Abstract
DNA binding proteins rapidly locate their specific DNA targets through a combination of 3D and 1D diffusion mechanisms, with the 1D search involving bidirectional sliding along DNA. However, even in nucleosome-free regions, chromosomes are highly decorated with associated proteins that may block sliding. Here we investigate the ability of the abundant chromatin-associated HMGB protein Nhp6A from Saccharomyces cerevisiae to travel along DNA in the presence of other architectural DNA binding proteins using single-molecule fluorescence microscopy. We observed that 1D diffusion by Nhp6A molecules is retarded by increasing densities of the bacterial proteins Fis and HU and by Nhp6A, indicating these structurally diverse proteins impede Nhp6A mobility on DNA. However, the average travel distances were larger than the average distances between neighboring proteins, implying Nhp6A is able to bypass each of these obstacles. Together with molecular dynamics simulations, our analyses suggest two binding modes: mobile molecules that can bypass barriers as they seek out DNA targets, and near stationary molecules that are associated with neighboring proteins or preferred DNA structures. The ability of mobile Nhp6A molecules to bypass different obstacles on DNA suggests they do not block 1D searches by other DNA binding proteins.
INTRODUCTION
DNA-binding proteins have to search over a vast excess of mixed polynucleotide sequences to find their functional targets, yet accomplish this within physiological timeframes. The search for targets typically involves both 3D diffusion in solution and 1D diffusion along DNA in a process known as facilitated diffusion (1–5). 1D diffusion along DNA has been characterized for a limited number of DNA-binding proteins using in vitro single-molecule fluorescence microscopy (6–12). In these studies, fluorescently-labeled individual proteins or protein complexes are tracked along extended DNA molecules from sequential images captured by a fluorescence microscope. Except for a few studies (13–16), the 1D movements of proteins have been evaluated on otherwise naked DNA. However, chromosomes in vivo are decorated with nucleosomes plus numerous chromatin-associated proteins that would likely place obstacles to 1D diffusion, thereby limiting sliding to intervals between neighboring proteins. Indeed, it has been shown that 1D movement of the Msh2-Msh6 DNA repair complex is effectively blocked by nucleosomes. On the other hand, the Mlh1-Pms1 DNA repair complex can bypass nucleosomes (13). In the present study, we investigate how the mobility of the most abundant non-histone chromatin protein in Saccharomyces cerevisiae is influenced by increasing densities of other proteins, each of which strongly deforms DNA structure.
Eukaryotic and bacterial architectural DNA-binding proteins (ADBPs) bind DNA with low sequence specificity and introduce large conformational changes into the DNA structure. These proteins are notable for the diversity of DNA transactions in which they participate, usually in collaboration with other more specialized DNA binding proteins in transcription, replication, recombination, and repair reactions, and in DNA packaging (17–22). Non-histone protein 6A (Nhp6A) is an abundant ADBP in S. cerevisiae (23,24) and is a member of the HMGB family of chromatin proteins that are ubiquitous throughout eukaryotes (25–27). Nhp6A is a 93 residue monomeric protein containing a single HMGB domain that binds within an expanded minor groove and introduces sharp bends into the DNA helix. It also contains an unstructured N-terminal arm rich in basic residues that stabilizes binding by wrapping around DNA primarily through the major groove (Figure 1A) (28,29). Yeast Nhp6A is present at ∼60 000 copies per haploid nucleus, and thus its levels approach those of nucleosomes (30). Yeast mutants lacking Nhp6A and its close paralog Nhp6B grow slowly at optimal growth temperatures (30°) and fail to grow at 38°, in part due to loss of SNR6 (snoRNA U6) expression (31,32). Nhp6A is primarily bound within intergenic nucleosome-free regions of chromatin, especially promotor-regulatory regions upstream of highly expressed pol II genes and within pol III genes (33). About 10% of yeast pol II genes exhibit significant changes in gene expression in nhp6a/b null mutants (33,34). Studies of individual genes have shown Nhp6A functions to facilitate looping between closely spaced UAS/enhancer and core promoter elements or by promoting assembly of complexes containing promoter-specific factors and the general transcription machinery (30,35,36). Examples of the latter include promoting assembly of pol II preinitiation complexes containing TBP, TFIIA, and TFIIB (30,37) and complexes at the SNR6 pol III gene containing TFIIIC, TFIIIB and TBP (32,38–40). Nhp6A also cooperates with MSH2 and MSH6 to selectively bind DNA mismatches in the initial steps of mismatch repair (41) and can function similarly to mammalian HMGB1/2 in assembling RAG1/2-RSS recombination complexes (42). In each of these cases, a 1D diffusion process whereby Nhp6A can bypass other proteins would enable the HMGB protein to embed itself within and stabilize the nucleoprotein complex by inducing changes in DNA structure. Nhp6A also helps recruit ATP-dependent SWI/SNF chromatin remodeling complexes to promoter regions and enables the yeast FACT chromatin modifying complex to facilitate translocation of transcription and replication elongation complexes through chromatin (36,43–48).
Figure 1.
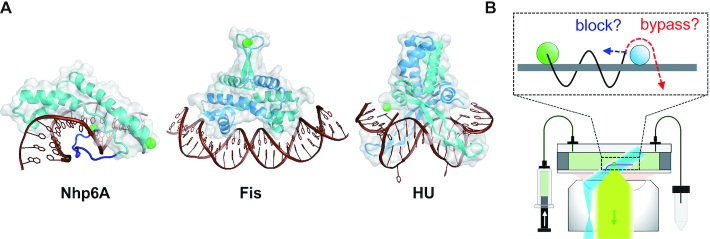
(A) ADBPs used for investigation of the effects of DNA-bound obstacles on 1D diffusion by Nhp6A. Structures of protein–DNA complexes from left to right: Nhp6A (PDB code: 1J5N), Fis (3IV5) and HU (1P71). Proteins are cyan or cyan/blue (dimer subunits) with transparent grey surfaces, and DNAs are brown. The Nhp6A N-terminal flexible arm is blue; patches of tandem basic residues are at residues 8–10 and 13–16. Green spheres are fluorescent labeling sites. (B) Fluorescence microscopy of fluorescent proteins (green dot) in a flow cell with DNA array (pink) using HILO illumination (blue) and fluorescence detection (light green). In inset, the sliding of Nhp6A (green circles) along DNA (grey) may be blocked by other ADBPs (cyan) or may bypass the ADBPs.
HU and Fis are the two most abundant DNA binding proteins in rapidly growing Escherichia coli (49). HU is most often a heterodimer of very similar 90 residue subunits that bind DNA through β-ribbon arms inserted into an expanded minor groove with additional dynamic DNA wrapping along the protein sides that introduce sharp bends (Figure 1A) (50,51). Fis is a homodimer of 98 residue subunits from E. coli that binds within the DNA major groove via helix-turn-helix motifs generating a bent DNA complex with obligatory changes in minor groove widths (Figure 1A) (52,53). Fis binds DNA the most selectively of the three proteins studied here, and in some genomic contexts, can function as a classical bacterial transcription factor or regulator of site-specific DNA recombination reactions (17,19,21,54–56).
Previously, we investigated the behavior of these three ADBPs on naked DNA using single-molecule fluorescence microscopy (12). We found that these ADBPs support 1D diffusion along DNA while maintaining continuous DNA contact, although Fis most often associates with DNA in a stationary mode. In this work, we tracked individual Nhp6A molecules traveling along DNA bound by increasing concentrations of ADBPs. We find that although the mobility of Nhp6A was impeded by protein obstacles, Nhp6A molecules could bypass obstacles with efficiencies that depended on the identity of the block. We present a potential mechanism for obstacle bypass that critically involves the N-terminal basic arm.
MATERIALS AND METHODS
Preparation of fluorescently-labeled proteins
Native or proteins containing one or two cysteines for labeling were expressed and purified without tags as described previously (12). Two versions of Nhp6A for labeling were employed: Nhp6A-A93C contains a cysteine substituted for the C-terminal alanine residue, and Nhp6A-2cys contains cysteines introduced at residue 2 and the C-terminal end. Fis-Q21C contains a cysteine near the tip of the N-terminal β-hairpin arm motif. The HU β subunit, containing a cysteine added to its C-terminal end, and HU α subunit were co-expressed in E. coli to form HU heterodimers. The proteins were labeled by Atto488 (ATTO-TEC) using maleimido chemistry as reported previously (12).
Single-molecule fluorescence microscopy
We used an inverted fluorescence microscope (IX-73; Olympus) with a total internal reflection fluorescence unit (IX3-RFAEVAW; Olympus) (12). Briefly, a 488-nm laser was illuminated into an objective lens with N.A. = 1.49 using a highly inclined thin illumination geometry at a laser power of 5 mW. Fluorescence collected by the objective lens was detected using an EM-CCD camera (iXon Ultra 888; Andor). Arrays of phage λ DNA (New England Biolabs) were constructed on the glass surface inside the flow cell using the DNA garden method described in Igarashi et al. (57). Atto488-labeled Nhp6A at 0.01−0.02 nM plus non-labeled ADBPs at various concentrations in a solution containing 20 mM HEPES, 150 mM KGlu, 1 mM EDTA, 1 mM DTT, 2 mM trolox, and 0.1 mg/ml BSA at pH 7.2 were introduced into the flow cell using a syringe pump. Images (300 × 800 pixels ROI) were taken at 44-ms intervals at a flow rate of 0.6 ml/min at 22°C.
The fluorescent spots of single Nhp6A molecules were tracked from sequential images using ImageJ software with the plugin ‘Particle track and analysis’. To remove non-specific adsorption of the fluorescent molecules on the flow cell surface, we selected trajectories using our in-house program (9) with some modifications. We selected traces with local mean square displacements (MSD) larger than 616 nm2 for the perpendicular axis against the stretched DNA, corresponding to the spatial resolution of the adsorbed molecules in this experimental set up. Also, we selected trajectories with at least 10 consecutive points. Displacements were calculated from all pairs of positions of a molecule at time intervals of 176 ms for all trajectories. For the fitting of displacement distribution analysis, we used the equation:
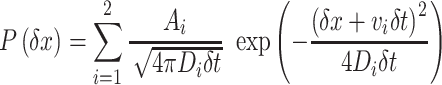 |
(1) |
where δt, δx, P(δx), Ai, vi and Di represent time interval, displacement in the time interval, the occurrence of δx, amplitude of the ith mode, drift velocity of the ith mode, and diffusion coefficient of the ith mode, respectively (9,12).
Estimation of the average distances between neighboring molecules
To determine the average distance between neighboring ADBP molecules, we calculated the number of protein molecules bound to the λ DNA. To this end, we measured the fluorescence intensity of labeled proteins bound to individual λ DNAs within the array under the same conditions as used for tracking except that the laser power was reduced to 0.3 or 0.6 mW to minimize photobleaching. Fifty sequential images were taken at 70 ms intervals. Time-averaged fluorescence intensities of single DNAs were obtained after subtraction of the background intensity. Twenty - 30 DNA molecules with at least 14 μm of end to end distance, corresponding to ≥85% extension of λ DNA (16.5 μm) under buffer flow (57), were used for the analysis. Similarly, we obtained the average fluorescence intensity of individual protein molecules bound to λ DNA from the single-molecule data. The average number of DNA-bound molecules was calculated by dividing the average fluorescence intensity of the single DNA by that of the single protein molecule after correcting for the excitation laser power and exposure time. A linear relation between the fluorescence intensity and the number of molecules was assumed. Finally, the average distance between neighboring protein molecules was obtained by dividing the DNA length by the average number of DNA-bound molecules.
Molecular dynamics simulation
To investigate the behavior of Nhp6A binding to DNA in the presence of a stable Fis dimer at the residue-level scale, we performed coarse-grained (CG) molecular dynamics (MD) simulations. We constructed a system consisting of one Nhp6A, one Fis dimer and one double-stranded DNA that included the 15-bp consensus motif for Fis. The initial structure of the simulations was built by appending 50-bp linear DNA to each end of the central 15-bp DNA structure in the Fis-specific DNA complex taken from PDB entry 3IV5 (52). PDB structure 1J5N (29) was used for Nhp6A modeling.
To model the interactions inside the proteins, we employed the 1-bead-per-amino-acid AICG2+ potentials (58). For DNA, we used the 3SPN.2C model (59), in which each nucleotide is represented by three beads corresponding to phosphate, sugar, and base. For non-specific interactions between protein and DNA, we considered excluded volume and electrostatic interactions. We used Go-type structure-based interactions between Fis and DNA to constrain the binding (60). For Nhp6A, we employed a variation of the recently developed PWMcos model to enable the recognition of DNA bases in the minor groove (61). More details of the CG models can be found in the Supplementary Methods.
All simulations were conducted by Langevin dynamics using the CafeMol package (62). The temperature was set to T = 300 K, and the ionic concentration was IC = 200 mM in all simulations reported here. The distances between the center-of-mass of Nhp6A and DNA were constrained to be smaller than 200 Å. Fifty independent simulations of 108 MD steps were conducted.
Two thousand Nhp6A-DNA structures were randomly chosen from the MD simulations, and for each structure ( ), a vector was assigned to describe the DNA-binding interface of Nhp6A:
), a vector was assigned to describe the DNA-binding interface of Nhp6A:
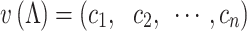 |
(2) |
where n = 93 (the number of residues in Nhp6A), and ci is 1 if residue i is within 10 Å of DNA, otherwise 0. A distance matrix based on these vectors was then constructed, and the DBSCAN method (63) was used to classify all the structures into two clusters. By direct visualization of representative structures in these two clusters, we found they corresponded to two binding modes: one with the HMGB domain inserted into the minor groove and the other with only the N-terminal flexible arm contacting DNA. We then classified each structure in the simulations with respect to the features of these two clusters, using the support vector classification method (64).
RESULTS
Experimental set up to examine the effect of obstacles on 1D diffusion by Nhp6A
To investigate how sliding dynamics is affected by obstacles bound to DNA, we measured the movement of individual fluorescently-labeled (Atto488) Nhp6A proteins along DNA bound by varying densities of non-labeled proteins using single-molecule fluorescence microscopy. For obstacles bound to DNA, we used three well-characterized ADBPs that exhibit different DNA binding mechanisms and dynamics: two abundant bacterial proteins Fis and HU, as well as unlabeled Nhp6A. The dynamics of each of these proteins associating with otherwise naked DNA has been previously characterized by single molecule assays (12). Nhp6A molecules bind and slide along individual DNA molecules at an input solution concentration of 0.01–0.1 nM, and Fis and HU bind beginning ∼0.04 and ∼0.25 nM, respectively. Each of these proteins form stable complexes with DNA whose lifetimes are much greater than the imaging times employed here (our unpublished results, (65–67)). Highly inclined and laminated optical sheet (HILO) illumination with a 488-nm laser was used for detecting Atto488-labeled Nhp6A proteins bound to DNA (Figure 1B) (12). The DNA garden method, in which arrayed λ DNA molecules (48.5 kb) are extended in a flow cell, was used for high throughput data collection (57).
1D diffusion of Nhp6A on DNA in the presence of Fis
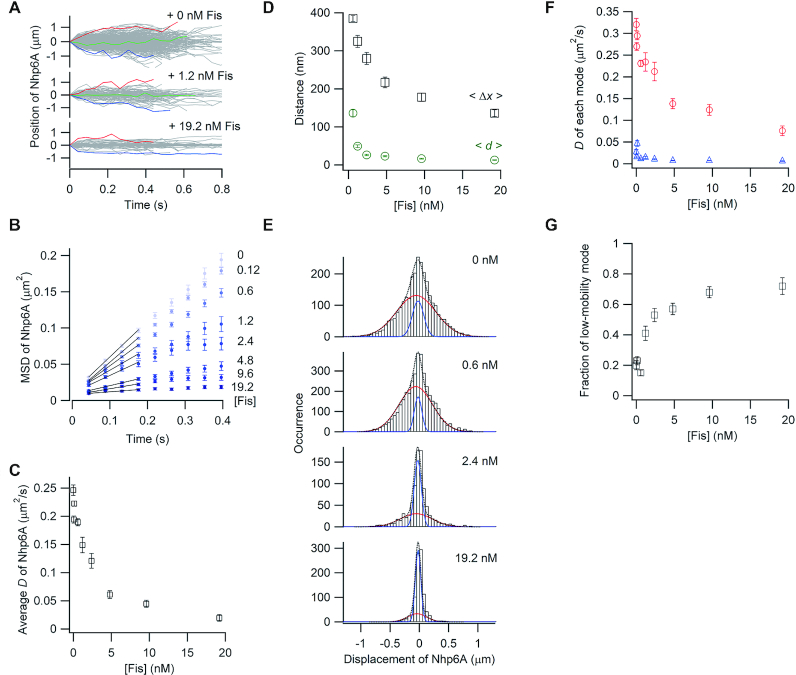
Labeled Nhp6A (0.01–0.02 nM) was introduced into the flow cell together with various concentrations of non-labeled Fis, and the 1D movement of the labeled Nhp6A on DNA was visualized. We have shown previously that most (∼90%) Fis molecules under the conditions used (150 mM potassium glutamate) associate with DNA in a near stationary mode with the remaining fraction undergoing slow and sometimes interrupted sliding (12). We obtained 70–855 single-molecule traces of Nhp6A with at least 11 consecutive data points in the presence of Fis concentrations ranging from 0 to 19.2 nM.
The movement of Nhp6A molecules along DNA was suppressed as the Fis concentration increased (Figure 2A). For quantitative analysis, the MSDs of Nhp6A molecules were plotted as a function of time (Figure 2B). MSD plots of Nhp6A in the presence of 0−0.6 nM of Fis increase linearly with time, whereas plots in the presence of ≥1.2 nM Fis deviate from linearity, and at high Fis concentrations, reflect little mobility. The deviation of the MSD plots suggests that Nhp6A diffuses along DNA bound by Fis at the shorter time scales but that sliding of Nhp6A is restricted by neighboring Fis molecules bound to DNA at the larger time scales. The average diffusion coefficient of Nhp6A, D, obtained by fitting four initial data points of the MSD plots with a linear function with 2D of the slope, decreased 12.5-fold as the Fis concentration increased to 19.2 nM (Figure 2C). These results demonstrate that Fis bound to DNA impedes the 1D sliding of Nhp6A on DNA.
Figure 2.
Single-molecule tracking of fluorescent Nhp6A along DNA bound by non-fluorescent Fis. (A) Single-molecule traces of Nhp6A in the presence of different concentrations of Fis. (B) MSD plots of Nhp6A with 0–19.2 nM Fis. (C) Fis concentration dependence of average diffusion coefficients (D) for Nhp6A. (D) Fis concentration dependence of average travel distances of Nhp6A at 0.4 s intervals (<Δx>, black square) and average spacings between neighboring Fis molecules (<d>, green circles). The error bars of <Δx> were determined from the fitting error of D. The errors of <d> represent standard errors calculated from the fluorescence intensity of at least 20 DNAs bound by fluorescently-labeled Fis. (E) Displacement distributions of Nhp6A at 0.176 s intervals with 0–19.2 nM Fis. Black-dashed curves are the best fitted curves with double Gaussian functions. Red and blue curves are the best fitted curves of the high-mobility and low-mobility modes, respectively. (F) Fis concentration dependence of diffusion coefficients in two modes of Nhp6A. Red and blue represent the diffusion coefficients of the high-mobility and low-mobility modes, respectively. (G) Fis concentration dependence of the fraction of the low-mobility mode of Nhp6A. The error bars in panels C, F and G represent the fitting errors.
We next compared the travel distance of Nhp6A to the distance between neighboring Fis molecules bound to DNA. If the sliding of Nhp6A is restricted to between neighboring Fis molecules, the travel distance of Nhp6A should correspond to the average spacing between Fis molecules. Under these non-equilibrium binding conditions, Fis associates randomly throughout the λ genome (Supplementary Figure S1, (12,65)). The average number of DNA-bound Fis dimers under different concentrations of Fis was estimated by dividing the total fluorescence intensity of Atto488-labeled Fis bound to single DNA molecules by the average fluorescence intensity of an individual Fis molecule (see Materials and Methods). The average distance between two neighboring Fis molecules, <d>, was obtained by dividing the total DNA length by the average number of DNA-bound Fis molecules. For comparison, we plotted the average travel distance of Nhp6A over 0.4 s, <Δx>, and the average spacing between Fis molecules, <d>, against the Fis concentration (Figure 2D). <Δx> values in the presence of ≥ 0.6 nM of Fis were 2.8–11 times larger than <d>. For example, Nhp6A molecules traveled an average distance of 178 nm (∼525 bp) in 0.4 s when the average spacing of Fis molecules at 9.6 nM is 16 nm (∼47 bp) and an average distance of 136 nm (∼400 bp) in 0.4 s when the average spacing of Fis molecules at 19 nM is 13 nm (∼38 bp). These results suggest that Nhp6A molecules can bypass Fis dimers on DNA to extend their travel distance well beyond the average spacing between neighboring Fis proteins.
To more closely examine how each molecule travels along DNA bound by Fis, we analyzed the displacement distribution of Nhp6A at 176 ms time intervals obtained from the single-molecule traces (Figure 2E). If Nhp6A has a single diffusional motion, a Gaussian function should be observed (9,10,68). Because the distributions in 0.6, 1.2, 2.4 nM Fis showed clear deviations from a bell-shaped Gaussian function, especially in the distribution tails, we fit the distributions with the sum of two Gaussian functions in all Fis concentrations (Equation 1). We refer to the two distributions as the high-mobility and low-mobility modes of 1D diffusion. The diffusion coefficients of the high-mobility mode decreased from 0.325 to 0.075 μm2/s as the non-labeled Fis concentration increased from 0 to 19 nM (Figure 2F). In contrast, the diffusion coefficients of the low-mobility mode (0.035 μm2/s) did not vary with the non-labeled Fis concentration (Figure 2F). The fraction of the low-mobility population increased from 0.20 to 0.72 as the non-labeled Fis concentration increased to 19 nM (Figure 2G). Considering that the displacement in the low-mobility mode was comparable to the spatial resolution of molecules on DNA (27 nm), the low-mobility mode at high Fis concentrations likely represents near stationary Nhp6A molecules blocked by adjacent Fis molecules. In contrast, Nhp6A proteins in the high-mobility mode likely reflect molecules bypassing Fis proteins bound to DNA, given that the average travel distance can be considerably greater than the average distance between two neighboring Fis molecules (Figure 2D). Individual trajectories in the presence of 2.4 nM Fis show that Nhp6A molecules can switch between low to high mobility modes or remain in a given mode over the time frame of the trajectory (Supplementary Figure S2).
In summary, we observe that the 1D diffusion of Nhp6A on DNA is retarded by bound Fis proteins by means of stabilizing the low-mobility mode, but that some Nhp6A molecules can slide past Fis and continue traveling on DNA.
1D diffusion of Nhp6A on DNA in the presence of HU
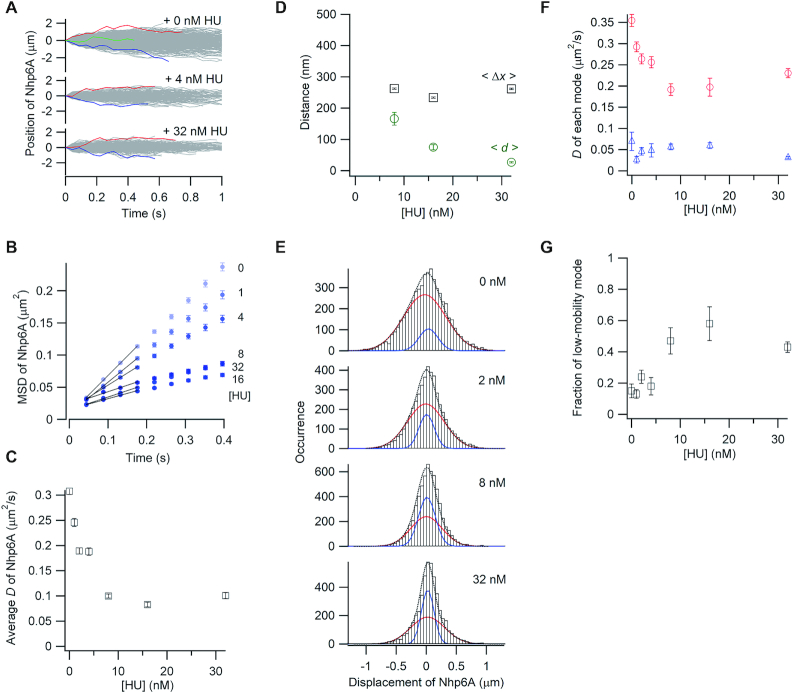
We explored the effects of a different obstacle, the bacterial nucleoid-associated protein HU, on 1D diffusion of Nhp6A. HU generates very different structures on DNA as compared with Fis (Figure 1A), and unlike Fis, most HU dimers associate with naked DNA in a mobile mode (D = 0.492 ± 0.007 μm2/s) under the conditions employed here (12). As shown in Figure 3A, DNA sliding by labeled Nhp6A molecules was retarded by addition of unlabeled HU. MSD plots of Nhp6A in the presence of 0−4 nM HU linearly increase over the time range, whereas the MSD plots deviate in the presence of ≥8 nM HU, reflecting decreasing travel distances (Figure 3B). The average diffusion coefficient of Nhp6A was reduced 3-fold, but then became constant as the concentration of HU increased above 8 nM (Figure 3C). The average travel distances of Nhp6A over a 0.4 s interval (<Δx>) increased up to about 9.8-fold greater than the spacing (<d>) between neighboring HU molecules as the HU concentration increased (Figure 3D). Notably, travel distances remained constant at about 250 nm (∼735 bp) per 0.4 s between 8 and 32 nM HU, as the HU binding density increased from about one dimer every 489 bp to dimers spaced every 79 bp on average. Thus, mobile Nhp6A molecules can bypass HU proteins on DNA.
Figure 3.
Single-molecule tracking of fluorescent Nhp6A along DNA bound by non-fluorescent HU. (A) Single-molecule traces of Nhp6A in the presence of different concentrations of HU. (B) MSD plots of Nhp6A with 0–32 nM HU. (C) Fis concentration dependence of average diffusion coefficients (D) for Nhp6A. (D) HU concentration dependence of average travel distances of Nhp6A at 0.4 s intervals (<Δx>, black squares) and average spacings between neighboring HU molecules (<d>, green circles). (E) Displacement distributions of Nhp6A at 0.176 s intervals with 0–32 nM HU. Black-dashed curves are the best fitted curves with double Gaussian functions. Red and blue curves are the best fitted curve of the high-mobility and low-mobility modes, respectively. (F) HU concentration dependence of diffusion coefficients in two modes of Nhp6A. Red circles and blue triangles represent the diffusion coefficients of the high-mobility and low-mobility modes, respectively. (G) HU concentration dependence of the fraction of the low-mobility mode of Nhp6A. The error bars in panels B, C, D, F, and G are described in the caption of Figure 2.
The displacement distributions were fit with the sum of two Gaussian functions (Figure 3E). The diffusion coefficient of the high-mobility mode decreased from 0.37 to about 0.21 μm2/s as the non-labeled HU concentration increased, whereas the diffusion coefficient of the low-mobility mode did not change with respect to the HU concentration (Figure 3F). The fraction of Nhp6A molecules in the low-mobility mode increased up to 3-fold by addition of HU (Figure 3G). The increasing fraction of non-mobile Nhp6A molecules combined with the reduction of mobility of the high-mobility fraction is consistent with HU blocking the 1D movement of Nhp6A on DNA. Nevertheless, over half of the Nhp6A molecules remain mobile in the presence of high concentrations of HU and are able to traverse distances that are greater than the average spacing of HU molecules.
1D diffusion of Nhp6A on DNA in the presence of Nhp6A
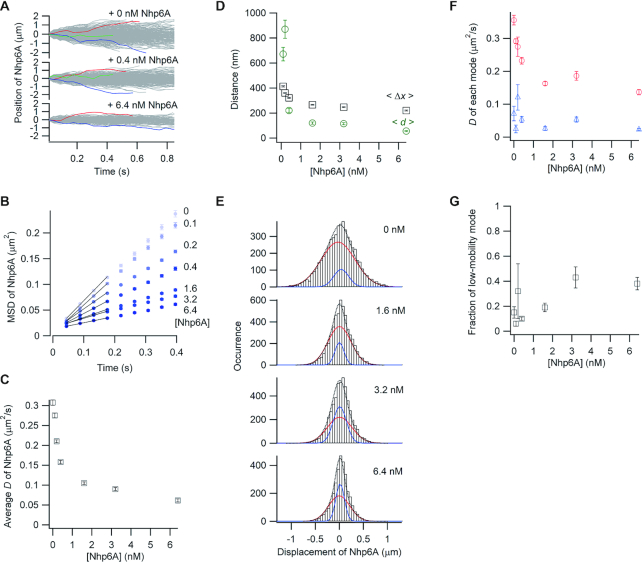
We next probed the movements of labeled Nhp6A along DNA bound with increasing amounts of non-labeled Nhp6A. Regions of high density binding by Nhp6A are found within promoter segments of about 12% of yeast pol II genes, and these genes are typically among the most highly transcribed (33). As observed with the bacterial proteins, the mobility of the labeled Nhp6A proteins in single-molecule traces decreased with the addition of non-labeled Nhp6A (Figure 4A). The average diffusion coefficient of the labeled Nhp6A decreased from 0.31 to 0.11 μm2/s as the total Nhp6A concentration increased to 1.6 nM, but additional Nhp6A (up to 6.4 nM) had smaller effects (Figure 4B and C). The average travel distances of labeled Nhp6A molecules over 0.4 s (<Δx>) in the presence of 0.4–6.4 nM non-labeled Nhp6A were 1.5- to 3.9-fold larger than the average distance (<d>) between neighbor Nhp6A molecules (Figure 4D). Thus, at 6.4 nM, Nhp6A molecules were bound on average every 56 nm (∼165 bp), but most of the labeled molecules traveled an average distance of 220 nm (∼647 bp).
Figure 4.
Single-molecule tracking of fluorescent Nhp6A along DNA bound by non-fluorescent Nhp6A. (A) Single-molecule traces of Nhp6A in the presence of different concentrations of non-fluorescent Nhp6A. b) MSD plots of Nhp6A with 0–6.4 nM non-fluorescent Nhp6A. (C) Non-fluorescent Nhp6A concentration dependence of average diffusion coefficients (D) for Nhp6A. (D) Non-fluorescent Nhp6A concentration dependence of average travel distances of Nhp6A at 0.4 s intervals (<Δx>, black squares) and average spacings between neighboring non-fluorescent Nhp6A molecules (<d>, green circles). (E) Displacement distributions of Nhp6A at 0.176 s intervals with 0–6.4 nM non-fluorescent Nhp6A. Black-dashed curves are the best fitted curves with double Gaussian functions. Red and blue curves are the best fitted curve of the high-mobility and low-mobility modes, respectively. (F) Non-fluorescent Nhp6A concentration dependence of diffusion coefficients in two modes of Nhp6A. Red and blue represent the diffusion coefficients of the high-mobility and low-mobility modes, respectively. (G) Non-fluorescent Nhp6A concentration dependence of the fraction of the low-mobility mode of Nhp6A. The error bars in panels B, C, D, F and G are described in the caption of Figure 2.
As with the Fis and HU blocks, the displacement distributions of the labeled Nhp6A molecules in the presence of increasing non-labeled Nhp6A were fit with the sum of two Gaussian functions (Figure 4E). The diffusion coefficient of the high-mobility mode decreased up to 2.6-fold as the total Nhp6A concentration increased, whereas the diffusion coefficient of the low-mobility mode was not changed (Figure 4F). The low-mobility population increased about 4-fold under the highest Nhp6A concentration tested, but ∼60% of the labeled molecules remained in the high-mobility mode (Figure 4G).
DISCUSSION
In this work, we investigate how Nhp6A molecules traveling along DNA negotiate the densely packed chromosome landscape. An alternative way to frame the issue is: Do Nhp6A molecules, which often bind at high density over nucleosome-free regulatory regions, impede the 1D search of sequence-specific binding proteins? We measured the mobility of Nhp6A molecules as they traversed along DNA bound by three model proteins that exhibit different modes of DNA binding. Each protein retarded the mobility of Nhp6A to varying extents, but Nhp6A molecules were able to bypass each of the protein obstacles, albeit with different efficiencies. The E. coli Fis protein, which forms the most stable DNA complex whereby most of the dimers associate in a low mobility or stationary mode, was the most effective block to Nhp6A sliding. The bacterial HU or other Nhp6A proteins, which bind otherwise naked DNA largely in a mobile mode, functioned as less effective blocks.
Nhp6A proteins on increasingly crowded DNA molecules partition into two populations: a low-mobility mode population in which the molecules exhibit little, if any, mobility within the resolution of the imaging, and a high-mobility mode in which molecules continued to traverse along the DNA decorated with obstacles. The low-mobility population probably includes Nhp6A molecules trapped and potentially sliding between closely-spaced neighboring proteins and molecules transiently associated with a blocking protein, as observed in two-color imaging experiments (Supplementary Figure S3a). The percentage of labeled Nhp6A molecules in the low-mobility mode increased with obstacle density, but for HU and Nhp6A blocks, plateaued at ≤50% of the population. 1D diffusion coefficients of molecules in the high-mobility mode decreased with obstacle density, but again with HU and Nhp6A, were only reduced to about half the coefficient measured on naked DNA. At high obstacle densities, average travel distances were measured to be up to 11-fold (Fis) greater than the mean separation between obstacles, implying multiple obstacles could be bypassed. Two-color imaging experiments have revealed examples of Nhp6A proteins moving across Fis proteins bound to DNA in a manner consistent with direct bypass of an obstacle (Supplementary Figure S3b).
Nhp6A, like other HMGB proteins, functions in part by assisting other effector proteins in the assembly of higher-order nucleoprotein complexes that promote diverse reactions. In this role, Nhp6A molecules would need to be able to seek out DNA positions embedded between other bound proteins and then through altered DNA conformation (e.g. local DNA bending or longer range looping), or perhaps through direct or indirect interactions with effector proteins, become stably associated within the nucleoprotein complex. Previous studies have shown that Nhp6A molecules form much more stable complexes with naked DNA containing kinks (e.g. cisplatin crosslinks) or tight loops (e.g. <100 bp microcircles) and can facilitate interactions between enhancer and reaction sites through DNA looping (24,30,69–71). Both mobile and stationary binding modes would be needed to function in its nucleoprotein assembly role. These binding modes would also enable Nhp6A to localize the FACT and SWI/SNF chromatin modifying complexes to appropriate nucleosomal targets (36,46,47). Nhp6A has been shown to bind nucleosomal DNA with lower but physiologically significant affinities (72), and may be able to slide along DNA wrapped around nucleosomes using the mechanism described below.
Obstacle bypass mechanisms
DNA-bound proteins would be expected to sterically block each other's movement, especially if the colliding proteins remained bound within the grooves of the DNA duplex. Several mechanisms have been proposed that could enable mobile proteins to bypass an obstacle. The bypassing molecule may have an intrinsic feature that allows it to circumvent blocks while maintaining continuous DNA contact (discussed for Nhp6A below). Alternatively, the bypassing protein could actively dissociate the blocking protein from DNA upon collision. Although eviction of proteins from DNA has been clearly demonstrated for energy-consuming translocases (15,73), to our knowledge it has not been reported for conventional DNA binding proteins when simultaneously bound to DNA; by contrast, closely-associated homotypic or heterotypic proteins have been reported to cooperatively stabilize each other by indirect mechanisms (74,75). On the other hand, there is compelling evidence that homotypic or heterotypic protein from solution can actively replace an existing protein bound to DNA by a process known as protein-facilitated dissociation (65,67,76,77). To test whether dissociation can occur when proteins on DNA collide we measured bulk lifetimes of DNA-bound labeled-Fis proteins in the presence of Nhp6A and bulk lifetimes of DNA-bound labeled-Nhp6A proteins in the presence of Fis. We observed no evidence of Nhp6A evicting Fis or dissociation of Nhp6A upon collision with Fis over timeframes much greater than those used to observe obstacle bypass in this study (Supplementary Figure S4). Mobile proteins could also potentially push the blocking molecule along DNA, but this mechanism seems unlikely without energy consumption and in light of the travel distances measured in the presence of high obstacle densities. Short-range DNA hopping (13,16,78,79) or intramolecular transfer via DNA looping are also potential mechanisms that could be used to evade obstacles. Although such mechanisms cannot be ruled out here, we note that the DNA molecules in our experiments are near fully extended (∼94% (57)) under hydrodynamic forces that would inhibit looping and that bypassing occurs bidirectionally on DNA under flow. Short-range hopping, involving total release from the DNA coulombic field followed by rapid rebinding, would likely be inefficient and unidirectional under the flow forces used in our in vitro experiments.
Mechanism of obstacle bypass by Nhp6A
The ability of Nhp6A molecules to evade DNA complexes with variable protein structures and DNA conformations implies a common mechanism for bypassing barriers. Each of the protein-DNA complexes tested would be expected to sterically block sliding of Nhp6A when its HMGB domain is bound within the minor groove, but in each complex, a continuous surface on the unbound side of the DNA molecule remains exposed. A clue into the mechanism of obstacle bypass comes from the observation that an Nhp6A mutant lacking the basic N-terminal arm is essentially immobile when bound to DNA (Supplementary Figure S5). Moreover, mutations that destabilize binding of the HMGB domain to the floor of the minor groove result in faster travel rates. These findings imply that Nhp6A may move along the surface of DNA with the protein connected to the phosphate backbone through its N-terminal disordered segment, which contains two patches of 3–4 tandem arginines and lysines. As described below, MD simulations provide support for sliding of Nhp6A along the DNA surface with continuous dynamic connections between the flexible basic arm and the phosphodiester backbone, as well as a paused mode with the HMGB domain bound within the minor groove and the DNA bent as in the NMR-derived Nhp6A-DNA structure. This mobile mode provides a mechanism whereby Nhp6A molecules could maintain continuous contact with DNA as it slides by an obstacle as long as an exposed surface of DNA is available. A recent study employing time-resolved FRET is also consistent with two binding states, with the DNA in one state being unbent (80). Also, force microscopy of Nhp6A-bound DNA suggests the presence of a loosely bound state that may correspond to the high mobility mode (81).
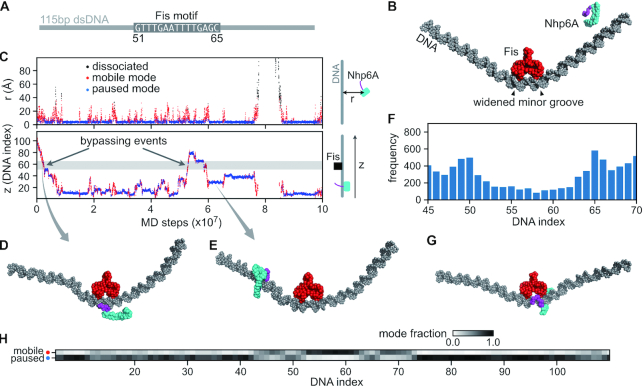
MD simulations were performed on Nhp6A binding to a 115 bp DNA segment containing a stationary Fis dimer bound at its center (position 51–65) (Figure 5A and B). In these simulations, Nhp6A bound Fis-free DNA and slid along the DNA switching between a paused mode where the HMGB domain is statically bound within the minor groove and a mobile mode where Nhp6A is dynamically associated with the DNA backbone through its basic N-terminal arm (Figure 5C–E, and Supplementary movie S1). Occasionally, mobile Nhp6A proteins traveled along the exposed DNA segment opposite to the Fis-bound interface, thereby bypassing Fis but maintaining continuous DNA contact with their flexible N-terminal arms (Figure 5D, Supplementary Figure S6). We noticed that the frequency of Nhp6A binding was moderately enhanced near the edges of the Fis binding motif (around positions 50 and 65) where Fis binding widens the minor groove (Figure 5F–H). Because the HMGB domain binds within an expanded minor groove (29), the local overrepresentation of Nhp6A in the paused mode at these positions suggests Fis may enhance Nhp6A binding through DNA conformation. The enhanced binding of Nhp6A to the ends of the Fis binding region resembles targeting of the phage λ Xis protein to the end of the Fis site within the λ attR recombination locus by Fis-induced changes to minor groove widths (82). Some MD trajectories also show that Nhp6A bypassed Fis by transiently dissociating and re-binding to DNA (DNA hopping, Supplementary Figure S6d–f). Overall, the MD simulations support the bypass of Fis-bound complexes by Nhp6A molecules in the mobile mode and suggest a mechanism for indirect stabilization of paused Nhp6A molecules via local DNA structural changes induced by Fis.
Figure 5.
Molecular dynamics (MD) simulations of Nhp6A bypassing a stationary Fis dimer. (A) A 115 bp dsDNA was used in the Nhp6A+Fis+DNA coarse-grained MD simulations. The central 15 bp was the Fis consensus motif (from DNA index 51–65), and the other regions had random sequences. (B) Initial structure of the MD simulations. Nhp6A was placed near one end of the DNA. The N-terminal arm of Nhp6A is shown in purple and the folded HMGB domain is in cyan. (C) A representative MD trajectory of Nhp6A sliding on DNA. Top panel: time series of distance from center-of-mass of Nhp6A HMGB domain to DNA (r); Bottom panel: Nhp6A binding position on DNA (z). In both panels the dots are colored by their corresponding binding status: mobile (red) or paused (blue) binding modes or dissociated from DNA (black). The grey region in the bottom panel represents the Fis binding motif (51–65). (D) A representative structure of Nhp6A using its N-terminal arm to contact DNA during Fis bypass. (E) A representative structure of Nhp6A binding to a Fis-free region in the paused mode. f) Nhp6A binding frequency to the region 45–70 on DNA in 50 independent MD simulations. (G) A representative structure of Nhp6A binding to the widened minor groove at DNA index 65. h) Normalized fraction of the two binding modes of Nhp6A along the DNA.
Other DNA binding proteins may use similar mechanisms for bypassing obstacles. The most conserved and abundant nucleoid-associated protein family in eubacteria are the HU/IHF-proteins. These proteins are distributed throughout the bacterial chromosome and function as nucleoprotein assembly factors by inducing or stabilizing DNA bends and in chromosome compaction (17,51,83). The HU dimer has two long flexible β-ribbon arms that are rich in basic residues and emanate from a highly basic patch on the protein surface (50). Like Nhp6A, MD simulations have modeled HU interacting with DNA in both sliding and paused modes, with the DNA highly bent only in the paused mode (84). The sliding mode, in which HU remains connected to the DNA backbone surface through dynamic electrostatic interactions, suggests a mechanism similar to that proposed for Nhp6A for bypassing obstacles while maintaining continuous contact with an exposed DNA surface. Indeed, our experiments have shown that labeled HU proteins can bypass high densities of unlabeled HU molecules with properties very similar to those measured for Nhp6A (Supplementary Figure S7).
Many eukaryotic DNA binding proteins contain nuclear localization sequences that are unstructured and rich in basic residues and may also function in DNA sliding (85). A well-studied example is the p53 transcription factor, which utilizes its C-terminal unstructured basic tail to search along DNA for its target (10,86). We predict that the p53 tetramer would be able to bypass barriers by sliding along the DNA surface through electrostatic connections with its C-terminal tails. As up to 70% of DNA binding proteins within the human proteome contain intrinsically-disordered tails (87), this mechanism of obstacle avoidance may be common.
Supplementary Material
ACKNOWLEDGEMENTS
K.K., E.M. and R.C.J. thank Prof. Margot E. Quinlan (UCLA) for use of her fluorescence microscope in early experiments, and Dr Yuji Itoh and Dwiky R.G. Subekti (Tohoku Univ.) for helpful support throughout the project.
Contributor Information
Kiyoto Kamagata, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan; Graduate School of Life Sciences, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan; Department of Chemistry, Faculty of Science, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Kana Ouchi, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan; Graduate School of Life Sciences, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Cheng Tan, Computational Biophysics Research Team, RIKEN Center for Computational Science, 7-1-26 Minatojima-minamimachi, Chuo-ku, Kobe, Hyogo 650-0047, Japan.
Eriko Mano, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Sridhar Mandali, Department of Biological Chemistry, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA 90095-1737 USA.
Yining Wu, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan; Department of Chemistry, Faculty of Science, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Shoji Takada, Department of Biophysics, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan.
Satoshi Takahashi, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan; Graduate School of Life Sciences, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan; Department of Chemistry, Faculty of Science, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Reid C Johnson, Department of Biological Chemistry, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA 90095-1737 USA; Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MEXT/JSPS KAKENHI [16KK0157 to K.K.]; NIGMS grant [GM038509 to R.C.J.]. Funding for open access charge: NIH [GM038509].
Conflict of interest statement. None declared.
REFERENCES
- 1. Von Hippel P.H., Berg O.G.. Facilitated target location in biological systems. J. Biol. Chem. 1989; 264:675–678. [PubMed] [Google Scholar]
- 2. Hammar P., Leroy P., Mahmutovic A., Marklund E.G., Berg O.G., Elf J.. The Lac repressor displays facilitated diffusion in living cells. Science. 2012; 336:1595–1598. [DOI] [PubMed] [Google Scholar]
- 3. Halford S.E., Marko J.F.. How do site-specific DNA-binding proteins find their targets. Nucleic Acids Res. 2004; 32:3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tafvizi A., Mirny L.A., van Oijen A.M.. Dancing on DNA: kinetic aspects of search processes on DNA. ChemPhysChem. 2011; 12:1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamagata K., Murata A., Itoh Y., Takahashi S.. Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging. J. Photochem. Photobiol. C Photochem. Rev. 2017; 30:36–50. [Google Scholar]
- 6. Wang F., Redding S., Finkelstein I.J., Gorman J., Reichman D.R., Greene E.C.. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013; 20:174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelson S.R., Dunn A.R., Kathe S.D., Warshaw D.M., Wallace S.S.. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2091–E2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murata A., Ito Y., Kashima R., Kanbayashi S., Nanatani K., Igarashi C., Okumura M., Inaba K., Tokino T., Takahashi S. et al.. One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca(2+) or Mg(2+) at millimolar concentrations. J. Mol. Biol. 2015; 427:2663–2678. [DOI] [PubMed] [Google Scholar]
- 10. Murata A., Itoh Y., Mano E., Kanbayashi S., Igarashi C., Takahashi H., Takahashi S., Kamagata K.. One-dimensional search dynamics of tumor suppressor p53 regulated by a disordered C-terminal domain. Biophys. J. 2017; 112:2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuculis L., Abil Z., Zhao H., Schroeder C.M.. TALE proteins search DNA using a rotationally decoupled mechanism. Nat. Chem. Biol. 2016; 12:831–837. [DOI] [PubMed] [Google Scholar]
- 12. Kamagata K., Mano E., Ouchi K., Kanbayashi S., Johnson R.C.. High free-energy barrier of 1D diffusion along DNA by architectural DNA-binding proteins. J. Mol. Biol. 2018; 430:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gorman J., Plys A.J., Visnapuu M.L., Alani E., Greene E.C.. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 2010; 17:932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stigler J., Camdere G.O., Koshland D.E., Greene E.C.. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell. Rep. 2016; 15:988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Terakawa T., Redding S., Silverstein T.D., Greene E.C.. Sequential eviction of crowded nucleoprotein complexes by the exonuclease RecBCD molecular motor. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E6322–E6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheon N.Y., Kim H.S., Yeo J.E., Schärer O.D., Lee J.Y.. Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 2019; 47:8337–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson R.C., Johnson L.M., Schmidt J.W., Gardner J.F.. Higgins N.P. The Bacterial Chromosome. 2005; Washington, D.C.ASM Press; 65–132. [Google Scholar]
- 18. Bianchi M.E., Agresti A.. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005; 15:496–506. [DOI] [PubMed] [Google Scholar]
- 19. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 20. Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta. 2010; 1799:101–113. [DOI] [PubMed] [Google Scholar]
- 21. Browning D.F., Grainger D.C., Busby S.J.. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010; 13:773–780. [DOI] [PubMed] [Google Scholar]
- 22. Reeves R. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 2010; 1799:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolodrubetz D., Burgum A.. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem. 1990; 265:3234–3239. [PubMed] [Google Scholar]
- 24. Paull T.T., Johnson R.C.. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 1995; 270:8744–8754. [DOI] [PubMed] [Google Scholar]
- 25. Landsman D., Bustin M.. A signature for the HMG-1 box DNA-binding proteins. Bioessays. 1993; 15:539–546. [DOI] [PubMed] [Google Scholar]
- 26. Bianchi M.E., Beltrame M.. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 1998; 63:1573–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas J.O., Travers A.A.. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001; 26:167–174. [DOI] [PubMed] [Google Scholar]
- 28. Allain F.H., Yen Y.M., Masse J.E., Schultze P., Dieckmann T., Johnson R.C., Feigon J.. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999; 18:2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Masse J.E., Wong B., Yen Y.M., Allain F.H., Johnson R.C., Feigon J.. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002; 323:263–284. [DOI] [PubMed] [Google Scholar]
- 30. Paull T.T., Carey M., Johnson R.C.. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996; 10:2769–2781. [DOI] [PubMed] [Google Scholar]
- 31. Costigan C., Kolodrubetz D., Snyder M.. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1994; 14:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kruppa M., Moir R.D., Kolodrubetz D., Willis I.M.. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell. 2001; 7:309–318. [DOI] [PubMed] [Google Scholar]
- 33. Dowell N.L., Sperling A.S., Mason M.J., Johnson R.C.. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev. 2010; 24:2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreira J.M., Holmberg S.. Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J. 2000; 19:6804–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fragiadakis G.S., Tzamarias D., Alexandraki D.. Nhp6 facilitates Aft1 binding and Ssn6 recruitment, both essential for FRE2 transcriptional activation. EMBO J. 2004; 23:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stillman D.J. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010; 1799:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biswas D., Imbalzano A.N., Eriksson P., Yu Y., Stillman D.J.. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol. Cell. Biol. 2004; 24:8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopez S., Livingstone-Zatchej M., Jourdain S., Thoma F., Sentenac A., Marsolier M.C.. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol. 2001; 21:3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin M.P., Gerlach V.L., Brow D.A.. A novel upstream RNA polymerase III promoter element becomes essential when the chromatin structure of the yeast U6 RNA gene is altered. Mol. Cell. Biol. 2001; 21:6429–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eriksson P., Biswas D., Yu Y., Stewart J.M., Stillman D.J.. TATA-binding protein mutants that are lethal in the absence of the Nhp6 high-mobility-group protein. Mol. Cell. Biol. 2004; 24:6419–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Labazi M., Jaafar L., Flores-Rozas H.. Modulation of the DNA-binding activity of Saccharomyces cerevisiae MSH2-MSH6 complex by the high-mobility group protein NHP6A, in vitro. Nucleic. Acids. Res. 2009; 37:7581–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai Y., Wong B., Yen Y.M., Oettinger M.A., Kwon J., Johnson R.C.. Determinants of HMGB proteins required to promote RAG1/2-recombination signal sequence complex assembly and catalysis during V(D)J recombination. Mol. Cell. Biol. 2005; 25:4413–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., Stillman D.J.. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001; 20:3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brewster N.K., Johnston G.C., Singer R.A.. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 2001; 21:3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rhoades A.R., Ruone S., Formosa T.. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 2004; 24:3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hepp M.I., Alarcon V., Dutta A., Workman J.L., Gutiérrez J.L.. Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast high mobility group box (HMGB) proteins. Biochim. Biophys. Acta. 2014; 1839:764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hepp M.I., Smolle M., Gidi C., Amigo R., Valenzuela N., Arriagada A., Maureira A., Gogol M.M., Torrejón M., Workman J.L. et al.. Role of Nhp6 and Hmo1 in SWI/SNF occupancy and nucleosome landscape at gene regulatory regions. Biochim. Biophys. Acta Gene Regul. Mech. 2017; 1860:316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kurat C.F., Yeeles J.T.P., Patel H., Early A., Diffley J.F.X.. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell. 2017; 65:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishihama A., Kori A., Koshio E., Yamada K., Maeda H., Shimada T., Makinoshima H., Iwata A., Fujita N.. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J. Bacteriol. 2014; 196:2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Swinger K.K., Lemberg K.M., Zhang Y., Rice P.A.. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003; 22:3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Swinger K.K., Rice P.A.. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004; 14:28–35. [DOI] [PubMed] [Google Scholar]
- 52. Stella S., Cascio D., Johnson R.C.. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010; 24:814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hancock S.P., Stella S., Cascio D., Johnson R.C.. DNA sequence determinants controlling affinity, stability and shape of DNA complexes bound by the nucleoid protein Fis. PLoS One. 2016; 11:e0150189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Finkel S.E., Johnson R.C.. The Fis protein: it's not just for DNA inversion anymore. Mol. Microbiol. 1992; 6:3257–3265. [DOI] [PubMed] [Google Scholar]
- 55. Johnson R.C. Site-specific DNA inversion by serine recombinases. Microbiol. Spectrum. 2015; 3:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Landy A. The λ Integrase site-specific recombination pathway. Microbiol. spectrum. 2015; 3:Mdna3–0051–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Igarashi C., Murata A., Itoh Y., Subekti D.R.G., Takahashi S., Kamagata K.. DNA garden: a simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins. Bull. Chem. Soc. Jpn. 2017; 90:34–43. [Google Scholar]
- 58. Li W., Wang W., Takada S.. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10550–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Freeman G.S., Hinckley D.M., Lequieu J.P., Whitmer J.K., de Pablo J.J.. Coarse-grained modeling of DNA curvature. J. Chem. Phys. 2014; 141:165103. [DOI] [PubMed] [Google Scholar]
- 60. Kenzaki H., Takada S.. Partial unwrapping and histone tail dynamics in nucleosome revealed by coarse-grained molecular simulations. PLoS Comput. Biol. 2015; 11:e1004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tan C., Takada S.. Dynamic and structural modeling of the specificity in protein-DNA interactions guided by binding assay and structure data. J. Chem. Theory Comput. 2018; 14:3877–3889. [DOI] [PubMed] [Google Scholar]
- 62. Kenzaki H., Koga N., Hori N., Kanada R., Li W., Okazaki K., Yao X.Q., Takada S.. CafeMol: a coarse-grained biomolecular simulator for simulating proteins at work. J. Chem. Theory Comput. 2011; 7:1979–1989. [DOI] [PubMed] [Google Scholar]
- 63. Ester M., Kriegel H.-P., Sander J., Xu X.. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996; 96:226–231. [Google Scholar]
- 64. Smola A.J., Schölkopf B.. A tutorial on support vector regression. Stat. Comput. 2004; 14:199–222. [Google Scholar]
- 65. Graham J.S., Johnson R.C., Marko J.F.. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011; 39:2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Giuntoli R.D., Linzer N.B., Banigan E.J., Sing C.E., de la Cruz M.O., Graham J.S., Johnson R.C., Marko J.F.. DNA-segment-facilitated dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response. J. Mol. Biol. 2015; 427:3123–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kamar R.I., Banigan E.J., Erbas A., Giuntoli R.D., Olvera de la Cruz M., Johnson R.C., Marko J.F.. Facilitated dissociation of transcription factors from single DNA binding sites. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3251–E3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Subekti D.R.G., Murata A., Itoh Y., Fukuchi S., Takahashi H., Kanbayashi S., Takahashi S., Kamagata K.. The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements. Biochemistry. 2017; 56:4134–4144. [DOI] [PubMed] [Google Scholar]
- 69. Yen Y.M., Wong B., Johnson R.C.. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem. 1998; 273:4424–4435. [DOI] [PubMed] [Google Scholar]
- 70. Wong B., Masse J.E., Yen Y.M., Giannikopoulos P., Feigon J., Johnson R.C.. Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein Nhp6A. Biochemistry. 2002; 41:5404–5414. [DOI] [PubMed] [Google Scholar]
- 71. Becker N.A., Maher L.J. 3rd. High-resolution mapping of architectural DNA binding protein facilitation of a DNA repression loop in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7177–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ruone S., Rhoades A.R., Formosa T.. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 2003; 278:45288–45295. [DOI] [PubMed] [Google Scholar]
- 73. Lee J.Y., Finkelstein I.J., Arciszewska L.K., Sherratt D.J., Greene E.C.. Single-molecule imaging of FtsK translocation reveals mechanistic features of protein-protein collisions on DNA. Mol. Cell. 2014; 54:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim S., Brostromer E., Xing D., Jin J.S., Chong S.S., Ge H., Wang S.Y., Gu C., Yang L.J., Gao Y.Q. et al.. Probing allostery through DNA. Science. 2013; 339:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jolma A., Yin Y., Nitta K.R., Dave K., Popov A., Taipale M., Enge M., Kivioja T., Morgunova E., Taipale J.. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015; 527:384–388. [DOI] [PubMed] [Google Scholar]
- 76. Chen T.Y., Cheng Y.S., Huang P.S., Chen P.. Facilitated unbinding via multivalency-enabled ternary complexes: new paradigm for protein-DNA interactions. Acc. Chem. Res. 2018; 51:860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Erbas A., Marko J.F.. How do DNA-bound proteins leave their binding sites? The role of facilitated dissociation. Curr. Opin. Chem. Biol. 2019; 53:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Marcovitz A., Levy Y.. Obstacles may facilitate and direct DNA search by proteins. Biophys. J. 2013; 104:2042–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Saito M., Terakawa T., Takada S.. How one-dimensional diffusion of transcription factors are affected by obstacles: coarse-grained molecular dynamics study. Mol. Simul. 2017; 43:1315–1321. [Google Scholar]
- 80. Sarangi M.K., Zvoda V., Holte M.N., Becker N.A., Peters J.P., Maher L.J., Ansari A.. Evidence for a bind-then-bend mechanism for architectural DNA binding protein yNhp6A. Nucleic Acids Res. 2019; 47:2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McCauley M.J., Rueter E.M., Rouzina I., Maher L.J. 3rd, Williams M.C.. Single-molecule kinetics reveal microscopic mechanism by which High-Mobility Group B proteins alter DNA flexibility. Nucleic Acids Res. 2013; 41:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hancock S.P., Cascio D., Johnson R.C.. Cooperative DNA binding by proteins through DNA shape complementarity. Nucleic Acids Res. 2019; 47:8874–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Prieto A.I., Kahramanoglou C., Ali R.M., Fraser G.M., Seshasayee A.S., Luscombe N.M.. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012; 40:3524–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tan C., Terakawa T., Takada S.. Dynamic coupling among protein binding, sliding, and DNA bending revealed by molecular dynamics. J. Am. Chem. Soc. 2016; 138:1520–5126. [DOI] [PubMed] [Google Scholar]
- 85. Mangel W.F., McGrath W.J., Xiong K., Graziano V., Blainey P.C.. Molecular sled is an eleven-amino acid vehicle facilitating biochemical interactions via sliding components along DNA. Nat. Commun. 2016; 7:10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tafvizi A., Huang F., Fersht A.R., Mirny L.A., van Oijen A.M.. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vuzman D., Levy Y.. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012; 8:47–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.