Abstract
Fungi have developed the ability to overcome extreme growth conditions and thrive in hostile environments. The model fungus Aspergillus nidulans tolerates, for example, ambient alkalinity up to pH 10 or molar concentrations of multiple cations. The ability to grow under alkaline pH or saline stress depends on the effective function of at least three regulatory pathways mediated by the zinc-finger transcription factor PacC, which mediates the ambient pH regulatory pathway, the calcineurin-dependent CrzA and the cation homeostasis responsive factor SltA. Using RNA sequencing, we determined the effect of external pH alkalinization or sodium stress on gene expression. The data show that each condition triggers transcriptional responses with a low degree of overlap. By sequencing the transcriptomes of the null mutant, the role of SltA in the above-mentioned homeostasis mechanisms was also studied. The results show that the transcriptional role of SltA is wider than initially expected and implies, for example, the positive control of the PacC-dependent ambient pH regulatory pathway. Overall, our data strongly suggest that the stress response pathways in fungi include some common but mostly exclusive constituents, and that there is a hierarchical relationship among the main regulators of stress response, with SltA controlling pacC expression, at least in A. nidulans.
Keywords: Aspergillus nidulans, alkaline pH stress, cation stress, fungi, PacC/Rim101, signal transduction, SltA, transcriptional control
Data Summary
The RNA sequencing data analysed in this work were deposited and are freely available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject ID PRJNA625291. All supporting data, code and protocols have been provided within the article or in the Supplementary Material.
Impact Statement.
Concurrently with their extended use in biotechnology, more than a thousand fungal species are known to act as human, animal or plant pathogens. To adapt to and thrive in host and natural environments, fungi have developed an array of stress response pathways. Here we used an RNA sequencing approach to analyse the transcriptional response of the filamentous fungus Aspergillus nidulans, one of the main reference systems within the subphylum Pezizomycotina, to alkaline pH and sodium stress. Sequencing was carried out in a wild-type genetic background and its transcriptional patterns were compared to those of a null sltA strain. SltA is one of the master regulators of alkaline pH and cation stress responses. Thus, this study contributes to our understanding of the level of overlap between stress response pathways, the general regulators that could be involved in those responses, and the level of dependence of each response mechanism on SltA activity, not only in A. nidulans, but also in other medically or industrially important Pezizomycotina species.
Background
The survival of organisms is subject to their ability to adapt to and overcome diverse challenges or stresses in the environment. The most general responses are produced by coordinating the regulation of gene expression and those mechanisms controlling the proper localization of proteins in the cell. Gene expression can be enhanced or repressed by the activity of transcription factors, which recognize specific consensus sequences at gene promoters through their DNA-binding domains. Concurrently, the activity of a transcription factor can be modulated by other regulatory elements or by post-translational modifications that may occur via elements belonging to a signalling cascade activated by a given ambient stimulus.
The model organism Aspergillus nidulans is capable of surviving a variety of ambient stresses, such as a wide range of pH values or high extracellular concentrations of monovalent and divalent cations [1]. Tolerance to ambient alkalinity has been intensively studied in this fungus [2–4]. Previous work led to the identification and functional characterization of three zinc-finger transcription factors that coordinate a response to ambient alkaline pH: PacC, regulator of the ambient pH regulatory pathway [2]; the calcineurin-dependent transcription factor CrzA; and the cation homeostasis transcription factor SltA [5]. The absence of any of these transcription factors inhibits vegetative growth of A.nidulans at alkaline pH [5].
PacC is a 674-amino-acid transcription factor with an N-terminally located DNA-binding domain consisting of three classical Cys2His2 zinc-fingers and C-terminally located key regulatory regions required for its signalling function [2, 6–8]. PacC can be found in three forms in the cell, depending on extracellular pH conditions [9]. At extremely acidic pH (4 or lower), the full-length form of 72 kDa is mainly detected, which is in a closed conformation to prevent its proteolysis [8]. At extracellular alkaline pH, PacC is activated through two proteolytic steps. The first step is pH-dependent and signalled by the Pal pathway [9]. PalH is the pH sensor protein at the plasma membrane, and is stabilized by PalI, another membrane-embedded protein [10]. PalF, an arrestin-like protein [11], is in a complex with PalH at the plasma membrane. At alkaline pH, PalH is phosphorylated and PalF is ubiquitylated, and then the Vps23 homologue, an element of the ESCRT-I complex, is recruited and possibly also favours the polymerization of the rest of the complex [12–14]. Subsequently, ESCRT-III and probably ESCRT-II complexes polymerize at the same plasma membrane foci. Finally in the PacC signalling cascade, Vps32, an ESCRT-III element, recruits PalC and PalA [12, 15], and the latter interacts with the PacC72kDa form [16]. Then, the putative calpain protease PalB joins the complex through interaction with the ESCRT-III component Vps24 and cleaves the PacC72kDa form at a specific site, rendering the intermediate form of PacC53kDa [17]. The second proteolytic step is pH-independent and carried out by the proteasome, resulting in the active form of PacC, PacC27kDa [18]. PacC plays a dual function at alkaline pH by repressing those genes expressed at acidic pH (acid-expressed genes) and positively regulating those genes whose function is required at alkaline pH (alkaline genes) [19, 20]. An example of this dual function is the opposite regulation of acid and alkaline phosphatases [21] or different xylanases [22]. This pH-dependent regulation of gene expression is possible due to the direct binding of PacC to 5′-GCCARG-3′ sequences at target promoters [2, 19, 20].
In contrast to PacC and CrzA, which can be found in most ascomycetes, SltA is a transcription factor that is exclusive to the subphylum Pezizomycotina [5, 23]. Only two elements have been described in the Slt pathway so far: SltA itself and its signalling protease SltB, also specific to Pezizomycotina [24]. SltA is a 698-amino-acid protein with three classical Cys2His2 zinc-fingers located between residues 416 and 500 that bind to a 5′-AGGCA-3′ target sequence [5, 25]. Initially, sltA was identified by studying the loss-of-function mutation sltA1, which truncates the protein at amino acid 502 and confers A. nidulans sensitivity to NaCl, UV radiation, MNNG, 4NQO and arginine [26]. Further studies showed that he absence of SltA function causes sensitivity to elevated extracellular concentrations of other cations, such as potassium, lithium, caesium or magnesium, but not to calcium, and extreme sensitivity or complete inhibition of colonial growth under ambient alkaline pH [5]. Similarly to PacC, SltA can be found simultaneously in the cell in three forms: a full-length form of 78 kDa and two versions of the proteolysed form of 32 kDa, one of which is phosphorylated [23]. This single proteolytic step that mediates the conversion of the 78 kDa form into the 32 kDa one is driven by the 1272-amino-acid protein SltB, a serine protease with a large prodomain similar to a pseudo-kinase domain [23, 24]. SltB is essential for SltA proteolysis, but SltA transcriptional activity is also necessary for sltB expression [23].
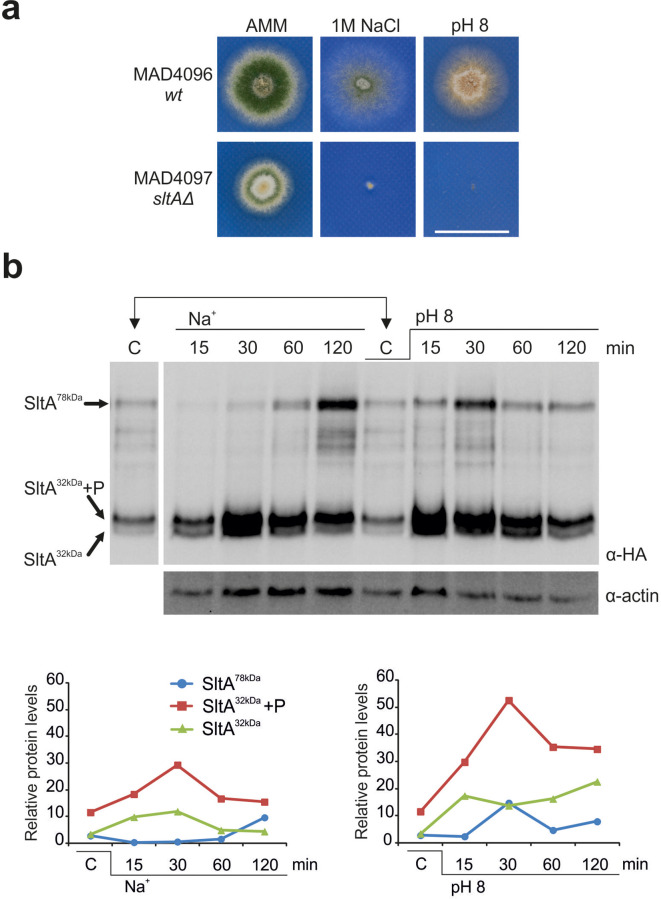
Despite the abundant genetic and molecular information available regarding these regulatory systems, only sparse data are available concerning the transcriptional response of A. nidulans to ambient pH alkalinization or the stress caused by an elevated concentration of sodium. When cultured on solid medium, inhibition of asexual sporulation at pH 8 and less dense growth in high sodium are observed in the wild-type strain, indicating differential effects of these stresses on colonial morphology (Fig. 1a). As reported previously [5, 23], the absence of SltA function causes defects in colony growth on standard minimal medium but renders both alkaline pH and high sodium stresses lethal.
Fig. 1.
Phenotype of the sltAΔ strain and levels of the different SltA forms under cation or alkaline pH stresses. (a) Colonial growth of wild-type (WT) and sltAΔ strains on AMM, AMM supplemented with 1.0 M NaCl or an alkalinized medium (addition of 100 mM Na2HPO4; pH 8) after 48 h of culture at 37 °C. Scale bar, 2 cm. (b) Immunodetection of SltA::HA3 in protein extracts obtained from mycelial samples grown in AMM (C, control), or at 15, 30, 60 and 120 min after the addition of 1.0 M NaCl or 100 mM Na2HPO4 (pH 8). α-actin was used as control and to quantify the levels of the different SltA forms detected. Bottom: charts showing the relative levels of SltA forms. Values were obtained by calculating the ratio between SltA and α-actin band intensities.
To determine the set of genes up- or downregulated under the above-mentioned stress conditions, the role of SltA in the corresponding homeostasis mechanisms and the existence of a transcriptional dependence of other regulators (PacC, CrzA or other unknown transcription factors) on SltA, we carried out RNA sequencing experiments on wild-type and sltAΔ samples followed by multiple comparisons of the transcriptomes. Our results show that there is a low degree of overlap between the transcriptional responses of A. nidulans to medium alkalinization or sodium stress, suggesting that each stress response pathway is largely distinct. In both cases, the expression of specific subsets of genes remains independent of SltA activity. Other subsets positively depend on SltA, including the PacC-dependent ambient pH regulatory pathway, suggesting that there is a hierarchical relationship among the main regulators of stress response in fungi.
Methods
Strains, oligonucleotides and culture conditions
The oligonucleotides used in this study are shown in Table S1 (available in the online version of this article), while the genotypes of the strains used are shown in Table S2. Strain MAD6669 was obtained by transformation of strain MAD1427 using a fusion PCR fragment as described previously [23], allowing the tagging of SltA with three copies of haemagglutinin epitope (HA3). The construct contained the riboB gene from Aspergillus fumigatus (riboBAf), which was used as the selection marker in the transformation.
The sensitivity of the null sltA mutant to sodium stress or medium alkalinity was compared to that of the reference wild-type strain by culturing both strains on adequately supplemented solid Aspergillus minimal medium (AMM) [27], AMM supplemented with 1.0 M NaCl (sodium stress) or AMM alkalinized to pH 8 by the addition of 100 mM Na2HPO4 [5, 24].
Samples for RNA extraction were cultured as follows (see also the diagram in Fig. 2a). Four independent culture sets were prepared for each strain (wild-type and null sltA) and condition (unstressed, 1 M sodium chloride and pH 8). Conidia (106 conidia ml−1) of each of the strains were inoculated into 100 ml Erlenmeyer flasks containing 25 ml of adequately supplemented standard liquid AMM. After 18 h of culture at 37 °C and 250 r.p.m., NaCl (1.0 M final concentration) or Na2HPO4 (100 mM final concentration) were added to each set, and the third flask of each set was considered to be the unstressed control condition. Cultures were further incubated for 60 min. Then, mycelia were collected and two sets were combined to minimize variations. The pH of filtered media was monitored and both control and NaCl samples showed an approximate final pH value of 4, while the sample containing Na2HPO4 showed a pH of 8. Mycelia of two combined samples were processed as duplicates.
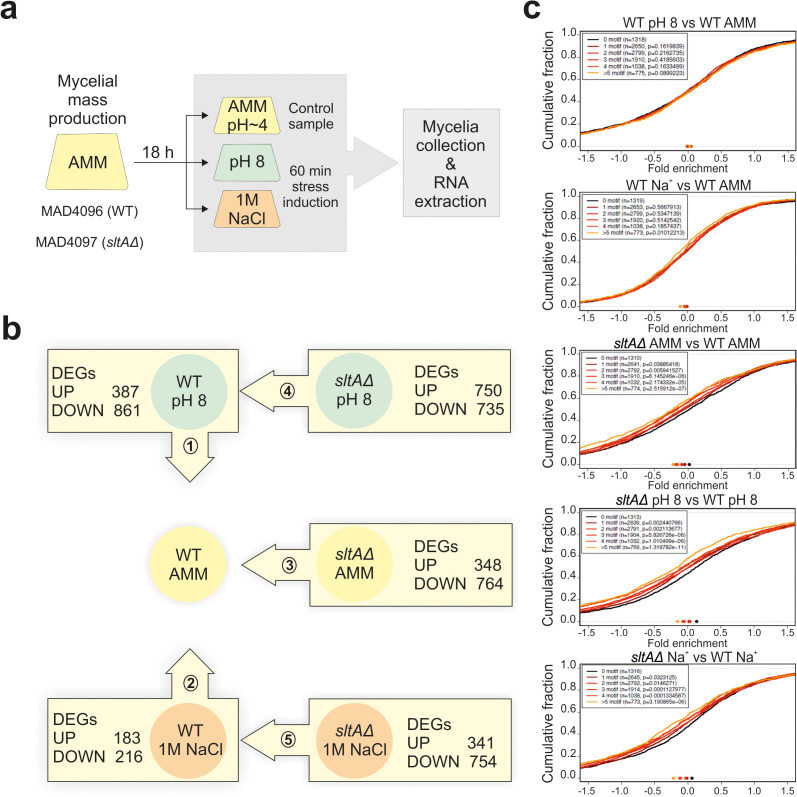
Fig. 2.
Experimental design and number of significantly deregulated genes in each RNA sequencing comparison. (a) Experimental design for the RNA sequencing experiment carried out in this work. Spores of wild-type and sltAΔ strains were collected and incubated (106 conidia ml−1) in AMM for 18 h at 37 °C. Then, Na2HPO4 (100 mM) or NaCl (1.0 M), final concentrations in both cases, were added, followed by an incubation of 60 min. Cultures grown in AMM were used as controls. Samples (duplicates; see the Methods section) were then filtered and processed for RNA extraction and sequencing. (b) Diagram representing the five transcriptional comparisons carried out in this work. Arrows indicate the direction of the comparison and boxes indicate the number of significantly up- and downregulated genes in each transcriptional comparison. (c) Cumulative distribution fraction plots correlating, in each of the five transcriptomic comparisons, log2FC values with the number of SltA target sites in gene promoters. Groups of promoters (1000 bp upstream of the starting ATG) with 0, 1, 2, 3, 4 and >5 SltA target motifs were differentiated and plotted. The points in the lower part of each graph indicate when 50 % of the genes are reached. The P value for each plot is included.
Samples for protein extraction were cultured as above in liquid AMM for 18 h. After the addition of 1.0 M NaCl or 100 mM Na2HPO4, mycelial samples were collected at 15, 30, 60 and 120 min and processed according to the alkaline lysis procedure.
RNA extraction
Mycelia cultivated under the conditions described in the previous section were harvested by filtration using Miracloth (Calbiochem, Merck-Millipore, Darmstadt, Germany). Mycelial samples of approximately 300 mg were frozen and ground in liquid nitrogen before the addition of 1 ml per sample of TRIreagent (Fluka, Sigma-Aldrich Quimica SL, Madrid, Spain) and further processing according to the manufacturer’s protocol. After TRIreagent addition, samples were incubated for 5 min at room temperature (RT). Then, chloroform (0.2 ml per sample) was added, mixed vigorously, incubated for 15 min at RT and centrifuged at 12 000 g and 4 °C for 15 min. After collection of the aqueous (upper) phase, 2-propanol (0.5 ml per sample) was added to precipitate RNA, followed by a new centrifugation at 12 000 g and 4 °C for 10 min. Ethanol (75 %; 1 ml) was added to wash the RNA, followed by centrifugation at 7500 g and 4 °C for 5 min., air drying and the addition of 200 µl of RNA-free milliQ water. A duplicate RNA extraction was performed for each mycelial sample. Total RNA samples were frozen at −80 °C until sequencing or use in quantitative PCR (qPCR).
Library construction and RNA sequencing
Library construction and RNA sequencing were carried out in Stabvida (Caparica, Portugal). Quality control of the total RNA samples was based on an RNA integrity number (RIN) equal to or higher than 6.5. Samples (input of ≥1 µg total RNA, free of contaminating DNA) were directed for library preparation using the Kapa Stranded mRNA Library Preparation kit (Roche Diagnostics Corporation, IN, USA) and sequencing in an lllumina HiSeq 4000 platform, using 150 bp paired-end sequencing reads. The RNA sequencing data analysed in this work were deposited and are freely available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject ID PRJNA625291.
Bioinformatic analyses
Adapters were trimmed using cutadapt version 2.5 (https://cutadapt.readthedocs.io/en/stable/ [28]). Reads were aligned to an Aspergillus genome reference file downloaded from the FungiDB database (FungiDB_36_AnidulansFGSCA4 [29]) using STAR aligner (version 2.5.2a [30]) and allowing no more than two mismatches. Counts for genes were generated using featureCounts version 1.4.6 [31] in the Rsubread package. The TPM calculation was performed using Rsem version 1.2.31 [32], while sample distance or PCA plots, as well as differential expression analyses, were carried out using the DESeq package [33]. Cumulative distribution fraction plots were generated using R version 3.3.3 [34].
Domain analyses of protein sequences were performed using Interpro [35]. Lists of DEGs were initially analysed using the FungiDB database (https://fungidb.org/fungidb/), including the distribution of DEGs by chromosomes, or the presence of signal peptides and/or transmembrane domains in the corresponding polypeptides. Fungifun (https://elbe.hki-jena.de/fungifun/) was used for GO enrichment analyses [36], while Interpro domain-enrichment analyses were carried out at the ShinnyGO website (http://bioinformatics.sdstate.edu/go/ [37]). Heatmaps showing the expression changes of DEGs were generated with Heatmapper (http://www.heatmapper.ca/ [38]), using average linkage clustering and Pearson distance measurement methods. Expression clusters were inferred and the list of genes in each one was also obtained also from Heatmapper. GraphPad was used to plot the expression pattern of genes in each cluster as well as ShinnyGO and Fungifun results, and to obtain the correlation coefficients between RNA sequencing duplicates. Venn diagrams were generated and downloaded with InteractiveVenn (http://www.interactivenn.net/ [39]).
Quantitative PCR
The SuperScript First-strand Synthesis System for RT-PCR (Invitrogen) was used to synthesize cDNA, following the manufacturer’s instructions and, after having eliminated traces of genomic DNA (DNAg) using deoxyribonuclease I, amplification grade (Invitrogen). Two micrograms of total RNA were treated with 1 unit of DNAseI (Invitrogen) for 15 min at RT (reaction volume of 10 µl). DNAg digestion was stopped by the addition of 1 µl of EDTA and incubation for 10 min at 65 °C. Total RNA concentration in samples was determined using a NanoDrop ND-1000. First-strand cDNA reaction consisted of 1 µg of DNAseI-treated RNA, 1 µl of 10 mM dNTPs mixture, 1 µl of oligonucleotide oligo(dT)12–18 and milliQ water up to a final volume of 10 µl. Samples were incubated at 65 °C for 5 min and then immediately transferred into ice for 1 min. After ice incubation, a mixture consisting of 2 µl of RT buffer 10×, 4 µl of MgCl2 25 mM, 2 µl of DTT 0.1 M and 1 µl of the enzyme RNAseOUT (40 U l−1) was added. After centrifugation at maximum speed for 20 s, samples were incubated at 42 °C for 2 min. Then, 1 µl of the retrotranscriptase SuperScript II RT was added to each reaction and incubated at 42 °C for 50 min, followed by a second incubation at 70 °C for 15 min, allowing it to cool on ice for 2–3 min at the end of the process. Finally, 1 µl of the enzyme RNAseH was added per sample and the samples were incubated at 37 °C during 20 min. The amount of cDNA obtained was measured in a NanoDrop ND-1000. Samples were stored at −20 °C until use.
Once the cDNA samples of the strains of interests were generated, dilutions were made at the final concentrations of 100 and 10 ng µl−1 in water PCR probe. Each reaction mixture contained 1 µl of the corresponding dilution, 0.3 µl (300 nmol) of each of the relevant oligonucleotides (Table S1), 8.4 µl of PCR probe water and 10 µl of the 2× commercial reaction mixture (SYBR Green Master Mix, Bio-Rad). A LightCycler96 (Roche) device was used and the PCR conditions applied were: preincubation at 95 °C for 5 min and 40 cycles of 95 °C for 10 s plus 65 °C for 30 s. To obtain the melting curve, a cycle was used in which the temperature was lowered from 95–65 °C at a speed of 4.4 °C 10 s−1.
Protein extraction and immunodetection
Protein extracts for immunodetection were obtained through the alkaline lysis protocol [40]. Briefly, approximately 6 mg of lyophilized mycelium was resuspended in 1 ml lysis buffer (0.2M NaOH, 0.2 % β-mercaptoethanol). After trichloroacetic acid (TCA) precipitation, 100 µL Tris-base (1 M) and 200 µl of loading buffer [62.5 mM Tris/HCl pH=6.8, 2 % SDS (p/v), 5 % β-mercaptoethanol (v/v), 6 M urea and 0.05 % bromophenol blue (p/v)] were added. Samples were then loaded and proteins separated on SDS-polyacrylamide gels (%10), and electro-transferred to nitrocellulose filters (Trans Blot Turbo transfer packs) using the Trans Blot Turbo system (BioRad) following the manufacturer’s instructions. HA3-tagged proteins were detected using a monoclonal rat α-HA3 (1 : 1000; clone 3F10; Sigma-Aldrich). Actin, used as loading control, was detected using monoclonal mouse α-actin (1 : 5000; clone C4; MP Biomedicals). Peroxidase-conjugated goat anti-rat IgG +IgM (1 : 4000; Southern Biotech) or anti-mouse (1 : 4000; Jackson ImmunoResearch Laboratories) were used as secondary antibodies. Western blots were developed using the Amersham Biosciences ECL kit and chemiluminescence was detected using a Chemidoc image system driven by Image Lab Touch software (version 2.2.0.08; BioRad). Images were processed to a minimum using Image Lab software (version 6.0; BioRad).
Results
Modification of the proteolytic pattern of SltA in response to sodium or alkaline pH stress
To establish the timing for sample collection, immunodetection experiments were carried out with protein samples of a strain expressing the fusion protein SltA::HA3 (Fig. 1b) [23]. As expected, the three main forms of SltA were immunodetected in mycelia grown under standard culture conditions (AMM): the full-length form of 78 kDa, SltA78kDa, and a proteolysed version of 32 kDa, SltA32kDa, also found in a phosphorylated state, SltA32kDa-P (C, control, in Fig. 1b) [23]. Distinctive patterns of SltA 78 kDa and 32 kDa forms have been repeatedly observed (Figs 1b and S1). The addition of 1.0 M sodium (as NaCl) or ambient pH alkalinization (pH 8) by the addition of 100 mM Na2HPO4 caused differential effects on the relative quantities, and probably on the total quantity, of these SltA forms. The presence of 1.0 M Na+ in the medium induced a reduction in the levels of SltA78kDa in the first 30 min, followed by an accumulation over time. In contrast, medium alkalinization caused a transient increase in the levels of SltA78kDa at 30 min. Both stresses induced a peak of SltA32kDa-P, the proteolysed and phosphorylated form, at 30 min (Fig. 1b). However, the 60 min time repeatedly showed a coincidence in the pattern of SltA forms between both conditions, offering a comparable time point for further experimental analysis of gene expression in these abiotic stresses. Thus, to investigate the effects of these stresses and the role of SltA on the pattern of gene expression, we decided to extract and sequence RNA samples after 60 min of induction of sodium or alkaline pH stress.
Transcriptional profiles under alkaline pH or sodium stress
Since growth of the sltAΔ strain under sodium or alkaline pH stress was completely inhibited on solid medium (Fig. 1a), sample collection was carried out from liquid cultures after addition of the compound causing stress (Fig. 2a; see also the Methods section). Mycelial samples cultured in AMM were used as controls. Importantly, mycelial cultures had a pH value of approximately 4 before the addition of sodium or the shift to pH 8. Both control and sodium stress-induced cultures maintained the same acidic pH at the time of sample collection. RNA was extracted in biological duplicates and sequenced (see the Methods section). PCA and sample distance plots showed that sample duplicates clustered together, with a higher similarity between sodium stress and control samples than those corresponding to alkaline pH stress (Fig. S2a, b). The correlation values obtained followed the general guidelines for biological duplicates, since in all cases they were between 0.94 and 0.97 (higher than 0.90; https://www.encodeproject.org/).
Five transcriptional comparisons were carried out with expression data (Fig. 2b; see also the dispersion of log fold change values in the MA plots of Fig. S2c). First, the transcriptomes of the reference wild-type strain grown in pH 8 medium versus standard (pH 4) AMM were compared (number 1 in Fig. 2b). The expression of 1248 genes showed a log2FC value higher than 2 or lower than −2, and were considered as significantly deregulated. This represents nearly 10 % of the genes annotated in the 36th version of the A. nidulans genome (https://fungidb.org/common/downloads/release-36/AnidulansFGSCA4/fasta/data/). Of these, 387 genes were found to be upregulated (log2FC >2) and 861 genes were found to be downregulated (log2FC <−2) at pH 8. The second analysis compared the transcriptome of the wild-type strain grown in AMM supplemented with 1.0 M sodium against the control condition (number 2 in Fig. 2b), finding 183 genes up- and 217 genes downregulated. The third set of comparisons used the transcriptomes of the null sltA and the reference wild-type strains in the three culture conditions tested (numbers 3: AMM; 4: pH 8; and 5 : 1.0 M sodium in Fig. 2b), rendering 348, 750 and 341 upregulated genes, and 764, 735 and 754 downregulated genes, respectively. The number of differentially expressed genes (DEGs) in each transcriptional comparison established the framework for further analyses.
Finally, and considering the affinity of SltA for the target sequence 5′-AGGCA-3′ [5], cumulative distribution fraction plots were drawn with the aim of correlating in each of the above-described transcriptomic comparisons log2FC values with the number of SltA target sites in gene promoters (groups of promoters with 0, 1, 2, 3, 4 and >5 SltA target motifs in the first 1000 bp upstream of the starting ATG were differentiated) (Fig. 2c). The general results suggest that mainly the response to pH 8, but also that to NaCl, does not exclusively depend on the number of putative SltA-binding sites on target promoters (first two plots in Fig. 2c). In all probability, this occurs because SltA-independent regulatory mechanisms are also required for alkaline pH and sodium homeostasis. They also suggest that an increase in the number of SltA target sequences in a given promoter correlates with a stronger dependence on the positive regulatory activity of SltA (lower log2FC values) in any of the three culture conditions tested (sltAΔ vs wild-type comparisons in Fig. 2c).
Alkaline pH causes a deregulation of genes coding for membrane transporters, with different levels of dependence on SltA activity
In the wild-type strain, there were twice as many downregulated genes at pH 8 compared to upregulated genes (861 vs 387 genes), suggesting that adaptation to medium alkalinization after a shift from an acid culture involves the induction but mainly the repression of specific sets of genes. Analyses of the lists of significantly up- and downregulated genes (Table S3; see also a specific bioinformatic analysis of genes with the Top100 log2FC values in Tables S4 and S5) confirmed the previously described upregulation of ‘alkaline-expressed genes’ such as pacC/An2855, aflR/An7820 and enaA/An6642 [2, 5, 41, 42], and the downregulation of ‘acid-expressed genes’ such as palF/An1844 (see below) [43]. These results support the reliability of the expression values obtained by RNA sequencing. Nevertheless, the expression of sltA/An2919 itself was high and was not significantly altered in the wild-type strain, suggesting that the transcription factor plays important roles in the three culture conditions studied.
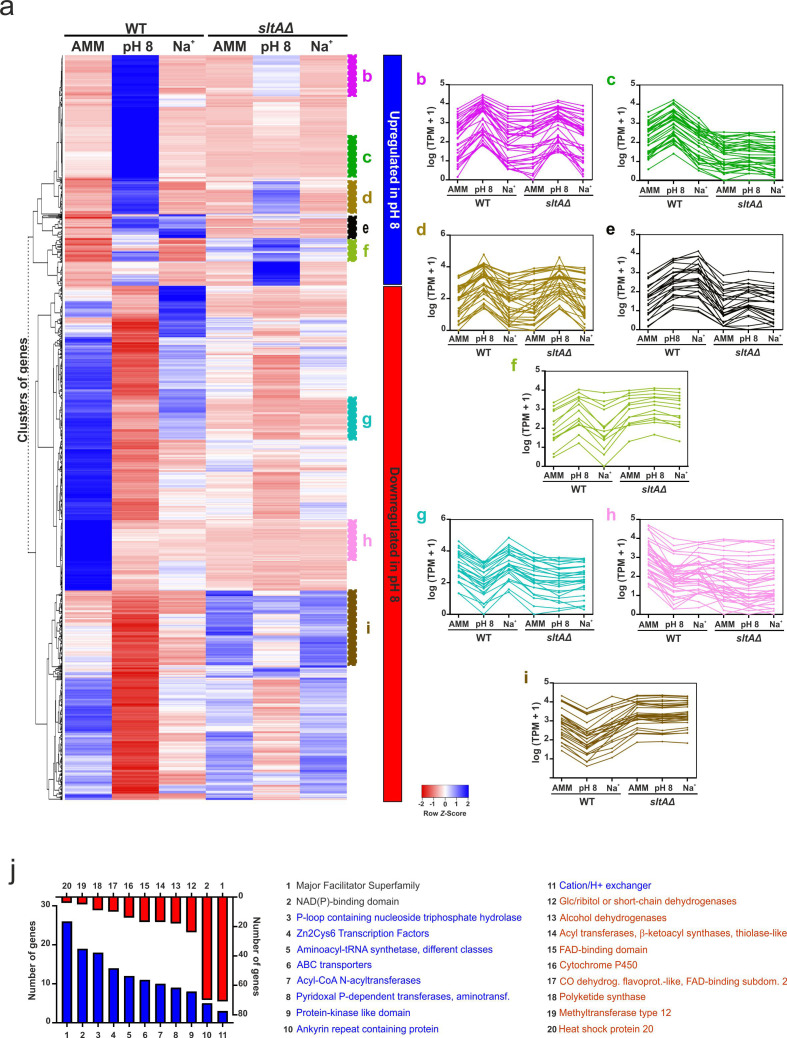
The heatmap in Fig. 3a includes all the genes up- and downregulated at pH 8 in the reference wild-type strain, but was built considering their row Z scores for all conditions and genetic backgrounds tested. The upper clade in the tree contains genes that are upregulated and the lower clade those downregulated at pH 8 (blue and red, respectively). Multiple gene expression patterns could be differentiated. Highlighted are five main expression clusters among upregulated genes and three among downregulated genes at alkaline pH. Those clusters of gene expression also uncovered different levels of dependence on SltA activity. Clusters B to D (Fig. 3b–d) correspond to those genes upregulated in the wild-type strain at pH 8. We differentiated three patterns of expression upon SltA activity: (i) the transcription of those in (b) is partially dependent on SltA (Fig. 3b), (ii) the transcription of those in (c) is totally dependent on SltA (Fig. 3c); and (iii) the transcription of those in (d) is apparently independent of SltA activity (Fig. 3d). These results suggest that the response to alkaline pH implies SltA-dependent and SltA-independent mechanisms and, in some cases, most probably the participation of additional regulators in cooperation with SltA.
Fig. 3.
Transcriptional response of A. nidulans to alkaline pH. (a) Heatmap showing the changes in expression of the genes significantly upregulated (blue; upper clade of the tree) and downregulated (red; bottom clade) at pH 8, for all six conditions and backgrounds tested. (b–i) Multiple expression clusters can be differentiated. (b) Genes upregulated at pH 8 and partially dependent on SltA at pH 8. (c) Genes upregulated at pH 8 and downregulated in the sltAΔ background under all conditions tested. (d) Genes upregulated at pH 8, almost independently of SltA at pH 8. (e) Genes upregulated both at pH 8 and in medium supplemented with 1M Na+; SltA-dependent. (f) Genes upregulated at pH 8 and also upregulated in the sltAΔ background under all conditions tested. (g) Genes downregulated at pH 8 but not in culture medium supplemented with 1.0 M Na+, and in a SltA-dependent manner. (h) Genes downregulated at pH 8 and in the sltAΔ background under all conditions tested. (i) Genes downregulated at pH 8 but upregulated in the sltAΔ background under all conditions tested. (j) Bar plot showing the number of DEGs coding for proteins predicted to contain significantly enriched Interpro domains. Blue indicates significantly upregulated genes at pH 8, while downregulated genes are highlighted in red. Genes coding for major facilitator superfamily transporters are the Top1 up- and downregulated gene family.
Cluster E (Fig. 3e) includes genes that are upregulated both at pH 8 and in medium supplemented with 1.0 M Na+. This result showed that the responses to these two types of abiotic stress may partially overlap. The expression of genes in cluster E is SltA-dependent under all conditions tested. To extend the diversity of gene expression patterns at pH 8, we show a small cluster (Fig. 3f) in which most genes are upregulated at pH 8 in the wild-type strain and also in the sltAΔ strain under all conditions tested.
Among downregulated genes at pH 8, we point to three interesting clusters. There are those genes whose expression is downregulated at pH 8 but not under 1.0 M Na+ stress (cluster G in Fig. 3g), and there are those whose expression is downregulated under both stress conditions (cluster H in Fig. 3h). Notably, SltA activity is required for the expression of those genes in any condition. The third cluster shows an opposed effect of lacking SltA activity, since in the wild-type strain these genes are downregulated at pH 8 but upregulated in the sltAΔ background under all conditions tested (cluster I in Fig. 3i). The overall data suggest that besides acting as an inducer, SltA may have a role as a repressor of transcription.
Finally, we analysed the enrichment of Interpro domains within proteins coded by genes significantly up- or downregulated at pH 8 (Fig. 3j). In both groups, the MFS and NAD(P)-binding domain-containing proteins were the Top1 and Top2 families, suggesting that the response to pH 8 involves a significant reorganization of the plasma membrane (and other subcellular membranes) and its transporters. In this sense, the enrichment of ABC transporters and cation/H+ exchangers among proteins coded by genes upregulated at pH 8 is also of note. Other enriched Interpro domains were, for example, those related to primary and secondary metabolism among the downregulated set of genes and transcription factors (mainly Zn cluster-type TFs) in the upregulated set.
Transcriptional response to sodium stress
A similar approach was followed to analyse RNA sequencing data for the samples corresponding to sodium stress compared to the control condition. In this case, the number of significantly up- or downregulated genes is clearly lower than the figures described for pH 8, suggesting that sodium stress has a smaller impact on the transcriptome of A. nidulans. Similarly, a preliminary analysis of the lists of significantly up- and downregulated genes (Table S6; see also a specific bioinformatic analysis of Top100 genes in Tables S7 and S8) showed that among previously known SltA-dependent and/or stress response genes, only sB/An2730, coding the principal sulfate transporter, varied (upregulation) significantly in its expression under sodium stress (see e.g. the upregulation of pacC/An2855, enaA/An6642 or aflR/An7820, but not that of sB, at pH 8) (Tables S6 and S7).
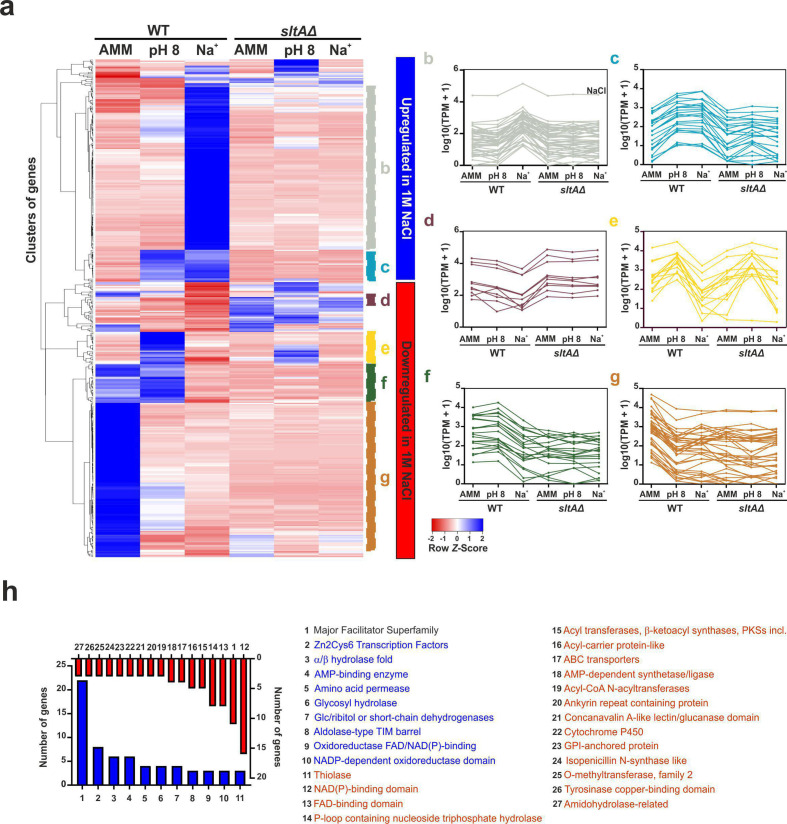
Six gene expression clusters are highlighted in the heatmap in Fig. 4a [see panels (b–g)], with two of them corresponding to the clade of upregulated genes and the remaining four corresponding to the clade of downregulated genes. Among the upregulated genes, we identified two clusters that were dependent on SltA activity and also activated by sodium stress. Cluster B shows the presence of multiple genes that are upregulated mainly or exclusively under sodium stress (Fig. 4b), while cluster C highlights genes that are upregulated both at pH 8 and under 1.0 M Na+ stress (Fig. 4c). Those genes would be common to both response mechanisms (see also Fig. 3e).
Fig. 4.
Transcriptional response of A. nidulans to sodium (1.0 M NaCl) stress. (a) Heatmap showing the changes in expression of the genes significantly upregulated (blue; upper clade) and downregulated (red; bottom clade) in medium supplemented with 1.0 M Na+, for all six conditions and backgrounds tested. (b–i) Again, multiple expression clusters can be differentiated. (b) Genes upregulated mainly or exclusively under sodium stress and in a SltA-dependent way. (c) Genes upregulated both at pH 8 and in medium supplemented with 1.0 M Na+; SltA-dependent. (d) Genes downregulated at pH 8 but mainly in 1.0 M Na+ but upregulated in the sltAΔ background under all conditions tested. (e) Genes upregulated at pH 8 but downregulated in medium supplemented with 1.0 M Na+ (different dependence patterns on SltA at pH 8). (f) Genes downregulated only in medium supplemented with 1.0 M Na+; SltA-dependent in standard minimal medium and at pH 8. (g) Genes expressed in AMM and downregulated for the rest of the conditions and backgrounds tested. (j) Bar plot showing the number of genes coding for proteins predicted to contain significantly enriched Interpro domains. Blue indicates significantly upregulated genes under Na+ stress, while downregulated genes are highlighted in red. As at pH 8, the genes coding for MFS transporters are the Top1 up- and downregulated gene family.
Among downregulated genes, cluster D contains genes that were downregulated at pH 8, though mainly in 1.0 M Na+, but were upregulated in the sltAΔ background under all conditions tested (Fig. 4d). Cluster E groups genes that were upregulated at pH 8 but downregulated in medium supplemented with 1.0 M Na+, with different patterns of dependence on SltA activity at pH 8 (independent or with positive dependence on SltA; Fig. 4e). Cluster F designates genes that were only downregulated in medium supplemented with 1.0 M Na+ and that were strongly dependent on the positive SltA activity in AMM and at pH 8 (Fig. 4f). Finally, the genes in cluster G were expressed in AMM and downregulated in the rest of conditions and backgrounds tested (Fig. 4g).
The significantly enriched Interpro domains predicted in the proteins encoded by genes with up- or downregulation under sodium stress (Fig. 4h) differed from those described in the case of pH 8 (see Fig. 3j for a comparison). However, other families, such as MFS (Top1, upregulated), NAD(P)-binding domain or Zn binuclear cluster-type TFs (Top2, upregulated), were common to the response to alkaline pH. Thus, we decided to analyse the sets and specific subsets of genes significantly deregulated at pH 8 and under sodium stress in greater depth.
Transcriptional responses under alkaline pH and sodium stress differ significantly
Only 43 genes were upregulated by both alkaline and salt stress from a total number of 387 (pH 8) and 183 (1.0 M Na+), respectively (only 3 when Top100 tables were compared; Fig. 5a). Those genes upregulated simultaneously included the ethanol regulon [44], which extends from An8977 to An8982 in ChrVII (only An8982 was not significantly upregulated in both conditions). This observation, as well as the upregulation of genes coding for MFS proteins annotated as solute : H+ symporters with a role in glycerol transport (such as An9168, the orthologue of slt1 of Saccharomyces cerevisiae, which is induced by osmotic shock [45]), may be related to an osmoregulation mechanism induced by pH 8 but primarily sodium stress. Indeed, in the yeast Pichia sorbitophila it has been seen that the ethanol regulon is induced by glycerol accumulated in response to sodium stress and that the main role of this system is osmoregulation, through accumulation of glycerol inside cells to compensate stress caused by alkaline pH or NaCl [46].
Fig. 5.
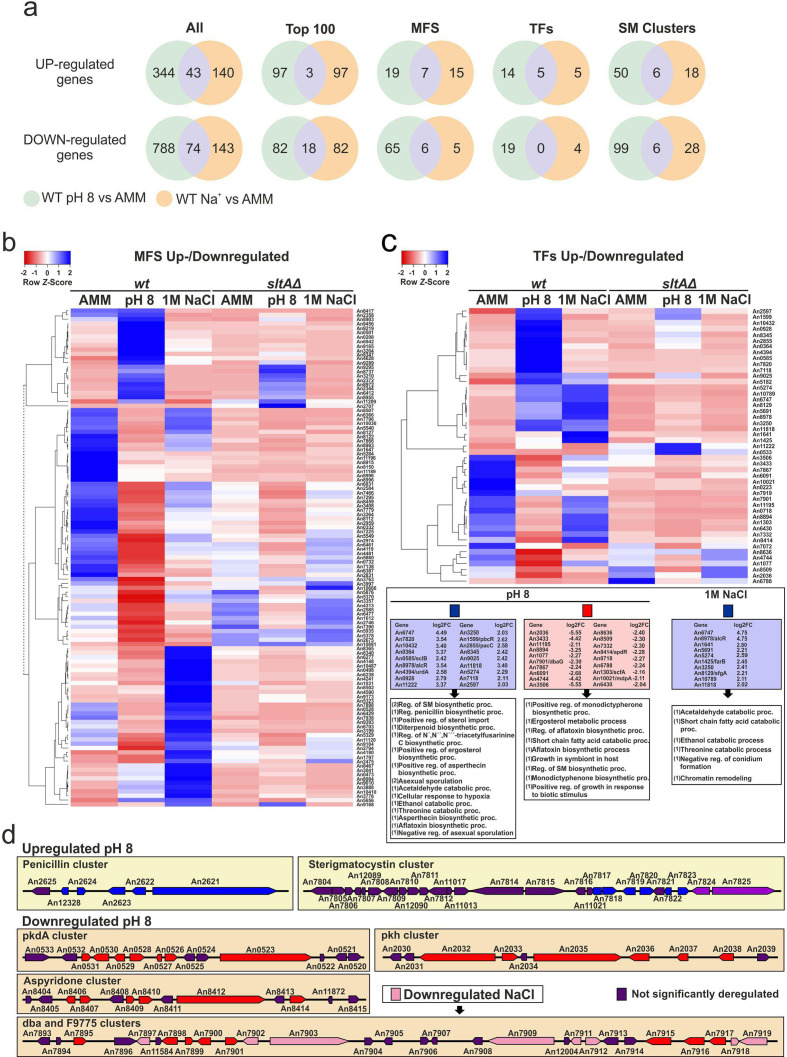
Differential transcriptional responses of A. nidulans to alkaline pH or cation stress. (a) Venn diagrams showing specific and common significantly deregulated genes when comparing the effect of medium alkalinization and high sodium stress. ‘All’ indicates the complete lists of DEGs. ‘Top100’ indicates analyses restricted to the first 100 DEGs of each comparison. The rest of the Venn diagrams focus on the up-/downregulation of genes coding for MFS, TFs and SM clusters. (b, c) Heatmaps showing the changes in expression of the genes that, coding for MFS transporters (b) or TFs (c), are significantly deregulated at pH 8 and/or in 1.0 M Na+, for all six conditions and backgrounds tested. The results suggest a very low degree of overlap between both responses regarding genes coding for these two types of proteins. (d) Diagrams showing SM clusters significantly deregulated exclusively at pH 8 or both at pH 8 and in 1.0 M Na+.
The number of simultaneously deregulated genes at pH 8 and 1.0 M Na+ increased to 74 in the sets of downregulated genes, from a total number of 861 (pH 8) and 217 (1.0 M Na+), respectively (only 18 when Top100 tables were compared; Fig. 5a). Again, we found little overlap when up- and downregulated genes coding for MFS or TFs were compared (Fig. 5a) or in the heatmaps shown in Fig. 5b, c (see e.g. genes coding for TFs that are upregulated both at pH 8 and 1.0 M Na+ in a SltA-dependent manner, such as An8978/alcR, coding for the TF of the ethanol regulon, An11818 or An6747). The differences in the transcriptional responses to alkaline pH and sodium stress were also supported by the analysis of those genes coding for proteins involved in secondary metabolite (SM) production (Fig. 5a). Panel (d) shows the deregulation of mainly different subsets of SM genes, such as the upregulation of penicillin and sterigmatocystin cluster genes at pH 8 [42, 47], or the downregulation of pkdA, aspyridone and pkh clusters at pH 8, with only the dba cluster also being downregulated under sodium stress [48–51]. Overall, the results strongly suggest that the transcriptional responses to sodium and alkaline pH stress differ significantly.
Deletion of SltA causes a profound reprogramming of gene expression, with specific subsets of DEGs depending on culture conditions
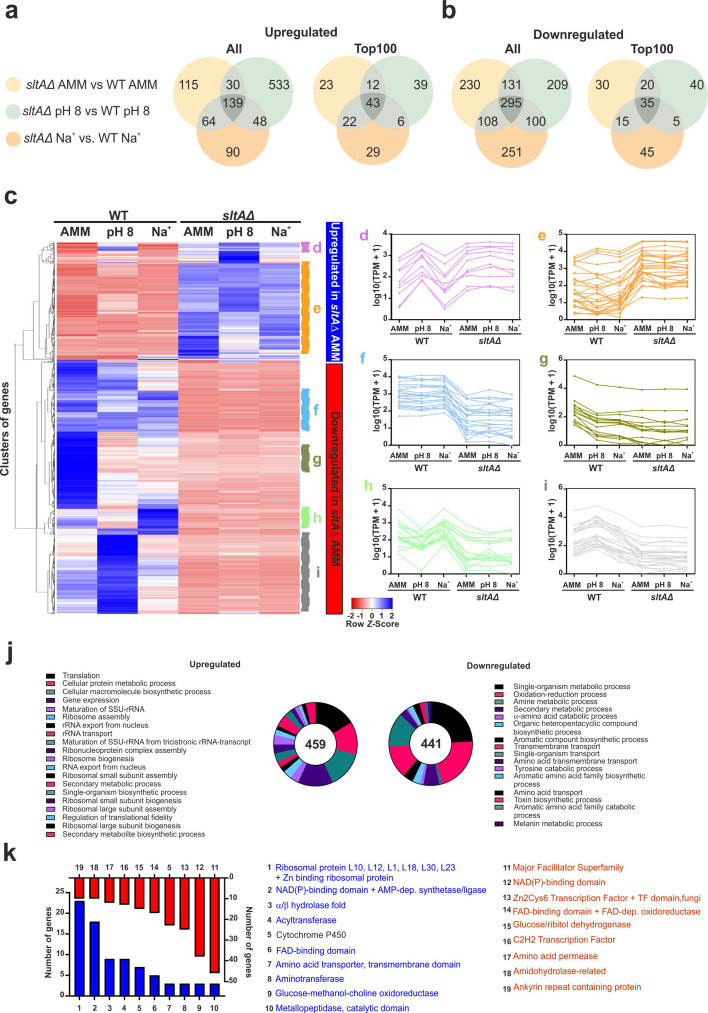
Next, we focused on the effect of sltA deletion on gene expression, based on three transcriptomic comparisons with the reference wild-type strain: under standard culture conditions (AMM), at pH 8 and after the addition of 1.0 M NaCl. We found that 139 genes were significantly upregulated and 295 were downregulated simultaneously in the three transcriptomic comparisons, with additional subsets of genes being deregulated in 2 or only 1 of the comparisons (Fig. 6a, b; see also Venn diagrams corresponding to Top100 lists of DEGs). These figures suggest that deletion of sltA has a general effect on gene expression independently of culture conditions, but also that there are most probably specific roles for SltA in transcriptional regulation at alkaline pH or sodium stress, as suggested earlier.
Fig. 6.
Comparison of sltAΔ and wild-type transcriptomes under standard culture conditions. (a,b) Venn diagrams showing the number of genes significantly upregulated (a) or downregulated (b) when comparing the sltAΔ strain and the reference wild-type under any of the three culture conditions analysed: AMM, AMM medium alkalinized to pH 8, or AMM supplemented with 1.0 M NaCl. Left: diagram corresponding to the complete lists of DEGs. Right: diagram corresponding to the Top100 DEGs. (c) Heatmap showing the changes in expression of the genes significantly upregulated (blue; upper clade) and downregulated (red; bottom clade) in the null sltA strain in AMM. Expression for all six conditions and backgrounds tested is shown. (d–i) Multiple expression clusters are differentiated. (d) Genes upregulated under all conditions in the sltAΔ strain and at pH 8 in the reference wild-type strain. (e) Genes with lower expression levels in the wild-type strain compared to the null sltA mutant, under all conditions. (f) Genes downregulated in the sltA mutant under all conditions tested. (g) Genes with higher expression levels in the wild-type strain in AMM but downregulated in the rest of samples. (h) Genes downregulated under all conditions in the sltAΔ strain and at pH 8 in the reference wild-type strain. (i) Genes upregulated at pH 8 in the wild-type strain but downregulated in the null sltA background under all conditions. (j) GO enrichment analyses of the genes up- (left) or downregulated (right) when comparing wild-type and ΔsltA samples in AMM. (k) Bar plot showing the number of DEGs coding for proteins predicted to contain significantly enriched Interpro domains. Blue indicates significantly upregulated genes in the sltAΔ strain in AMM, while downregulated genes are highlighted in red.
Again, multiple expression clusters could be differentiated from the expression patterns of significantly deregulated genes when sltAΔ and wild-type samples cultured in AMM were compared (Fig. 6c–i). In the tables of genes corresponding to this comparison (Table S9; see also a specific bioinformatic analysis of Top100 genes in Tables S10 and S11), we found that previously known SltA-related genes such as enaA/An6642, aflR/An7820 or brlA/An0973 were significantly downregulated and putative vacuolar calcium ATPases pmcA/An1189 and pmcB/An4920 were upregulated in the null sltA strain [5, 52, 53]. Cluster D corresponds to genes upregulated under all conditions in the sltAΔ strain and at pH 8 in the reference wild-type strain (Fig. 6d). Cluster E highlights genes with lower expression levels in the wild-type strain compared to the null sltA mutant (Fig. 6e), under all conditions, and probably corresponds to the subset of 139 genes mentioned in Fig. 6a (left). The remaining four expression clusters highlighted in the heatmap of Fig. 6 correspond to genes that were downregulated in the null sltA strain in AMM (lower clade of the heatmap). Cluster F shows genes that were downregulated in the sltA mutant under all conditions tested (Fig. 6f). Cluster G includes those genes with higher expression levels in the wild-type strain in AMM but downregulated in the rest of samples (Fig. 6g). Cluster H highlights those genes downregulated under all conditions in the sltAΔ strain and at pH 8 in the reference wild-type strain (Fig. 6h). Finally, cluster I groups genes were upregulated at pH 8 in the wild-type strain but downregulated in the null sltA background under all conditions (Fig. 6i).
GO enrichment analyses showed that most GO terms among upregulated genes were related to ribosome biogenesis and translation (Fig. 6j), suggesting that deletion of sltA caused a ribosomal stress that would correlate with the vegetative growth and developmental phenotypes of the null strain on AMM (see Fig. 1a). We also found that a majority of GO terms related to primary and mainly secondary metabolism among deregulated genes. A hypothetical deregulation of secondary metabolism would correlate with an induction of ribosomal stress [50, 51]. These results were confirmed by the Interpro domains enriched in the proteins coded by DEGs (Fig. 6k). Of note is, again, the downregulation of MFS transporter-coding genes, those coding for NAD(P)-binding proteins and those coding for TFs, suggesting that the previously mentioned deregulation of these groups of genes depends, at least partially, on SltA.
In the tables of genes corresponding to the comparison between sltAΔ and wild-type strains at pH 8 (Table S12; see also a specific bioinformatic analysis of Top100 genes in Tables S13 and S14) or sodium stress (Tables S15–S17), we found that previously known SltA-regulated genes such as vcxA/An0471, pmcA/An1189 and pmcB/An4920 were significantly upregulated, while others such as sltB/An6132, aflR/An7820, enaA/An6642 and brlA/An0973 were downregulated [5, 24, 52, 53]. The clusters inferred from the heatmaps in Figs S3a and S4a confirmed most of the gene expression patterns described above for the comparison of the same strains in AMM (see Figs S3b–h and S4b–j). In this case, and as described for the wild-type strain, genes coding for MFS transporters, NAD(P)-binding domain proteins and TFs (Zn2Cys6 and C2H2 types) were among most significantly deregulated ones (Figs S3h–k). Overall, the results suggest that SltA has a broad positive role in the control of genetic pathways.
SltA function is required for proper expression of pacC and palF genes
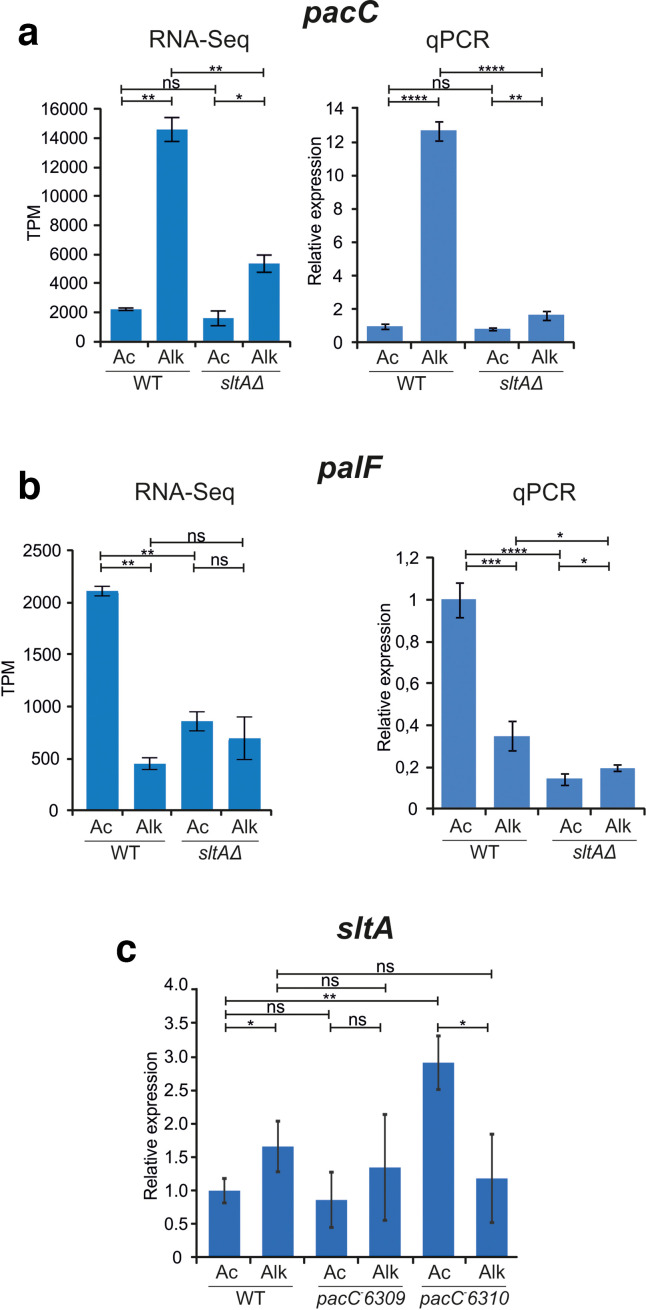
The RNA sequencing data agreed with previous results that showed that pacC is an alkaline-expressed gene and palF is an acid-expressed gene [2, 43]. In this opposed regulation PacC has been suggested to play an important role in an auto-regulatory model of the ambient pH regulatory system. However, the expression levels of pacC and palF were markedly altered in the null sltA background. RNAseq data evidenced that upregulation of pacC expression caused by medium alkalinization was lost in the sltA mutant (Fig. 7a) without being affected at those levels found at acid pH (AMM). In contrast, palF expression was notably reduced at acid pH, close to those levels found at alkaline pH (Fig. 7b). To verify this finding, independent biological repetitions were performed and pacC and palF expression was determined by means of quantitative PCR. The results of qPCRs, using α-tubulin coding gene benA as a control, were similar to those found in RNA sequencing experiments (Fig. 7a, b, compare RNA-seq and qPCR charts). Absence of SltA activity caused deregulation of these principal genes in the ambient pH regulatory system.
Fig. 7.
Analyses of pacC and palF expression levels under alkaline pH stress and the effect of lacking SltA activity. Confirmation of RNA sequencing data in new biological samples using quantitative PCR. (a) Charts showing RNA sequencing data (TPM values) and relative expression levels of pacC in wild-type and sltAΔ strains, under control AMM (Ac) and pH 8 (Alk) conditions. (b) As in (a) but for palF gene expression levels. The strains used in qPCR experiments were MAD6669 (reference wild-type strain) and MAD3816 (sltAΔ). (c) Chart showing relative expression levels of sltA in wild-type and pacC-6310 mutant backgrounds, under the same conditions as in (a, b). Error bars correspond to standard deviations of two replicates in the case of sequencing results and three replicates in the case of qPCR results. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001; ns, not significant (P>0.05).
Finally, to determine whether the expression of sltA and pacC was interdependent, we analysed the relative expression of the former in pacC−6309 and pacC−6310 mutants, suppressors of pacCc63 allele, which express truncated non-functional versions of PacC (Met5fs and His164fs, respectively) [54]. qPCR results did not show any downregulation (but a slight upregulation) of sltA in these pacC − mutant backgrounds (Fig. 7c), suggesting that the expression of both genes is not interdependent.
Discussion
Tolerance to abiotic stress conditions has served as a basis for the classification of micro-organisms. The capacity to grow at a given pH results in differentiating acidophiles, neutrophiles and alkaliphiles, while those organisms able to thrive when alkali cations, such as sodium, are present in high quantities are known as halo-tolerants (recently reviewed in [55]). Of course, tolerance relies on genomes coding for subsets of proteins enabling adaptation to those abiotic stress conditions and also the induction of regulatory mechanisms that coordinate the scheduled expression of those resistance genes [56]. A. nidulans has served as an important reference system to investigate these abiotic stress-responsive pathways (recently reviewed in [1]).
In this work, we have focused on studying the transcriptional response to alkalinization of the extracellular medium and to high concentrations of extracellular sodium. Both stress conditions compromise the colonial growth capacity of A. nidulans, particularly when the function of the transcriptional factor SltA is lost (Fig. 1a) [5, 23]. Here we show that over time A. nidulans modulates the relative amounts of the three main forms of SltA, SltA78kDa as well as phosphorylated and non-phosphorylated SltA32kDa forms, differently, after the induction of each stress condition (Fig. 1b). This observation suggests that all forms of SltA may play a role in controlling gene expression, and that specific relative amounts are required to coordinate the adequate transcriptional response to each stimulus. To determine this transcriptional response, we concentrated at the time point of 60 min after the induction of stress, because this time point consistently showed a similar distribution of SltA forms in two markedly different response patterns to both stresses (Fig. 1b).
Comparisons of RNA sequencing results in the wild-type strain revealed important transcriptomic differences between the responses to medium alkalinization and sodium stress. The PCA plot (Fig. S1) denotes that ambient pH alkalinization causes a major modification of the transcriptional pattern, while the addition of sodium has a more specific effect. Subsequent comparisons supported the above conclusion, with the observation that only a few genes are simultaneously deregulated in the wild-type strain when each of the two types of stress is induced (Fig. 5a). Among upregulated genes, 43 are common but 344 are specific to alkaline pH response and 140 are specific to high sodium response. By contrast, 74 genes are downregulated in both conditions and 788 and 143 genes are specifically downregulated at pH 8 and under high sodium, respectively. Ethanol regulon and glycerol/H+ symporters are part of this common response. The accumulation of osmolytes such as glycerol or trehalose is a common mechanism of osmoregulation [57]. Although the anabolic pathway of glycerol (or the expression of vosA/An1959, which codes for a transcriptional regulator of trehalose synthesis [58]) is not found in the list of DEGs, the upregulation of the ethanol regulon and glycerol/H+ symporters suggests that intracellular accumulation of glycerol is probably a primary response to these abiotic stresses in A. nidulans, and one that is more prominent in the case of high sodium concentrations.
Intracellular production of glycerol as an osmolyte is regulated by the mitogen-activated protein kinase cascade HOG through the transcription factor MsnA/An1652 [41, 59, 60]. In the wild-type strain, none of the genes encoding elements of this pathway (reviewed in [56, 61, 62]) are included in the DEG lists of this work. This is interesting because the absence of any of the elements of the MAP kinase cascade, sskB/An10153 (MAPKKK), pbsA (alias pbsB)/An0931 (MAPKK) and hogA/sakA/An1017 (MAPK), cause sensitivity to high NaCl [61]. The work of Han and Prade [41] showed a transient upregulation of pbsA and hogA after exposure to 1 M sodium. Our results agree with their observation that expression of both genes decreased after a peak at 30 min and led us to conclude that this key signalling pathway in osmoregulation is not subjected to transcriptional up- or downregulation after 60 min of the response to alkaline or sodium stress. Which, then, is the main transcriptional response observed in the wild-type? At alkaline pH, among the most upregulated genes are those coding for sodium ATPases, enaA/AN6642 and enaB/AN1628, the Pi/Na+ symporter Pho89-like/AN8956, and two sodium/H+ antiporters (AN5035 and AN4131) of the Nha1 family [5, 41, 63]. The upregulation of these ion transporters could reflect the need for sodium and phosphate detoxification or for the accumulation of protons to compensate for intracellular alkalinization caused by the addition of the buffer. However, this response is specific to alkaline pH because these genes are not significantly deregulated in the response to sodium. In contrast, sodium generates a more general response: the previously described activation of ethanol regulon and a deregulation of genes related to primary metabolism of carbon and nitrogen sources. This effect on primary metabolisms has previously been described as a general response to environmental stress [56, 62].
What is the role of SltA in transcription? The absence of SltA causes evident changes in the pattern of gene expression compared to the wild-type strain, both in the absence or presence of alkaline pH/sodium stress conditions. This is well illustrated by the PCA plot in Fig. S1a, in which data from the null strain do not cluster with and are distant from those of the reference strain. The need for SltA activity under non-stressing conditions is well supported by the vegetative growth and developmental phenotype of the null strain, and also the detection of the three forms of SltA in the wild-type background (Fig. 1b). The increase of full-length SltA form at early time-points after alkaline pH stress induction suggests either a role for this form or the effect of a pH-dependent upregulation of sltA expression which is not observed in our RNA sequencing data at 60 min. In contrast, the truncated 32 kDa form predominates at high sodium concentrations. These observations might explain previous results in which the sole expression of a form mimicking the SltA32kDa truncated version suffices to enable wild-type growth rates in sodium stress, but not at alkaline pH [23]. Thus, SltA78kDa may play a role in the response to alkalinity and SltA32kDa under both stress conditions. In addition, these changes in the patterns of SltA forms in response to pH 8 and sodium stress predict differential expression levels of sltA, but our RNA sequencing data showed no major changes at 60 min. Interestingly, sltA promoter contains four putative SltA-binding sites within 1.5 kb from ATG, two very close to the possible transcription start point, suggesting the possibility of autorregulation. Thus, extensive and specific studies on the mechanisms affecting sltA expression levels will be required (see also below).
More than a thousand genes are likely to be significantly deregulated in each of the three wild-type versus sltAΔ comparisons (i.e. 1112 genes under standard, non-stressing, culture conditions; Fig. 2b) and may require the direct or indirect activity of SltA. Among them, we found genes that are up- and downregulated, confirming our previous hypothesis that SltA may play a dual role as a repressor and as an activator [5, 52]. RNA sequencing data confirm that SltB, coding for the protease that cleaves SltA, is among the genes that are positively dependent on SltA activity [23]. Other known targets of SltA are also present among DEGs (see the Results section). However, the most representative GO classes upregulated in the null sltA strain under non-stressing conditions are related to ribosome biogenesis and translation (Fig. 6j, k), indicating that the absence of SltA forces a mis-scheduled regulation of the expression of these genes. The deregulation of the expression of ribosome-forming or regulatory proteins has been related to a general response to ambient stress [56]. It must therefore be assumed that part of the deregulation observed here may be due to an indirect effect of the absence of SltA. Using other means to establish the genes that are regulated directly by SltA itself or by transcriptional factor cascades regulated by SltA will help to provide a definitive picture. The presence of SltA targets in the promoters will help us to determine the range of action of SltA.
Several transcriptional regulators are subject to SltA control. Expression of msnA/An1652, a transducer of the HOG osmoregulatory pathway [41], is significantly downregulated in the null sltA background. In S. cerevisiae, ALD2 gene, coding for cytoplasmic aldehyde dehydrogenase [64], is under regulation of Msn1p, the MsnA homologue. In concordance, the homologue of ALD2 in A. nidulans, AN9034, is strongly downregulated in the null sltA background. Predictably, the product of AN9034 converts acetaldehyde in acetyl-CoA and its downregulation in the null sltA strain might cause the intracellular elevation of toxic aldehydes, as has been described in yeast [64], contributing to the reduced viability of the sltAΔ mutant under the stress conditions analysed in this work. Other transcription factor-coding genes deregulated in the sltA deletion strain belong to well-known SM biosynthetic clusters [65], evidencing regulation by ambient stress and/or in concert with SltA regulation. As previously known, the penicillin cluster is one of this alkaline pH-activated SM pathways [47], but only acvA/An2622 (ACV-peptide synthase) and ipnA/An2621 (isopenicillin N synthase), the genes coding for the first two enzymes, are among the Top100 upregulated genes at alkaline pH (Fig. 5d). The penicillin pathway is not under SltA regulation, but certainly SltA plays an important role in the regulation of the aflatoxin pathway [53], probably by positively regulating the expression of the zinc binuclear cluster-coding gene aflR/An7820 (Tables S9 and S12). Other deregulated SM pathways at alkaline pH are the pkh cluster, most likely through downregulation of the predicted specific regulator AN2036, the pkdA cluster, the aspyridone pathway (probably via downregulating the specific transcription factor apdR/An8414) or upregulation of the terrequinone cluster, via upregulation of tdiB/An8514 and tdiC/An8518 genes. Several genes of contiguous clusters dba and F9775 are deregulated under sodium stress (Fig. 5d). Thus, abiotic stresses and SltA are key regulators of several SM pathways [65].
In particular, this investigation opens an avenue for defining the hierarchy among the main transcriptional regulators controlling the response to ambient pH. PacC, the transcription factor regulating ambient pH response, is upregulated at alkaline pH, as has been shown previously (Figs 5c and 7) [2, 43]. The absence of SltA function strongly reduces the upregulation of pacC/An2855 at alkaline pH, and this has been confirmed using an alternative null sltA strain and through qPCR analyses (Fig. 7a). Transcriptional upregulation of pacC has been proposed as a key step in the PacC-Pal ambient pH pathway, and thus the fact that pacC transcription levels remain close to those determined at acid pH in the null sltA mutant suggests that there is a significant mismatch in the PacC signalling process. In agreement with this idea, the acid-expressed gene palF/An1844, coding for the signalling arrestin-like protein [43], is also not properly upregulated at acid pH in the sltA mutant. Overall, these data suggest an important role of SltA in modulating signalling in the PacC/Pal pathway in response to alkaline pH. Recently, PacX/An0766, a negative regulator, was included in this pathway [43]. Here, SltA is proposed to act as a positive regulator of pacC and palF expression. The presence of three putative SltA-binding sites at the pacC promoter (first 1000 bp; not shown) suggests a direct role of SltA through direct interaction with those sites. Furthermore, here we have described a correlation between the number of SltA target sites in a promoter and an increase in the fold change values of wild-type versus sltAΔ samples, although this is clearly not the only requirement for a direct transcriptional dependence on SltA activity, since there are multiple genes with multiple SltA sites in their promoters, but with log2FC values between −1 and 1. On the other hand, no SltA-binding sites are found in the palF promoter, suggesting an indirect function or recognition of alternative target sites. Future work will focus on delimiting the role of SltA on the ambient pH regulatory pathway and whether corregulation with PacX/An0766 occurs, since both SltA and PacX are transcription factors specific to the subphylum Pezizomycotina [5, 23, 43]. In the context of ancient gene rewiring phenomena [66], both factors may have been recruited for new functions in ancestors of Aspergillus species. Deciphering the roles of SltA in distant Pezizomycotina species would contribute to our understanding these genetic processes and dissection of the mechanisms of abiotic stress response in fungi.
Conclusions
Our data suggest that alkaline pH and sodium stress responses in Aspergillus nidulans are largely distinct, with only a small fraction of common elements. Adaptation to alkaline pH apparently requires a more general transcriptional response, while the response is more specific in the case of sodium stress. Both stress responses probably include a general reorganization of lipid bilayers and their transporters, but probably using different subsets of proteins, suggesting that the study of each response will require specific experimental approaches in the future. The transcription factor SltA plays a key role in both responses, and also the control of, for example, SM and development. Furthermore, there is a dependence of pacC expression on SltA activity, but not in the opposite way, suggesting that there is a hierarchical relationship between these two transcription factors. Future studies will elucidate how this dependence is established in A. nidulans.
Supplementary Data
Funding information
Work at CIB-CSIC was funded by MINECO (BFU2015-66806-R to E. A. E) and MICIU/AEI (RTI2018-094263-B-100) to E. A. E (both partially supported by FEDER, EU). Work at the UPV/EHU lab was funded by UPV/EHU grants PPGA19/08 and GIU19/014 to O. E and the Basque Government grant Elkartek19/72 (to Professor María Teresa Dueñas). I. P and E. R. held research contracts associated with grants RTI2018-094263-B-100 and BFU2015-66806-R, respectively. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgements
We thank Irene Tomico for sharing strain MAD6669. We acknowledge the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI) for enabling this open access publication through a Publish and Read deal with the Microbiology Society.
Author contributions
E. A. E. conceived and supervised the experiments. I. P. and E. R. performed experimental work. A. G. carried out trimming and mapping of sequencing reads. Bioinformatic analyses of the sequencing results were performed by O. E., I. P. and E. A. E. Original draft manuscript written by O. E., I. P and E. A. E. All authors contributed to the improvement of the text and figures.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMM, Aspergillus minimal medium; Chr, chromosome; DEG, differentially expressed gene; FC, fold change; GO, gene ontology; MA plot, log ratio (M) and mean average (A) plot; MFS, major facilitator superfamily; MNNG, N-methyl-N'-nitro-N-nitrosoguanidine; 4NQO, 4-nitroquinoline-1-oxide; PCA, principal component analysis; qPCR, quantitative PCR; RIN, RNA integrity number; RT-PCR, reverse transcriptase PCR; SM, secondary metabolism; TF, transcription factor.
All supporting data, code and protocols have been provided within the article or through supplementary data files. 17 supplementary tables and four supplementary figures are available with the online version of this article.
The RNA sequencing data analysed in this work were deposited and are freely available in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA625291.
References
- 1.Etxebeste O, Espeso EA. Aspergillus nidulans in the post-genomic era: a top-model filamentous fungus for the study of signaling and homeostasis mechanisms. Int Microbiol. 2020;23:5–22. doi: 10.1007/s10123-019-00064-6. [DOI] [PubMed] [Google Scholar]
- 2.Tilburn J, Sarkar S, Widdick DA, Espeso EA, Orejas M, et al. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. Embo J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arst HN, Peñalva MA. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 2003;19:224–231. doi: 10.1016/s0168-9525(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 4.Peñalva MA, Tilburn J, Bignell E, Arst HN. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Spielvogel A, Findon H, Arst HN, Araújo-Bazán L, Hernández-Ortíz P, et al. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans . Biochem J. 2008;414:419-29. doi: 10.1042/BJ20080344. [DOI] [PubMed] [Google Scholar]
- 6.Orejas M, Espeso EA, Tilburn J, Sarkar S, Arst HN, et al. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 7.Espeso EA, Tilburn J, Sánchez-Pulido L, Brown CV, Valencia A, et al. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol. 1997;274:466–480. doi: 10.1006/jmbi.1997.1428. [DOI] [PubMed] [Google Scholar]
- 8.Espeso EA, Roncal T, Díez E, Rainbow L, Bignell E, et al. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. Embo J. 2000;19:719–728. doi: 10.1093/emboj/19.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Díez E, Álvaro J, Espeso EA, Rainbow L, Suárez T, et al. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. Embo J. 2002;21:1350–1359. doi: 10.1093/emboj/21.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink H-J, et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell. 2007;6:2365 LP–2375. doi: 10.1128/EC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herranz S, Rodríguez JM, Bussink H-J, Sánchez-Ferrero JC, Arst HN, et al. Arrestin-Related proteins mediate pH signaling in fungi. Proc Natl Acad Sci U S A. 2005;102:12141 LP–12146. doi: 10.1073/pnas.0504776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galindo A, Calcagno-Pizarelli AM, Arst HN, Peñalva Miguel Ángel. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J Cell Sci. 2012;125:1784–1795. doi: 10.1242/jcs.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucena-Agell D, Hervás-Aguilar A, Múnera-Huertas T, Pougovkina O, Rudnicka J, et al. Mutational analysis of the Aspergillus ambient pH receptor PalH underscores its potential as a target for antifungal compounds. Mol Microbiol. 2016;101:982–1002. doi: 10.1111/mmi.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peñalva MA, Lucena-Agell D, Arst HN. Liaison alcaline: PalS entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol. 2014;22:49–59. doi: 10.1016/j.mib.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Galindo A, Hervás-Aguilar A, Rodríguez-Galán O, Vincent O, Arst HN, et al. Palc, one of two BRO1 domain proteins in the fungal pH signalling pathway, localizes to cortical structures and binds Vps32. Traffic. 2007;8:1346–1364. doi: 10.1111/j.1600-0854.2007.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent O, Rainbow L, Tilburn J, Arst HN, Peñalva MA. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol Cell Biol. 2003;23:1647 LP–1655. doi: 10.1128/mcb.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peñas MM, Hervás-Aguilar A, Múnera-Huertas T, Reoyo E, Peñalva MA, et al. Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot Cell. 2007;6:960–970. doi: 10.1128/EC.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hervás-Aguilar A, Rodríguez JM, Tilburn J, Arst HN, Peñalva MA. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J Biol Chem. 2007;282:34735–34747. doi: 10.1074/jbc.M706723200. [DOI] [PubMed] [Google Scholar]
- 19.Espeso EA, Peñalva MA. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J Biol Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 20.Espeso EA, Arst HN. On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol Cell Biol. 2000;20:3355 LP–3363. doi: 10.1128/mcb.20.10.3355-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caddick MX, Brownlee AG, Arst HN. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans . Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 22.MacCabe AP, Orejas M, Pérez-González JA, Ramón D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J Bacteriol. 1998;180:1331 LP–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellado L, Arst HN, Espeso EA. Proteolytic activation of both components of the cation stress-responsive Slt pathway in Aspergillus nidulans . Mol Biol Cell. 2016;27:2598–2612. doi: 10.1091/mbc.E16-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellado L, Calcagno-Pizarelli AM, Lockington RA, Cortese MS, Kelly JM, et al. A second component of the SltA-dependent cation tolerance pathway in Aspergillus nidulans . Fungal Genet Biol. 2015;82:116–128. doi: 10.1016/j.fgb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saloheimo A, Aro N, Ilmén M, Penttilä M. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei . J Biol Chem. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- 26.O’Neil JD, Bugno M, Stanley MS, Barham-Morris JB, Woodcock NA, et al. Cloning of a novel gene encoding a C2H2 zinc finger protein that alleviates sensitivity to abiotic stresses in Aspergillus nidulans. Mycol Res. 2002;106:491–498. [Google Scholar]
- 27.Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans . Biochim Biophys Acta - Enzymol Biol Oxid. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 29.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, et al. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2012;40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. Star: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y, Smyth GK, Shi W. featureCounts: an efficient General purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team R: A language and environment for statistical computing. Austria: R Found Stat Comput Vienna; 2107. [Google Scholar]
- 35.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2018;gky1100-gky1100. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. FungiFun2: a comprehensive online resource for systematic analysis of gene Lists from fungal species. Bioinformatics. 2015;31:445–446. doi: 10.1093/bioinformatics/btu627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SX G, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernández-Ortiz P, Espeso EA. Phospho-regulation and nucleocytoplasmic trafficking of CrzA in response to calcium and alkaline-pH stress in Aspergillus nidulans . Mol Microbiol. 2013;89:532–551. doi: 10.1111/mmi.12294. [DOI] [PubMed] [Google Scholar]
- 41.Han K-H, Prade RA. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans . Mol Microbiol. 2002;43:1065–1078. doi: 10.1046/j.1365-2958.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- 42.Delgado-Virgen F, Guzman-de-Peña D. Mechanism of sterigmatocystin biosynthesis regulation by pH in Aspergillus nidulans . Braz J Microbiol. 2009;40:933–942. doi: 10.1590/S1517-838220090004000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bussink H-J, Bignell EM, Múnera-Huertas T, Lucena-Agell D, Scazzocchio C, et al. Refining the pH response in Aspergillus nidulans: a modulatory triad involving PacX, a novel zinc binuclear cluster protein. Mol Microbiol. 2015;98:1051–1072. doi: 10.1111/mmi.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felenbok B, Sequeval D, Mathieu M, Sibley S, Gwynne DI, et al. The ethanol regulon in Aspergillus nidulans: characterization and sequence of the positive regulatory gene alcR. Gene. 1988;73:385–396. doi: 10.1016/0378-1119(88)90503-3. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, et al. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:2068–2076. doi: 10.1091/mbc.e04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lages F, Lucas C. Characterization of a glycerol/H+ symport in the halotolerant yeast Pichia sorbitophila. Yeast. 1995;11:111–119. doi: 10.1002/yea.320110203. [DOI] [PubMed] [Google Scholar]
- 47.Espeso EA, Tilburn J, Arst HN, Peñalva MA. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. Embo J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahuja M, Chiang Y-M, Chang S-L, Praseuth MB, Entwistle R, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans . J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oiartzabal-Arano E, Garzia A, Gorostidi A, Ugalde U, Espeso EA, et al. Beyond asexual development: modifications in the gene expression profile caused by the absence of the Aspergillus nidulans transcription factor FlbB. Genetics. 2015;199:1127–1142. doi: 10.1534/genetics.115.174342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerke J, Braus GH. Manipulation of fungal development as source of novel secondary metabolites for biotechnology. Appl Microbiol Biotechnol. 2014;98:8443-55. doi: 10.1007/s00253-014-5997-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Findon H, Calcagno-Pizarelli A-M, Martínez JL, Spielvogel A, Markina-Iñarrairaegui A, et al. Analysis of a novel calcium auxotrophy in Aspergillus nidulans . Fungal Genet Biol. 2010;47:647–655. doi: 10.1016/j.fgb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shantappa S, Dhingra S, Hernández-Ortiz P, Espeso EA, Calvo AM. Role of the zinc finger transcription factor SltA in morphogenesis and sterigmatocystin biosynthesis in the fungus Aspergillus nidulans . PLoS One. 2013;8:e68492. doi: 10.1371/journal.pone.0068492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernández-Martínez J, Brown CV, Díez E, Tilburn J, Arst HN, et al. Overlap of nuclear localisation signal and specific DNA-binding residues within the zinc finger domain of PacC. J Mol Biol. 2003;334:667–684. doi: 10.1016/j.jmb.2003.09.072. [DOI] [PubMed] [Google Scholar]
- 55.Coker JA. Recent advances in understanding extremophiles [version 1; peer review: 2 approved] F1000 Research. 2019;8 doi: 10.12688/f1000research.20765.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 2007;24:961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 57.Ianutsevich EA, Tereshina VM. Combinatorial impact of osmotic and heat shocks on the composition of membrane lipids and osmolytes in Aspergillus niger. Microbiology. 2019;165:554–562. doi: 10.1099/mic.0.000796. [DOI] [PubMed] [Google Scholar]
- 58.Ni M, Yu J-H. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans . PLoS One. 2007;2:e970. doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawasaki L, Sánchez O, Shiozaki K, Aguirre J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol Microbiol. 2002;45:1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x. [DOI] [PubMed] [Google Scholar]
- 60.Klein M, Swinnen S, Thevelein JM, Nevoigt E. Glycerol metabolism and transport in yeast and fungi: established knowledge and ambiguities. Environ Microbiol. 2017;19:878–893. doi: 10.1111/1462-2920.13617. [DOI] [PubMed] [Google Scholar]
- 61.Furukawa K, Hoshi Y, Maeda T, Nakajima T, Abe K. Aspergillus nidulans hog pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol Microbiol. 2005;56:1246–1261. doi: 10.1111/j.1365-2958.2005.04605.x. [DOI] [PubMed] [Google Scholar]
- 62.Miskei M, Karányi Z, Pócsi I. Annotation of stress-response proteins in the aspergilli. Fungal Genet Biol. 2009;46 Suppl 1:S105–S120. doi: 10.1016/j.fgb.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 63.Harris SD, Turner G, Meyer V, Espeso EA, Specht T, et al. Morphology and development in Aspergillus nidulans: a complex puzzle. Fungal Genet Biol. 2009;46 Suppl 1:S82–S92. doi: 10.1016/j.fgb.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 64.Aranda A, del Olmo Ml Marcel lí, l delOM. Response to acetaldehyde stress in the yeast Saccharomyces cerevisiae involves a strain-dependent regulation of several ALD genes and is mediated by the general stress response pathway. Yeast. 2003;20:747–759. doi: 10.1002/yea.991. [DOI] [PubMed] [Google Scholar]
- 65.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae . BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Etxebeste O, Otamendi A, Garzia A, Espeso EA, Cortese MS. Rewiring of transcriptional networks as a major event leading to the diversity of asexual multicellularity in fungi. Crit Rev Microbiol. 2019;45:548–563. doi: 10.1080/1040841X.2019.1630359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.