerol. Samples were then stained with 2% uranyl acetate, blotted with filter paper, and air dried. Electron micrographs of uncrosslinked BRCA2–DSS1–ssDNA complexes were collected on a TVIPS 4k × 4k FastScan CCD camera with 10 μm pixels on a JEOL-2100F transmission electron microscope operating at 200 kV and × 40 000 nominal magnification, corresponding to a sampling of 2.0 Å/pixel. Images were collected using 1.0–3.5 μm under focus and low dose (25–30 e/Å2) configuration using EM-MENU software (TVIPS). Electron micrographs of crosslinked BRCA2-DSS1-ssDNA complex were collected on Direct Electron DE-20 detector on a JEOL-2100F transmission electron microscope operating at 200 kV and × 40 000 nominal magnification, corresponding to a sampling of 1.48 Å/pixel. Images were collected using 1.0–3.5 μm under focus and low dose (25–50 e/Å2) configuration using SerialEM (35).
Sucrose gradient centrifugation
A 5-ml 10–35% sucrose gradient was prepared by mixing equal volumes of 10% and 35% sucrose in buffer S containing 20 mM HEPES (pH 7.5), 1 mM DTT, 100 mM NaCl, 5% glycerol using a gradient maker (JULE, INC). About 2.5–5 μg of BRCA2 (0.5 nM) alone, with either 28.7 μM in nucleotides/0.287 μM molecule of 5′-biotinylated oligo (Supplementary Table S1), 0.64 μM of DSS1, or both were incubated in buffer R containing 20 mM HEPES (pH 7.5), 1 mM DTT and 100 mM NaCl, at 37°C for 30 min and loaded onto the top of the sucrose gradient in a 5-ml ultraclear centrifugation tube (Beckman). Gradients were centrifuged at 45 000 rpm for 16 h in a Beckman SW55-Ti rotor at 4°C. Gradients were freshly fractionated in a top to bottom fashion by manually pipetting 200 μl solution as each fraction. 3 μl of pooled fractions from each condition were used for negative staining. We collected about 50 images each for BRCA2, BRCA2–DSS1 and BRCA2–ssDNA, and 686 images for BRCA2 + DSS1 + ssDNA, from different squares and areas of the grids with identical imaging settings. For analysis of the largest diameters, 100 particles were measured manually for each condition. 12 μl of each fraction was used to analyze the level of BRCA2 by immunoblots, and 6 μl for dot blots to test the levels of DNA or DSS1. BRCA2 was detected using anti-BRCA2 (H-300, Santa Cruz Biotechnology), anti-MBP (NEB) or anti-FLAG (Sigma) antibodies. The DSS1 dot blot was first crosslinked with 2% glutaraldehyde for 45 min before blotting. Anti-DSS1 (Santa Cruz Biotechnology) was used as the primary antibody. The 5′-biotinylated oligo was crosslinked with 120 mJ/cm2 of UV at 254 nm for 12 s before blotting, and an anti-Streptavidin antibody (HRP) (Abcam) was used to recognize the biotin-cojugated oligonucleotide. Globular protein standards were made as 20 mg/ml stocks in buffer R. Five μl BSA, 5 μl ovalbumin, 5 μl aldolase, 15 μl catalase and 25 μl thyroglobulin were mixed and loaded on top of the gradients for identical centrifugation process as BRCA2 species. Protein standards were analyzed by SDS-PAGE and Denville Blue (Denville Scientific) staining. The relative amount of protein and DNA in each fraction were determined through densitometry using ImageQuant (GE).
Gel filtration analysis
About 2.5 μg of BRCA2, alone or with 28.7 μM (in nucleotides) of 5′-biotinylated oligo (Supplementary Table S1) and 0.64 μM of DSS1, were incubated in buffer R containing 20 mM HEPES (pH 7.5), 1 mM DTT and 100 mM NaCl, at 37°C for 20 min and loaded onto the top of a superose 6 increase column, pre-equilibrated with buffer T containing 10 mM Tris–HCl (pH 7.5), 1 mM TECP, 1 mM Mg(OAc)2 and 140 mM NaCl. Fractions were collected and concentrated before analysis by SDS-PAGE and then Denville Blue (Denville Scientific) staining. Simultaneously, 5 μl of concentrated fractions (Supplementary Figure S3B) was applied to glow-discharged continuous carbon copper grids and incubated for 1 min. Samples were then stained with 2% uranyl acetate, blotted with filter paper, air dried, and then imaged with a JEOL-1230 using a Tietz 2k × 2k CCD at the indicated magnification (Supplementary Figure S3C,D). Globular protein standards were prepared as 20 mg/ml stocks in buffer R. Ten microliters ovalbumin, 10 μl aldolase, 15 μl catalase and 25 μl thyroglobulin were mixed and loaded on top of the superose 6 increase column for the identical separation process as the BRCA2 species. Protein standards were analyzed by SDS-PAGE and then Denville Blue (Denville Scientific) staining. The relative amount of protein in each fraction were determined through densitometry using ImageQuant (GE).
Image processing
For BRCA2–DSS1–ssDNA complexes without crosslinking, a total of 686 micrographs were collected, and a total of 35 985 particles were boxed and extracted using EMAN2 (36). The particles were phased flipped, CTF corrected and filtered using a high-pass filter of 250 Å and a Gaussian low-pass filter of 10 Å. The filtered particle set was then classified using Sparx's 2D clustering with ISAC (37), from which 369 stable classes, representing 19 968 particles, were obtained. Best classes with unique views were selected to generate 15 initial models using Sphire's ab initio 3D model generator, VIPER. Each model was manually compared using its projections to the class averages from which they were derived and the model with the best matching was chosen as the initial model for refinement through Sphire (38). Iterative 3D reconstructions were perfomed in Sphire using MERIDIEN from two independently refined half datasets and reached an average resolution of 17.1 Å as indicated by Fourier Shell Correlation (FSC) criterion at 0.5. The class averages generated in Sphire from the final data subset were then compared to back-projections of the reconstructed model using EMAN2’s classvsproj.py to assess that the reconstructed model encompassed the different views observed in the class averages. Similarly, for BRCA2–DSS1–ssDNA complexes with mild crosslinking, a total of 1359 micrographs with 80 985 particles were extracted and processed in the same way through Sphire. The average resolution reached 14.6 Å, as indicated by FSC criterion at 0.5. All figures containing 3D density maps were generated using CHIMERA (39).
Immuno nanogold and Fab fragment staining
Nanogold labeling was set up with BRCA2–DSS1–ssDNA complexes, isolated from fractions #15-17 of the 10–35% sucrose gradient. One to 10 μl of peak fraction was incubated with 2 to 4 μl of 1:20 or 1: 50 diluted Ni-NTA nanogold (Nanoprobes) or streptavidin-conjugated nanogold (Cytodiagnostics) in buffer R containing 20 mM HEPES (pH 7.5) and 100 mM NaCl for an hr at room temperature. Either 3 or 5 μl of the mixture was applied to glow-discharged continuous carbon copper grids and incubated for 2 min before staining with 2% uranyl acetate.
Anti-MBP Fab fragments were produced through the Pierce Fab micro preparation kit (Thermo Fisher Scientific). Crosslinked BRCA2–DSS1–ssDNA complexes were incubated with 1 μl of purified Fab fragment in buffer R containing 20 mM HEPES (pH7.5) and 100 mM NaCl for an hr at room temperature. Either 3 or 5 μl of the mixture was applied to glow-discharged copper grids with a layer of continuous carbon and incubated for 2 min before staining with 2% uranyl acetate.
Pull-down assay
Glutathione pull-down assays using purified proteins were performed in binding buffer containing 50 mM HEPES–NaOH (pH 7.8), 4 mM Mg(OAc)2, 150 mM NaCl, 1 mM TCEP, 10% glycerol, 100 μg/ml BSA and 0.1% NP-40. For assays using GST-tagged full-length BRCA2 as a bait, in Figure 4C, a varying amount of MBP-BRCA2NT at 0.5, 1.5, 3, 5 and 6 nM was mixed with 0.65 nM GST-BRCA2-FLAG-His10 which was purified as described previously (9) in 100 μl binding buffer. In Figure 5A, 15 nM MBP-BRCA2CT was mixed with 1.5 μM DSS1 and then 1.5 nM GST-BRCA2-FLAG-His10 in 100 μl binding buffer. The mixtures were incubated at 22°C for 1 h. Equilibrated and BSA-treated glutathione agarose resin (Thermo Fisher Scientific) were added into the mixture and incubated for another hour. The beads and supernatant were separated by centrifugation and the beads were washed three times with 500 μl binding buffer. The resulting protein complexes were eluted by boiling at 55°C for 10 min in 15 μl SDS-PAGE loading buffer and analyzed by 4–20% SDS-PAGE gradient gel. Separated proteins were then transferred onto PVDF membranes for immunoblotting. GST-BRCA2-FLAG-His10 and GST control (GE Healthcare) proteins were detected using an anti-GST antibody (GE Healthcare), and MBP-BRCA2CT and MBP-BRCA2NT were blotted by an anti-MBP antibody (New England Biolabs).
For assays using GST-tagged BRCA2NT as a bait, typically 100 nM MBP-BRCA2CT or MBP-BRCA2NT, unless indicated otherwise in the figure legends, were mixed with additional factors as indicated in the figure legends and then with 30 nM GST-BRCA2NT in 100 μl binding buffer. The mixtures were then conducted similarly as reactions for GST-tagged full-length BRCA2. GST control (GE Healthcare) protein was detected using an anti-GST antibody (GE Healthcare), GST-BRCA2NT protein was visualized by an anti-BRCA2 (ab123491, Abcam) or anti-GST (GE Healthcare) antibodies, and MBP-BRCA2CT or MBP-BRCA2NT were blotted by an anti-MBP antibody (New England Biolabs).
To quantify the amount of bound proteins, the input signals of samples were first normalized to the input signal of GST control to obtain the normalized ratio. And then, the percentage of bound proteins was calculated as the following formula: %bound protein = [(pulled-down signal of sample/normalized ratio) – pulled-down signal of GST control)]/normalized Input signal × 10%.
RESULTS
An N- and C-terminally tagged BRCA2 construct is functional In Vivo
Purification of full-length human BRCA2 protein faces significant technical challenges, complicating its in vitro biochemical and structural characterization. To improve protein expression and purification, we re-engineered a full-length human BRCA2 construct for human cell expression, with two consecutive MBP tags at the N-terminus (8) and a tandem FLAG-His10 tag at the C-terminus (Figure 1A) (9). To test whether this tagged BRCA2 is functional in vivo, we transfected the hamster V-C8 brca2−/− cell line (40) and established a stable cell line expressing 2 × MBP-BRCA2-FLAG-His10. As shown in Figure 1B, the immunoprecipitated BRCA2 from the stable clone #14 contains the MBP tag as detected by immunoblots. The same band with the size corresponding to the full-length construct is also recognized by another BRCA2 antibody towards a different region (Figure 1A, B). Clone #14 shows full resistance to mitomycin C, very similar to wild type V79 hamster cells. In contrast, untransfected brca2−/− V-C8 cells and V-C8 cells transfected with an empty vector display nearly identical severe sensitivity to mitomycin C (Figure 1C). After confirming that the tagged BRCA2 is functional in vivo, we generated a stable HEK293 Flp-In cell line expressing tetracycline-inducible 2× MBP-BRCA2-FLAG-His10, and we will refer it as BRCA2 for simplicity hereafter (41). As shown in Figure 1D, full-length human BRCA2 with >90% purity can be obtained after sequential affinity chromatography selecting for the MBP and FLAG tags. The presence of both tags in the purified BRCA2 protein was also documented by immunoblotting using the respective antibodies (for example, see Supplementary Figure S2C). This improved BRCA2 purification enables us to obtain ∼100 μg of highly purified protein without apparent contaminations from about 180 15-cm plates of HEK293 culture. The twenty-fold increase in protein yield compared to our previous yeast expression system enables us to further investigate the biochemical and structural features of BRCA2 (9).
Figure 1.
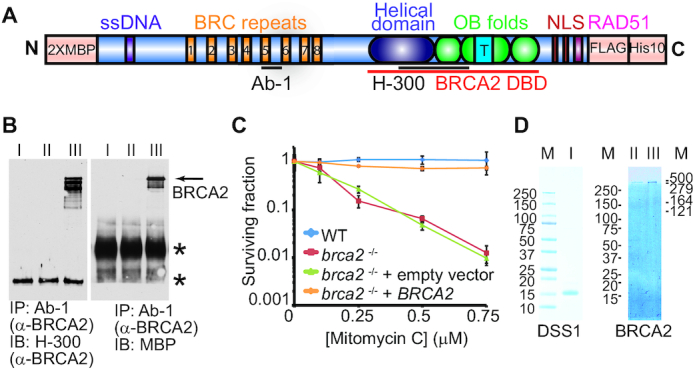
The tagged full-length human BRCA2 is functional in vivo. (A) Scheme of 2× MBP-BRCA2-FLAG-His10 construct. The BRCA2 purification construct was engineered to have 2× MBP at the N-terminus and FLAG-His10 at the C-terminus. (B) Expression of 2× MBP-BRCA2-FLAG-His10 in established stable clone (#14) was verified by immunoprecipitation and immunoblots. BRCA2 from cellular lysate was immunoprecipitated with anti-BRCA2 (Ab-1) antibody and blotted with either a different anti-BRCA2 (H-300) or anti-MBP antibodies. I. brca2−/- mutant V-C8 cells. II. V-C8 cells transfected with empty vector. III. V-C8 cells transfected with purification vector coding 2 × MBP-BRCA2-FLAG-His10 construct, clone #14. Clone #14 was used in the clonogenic survival assay to assess complementation in C. The stars (*) indicate crossreacting proteins. (C) Mitomycin C survival curve of BRCA2 wild type V79 cells, brca2−/- V-C8 cells, and V-C8 cells transfected with control or purification vectors. Plotted are means ± standard deviation from n = 3. (D) Purified human DSS1 (I) and full-length human BRCA2 visualized on a 4–15% gradient SDS-PAGE gel stained with Denville Blue. II, FLAG fraction of BRCA2. III, the final pool of BRCA2. M, protein standards.
BRCA2 particles are a mixture of monomers and multimers
We used transmission electron microscopy (TEM) to examine negatively stained BRCA2, which shows a high level of heterogeneity. The structures of BRCA2 span from small degradation products to large multimers (Supplementary Figure S1A), consistent with the previous observation that BRCA2 particles show a wide range of particle sizes (1,3). Full-length BRCA2 appears to be fragile in vitro, inferred from the degradation products observed on grids, possibly induced by the acidity of the uranyl acetate staining. Interestingly, the incubation with DSS1 or ssDNA decreases the extent of BRCA2 aggregation, and smaller protein particles were observed instead (Supplementary Figure S1A).
We developed a mild crosslinking condition to stabilize BRCA2 species and improve the structural appearance during negative staining, in order to further investigate BRCA2 multimerization (Figure 2A). We measured the side-to-side distance of the BRCA2 particles and used the largest dimension to compare their sizes. Alone, BRCA2 particles show a wide distribution in the largest dimensions ranging from 120 to 1257 Å, with an average of 324 ± 128 Å (Figure 2B). As a reference, the largest dimension of the published dimeric structure of BRCA2 is 250 Å, and the molecule assumes an elongated and hollow conformation (3), as shown in Figure 3F. Our construct adds a 90-kDa 2× MBP tag, and dimeric BRCA2 is expected to have the largest dimension at ∼300 Å, assuming a similar conformation (Figure 2A). Particles smaller than 120 Å were excluded in the analysis as degradation products. Thus, most of the BRCA2 particles show sizes consistent with dimers and multimers but display significant structural heterogeneity (Figure 2A, B). These results are consistent with the previous reports that described the heterogeneous nature of BRCA2 in vitro and in vivo (1–3).
Figure 2.
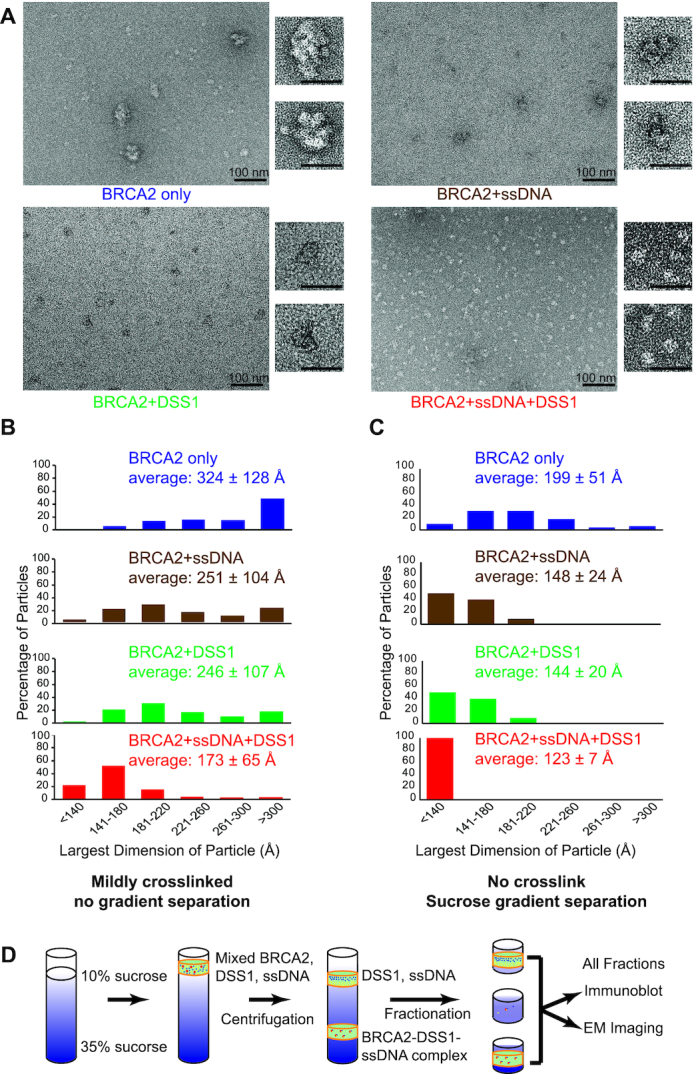
DSS1 and ssDNA counteract the self-association of BRCA2 synergistically. (A) Representative micrograph overviews of negative stained BRCA2, BRCA2-ssDNA, BRCA2-DSS1 and BRCA2–DSS1–ssDNA complexes. For complex formation in A–C, 0.5 nM BRCA2 was incubated with either 28.7 μM in nucleotides/0.287 μM molecule of 5′-biotinylated oligo, or 0.64 μM of DSS1, or both DNA and DSS1. Enlarged views of single particles were shown on the right next to the overview. Scale bar: 100 nm in overviews and 50 nm in enlarged panels. (B) Distribution of the largest dimensions of the crosslinked particles in (A). (C) Distribution of the largest dimensions of the uncrosslinked particles after sucrose gradient separation, as illustrated in (D). The average diameter ± standard deviation was shown for each condition in (B) and (C).
Figure 3.
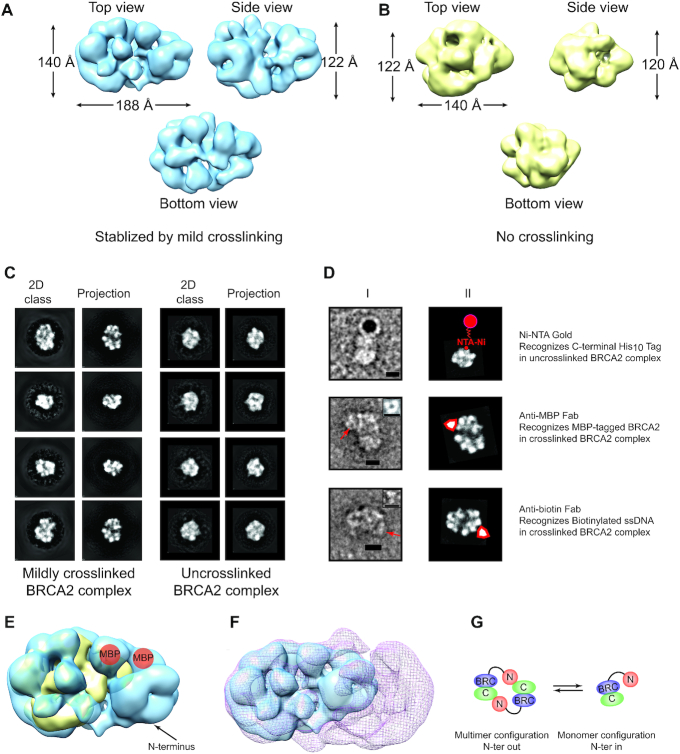
3D reconstruction of the monomeric BRCA2-DSS1-ssDNA complex. Top, side and bottom views of the 3D reconstruction of crosslinked (EMD-21998) (A) or uncrosslinked (EMD-20348) (B) BRCA2–DSS1–ssDNA complexes. (C) Representative reference-free 2D class averages of BRCA2–DSS1–ssDNA complex compared to forward reprojections of the 3D structure in A (crosslinked) or B (uncrosslinked). (D) Top: Ni-NTA nanogold labeling against C-terminal His10 tag; Middle: anti-MBP Fab labeling MBP-tagged BRCA2; Bottom: anti-biotin Fab labeling biotinylated ssDNA. I, Raw particles with nanogold or Fab. II, Matching reference-free 2D class averages with the same orientation as the particle with an illustration of nanogold or Fab. Free Fab particles shown in small inserts. Scale bar, 10 nm. (E) Comparison of the crosslinked BRCA2 complex (EMD-21998, blue) versus uncrosslinked complex (EMD-20348, yellow). Mapped MBP tag from D is shown in orange, and N-terminus is labeled. (F) Comparison of the monomeric BRCA2–DSS1–ssDNA complex (EMD-21998, blue, crosslinked) versus dimeric BRCA2 structure (EMD-2779, red mesh) (3). (G) Structural comparisons indicate that the N-to-C terminal contact is disrupted during the dimer-to-monomer transition, accompanied by a significant structural rearrangement.
Binding of DSS1 and ssDNA synergistically stabilizes the monomeric state of BRCA2
Next, we asked whether DSS1 and ssDNA could modulate the self-association of BRCA2. A large number of micrographs were randomly collected from different areas across the grids with identical imaging settings to ensure fair presentation of the data with no bias. We noticed that particles from different complexes were stained differently with uranyl acetate, leading to varying contrast in the micrographs and difficulty in showing morphological changes (Figure 2A & Supplementary Figure S1B). However, both BRCA2–DSS1 and BRCA2–ssDNA particles show a clear shift towards smaller sizes, and the majority of these particles have a largest dimension of <300 Å (Figure 2A, B). The sizes are more consistent with the monomeric form of BRCA2 in different conformations and not compatible with the dimeric form of BRCA2. In conclusion, both DSS1 and ssDNA inhibit multimerization of BRCA2, consistent with previous AFM studies showing that incubation with ssDNA appeared to shift branched BRCA2 multimers to smaller forms (1).
The simultaneous addition of both DSS1 and ssDNA further decreases the size of BRCA2 particles, suggesting a synergistic action in stabilizing BRCA2 monomeric species (Figure 2A, B). Indeed, these particles of the BRCA2-DSS1-ssDNA complex show significant improvement in structural homogeneity, illustrated by the micrograph overview and the enlarged particles (Figure 2A). The apparent shift in the sizes of BRCA2 particles when purified DSS1 is added (Figure 2A, B), along with the morphology changes in BRCA2–DSS1 or BRCA2–DSS1–ssDNA particles, suggest that our purified BRCA2 protein does not contain endogenous DSS1 from mammalian cells through the purification process. Furthermore, there is no detectable DSS1 immunoblot signal in the conditions without added DSS1 (BRCA2 only or BRCA2 with ssDNA in Supplementary Figure S1D).
DSS1 and ssDNA inhibit self-interactions of BRCA2 without crosslinking
To exclude the possibility that the mild crosslinking might affect how BRCA2 multimers behave in the presence of DSS1 and ssDNA, we developed another strategy to stabilize BRCA2 complexes and avoid crosslinking by using sucrose gradient separation. An excess of DSS1 and ssDNA were used to ensure complex formation, and free DSS1 and ssDNA were effectively removed by a continuous 10–35% sucrose gradient, along with small degradation products and large aggregates (Figure 2D). Immunoblots with different antibodies were used to identify each component and confirm BRCA2 integrity through the entire gradients (Supplementary Figure S1C,D). Sucrose gradients are limited in their resolution (Supplementary Figure S2A), causing a broad distribution of full-length BRCA2 with two major overlapping peaks (Supplementary Figure S1B), identified by the immunoblot signal with anti-BRCA2 antibodies (Supplementary Figure S1C). Different concentrations and ranges do not change this wide distribution profile in the sucrose gradient significantly. We prepared every fraction containing full-length BRCA2 for negative stain and collected images to detect particles. Only fractions 15–17 contain a large number of visible particles with a variety of sizes (Supplementary Figure S1B). The position of fractions 15–17 is consistent with a monomeric form of BRCA2 relying on globular protein standards (Supplementary Figure S2A), but the low resolution of this approach does not allow a firm conclusion. Later fractions show large protein aggregates on the grids with very few visible particles which were not suitable for further analysis.
In order to compare BRCA2 species formed with or without DSS1 and ssDNA, we pooled fractions 15–17 across all conditions and analyzed them in more detail. We measured the side-to-side distance of the BRCA2 particles and compared the largest dimension in the same way as crosslinked samples. Figure 2C shows the same trend that BRCA2 particles are decreasing in sizes with the addition of DSS1 and ssDNA. The synergy of DSS1 and ssDNA in stabilizing the monomeric form of BRCA2 is also observed with uncrosslinked protein complexes. More importantly, the uncrosslinked BRCA2-DSS1-ssDNA complex also shows improved homogeneity and display similar structural features to the crosslinked samples.
We also used gel filtration chromatography to analyze the effect of DSS1 and ssDNA on BRCA2 multimerization. Alone in solution, BRCA2 forms large multimers and aggregates, and most of the protein was probably lost on the chromatography resin. In sharp contrast, the same amount of BRCA2 could be detected after the gel filtration chromatography in the presence of DSS1 and ssDNA by both A280 absorbance and SDS-PAGE gel with Denville Blue staining. This elution position is consistent with a monomeric BRCA2 if following globular protein standards (Supplementary Figure S3A, B). Single particles of similar morphology were observed in a simple Tris-based buffer system with minimum salt and no glycerol, as another safeguard against staining artifacts (Supplementary Figures S1A & S3C, D). Without crosslinking or sucrose separation, the BRCA2–DSS1–ssDNA complex in the presence of minimum salt shows significant heterogeneity (Figure 2A vs. Supplementary Figures S1A & S3C, D). It appears that the BRCA2 complex occupies a range of different oligomeric states in solution and crosslinking promotes formation of monomers. For the complex without crosslinking, sucrose gradients separate the aggregates from the monomers and thus remove the heterogeneity.
3D structure of monomeric BRCA2–DSS1–ssDNA complex
The low density of these particles presents a technical obstacle for high-resolution structural determination using cryo-EM (Figure 2A). Instead, we performed single-particle 3D reconstruction on negatively stained BRCA2–DSS1–ssDNA complexes with or without crosslinking independently (Figure 3A, B). As expected, the sucrose-gradient-prepared BRCA2–DSS1–ssDNA model without crosslinking has a smaller volume and reached a resolution of 17.1 Å, losing density at regions with high flexibility (Supplementary Figure S4B). The density of the flexible regions is recovered after crosslinking, and the resolution is improved to 14.6 Å (Supplementary Figure S4A). The difference in 3D volume in crosslinked vs. uncrosslinked BRCA2–DSS1–ssDNA complex is consistent with the particle size distribution (Figure 2B, C). As shown in Figure 3C, there is a strong match between the reference-free 2D classes and the forward reprojections of the 3D reconstructed model. We also made multiple 3D sorting attempts and were not successful in obtaining 3D classes for both crosslinked and uncrosslinked BRCA2 complexes. We suspect that it could be caused by the negative staining process and the limited resolution.
To confirm that the particles are BRCA2, we were able to use Ni-NTA nanogold with a 5 nm diameter to label the C-terminal region of BRCA2 through the His10 tag (Figure 3D). We were also able to use purified anti-MBP Fab fragments to confirm the presence of the MBP-tag in the BRCA2–DSS1–ssDNA particles, as well as the anti-biotin Fab fragment to confirm the presence of the biotinylated ssDNA (Figure 3D). The positions of the MBP-tag at the N-terminus, the ssDNA-binding domain, and the C-terminus are consistent in the structural comparisons between the uncrosslinked and crosslinked BRCA2–DSS1–ssDNA complex, assigning the N-terminus of BRCA2 as the flexible domain (Figure 3E). We further compared the monomeric BRCA2–DSS1–ssDNA complex with the published dimeric structure (∼250 Å × 135 Å × 120 Å, EMD-2779), which supported our conclusion that the N-to-C terminal interactions were disrupted during the dimer-to-monomer transitions (Figure 3F, G). The transition is similar to a process when two clasped hands separate into two fists, and the stretched N-terminus at the dimer interface would be the two thumbs folding backward.
Both N- and C-terminus of BRCA2 are prime regions for BRCA2 self-association
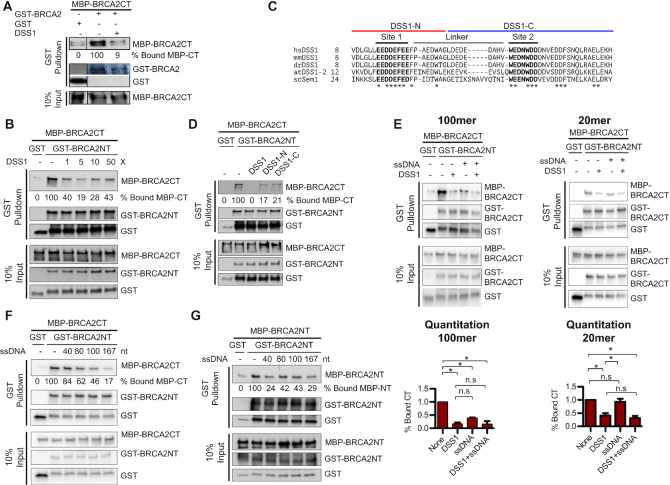
The mapped positions of the N- and C-terminus in the BRCA2 complex (Figure 3D), along with the structural comparisons (Figure 3E, F), lead to the conclusion that N- and C-terminus of BRCA2 interact in the dimer (Figure 3G). There are limitations to use the match between 3D projections and 2D class averages as the only model validation tool. Thus, we decided to test and validate the structural findings with biochemical studies of BRCA2 truncation mutants, focusing on the two termini. First, we constructed and purified an MBP-tagged N-terminal fragment of BRCA2 containing the first 714 amino acids (MBP-BRCA2NT) (Figure 4A, Supplementary Figure S5E). BRCA2NT was tested in protein pulldown assays and showed a saturable and robust interaction with a fixed amount of GST-tagged full-length BRCA2 (Figure 4B, C). Next, we constructed and purified an MBP-tagged C-terminal fragment of BRCA2 containing the last 1,119 amino acids (MBP-BRCA2CT), along with the GST-tagged N-terminal fragment of BRCA2 (GST-BRCA2NT) (Figure 4A and Supplementary Figure S5E, F). As shown in Figure 4D, GST-BRCA2NT interacts strongly with MBP-BRCA2CT in a stoichiometric manner, providing solid support for the N-to-C terminal interaction model (Figure 3G). We also detected interactions between GST-tagged BRCA2NT and MBP-tagged BRCA2NT (Figure 4E), indicating a more complex pattern of self-interaction. The protein interaction of the N-to-C termini is ∼6-fold stronger than the one between the N-termini (Figure 4D, E and Supplementary Figure S5I).
Figure 4.
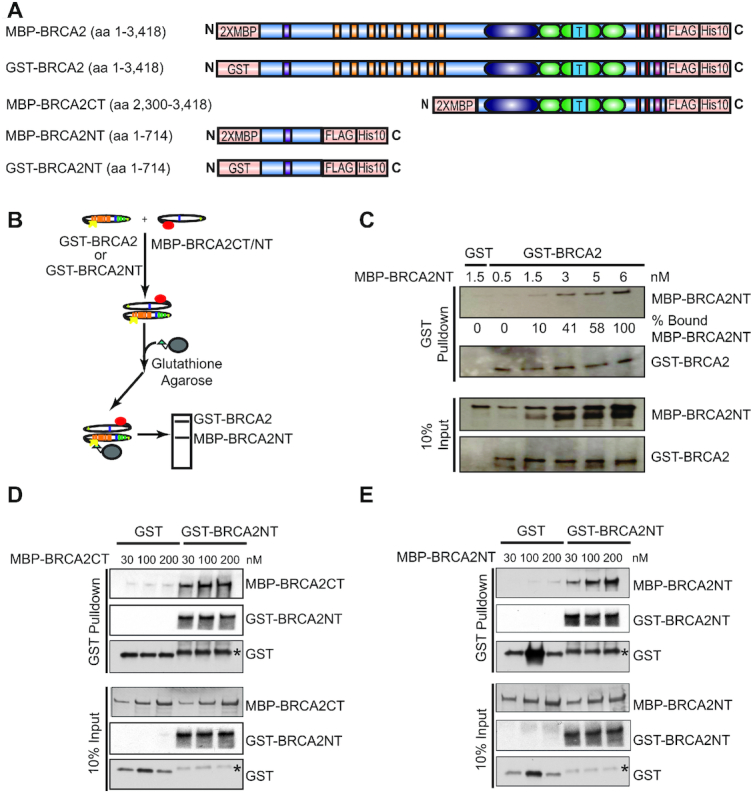
The N- and C-terminus of BRCA2 are prime regions for BRCA2 self-interactions. (A) Constructs of wild type and truncated mutant BRCA2 proteins tested in pulldown interaction assay shown in B. (B) Scheme of protein interaction pulldown assay. (C) Protein interactions between 0.65 nM full-length BRCA2 (GST-BRCA2) and increasing amount of BRCA2 N-terminus (MBP-BRCA2NT) at 0.5, 1.5, 3, 5 and 6 nM. (D) Protein interactions between 30 nM GST-tagged N-terminus of BRCA2 (GST-BRCA2NT) and MBP-tagged C-terminus of BRCA2 (MBP-BRCA2CT) at 30, 100 and 200 nM. (E) Protein interactions between 30 nM GST-tagged N-terminus of BRCA2 (GST-BRCA2NT) and MBP-tagged N-terminus of BRCA2 (MBP-BRCA2NT) at 30 nM, 100 nM and 200 nM. Asterisks in D and E indicate degraded GST-BRCA2NT fragments.
Three putative self-interaction regions (SIRs) involved in BRCA2 self-association
To identify the regions responsible for the self-interactions of BRCA2, we decided to probe the potential role of coiled coils, a motif involved at the protein dimerization interfaces of several DNA repair proteins (42–44). Bioinformatic analysis indicates many sites as potential coiled coils across the BRCA2 sequence (Supplementary Figure S5A). Two predicted coiled coils drew our attention, one from residue 331 to residue 351 in the N-terminus and one from residue 2845 to residue 2964 in the C-terminal OB2 region (Supplementary Figure S5A, B). We created a series of small N-terminal truncation mutants with GST tags, ranging from 100 to 230 amino acids in size (Supplementary Figure S5D,E). We screened them in the pulldown assays for their interactions with the full-length N-terminal fragment containing 714 amino acids (MBP-BRCA2NT, Supplementary Figure S5G). Fragment N7 (aa 250–370) showed an interaction with BRCA2NT, and interestingly, this fragment contains both the secondary DNA binding site (aa 265–349) (11) and the putative coiled coil (aa 331–351). The coiled coil could serve as a SIR of BRCA2, and its proximity to the DNA binding site provides a rationale for the inhibition from ssDNA on BRCA2 self-association. Fragment N9 (aa 481–714) showed a stronger interaction with BRCA2NT, compared to fragment N7. We performed a SMART domain prediction analysis of fragment N9 and discovered a second potential SIR for BRCA2 self-interaction from residue 501 to 620. This short stretch shares a structural similarity with the N-terminal lobe of human histone chaperone FACT subunit Spt16, an interacting domain for its hetero-protein association (Supplementary Figure S5C) (45).
To identify the C-terminal SIR of BRCA2, we constructed a small BRCA2 fragment (aa 2780–3080) in the OB2 region carrying the putative coiled coil ranging from aa 2845 to aa 2964, as an MBP fusion protein (Supplementary Figure S5D, E). This OB2 fragment showed a strong interaction with fragment N9, but no detectable interaction with fragment N7 (Supplementary Figure S5H). Overall, we concluded that there are at least three SIRs involved in BRCA2 self-association, two at the N-terminus and one at the C-terminus, as suggested by the structural findings (Figure 3E–G). The N9 and OB2 interaction might dominate the N-to-C terminus interaction, while the N7 and N9 interaction could explain the observed weaker N-to-N terminus interaction. Fragment N3 contains the PALB2-binding domain (46) but showed no detectable association with either the full-length N-terminus (BRCA2NT) or the OB2 fragment (Supplementary Figure S5G, H).
DSS1 inhibits the N-to-C terminal interactions of BRCA2
The imaging studies have shown that DSS1 and ssDNA inhibit BRCA2 multimerization and synergistically stabilize the monomeric state of BRCA2 (Figure 2). The protein pulldown assays confirmed and supported an N-to-C terminal self-interaction of BRCA2 (Figure 4 & Supplementary Figure S5). To gain more mechanistic insights, we tested the effects of DSS1 and ssDNA in the protein pulldown assays with the BRCA2 fragments. As expected, the addition of DSS1 almost completely blocks the self-interaction of BRCA2 between full-length BRCA2 and the C-terminal one-third fragment of BRCA2 (BRCA2CT) with an 11-fold reduction to almost background level (Figure 5A). We decided to focus on the interactions between the BRCA2NT and BRCA2CT fragments for further exploration. We showed that DSS1 also inhibits this interaction in a concentration-dependent manner, and even an equimolar amount of DSS1 significantly reduced the N-C interaction to 40% (Figure 5B). In conclusion, the N-to-C terminal self-interaction of BRCA2 is sensitive to DSS1 inhibition, suggesting that the binding of DSS1 could block the OB2 SIR in its proximity.
Figure 5.
DSS1 and ssDNA disrupt BRCA2 self-association. (A) DSS1 inhibits the interactions between the C-terminus of BRCA2 (MBP-BRCA2CT) and full-length BRCA2 (GST-BRCA2). The reactions contained 1.5 nM GST-BRCA2, 15 nM MBP-BRCA2CT, and 1.5 μM DSS1. (B) A stoichiometric amount of DSS1 inhibits the self-interaction between the N-terminus of BRCA2 (GST-BRCA2NT) and C-terminus of BRCA2 (MBP-BRCA2CT). The reactions contained 30 nM GST-BRCA2NT and 100 nM MBP-BRCA2CT, with 0, 0.1, 0.5, 1 and 5 μM DSS1. (C) Sequence alignment of DSS1 proteins from different species. Conserved acidic regions 1 and 2 and the poorly conserved linker are labeled. Asterisks indicated the conserved residues in all shown species. The sequences of DSS1 halves (DSS1-N and DSS1-C) are illustrated. (D) DSS1 halves partially inhibit the self-interaction between the N-terminus of BRCA2 (GST-BRCA2NT) and C-terminus of BRCA2 (MBP-BRCA2CT). The reactions contained 30 nM GST-BRCA2NT, 100 nM MBP-BRCA2CT, and 0.5 μM DSS1, or 1 μM DSS1-N, or 1 μM DSS1-C. (E) Both DSS1 and ssDNA inhibits the N-to-C terminal interaction, and the inhibition from ssDNA has a requirement in length. The reactions contained 30 nM GST-BRCA2NT, 100 nM MBP-BRCA2CT, and/or 0.5 μM DSS1, 0.5 μM 100mer (olWDH1734), or 0.5 μM 20mer (olWDH1728). Quantitation is shown below. Values are expressed as mean and SD (n = 3). P values were determined with a two-tailed t-test. *P<0.05, n.s: no significance. (F) The inhibition of ssDNA on the self-interaction between the N- and C-terminus of BRCA2 increases with its length. (G) The inhibition of ssDNA on the N-to-N interaction of BRCA2 also depends on its length. The reactions contained 30 nM GST-BRCA2NT, 100 nM MBP-BRCA2CT in F or 100 nM MBP-BRCA2NT in G, and 0.5 μM ssDNA with indicated length.
The yeast DSS1 homolog, Sem1, has been shown to function as a molecular tether to sequester two proteins, Rpn3 and Rpn7, into a complex through its two acidic stretches during early proteasome assembly (47). Similarly, the two acidic stretches of DSS1 bind the same molecule of BRCA2 at two different interfaces. The N-terminal stretch of DSS1 spans the helical domain and OB1 while the C-terminal stretch winds through OB2 and OB1 to touch the helical domain (12). We synthesized two separate DSS1 halves containing one acidic stretch each, DSS1-N and DSS1-C (Figure 5C, Supplementary Table S1), to test the hypothesis whether molecular tethering is key for DSS1 to stabilize the monomeric state of BRCA2. To our surprise, both DSS1 halves partially inhibit the self-interaction of BRCA2 to a large extent, but not achieving a complete inhibition as full-length DSS1 (Figure 5D). Thus, eliminating the tethering still allows partial inhibition, indicating that while there is a second mechanism of inhibition at work, tethering is required for complete inhibition. We propose that DSS1 might function as a molecular chaperone to facilitate the rearrangement of BRCA2, disrupting its self-association during the process.
DNA inhibits both the N-to-C and the N-to-N terminal interactions of BRCA2
The addition of DNA usually complicates protein pulldown interaction assays by providing a common binding site for DNA-binding proteins leading to a pulldown signal based on independent binding to the same DNA molecule. For this reason, we used short DNA, and the addition of a 100mer ssDNA oligo dramatically decreases the interaction between BRCA2NT and BRCA2CT, corroborating the EM imaging result that self-interactions of BRCA2 are inhibited by ssDNA (Figure 5E). Furthermore, the presence of both DSS1 and ssDNA further decreases the level of interaction to the background level (Figure 5E). In conclusion, the N- and C-terminal interactions of BRCA2 are sensitive to the inhibition from both DSS1 and ssDNA with an additive effect.
Since the ssDNA inhibition of N-to-C terminal BRCA2 interaction seems to depend on its length, we tested ssDNA oligos with various length ranging from 20 to 167 nucleotides (Supplementary Table S1) in the pulldown assays between BRCA2NT and BRCA2CT. While the 20mer had no effect on interaction (Figure 5E), 40–167mers decreased the interaction between the N- and C-terminus of BRCA2 gradually with increasing length (Figure 5F). We also tested these oligos in the pulldown interactions assays between GST-tagged BRCA2NT and MBP-tagged BRCA2NT, considering the secondary DNA-binding site in the N-terminus of BRCA2 (11). A similar but not identical length dependence was identified for disruption of the N–N self-interaction (Figure 5G). DSS1 was not tested for the lack of DSS1 interaction sites at the N-terminus of BRCA2 (12). Overall, ssDNA inhibits both the N-to-C and the N-to-N interaction.
DISCUSSION
The bioinformatic analysis predicts that BRCA2 has a significant amount of intrinsically disordered regions across the N-terminus, the central domain, and the extreme C-terminus (Supplementary Figure S6A, B). These disordered regions could provide an amply exposed surface as potential loops to interact with different protein and nucleic acid partners (Supplementary Figure S6C, D). Reorganization of these disordered regions upon ligand binding might provide the structural plasticity needed to regulate BRCA2 function. In this study, we show that binding to DSS1 and ssDNA blocks BRCA2 multimerization and stabilizes the monomeric state of BRCA2 through major structural rearrangements. Negative stained EM imaging has served as a useful tool, but there are limitations to quantitate the solution behavior of BRCA2 in response to DSS1 and ssDNA through still images.
Self-association of BRCA2 has been observed in vivo through fluorescence bleaching experiments, demonstrating that BRCA2-GFP could exist as monomers, dimers, and larger multimers (2). In vitro, we observed a highly heterogeneous population of BRCA2 multimers in solution when no binding partner is present (Figure 2A, B), consistent with previous observations that negative stained BRCA2 particles show different sizes (3). We have discovered two kinds of self-interactions of BRCA2, leading to BRCA2 multimers in different configurations (Figure 6). The major population of BRCA2 multimers is featuring an N-to-C terminal, or head-to-tail, configuration, and they are sensitive to both DSS1 and ssDNA disruption. A small population of BRCA2 multimers could have an N-to-N terminal, or head-to-head, configuration, and they are only sensitive to ssDNA disruption and not to DSS1. Three SIRs were identified in this study, and two of them are putative coiled coils in proximity to DNA- or DSS1-binding sites. The two configurations of self-interaction and the existence of multiple SIRs could provide an explanation for the large structures of BRCA2 alone (Figure 6) observed in this study and previous scanning force microscopy study (1). However, these BRCA2 multimers with irregular structures (Figure 2A) are less likely to be the nucleating configuration for RAD51 to initiate a uni-directional filament growth in vivo.
Figure 6.
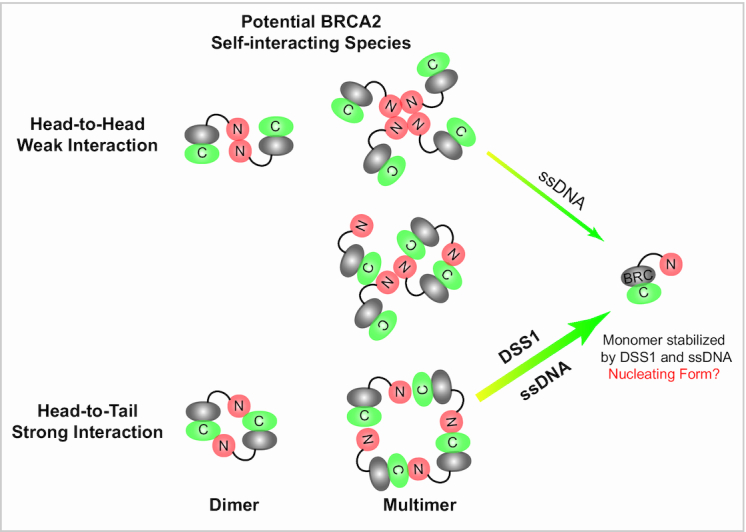
DSS1 and ssDNA disrupt BRCA2 multimers and stabilize BRCA2 monomers. The majority of BRCA2 multimers (including dimers) have a head-to-tail configuration featuring an N-to-C terminus interaction. Two SIRs at the N-terminus of BRCA2 form contacts with the third SIR on OB2 at the C-terminus of BRCA2. DSS1 and ssDNA could block this self-interaction synergistically and efficiently. A small population of BRCA2 multimers (including dimers) has a head-to-head configuration featuring an N-to-N terminus interaction. ssDNA counteracts this self-interaction, potentially by blocking the two SIRs at the N-terminus of BRCA2 in the proximity to the secondary N-terminal DNA-binding site. Large multimers of BRCA2 could be formed with both N-to-C and N-to-N configurations.
The transition between monomers and multimers might represent a regulatory step of BRCA2 activation in recombinational repair (Figure 6). The observation that BRCA2 binds ssDNA as a monomer suggests that the dimeric BRCA2 bound by RAD51 might represent a different form than the one active in nucleation (3). In the absence of genotoxins, almost all nuclear RAD51 is associated with BRCA2 based on their diffusive patterns (2). However, some BRCA2 foci are not overlapping with RAD51 foci without ionizing radiation treatment, visualized by traditional confocal microscopy (1). Upon ionizing radiation treatment, the proportion of overlapping RAD51 and BRCA2 foci increases over time as detected by super-resolution microscopy (1). We propose that the binding of DSS1 stabilizes the monomeric BRCA2–RAD51 complex into a pre-nucleation conformation. The recruitment of monomeric BRCA2–RAD51–DSS1 complex onto ssDNA enables a nucleation conformation of BRCA2 and thus initial RAD51 cluster formation at the damaged site. The presence of dominant-negative mutations, such as in RAD51 (48,49), provides genetic evidence for a functional multimer. Conversely, the absence of BRCA2 mutations that act in a dominant-negative fashion is consistent with a monomer being the functional unit for BRCA2.
A critical observation in U. maydis showed that Dss1 blocks the self-association of Brh2 (BRCA2 homolog) and associates with monomeric Brh2 (20). It supports an earlier proposal that Dss1 activates the Brh2-Rad51 complex (50). Dss1 also inhibits Brh2-DNA binding and Brh2-Rad51 binding (23,25,27). However, Brh2 has only one BRC repeat capable of interacting with a single Rad51 molecule, but nucleation of the human RAD51 filament requires a RAD51 dimer (51,52). Consequently, the molecular mechanisms of fungal Brh2 and mammalian BRCA2 could be different. Fungal Dss1 was shown to inhibit the Brh2–Rad51 interaction (27), but the loss of mammalian DSS1 does not affect either the protein level or the interaction of BRCA2 and RAD51 (18). Here, we showed that DSS1 blocks the N-to-C terminal interaction of BRCA2 in the multimeric state and thus promotes the monomeric state of BRCA2. Unlike Sem1, the yeast DSS1 homolog, DSS1 does not fully require physical tethering of the two binding sites on BRCA2 intra-molecularly to stabilize its monomeric form.
DSS1 was found to act as a DNA mimic, displacing RPA to allow BRCA2 binding to ssDNA (28). Recently, an unexpected interaction between RAD52 and DSS1 was discovered, which also modulates RAD52 oligomeric state and increases its DNA binding and single strand annealing activities (53). In this new role, DSS1 does not function as a DNA mimic and independent of RPA (53). Our experiments were conducted in the absence of RPA, and the synergistic manner by which DSS1 blocks BRCA2 self-association in conjunction with ssDNA suggests an additional function of DSS1 besides acting as a DNA mimic. Thus, we propose that DSS1 functions as a molecular chaperone of BRCA2 rearrangement towards a stable monomeric form. This rearrangement of BRCA2 could result in proximity between the BRCA2 C-term-DBD and BRC repeats, relating DNA-binding to RAD51-interaction. This structural role of DSS1 is consistent with previous observations that DSS1 stimulates BRCA2 in RAD51 recruitment onto RPA-covered ssDNA and RAD51-catalyzed DNA strand exchange (9,28). It also provides an explanation for the observations that DSS1 binding in cells is only essential when the BRCA2 C-termDBD is present (21). The absence of BRCA2 C-termDBD possibly limits multimer formation and thus the DSS1 regulation. Therefore, we speculate that monomeric BRCA2 in complex with DSS1 and ssDNA could be the active form in recombinational DNA repair as well as replication fork stabilization.
In summary, we revealed the nature of self-association for BRCA2 by pinpointing two configurations of BRCA2 multimers, N-to-C (head-to-tail) and N-to-N (head-to-head). The N-to-C interaction is dominant and sensitive to the inhibition from DSS1 and ssDNA, while the N-to-N interaction is minor and sensitive to ssDNA. Binding of DSS1 and ssDNA leads to a synergistic rearrangement of BRCA2 and stabilizes the monomeric state of BRCA2 potentially through improved folding at disordered regions. Our study demonstrates that self-association and protein conformation of BRCA2 are influenced by its binding partners, revealing a new regulatory strategy in recombinational DNA repair.
DATA AVAILABILITY
The density maps of BRCA2–DSS1–ssDNA with (EMD-21998) or without crosslinking (EMD-20348) have been deposited in the Electron Microscopy Data Bank.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Qing Zhong (UT Southwestern Medical Center) for providing the HEK293 Flp-In expression system. We thank Robert Glaeser and Bong-Gyoon Han (UC Berkeley) for providing streptavidin crystal affinity grids. We also thank Stephen Kowalczykowski for providing the V79 and V-C8 cell lines, and the phCMV1 vector. We thank Henning Stahlberg (University of Basel) and his laboratory for providing training and technical advice in EM data collection and image processing. We thank Farideh Zakeri, Rupleen Kaur, Nikan Mofakham Fini, Sukhman Chatrath, and Cindy Feng for cell culture and imaging support. We thank all members of the Heyer laboratory, Xiaodong Zhang (Imperial College London) and Claire Wyman (Erasmus University, Rotterdam) for providing critical comments.
Author contributions: J.L. and W.-D.H. designed research; H.P.L. and M.B. performed BRCA2 mutant studies; X.M. and J.L. performed biochemical and cellular studies on wild type BRCA2; J.V. and J.L. collected and analyzed EM data with the help of F.G.; J.L. and W.D.H. wrote the paper.
Contributor Information
Hang Phuong Le, Department of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA 95616–8665, USA.
Xiaoyan Ma, Department of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA 95616–8665, USA.
Jorge Vaquero, Department of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA 95616–8665, USA.
Megan Brinkmeyer, Department of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA 95616–8665, USA.
Fei Guo, Department of Molecular and Cellular Biology, University of California, Davis, Davis, CA 95616–8665, USA.
Wolf-Dietrich Heyer, Department of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA 95616–8665, USA; Department of Molecular and Cellular Biology, University of California, Davis, Davis, CA 95616–8665, USA.
Jie Liu, Department of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA 95616–8665, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [P30 CA93373, CA92267 to W.D.H., GM58015 to W.D.H., CA187561 to J.L.]; Placer Breast Cancer Foundation; Sorenson Foundation. Funding for open access charge: National Cancer Institute [CA187561].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sanchez H., Paul M.W., Grosbart M., van Rossum-Fikkert S.E., Lebbink J.H.G., Kanaar R., Houtsmuller A.B., Wyman C.. Architectural plasticity of human BRCA2-RAD51 complexes in DNA break repair. Nucleic Acids Res. 2017; 45:4507–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reuter M., Zelensky A., Smal I., Meijering E., van Cappellen W.A., de Gruiter H.M., van Belle G.J., van Royen M.E., Houtsmuller A.B., Essers J. et al.. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J. Cell Biol. 2014; 207:599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shahid T., Soroka J., Kong E.H., Malivert L., McIlwraith M.J., Pape T., West S.C., Zhang X.. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat. Struct. Mol. Biol. 2014; 21:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. King M.C., Marks J.H., Mandell J.B. New York Breast Cancer Study, G. . Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003; 302:643–646. [DOI] [PubMed] [Google Scholar]
- 5. Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G.. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995; 378:789–792. [DOI] [PubMed] [Google Scholar]
- 6. Prakash R., Zhang Y., Feng W., Jasin M.. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015; 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roy R., Chun J., Powell S.N.. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012; 12:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jensen R.B., Carreira A., Kowalczykowski S.C.. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010; 467:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu J., Doty T., Gibson B., Heyer W.D.. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010; 17:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C.. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010; 17:1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. von Nicolai C., Ehlen A., Martin C., Zhang X., Carreira A.. A second DNA binding site in human BRCA2 promotes homologous recombination. Nat. Commun. 2016; 7:12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang H.J., Jeffrey P.D., Miller J., Kinnucan E., Sun Y.T., Thoma N.H., Zheng N., Chen P.L., Lee W.H., Pavletich N.P.. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002; 297:1837–1848. [DOI] [PubMed] [Google Scholar]
- 13. Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R.. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002; 420:287–293. [DOI] [PubMed] [Google Scholar]
- 14. Wong A.K.C., Pero R., Ormonde P.A., Tavtigian S.V., Bartel P.L.. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997; 272:31941–31944. [DOI] [PubMed] [Google Scholar]
- 15. Chen P.L., Chen C.F., Chen Y., Xiao J., Sharp Z.D., Lee W.H.. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:5287–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carreira A., Kowalczykowski S.C.. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10448–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oliver A.W., Swift S., Lord C.J., Ashworth A., Pearl L.H.. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009; 10:990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gudmundsdottir K., Lord C.J., Witt E., Tutt A.N., Ashworth A.. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004; 5:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kojic M., Yang H., Kostrub C.F., Pavletich N.P., Holloman W.K.. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell. 2003; 12:1043–1049. [DOI] [PubMed] [Google Scholar]
- 20. Zhou Q., Kojic M., Cao Z., Lisby M., Mazloum N.A., Holloman W.K.. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol. Cell. Biol. 2007; 27:2512–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siaud N., Barbera M.A., Egashira A., Lam I., Christ N., Schlacher K., Xia B., Jasin M.. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet. 2011; 7:e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeyasekharan A.D., Liu Y., Hattori H., Pisupati V., Jonsdottir A.B., Rajendra E., Lee M., Sundaramoorthy E., Schlachter S., Kaminski C.F. et al.. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat. Struct. Mol. Biol. 2013; 20:1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Q., Kojic M., Holloman W.K.. Dss1 release activates DNA binding potential in Brh2. Biochemistry. 2012; 51:9137–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li J., Zou C., Bai Y., Wazer D.E., Band V., Gao Q.. DSS1 is required for the stability of BRCA2. Oncogene. 2006; 25:1186–1194. [DOI] [PubMed] [Google Scholar]
- 25. Zhou Q., Mazloum N., Mao N., Kojic M., Holloman W.K.. Dss1 regulates interaction of Brh2 with DNA. Biochemistry. 2009; 48:11929–11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marston N.J., Richards W.J., Hughes D., Bertwistle D., Marshall C.J., Ashworth A.. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol. Cell. Biol. 1999; 19:4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Q., Holloman W.K.. Dss1 regulates Association of Brh2 with Rad51. Biochemistry. 2017; 56:3318–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao W., Vaithiyalingam S., San Filippo J., Maranon D.G., Jimenez-Sainz J., Fontenay G.V., Kwon Y., Leung S.G., Lu L., Jensen R.B. et al.. Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA Mimicry. Mol. Cell. 2015; 59:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Faza M.B., Kemmler S., Jimeno S., Gonzalez-Aguilera C., Aguilera A., Hurt E., Panse V.G.. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J. Cell Biol. 2009; 184:833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ellisdon A.M., Dimitrova L., Hurt E., Stewart M.. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat. Struct. Mol. Biol. 2012; 19:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilmes G.M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., Cagney G., Collins S.R., Whitworth G.B., Kress T.L., Weissman J.S. et al.. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008; 32:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhatia V., Barroso S.I., Garcia-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 33. Gudmundsdottir K., Lord C.J., Ashworth A.. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007; 26:7601–7606. [DOI] [PubMed] [Google Scholar]
- 34. Wei S.J., Williams J.G., Dang H., Darden T.A., Betz B.L., Humble M.M., Chang F.M., Trempus C.S., Johnson K., Cannon R.E. et al.. Identification of a specific motif of the DSS1 protein required for proteasome interaction and p53 protein degradation. J. Mol. Biol. 2008; 383:693–712. [DOI] [PubMed] [Google Scholar]
- 35. Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005; 152:36–51. [DOI] [PubMed] [Google Scholar]
- 36. Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J.. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- 37. Hohn M., Tang G., Goodyear G., Baldwin P.R., Huang Z., Penczek P.A., Yang C., Glaeser R.M., Adams P.D., Ludtke S.J.. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 2007; 157:47–55. [DOI] [PubMed] [Google Scholar]
- 38. Moriya T., Saur M., Stabrin M., Merino F., Voicu H., Huang Z., Penczek P.A., Raunser S., Gatsogiannis C.. High-resolution single particle analysis from electron cryo-microscopy images using SPHIRE. J. Vis. Exp. 2017; 123:55448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 40. Kraakman-van der Zwet M., Overkamp W.J.I., van Lange R.E.E., Essers J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P.P.W., Errami A., Tan R.T.L. et al.. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002; 22:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O’Gorman S., Fox D.T., Wahl G.M.. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991; 251:1351–1355. [DOI] [PubMed] [Google Scholar]
- 42. Itakura E., Sawada I., Matsuura A.. Dimerization of the ATRIP protein through the coiled-coil motif and its implication to the maintenance of stalled replication forks. Mol. Biol. Cell. 2005; 16:5551–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X.. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 2009; 19:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park Y.B., Hohl M., Padjasek M., Jeong E., Jin K.S., Krezel A., Petrini J.H., Cho Y.. Eukaryotic Rad50 functions as a rod-shaped dimer. Nat. Struct. Mol. Biol. 2017; 24:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stuwe T., Hothorn M., Lejeune E., Rybin V., Bortfeld M., Scheffzek K., Ladurner A.G.. The FACT Spt16 ‘peptidase’ domain is a histone H3-H4 binding module. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8884–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M.. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006; 22:719–729. [DOI] [PubMed] [Google Scholar]
- 47. Tomko R.J. Jr., Hochstrasser M.. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol. Cell. 2014; 53:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K. et al.. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015; 59:478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ameziane N., May P., Haitjema A., van de Vrugt H.J., van Rossum-Fikkert S.E., Ristic D., Williams G.J., Balk J., Rockx D., Li H. et al.. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat. Commun. 2015; 6:8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kojic M., Zhou Q., Lisby M., Holloman W.K.. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol. Cell. Biol. 2005; 25:2547–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hilario J., Amitani I., Baskin R.J., Kowalczykowski S.C.. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Nat. Acad. Sci. U.S.A. 2009; 106:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Candelli A., Holthausen J.T., Depken M., Brouwer I., Franker M.A., Marchetti M., Heller I., Bernard S., Garcin E.B., Modesti M. et al.. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15090–15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stefanovie B., Hengel S.R., Mlcouskova J., Prochazkova J., Spirek M., Nikulenkov F., Nemecek D., Koch B.G., Bain F.E., Yu L. et al.. DSS1 interacts with and stimulates RAD52 to promote the repair of DSBs. Nucleic Acids Res. 2020; 48:694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The density maps of BRCA2–DSS1–ssDNA with (EMD-21998) or without crosslinking (EMD-20348) have been deposited in the Electron Microscopy Data Bank.