Abstract
Centromeres are genomic regions essential for faithful chromosome segregation. Transcription of noncoding RNA (ncRNA) at centromeres is important for their formation and functions. Here, we report the molecular mechanism by which the transcriptional regulator ZFAT controls the centromeric ncRNA transcription in human and mouse cells. Chromatin immunoprecipitation with high-throughput sequencing analysis shows that ZFAT binds to centromere regions at every chromosome. We find a specific 8-bp DNA sequence for the ZFAT-binding motif that is highly conserved and widely distributed at whole centromere regions of every chromosome. Overexpression of ZFAT increases the centromeric ncRNA levels at specific chromosomes, whereas its silencing reduces them, indicating crucial roles of ZFAT in centromeric transcription. Overexpression of ZFAT increases the centromeric levels of both the histone acetyltransferase KAT2B and the acetylation at the lysine 8 in histone H4 (H4K8ac). siRNA-mediated knockdown of KAT2B inhibits the overexpressed ZFAT-induced increase in centromeric H4K8ac levels, suggesting that ZFAT recruits KAT2B to centromeres to induce H4K8ac. Furthermore, overexpressed ZFAT recruits the bromodomain-containing protein BRD4 to centromeres through KAT2B-mediated H4K8ac, leading to RNA polymerase II-dependent ncRNA transcription. Thus, ZFAT binds to centromeres to control ncRNA transcription through the KAT2B–H4K8ac–BRD4 axis.
INTRODUCTION
The centromere is a unique chromosomal region essential for the accurate segregation of sister chromatids into daughter cells (1). The kinetochore complex, which is built upon the centromere, mediates the attachment of each chromosome to the spindle microtubules during mitosis. The functional centromere is epigenetically defined by the specific incorporation of the histone H3 variant CENP-A (2–4). The centromere chromatin is composed of interspersed canonical H3 nucleosomes and nucleosomes containing CENP-A (5–7).
The eukaryotic centromere, which mostly consists of species-specific repetitive DNA sequences that lack protein-coding genes, had long been thought to be a transcriptionally inactive region. However, recent studies in various organisms have demonstrated that centromeric repeat sequences are transcribed into noncoding RNA (ncRNA). RNA polymerase II (RNAPII) was detected at the centromere in yeast, fly and humans (8–12). Furthermore, transcripts derived from centromeric DNA were identified in various species from yeast to humans (10–18). These centromeric transcripts have been thought to play crucial roles in the formation and functions of centromeres through the association with CENP-A (16,18,19), CENP-C (12,20,21), Aurora B (13,22,23) and Shugoshin 1 (24). Furthermore, the process of centromeric transcription has been thought to mediate chromatin remodeling at the centromeres, which is required for the assembly of CENP-A (8,9,25). These reports demonstrate that RNAPII-mediated centromeric transcription and its ncRNA products play crucial roles in chromosome segregation. However, there is limited understanding regarding the regulation of this process at the molecular level.
ZFAT is a nuclear protein harboring an AT-hook domain and 18-repeats of C2H2 zinc-finger domains (26–28). It regulates mRNA transcription by binding to the proximal region of transcription start sites in ZFAT-target genes (29). Zfat-deficient mice exhibited embryonic lethality by embryonic day 8.5 (30). Furthermore, T-cell specific deletion of the Zfat gene in mice resulted in a marked reduction in the number of T cells (31–33). Therefore, ZFAT has been thought to be a transcriptional regulator essential for embryonic development and T-cell homeostasis.
Here, we report crucial roles of ZFAT in centromeric ncRNA transcription in human and mouse cells. ZFAT was bound to centromeres through a specific 8-bp DNA sequence that is highly conserved and widely distributed at whole centromere regions of every chromosome. Overexpression of ZFAT caused a marked increase in the levels of centromeric ncRNA, whereas silencing of ZFAT reduced them, indicating crucial roles of ZFAT in centromeric ncRNA transcription. ZFAT induced acetylation at the lysine 8 in histone H4 (H4K8ac) at centromeres by recruiting the histone acetyltransferase KAT2B, leading to the accumulation of the bromodomain-containing protein BRD4 at centromeres. Therefore, we propose that ZFAT binds to centromeres to control ncRNA transcription through the KAT2B–H4K8ac–BRD4 axis.
MATERIALS AND METHODS
Cell culture
HEK293, HeLa, NIH3T3 and HT1080 cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Wako Pure Chemical Industries), supplemented with 10% fetal calf serum and penicillin/streptomycin. For inhibition of RNAPII, α-amanitin (Wako Pure Chemical Industries, 010-22961) was used at a final concentration of 1 μM. For inhibition of BRD4, JQ1 (Sigma-Aldrich, SML1524) was used at a final concentration of 0.5 μM.
Mice
Mouse thymocytes and splenic CD4+ T cells were prepared as previously described (32,33). All animal experiments followed the guidelines established by the Institutional Animal Care and Use Committee of Fukuoka University in accordance with approved protocols.
Constructs
The expression vectors and primers used for cloning and mutagenesis in this study are listed in Supplementary Tables S1 and S2. The expression vectors for mouse Zfat were previously described (26,29). The previously described cDNA for human ZFAT (27) was cloned into plasmid DNA for expression in cultured mammalian cells. The cDNA for human BRD4 (FHC11882) was purchased from Promega and cloned into an EGFP-C3 vector. The cDNA for mouse KAT2B was amplified from reverse transcription products obtained from thymocytes of C57BL/6 mice and cloned into an pcDNA3 (Invitrogen) or MSCVpuro (Clontech) vector with a FLAG-tag at the N-terminus. A QuickChange Site-Directed Mutagenesis kit (Takara Bio) was used for mutagenesis. All expression vectors were verified by DNA sequencing.
Chromatin immunoprecipitation (ChIP) and high-throughput sequencing (ChIP-seq), and ChIP-quantitative PCR (ChIP-qPCR)
The antibodies used for ChIP and the primers used for qPCR are listed in Supplementary Tables S2 and S3.
For the ChIP-seq analysis of HA-hZFAT using an anti-HA antibody, HEK293 cells were transfected with the empty vector or the expression vector for HA-hZFAT using Lipofectamine 3000 (Invitrogen). ChIP-seq analysis was performed as previously described (29). After trimming adaptor sequences and low-quality sequences, approximately nine million sequence reads (only Read 1) were mapped to a human reference genome set (hg38) using the Burrows-Wheeler Aligner (ver0.6.2). The reference genome sequence set used consists of the sequences of the autosomal (1–22) and sex (X, Y) chromosomes, and the mitochondrial DNA from the GRCh38 assembly (GenBank assembly accession: GCA_000001405.15), the human gamma herpesvirus 4 (Epstein-Barr virus) genome sequence (GenBank accession: NC_007605), and the decoyJRGv1 sequence (available at https://jmorp.megabank.tohoku.ac.jp/dj1/datasets/tommo-jrgv1/files/decoyJRGv1.fasta.zip?download=true). The resultant bam files were de-duplicated using the samtools-1.6 (https://sourceforge.net/projects/samtools/files/samtools) and picard-tools-2.8.1 (https://broadinstitute.github.io/picard/). To visualize the mapping results in bam files, the count function of IGVTools (https://software.broadinstitute.org/software/igv/igvtools) was used to generate .tdf files from the de-duplicated bam files. The .tdf files were visualized using the Integrative Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/). The de-duplicated bam files were analyzed using the MACS2 algorithm (https://github.com/taoliu/MACS/) to identify binding regions. Genomic feature annotation of the ZFAT-binding sites was performed using ChIPseeker annotating tools with ‘TxDb.Hsapiens.UCSC.hg38.knownGene’ as an annotation data source (34). Cells transfected with the empty vector were used as a control. ChIP-seq data has been deposited in NCBI GEO as GSE134612.
ChIP-qPCR was similarly performed with minor modifications. The modifications were as follows. HEK293 cells were transfected with the expression vector for EGFP or HA-hZFAT-EGFP using Lipofectamine 3000. After 24 h, the cells were crosslinked with formaldehyde, and EGFP+ cells were sorted using FACSAria II (BD Biosciences). The cells were lysed in RIPA buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) and sonicated using a Bioruptor (Cosmo Bio) for 15 cycles of 1 min with 30 s on/off without MNase treatment. After the ChIP reaction, the beads were serially washed with RIPA, RIPA containing 500 mM NaCl, and RIPA containing 250 mM LiCl and TE buffers (10 mM Tris–HCl, pH 8.0, 1 mM EDTA). qPCR was performed using TB Green Premix Ex Taq GC (Perfect Real Time) (Takara Bio) with ABI PRISM 7900HT (Applied Biosystems).
RNA-sequencing (RNA-seq) analysis
HT1080 cells were transfected with expression vectors for HA-hZFAT-EGFP or EGFP. After 24 h, EGFP+ cells were sorted using FACSAria II, and total RNA was extracted using the TRIZol reagent (Life Technologies). Libraries for RNA-seq were prepared using NEBNext rRNA Depletion Kit (New England Biolabs, #E6310L) and NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs, #E7760S) using 750 ng of total RNA as a starting material by following the manufacturer's instructions with 6 cycles of PCR amplification. Paired-end sequencing (76 bp × 2) was performed on the NextSeq550 system (Illumina). Approximately 30 million read pairs per library were trimmed for adaptor sequence and low-quality sequence, and mapped to the human reference genome sequence set described in the ChIP-seq section using HISAT2 v2.1.0 (https://ccb.jhu.edu/software/hisat2/manual.shtml) with the following options: -p, –dta, -rna-stranness RF. The resultant .sam files were converted to .bam files and de-duplicated using samtools-1.6 and picard-tools-2.8.1. The resultant bam files were subjected to StringTie ver. 2.0.6 (https://ccb.jhu.edu/software/stringtie/) to determine read counts for each gene described in the gtf file for NCBI RefSeq genes (GRCh38/hg38) with the following options: -A -B -C -e -G –rf. The gtf file was obtained using the Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables). RNA-seq data has been deposited in NCBI GEO as GSE145651.
Count of reads mapped to centromeric α-satellite DNA regions
The RepeatMasker annotation for the GRCh38/hg38 genome was obtained in bed format using the Table Browser, and used to generate a bed file for 1,653 genomic intervals annotated as α-satellite DNA regions on autosomal (1–22) and sex (X and Y) chromosomes. The read counts mapped in each of the 1653 α-satellite DNA intervals were counted for de-duplicated bam files using the multicov function of bedtools v2.26.0 (https://bedtools.readthedocs.io/en/latest/). The sum of the read counts mapped to the α-satellite DNA regions was calculated for each chromosome. The mapped read counts were normalized as read per million reads (RPM).
siRNA transfection
The siRNAs against KAT2B, BRD4 and ZFAT, and the negative control siRNA were purchased from Thermo Scientific. Sequences for siRNA are shown in Supplementary Table S2. The cells were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the reverse transfection protocol provided by the manufacturer. Briefly, cells were seeded with siRNA (20 pmol)–Lipofectamine RNAiMAX (5 μl) complexes in six-well plates at a density of 2 × 105 cells per well. After 48 h, the cells were transfected with plasmid DNAs using Lipofectamine 3000 (Invitrogen) and incubated for a further 24 h. Following incubation, the cells were subjected to further analyses.
Immunofluorescence microscopy
Cells were seeded onto 12-mm diameter glass coverslips coated with 0.1 mg/ml poly-l-lysine (Sigma-Aldrich, P5899), placed in 24-well plates, and transfected with plasmid DNAs using Lipofectamine 3000. After 24 h, the transfected cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature or ice-cold 100% methanol for 15 min at –20°C, washed thrice with PBS, permeabilized/blocked with 5% FBS or 5% nonfat dry milk in PBS containing 0.3% Triton X-100 for 30 min at room temperature, and subsequently incubated with primary antibodies. The antibodies used for immunostaining are listed in Supplementary Table S3. Following incubation, the cells were washed thrice with PBS, permeabilized/blocked with 5% FBS or 5% nonfat dry milk in PBS containing 0.3% and subsequently incubated with secondary antibodies conjugated with fluorescent dyes for 1 h at room temperature. The cells were washed thrice with PBS, permeabilized/blocked with 5% FBS or 5% nonfat dry milk in PBS containing 0.3%, stained with DAPI, mounted using Fluorescence Mounting Medium (Dako), and examined using a TCS SP5 laser-scanning confocal microscope (Leica Microsystems). To determine proportion of cells with centromeric ZFAT foci, cells were counted as ZFAT foci positive cells when >80% of the detected ZFAT foci were colocalized with CENP-A foci.
Quantitative RT-PCR (qRT-PCR)
Cells were transfected with the expression vector for EGFP or HA-hZFAT-EGFP using Lipofectamine 3000. After 24 h, EGFP+ cells were sorted using FACSAria II. Total RNA was extracted using the TRIZol reagent (Life Technologies). cDNA was synthesized using the ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). The RNA expression levels were determined in both reactions with and without reverse transcriptase, and are shown as the relative values to those of β-actin. qPCR was performed using the Thunderbird SYBR qPCR Mix (Toyobo) with ABI PRISM 7900HT (Applied Biosystems) according to the instructions provided by the manufacturer. The primers used for qPCR are listed in Supplementary Table S2.
Immunoblotting and co-immunoprecipitation
Immunoblotting was performed as previously described (35,36) using the antibodies shown in Supplementary Table S3. Co-immunoprecipitation was performed as previously described (37).
Flow cytometry
Cell sorting and flow cytometry analysis were performed using FACSAria II as previously described (33). Flow cytometry data were analyzed using the FlowJo software (Tomy Digital Biology).
Luciferase reporter assay
Luciferase reporter assay was performed as previously described (29). Mouse Zfat cDNA was subcloned into the pVP16-AD vector (Clontech) to express Zfat fused with the transcription activation domain (AD) of herpes simplex virus VP16. The minimal adenovirus E1b promoter sequence (GGGTATATAATG, E1bTATA) was inserted into the pGL4.14 vector (Promega) at the HindIII site directly preceding firefly luciferase (pGL4.14-E1bTATA). The oligonucleotides shown in Supplementary Table S2 were annealed and then inserted into the pGL4.14-E1bTATA vector to produce the reporter constructs containing a part of α- or minor satellite DNA. The nucleotide sequence for the ZFAT box in the reporter construct containing CENP-B box was substituted to ‘TCCTCGTA’. HEK293 cells were seeded at 1 × 104 cells on 96-well plates and transfected with 2 ng of pRL-SV40 vector (Promega), 50 ng of pGL4.14-E1bTATA reporter constructs, and 50 ng of pVP16-AD or pVP16-AD-Zfat vector using Lipofectamine 3000 (Invitrogen). After 24 h, the cells were lysed and analyzed using the Dual Glo Luciferase Assay system and Glomax luminometer (both from Promega), according to the manufacturer's protocol. The firefly luciferase activity was normalized against the corresponding Renilla luciferase activity, and the relative luciferase activity was calculated as the change relative to the reporter in the presence of pVP16-AD. In all of the reporter assays, we obtained consistent results from at least three independent experiments.
Statistical analysis
The data were expressed as the mean ± standard deviation. The statistical analyses were performed using an unpaired two-tailed Student's t-test. A P < 0.05 denoted statistically significant difference.
RESULTS
ZFAT binds to centromeres of every chromosome in human and mouse cells
We previously reported ZFAT-binding sites on the mouse genome DNA, determined by ChIP-seq analysis of mouse Zfat (mZfat) (29). To compare the genomic binding profiles of ZFAT between human and mouse, we attempted to determine the ZFAT-binding sites on the human genome by ChIP-seq analysis of human ZFAT (hZFAT). However, the anti-mZfat antibody, which was used in the ChIP-seq analysis of mZfat, was not suitable for the hZFAT ChIP-seq analysis. Mouse and human ZFAT showed 91% amino acid sequence identity in the C-terminal region used for generation of the anti-mZfat antibody (Supplementary Figure S1A). However, the anti-mZfat antibody specifically recognized mZfat, but not hZFAT, in immunoprecipitation and immunoblotting analyses (Supplementary Figure S1B). Therefore, we established HEK293 human cells stably expressing hZFAT tagged with HA (HA-hZFAT), and performed immunoprecipitation of HA-hZFAT using an anti-HA antibody. Ectopic HA-hZFAT was overexpressed over 50-fold higher than endogenous ZFAT through the CAG promoter (Supplementary Figure S1C). In the ChIP-seq analysis of HA-hZFAT, majority (92%) of the hZFAT-binding sites was found to be located at α-satellite DNA regions (Figure 1A and Supplementary Figure S2A). On the other hand, at non-α-satellite DNA regions, hZFAT was mainly bound to the gene promoters, which correspond to the mZfat-binding sites previously identified through the mZfat ChIP-seq analysis (Supplementary Figures S2A and S2B), indicating the efficacy of the HA-hZFAT ChIP-seq analysis.
Figure 1.
ZFAT binds to every chromosome centromere. (A,B) ChIP-seq analysis of ZFAT at the α-satellite DNA region using an anti-HA antibody in HEK293 cells transfected with HA-hZFAT expression vector or empty vector. (A) Results of the α-satellite DNA region of the chromosome 4 are shown as representative data. Chr4 row shows the structure of human chromosome 4. Red rectangle indicates chromosome region where the results of ZFAT ChIP-seq analysis are shown. HA-hZFAT row presents the result of the ZFAT ChIP-seq analysis in cells transfected with HA-hZFAT expression vector using an anti-HA antibody. Control vector row presents the result of the ChIP-seq analysis in cells transfected with empty vector using an anti-HA antibody as the negative control. Values in the panels represent the ranges of mapped read. ZFAT peak summits row shows the position of ZFAT peak summit identified by MACS analysis. Satellite and Gene rows show the position of satellite DNA and genes, respectively. Results of every chromosome are shown in Supplementary Figure S3. (B) The reads mapped in the α-satellite DNA regions are counted as described in the Materials and Methods. The sum of mapped reads is calculated, and normalized as read per million reads (RPM). The centromeric ZFAT levels at each chromosome centromere are shown as the ratio of the values in cells expressing HA-hZFAT against those in cells transfected with empty vector. (C) ChIP-qPCR analysis of ZFAT at human repetitive DNA sequences using an anti-HA antibody or control IgG in HT1080 cells transiently expressing HA-hZFAT-EGFP or EGFP. The results are shown as the values for relative recovery rate of immunoprecipitated DNA. (D) ChIP-qPCR analysis of mZfat at minor and major satellite DNA sequences in NIH3T3 cells using an anti-ZFAT antibody or control IgG. The mZfat protein levels are shown as the relative values to control IgG. Primers for tubulin were used as the negative control. (E) Immunofluorescence images of endogenous hZFAT and CENP-B in HT1080 cells using anti-ZFAT and anti-CENP-B antibodies. (F) Intensity profile of the indicated cross section in the image in (E) (white line). (G) Immunofluorescence images of ZFAT and CENP-A in HEK293 cells transiently expressing hZFAT-HA or mZfat-HA. (H) Percentage of cells containing centromeric ZFAT foci in cells expressing ZFAT-HA, determined in (G). (C, D, H) Data represent the mean ± SD of three independent experiments. (E, G) Data are representative of three independent experiments. Scale bar, 5 μm.
Binding of ZFAT to α-satellite DNA was observed at every chromosome, except for the Y chromosome (Figure 1B and Supplementary Figure S3). HEK293 cells do not have the Y chromosome because they are of female origin. To examine whether ZFAT binds to α-satellite DNA at the Y chromosome, ChIP-quantitative PCR (qPCR) analysis of overexpressed hZFAT was performed using primers for centromeric α-satellite DNA and other repetitive DNA sequences in HT1080 cells, which are of male origin (Supplementary Figure S4A). Binding of HA-hZFAT-EGFP was observed at α-satellite DNA of the Y chromosome, as well as of the chromosome 21, but not at the genomic region for 5S ribosomal RNA (Figure 1C). Furthermore, the binding of overexpressed hZFAT at α-satellite DNA was also validated in HEK293 cells by ChIP-qPCR analysis. Binding of hZFAT was specifically observed at the centromeric α-satellite DNA, but not at other repetitive DNA sequences, in cells expressing HA-hZFAT-EGFP (Supplementary Figure S4B). These results suggest that ZFAT binds to centromeres of every chromosome, including the Y chromosome.
As DNA sequences for mouse centromeric regions have not fully been reported in the mouse genome assembly (mm10), it is unknown that mZfat binds to centromeres in mouse cells. To address whether endogenous ZFAT binds to mouse centromeres, we performed ChIP-qPCR analysis of mZfat in NIH3T3 mouse cells using the anti-mZfat antibody. Centromeric and pericentromeric regions at the mouse chromosomes consist of distinct repetitive DNA sequences, called minor and major satellite DNAs, respectively. Binding signals of endogenous mZfat were detected at both minor and major satellite DNAs (Figure 1D), suggesting that endogenous mZfat binds to both centromeres and pericentromeres in mouse cells.
The localization of ZFAT was examined through immunofluorescence analysis using an anti-hZFAT antibody. Endogenous hZFAT formed foci within the nucleus in HT1080 cells (Figure 1E). A part of ZFAT foci were completely colocalized with CENP-B foci, but there were also several ZFAT foci, which were only partially colocalized with CENP-B foci (Figure 1E and F). Furthermore, the centromeric localization of hZFAT was also examined in HEK293 cells overexpressing hZFAT-HA. hZFAT-HA showed the formation of foci, which were partially colocalized with CENP-A (Figure 1G and H). The hZFAT foci were larger than those of CENP-A, suggesting that hZFAT binds to centromeres and the surrounding regions. Furthermore, formation of the ZFAT foci was also observed in other human and mouse cell lines (Supplementary Figure S4C). Interestingly, mZfat-HA, which was ectopically overexpressed in human cells, showed formation of foci at centromeres (Figure 1G and H, and Supplementary Figure S4C). Similarly, ectopically overexpressed hZFAT-HA formed centromeric foci in mouse cells (Supplementary Figure S4C). These results suggest that the centromeric binding of ZFAT is conserved between human and mouse. Collectively, these results indicate that ZFAT binds to centromeres of every chromosome in human and mouse cells.
ZFAT binds to a specific 8-bp DNA sequence named ZFAT box, which is widely distributed at every chromosome centromere
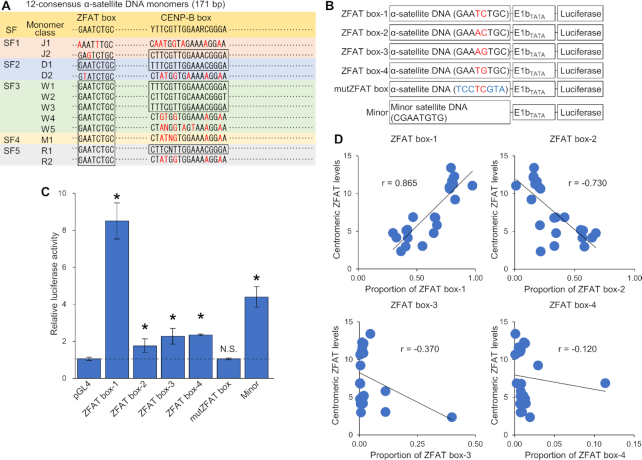
We previously reported that ZFAT was directly bound to specific nucleotide sequences on the mouse genome DNA, and identified the GAA(T/A)(C/G)TGC sequence as ZFAT-binding motif (29). As shown in Supplementary Figure S3, every human chromosome harbored a large number of the ZFAT-binding motif at whole α-satellite DNA regions. Based on sequence analysis of hundreds of α-satellite DNA monomers, 12 consensus monomers (J1, J2, D1, D2, W1, W2, W3, W4, W5, M1, R1 and R2) have been designated (Figure 2A and Supplementary Figure S5) (38–40). While only half of 12 consensus monomers contained CENP-B box that is the only sequence element known at centromeres, the ZFAT-binding motif was conserved in 9 of 12 consensus monomers (Figure 2A and Supplementary Figure S5). These results suggest that a specific 8-bp DNA sequence for the ZFAT-binding motif is highly conserved and widely distributed at whole centromere regions of every chromosome. Thus, we named this 8-bp DNA sequence ‘ZFAT box’.
Figure 2.
ZFAT binds to a specific 8-bp DNA sequence conserved in α-satellite DNA. (A) Schematic diagram of 12-consensus α-satellite DNA monomers. Suprachromosomal family (SF) classification for monomers are shown. Nucleotide sequences for the ZFAT box and CENP-B box are presented at the top. Nucleotides different from ZFAT or CENP-B box are colored in red in each monomer. The entire nucleotide sequences for 12-consensus α-satellite DNA monomers are shown in Supplementary Figure S5. (B) Schematic diagram of constructs used in (C). Nucleotide sequences of the ZFAT box are shown. Different nucleotides among the ZFAT box-1-4 are colored in red. Nucleotides substituted in the ZFAT box are colored in blue. The entire nucleotide sequences for each reporter constructs are described in Supplementary Table S2. (C) Luciferase assay to characterize the ZFAT box in α- or minor satellite DNA regions. The luciferase activity is expressed as the relative values to those in cells transfected with a combination of pVP16-AD and each reporter construct. Data represent the mean ± SD based on three replicates from three independent experiments. Values in cells transfected with the reporter constructs containing ZFAT box are compared with those in cells transfected with the empty pGL4 vector. *P < 0.05, N.S.; not significant (P > 0.05). (D) Correlation between the centromeric ZFAT levels and proportion of ZFAT boxes at each chromosome. Proportion of each ZFAT box to the total number of all ZFAT boxes is determined, as shown in Supplementary Figure S6. Values of correlation coefficient (r) are shown in the panels.
To confirm the binding of ZFAT to the ZFAT box within α-satellite DNA, the luciferase reporter assay was performed in HEK293 cells. We used mZfat fused with the transcription activation domain (AD) of herpes simplex virus VP16 (AD-Zfat). Furthermore, the minimal adenovirus E1b promoter sequence (E1bTATA) was inserted directly preceding the luciferase-coding sequence at a reporter construct (Figure 2B). In our assay system, luciferase is expressed through the E1bTATA promoter activated by the AD fused with mZfat. Therefore, the luciferase activities mostly reflect the ZFAT-binding levels at a reporter construct in cells (29). A part of α-satellite DNA, a 50-bp nucleotide sequence containing the ZFAT box, was cloned in front of the E1bTATA sequence in the reporter construct (Figure 2B). The reporter construct was transiently transfected into HEK293 cells along with a construct expressing AD-Zfat. AD-Zfat strongly induced luciferase activity through the reporter construct containing α-satellite DNA with the ZFAT box-1, whereas luciferase activity was slightly observed in cells transfected with the reporter constructs containing the ZFAT box-2, 3 and 4 (Figure 2C). Furthermore, AD-Zfat hardly induced luciferase expression through the reporter construct containing the mutated ZFAT box (Figure 2C). These results suggest that ZFAT binds to the ZFAT box within α-satellite DNA, and shows a binding preference for the ZFAT box-1.
Binding levels of ZFAT at α-satellite DNA regions were different in each chromosome (Figure 1B and Supplementary Figure S3). To explore whether the different levels of ZFAT-binding in each chromosome were due to selectivity of ZFAT to the ZFAT boxes, we examined the number of ZFAT box per million base pairs at centromeres of every chromosome. Majority of the ZFAT box present in centromeres was the ZFAT box-1 and 2 at every chromosome, but their proportions to the total number of all ZFAT boxes were different in each chromosome (Supplementary Figure S6). We further examined correlation on each chromosome between the centromeric ZFAT levels and proportions of the ZFAT boxes. Of the 4-types of ZFAT boxes, the proportion of ZFAT box-1 was positively correlated with the centromeric ZFAT levels at each chromosome (Figure 2D). These results were consistent with the binding selectivity of ZFAT to ZFAT boxes (Figure 2C), suggesting that the binding preference of ZFAT for ZFAT box-1 participates in the different ZFAT-binding levels at each chromosome centromere.
We investigated whether mouse minor satellite DNA contained the ZFAT box, and found a nucleotide sequence (CGAATGTGT) with one nucleotide substitution against the ZFAT box (GAATGTGC) within minor satellite DNA. We previously reported that mZfat was strongly bound to the modified ZFAT-binding motif (CGAATGTG) at promoter region of the Brpf1 gene, which is perfectly matched with the nucleotide sequence of minor satellite DNA (29). Indeed, AD-Zfat significantly induced luciferase expression through the reporter constructs containing a part of minor satellite DNA with the CGAATGTG sequence (Figure 2C), suggesting that the CGAATGTG sequence at minor satellite DNA functions as the ZFAT-binding site in mouse cells. These results suggest that ZFAT binds to a specific 8-bp DNA sequence named ZFAT box, which is widely distributed at every chromosome centromere, in human and mouse cells.
A cluster of basic amino acids in the ZFAT N-terminal region is required for its centromeric binding
To determine hZFAT regions involved in binding to the centromeres, we examined the centromeric localization of the hZFAT deletion mutants through immunofluorescence analysis. The deletion mutants without the C-terminal half of ZFAT showed formation of the foci that were colocalized with CENP-A (ZF-ΔC-2 and -3), whereas the formation of centromeric ZFAT foci was inhibited by deletion of the N-terminal region of ZFAT (1–135 amino acids) (ZF-ΔN-1, Supplementary Figures S7A and S7B). These findings indicate that the N-terminal region, but not zinc-finger domains in the C-terminal region, of ZFAT is involved in its centromeric binding. The N-terminal region of ZFAT contains a cluster of basic amino acids, as well as an AT-hook domain which is known to bind to DNA, both of which are highly conserved in ZFAT amino acid sequences among vertebrates (Supplementary Figure S7C). The formation of foci of ZFAT was abolished by deleting a cluster of basic amino acids in the N-terminal region (Δ71–83), whereas mutations in the AT-hook domain (GRP/AAA) did not affect the formation of foci (Supplementary Figures S7A and S7B). These results suggest that a cluster of basic amino acids at 71–83 residues, but not the AT-hook domain, in the ZFAT N-terminal region is required for its centromeric binding.
ZFAT plays crucial roles in the transcription of ncRNA at specific chromosome centromeres
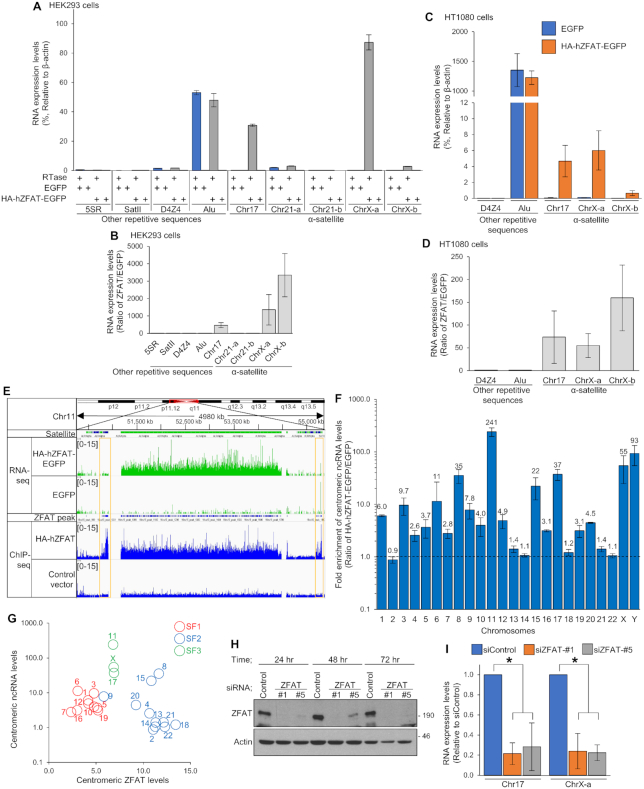
We previously reported that mZfat regulated the mRNA transcription of particular genes by binding to their promoter regions (29). To elucidate the role of ZFAT at centromeres, we examined the expression levels of centromeric ncRNAs in HEK293 cells overexpressing HA-hZFAT-EGFP by quantitative RT-PCR (qRT-PCR) analysis using primers for centromeric α-satellite RNA and other repetitive sequences (Supplementary Figure S4A). Notably, the expression levels of centromeric ncRNAs from the chromosomes 17 and X, but not from the chromosome 21, were markedly increased by the overexpression of hZFAT (Figure 3A). Since the centromeric ncRNAs were expressed at very low levels in control cells, the ectopic overexpression of hZFAT resulted in hundreds- to thousands-fold increase in the levels of centromeric ncRNA from the chromosomes 17 and X, compared with those observed in control cells (Figure 3B). Furthermore, a marked increase in the expression levels of centromeric ncRNA from the chromosomes 17 and X caused by the ectopic overexpression of ZFAT was also observed in HT1080 cells (Figure 3C and D). These results suggest that overexpression of ZFAT increases the centromeric ncRNA transcription at specific chromosomes in human cells.
Figure 3.
ZFAT plays crucial roles in the ncRNA transcription at specific chromosome centromeres. (A, C) qRT-PCR analysis of RNA derived from human repetitive sequences in HEK293 (A) and HT1080 (C) cells expressing EGFP or HA-hZFAT-EGFP. The RNA expression levels were determined in both reactions with and without reverse transcriptase (RTase), and are shown as the relative values to those of β-actin. (B, D) The RNA expression levels determined in (A and C) are shown as the ratio of HA-hZFAT-EGFP-expressing cells against EGFP-expressing cells. (E) Combined results of the RNA-seq analysis of HT1080 cells expressing HA-hZFAT-EGFP or EGFP, and the ChIP-seq analysis of hZFAT in HEK293 cells transfected with HA-hZFAT expression vector or control vector, at the α-satellite DNA region of the chromosome 11. Regions showing ZFAT-binding, but not the increased ncRNA levels, are surrounded by orange boxes. (F) RNA-seq analysis of HT1080 cells expressing HA-hZFAT-EGFP or EGFP. Fold enrichment of centromeric ncRNA levels is shown as the ratio of HA-hZFAT-EGFP expressing cells against EGFP-expressing cells. The Y-axis is represented in log scale. (G) Correlation between the centromeric ncRNA and ZFAT levels at each chromosome. Dots for each chromosome are color-coded based on suprachromosomal family (SF) classification. (H) Immunoblotting analysis of ZFAT using an anti-ZFAT antibody in HT1080 cells transfected with siRNAs for control or ZFAT (#1 and #5). Data are representative of three independent experiments. (I) qRT-PCR analysis of centromeric ncRNA in HT1080 cells transfected with siRNAs for control or ZFAT (#1 and #5). Reverse transcription was performed in both reactions with and without RTase, and background values (no RTase) are subtracted. The RNA expression levels are normalized to those of β-actin. (A–D, F, I) Data represent the mean ± SD of three independent experiments. *P < 0.05.
To investigate the chromosomal specificity of ZFAT in the centromeric ncRNA transcription, we performed RNA-seq analysis in HT1080 cells overexpressing HA-hZFAT-EGFP or EGFP. As shown in Figure 3E, an increase in the expression levels of centromeric ncRNA was observed in cells overexpressing HA-hZFAT-EGFP. The regions where ncRNA expression was induced by ZFAT mostly coincided with the regions where ZFAT-binding was observed (Figure 3E). However, there were also narrow regions showing the ZFAT-binding, but not the increased ncRNA levels (Figure 3E). Interestingly, the effects of ZFAT overexpression on the centromeric ncRNA levels greatly varied between chromosomes (Figure 3F). The centromeric ncRNA levels markedly increased at the chromosomes 8, 11, 15, 17, X and Y in HA-hZFAT-EGFP-expressing cells (Figure 3F). In contrast, the ectopic overexpression of ZFAT did not significantly affect the centromeric ncRNA levels at the chromosomes 2, 13, 14, 18, 21 and 22 although the centromeric binding of ZFAT at these chromosomes was observed (Figures 1B and 3F). Consistent with the results of qRT-PCR analysis, an increase in the centromeric ncRNA levels by the expression of hZFAT was observed at the chromosomes 17 and X, but not at the chromosome 21, in the RNA-seq analysis. These results suggest that ZFAT plays roles in centromeric ncRNA transcription at specific chromosomes.
Twelve consensus α-satellite monomers have been classified into five suprachromosomal families (SFs) that are defined by sequence homology and linear order of the monomers that create a higher-order repeat (HOR) (Figure 2A) (38,39,41,42). The centromeres of each chromosome contain specific SFs (Supplementary Figure S6) (38,42). We examined correlation on each chromosome between the SF classification, and the centromeric ncRNA and ZFAT-binding levels. Interestingly, chromosomes with the same SF classification showed similar patterns of correlation between the centromeric ncRNA and ZFAT levels (Figure 3G). Chromosomes containing SF1 monomers showed the low ZFAT-binding levels and slightly high ncRNA levels at centromeres. In contrast, at many chromosomes containing SF2 monomers, except for the chromosomes 8, 9 and 15, the centromeric ZFAT levels were high, but their ncRNA levels were low. Importantly, all chromosomes containing SF3 monomers (chromosomes 11, 17 and X) showed the clearly high centromeric ncRNA levels despite of their moderate ZFAT-binding levels. Furthermore, we observed the obviously high centromeric ncRNA levels at the Y chromosome that uniquely contains SF4 monomers although the levels of centromeric ZFAT-binding were not determined in the hZFAT ChIP-seq analysis (Figure 3F and Supplementary Figure S6). These results suggest that the ZFAT-binding levels and its effects on ncRNA levels are different in each chromosome group containing specific SFs. Nucleotide sequences or linear order of α-satellite monomers in the HOR may play an important role in the ZFAT-regulated centromeric ncRNA transcription.
To examine the involvement of endogenous ZFAT on centromeric ncRNA transcription, we quantitated the centromeric ncRNA levels in HT1080 cells treated with siRNA against hZFAT. Two siRNAs targeting different hZFAT sequences resulted in a drastic decrease in the expression levels of hZFAT protein, determined by immunoblotting analysis using an anti-ZFAT antibody (Figure 3H). qRT-PCR analysis showed that the depletion of ZFAT significantly reduced the levels of centromeric ncRNA in HT1080 cells (Figure 3I). These results suggest that ZFAT plays crucial roles in the transcription of ncRNA at the centromeres of, at least, chromosomes 17 and X.
ZFAT induces acetylation at the lysine 8 in histone H4 at centromeres
We previously reported that mZfat was involved in histone acetylation on the mouse genome DNA (29). Histone modifications, such as acetylation and methylation, are important for transcriptional regulation. To elucidate the role of ZFAT in centromeric ncRNA transcription, we examined the effects of ZFAT overexpression on histone modification patterns in HEK293 cells by immunofluorescence analysis. Interestingly, overexpressed ZFAT specifically induced the foci formation of histone H4 acetylated at the lysine 8 (H4K8ac), compared with that observed in neighboring non-transfected cells (Figure 4A and B, and Supplementary Figure S8A). In contrast, other histone modification patterns examined in this study were apparently unaffected by the expression of ZFAT (Supplementary Figure S8A). The H4K8ac foci induced by overexpressed ZFAT were colocalized with the foci of ZFAT and CENP-A (Figure 4A). The formation of H4K8ac foci caused by the overexpression of ZFAT was also observed in other human cell lines (Supplementary Figure S8B). Furthermore, ChIP-qPCR analysis using an anti-H4K8ac antibody revealed that the overexpression of ZFAT resulted in a significant increase in the levels of H4K8ac at centromeric α-satellite DNA, but not at other repetitive sequences, compared with those in cells expressing EGFP (Figure 4C). In contrast, the acetylation levels of lysine 5, lysine 12 and lysine 16 in histone H4 at centromeric α-satellite DNA were unaffected by the expression of ZFAT (Supplementary Figure S8C). Thus, these results indicate that ZFAT specifically induces H4K8ac at centromeres.
Figure 4.
ZFAT induces centromeric H4K8ac and recruits KAT2B to centromeres. (A) Immunofluorescence images of ZFAT, CENP-A, H4K8ac and DNA (DAPI) in HEK293 cells transiently expressing HA-hZFAT-EGFP. (B) Percentage of cells containing centromeric H4K8ac foci in cells expressing EGFP or HA-hZFAT-EGFP, determined in (A). (C) ChIP-qPCR analysis of H4K8ac at human repetitive sequences using an anti-H4K8ac antibody or control IgG in HEK293 cells expressing HA-hZFAT-EGFP or EGFP. The H4K8ac levels are shown as the ratio of HA-ZFAT-EGFP-expressing cells against EGFP-expressing cells. Values in cells expressing HA-hZFAT-EGFP are compared with those in cells expressing EGFP. *P < 0.05, N.S.; not significant (P > 0.05). (D) Immunofluorescence images of ZFAT and KAT2B in HEK293 cells expressing HA-hZFAT-EGFP. (E) Percentage of cells containing KAT2B foci in cells expressing EGFP or HA-hZFAT-EGFP, determined in (D). (F) Co-immunoprecipitation (Co-IP) analysis of mZfat and KAT2B using an anti-mZfat antibody or control IgG (IgG) in mouse thymocytes and splenic CD4+ T cells. (G) Co-IP analysis of hZFAT-HA and FLAG-KAT2B using anti-HA and anti-FLAG antibodies in HEK293 cells transfected with indicated expression vectors. (H) Immunofluorescence images of ZFAT, KAT2B, CENP-A and DNA (DAPI) in HEK293 cells expressing HA-hZFAT-EGFP and FLAG-KAT2B. (I) Percentage of cells containing centromeric FLAG-KAT2B foci in cells expressing EGFP or HA-hZFAT-EGFP, determined in (H). (J) ChIP-qPCR analysis of KAT2B at human repetitive DNA sequences using an anti-KAT2B antibody or control IgG in HEK293 cells expressing HA-hZFAT-EGFP or EGFP. The KAT2B protein levels are shown as the relative values to control IgG. (A, D, F, G, H) Data are representative of three independent experiments. (B, C, E, I, J) Data represent the mean ± SD of three independent experiments. *P < 0.05. (A, D, H) Scale bar, 5 μm. Enlarged images of the region enclosed by a white dotted line are shown. (F, G) WCL; whole cell lysate. IP; immunoprecipitated.
ZFAT recruits the histone acetyltransferase KAT2B to centromeres
The levels of histone acetylation are tightly regulated by the balance between histone acetyltransferases (KATs) and histone deacetylases. To elucidate the mechanism through which ZFAT induces centromeric H4K8ac, we examined the effects of ZFAT overexpression on the distribution patterns of KATs in HEK293 cells by immunofluorescence analysis. Intriguingly, the overexpression of ZFAT caused the foci formation of KAT2B in the proximity of the ZFAT foci (Figure 4D and E). In contrast, the distribution patterns of other HATs examined in this study were apparently unaffected by the overexpression of ZFAT, compared with those in cells expressing EGFP (Supplementary Figure S9). KAT2B was reported to possess acetyltransferase activity for different sites of histones H3 and H4 (43–46). To examine whether ZFAT recruits KAT2B to centromeres, we investigated the interaction between mZfat and KAT2B by Co-IP analysis. As shown in Figure 4F, endogenous mZfat was bound to endogenous KAT2B in the thymocytes and splenic CD4+ T cells obtained from mice. Similarly, ectopically overexpressed hZFAT-HA was bound to FLAG-tagged KAT2B (FLAG-KAT2B) in HEK293 cells (Figure 4G). These results indicate that ZFAT interacts with KAT2B. Furthermore, we examined the mechanism of interaction between ZFAT and KAT2B by Co-IP analysis of FLAG-KAT2B and the deletion mutants of hZFAT-HA (Supplementary Figures S10A and S10B). Although the deletion mutants of the N-terminal regions of ZFAT (ZF-ΔN-1, 2 and 3) retained the ability to interact with KAT2B, the deletion of the C-terminal regions of ZFAT markedly diminished the interaction with KAT2B (ZF-ΔC-1, 2 and 3, Supplementary Figures S10A and S10B). These results indicate that the C-terminal region of ZFAT is involved in the interaction with KAT2B. Importantly, co-expression of HA-hZFAT-EGFP and FLAG-KAT2B resulted in the formation of KAT2B foci, which were colocalized with the foci of ZFAT and CENP-A (Figure 4H and I). Furthermore, ChIP-qPCR analysis using an anti-KAT2B antibody revealed that the overexpression of ZFAT resulted in a significant increase in the levels of KAT2B protein at centromeric α-satellite DNA, but not at other repetitive sequences (Figure 4J). Thus, these results suggest that ZFAT recruits KAT2B to centromeres through its C-terminal region.
ZFAT induces centromeric H4K8ac by recruiting KAT2B to promote the transcription of ncRNA
To explore the involvement of KAT2B in ZFAT-induced H4K8ac at centromeres, we used siRNA for the depletion of KAT2B in HEK293 cells overexpressing HA-hZFAT-EGFP, and examined the levels of H4K8ac by ChIP-qPCR analysis. Loss of KAT2B expression was confirmed by immunoblotting analysis using an anti-KAT2B antibody (Figure 5A). Depletion of KAT2B substantially suppressed the overexpressed ZFAT-induced increase in the levels of H4K8ac at centromeric α-satellite DNA (Figure 5B), indicating that KAT2B is required for the ZFAT-induced H4K8ac at centromeres. Furthermore, we examined the ability of the ZFAT deletion mutants to induce the formation of H4K8ac foci through immunofluorescence analysis. The ZFAT C-terminal deletion mutants, which did not interact with KAT2B, failed to induce the formation of H4K8ac foci (ZF-ΔC-2 and -3, Figure 5C and D) although they retained the ability to form the centromeric foci (Supplementary Figures S7A and S7B). These results indicate that the ZFAT C-terminal region, which is involved in the interaction with KAT2B, is required for the ability of ZFAT to induce centromeric H4K8ac. Collectively, these results suggest that ZFAT induces centromeric H4K8ac by recruiting KAT2B.
Figure 5.
ZFAT induces centromeric H4K8ac by recruiting KAT2B to promote the transcription of ncRNA. (A) Immunoblotting analysis of ZFAT (HA) and KAT2B in HEK293 cells transfected with indicated expression vectors and siRNAs. N.S.; non-specific band. (B) ChIP-qPCR analysis of H4K8ac at centromeric α-satellite DNA using an anti-H4K8ac antibody or control IgG in HEK293 cells transfected with indicated expression vectors and siRNAs. The H4K8ac levels are shown as the ratio of HA-hZFAT-EGFP-expressing cells against EGFP-expressing cells. (C) Immunofluorescence images of ZFAT (HA) and H4K8ac in HEK293 cells expressing the full-length or deletion mutants of hZFAT-HA. Scale bar, 5 μm. (D) Percentage of cells containing centromeric H4K8ac foci in cells expressing the full length or deletion mutants of hZFAT-HA, determined in (C). (E) qRT-PCR analysis of centromeric ncRNA in HEK293 cells transfected with indicated expression vectors and siRNAs. The RNA expression levels are shown as the relative values against those in cells transfected with control siRNA and HA-hZFAT-EGFP expression vector. (F) qRT-PCR analysis of centromeric ncRNA in HEK293 cells expressing EGFP, or the full-length or deletion mutants of hZFAT. The RNA expression levels are shown as the relative values against those in cells expressing EGFP-hZFAT. (A, C) Data are representative of three independent experiments. (B, D, E, F) Data represent the mean ± SD of three independent experiments. *P < 0.05. (E, F) The RNA expression levels are normalized to those of β-actin.
To elucidate the role of KAT2B-mediated H4K8ac in the centromeric transcription of ncRNA, we examined the effects of KAT2B knockdown on the levels of centromeric ncRNA in HEK293 cells overexpressing HA-hZFAT-EGFP. Depletion of KAT2B significantly inhibited the overexpressed ZFAT-induced increase in the levels of centromeric ncRNA, (Figure 5E), demonstrating that KAT2B is required for the ZFAT-regulated centromeric transcription of ncRNA. Furthermore, the ZFAT C-terminal deletion mutants, which did not induce the formation of H4K8ac foci, failed to cause an increase in the levels of centromeric ncRNA (Figure 5F). These results suggest that ZFAT controls centromeric ncRNA transcription through KAT2B-mediated H4K8ac.
ZFAT recruits BRD4 to centromeres through KAT2B-mediated H4K8ac to promote the transcription of ncRNA
Histone acetylation has the ability to recruit particular proteins, such as bromodomain-containing proteins that modulate transcription (47). Through immunofluorescence analysis, we found that the overexpression of ZFAT resulted in the formation of foci of the bromodomain-containing protein BRD4, compared with that observed in neighboring non-transfected cells (Figure 6A). The overexpressed ZFAT-induced BRD4 foci were colocalized with the foci of ZFAT and CENP-A (Figure 6B and C). Furthermore, we performed ChIP-qPCR analysis of BRD4 in HEK293 cells transiently overexpressing HA-hZFAT-EGFP or EGFP. The overexpression of ZFAT resulted in a significant increase in the levels of BRD4 protein at centromeric α-satellite DNA, but not at other repetitive sequences (Figure 6D). These results indicate that ZFAT recruits BRD4 to centromeres.
Figure 6.
ZFAT recruits BRD4 to centromeres. (A) Immunofluorescence images of ZFAT (HA) and BRD4 in HEK293 cells transiently expressing hZFAT-HA. The arrowhead indicates a transfected cell, as determined by the anti-HA antibody signal. (B) Immunofluorescence images of ZFAT, BRD4, CENP-A and DNA (DAPI) in HEK293 cells transiently expressing HA-hZFAT-EGFP. Enlarged images of the region enclosed by a white dotted line are shown. (C) Percentage of cells containing centromeric BRD4 foci in cells expressing EGFP or HA-hZFAT-EGFP, determined in (B). (D) ChIP-qPCR analysis of BRD4 at human repetitive DNA sequences using an anti-BRD4 antibody or control IgG in HEK293 cells expressing HA-hZFAT-EGFP or EGFP. The BRD4 protein levels are shown as the relative values to control IgG. (A, B) Scale bar, 5 μm. Data are representative of three independent experiments. (C, D) Data represent the mean ± SD of three independent experiments. *P < 0.05.
BRD4 contains two bromodomains, BD1 and BD2, which bind to acetylated lysines in histones H3 and H4, in the N-terminal region (Figure 7A) (47). To elucidate the mechanism through which ZFAT recruits BRD4 to centromeres, we examined the ZFAT-induced foci formation of the BRD4 deletion mutants through immunofluorescence analysis (Figure 7A–C). Deletion of BD1 (EGFP-ΔBD1) considerably suppressed the ZFAT-induced foci formation of BRD4. Furthermore, the double deletion of BD1 and BD2 (EGFP-ΔBD1+2) completely prevented the foci formation of BRD4 caused by the expression of ZFAT, indicating that the BD1 and BD2 domains are indispensable for the ZFAT-induced recruitment of BRD4 to centromeres. On the other hand, ZFAT weakly induced foci formation of EGFP containing only the BD1 domain (nlsEGFP-BD1). Furthermore, the construct expressing EGFP containing both BD1 and BD2 domains (EGFP-BD1+2) showed significant ability for foci formation, indicating that the BD1 and BD2 domains are sufficient for the ZFAT-induced recruitment of BRD4 to centromeres. Next, we examined the effects of the BRD4 inhibitor JQ1, which specifically inhibits the interaction between the BRD4 bromodomains and acetylated lysines, on the BRD4 localization in cells expressing HA-hZFAT-EGFP. Treatment with JQ1 completely blocked the overexpressed ZFAT-induced foci formation of BRD4 without affecting the ZFAT foci (Figure 7D and E). These results indicate that ZFAT recruits BRD4 to centromeres through interaction between BRD4 bromodomains and acetylated lysines.
Figure 7.
ZFAT recruits BRD4 to centromeres through histone acetylation. (A) Schematic diagram of the BRD4 constructs used in (B). (B) Immunofluorescence images in HEK293 cells transiently expressing mCherry-hZFAT, and the full-length or deletion mutants of EGFP-BRD4. Enlarged images of the region enclosed by a white dotted line are shown. nls; nuclear localization signal. (C) Percentage of cells containing centromeric BRD4 foci in cells transfected with indicated expression vectors, determined in (B). (D) Immunofluorescence images of ZFAT (EGFP) and BRD4 in HEK293 cells expressing EGFP-hZFAT, treated with JQ1 or vehicle (Control). (E) Percentage of cells containing centromeric BRD4 foci in cells expressing EGFP-hZFAT, treated with JQ1 or vehicle, determined in (D). (B, D) Data are representative of three independent experiments. Scale bar, 5 μm. (C, E) Data represent the mean ± SD of three independent experiments.
To examine the role of KAT2B-mediated H4K8ac in the ZFAT-induced recruitment of BRD4 to centromeres, we used siRNA for the depletion of KAT2B in HEK293 cells overexpressing HA-hZFAT-EGFP. We examined the levels of centromeric BRD4 by ChIP-qPCR analysis. Depletion of KAT2B significantly suppressed the overexpressed ZFAT-induced increase in the levels of BRD4 at centromeric α-satellite DNA (Figure 8A), indicating that KAT2B is required for the ZFAT-induced recruitment of BRD4. Furthermore, the deletion mutants of ZFAT, which did not interact with KAT2B nor induce centromeric H4K8ac (ZF-ΔC-2 and -3), failed to induce the foci formation of BRD4 at centromeres (Figure 8B and C). These results suggest that KAT2B-mediated H4K8ac is involved in the ZFAT-induced recruitment of BRD4 to centromeres. Next, we examined the effects of ZFAT knockdown on the expression levels of KAT2B and BRD4 through immunoblotting analysis (Figure 8D). Depletion of ZFAT did not affect the expression levels of KAT2B or BRD4, suggesting that ZFAT is not involved in the regulation of KAT2B or BRD4 expression. Thus, these results show that ZFAT recruits BRD4 to centromeres through interaction between the BRD4 bromodomains and KAT2B-mediated H4K8ac.
Figure 8.
ZFAT recruits BRD4 to centromeres through KAT2B-mediated H4K8ac to promote the transcription of ncRNA. (A) ChIP-qPCR analysis of BRD4 at human repetitive DNA sequences using an anti-BRD4 antibody or control IgG in HEK293 cells transfected with indicated expression vectors and siRNAs. The BRD4 protein levels are shown as the relative values to control IgG. (B) Immunofluorescence images of ZFAT (HA) and BRD4 in HEK293 cells expressing the full-length or deletion mutants of hZFAT-HA. Enlarged images of the region enclosed by a white dotted line are shown. Scale bar, 5 μm. (C) Percentage of cells containing centromeric BRD4 foci in cells expressing the full length or deletion mutants of hZFAT-HA, determined in (B). (D) Immunoblotting analysis of ZFAT, KAT2B and BRD4 in HEK293 cells transfected with siRNAs for control or ZFAT (#1 and #5). (E) qRT-PCR analysis of centromeric ncRNA in HEK293 cells expressing EGFP or HA-hZFAT-EGFP (ZFAT), treated with JQ1 or vehicle. The RNA expression levels are shown as the relative values against those in vehicle-treated cells expressing HA-hZFAT-EGFP. (F) Immunoblotting analysis of ZFAT (HA) and BRD4 in HEK293 cells transfected with indicated expression vectors and siRNAs. N.S.; non-specific band. (G) qRT-PCR analysis of centromeric ncRNA in HEK293 cells transfected with indicated expression vectors and siRNAs. The RNA expression levels are shown as the relative values against those in cells transfected with control siRNA and HA-hZFAT-EGFP expression vector. (H) qRT-PCR analysis of centromeric ncRNA in HEK293 cells expressing EGFP or HA-hZFAT-EGFP (ZFAT), treated with α-amanitin or vehicle for 3 h. The RNA expression levels are shown as the relative values against those in vehicle-treated cells expressing HA-hZFAT-EGFP. (B, D, F) Data are representative of three independent experiments. (A, C, E, G, H) Data represent the mean ± SD of three independent experiments. *P < 0.05. (E, G, H) The RNA expression levels are normalized to those of β-actin.
Although BRD4 regulates the mRNA transcription of numerous genes through RNAPII (48–50), the role of BRD4 at centromeres has not been explored. Therefore, we examined the involvement of BRD4 and RNAPII in the ZFAT-regulated centromeric ncRNA transcription. Treatment with the BRD4 inhibitor JQ1 significantly suppressed the overexpressed ZFAT-induced increase in the centromeric ncRNA levels (Figure 8E). Furthermore, siRNA-mediated knockdown of BRD4 significantly reduced the levels of centromeric ncRNA in HEK293 cells overexpressing HA-hZFAT-EGFP (Figure 8F and G). The RNAPII inhibitor α-amanitin significantly inhibited the overexpressed ZFAT-induced increase in the levels of centromeric ncRNA (Figure 8H). These results suggest that ZFAT promotes the RNAPII-dependent centromeric ncRNA transcription by recruiting BRD4.
ZFAT is involved in chromosome segregation
Centromeric ncRNA transcription is important for chromosome segregation (51–54). To elucidate the roles of ZFAT in chromosome segregation, we used siRNA for the depletion of ZFAT, and examined its effects on chromosome segregation. Immunofluorescence analysis using an anti-α-tubulin antibody revealed that abnormal spindle morphology was observed in cells transfected with siRNAs for ZFAT, but not in cells transfected with control siRNA (Figure 9A and B), indicating that loss of ZFAT results in chromosome segregation error. These results suggest that ZFAT plays important roles in chromosome segregation. Elucidating functional significance of the ZFAT-regulated centromeric transcription in chromosome segregation will be addressed in future studies.
Figure 9.

ZFAT is involved in chromosome segregation. (A) Immunofluorescence images of α-tubulin and DAPI in HT1080 cells transfected with indicated siRNAs. Data are representative of three independent experiments. Scale bar, 5 μm. (B) Percentage of cells with abnormal spindle morphology in mitotic cells transfected with indicated siRNAs, determined in (A). Data represent the mean ± SD of three independent experiments. (C) Model of ZFAT-regulated centromeric ncRNA transcription through the KAT2B–H4K8ac–BRD4 axis.
DISCUSSION
Recent studies have shown that centromeric transcription and its ncRNA products play important roles in the formation and functions of centromeres (51–54). On the other hand, the regulatory mechanism of this process at the molecular level remained unknown. In this study, we identified ZFAT as a crucial transcriptional regulator for centromeric ncRNA, and defined the molecular mechanism of ZFAT-regulated centromeric transcription. ZFAT recruits KAT2B to the centromeres for the induction of H4K8ac, leading to the recruitment of BRD4 for the activation of RNAPII-dependent ncRNA transcription. Furthermore, we showed that ZFAT binds to centromeres through the ZFAT box. Thus, we propose that ZFAT binds to the centromere to control ncRNA transcription at specific chromosomes through the KAT2B-H4K8ac-BRD4 axis (Figure 9C). These findings would lead to a better understanding of the functional significance of centromeric ncRNA transcription.
ZFAT-binding domains at α-satellite DNA regions
Here we found that ZFAT binds to α-satellite DNA regions through the ChIP-seq analysis of ectopically overexpressed ZFAT. Human centromeres are built on arrays of a 171-bp α-satellite DNA monomer, which is tandemly arranged in a head-to-tail fashion to form a HOR (55–57). Sequence assembly across α-satellite DNA regions at human centromeres is difficult due to high sequence similarities with the HOR and variability between individuals. Nevertheless, in the latest hg38 human genome assembly, centromere regions have been filled with so-called ‘reference models’, which are somewhat arbitrary representations of α-satellite DNA regions. Reference models are not real DNA sequences like traditional GenBank contigs, but are collections of all reads from whole-genome shotgun sequencing that match a certain HOR (58,59). These models are the most prevailing and informative sequences that are available at the moment as references for α-satellite DNA regions.
In mapping of reads to highly repetitive sequences, reads, which show the equal mapping probability to multiple regions due to their high sequence similarities, are randomly assigned to somewhere in the repetitive sequences, as Nechemia-Arbely et al. described (60). The alignments are distributed across the array when there are numerous such reads in ChIP-seq data, as we observed in the hZFAT ChIP-seq analysis (Figure 1A). Therefore, it is difficult to distinguish that ZFAT binds to a whole or certain limited region at centromeres through ChIP-seq analysis. On the other hand, the binding of mZfat is observed not only at centromeres but also at pericentromeres, determined by the ChIP-qPCR analysis of endogenous mZfat (Figure 1D). Furthermore, immunofluorescence analysis of ZFAT revealed that ZFAT forms foci, which are partially colocalized with those of CENP-A or CENP-B (Figure 1E and G). These results suggest that ZFAT binds to centromeres and the surrounding regions including pericentromeres. To determine the exact ZFAT-binding domains at centromeres, further progress in mapping algorithm for ChIP-seq analysis and determination of exact DNA sequences of centromere regions are required.
ZFAT is a novel transcriptional regulator of centromeric ncRNA
Studies in yeast have identified various molecules involved in centromeric transcription, including Cbf1, Dig1, Ste12 and Ams2 (17,61). In contrast, the molecular mechanism of centromeric transcription in mammalian cells remains to be elucidated. The CTDP1 (C-terminal domain, RNA polymerase II, polypeptide A phosphatase, subunit 1) is reported to be the only transcription factor localizing at the centromere in human and mouse cells (10). Recently, the mitotic kinase Bub1 was reported to recruit RNAPII to the centromere through histone H2A phosphorylation in human cells (24). However, the mechanism through which the transcriptional activity of RNAPII at the centromere is regulated remains unknown. We showed that ZFAT recruits BRD4 to the centromere through the KAT2B-mediated H4K8ac (Figures 7 and 8). Importantly, BRD4 activates RNAPII to regulate the mRNA transcription of numerous genes (48–50). Thus, ZFAT-recruited BRD4 could activate the transcriptional activity of RNAPII at the centromere.
Here, we showed that ZFAT expression induces centromeric ncRNA transcription only at specific chromosomes although the centromeric binding of ZFAT is observed at every chromosome (Figures 1B and 3F). Furthermore, there are centromeric regions to which ZFAT binds but does not induce ncRNA transcription (Figure 3E). Thus, only centromeric binding of ZFAT is not sufficient for the induction of ncRNA transcription. Interestingly, chromosome groups containing specific SFs have a unique correlation between the centromeric ZFAT-binding and ncRNA levels (Figure 3G). Chromosomes containing SF1 or SF2 monomers show the relatively low centromeric ncRNA levels. On the other hand, all chromosomes containing SF3 monomers (chromosomes 11, 17 and X) show the clearly high centromeric ncRNA levels (Figure 3G). The SF3, which consists of W1-5 monomers, represents a pentameric HOR configuration (41). The unique nucleotide sequences or linear order of W1-5 monomers in the HOR may be responsible for the significant increase in centromeric ncRNA levels caused by ZFAT overexpression. Furthermore, we also observed the high centromeric ncRNA levels at the Y chromosome in cells overexpressing hZFAT (Figure 3F). The Y chromosome uniquely contains SF4 monomers at centromeres (38). Interestingly, the chromosome 15, which is classified into SF2, shows the obviously higher centromeric ncRNA levels, compared with those of many other chromosomes containing SF2 monomers (Figure 3G). It has been reported that the chromosome 15 centromeres also contain quite many SF4 monomers although they mainly consist of the HOR containing SF2 monomers (42), suggesting that SF4 monomers may contribute to the significantly high levels of centromeric ncRNA at the chromosomes 15 and Y. Elucidating sequence elements in SF3 and SF4 monomers involved in the centromeric ncRNA transcription will be addressed in future studies.
Several studies have shown that RNAPII localizes at centromeres at M-phase (10,12). The RNAPII foci at mitotic centromeres can be detected through immunofluorescence analysis probably because most of the non-centromeric regions are transcriptionally inactive during mitosis. In contrast, at interphase, RNAPII actively transcribes throughout the genome, making it difficult to observe the centromeric RNAPII foci at interphase, even when centromeric transcription is active. Indeed, Bobkov et al. have shown that ectopically expressed RNAPII is detected at centromeres at G1-phase in fly cells (8). Furthermore, McNulty et al. have shown that the centromeric transcript levels are not different between M and G1-phases in human cells (16). These reports suggest that centromeres are transcribed at interphase, as well as during mitosis. Our conclusions in this study are based on experiments using asynchronized cells, suggesting that ZFAT controls the centromeric transcription at interphase. Elucidating roles of ZFAT in centromeric transcription at each cell-cycle phase will be addressed in future studies.
Here, we show that endogenous ZFAT binds to the centromeres (Figure 1D and E), and interacts with endogenous KAT2B protein (Figure 4F). Furthermore, siRNA-mediated depletion of endogenous ZFAT significantly reduces the centromeric ncRNA levels (Figure 3I), and results in chromosome segregation error (Figure 9A and B). These results support that endogenous ZFAT binds to centromeres to control ncRNA transcription through KAT2B. On the other hand, we show that H4K8ac and BRD4 are involved in centromeric ncRNA transcription through ectopic overexpression of ZFAT (Figures 4–8). Thus, there is a possibility that non-physiological effects caused by overexpressed ZFAT may change the H4K8ac levels and BRD4 localization at the centromeres. Further verification of the involvement of H4K8ac and BRD4 in the ZFAT-regulated centromeric ncRNA transcription awaits future studies.
Histone acetylation plays important roles in the formation and functions of centromeres through the transcription of ncRNA
Centromere nucleosomes show unique histone methylation patterns, including dimethylation of lysine 4 (H3K4me2) and lysine 36 (H3K36me2) (7,62). On the other hand, the acetylation of histone H3 and H4 is in low abundance at centromeres in human and fly cells (7). However, it has been reported that the acetylation of histone H3 at HAC centromeres is involved in the formation and function of centromeres (63–66). Furthermore, it was recently reported that acetylation of histone H4 facilitated efficient establishment of centromeres in Caenorhabditis elegans (67). In this study, we define the molecular mechanism of histone acetylation at centromeres where ZFAT specifically induces the acetylation at a particular lysine residue in histone H4 by recruiting KAT2B (Figures 4 and 5). Furthermore, ZFAT-induced histone H4 acetylation is required for centromeric ncRNA transcription (Figure 5). Therefore, histone acetylation plays important roles in the formation and functions of centromeres through ncRNA transcription.
H4K8ac may be broadly involved in ncRNA transcription at both centromeres and pericentromeres
The lysine 8 in histone H4 is one of the four acetylatable lysines at the N-terminus. It was previously reported that heat shock in human cells triggered the enrichment of H4K8ac at stress bodies, which were formed on the pericentromeric heterochromatin domains (68). This induced the accumulation of heterogenous RNA molecules containing the subclass of satellite III sequences found in pericentromeric heterochromatin domains (68). Furthermore, BRD4 is also recruited to the stress bodies in response to heat shock (69). Thus, H4K8ac may be broadly involved in ncRNA transcription through the recruitment of BRD4 not only at centromeres but also at pericentromeres. Given that ZFAT binds to both centromeres and pericentromeres (Figure 1D), ZFAT may play a role in ncRNA transcription at pericentromeres through the KAT2B–H4K8ac–BRD4 axis.
DATA AVAILABILITY
ChIP-seq and RNA-seq data have been deposited in NCBI GEO as GSE134612 and GSE145651, respectively.
Supplementary Material
Contributor Information
Shuhei Ishikura, Department of Cell Biology, Faculty of Medicine, Fukuoka University, Fukuoka 814-0180, Japan; Central Research Institute for Advanced Molecular Medicine, Fukuoka University, Fukuoka 814-0180, Japan.
Kazuhiko Nakabayashi, Department of Maternal-Fetal Biology, National Research Institute for Child Health and Development, Tokyo 157-8535, Japan.
Masayoshi Nagai, Central Research Institute for Advanced Molecular Medicine, Fukuoka University, Fukuoka 814-0180, Japan.
Toshiyuki Tsunoda, Department of Cell Biology, Faculty of Medicine, Fukuoka University, Fukuoka 814-0180, Japan; Central Research Institute for Advanced Molecular Medicine, Fukuoka University, Fukuoka 814-0180, Japan.
Senji Shirasawa, Department of Cell Biology, Faculty of Medicine, Fukuoka University, Fukuoka 814-0180, Japan; Central Research Institute for Advanced Molecular Medicine, Fukuoka University, Fukuoka 814-0180, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for the FCAM from MEXT-Supported Program for the Strategic Research Foundation at Private Universities [S0801082 to S.S.]; JSPS KAKENHI [20K07317 to S.I.]; Fukuoka University [925]. Funding for open access charge: Fukuoka University [925].
Conflict of interest statement. None declared.
REFERENCES
- 1. Muller S., Almouzni G.. Chromatin dynamics during the cell cycle at centromeres. Nat. Rev. Genet. 2017; 18:192–208. [DOI] [PubMed] [Google Scholar]
- 2. Henikoff S., Ahmad K., Platero J.S., van Steensel B.. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McKinley K.L., Cheeseman I.M.. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016; 17:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vafa O., Sullivan K.F.. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 1997; 7:897–900. [DOI] [PubMed] [Google Scholar]
- 5. Blower M.D., Sullivan B.A., Karpen G.H.. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002; 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C.C., Mellone B.G.. Chromatin assembly: journey to the CENter of the chromosome. J. Cell Biol. 2016; 214:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sullivan B.A., Karpen G.H.. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004; 11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bobkov G.O.M., Gilbert N., Heun P.. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol. 2018; 217:1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Catania S., Pidoux A.L., Allshire R.C.. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLos Genet. 2015; 11:e1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H., Wong L.H.. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl Acad. Sci. U.S.A. 2012; 109:1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi E.S., Stralfors A., Castillo A.G., Durand-Dubief M., Ekwall K., Allshire R.C.. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011; 286:23600–23607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosic S., Kohler F., Erhardt S.. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014; 207:335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blower M.D. Centromeric transcription regulates Aurora-B localization and activation. Cell Rep. 2016; 15:1624–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bouzinba-Segard H., Guais A., Francastel C.. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl Acad. Sci. U.S.A. 2006; 103:8709–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carone D.M., Longo M.S., Ferreri G.C., Hall L., Harris M., Shook N., Bulazel K.V., Carone B.R., Obergfell C., O’Neill M.J. et al.. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009; 118:113–125. [DOI] [PubMed] [Google Scholar]
- 16. McNulty S.M., Sullivan L.L., Sullivan B.A.. Human centromeres produce Chromosome-Specific and Array-Specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell. 2017; 42:226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohkuni K., Kitagawa K.. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 2011; 21:1695–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topp C.N., Zhong C.X., Dawe R.K.. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quenet D., Dalal Y.. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife. 2014; 3:e03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du Y., Topp C.N., Dawe R.K.. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010; 6:e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L., Hannan R., Saffery R., Shaw M.L., Williams E. et al.. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007; 17:1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferri F., Bouzinba-Segard H., Velasco G., Hube F., Francastel C.. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009; 37:5071–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ideue T., Cho Y., Nishimura K., Tani T.. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells. 2014; 19:528–538. [DOI] [PubMed] [Google Scholar]
- 24. Liu H., Qu Q., Warrington R., Rice A., Cheng N., Yu H.. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell. 2015; 59:426–436. [DOI] [PubMed] [Google Scholar]
- 25. Chen C.C., Bowers S., Lipinszki Z., Palladino J., Trusiak S., Bettini E., Rosin L., Przewloka M.R., Glover D.M., O’Neill R.J. et al.. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-Mediated transcription. Dev. Cell. 2015; 34:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koyanagi M., Nakabayashi K., Fujimoto T., Gu N., Baba I., Takashima Y., Doi K., Harada H., Kato N., Sasazuki T. et al.. ZFAT expression in B and T lymphocytes and identification of ZFAT-regulated genes. Genomics. 2008; 91:451–457. [DOI] [PubMed] [Google Scholar]
- 27. Shirasawa S., Harada H., Furugaki K., Akamizu T., Ishikawa N., Ito K., Ito K., Tamai H., Kuma K., Kubota S. et al.. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum. Mol. Genet. 2004; 13:2221–2231. [DOI] [PubMed] [Google Scholar]
- 28. Tochio N., Umehara T., Nakabayashi K., Yoneyama M., Tsuda K., Shirouzu M., Koshiba S., Watanabe S., Kigawa T., Sasazuki T. et al.. Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT. J. Struct. Funct. Genomics. 2015; 16:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishikura S., Tsunoda T., Nakabayashi K., Doi K., Koyanagi M., Hayashi K., Kawai T., Tanaka Y., Iwaihara Y., Luo H. et al.. Molecular mechanisms of transcriptional regulation by the nuclear zinc-finger protein Zfat in T cells. Biochim. Biophys. Acta. 2016; 1859:1398–1410. [DOI] [PubMed] [Google Scholar]
- 30. Tsunoda T., Takashima Y., Tanaka Y., Fujimoto T., Doi K., Hirose Y., Koyanagi M., Yoshida Y., Okamura T., Kuroki M. et al.. Immune-related zinc finger gene ZFAT is an essential transcriptional regulator for hematopoietic differentiation in blood islands. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14199–14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doi K., Fujimoto T., Okamura T., Ogawa M., Tanaka Y., Mototani Y., Goto M., Ota T., Matsuzaki H., Kuroki M. et al.. ZFAT plays critical roles in peripheral T cell homeostasis and its T cell receptor-mediated response. Biochem. Biophys. Res. Commun. 2012; 425:107–112. [DOI] [PubMed] [Google Scholar]
- 32. Ishikura S., Ogawa M., Doi K., Matsuzaki H., Iwaihara Y., Tanaka Y., Tsunoda T., Hideshima H., Okamura T., Shirasawa S.. Zfat-deficient CD4(+) CD8(+) double-positive thymocytes are susceptible to apoptosis with deregulated activation of p38 and JNK. J. Cell. Biochem. 2015; 116:149–157. [DOI] [PubMed] [Google Scholar]
- 33. Ogawa M., Okamura T., Ishikura S., Doi K., Matsuzaki H., Tanaka Y., Ota T., Hayakawa K., Suzuki H., Tsunoda T. et al.. Zfat-deficiency results in a loss of CD3zeta phosphorylation with dysregulation of ERK and Egr activities leading to impaired positive selection. PLoS One. 2013; 8:e76254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 35. Ishikura S., Iwaihara Y., Tanaka Y., Luo H., Nishi K., Doi K., Koyanagi M., Okamura T., Tsunoda T., Shirasawa S.. The nuclear zinc finger protein Zfat maintains FoxO1 protein levels in peripheral T cells by regulating the activities of autophagy and the Akt signaling pathway. J. Biol. Chem. 2016; 291:15282–15291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwaihara Y., Ishikura S., Doi K., Tsunoda T., Fujimoto T., Okamura T., Shirasawa S.. Marked reduction in FoxO1 protein by its enhanced proteasomal degradation in Zfat-deficient peripheral T-Cells. Anticancer Res. 2015; 35:4419–4423. [PubMed] [Google Scholar]
- 37. Nishi K., Iwaihara Y., Tsunoda T., Doi K., Sakata T., Shirasawa S., Ishikura S.. ROS-induced cleavage of NHLRC2 by caspase-8 leads to apoptotic cell death in the HCT116 human colon cancer cell line. Cell Death. Dis. 2017; 8:3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McNulty S.M., Sullivan B.A.. Alpha satellite DNA biology: finding function in the recesses of the genome. Chromosome Res. 2018; 26:115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Romanova L.Y., Deriagin G.V., Mashkova T.D., Tumeneva I.G., Mushegian A.R., Kisselev L.L., Alexandrov I.A.. Evidence for selection in evolution of alpha satellite DNA: the central role of CENP-B/pJ alpha binding region. J. Mol. Biol. 1996; 261:334–340. [DOI] [PubMed] [Google Scholar]
- 40. Rosandic M., Paar V., Basar I., Gluncic M., Pavin N., Pilas I.. CENP-B box and pJalpha sequence distribution in human alpha satellite higher-order repeats (HOR). Chromosome Res. 2006; 14:735–753. [DOI] [PubMed] [Google Scholar]
- 41. Alexandrov I.A., Medvedev L.I., Mashkova T.D., Kisselev L.L., Romanova L.Y., Yurov Y.B.. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 1993; 21:2209–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shepelev V.A., Uralsky L.I., Alexandrov A.A., Yurov Y.B., Rogaev E.I., Alexandrov I.A.. Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom Data. 2015; 5:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y., Ge K.. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011; 30:249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Agalioti T., Chen G., Thanos D.. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002; 111:381–392. [DOI] [PubMed] [Google Scholar]
- 45. Ito K., Barnes P.J., Adcock I.M.. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol. 2000; 20:6891–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schiltz R.L., Mizzen C.A., Vassilev A., Cook R.G., Allis C.D., Nakatani Y.. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999; 274:1189–1192. [DOI] [PubMed] [Google Scholar]
- 47. Taniguchi Y. The Bromodomain and Extra-Terminal Domain (BET) Family: Functional anatomy of BET paralogous proteins. Int. J. Mol. Sci. 2016; 17:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A., Rosenfeld M.G.. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013; 155:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanno T., Kanno Y., LeRoy G., Campos E., Sun H.W., Brooks S.R., Vahedi G., Heightman T.D., Garcia B.A., Reinberg D. et al.. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 2014; 21:1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K.. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005; 19:523–534. [DOI] [PubMed] [Google Scholar]
- 51. Chan F.L., Wong L.H.. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 2012; 40:11178–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hall L.E., Mitchell S.E., O’Neill R.J.. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res. 2012; 20:535–546. [DOI] [PubMed] [Google Scholar]
- 53. Perea-Resa C., Blower M.D.. Centromere Biology: transcription goes on stage. Mol. Cell. Biol. 2018; 38:e00263–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Talbert P.B., Henikoff S.. Transcribing centromeres: noncoding RNAs and kinetochore assembly. Trends Genet. 2018; 34:587–599. [DOI] [PubMed] [Google Scholar]
- 55. Choo K.H., Vissel B., Nagy A., Earle E., Kalitsis P.. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991; 19:1179–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mitchell A.R., Gosden J.R., Miller D.A.. A cloned sequence, p82H, of the alphoid repeated DNA family found at the centromeres of all human chromosomes. Chromosoma. 1985; 92:369–377. [DOI] [PubMed] [Google Scholar]
- 57. Schueler M.G., Sullivan B.A.. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 2006; 7:301–313. [DOI] [PubMed] [Google Scholar]
- 58. Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., Kent W.J.. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014; 24:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al.. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015; 43:D670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nechemia-Arbely Y., Miga K.H., Shoshani O., Aslanian A., McMahon M.A., Lee A.Y., Fachinetti D., Yates J.R. 3rd, Ren B., Cleveland D.W.. DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat. Cell Biol. 2019; 21:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen E.S., Saitoh S., Yanagida M., Takahashi K.. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003; 11:175–187. [DOI] [PubMed] [Google Scholar]
- 62. Bergmann J.H., Rodriguez M.G., Martins N.M., Kimura H., Kelly D.A., Masumoto H., Larionov V., Jansen L.E., Earnshaw W.C.. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011; 30:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bergmann J.H., Jakubsche J.N., Martins N.M., Kagansky A., Nakano M., Kimura H., Kelly D.A., Turner B.M., Masumoto H., Larionov V. et al.. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012; 125:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Molina O., Vargiu G., Abad M.A., Zhiteneva A., Jeyaprakash A.A., Masumoto H., Kouprina N., Larionov V., Earnshaw W.C.. Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat. Commun. 2016; 7:13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ohzeki J., Bergmann J.H., Kouprina N., Noskov V.N., Nakano M., Kimura H., Earnshaw W.C., Larionov V., Masumoto H.. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012; 31:2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ohzeki J., Shono N., Otake K., Martins N.M., Kugou K., Kimura H., Nagase T., Larionov V., Earnshaw W.C., Masumoto H.. KAT7/HBO1/MYST2 regulates CENP-A chromatin assembly by antagonizing Suv39h1-Mediated centromere inactivation. Dev. Cell. 2016; 37:413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhu J., Cheng K.C.L., Yuen K.W.Y.. Histone H3K9 and H4 acetylations and transcription facilitate the initial CENP-A(HCP-3) deposition and de novo centromere establishment in caenorhabditis elegans artificial chromosomes. Epigenet. Chromatin. 2018; 11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rizzi N., Denegri M., Chiodi I., Corioni M., Valgardsdottir R., Cobianchi F., Riva S., Biamonti G.. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell. 2004; 15:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Col E., Hoghoughi N., Dufour S., Penin J., Koskas S., Faure V., Ouzounova M., Hernandez-Vargash H., Reynoird N., Daujat S. et al.. Bromodomain factors of BET family are new essential actors of pericentric heterochromatin transcriptional activation in response to heat shock. Sci. Rep. 2017; 7:5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data have been deposited in NCBI GEO as GSE134612 and GSE145651, respectively.