Abstract
Autophagy is a highly conserved catabolic process that degrades cytosolic proteins and organelles via formation of autophagosomes that fuse with lysosomes to form autolysosomes, whereby autophagic cargos are degraded. Numerous studies have demonstrated that autophagy plays a critical role in the regulation of liver physiology and homeostasis, and impaired autophagy leads to the pathogenesis of various liver diseases such as viral hepatitis, alcohol associated liver diseases (AALD), non-alcoholic fatty liver diseases (NAFLD), and liver cancer. Recent evidence indicates that autophagy may play a dual role in liver cancer: inhibiting early tumor initiation while promoting progression and malignancy of already formed liver tumors. In this review, we summarized the progress of current understanding of how hepatic viral infection, alcohol consumption and diet-induced fatty liver diseases impair hepatic autophagy. We also discussed how impaired autophagy promotes liver tumorigenesis, and paradoxically how autophagy is required to promote the malignancy and progression of liver cancer. Understanding the molecular mechanisms underlying how autophagy differentially affects liver cancer development and progression may help to design better therapeutic strategies for prevention and treatment of liver cancer.
Keywords: Alcohol, mTOR, Nrf2, p62, TFEB
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved catabolic process for maintaining cellular homeostasis under both basal and stress conditions. Autophagy facilitates the clearance or turnover of long-lived or misfolded proteins, insoluble protein aggregates, invading microbes and damaged or excess organelles [1-3]. Autophagy has generally been thought to play dual roles in tumorigenesis and cancer progression. Autophagy serves as a tumor suppressor to inhibit tumorigenesis by removing damaged organelles and reducing oxidative stress to mitigate DNA damage and genome instability. However, after the cells have transformed and become cancerous, they can use autophagy as a survival mechanism against the harsh environment characterized by a lack of both oxygen and nutrients. As a result, blocking autophagy in combination with other chemotherapeutic drugs is believed to be a promising therapeutic strategy to treat various cancers [4].
Liver cancer is the third leading cause of cancer-related deaths worldwide and accounts for more than 700,000 deaths annually [5,6]. According to a recent study, the mortality rate of hepatocellular carcinoma (HCC) in the USA increased approximately 40% between 2000 and 2016 [7]. As a multistage disease, HCC development is linked to many environmental risk factors such as alcohol associated liver disease (AALD), hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, aflatoxin B1 and obesity-related nonalcoholic fatty liver diseases (NAFLD) [8]. Interestingly, among the high-risk factors of HCC, impaired hepatic autophagy has been shown to contribute to the pathogenesis of AALD, NAFLD as well as HBV and HCV hepatitis [9-14], suggesting that defective autophagy may be a common mechanism for HCC development. In this review, we briefly discuss the current understanding of the role and mechanisms of autophagy in the pathogenesis of liver cancer.
DUAL ROLES OF AUTOPHAGY IN LIVER TUMORIGENESIS AND CANCER PROGRESSION
As mentioned above, autophagy may play dual roles in cancer depending on the stage of tumor development. Autophagy serves as a tumor suppressor to inhibit tumorigenesis likely before the normal cells are transformed into cancer cells. Autophagy also promotes cancer progression and malignancy in already transformed cancer cells. Below we will discuss in detail the paradoxical role of autophagy in tumor initiation and progression. Understanding the complex dual roles of autophagy in cancer has very important implications as it will help to redefine the context in which autophagy manipulation can act as tumor prevention or treatment/therapy. For instance, agents that increase autophagy activity may be used to prevent tumorigenesis whereas agents that inhibit autophagy may be used to treat existing cancers.
AUTOPHAGY INHIBITS LIVER TUMORIGENESIS
The first direct evidence to support autophagy as a tumor suppressor came from the pioneering work of Dr. Beth Levine’s group, who observed spontaneous tumor occurrence in multiple tissues, including the liver, in aged beclin1 heterozygous mice [15,16]. This result was later confirmed by Yue et al. [17]. The beclin1 heterozygous mice are also highly susceptible to HCC upon HBV infection [16]. Moreover, beclin1 is frequently found mono-allelically deleted in many human cancers such as prostate, ovarian and breast cancers [15] suggesting that Beclin1 inhibits tumorigenesis and acts as a haploinsufficient tumor suppressor. Further studies show that decreased expression of beclin1 is correlated with HCC grade, suggesting its possibility as a HCC prognostic bio-marker [18]. However, beclin1 has a BH3 domain and interacts with Bcl-2 protein, a critical regulator of cell death. Moreover, beclin1 homozygous knockout (KO) mice are embryonic lethal. This is in great contrast to the phenotypes of homozygous deletion of other Atgs such as atg5 or atg7 in mice, which die at neonatal stage with normal embryo development [19,20]. Therefore, it has been questioned whether the tumor suppressive function of Beclin1 was directly due to its inhibition of autophagy or its non-autophagic functions. Subsequent studies from Komatsu’s and Mizushima’s group as well as ours show that liver-specific atg5 or atg7 KO mice also develop spontaneous liver tumors [21-23], confirming that autophagy is indeed a bona fide tumor suppressor. The tumor suppressive function of autophagy is likely due to its ability to remove cellular protein aggregates and damaged organelles. The failure to remove damaged organelles such as mitochondria and protein aggregates in a timely manner can lead to a cascade of increased oxidative and metabolic stress, genome instability, and ultimately, cell malignant transformation [24-28]. Indeed, liver-specific atg5 or atg7 KO mice have increased hepatic oxidative DNA damage, accumulation of protein aggregates and damaged mitochondria as well as an increased number of peroxisomes and endoplasmic reticulum (ER) content, increased apoptosis, compensatory hepatocyte proliferation, hepatomegaly, increased ductular reaction, inflammation, and fibrosis, all of which can result in the development of liver tumors [21,23,29-32]. Several signaling pathways have been identified that may contribute to the liver pathogenesis and tumorigenesis in autophagy-defective liver including high mobility group box 1 (HMGB1), yes-associated protein (Yap), mechanistic target of rapamycin (mTOR), and p62-nuclear factor (erythroid-derived 2)-like 2 (Nrf2) pathway [32-35]. Among these signaling pathways in autophagy-defective liver, the p62-Nrf2 pathway seems to be the central player and has a more profound impact on liver pathogenesis and tumorigenesis than HMGB1, Yap, or mTOR, is discussed in detail later.
HMGB1 is a non-histone DNA binding protein and can be released into the extracellular space either passively during cell death or actively following cytokine stimulation. Once released, HMGB1 acts as a damage-associated molecular pattern (DAMP) molecule to activate immune cells [36,37], which is associated with liver injury, sepsis, inflammation, and fibrosis [38,39]. Deletion of hmgb1 in liver-specific atg7 KO mice markedly attenuated ductular reaction and liver tumorigenesis but did not improve liver injury, hepatomegaly, inflammation or fibrosis in these mice [33].
Yap is a key component of the Hippo signaling pathway and acts as a transcriptional co-activator. Yap binds to the transcriptional enhanced associate domain (TEAD) family of transcription factors and regulates the expression of a set of genes for cell proliferation, anti-apoptosis and “stemness [40,41]”. Lee et al. [34] found that liver-specific atg7 KO mice have increased hepatic cytoplasmic and nuclear Yap protein accumulation and increased expression of Yap target genes. They further found that Yap colocalizes with GFP-LC3 positive autophagosomes and lysotracker positive lysosomes in hepatocytes. Moreover, Yap protein increases when autophagy is inhibited either pharmacologically or genetically, suggesting that Yap protein may be a novel autophagy substrate. Deletion of yap in liver-specific-atg7 KO mice leads to a decrease in hepatocyte size, hepatomegaly, inflammation, ductular reaction, progenitor cell expansion and fibrosis. More importantly, the number and size of liver tumors decreases but are not eliminated in yap/atg7 double KO mice [34].
mTOR is a key cellular nutrient and energy sensor that regulates the cellular anabolic pathway by increasing protein and lipid synthesis to meet the demand for cell proliferation. Mammalian cells have two mTOR complexes: mTOR complex1 (mTORC1) and mTORC2, in which mTORC1 is rapamycin-sensitive but mTORC2 is relatively rapamycin resistant. Both mTORC1 and mTORC2 share several common components such as the mTOR kinase but also have distinct subunits. For instance, mTORC1 has the scaffold protein Raptor whereas mTORC2 contains Rictor [42]. As cancer cells have a higher proliferation rate, it is not surprising that approximately 50% human cancers, including HCC, have aberrant mTOR activation [43]. Increased mTOR activation in HCC is mainly attributed to loss of function mutations of phosphatase and tensin homolog (PTEN), tuberous sclerosis complex 1 (TSC1) or TSC2 [44,45]. Mice with liver-specific deletion of pten or tsc1 have persistent hepatic mTOR activation, hepatomegaly, and develop spontaneous tumors [46]. Therefore, targeting mTOR has been an attractive approach for treating various cancers. However, results from clinical trials using rapamycin and its pharmacological analogs are not promising as only minimal beneficial effects are observed [47,48]. We recently demonstrated that genetic ablation of mtor has a dual role in the liver pathogenesis and tumorigenesis of L-atg5 KO mice. Deletion of mtor decreases hepatomegaly, cell death and inflammation but not fibrosis in young (2 months-old) L-atg5 KO mice. However, these beneficial effects are gradually lost in older (6–12 months-old) L-atg5/mtor DKO mice as they all develop tumors at 12 months-old. Moreover, more than 50% of L-atg5/mtor DKO mice develop spontaneous tumors but none of the L-atg5 KO mice have tumors at 6 months-old. These results suggest that ablation of mtor exacerbates tumorigenesis in autophagy defective mouse livers. Mechanistically, we found that L-atg5/mtor DKO mice have increased protein kinase B (PKB, also known as AKT) activation compared with L-atg5 KO, suggesting that the early development of liver tumors could be mediated by compensatory AKT activation in the absence of both mTOR and autophagy. It is possible that pharmacological inhibiting both mTOR and AKT may have increased beneficial effects compared to targeting mTOR alone for treating HCC.
Taken together, it seems that HMGB1, Yap, and mTOR all contribute to tumorigenesis in autophagy defective livers although ablation of individual proteins cannot completely eliminate liver tumors, which is in contrast with the ablation of Nrf2 that completely abolishes liver tumors in autophagy defective livers (see below discussion).
AUTOPHAGY PROMOTES MALIGNANT TUMOR PROGRESSION
Perhaps one of the most intriguing findings from the liver-specific atg5 or atg7 KO mice is that these hepatic autophagy defective mice develop benign adenomas but not malignant HCC [21,23]. Notably, liver-specific atg5 KO mice fail to develop HCC even after they were exposed to the hepatic carcinogen diethylnitrosamine (DEN) [29]. These data may suggest that during the late tumor progression stage, autophagy may be required for benign tumors to progress to malignant tumors. One possible mechanism is that loss of hepatic autophagy leads to the induction of several tumor suppressor genes, such as p53, p21, p27, p16, and PTEN [29,32]. Increased p53 in autophagy deficient hepatoma cells negatively regulates Nanog homeobox (NANOG) and cancer stem cell (CSC) proliferation, which may suppress tumor progression [49]. Mechanistically, it is known that mitophagy, a selective autophagy for mitochondrial removal, positively regulates hepatic CSC by suppressing the tumor suppressor p53. When mitophagy is induced, p53 colocalizes with mitochondria and is removed in a mitophagy-dependent manner. However, when mitophagy or autophagy is inhibited, mitochondrial p53 is phosphorylated by PTEN-induced kinase 1 (PINK1) and translocated into the nucleus. Once in the nucleus, p53 binds to the NANOG promoter to prevent OCT4 and SOX2 transcription factors from activating the expression of NANOG, a transcription factor critical for maintaining the stemness and self-renewal ability of CSCs, resulting in the reduction of hepatic CSC populations. These results demonstrate that autophagy, and more likely mitophagy, controls the activities of p53 to maintain hepatic CSC and provides an explanation as to why autophagy is required to promote hepatocarcinogenesis [49,50]. However, it remains to be studied whether further deletion of tumor suppressor genes such as Trp53 or pten in liver-specific atg5 or atg7 KO mice would promote the progression of benign tumors to malignant tumors.
Increasing evidence has demonstrated that autophagy is robustly activated in tumor cells including HCC under a multitude of stressors, such as starvation, growth factor deprivation, hypoxia, proteasome inhibition and therapeutic agents [51]. Inducible autophagy constitutes an important pro-survival mechanism for cancer cells by removing toxic oxygen radicals or damaged misfolded proteins, relieving ER stress, maintaining mitochondrial function, sustaining metabolism, and preventing diversion of tumor progression to benign oncocytomas [51,52-56]. It is reported that autophagosome formation is induced in hypoxic tumor regions and that Beclin1 deletion leads to tumor cell death specifically in these hypoxic regions [57,58]. Moreover, inhibition of autophagy restores the sensitivity of hepatoma cells to chemotherapy, suggesting that autophagy plays a pro-survival role in chemotherapeutic agent-induced cell death [57]. In many other cancer cells, such as pancreatic cancer, elevated basal autophagy is required for continued cell growth and maintaining the intracellular amino acid pool, suggesting that tumor cells have evolved to rely on autophagy even under basal conditions [59,60]. In KrasG12D- or BrafV600E-driven lung cancer, KrasG12D-driven pancreatic ductal adenocarcinoma (PDAC), or pten–/–-driven prostate cancer, deletion of atg5 or atg7 decreases tumor progression [61-64], further supporting the notion that functional autophagy is required for tumor progression.
While cancer cell autophagy is important for cancer cell survival, emerging evidence now suggests that host autophagy also plays a critical role in tumor growth. It should be noted that tumors obtain their nutrient supply from the host and thus whole-body metabolism and the tumor microenvironment are critical for tumor growth. To address host autophagy in metabolism and its impact on tumor growth, Dr. Eileen White’s group established an animal model that allograft autophagy-competent cancer cell lines onto autophagy wild-type and autophagy-deficient (inducible wholebody atg7 deleted) host mice [65]. Loss of host autophagy markedly impairs growth of multiple different allografted tumors due to decreased levels of circulating arginine. Arginine is a non-essential amino acid, which has to be obtained from the diet, de novo synthesis or protein turnover. Arginine is also important for mTOR activation to promote tumor cell growth. Interestingly, some human cancer cells have silenced the expression of the enzyme argininosuccinate synthase (ASS1) for arginine synthesis that render these cancer cells sensitive to arginine deficiency [66]. Circulating arginine is generally broken down by arginase I (ARG1), an enzyme predominantly expressed in liver and released from hepatocytes into circulation. Host-atg7 KO mice have increased liver injury (similar to L-atg7 KO mice) resulting in increased release of ARG1 into circulation. Dietary supplementation of arginine for host-atg7 KO mice partially restored circulating arginine levels and tumor growth. Ablation of host autophagy by conditional induction of systemic expression of a dominant-negative ATG4b in mice with KrasG12D- and Trp53–/+-driven PDAC also leads to inhibition of PDAC progression [67]. These results indicate that systemic inhibition of autophagy may be beneficial for cancer treatment at multiple levels by targeting both cancer cells and host cells.
The tumor microenvironment is an integral part of the tumor, which is composed of cancerous and noncancerous cells (e.g., immune cells, stromal cells, endothelial cells, and adipocytes) as well as various mediators (e.g., cytokines, chemokines, growth factors, and humoral factors). The tumor microenvironment plays a critical role in cancer cell adaptation, growth and progression as well as resistance to therapies. Autophagy in the tumor microenvironment can provide the cancer cells/tumor cells with amino acids, extracellular matrix molecules and cytokines (e.g., interleukin-6) to promote tumor growth [68,69]. In addition, autophagy may also affect the anti-tumor T cell responses and reduce tumor growth in syngeneic host mice. In PDAC, autophagy selectively degrades major histocompatibility complex class I (MHC-I) via autophagy receptor protein NBR1, which results in decreased surface levels of MHC-1 on PDAC cells and reduced CD8+ T cells-mediated antitumor immunity. Inhibition of autophagy in PDAC cells leads to increased antigen presentation, enhanced CD8+ T cell proliferation and activation that results in increased tumor cell killing and decreased tumor size [70,71].
Taken together, it is evident that autophagy plays a complex dual role in cancer development. Autophagy acts as a tumor suppressor during the early stage of tumorigenesis but promotes tumor malignancy and progression in existing tumors. Autophagy in both the tumor cells themselves and the host as well as the surrounding microenvironment promotes tumorigenesis and cancer progression.
ROLE OF P62 AND NRF2 IN LIVER TUMORIGENESIS
p62 is considered a cargo adaptor for selective autophagy due to its ability to bind directly to LC3 via its LC3 interaction region (LIR) [72,73]. However, more evidence now supports that p62 also serves as a multifunctional signaling hub within the cell [74-76]. In response to stress, p62 can bind to various partners through its multiple domains that are involved in various signaling pathways [75,77]. For example, the Phox/Bemp1 (PB1) domain at the N terminal of p62 can self-oligomerize to form homo-oligomers and hetero-oligomers, and can interact with the atypical protein kinase Cζ (PKCζ) [78,79]. The ZZ zinc finger region, and TB domains interact with multiple proteins that are related to p62-mediated nuclear factor-κB (NF-κB) activation [75]. The region between the ZZ and TB domain binds to Raptor, a subunit of the mTORC1 complex, leading to mTORC1 activation [80]. The LIR domain at the C terminal region of p62 binds to LC3 and triggers the selective-autophagy pathway [72,75]. Moreover, the KIR domain next to the LIR domain binds to Keap1 and transports it into the autophagosome for degradation, which leads to the activation of the non-canonical Keap1-Nrf2 pathway [81-84]. Both accumulated p62- and Keap1-positive aggregates have been found in autophagy-deficient mice (L-atg7 KO mice), causing persistent activation of Nrf2 and increasing the expression of Nrf2 target genes [22]. In addition, the ubiquitin-associated (UBA) domain binds to both mono- and polyubiquitinated proteins, which may explain why p62 is often found in protein aggregates and inclusion bodies [75,85].
As discussed above, p62 accumulation in the liver occurs in various types of liver diseases, including HCC [86-89], intrahepatic cholangiocarcinoma (CCA) [90], HCV and HBV infection [89,91], AALD [92], and non-alcoholic steatohepatitis [93,94]. Ectopic expression of p62 in mouse liver is sufficient to induce HCC in mice [87]. Moreover, loss of p62 in the human HCC cell line abrogates anchorage-independent growth, and whole-body depletion of p62 in mice restricts the size of hepatocellular adenomas in L-atg7 KO mice and L-tsc1 KO mice [21,22,87]. The mechanism by which hepatic p62 promotes HCC is generally thought to be due to its activation of Nrf2 via the noncanonic p62-Keap1-Nrf2 pathway, of which phosphorylated p62 further strengthens the binding affinity between p62 and Keap1 for Nrf2 activation [21,22,87,89]. Indeed, we found that deletion of Nrf2 in L-atg5 KO mice abolished hepatomegaly, liver injury and liver tumorigenesis [23]. Tumors grow in a relatively hash environment, often associated with hypoxia and limited nutrients, Nrf2 activation promotes the expression of genes for detoxification and antioxidant enzymes [95]. Moreover, p62-mediated Nrf2 activation also directs glucose to the glucuronate pathway, and glutamine towards glutathione synthesis which provides HCC cells with tolerance to anti-cancer drugs and increases their ability to proliferate [89].
In great contrast to p62’s role in hepatocytes, p62 in hepatic stellate cells (HSC) directly interacts with vitamin D receptor (VDR) and retinoid X receptor (RXR) to promote their heterodimerization resulting in the inhibition of HSC activation [88]. Whole body and HSC-specific p62 KO mice have increased HSC activation, enhanced liver inflammation and fibrosis, as well as HCC progression in response to high fat diet and hepatic carcinogen DEN [88]. Furthermore, decreased p62 protein levels have been observed in human HCC stroma cells. These findings suggest that p62 may have a cell-type specific role in HCC development. In hepatocytes, p62 may promote HCC development via Nrf2 activation whereas p62 in HSC may inhibit HSC activation and HCC progression via activation of VDR and RXR. Therefore, for clinical treatment purposes it is important to monitor p62 expression in different cell types. However, it remains a challenge to design cell type specific p62 targeting therapies for treating HCC.
As discussed above, genetic deletion of HMGB1 or Yap improved pathogenesis and tumorigenesis but did not eliminate tumor development in hepatic autophagy deficient mice [33,34]. However, deletion of Nrf2 completely abolished tumorigenesis in L-atg5 KO and L-atg7 KO mice [23,33]. These findings suggest that persistent activation of Nrf2 plays a more critical role than HMGB1 and Yap in autophagy-deficiency-induced liver tumorigenesis. Inhibition of Nrf2 may be a promising approach for treating HCC.
ROLE OF TFEB IN LIVER TUMORIGENESIS
The lysosome is the terminal component of autophagy and contains more than 50 acid hydrolases. Recent evidence suggests that transcriptional regulation of genes for lysosomal biogenesis and autophagy plays a critical role in autophagy [96]. Transcription factor EB (TFEB) is a basic helix-loop-helix leucine zipper transcription factor belonging to the microphthalmia/transcription factor E (MiT/TFE) family of transcription factors, which can bind to a specific gene sequence called the coordinated lysosomal expression and regulation (CLEAR) gene network motif [96]. TFEB is a master regulator for transcription of lysosomal biogenesis and autophagy genes [97,98]. TFEB is mainly regulated at the posttranslational level via phosphorylation of specific amino acid residues. Several kinases including the extracellular signal-regulated kinase 2 (ERK2) [98], mTORC1 [97], AKT [99], GSK3β [100], and protein kinase Cβ (PKCβ) [96] have been reported to phosphorylate TFEB. TFEB is phosphorylated at Ser142 by ERK2 and at both Ser142 and Ser211 by mTORC1, which promotes TFEB binding with the cytosolic chaperone 14-3-3 and sequesters TFEB in the cytosol, resulting in its inactivation [96,98]. In addition, STIP1 homology and U-box containing protein 1 (STUB1), a chaperone-dependent E3 ubiquitin ligase, interacts with phosphorylated TFEB resulting in TFEB ubiquitination and proteasomal degradation [101]. Conversely, calcineurin, a Ca2+-dependent phosphatase, dephosphorylates TFEB and increases TFEB nuclear translocation and activation [102]. In addition to phosphorylation and ubiquitination, TFEB can also be regulated by acetylation. General control non-repressed protein 5 (GCN5), a histone acetyltransferase, acetylates TFEB at multiple sites resulting in decreased TFEB transcription activity and lysosomal biogenesis [103]. In contrast, SIRT1, an NAD+-dependent deacetylase, deacetylates TFEB at lysine 116 leading to increased TFEB-mediated lysosomal gene expression [104].
TFEB and other members of the MiT-TFE family of transcription factors exhibit oncogenic features. Previous studies have revealed that the aberrant expression of several MiT-TFE family members is associated with different types of human cancers, such as renal cell carcinomas (RCC), alveolar sarcomas [105], non-small cell lung cancer, melanomas [106], and PDAC [59]. Chromosomal translocations and fusions of TFEB and TFE3 with other partner genes cause a particular type of RCC, referred to as translocation-RCC [107]. These fusions consistently preserve the TFEB/TFE3 open reading frame and always include the DNA-binding domains. While a variety of genes can be fused to TFEB or TFE3, a major consequence of the translocation with respect to oncogenic activity is a massive increase in the expression levels of TFEB or TFE3 protein [108,109]. In addition, altered TFEB expression and/or activity is associated with PDAC and non-small cell lung cancer, where they appear to support tumor growth via the induction of autophagy [59]. Interestingly, tumors in which MiT-TFE genes are amplified or overexpressed show an induction of RagD, a direct transcriptional target of TFEB, that resulted in mTORC1 hyperactivation [110]. The RagD GTPase is important for efficient mTORC1 recruitment to the lysosomal surface [111]. Silencing of MiT-TFE genes in primary cultured cells obtained from tumors resulted in a significant reduction of the hyperproliferative phenotype. Furthermore, xenotransplantation experiments performed using a melanoma cell line showed that silencing of RagD significantly reduced tumor growth [110]. These findings suggest that one of the mechanisms accounting for the oncogenic function of TFEB is likely due to RagD-mediated mTORC1 activation.
Birt-Hogg-Dubé syndrome is characterized by benign skin tumors, pulmonary and kidney cysts and RCC, which is caused by mutations in the tumor suppressor gene folliculin (FLCN) [112]. FLCN and its interacting partner protein FNIP2 act as a positive component of the mTORC1 pathway by promoting the binding of mTORC1 to the Rag heterodimer via its GTPase-activating protein (GAP) activity for RagC and RagD resulting in mTORC1 activation in cultured cells [113]. However, loss of FLCN in kidney cancers leads to hyperactivation of mTORC1 [114]. In a recent study, using a kidney-specific FLCN KO mouse model that mimics Birt-Hogg-Dubé syndrome, Napolitano et al found that TFEB is constitutively activated despite the hyperactivity of mTORC1 in kidney-specific FLCN KO mice [115]. They further found that mTORC1 can phosphorylate different substrates via a substrate-specific mechanism, which is mediated by Rag GTPases. Unlike other substrates of mTORC1, such as S6K and 4E-BP1, TFEB is strictly dependent on the amino-acid-mediated activation of RagC and RagD GTPases but is insensitive to Ras homologue enriched in brain (Rheb) activity induced by growth factors. Depletion of TFEB in kidneys of FLCN KO mice fully normalized mTORC1 activity and rescued the disease phenotype, suggesting activation of TFEB is the major driver for kidney cysts and cancer in FLCN KO mice [115].
Despite the above-mentioned evidence linking TFEB to several cancers, whether TFEB inhibits or promotes HCC remains elusive. Notably, TFEB activity is impaired in both AALD and NALFD [14,116,117], two risk factors for HCC, implicating a potential role of TFEB in HCC. The liver is the most active metabolic organ and is frequently exposed to xenobiotics and metabolites that are often toxic which can lead to acute or chronic liver injury. To deal with various toxic insults, the liver has evolved a remarkable regenerative capacity to help with recovery from injury [118]. The high regeneration capacity is largely attributable to the ability of its differentiated epithelial cells, hepatocytes and biliary epithelial cells, to proliferate after injury. However, when the proliferation of one subtype of cells in the liver is compromised and unable to make its own histologic recovery, the other cell types will step in and transdifferentiate to the lost cell type [118]. While it remains to be a controversial hot topic, increasing evidence indicates that hepatocytes can trans-differentiate to cholangiocytes in response to biliary injury and cholangiocytes can trans-differentiate to hepatocytes for the loss of injured hepatocytes [119,120].
In a recent study, Pastore et al reported that TFEB can drive liver progenitor cell (LPC) differentiation and promote the formation of ductular structures [121]. Among the different cell types in the liver, TFEB is highly expressed in biliary cells. TFEB drives the differentiation of LPC into the progenitor/cholangiocyte lineage while inhibiting hepatocyte differentiation during the mouse liver development or upon regeneration in response to injury. Tfeb transgenic mice exhibit an increase in the number of bile ductules after injury whereas L-tfeb KO mice exhibit a decrease. Transcriptomic and CHIP studies show that Sox9, a marker of precursor and biliary cells, is a direct TFEB target and a primary mediator of its effects on liver cell fate. Interestingly, results from lineage tracing experiments show that overexpression of TFEB in hepatocytes can promote trans-differentiate to cholangiocytes. Tfeb transgenic mice at 6-months-old either die or have to be euthanized due to poor health conditions. These mice have increased fibrosis and develop liver cysts which mimic a polycystic liver and develop CCA-like phenotype [121]. These findings suggest that TFEB may play a crucial role in determining liver cell fate during development and regeneration, and may also contribute to CCA, which seems independent of its functions in lysosomal biogenesis.
ONCOGENES AND TUMOR SUPPRESSORS
In general, oncogenes such as class-I phosphatidylinositol 3-kinase (PI3K) and AKT suppress autophagy via activating mTORC1 whereas tumor suppressor genes such as tsc1, pten, lkb, and ampk activate autophagy through inhibition of mTORC1 (Fig. 1) [122]. Class I- PI3Ks are activated by growth factor receptor tyrosine kinases (RTKs) that catalyze the formation of lipid secondary messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) from phosphatidylinositol-4,5-bisphosphate (PIP2). PIP3 then binds with AKT via its pleckstrin homology (PH) domain to recruit AKT to the plasma membrane, where AKT is activated by 3-phosphoinositide dependent kinase (PDK1) at threonine 308 and serine 473 by the rapamycin-insensitive mTORC2 [123,124]. The tumor suppressor PTEN reverses the effects of PI3K by dephosphorylating PIP3 which in turn inhibits AKT activation. However, loss of function mutations in PTEN results in AKT activation in a wide spectrum of human cancers [125]. Activated AKT directly phosphorylates TSC2 and inactivates TSC1-TSC2 complex, which has GAP activity towards the small G-protein Rheb, and in turn activates mTORC1 [126,127]. In contrast, the AMP-activated protein kinase (AMPK) phosphorylates TSC2 and enhances TSC1-TSC2 activity resulting in mTORC1 inhibition in response to energy starvation [128]. Alternatively, AMPK can be phosphorylated and activated by serine-threonine kinase liver kinase B1 (LKB1) [129,130], a tumor suppressor gene that is responsible for the inherited cancer disorder Peutz-Jeghers syndrome [131]. Together, AMPK negatively regulates mTORC1 to trigger the catabolic autophagy process to generate more energy substrates, whereas, AKT positively regulates mTORC1 to promote the anabolic process and shutdown autophagy. L-pten KO mice have increased AKT and mTORC1 activation, impaired hepatic autophagy and develop hepatomegaly, steatohepatitis and spontaneous HCC [132,133]. L-tsc1 KO mice have constitutive mTORC1 activation, impaired autophagy and develop spontaneous HCC [134]. Therefore, oncogenes inhibit whereas tumor suppressor genes activate autophagy resulting in either the promotion or suppression of HCC in the liver.
Figure 1.
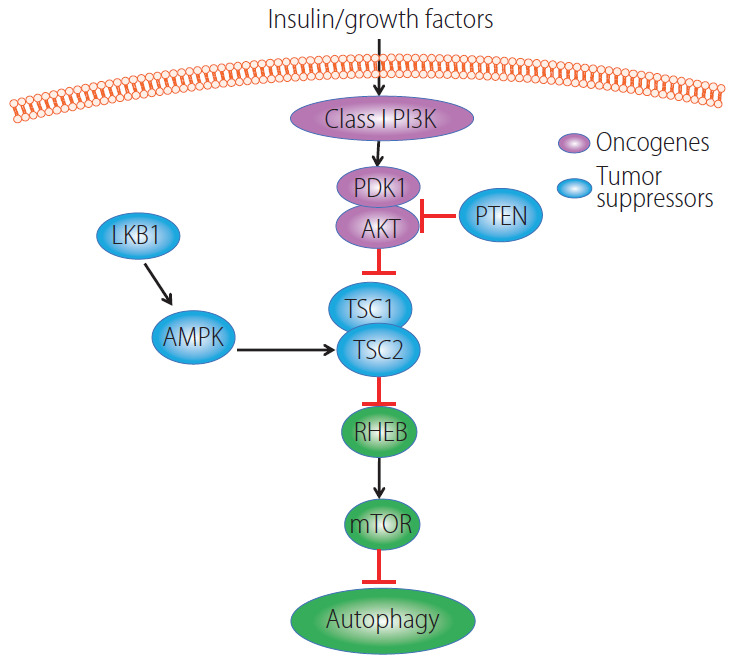
Regulation of autophagy by oncogenes and tumor suppressor genes. In the presence of insulin or growth factors, AKT is activated due to increased PI3K activity and binding with PDK1. AKT is negatively regulated by PTEN. Activated AKT phosphorylates TSC2 resulting in decreased TSC1-TSC2 complex activity leading to enhanced GTP-bound form of Rheb to stimulate mTORC1 activation. LKB1 phosphorylates and activates AMPK, and activated AMPK increased TSC2 phosphorylation on a different site of AKT resulting in increased TSC1-TSC2 complex activity which leads to decreased mTORC1 activation. Increased mTORC1 activity negatively regulates autophagy. PI3K, phosphatidylinositol 3-kinase; PDK1, phosphoinositide dependent kinase; AKT, protein kinase B; LKB1, serine-threonine kinase liver kinase B1; PTEN, phosphatase and tensin homolog; AMPK, AMP-activated protein kinase; TSC, tuberous sclerosis complex; RHEB, Ras homologue enriched in brain; mTOR, mechanistic target of rapamycin; mTORC1, mechanistic target of rapamycin complex 1.
SUMMARY
Since the identification of the first Atg in yeast in the 1990s, significant progress has been made in the understanding of the molecular mechanisms underlying the process of autophagy as well as the vital physiological role of autophagy in human diseases. In regards to the role of autophagy in cancer, it is now clear that autophagy exerts both tumor-suppressive and tumor-promoting roles in a context dependent manner. In the early phase of tumor initiation, autophagy acts as a tumor suppressor by removing damaged organelles and proteins to guard against genome instability. In the late phase of tumor development and progression, transformed cancer cells and stromal cells use autophagy to generate nutrients within the hash tumor microenvironment to increase their chances of survival. Viral infection, AALD and NAFLD, all lead to impaired autophagy and promotion of HCC. Therefore, different approaches to manipulate autophagy should be considered for the prevention/treatment of HCC. For preventative purposes, pharmacological activation of autophagy to correct virus or alcohol and diet-impaired hepatic autophagy may be beneficial. However, for treating existing HCC, pharmacological inhibition of autophagy or Nrf2 in combination with other chemotherapy drugs should be a more efficient strategy.
Acknowledgments
This work was partially supported by NIH grants: R37 AA020518, R01 DK102142, U01 AA024733 and P20GM103549 & P30GM118247.
Abbreviations
- AALD
alcohol associated liver disease
- AKT
protein kinase B
- AMPK
AMP-activated protein kinase
- ARG1
arginase I
- ASS1
argininosuccinate synthase
- CCA
cholangiocarcinoma
- CLEAR
coordinated lysosomal expression and regulation
- CSC
cancer stem cell
- DAMP
damage-associated molecular pattern
- DEN
diethylnitrosamine
- ER
endoplasmic reticulum
- ERK2
extracellular signal-regulated kinase 2
- FLCN
folliculin
- GAP
GTPase-activating protein
- GCN5
general control non-repressed protein 5
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HMGB1
high mobility group box 1
- HSC
hepatic stellate cells
- KO
knockout
- LIR
LC3 interaction region
- LKB1
serine-threonine kinase liver kinase B1
- LPC
liver progenitor cell
- MHC
major histocompatibility complex
- MiT
microphthalmia
- mTOR
mechanistic target of rapamycin
- mTORC1
mechanistic target of rapamycin complex 1
- NAFLD
nonalcoholic fatty liver diseases
- NANOG
Nanog homeobox
- NF-κB
nuclear factor-κB
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- PB1
phox/bemp1
- PDAC
pancreatic ductal adenocarcinoma
- PDK1
phosphoinositide dependent kinase
- PH
pleckstrin homology
- PI3K
phosphatidylinositol 3-kinase
- PINK1
PTEN-induced kinase 1
- PIP2
phosphatidylinositol-4,5-bisphosphate
- PIP3
phosphatidylinositol-3,4,5-trisphosphate
- PKC
protein kinase C
- PTEN
phosphatase and tensin homolog
- RCC
renal cell carcinomas
- Rheb
ras homologue enriched in brain
- RTKs
receptor tyrosine kinases
- RXR
retinoid X receptor
- STUB1
STIP1 homology and U-box containing protein 1
- TEAD
transcriptional enhanced associate domain
- TFE
transcription factor E
- TFEB
transcription factor EB
- TSC
tuberous sclerosis complex
- UBA
ubiquitin-associated
- VDR
vitamin D receptor
- Yap
yes-associated protein
Footnotes
Authors’ contributions
Conceptualization, W.X.D., X.J.C., and H.Q.; Writing and Original Draft Preparation, W.X.D., X.J.C., H.Q., and S.G.W., Writing, Review and Editing, W.X.D., X.J.C., H.Q., S.G.W., and S.F.
Conflicts of Interest
The authors have no conflicts to disclose.
REFERENCES
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 7.Xu J. Trends in liver cancer mortality among adults aged 25 and over in the United States, 2000-2016. NCHS Data Brief. 2018;(314):1–8. [PubMed] [Google Scholar]
- 8.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis:from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 9.Chao X, Ding WX. Role and mechanisms of autophagy in alcohol-induced liver injury. Adv Pharmacol. 2019;85:109–131. doi: 10.1016/bs.apha.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X, McKeen T, Zhang J, Ding WX. Role and mechanisms of mitophagy in liver diseases. Cells. 2020;9:837. doi: 10.3390/cells9040837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sir D, Ann DK, Ou JH. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy. 2010;6:548–549. doi: 10.4161/auto.6.4.11669. [DOI] [PubMed] [Google Scholar]
- 13.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao X, Wang S, Zhao K, Li Y, Williams JA, Li T, et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879. doi: 10.1053/j.gastro.2018.05.027. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 16.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao XL, et al. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer. 2014;14:327. doi: 10.1186/1471-2407-14-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, et al. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding WX, Ni HM, Gao W, Chen X, Kang JH, Stolz DB, et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol Cancer Ther. 2009;8:2036–2045. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 26.Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2010;584:1427–1435. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vessoni AT, Filippi-Chiela EC, Menck CF, Lenz G. Autophagy and genomic integrity. Cell Death Differ. 2013;20:1444–1454. doi: 10.1038/cdd.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, et al. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025–1034. doi: 10.1038/cdd.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, et al. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–450. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Ni HM, Guo F, Ding Y, Shi YH, Lahiri P, et al. Sequestosome 1/p62 protein is associated with autophagic removal of excess hepatic endoplasmic reticulum in mice. J Biol Chem. 2016;291:18663–18674. doi: 10.1074/jbc.M116.739821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni HM, Chao X, Yang H, Deng F, Wang S, Bai Q, et al. Dual roles of mammalian target of rapamycin in regulating liver injury and tumorigenesis in autophagy-defective mouse liver. Hepatology. 2019;70:2142–2155. doi: 10.1002/hep.30770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khambu B, Huda N, Chen X, Antoine DJ, Li Y, Dai G, et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagydeficient livers. J Clin Invest. 2018;128:2419–2435. doi: 10.1172/JCI91814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YA, Noon LA, Akat KM, Ybanez MD, Lee TF, Berres ML, et al. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat Commun. 2018;9:4962. doi: 10.1038/s41467-018-07338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Ni HM, Ding WX. Emerging Players in autophagy deficiency-induced liver injury and tumorigenesis. Gene Expr. 2019;19:229–234. doi: 10.3727/105221619X15486875608177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinotti S, Patrone M, Ranzato E. Emerging roles for HMGB1 protein in immunity, inflammation, and cancer. Immunotargets Ther. 2015;4:101–109. doi: 10.2147/ITT.S58064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 38.Arriazu E, Ge X, Leung TM, Magdaleno F, Lopategi A, Lu Y, et al. Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut. 2017;66:1123–1137. doi: 10.1136/gutjnl-2015-310752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 40.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 42.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 43.Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60:855–865. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu TH, Huang CC, Lin PR, Chang HW, Ger LP, Lin YW, et al. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer. 2003;97:1929–1940. doi: 10.1002/cncr.11266. [DOI] [PubMed] [Google Scholar]
- 45.Ho DWH, Chan LK, Chiu YT, Xu IMJ, Poon RTP, Cheung TT, et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication. Gut. 2017;66:1496–1506. doi: 10.1136/gutjnl-2016-312734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenerson HL, Yeh MM, Kazami M, Jiang X, Riehle KJ, McIntyre RL, et al. Akt and mTORC1 have different roles during liver tumorigenesis in mice. Gastroenterology. 2013;144:1055–1065. doi: 10.1053/j.gastro.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraham RT, Gibbons JJ. The mammalian target of rapamycin signaling pathway: twists and turns in the road to cancer therapy. Clin Cancer Res. 2007;13:3109–3114. doi: 10.1158/1078-0432.CCR-06-2798. [DOI] [PubMed] [Google Scholar]
- 48.Chiang GG, Abraham RT. Targeting the mTOR signaling network in cancer. Trends Mol Med. 2007;13:433–442. doi: 10.1016/j.molmed.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan ST, et al. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell. 2017;68:281–292. doi: 10.1016/j.molcel.2017.09.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian H, Chao X, Ding WX. A PINK1-mediated mitophagy pathway decides the fate of tumors-to be benign or malignant? Autophagy. 2018;14:563–566. doi: 10.1080/15548627.2018.1425057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barnard RA, Regan DP, Hansen RJ, Maycotte P, Thorburn A, Gustafson DL. Autophagy inhibition delays early but not late-stage metastatic disease. J Pharmacol Exp Ther. 2016;358:282–293. doi: 10.1124/jpet.116.233908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 57.Song J, Qu Z, Guo X, Zhao Q, Zhao X, Gao L, et al. Hypoxiainduced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131–1144. doi: 10.4161/auto.5.8.9996. [DOI] [PubMed] [Google Scholar]
- 58.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-rasinduced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 64.Santanam U, Banach-Petrosky W, Abate-Shen C, Shen MM, White E, DiPaola RS. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 2016;30:399–407. doi: 10.1101/gad.274134.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, et al. Autophagy maintains tumour growth through circulating arginine. Nature. 2018;563:569–573. doi: 10.1038/s41586-018-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dillon BJ, Prieto VG, Curley SA, Ensor CM, Holtsberg FW, Bomalaski JS, et al. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer. 2004;100:826–833. doi: 10.1002/cncr.20057. [DOI] [PubMed] [Google Scholar]
- 67.Yang A, Herter-Sprie G, Zhang H, Lin EY, Biancur D, Wang X, et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018;8:276–287. doi: 10.1158/2159-8290.CD-17-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Endo S, Nakata K, Ohuchida K, Takesue S, Nakayama H, Abe T, et al. Autophagy is required for activation of pancreatic stellate cells, associated with pancreatic cancer progression and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;152:1492–1506. doi: 10.1053/j.gastro.2017.01.010. e24. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature. 2020;581:100–105. doi: 10.1038/s41586-020-2229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamamoto K, Venida A, Perera RM, Kimmelman AC. Selective autophagy of MHC-I promotes immune evasion of pancreatic cancer. Autophagy. 2020;16:1524–1525. doi: 10.1080/15548627.2020.1769973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 73.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 74.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 75.Manley S, Williams JA, Ding WX. Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med (Maywood) 2013;238:525–538. doi: 10.1177/1535370213489446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 77.Sánchez-Martín P, Komatsu M. p62/SQSTM1 - steering the cell through health and disease. J Cell Sci. 2018;131:jcs222836. doi: 10.1242/jcs.222836. [DOI] [PubMed] [Google Scholar]
- 78.Jakobi AJ, Huber ST, Mortensen SA, Schultz SW, Palara A, Kuhm T, et al. Structural basis of p62/SQSTM1 helical filaments, their presence in p62 bodies and role in cargo recognition in the cell. Nat Commun. 2020;11:440. doi: 10.1038/s41467-020-14343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 80.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katsuragi Y, Ichimura Y, Komatsu M. Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Curr Opin Toxicol. 2016;1:54–61. [Google Scholar]
- 82.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 83.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 84.Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Isogai S, Morimoto D, Arita K, Unzai S, Tenno T, Hasegawa J, et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem. 2011;286:31864–31874. doi: 10.1074/jbc.M111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taniguchi K, Yamachika S, He F, Karin M. p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett. 2016;590:2375–2397. doi: 10.1002/1873-3468.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duran A, Hernandez ED, Reina-Campos M, Castilla EA, Subramaniam S, Raghunandan S, et al. p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell. 2016;30:595–609. doi: 10.1016/j.ccell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aishima S, Fujita N, Mano Y, Iguchi T, Taketomi A, Maehara Y, et al. p62+ hyaline inclusions in intrahepatic cholangiocarcinoma associated with viral hepatitis or alcoholic liver disease. Am J Clin Pathol. 2010;134:457–465. doi: 10.1309/AJCP53YVVJCNDZIR. [DOI] [PubMed] [Google Scholar]
- 91.Xiang X, Qin HG, You XM, Wang YY, Qi LN, Ma L, et al. Expression of P62 in hepatocellular carcinoma involving hepatitis B virus infection and aflatoxin B1 exposure. Cancer Med. 2017;6:2357–2369. doi: 10.1002/cam4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Menk M, Graw JA, Poyraz D, Möbius N, Spies CD, von Haefen C. Chronic alcohol consumption inhibits autophagy and promotes apoptosis in the liver. Int J Med Sci. 2018;15:682–688. doi: 10.7150/ijms.25393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fukushima H, Yamashina S, Arakawa A, Taniguchi G, Aoyama T, Uchiyama A, et al. Formation of p62-positive inclusion body is associated with macrophage polarization in non-alcoholic fatty liver disease. Hepatol Res. 2018;48:757–767. doi: 10.1111/hepr.13071. [DOI] [PubMed] [Google Scholar]
- 94.González-Rodríguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bartolini D, Dallaglio K, Torquato P, Piroddi M, Galli F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54–71. doi: 10.1016/j.trsl.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 96.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18:1065–1077. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- 101.Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017;36:2544–2552. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y, Huang Y, Liu J, Zhang J, Xu M, You Z, et al. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020;21:e48335. doi: 10.15252/embr.201948335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li Z, et al. Deacetylation of TFEB promotes fibrillar Aβ degradation by upregulating lysosomal biogenesis in microglia. Protein Cell. 2016;7:417–433. doi: 10.1007/s13238-016-0269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanaka M, Homme M, Yamazaki Y, Shimizu R, Takazawa Y, Nakamura T. Modeling alveolar soft part sarcoma unveils novel mechanisms of metastasis. Cancer Res. 2017;77:897–907. doi: 10.1158/0008-5472.CAN-16-2486. [DOI] [PubMed] [Google Scholar]
- 106.Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mendel L, Ambrosetti D, Bodokh Y, Ngo-Mai M, Durand M, Simbsler-Michel C, et al. Comprehensive study of three novel cases of TFEB-amplified renal cell carcinoma and review of the literature:Evidence for a specific entity with poor outcome. Genes Chromosomes Cancer. 2018;57:99–113. doi: 10.1002/gcc.22513. [DOI] [PubMed] [Google Scholar]
- 108.Argani P, Lui MY, Couturier J, Bouvier R, Fournet JC, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 109.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 110.Di Malta C, Siciliano D, Calcagni A, Monfregola J, Punzi S, Pastore N, et al. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science. 2017;356:1188–1192. doi: 10.1126/science.aag2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmidt LS, Linehan WM. FLCN: the causative gene for Birt-Hogg-Dubé syndrome. Gene. 2018;640:28–42. doi: 10.1016/j.gene.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baba M, Furihata M, Hong SB, Tessarollo L, Haines DC, Southon E, et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst. 2008;100:140–154. doi: 10.1093/jnci/djm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Napolitano G, Di Malta C, Esposito A, de Araujo MEG, Pece S, Bertalot G, et al. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Nature. 2008 Jul 1; doi: 10.1038/s41586-020-2444-0. doi: 10.1038/s41586-020-2444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chao X, Ni HM, Ding WX. Insufficient autophagy: a novel autophagic flux scenario uncovered by impaired liver TFEB-mediated lysosomal biogenesis from chronic alcohol-drinking mice. Autophagy. 2018;14:1646–1648. doi: 10.1080/15548627.2018.1489170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Z, Qian Q, Li M, Shao F, Ding WX, Lira VA, et al. The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB. Autophagy. 2020 Jul 15; doi: 10.1080/15548627.2020.1788889. doi:10.1080/15548627.2020.1788889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Michalopoulos GK. The regenerative altruism of hepatocytes and cholangiocytes. Cell Stem Cell. 2018;23:11–12. doi: 10.1016/j.stem.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 119.Deng X, Zhang X, Li W, Feng RX, Li L, Yi GR, et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell. 2018;23:114–122. doi: 10.1016/j.stem.2018.05.022. e3. [DOI] [PubMed] [Google Scholar]
- 120.Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, et al. De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pastore N, Huynh T, Herz NJ, Calcagni’ A, Klisch TJ, Brunetti L, et al. TFEB regulates murine liver cell fate during development and regeneration. Nat Commun. 2020;11:2461. doi: 10.1038/s41467-020-16300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 124.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 125.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 126.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 127.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 128.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 129.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Mäkelä TP, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 131.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 132.Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113:1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ueno T, Sato W, Horie Y, Komatsu M, Tanida I, Yoshida M, et al. Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy. 2008;4:692–700. doi: 10.4161/auto.6085. [DOI] [PubMed] [Google Scholar]
- 134.Menon S, Yecies JL, Zhang HH, Howell JJ, Nicholatos J, Harputlugil E, et al. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci Signal. 2012;5:ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]


