Abstract
NBS1 is a critical component of the MRN (MRE11/RAD50/NBS1) complex, which regulates ATM- and ATR-mediated DNA damage response (DDR) pathways. Mutations in NBS1 cause the human genomic instability syndrome Nijmegen Breakage Syndrome (NBS), of which neuronal deficits, including microcephaly and intellectual disability, are classical hallmarks. Given its function in the DDR to ensure proper proliferation and prevent death of replicating cells, NBS1 is essential for life. Here we show that, unexpectedly, Nbs1 deletion is dispensable for postmitotic neurons, but compromises their arborization and migration due to dysregulated Notch signaling. We find that Nbs1 interacts with NICD-RBPJ, the effector of Notch signaling, and inhibits Notch activity. Genetic ablation or pharmaceutical inhibition of Notch signaling rescues the maturation and migration defects of Nbs1-deficient neurons in vitro and in vivo. Upregulation of Notch by Nbs1 deletion is independent of the key DDR downstream effector p53 and inactivation of each MRN component produces a different pattern of Notch activity and distinct neuronal defects. These data indicate that neuronal defects and aberrant Notch activity in Nbs1-deficient cells are unlikely to be a direct consequence of loss of MRN-mediated DDR function. This study discloses a novel function of NBS1 in crosstalk with the Notch pathway in neuron development.
INTRODUCTION
The DNA damage response (DDR), which includes cell cycle checkpoint activation, DNA repair, induction of senescence, apoptosis and transcription, safeguards genomic stability. It also has multifaceted functions in cellular processes and tissue homeostasis. Many key DDR and DNA repair molecules—including ATR, MRN, CHK1, TopBP1, BRCA1/2, RAD51, etc.—are essential for the life of cells and organisms, believed to be due to their crucial function in handling damages from replication stress and preventing cell death. Given the essentiality and the choice of cellular model systems applied to study DDR function, their role in non-dividing cells – neurons, for example – is largely masked.
The MRN complex consisting of MRE11, RAD50 and NBS1 (also known as Nibrin, p95), acts as a sensor of DNA double strand breaks (DSBs) to activate ATM-mediated DDR (1) and also resolves endogenous replication intermediates (2). This complex is recruited to damage sites through binding of MRE11 and RAD50 to damaged DNA, which is vital for activation of the protein kinase ATM that can phosphorylate many downstream substrates, including P53, CHK2, MDC1 and histone variant H2AX (3,4). MRE11 has both 3′-5′ exonuclease and ssDNA endonuclease activities, which are enhanced by RAD50 that holds the broken ends of DNA (5,6). Although the function of NBS1 in the assembly of MRN in DNA termini is less transparent, it is believed that the C-terminus of NBS1 interacts with MRE11 to facilitate its enzymatic activities in the MRN complex, to process broken DNA ends and mediate an essential step for repairing DSBs by non-homologous end joining (NHEJ) or homologous recombination (HR). Apart from its role in direct activation of ATM (7–9), NBS1 has been reported to activate ATR in response to single strand breaks (SSBs) or replication fork stalling (10–13). MRN is in the center of the DDR network and is essential for life; deletion of any component of MRN is lethal to cells and mice (2,14–16).
Mutations in genes encoding the MRN complex, or any other key DDR molecules, cause human genomic instability syndromes. These syndromes are characterized by many symptoms, amongst which neurological defects are common (17,18). For example, patients with Ataxia-Telangiectasia (A–T, mutations in ATM) or Ataxia-Telangiectasia-like disorder (A-TLD, mutations in MRE11) suffer from cerebellar degeneration and ataxia. Seckel Syndrome (mutations in ATR) or NBS (mutations in NBS1) patients present with microcephaly and intellectual disabilities (1,17,19). Although some of these neurological symptoms result from neuroprogenitor loss during brain development, others are likely due to dysfunction of postmitotic neuronal cells (17).
Brain development is strictly regulated by a concerted serial process of proliferation and differentiation of neuroprogenitors, migration of newborn neurons from origin to final destination, outgrowth of neurites from the soma, and synaptogenesis (20). In early embryonic brain development, neuroprogenitors located in the ventricular zone (VZ) of the neocortex undergo extensive expansion to establish the progenitor pool of the neocortex (21,22). The rapid proliferation of these neuroprogenitors generates a high level of DNA lesion encounters at replication forks, which request a robust DDR machinery (23,24). Neuroprogenitors are thus highly susceptible to defective DDR. The accumulation of damaged DNA in neuroprogenitors subsequently ceases proliferation and promotes apoptosis, resulting in neurodevelopmental disorders (17,23). In an attempt to delineate the function of NBS1 and the MRN complex responsible for neurological defect of human patients, we deleted Nbs1 in neural stem cells of the mouse central nervous system (CNS) (Nbs1-CNSΔ) and found cerebellar developmental defects due to a blockage of proliferation and increased apoptosis in proliferating neuroprogenitors. These defects are attributed to the loss of the DDR function of Nbs1 in activating the ATM-p53 axis (25–29). Considering the postnatal neural deficits of intellectual disabilities and cerebellar degeneration in NBS, A-TLD and ATR-Seckel patients (30), it is possible that DDR molecules—such as NBS1, MRE11, ATR—also effect an important role in postmitotic neuronal cells. To this end, RNA-sequencing databases of the developing mouse brain (31,32) reveals the expression of DDR genes NBS1, MRE11 and ATM in neurons, suggesting that these genes are biologically active in postmitotic neurons and may have functions in addition to their DDR role within replicating cells.
This study has endeavored to explore the biological function of the essential DDR molecule NBS1 during neuronal development and has found, unexpectedly, that NBS1 is not required for neuronal formation and survival; rather, it regulates postmitotic processes of neurons such as neurite arborization and neuronal migration, through modulating Notch activity. This novel function of NBS1 appears to be independent of its canonical DDR role, therein highlighting the physiological importance of this DDR molecule during neuronal development.
MATERIALS AND METHODS
Vector construction for shRNA knockdown and overexpression
The construction of shRNA expression vectors was performed as previously described (33). Briefly, oligonucleotides targeting the coding sequences and their complementary sequences were inserted into the vector under control of the human U6 promoter. U6 promoter-mediated shRNA expression cassettes were then sub-cloned into vectors with GFP or Tomato genes. All oligonucleotides contained the following hairpin loop sequence: ttcaagaga. The targeting sequences used were: shLuciferase: aatccctggtaatccgttg, shNbs1#1: gggccagccttgtacagaatt, shNbs1#2: gctccagtgaatatgaccacata, shMre11: ggactatagtggaggctttga, shRad50: gggcagacttaaagaagaaat, and shNotch1: gcagctatgagactgccaaag. For preparing shRNA-resisted GFP-rsNbs1, two fragments (F1 and F2) of Nbs1 were amplified with the following primers and digested with EcoRI/KpnI before insertion into pCAG-GFP (Plasmid #11150, Addgene, Cambridge, MA, USA). F1f (5′-cggaattcccgccaccatgtggaagctgctcc-3′) and F1r (5′-cacggttggccctgcggattacag-3′); F2f (5′-cacggaattaaagacaacgactcc-3′) and F2r (5′-gacggtaccgctcttctttttacattaggat-3′). For preparing shRNA-resisted GFP-Nbs1-N (1–330AA), GFP-rsNbs1 was used as a template and amplified by F1f and F3r (5′-gacGGTACCGtGcaCggctggccc-3′), PCR fragment was digested with EcoRI/KpnI before insertion into pCAG-GFP.
3xFlag-tagged full length (FL) Nbs1 (Nbs1FL) was constructed by amplifying Nbs1 cDNA using primers Nbs1-oligo15 (cggaattcatgtggaagctgctcc) and Nbs1-oligo13 (ccgctcgagttatcttctttttacattag), then inserted into the pCDNA3–3xFlag-A plasmid at the EcoRI/XhoI sites. Truncation fragments were created by Multi Site-Directed Mutagenesis Kit (QuickChange®, Agilent Technologies, Santa Clara, CA, USA) per manufacturer's instruction using, 3xFlag-Nbs1 as a template with the following primers – for deletion of N-terminal amino acids 24–330 (Nbs1Δ24–330): Nbs1-delN-5 (ccggcgtggagacagaattaaagacaacgactcc) and Nbs1-delN-3 (ctccacgccggccaaaagtcggtatggttctc); for deletion of mid-part amino acids 331 to 670 (Nbs1Δ331–670): Nbs1-delM-5 (ggccagccttgtaatctatgtgtaaatgaatgtgg) and Nbs1-delM-3 (acaaggctggccctgcggattacagtaattc); for deletion of C-terminal amino acids 671 to 752 (Nbs1Δ671–752): Nbs1 cDNA was amplified with Nbs1-oligo15 and Nbs1-delC-3 (gctctagatctggaggtggagttg), digested with EcoRI/XbaI before insertion into pCDNA3–3Flag-A. All constructs were confirmed by DNA sequencing.
Mice and genotyping
Nbs1-CNSΔ mice were generated as described previously (25). Inducible Nbs1 deletion mice were created as described in (26). All animals were maintained in the SPF facility and experiments conducted according to German animal welfare legislation. The genotypes were confirmed by PCR using primers, as follows. For Nbs1: exon6 (cagggcgacatgaaagaaaac), Intron5F (ataagacagtcaccactgcg) and LoxPtestR (aatacagtgactcctggagg); For Cre: Cre1 (cggtcgatgcaacgagtgatg) and Cre2 (ccagagacggaaatccatcgc).
mRNA isolation, RNA sequencing and semi-quantitative PCR
Total RNA was isolated by using Tri Reagent (T9424, Sigma-Aldrich, Munich, Germany) and used for library preparation by a TruSeq RNA Sample Prep Kit v2 (Illumina, Munich, Germany), per manufacturer's instruction. The libraries were sequenced with HiSeq2000 (Illumina) in single-read mode and RNA-seq reads of 50bp were mapped to the mouse genome (mm9) with TopHat2 (34). Differential expression analysis was performed by Cufflinks2 according to their protocol (35).
For real-time PCR, 1 μg of RNA was used for the synthesis of the first-strand cDNA by Affinity Script Multiple Temperature cDNA Synthesis Kit (200436, Agilent Technologies), per manufacturer's instruction. SYBR Green Master Mix (Invitrogen, Darmstadt, Germany) was used for real-time PCR reactions, each in triplicate, on a LightCycler 480II (Roche, Berlin, Germany). The relative differences in gene expression were calculated using the 2−ΔΔCt method (36) and normalized to untreated controls.
The following primers were used—for GAPDH: mGAPDH-F (gcacagtcaaggccgagaat) and mGAPDH-R (gccttctccatggtggtgaa); for Notch1: mNotch1-F (gctccgaggagatcaacgag) and mNotch1-R (ttgacatcaccctcacaccg); For Notch2: mNotch2-F (agcaggagcaggaggtgata) and mNotch2-R (tgggcgtttcttggactctc); mNotch3-F (gactgctcactgaacgtgga); for Notch3: mNotch3-R (cacaccggctgttgttgaag); for Notch4: mNotch4-F (acctgtgtgcctcagcccagt) and mNotch4-R (gggctgggactgacaagcgtc); for Hes1: mHes1-F (tcagcgagtgcatgaacga) and mHes1-R (tgcgcacctcggtgttaac); for Hes5: mHes5-F (cgcatcaacagcagcatagag) and mHes5-R (tggaagtggtaaagcagcttc); for Hey1: mHey1-F(cgtgagtgggatcagtgtgc) and mHey1-R (ctcgatgatgcctctccgtc); for Hey2: mHey2-F (ttctgtctctttcggccact) and mHey2-R (tttgtcccagtgcttgtctg); for p21: p21-F (gtcaggctggtctgcctccg) and p21-R (cggtcccgtggacagtgagcag).
Cells, cell culture, 4-OHT treatments, transfection and γ-irradiation treatment
The cell lines used in this study were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) high glucose, containing 10% fetal calf serum (FCS) and 1% penicillin/streptomycin. Mouse embryonic fibroblasts (MEFs) were isolated from wildtype or corresponding Nbs1 mutant mice and immortalized by a standard 3T3 protocol or by the shP19ARF-mediated protocol (a kind gift from Martin Eilers, Würzburg University, Germany). Primary MEFs were cultured at 37°C and 5% CO2 with 3% O2. For induced deletion of Nbs1, MEF cells were treated with 4-Hydroxytamoxifen (4-OHT, H6278, Sigma-Aldrich) for 4 days and harvested 2 (6dpo) or 4 days (8dpo) after withdrawal of the drug. MEFs were transfected using Amaxa Nucleofector Kit R (VCA-1001, Lonza, Cologne, Germany). Briefly, 1 × 106 cells MEFs were centrifuged and the cell pellet resuspended in 100 μl Nucleofector Solution mixture with 5 μg of plasmid DNA. The cell suspension was electroporated using Nucleofector I Device (Lonza). The GFP+ cells were sorted by flow cytometry 24 h after electroporation. The sorted cells were either used for protein extraction, mRNA isolation, or further cultured in the presence of 400 μg/ml of G418 (Invitrogen). HEK293T cells were transfected with polyethylenimine (PEI, Polyscience, Eppelheim, Germany). Neuro2A cells were transfected with GFP-shRNA expression plasmids in presence of PEI. Transfected cells were selected with 500 μg/ml G-418 (Invitrogen) 24 h post-transfection, for at least 24 h. Ionizing radiation (IR) of cells was completed using Cesium-137 from Gammacell 40 (GC40) Irradiator (MDS Nordion, Ottawa, Canada).
Primary neuron cultures and deletion of Nbs1
After euthanasing the pregnant female, mouse embryos (E15.5) were removed from the uterus. Embryos were decapitated and placed into a sterile Petri dish containing ice-cold GBSS (Gey's Balanced Salt Solution) with 0.5% glucose. Following removal of the skull and cerebral dura mater, dissected cerebral cortices were collected in ice-cold HBSS (Hank's Balanced Salt Solution) containing 0.05% glucose. The tissue was then incubated in 1× trypsin solution for 15 min at 37°C. Upon removing the supernatant, ice-cold plating medium (MEM with 0.5% glucose, 1 mM sodium pyruvate, 1% penicillin/streptomycin, 10 mM HEPES, 10% FCS, 1 mM l-glutamine and B-27 supplement (Thermo-Fisher Scientific, Karlsruhe, Germany) was added to inactivate the trypsin, the tissue re-suspended and the cell suspension filtrated through a nylon mesh. Cell density was determined and cells were seeded into laminin-poly-l-lysine-coated plates at a density of 1 × 105 cells/ml and cultured in plating medium. From the second day, cortical neurons were cultured in neuronal media (neuro-basal media (Invitrogen) containing B27 and 0.5 mM l-glutamine). For induced deletion of Nbs1, 1–2 days after culturing with neuronal media, cells were treated with 4-OHT for 4 days and harvested 2 (6dpo) or 4 days (8dpo) after withdrawal of the drug.
Neuro2A cell differentiation and drug treatment
Neuro2a cells were cultured in DMEM medium supplemented with 10% FCS. To induce differentiation, 24 h after transfection with Tomato-tagged-shRNA expression plasmid DNA, Neuro2A cells were cultured with DMEM supplemented with 2.5 mM Dibutyryladenosine 3′,5′-cyclic monophosphate (dbcAMP, Sigma-Aldrich), and/or DAPT inhibitors (10 μM of DAPT or L685, 458) for 24 h before fixation, for further analysis.
In utero electroporation (IUE)
In utero electroporation was performed as described previously (37). Briefly, 1 μg of plasmid DNA in PBS was injected into the lateral ventricle of E15.5 embryos and electroporated. Brains were isolated at postnatal day 16 (P16) or P26 and fixed in 4% PFA for cryosection, immunostaining and imaging.
Transwell migration assay
Neuronal transwell migration assays were performed using Neuro2A cell line and primary neuronal cells isolated from indicated embryos. 1.5 × 105 cells were transferred in 300 μl serum-free medium into the upper chamber of a 12-well chemotaxis insert (ThinCert™, 8 μm pores; Greiner-Bio-One GmbH, Frickenhausen, Germany). The chamber was placed in 700 μl medium containing 10% FCS and incubated in a tissue culture incubator for 20 h at 37°C with 5% CO2. Cells on the underside of the filter membrane were then fixed with 4% paraformaldehyde (15 min) and stained with DAPI solution for 5 min (1:1000 in 1× PBS), counted from five independent areas under a fluorescence microscope and normalized to the control. For Notch inhibitor treatment, 40 hr after transfection, cells were suspended in starvation medium together, with or without Notch inhibitors (10 μM of DAPT or L685, 458) and plated onto the transwell membrane.
Immunostaining and imaging
Immunocytochemistry was performed as described previously (37). Briefly, PFA-fixed cells were incubated with blocking solution (1% BSA, 5% goat serum and 0.4% Triton X-100 in PBS) for 1 h at room temperature and incubated with a primary antibody diluted in blocking solution at 4°C overnight, washed with PBS, followed by incubation with secondary antibodies for 2 h at room temperature. After washing three times with PBS, the coverslips were mounted on glass slides with DAPI-containing mounting medium (Invitrogen). For immunohistochemistry, brains were isolated and transferred into 4% PFA solution overnight at 4°C for fixation. Fixed tissue was cryoprotected 2–3 days with 30% (w/v) sucrose in PBS embedded in OCT compound (NEG-50™, Thermo-Fisher Scientific). Sections of 10 μm thickness were cut on a cryostat (Leica, Wetzlar, Germany), mounted on glass-slides and stored at −20°C until staining. Brain sections were washed three times with PBS, prior to incubation with the blocking solution for 1 h at room temperature and then incubated with primary antibodies at 4°C overnight. For immunofluorescence detection, the bound antibodies were visualized using fluorescent-dye conjugated secondary antibodies (Invitrogen). Samples were mounted with glass coverslips with ProLong Gold antifade (Invitrogen) mounting medium containing DAPI to counterstain for DNA. For TUNEL staining, brain sections were sub-boiled with an antigen retrieval buffer (10 mM sodium citrate, pH 6.0) in a microwave for 10 min. After 30 min at room temperature, the terminal deoxynucleotidyl transferase (TdT) (Fermentas, St. Leon-Rot, Germany) reaction was conducted per manufacturer's instruction. All images were acquired using a virtual microscope (BX61VS, Olympus, Tokyo, Japan) or a confocal microscope (LSM510, Zeiss, Jena, Germany).
The primary antibodies and respective dilutions are: rabbit anti-MAP2 (1:200, PRB-547C, BioLegend, San Diego, CA, USA); rabbit anti-CUX1/CDP (1:200, sc-13024, Santa Cruz); mouse anti-NeuN (1:200, MAB377, Millipore, Darmstadt, Germany); rat anti-Ctip2 (1:200, ab18465, Abcam). The following secondary antibodies were used: donkey anti-rabbit Cy2 (1:100, 711-225-152, Jackson ImmunoResearch Inc, PA, USA); donkey anti-mouse Cy2 (1:100, 715-225-150, Jackson ImmunoResearch Inc); sheep anti-mouse-Cy3 (1:500, Sigma-Aldrich); goat anti-rabbit IgG FITC (1:100, Sigma-Aldrich); sheep anti-rabbit-Cy3 (1:500, Sigma-Aldrich); goat anti-rabbit-Cy5 (1:500, Invitrogen); mouse anti-phospho-H2AX (ser139) (γ-H2AX) (1:100, 05-636, Upstate, New York, USA).
Dual luciferase reporter assay
To monitor Notch1 transcriptional activity Dual Luciferase® Reporter Assay (DLRA) system (Promega, Madison WI, USA) was used according to manufacturer's instructions and as described previously (38). The pGL4.20-12xCSL-luciferase plasmid was used as a reporter for Notch activity. Briefly, cells were transfected with the reporter construct and treated with inhibitors or IR. Cells were harvested 24 h after transfection by lysing them on culture plates for 15 min with 1× passive lysis buffer provided by the manufacturer (Promega). 10 μl of clarified cell lysate was used to measure the activity of Firefly luciferase after adding 50 μl of 1× LARII substrate. Renilla luciferase activity was measured by adding 50 μl of 1× Stop&Glo solution. Measurements were completed using a 96-well plate on the Tecan infinite M1000-pro plate reader (Tecan, Männedorf, Switzerland).
Subcellular fractionation
MEF cells were collected by scraping, washed with ice-cold PBS and pelleted at 4°C to resuspend the cells in sucrose-based lysis buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 10% glycerol, 0.1% Triton X-100 and 0.32 M sucrose, pH 7.9). The resuspension was incubated on ice for 5 min and centrifuged at 1300 g for 4 min at 4°C. The supernatant and pellet present crude cytoplasmic and nuclear fractions, respectively. The supernatant was cleared by centrifugation at 16 000 g for 10 min at 4°C to generate cytoplasmic fraction (S1). The pellet (P1) was resuspended with lysis buffer B (3 mM EDTA, 0.2 mM EGTA), incubated on ice for 10 min and centrifuged at 1700 g for 4 min at 4°C. The supernatant generates the soluble nuclear fraction (S2). The remaining pellet was washed with buffer B and centrifuged at 10 000 g for 1 min at 4°C to obtain chromatin-bound fraction (P2).
Immunoprecipitation and immunoblotting
Cells were lysed with RIPA buffer (50 mM Tris–HCl, pH 7.6, 10% glycerol, 0.15 M NaCl, 1.5mM MgCl2, 0.2 mM EDTA, pH 8.0, 1% NP-40, 1 mM DTT, 1 mM PMSF, 5 mg/ml leupeptin, 2 mg/ml aprotinin, 1 mM β-glycerophosphate, 1 mM Na3VO4 and 10 mM NaF). For immunoprecipitation, 2 μg of antibody was incubated with 1 mg of cell lysate, together with protein A sepharose™ CL-4B or protein G sepharose™ 4 fast flow (GE Healthcare, München, Germany) at 4°C overnight. Precipitates were washed with the NETN (100 mM NaCl, 20 mM Tris–Cl (pH 8.0), 0.5 mM EDTA, 0.5% (v/v) NP-40) buffer without protease inhibitors. Immunoblots on nitrocellulose or PVDF membrane were blotted with antibodies in TBST containing 5% non-fat dried milk (NFDM), ahead of incubation with horseradish peroxidase-conjugated secondary antibodies and detected by the ECL reagents (Amersham Biosciences, Buckinghamshire, UK). The following antibodies were used: mouse anti-NBS1 (1:1000, GTX70224, GeneTex, Irvine, CA, USA); rabbit anti-Nbs1 (1:1000; #3002, Cell Signaling, Danvers, MA, USA) or homemade serum (1:5000); mouse anti-FLAG M2 (1:5000, F-1804, Sigma-Aldrich); mouse anti-GFP (1:1000, #sc-9996, Santa Cruz, Heidelberg, Germany); mouse anti-β-Actin (1:5000, #T4026, Sigma-Aldrich); rabbit anti-Notch1 (1:1000, ab27526, Abcam, Cambridge, UK); rabbit anti-activated Notch1 (NICD) (1:1000, ab8925, Abcam); rabbit anti-RBPJ (1:1000, ab25949, Abcam); goat anti-Lamin B (1:1000, sc-6217, Santa Cruz); mouse anti-β-Tubulin (1:1000; #T4026, Sigma-Aldrich); rabbit anti-Mre11 (1:1000, NB100–142, Novus Biologicals, Littleton, CO, USA); mouse anti-Rad50 (1:1000, 05-525, Upstate, Darmstadt, Germany); rabbit anti-Histone H3 (1:1000, ab1791, Abcam); mouse anti-p53 (1:1000; #2524, Cell Signaling); rabbit anti-phopho-p53 (1:1000, 9284S, Cell Signaling).
ChIP-IP and PCR
Cells were treated or not with 10 Gy of IR and crosslinked with formaldehyde 1% for 10 min at room temperature, 1 hr after IR. Fixed cells were rinsed twice with PBS and resuspended in 250 μl (for two 150 mm dishes) of lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8, 0.1% Triton X-100, 0.1% sodium deoxycholate, 1% SDS, protease inhibitor) and incubated for 30 min on a rotator at 4°C. Lysate was sonicated for 10 min (30 s on/30 s off) in Diagenode water bath-sonicator and centrifuged at 14 000 rpm for 10 min.
The cleared supernatant was diluted 10 times in ChIP Dilution Buffer (1% Triton X-100, 2 mM EDTA pH 8, 20 mM Tris–HCl pH 8, 150 mM NaCl, Protease inhibitor). 600 μl of lysates were then incubated overnight with 2 μg of antibody at 4°C with rotation. 40 μl of Dynabeads (Protein G) were blocked overnight with 0.5 mg/ml BSA in PBS. Chromatin lysates were added and incubated with beads for 2 h, rotating at 4°C. The beads were washed 3 times with Washing Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA pH 8, 20 mM Tris–HCl pH 8, 150 mM NaCl), 1 time with High Salt Buffer (0.1% SDS, 1% Triton X- 100, 2 mM EDTA pH 8, 20 mM Tris–HCl pH 8, 500 mM NaCl) and 1 time with TE. ChIPed material was eluted by 30 min incubations at room temperature with 250 μl Elution Buffer (1% SDS, 150 mM NaCl, 5 mM DTT). Chromatin was reverse-crosslinked by adding 20 μl of NaCl 5M, incubated at 65°C for 16 h and DNA purified with PCR purification Kit (Qiagen). Purified DNA was subjected to quantitative PCR with the following primers to the promoter of Hes5: HES5-F2 ‘cctctggggagtgggagggaa’ and HES5-R2 ‘gccatgcctggagctctggag’.
CAPS analysis
Coevolution Analysis using Protein Sequences (CAPS) was performed as described previously (39). Briefly, orthologous protein sequences of NBS1 and NICD were exported from NCBI Homologene or OMA Orthology databases and aligned using M-Coffee. Multiple sequence alignment files were used as input for CAPS 2.0 (Coevolution Analysis using Protein Sequences) web-server to identify co-evolution between amino acid sites (39). The co-evolving sites between two proteins were used to predict the possibility of interaction between these proteins.
Measurement of neurite length
The length of neurites was measured as described previously (40), using NeuriteTracer, an ImageJ plugin for tracing neurites.
Statistical analysis
Statistical analysis methods are detailed in the figure legends.
RESULTS
NBS1 is required for proper neurite arbor formation of postmitotic neurons
To investigate whether NBS1 and other DDR molecules play a role in differentiating or postmitotic neurons, we analyzed RNA-sequencing data of the brain cortex of embryonic E14.5 and adult (10-month old) mice, which revealed that Nbs1 mRNA was similar in the brain cortexes of both (Supplementary Figure S1a). Yet, the mRNA levels of Mre11 and Rad50, as well as other DDR molecules, were reduced in the adult brain compared to the embryonic neocortex (Supplementary Figure S1a). Reverse transcription PCR (RT-PCR) further confirmed expression of Nbs1 in brain cortices of newborn and adult (3-month old) mice (Supplementary Figure S1b).
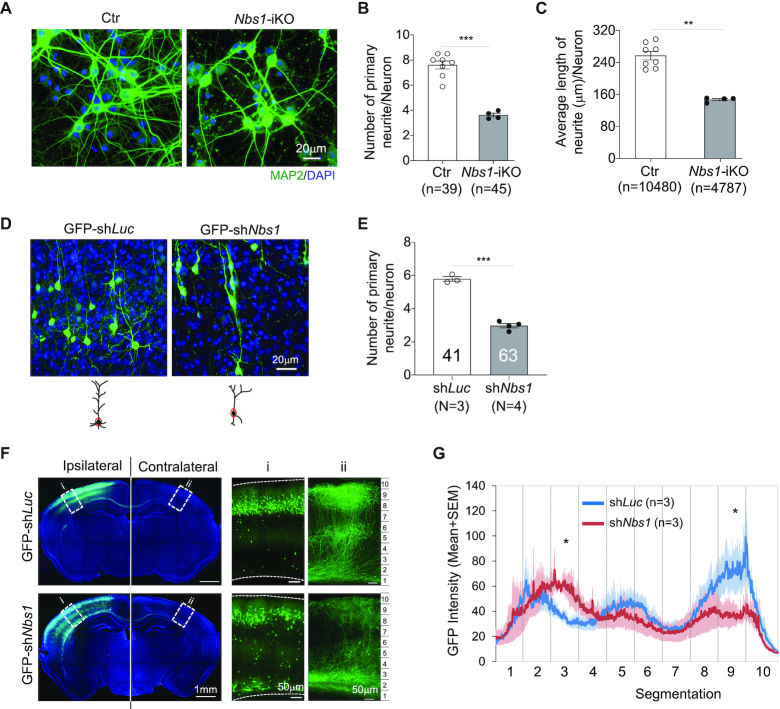
To investigate the function of NBS1 during neuronal development, we isolated primary neurons from E15.5 cortical plate of inducible Nbs1-deletion mice (Nbs1f/f-CreERT2, Nbs1-CER) (26) (Supplementary Figure S2a) and incubated them with 4-OHT for 4 days to induce Nbs1 deletion (thereafter Nbs1-iKO) (Supplementary Figure S2a and b). Staining of the culture with the neuronal marker NeuN at 8dpo revealed a similar number of neurons between control and Nbs1-iKO (Supplementary Figure S2c), indicating that deletion of Nbs1 does not compromise the viability of postmitotic neurons. To our surprise, the complexity of neurons was affected after Nbs1 deletion (Figure 1A). The number of primary neurites, as well as the average length of neurites per neuron, was significantly reduced in Nbs1-iKO neurons (Figure 1B and C). In further analyzing this neurite outgrowth defect in vivo and to test a cell-autonomous effect of Nbs1 deletion, we performed in utero electroporation (IUE) to knock down Nbs1 in developing neurons by introducing GFP-tagged shRNAs into the brain lateral ventricle (Supplementary Figure S3a and b). ShNbs1#1 had a higher knockdown efficiency than shNbs1#2 (Supplementary Figure S3b), which also correlated well with their biological effects (see below Figure 2A and B)—thus we used the former for most subsequent experiments. Electroporation was carried out at E15.5, at which stage neurons are generated to form layers II/III in the neocortex (41). Imaging analysis of cortical sections from postnatal day 26 (P26) mice revealed that the Nbs1 knockdown neurons (GFP-shNbs1) in the brain cortex exhibited a less complex morphology compared to shLuciferase (GFP-shLuc) controls (Figure 1D). The number of primary neurites per neuron was significantly reduced in GFP-shNbs1 samples (Figure 1E). Moreover, when measuring the axon projections of callosal projection neurons (CPN) after Nbs1 knockdown, we found the intensity of the GFP signal in the corpus callosal region of the contralateral cortex to be higher (segment 3), but significantly lower in the upper layer (segments 8) as compared to controls (Figure 1F and G); suggesting that neurite arborization of CPN is defective without Nbs1.
Figure 1.
The deletion of Nbs1 inhibits neurite outgrowth. (A) In vitro cultured neurons (8dpo) of control (Ctr) or Nbs1 inducible deletion (Nbs1-iKO) genotypes (see Supplementary Figure S2a) were stained with the dendritic marker MAP2. Nbs1-iKO: Nbs1-CER (Nbs1f/f-CreERT2), with 4-OHT treatment. Controls include all genotypes without 4-OHT treatment or Nb1+/+ or Nbs1f/+ with 4-OHT treatment. (B, C) The number (B) and the average length (C) of neurites per neuron were quantified from the indicated number of neurons (n) from four animals of each genotype. n, the number of neurons analyzed. Data are mean ± SEM. Welch's t-test (for B) or Mann–Whitney test (for C) was used for statistical analysis. **P < 0.01, ***P < 0.001. (D) GFP+ cells after IUE at E15.5 (see Supplementary Figure S3a) in layers II/III of the cortex of postnatal days 26 (P26) mice are shown. (E) The number of primary neurites per neuron was quantified from three to four animals of each treatment. N: number of animals analyzed. The number of neurons analyzed is depicted within the bar. Data are mean ± SEM. Welch's t-test was used for statistical analysis. ***P < 0.001. (F) The whole cross sections of P16 brains after IUE at E15.5 were imaged by confocal microscopy and the GFP signal in selected areas (i, ii) are shown enlarged on the right. (G) The average GFP intensity in control (shLuc) and Nbs1 knockdown (shNbs1) brain in area (ii) of the contralateral cortex were quantified. Three to five sections from each of three animals were analyzed for each condition. Data are presented as Mean ± SEM. The contralateral cortex region was equally divided into ten segments for statistical analysis by two-way ANOVA with Holm-Sidak's multiple comparison test. *P < 0.05.
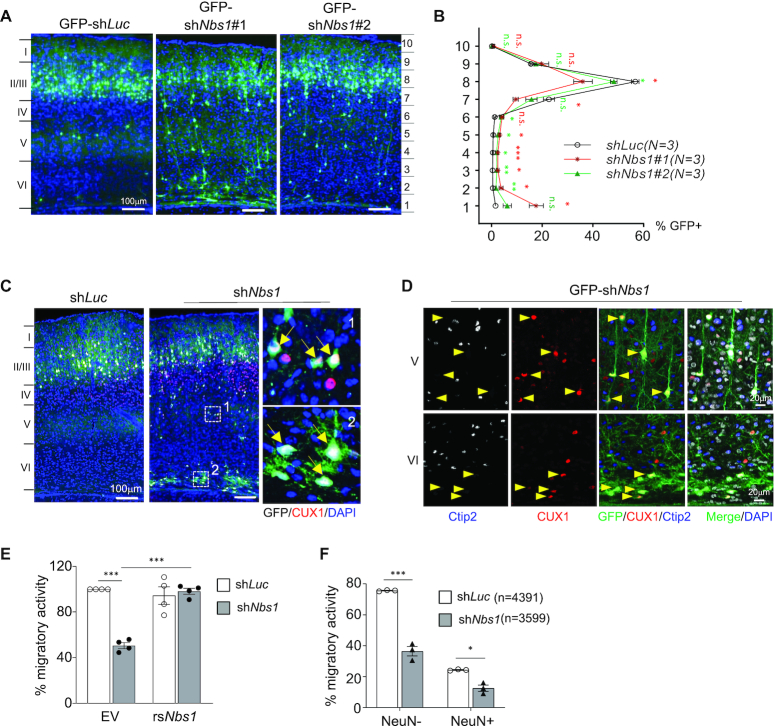
Figure 2.
Nbs1 deficiency inhibits neuronal migration. (A) Brain sections of P16 mice electroporated with indicating GFP-shRNA vectors at E15.5 are shown. GFP-shLuc targeting luciferase is control and GFP-shNbs1#1 or shNbs1#2 targets Nbs1. (B) The brain cortex from (A) was equally divided into ten segments and the percentage of GFP+ cells from each segment quantified based on three to five sections from each of three (N) animals of each condition (right). Data are Mean ± SEM. Two-way ANOVA with Holm-Sidak's multiple comparison test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001; n.s. = not significant. (C) Cortical sections were imaged after staining with CUX1, a marker for recently born neurons. The right panels are enlargements of the indicated areas of shNbs1 images. Arrows indicate GFP+CUX1+ cells. (D) The layers V and VI of the cortex (C) are shown after staining with CUX1 and early-born neuronal marker Ctip2. Arrowheads indicate GFP+CUX1+Ctip2– cells. (E) In vitro transwell migration of Neuo2A cells after co-transfection with shLuc or shNbs1 together with GFP-empty vector (EV) or GFP-shNbs1-resistant Nbs1 (rsNbs1) vector. The migratory activity was analyzed 24 h after plating on the membrane. Data are Mean ± SEM of minimum three independent experiments. Statistical analysis was performed using Two-way ANOVA followed by Tukey's post-hoc test. ***P < 0.001. (F) Transwell migration assay of primary neurons from E17.5 cortex of embryos that were electroporated with indicated shRNA expression vectors at E15.5. The migratory activity was calculated as GFP+, or GFP and NeuN double-positive (GFP+NeuN+) cells among the number of DAPI positive cells counted (n). Data are mean ± SEM of at least three embryos. Statistical analysis was performed using two-way ANOVA followed by Tukey's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown of Nbs1 inhibits neuron migration
Neuronal maturation is also associated with migration of newborn neurons to the destination. Next, the role of Nbs1 in neuronal migration was investigated, using IUE to knock down Nbs1 and the migratory capacity of neurons analyzed in situ from the cortical sections of P16 and P26 mice who received GFP-shNbs1 at E15.5. Almost all GFP positive (GFP+) cells in control (GFP-shLuc) reached layers II/III of the brain cortex (Figure 2A and B). In sharp contrast, many GFP+ cells in shNbs1-treated samples were found in layers IV/V/VI (Figure 2A and B), indicating defective neuronal migration. Immunofluorescent staining revealed that these GFP+ cells at layers IV/V of the shNbs1-treated cortex were positively labeled with CUX1, a marker for neurons of layers II/III (Figure 2C and D)—but not with Ctip2—a neuronal marker for the neurons of cortical layers V/VI (Figure 2D). Thus, neurons for layers II/III were eventually formed but could not reach the right destination in the absence of Nbs1, suggesting a role for Nbs1 in neuronal migration. To further explore whether the migration defect is due to a cell-autonomous defect, we performed a transwell migration assay using mouse brain neuroblastoma cells (Neuro2A)—a well-established cellular system. Nbs1 deletion by shRNA impaired migration of differentiated Neuro2A cells, which can be rescued by ectopic expression of an shRNA-resistant Nbs1 cDNA (Figure 2E). A similar degree of the migratory defect was observed using different pole size of the membrane in this assay (Supplementary Figure S3c), suggesting that the migration defect of Nbs1-deficient neural cells as an intrinsic property, not linked to the complexity of neurites. Furthermore, the transwell migration assay using primary cortical neurons isolated from brains 2 days after IUE-mediated transfection of GFP-shNbs1, also revealed that shNbs1 reduced the migratory activity of neurons (Figure 2F). Taken together, the migration phenotype of Nbs1-deleted neurons results from an intrinsic function of Nbs1.
Nbs1 deletion elevates expression of Notch target genes and Notch activity
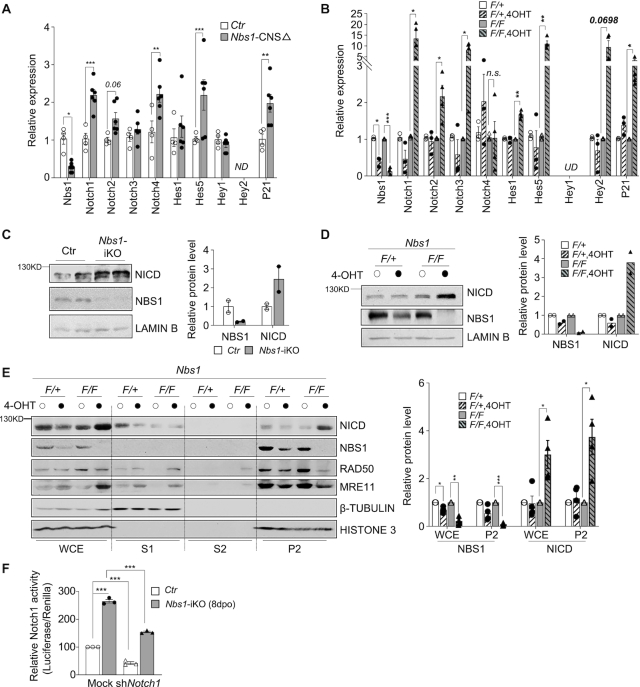
Notch signaling is a master regulator of brain development (42,43) and critical for such as neurite arborization, maturation, and migration of postmitotic neurons in neuronal development (44–46). We were moved to investigate whether Notch signaling is altered in Nbs1 knockout neurons. Interestingly, Nbs1 knockout upregulated the mRNA level of many genes of the Notch pathway, such as Notch1, Notch2 (P = 0.06), Notch4, Hes5 and P21, in neurons isolated from P26 Nbs1-CNSΔ mouse cerebellums (Figure 3A). We also found increased levels of Notch targets, including Notch1, Notch2, Notch3, Hes1, Hes5, Hey2 (P = 0.0698), and P21, in mouse fibroblast cells (MEFs) after Nbs1 depletion (Figure 3B). Of the Notch receptors, Notch1 is the most affected (Figure 3A and B). Western blotting further revealed a high level of NICD (Notch intracellular domain)—a proteolytic product of Notch receptors and a transcriptional co-activator for expression of Notch target genes (43,46)—in Nbs1-deleted primary neurons (Figure 3C), as well as in MEFs (Figure 3D, E and also below 5C); shRNA knockdown of Nbs1 also elevated NICD in MEF cells (Supplementary Figure S4a). These findings indicate a general regulation of Notch by Nbs1 in both cell types. Further mapping of the accumulation of NICD in Nbs1-deleted cells involved cell fractionation experiments, which detected the elevated NICD protein mainly in the nuclear fraction of the Nbs1-iKO MEF cells—where lowered Mre11 and Rad50 were detected as expected (Figure 3E). Consistent with an increased NICD level, Nbs1-deficient cells (by Nbs1-iKO or shNbs1) displayed a higher Notch activity (Figure 3F, Supplementary Figure S4b), which nevertheless can be suppressed by Notch1 knockdown (Figure 3F) or by the Notch inhibitor DAPT (Supplementary Figure S4b). All indicate an inhibitory function of Nbs1 in Notch signaling.
Figure 3.
The deletion of Nbs1 elevates Notch signaling. (A, B) Real-Time-quantitative-PCR (qPCR) analyses of Notch receptors and their target genes after Nbs1 deletion. (A) RNA was isolated from cerebellar neurons of P26 Nbs1-CNSΔ and control mice. Data are Mean ± SEM from two repeat experiments of 2–3 animals of each genotype. (B) RNA was from one Nbs1+/f-CreERT2 (+/F) and two independent Nbs1f/f-CreERT2 (F/F) MEF cell lines after treatment with or without 4OHT for 4 days and analyzed at 8dpo. Data are mean ± SEM of 2–3 independent experiments. Two-way ANOVA followed by uncorrected Fisher's LSD test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001; n.s.: not significant; UD: undetectable, ND: not detected. (C, D) Western blot analysis of Notch protein in primary neurons (C) or in 8dpo MEFs (D) with the indicated antibodies. The Notch1 antibody detects NICD at ∼110 kDa. Lamin B is a loading control. Data are mean ± SD of the relative protein level after quantifying two animals of each genotype (C), or two repeats of two independent cell lines of each genotype (D). (E) Western blot analysis of cell fractionations prepared from Nbs1-iKO MEFs (8dpo) using the indicated antibodies. The Notch1 antibody detects NICD (∼110 kDa). Histone 3 and β-Tubulin were used to control the nuclear and cytoplasmic fractions, respectively. WCE: whole-cell extract; S1: soluble cytoplasmic fraction; S2: soluble nuclear fraction; P2: chromatin-bound fraction. +/F: Nbs1+/f-CreERT2, heterozygous of Nbs1 alleles; F/F: Nbs1f/f-CreERT2, homozygous of Nbs1 floxed alleles. The mean ± SEM of the relative protein level is from four independent experiments (right panel). Two-way ANOVA followed by uncorrected Fisher's LSD test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001. (F) Notch activity in Nbs1-deleted MEF cells. Control or Nbs1-iKO MEFs were co-transfected with the luciferase reporters together with empty vector (EV) or shNotch1 at 7dpo. Dual-luciferase assay was carried out 24 h after transfection. Data are mean ± SEM of three independent experiments. Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis. ***P < 0.001.
Figure 5.
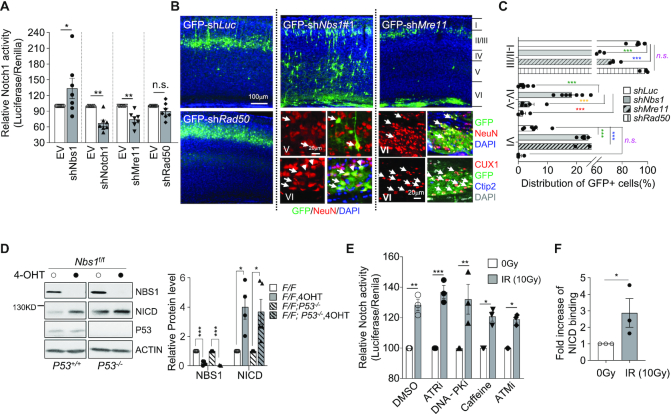
Distinct effects of MRN and DNA damage on Notch activity and neuronal development. (A) The effect of the MRN complex on Notch activity. The luciferase reporters were co-transfected with empty (EV) or indicated shRNA expression vectors into wild-type MEF cells. Dual-luciferase assay was carried out 24 h after transfection. Data are mean ± SEM of the relative activity from seven independent experiments. One-way ANOVA with Holm–Sidak's multiple comparison test was used for statistical analysis. *P < 0.05; **P < 0.01; n.s.: not significant. (B) Neuron migration after knockdown of the MRN complex. Cortical sections of P16 mice after IUE at E15.5 with indicated shRNA expression vectors are shown. High magnification of immunostained sections with neuron markers (NeuN, CUX1, Ctip2) are shown under respective images. Arrows mark the GFP+NeuN+ or GFP+CUX1+ cells. (C) The percentage of GFP+ cells in the indicated regions from 4∼7 sections of three animals of each treatment is shown (right panel). Layers of the cortex are marked I to VI in (B). Data are presented as mean ± SEM. Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis within marked corresponding layer group. *P < 0.05; **P < 0.01; ***P < 0.001; n.s.: not significant. (D) Western blot analysis of Nbs1-iKO MEFs at 8dpo, (see Supplementary Figure S2a) in the p53+/+ or p53−/− background using the indicated antibodies. The Notch1 antibody detects NICD (∼110 kDa). Actin serves as a loading control. The mean ± SEM of the relative protein level from four independent experiments is shown (right panel). Two-way ANOVA followed by uncorrected Fisher's LSD test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001. (E) Notch activity analysis was carried out using MEF cells transfected with the luciferase reporter, 24 h before treatment with different kinase inhibitor (1.6 μM ATRi, 1.0 μM DNA-PKi, 5 mM caffeine, 10 μM ATMi). Cells were exposed to 10 Gy of IR 1 h after drug incubation. Dual-luciferase assay was carried out 1 h after IR treatment. Data are mean ± SEM of the relative activity from three independent experiments. One-way ANOVA with Holm-Sidak's multiple comparison test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001. (F) ChIP quantitative real-time PCR detection of binding of NICD to the Hes5 promoter after MEF cells were treated with 10 Gy of IR. The Notch1 antibody was used for the ChIP assay and rabbit IgG was used as a negative control. All data shown as fold increase of binding activity after normalization first to the input, then the ratio of NICD to IgG in IR untreated control samples, were used to calculate the fold change. The mean ± SEM of fold changes from three experiments is shown. Welch's t-test was used for statistical analysis. *P < 0.05.
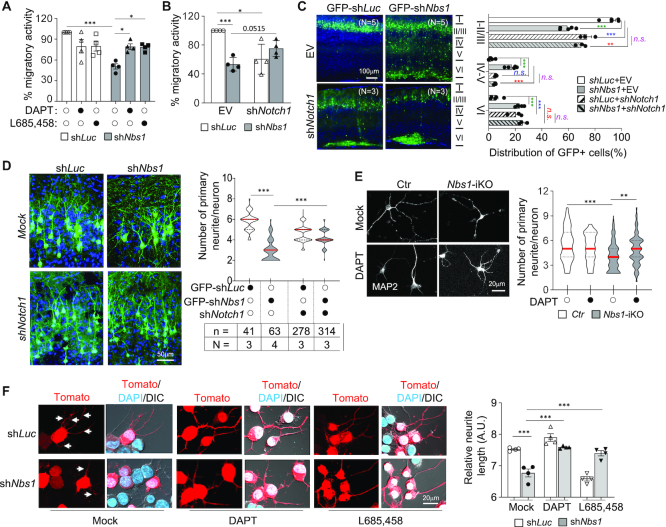
Inhibition of Notch signaling ameliorates the phenotypes of Nbs1-deficient neurons
Notch inhibitors were applied (DAPT and L685,458) in the transwell migration assay using Neuro2A cells after Nbs1 knockdown, to substantiate that the Notch pathway is responsible for neuronal arborization and migration defects in Nbs1-deficient cells. These inhibitors enhanced the migration activity of the Nbs1-knockdown (shNbs1) cells to the level of shLuc control cells (Figure 4A). Moreover, knockdown of Notch1 by shRNA largely—although not statistically significant (P = 0.0515)—corrected the migration defect in Nbs1-deficient cells (Figure 4B). We next applied IUE to examine the influence of Notch signaling on the migration of Nbs1 knockdown neurons in vivo. Notch knockdown ameliorated the migratory defects of shNbs1 (GFP+, shNbs1-shNotch1 double-knockdown) neurons in layers IV/V of the cortex, resulting in a similar migration pattern to Notch single-knockdown—which displayed a stack of GFP+ neurons in either layers II/III or VI of brain cortex (Figure 4C). These data indicate that Notch signaling is indeed responsible for the neuronal migration defects of Nbs1 deletion.
Figure 4.
Inhibition of Notch signaling rescues the migration and neurite defects of Nbs1-deficient neural cells. (A, B) Blockage of Notch signaling by inhibitors (10μM of DAPT or L685, 458) (A) or shRNA targeting Notch1 (B) improves migration deficiency of N2A cells caused by shNbs1 knockdown. The migratory activity was analyzed 24 h after drug treatment or shRNA transfection. Data are Mean ± SEM from four independent experiments. Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Rescue of the neuronal migration in vivo. Brain cortical sections from P16 mice after IUE with indicated plasmids at E15.5 were imaged by confocal microscopy. The distribution of GFP+ cells in indicated regions of the cortex was quantified (right). The mean ± SEM of the percentage of GFP+ cells in indicated regions from three to five animals is shown (right panel). N: number of mice analyzed. Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis within marked corresponding layer group. *P < 0.05; ** or ##P < 0.01; *** or ### or $$$P < 0.001; n.s.: not significant. (D) Rescue of the neurite outgrowth in vivo. Brain cortical sections from P26 mice after IUE with indicated plasmids at E15.5 were imaged and GFP+ neurons in layers II/III of the cortex are presented (up penal). The mean ± SEM were quantified from indicated animals of each treatment. N: number of mice analyzed; n: number of neurons analyzed. One-way ANOVA followed by Dunnett's multiple comparisons test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001. (E) Inhibition of Notch signaling by DAPT ameliorates the arborization defects of Nbs1-iKO neurons. Primary neurons were isolated from two Nbs1+/f-CreERT2 (+/F) and two Nbs1f/f-CreERT2 (F/F) E15.5 embryonic brains and cultured with 4-OHT for four days. Cells were then treated with or without 10 μM of DAPT for another 4 days before analysis at 8dpo. The number of primary neurites (marked by MAP2) per neuron from 41∼131 neurons (from at least two animals) are shown (right panel). The Mean ± SEM is shown. One-way ANOVA followed by Dunnett's multiple comparisons test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001. (F) Inhibition of Notch activity corrects neurite outgrowth defects of shNbs1 knockdown N2A. N2A cells were transfected with Tomato-tagged shRNA against luciferase (shLuc) or Nbs1 (shNbs1). Cells were induced to neuronal differentiation by 2.5 mM cAMP in the presence or absence of Notch inhibitors (10 μM of DAPT or L685, 458) 12 h after transfection. Cells were imaged 24 h after differentiation induction and represented imaged (left). Quantification of the total length of neurites and the areas of cell clusters was acquired by the IncuCyte3 system. Arrows mark the neurites. Data are presented as mean ± SEM of the relative neurite length (dividing total length by areas of cell cluster) from four experiments (right panel). Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis. ***P < 0.001.
Next we examined whether the abnormally high Notch activity is also responsible for the neurite outgrowth defects of Nbs1 knockout neuronal cells. After knocking down Notch1 in vivo by the IUE assay, we observed a significant improvement in the number of primary neurites of Nbs1-deficient neurons (Figure 4D). Moreover, the Notch inhibitor DAPT increased the number of primary neurites in Nbs1-deficient primary neurons in vitro (Figure 4E). This was further confirmed in differentiating Neuro2A cells after shNbs1 knockdown, followed by treatment with Notch inhibitors (Figure 4F). Consistent with the findings in primary neurons (Figure 1), depletion of Nbs1 dramatically impaired neurite outgrowth and reduced the average length of differentiated Neuro2A cells, which nevertheless could be restored by Notch inhibitors (Figure 4F). Combined, these genetic and pharmacological experiments demonstrate that Nbs1 regulates neuronal development through the Notch pathway.
Notch dysfunction and migration defects of Nbs1-deficient cells are not direct consequences of DDR
The classical function of NBS1 is to activate the DDR cascade through the assembly of the MRN complex at DSBs, failure of which provokes DNA damage to elicit the p53-mediated DDR cascade and apoptosis (1,9). In establishing whether the classical function of Nbs1 in the MRN complex might be responsible for alteration of Notch signaling, we knocked down other components of the MRN complex—namely Mre11 and Rad50—(Supplementary Figure S5a) and analyzed Notch activity and neuronal migration. While shRad50 had a negligible effect on Notch activity in MEF cells, shNbs1-transfected cells showed an expected higher Notch activity (Figure 5A). Intriguingly, shMre11 decreased Notch activity, similarly to shNotch1 (Figure 5A), whereas the ectopic expression of Mre11 enhanced Notch activity (Supplementary Figure S5b), phenotypically copying Nbs1-deficient cells. We extended our analysis to knocking down Mre11 and Rad50 in vivo by IUE and scored neuronal migration. Whereas Rad50 knockdown resulted in a similar distribution pattern of GFP+ cells as shLuc control, Mre11 depletion led to GFP+ cells located in either layers II/III or VI of brain cortex (Figure 5B and C). This was in great contrast to the patterns from the shNbs1 samples (Figure 2A–C). All GFP+ cells in the VZ region of the shMre11-treated cortex were positive for NeuN and CUX1, but negative for Ctip2 (Figure 5B). Of note, the Mre11 knockdown significantly compromised neuronal migration, reminiscent of Notch1 knockdown (see Figure 4C). The differing migratory patterns seem to correlate well with the distinct Notch activity influenced by the deficiency of the individual MRN subunit (see Figure 5A). Moreover, we did not observe obvious accumulation of DNA damage, as judged by γH2AX foci in shNbs1 knockdown neuronal cells (GFP+) in the cortex in vivo (Supplementary Figure S5c). TUNEL staining of shNbs1-transfected neuronal cells (GFP+) in IUE-treated brain slices did not detect an apoptosis increase (Supplementary Figure S5d). Overall, Nbs1-deletion-induced upregulation of Notch activity and neuronal defects are unlikely to be a direct consequence of the loss of MRN-mediated DDR.
DNA damage has been shown to alter Notch signaling via p53-mediated regulation of Notch gene expression (47–50). To further investigate whether the abnormal Notch activity in Nbs1-deficient cells is secondary to p53-related DDR, we next analyzed Notch expression in Nbs1-deficient cells in the presence or absence of p53. Western blot analysis showed that Nbs1 depletion increased the NICD level regardless of the p53 background (Figure 5D), suggesting that the upregulation of Notch signaling in Nbs1-deficient cells is likely independent of DDR involving p53. Moreover, acute DNA damage induced by ionizing radiation (IR, 1 h after 10 Gy treatment) elevated Notch activity (Figure 5E, DMSO treated), while concurrently enhancing the binding of NICD to the promoter of Notch target gene Hes5 (Figure 5F). Of note, inhibitors to ATM (ATMi and Caffeine) or other early DDR kinases, i.e. ATR and DNA-PK, did not repress IR-induced Notch activity (Figure 5E)—suggesting that IR-mediated Notch upregulation may involve pathways other than the DDR kinases of the PI3KK family, via an as yet unknown mechanism.
Nbs1 regulates Notch activity through direct interaction with NICD-RBPJ
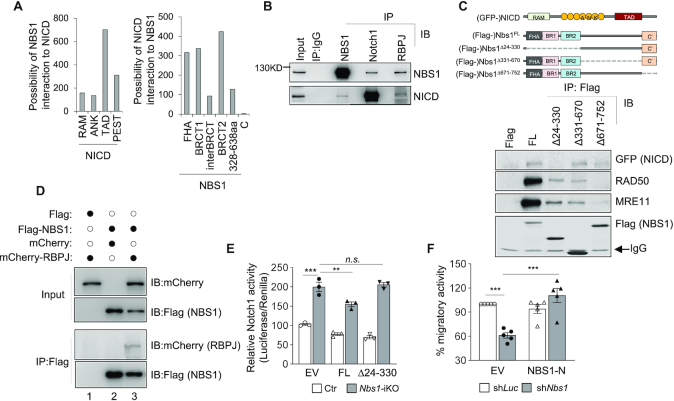
NICD–RBPJ association to displace co-repressor at the promoter region of Notch target genes is necessary for transcriptional activation (43). We were thus prompted to gain molecular understanding of how NBS1 participates in NICD-mediated transcription machinery. To this end, a co-evolution analysis utilising protein sequences (CAPS) (39) was applied to amino acid sequences of NBS1 and NICD from different species. CAPS analysis indicated that the FHA domain and both BRCT domains (BRCT1, BRCT2) in the N-terminus of NBS1 have high potential to interact with NICD, especially with the transactivation domain (TAD) of NICD (Figure 6A). An immunoprecipitation (IP) assay was conducted to determine whether NBS1 indeed interacts with NICD, which found that the Nbs1 antibody could precipitate NICD in HeLa cell lysates (Figure 6B). To further validate this interaction, GFP-tagged NICD (GFP-NICD) were co-transfected with Flag-tagged Nbs1 truncation mutants from deleting the N-terminal 24–330 amino acids (Nbs1Δ24–330); or deleting the mid part 331–670 amino acids (Nbs1Δ331–670); or deleting the C-terminal 671–752 amino acids (Nbs1Δ671–752) (Figure 6C, upper panel), into HEK293 cells. Full length (Nbs1FL), as well as Nbs1Δ331–670 and Nbs1Δ671–752 mutant Nbs1, could pull down NICD (Figure 6C). Consistent with the notion that the C-terminus of Nbs1 is necessary for interaction with other components of the MRN complex (1), the C-terminal mutant Nbs1Δ671–752 failed to pull down Mre11 and Rad50 (Figure 6C). Of note, the N-terminal mutant Nbs1Δ24–330 lost its interaction with NICD, while maintaining its association with Mre11–Rad50 (Figure 6C). Noticeably, IP experiments also detected an interaction of endogenous and ectopically expressed Nbs1 with RBPJ, the core component of the NICD-mediated transcription complex (Figure 6B and D). These data indicate a molecular link of Nbs1 with Notch signaling.
Figure 6.
NBS1 interacts with NICD-RBPJ and suppresses NICD activity. (A) Prediction of inter-relationship between NBS1 and NICD. Co-evolution analysis was carried out by using amino acid sequences of NBS1 and NICD of Notch1 from different species. The potential of functional interaction between NBS1 and different domains of NICD (left panel), or between NICD and different domains of NBS1 (right panel) were scored. FHA: forkhead-associated domain; BRCT: BRCA1 C-Terminus domain; RAM: RBPJ-associated molecule domain; ANK: ankyrin repeats domain; TAD: transactivation domain; PEST: proline (P), glutamic acid (E), serine (S), and threonine (T) rich domain; BRCT: BRCA1 C-terminal domain; C’: C-terminus. (B) Co-immunoprecipitation (Co-IP) analysis of endogenous interaction between Nbs1 and NICD or RBPJ in HeLa cells. Antibodies used for IP and Immunoblotting (IB) are indicated. The Notch1 antibody detects NICD (∼110 kDa). Representative images of four experiments are shown. (C) Identification of the binding domains between Nbs1 and NICD by Co-IP. Schematic structures of functional domains of Nbs1 and NICD (upper panel) are shown. HEK293 cells were co-transfected with Flag-tagged full length or truncated mutants of Nbs1 together with GFP-tagged NICD. IP (IP:Flag) samples were used for immunoblot analysis with indicated antibodies (lower panel). FL: full length; Δ24–330: deletion of N-terminal amino acids 24–330; Δ331–670: deletion of mid-part amino acids 331–670; Δ671–752: deletion of C-terminal amino acids 671–752. Representative images from four experiments are shown. (D) mCherry-tagged RBPJ was co-transfected with Flag-Nbs1 or GFP-NICD into U2OS cells and a Co-IP assay was performed using indicated antibodies. Representative images from two experiments are shown. (E) Notch activity assay of wildtype (Ctr) or Nbs1-iKO MEF cells (7dpo) after co-transfection with the luciferase reporter and the GFP-tagged empty vector (EV) or the full length Nbs1FL (FL) or N-terminal deletion mutant Nbs1Δ24–330 (Δ24–330) vector. Reporter activity was measured 24 hr after transfection. Data are from three independent experiments. Data are Mean ± SEM of relative Notch activity from three independent experiments. Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis. **P < 0.01; ***P < 0.001, n.s.: not significant. (F) Transwell migration assay of N2A cells after co-transfection with Tomato-tagged shRNA targeting luciferase (shLuc) or Nbs1 (shNbs1), together with GFP-empty vector (EV) or a GFP-shNbs1-resistant Nbs1-N terminus (Nbs1-N) plasmid. The migratory activity was analyzed 24 hr after plating cells on the membrane. Data are mean ± SEM of the relative migration activity from five independent experiments. Two-way ANOVA followed by Tukey's post-hoc test was used for statistical analysis. **P < 0.01; ***P < 0.001.
Notch auto-regulates expression of its target genes (51,52) and Nbs1 knockout increased both Notch activity and mRNA levels of Notch targets (see Figures 3, 5A and D). We investigated whether the interaction of Nbs1 and NICD regulates the Notch activity and thereby transcription. Indeed, ectopic expression of Nbs1FL repressed Notch activity, whereas the NICD interaction mutant Nbs1Δ24–330 failed to suppress Notch1 activity in Nbs1-deleted cells (Figure 6E) – positing that Nbs1-NICD interaction is necessary to repress NICD transcription activity. Furthermore, the N-terminal Nbs1 (Nbs11–330) was sufficient to correct the migration defect of Nbs1-disrupted Neuro2A cells (Figure 6F), as does full-length Nbs1 (see Figure 2E). Thus, via a direct interaction of its N-terminus with NICD, Notch activity and neuronal development are modulated by Nbs1.
DISCUSSION
Mutations in various DDR pathways are responsible for genomic instability disorders, of which neurological deficits such as developmental defects and neuronal degeneration, are common features (17,23,53). Malfunction of the DDR compromises the proliferation and survival of neuroprogenitors, which has been proposed as a major cause of brain developmental disorders in humans and mouse models (18,54). All components of the MRN complex are essential for proliferating cells. The current study shows that, unlike the effect in proliferating cells (2) and neuroprogenitors (25,55), Nbs1 deletion does not compromise the survival of differentiating and mature neurons. The dispensability of Nbs1 in postmitotic cells reflects a previous report showing that tissue-specific knockout of Rad50 in the murine liver (postmitotic hepatocytes) did not cause any obvious phenotype (56). Whether Mre11 is dispensable for non-dividing cells has not yet been studied. Intriguingly, it is unexpected that silencing or knocking out Nbs1 specifically compromises neurite outgrowth and the migration of neurons, which can explain the observation of the cortical layering defect in Nbs1-CNSΔ mice (57) and also the neurological deficits—e.g., microcephaly and intellectual disability—of NBS patients (30). The current study is interesting because it shows that if cells survive the elimination of the essential genes (e.g. Nbs1), for example in postmitotic neurons, novel functions of the proteins can be revealed.
We found that elevated expression of several Notch receptors and targets in Nbs1-deleted neural cells and also MEFs, of which Notch1 is the most affected. It is well-known that all Notch receptors function redundantly and have often overlapping contributions in many cellular, developmental and disease processes (58) (see review (59)). Notch signaling is a master regulator of a wide range of developmental processes, including the nervous development (42,60), where Notch-RBPJ is critically important for neurite outgrowth and neuronal migration (42,61). We show that Nbs1 is a regulatory component of the NICD-RBPJ-mediated transcriptional activity of Notch signaling. Nbs1 deletion upregulates the NICD protein level, as well as Notch activity in neurons and also other cell types tested—which may function to repress neurite outgrowth and neuronal migration, consistent with the instrumental function of Notch in neuronal differentiation, maturation, and migration (44–46). Nbs1 negatively regulates the Notch pathway in neuronal homeostasis; most likely achieved by its direct interaction with NICD. It is plausible that Nbs1 destabilizes or induces degradation of NICD once Nbs1 interacts with NICD; therefore, Nbs1 depletion results in an increase of NICD, which may also explain a limited amount of co-IP signals. Alternatively, the Nbs1-NICD engagement weakens occupancy of NICD-RBPJ at the target gene promoter, or impairs recruitment of transcriptional co-activators by NICD-RBPJ at the same site (43). In this regard, it is worth mentioning that NBS1 can interact with P300/CBP (62) and CtIP (63) – both are co-factors of the NICD-RBPJ transcriptional machinery. Notch signaling includes the interaction of signal-presenting (or ligand-presenting, or environment/niche) cells and signal-receiving cells (e.g. neurons in this case) (46,64). Our IUE experiments exclude a major contribution of the environment or ligand-presenting cells of the brain in Nbs1-mediated neuronal migration. The in vitro transwell assay of primary neurons as well as neural cells N2A all demonstrate an intrinsic migration defect in Nbs1-deficient cells. Consequently, the Nbs1-NICD function in migration and neurite outgrowth is cell-autonomous. Given the diverse functions of Notch signaling during neuronal maturation, migration and also in synaptic plasticity (see (46), references therein), our discovery of NBS1 in crosstalk with the Notch pathway adds a plausible explanation for the etiology of aforementioned neuronal deficits in NBS patients.
DNA damage may alter neuronal differentiation (65,66). However, for the following reasons the Notch-mediated neuronal arborization and migration defects in Nbs1-deficient neurons are unlikely to be a secondary consequence of the MRN-mediated DDR function: (i) knocking down of each of MRN components resulted in distinct phenotypes concerning Notch activity and neuronal migration patterns. It is interesting to note that Mre11 knockdown caused downregulation of Notch activity, which is opposite to Nbs1 knockdown and concurrently a defective neuronal migration, similar to the Notch1 knockdown. (ii) We did not observe an obvious accumulation of DNA damage (γ-H2AX foci) or increased DDR-mediated apoptosis (TUNEL+ cells) in Nbs1-deficient neurons. (iii) p53 deletion failed to repress the NICD level in Nbs1-deleted cells. (iv) Pharmacological blockage of Notch signaling by inhibitors, or shRNA knockdown of Notch1, which does not repair DDR function, can rescue both neurite outgrowth and the neuronal migration defects in Nbs1-deficient cells. Taken together, the function of NBS1 in promoting neuronal morphogenesis and migration through the Notch pathway is distinct from its classical MRN-mediated DDR function.
Yet we observed that both DNA damage (e.g. IR treatment) (Figure 5E) or Nbs1 depletion (Figures 3, 5A and D) consistently upregulate Notch activity – How both events are mechanistically connected is currently unclear. We speculate that DNA damage can induce assembly of the MRN complex at DSBs, which alleviates the NBS1 inhibitory interaction with NICD, then facilitates NICD binding to target promoters and upregulates Notch activity (Figure 5F)—which is however independent of early DDR kinases’ activity. In support of this hypothesis, we found that when Mre11 is knocked down, presumably unable to drag Nbs1 away from the Nbs1-NICD-RBPJ engagement, Notch activity is repressed. Also, when Mre11 is overexpressed, which presumably sequesters Nbs1 from its binding with NICD, Notch activity is increased (Supplementary Figure S4b).
The observation that IR and Nbs1 deletion induces Notch activity is interesting because high levels or stabilization of Notch can promote neuronal death in ischemic stroke, indicating that Notch signaling is important for cell fate of neurons (67). Also, Tip60, a regulator of ATM, acetylates upon UV treatment and inhibits transcriptional activity via direct interaction with NICD—suggesting that Notch is downstream of the DDR (50). However, Vermezovic et al. reported that Notch1 is a negative regulator (upstream) of the ATM activity in the DDR (68), because it can compete with FOXO3a and thereby compromises Tip60 binding to ATM (69). Whilst we cannot formally exclude the possibility that the ATM-NICD interaction might interfere with NBS1-NICD because DSBs can induce NBS1-ATM association, our findings rather indicate that NBS1’s function to suppress Notch activity is unlikely to be a downstream event of the canonical MRN-ATM-DDR cascade.
Under genotoxic stress, NBS1 together with MRE11 and RAD50 (in the MRN complex) regulates the DDR. The canonical DDR function of NBS1 protects neuroprogenitors from proliferation arrest and cell death (25), which is however spared from non-dividing neurons. In the absence of DNA replication and the presence of a negligible level of DSBs, namely in non-dividing cells, NBS1 serves as a scaffold, as it does for MRN, to engage other signaling pathway components – in this case, NICD-RBPJ. The current study demonstrates that NBS1, via its physical and functional interaction with Notch signaling, ensures proper neuronal maturation, migration, and neurite outgrowth. Our study strongly implies that the essential DDR genes in non-dividing cells and tissues—if expressed—may harbor diverse physiological functions other than those previously deemed to handle DNA damage derived from the replication stress and, that these non-canonical functions are highly relevant for the maintenance of tissue homeostasis in adult life under non-genotoxic and physiological conditions.
DATA AVAILABILITY
RNA sequencing data are deposited at NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE137506.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Andreas for his excellent assistance for the cytometry analysis. We also thank M. Rodriguez and P. Elsner for their excellent assistance in the maintenance of the animal colonies. Further thanks go to Yossi Shiloh for scientific discussion on the development of the project. We are grateful to Professor Xin Li for his help in statistical analyses. We thank Iree Chang for providing the luciferase vectors and discussion of Notch activity and Marco Groth for RNA sequencing. We are grateful to D. Weih and E. Stöckl for their editing of the manuscript. We are also grateful to many members of Wang laboratory for the helpful discussions. Z.-W.Z was a recipient of a fellowship from the Deutsche Forschungsgemeinschaft (DFG) and is supported by the Natural Science Foundation of Guangdong Province (2019A1515010881) and the Shenzhen Science and Technology Innovation Commission (JCYJ20190807154407467).
Author Contributions: Z.W.Z. conceived the project, performed the majority of the experiments, interpreted the data, and wrote the manuscript. M.K. performed immunofluorescent staining on brain section and Notch activity reporter assay. N.S. performed transwell membrane migration assay. G.Y. performed Notch activity reporter assay and studied the interaction between NBS1-NICD. M.D. studied the interaction between NBS1-BBPJ and ChIP-qPCR; T.R. performed primary neuron isolation and culture. K.S. performed co-evolution analysis. R.Q. and Y. N. performed some of the qPCR analyses. T.-L.L. contributed shRNA vectors and to the development of the project. C.K. and A.B. contributed to the scientific development and discussion; Z.Q.W. designed the experiments, supervised the project and wrote the manuscript.
Contributor Information
Zhong-Wei Zhou, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany; School of Medicine (Shenzhen), Sun Yat-Sen University, Guangzhou, China.
Murat Kirtay, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany.
Nadine Schneble, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany.
George Yakoub, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany.
Mingmei Ding, School of Medicine (Shenzhen), Sun Yat-Sen University, Guangzhou, China.
Tina Rüdiger, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany.
Kanstantsin Siniuk, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany.
Ruiqing Lu, School of Medicine (Shenzhen), Sun Yat-Sen University, Guangzhou, China.
Yi-Nan Jiang, School of Medicine (Shenzhen), Sun Yat-Sen University, Guangzhou, China.
Tang-Liang Li, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany; Institute of Aging Research, School of Medicine, Hangzhou Normal University, Hangzhou, China.
Christoph Kaether, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany.
Ari Barzilai, Department of Neurobiology, George S. Wise Faculty of Life Sciences and Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel.
Zhao-Qi Wang, Leibniz Institute on Aging – Fritz Lipmann Institute (FLI), Jena, Germany; Faculty of Biological Sciences, Friedrich-Schiller-University Jena, Jena, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG [WA2627/1-1, WA2627/5-1 to Z.-Q.W.]; Leibniz Association [SAW2014, SAW2015 to Z.-Q.W.], Germany; German-Israel Foundation (GIF) [I-1307-418.13/2015 to Z.-Q.W. and A.B.]; Natural Science Foundation of Guangdong Province supports Z.-W.Z.’s research [2019A1515010881]. Funding for open access charge: DFG and Institute core budget.
Conflict of interest statement. None declared.
REFERENCES
- 1. Stracker T.H., Petrini J.H.. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011; 12:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruhn C., Zhou Z.W., Ai H., Wang Z.Q.. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep. 2014; 6:182–195. [DOI] [PubMed] [Google Scholar]
- 3. Lee J.H., Paull T.T.. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004; 304:93–96. [DOI] [PubMed] [Google Scholar]
- 4. Shiloh Y., Ziv Y.. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013; 14:197–210. [PubMed] [Google Scholar]
- 5. Trujillo K.M., Yuan S.S., Lee E.Y., Sung P.. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998; 273:21447–21450. [DOI] [PubMed] [Google Scholar]
- 6. Paull T.T., Gellert M.. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998; 1:969–979. [DOI] [PubMed] [Google Scholar]
- 7. Falck J., Coates J., Jackson S.P.. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005; 434:605–611. [DOI] [PubMed] [Google Scholar]
- 8. Berkovich E., Monnat R.J. Jr., Kastan M.B.. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007; 9:683–690. [DOI] [PubMed] [Google Scholar]
- 9. Lee J.H., Paull T.T.. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005; 308:551–554. [DOI] [PubMed] [Google Scholar]
- 10. Jazayeri A., Balestrini A., Garner E., Haber J.E., Costanzo V.. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008; 27:1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobayashi M., Hayashi N., Takata M., Yamamoto K.. NBS1 directly activates ATR independently of MRE11 and TOPBP1. Genes Cells. 2013; 18:238–246. [DOI] [PubMed] [Google Scholar]
- 12. Lee J., Dunphy W.G.. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol. Biol. Cell. 2013; 24:1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shiotani B., Nguyen H.D., Hakansson P., Marechal A., Tse A., Tahara H., Zou L.. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 2013; 3:1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao Y., Weaver D.T.. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997; 25:2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dumon-Jones V., Frappart P.O., Tong W.M., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q.. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003; 63:7263–7269. [PubMed] [Google Scholar]
- 16. Luo G., Yao M.S., Bender C.F., Mills M., Bladl A.R., Bradley A., Petrini J.H.. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKinnon P.J. Genome integrity and disease prevention in the nervous system. Genes Dev. 2017; 31:1180–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lavin M.F. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 2008; 9:759–769. [DOI] [PubMed] [Google Scholar]
- 20. Stiles J., Jernigan T.L.. The basics of brain development. Neuropsychol. Rev. 2010; 20:327–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kennedy H., Douglas R., Knoblauch K., Dehay C.. Self-organization and pattern formation in primate cortical networks. Novartis Found. Symp. 2007; 288:178–194. [DOI] [PubMed] [Google Scholar]
- 22. Gupta A., Tsai L.H., Wynshaw-Boris A.. Life is a journey: a genetic look at neocortical development. Nat. Rev. Genet. 2002; 3:342–355. [DOI] [PubMed] [Google Scholar]
- 23. McKinnon P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 2009; 10:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rass U., Ahel I., West S.C.. Defective DNA repair and neurodegenerative disease. Cell. 2007; 130:991–1004. [DOI] [PubMed] [Google Scholar]
- 25. Frappart P.O., Tong W.M., Demuth I., Radovanovic I., Herceg Z., Aguzzi A., Digweed M., Wang Z.Q.. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat. Med. 2005; 11:538–544. [DOI] [PubMed] [Google Scholar]
- 26. Zhou Z., Bruhn C., Wang Z.Q.. Differential function of NBS1 and ATR in neurogenesis. DNA Repair (Amst.). 2012; 11:210–221. [DOI] [PubMed] [Google Scholar]
- 27. Assaf Y., Galron R., Shapira I., Nitzan A., Blumenfeld-Katzir T., Solomon A.S., Holdengreber V., Wang Z.Q., Shiloh Y., Barzilai A.. MRI evidence of white matter damage in a mouse model of Nijmegen breakage syndrome. Exp. Neurol. 2008; 209:181–191. [DOI] [PubMed] [Google Scholar]
- 28. Dar I., Yosha G., Elfassy R., Galron R., Wang Z.Q., Shiloh Y., Barzilai A.. Investigation of the functional link between ATM and NBS1 in the DNA damage response in the mouse cerebellum. J. Biol. Chem. 2011; 286:15361–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galron R., Gruber R., Lifshitz V., Lu H., Kirshner M., Ziv N., Wang Z.Q., Shiloh Y., Barzilai A., Frenkel D.. Astrocyte dysfunction associated with cerebellar attrition in a Nijmegen breakage syndrome animal model. J. Mol. Neurosci. 2011; 45:202–211. [DOI] [PubMed] [Google Scholar]
- 30. Chrzanowska K.H., Gregorek H., Dembowska-Baginska B., Kalina M.A., Digweed M.. Nijmegen breakage syndrome (NBS). Orphanet. J. Rare. Dis. 2012; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Molyneaux B.J., Goff L.A., Brettler A.C., Chen H.H., Hrvatin S., Rinn J.L., Arlotta P.. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015; 85:275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Telley L., Agirman G., Prados J., Amberg N., Fievre S., Oberst P., Bartolini G., Vitali I., Cadilhac C., Hippenmeyer S. et al.. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019; 364:eaav2522. [DOI] [PubMed] [Google Scholar]
- 33. Zhou Z., Sun X., Zou Z., Sun L., Zhang T., Guo S., Wen Y., Liu L., Wang Y., Qin J. et al.. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 2010; 20:1023–1033. [DOI] [PubMed] [Google Scholar]
- 34. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 37. Gruber R., Zhou Z., Sukchev M., Joerss T., Frappart P.O., Wang Z.Q.. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat. Cell Biol. 2011; 13:1325–1334. [DOI] [PubMed] [Google Scholar]
- 38. Huenniger K., Kramer A., Soom M., Chang I., Kohler M., Depping R., Kehlenbach R.H., Kaether C.. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell. Mol. Life Sci. 2010; 67:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fares M.A., McNally D.. CAPS: coevolution analysis using protein sequences. Bioinformatics. 2006; 22:2821–2822. [DOI] [PubMed] [Google Scholar]
- 40. Pool M., Thiemann J., Bar-Or A., Fournier A.E.. NeuriteTracer: a novel ImageJ plugin for automated quantification of neurite outgrowth. J. Neurosci. Methods. 2008; 168:134–139. [DOI] [PubMed] [Google Scholar]
- 41. Finlay B.L., Darlington R.B.. Linked regularities in the development and evolution of mammalian brains. Science. 1995; 268:1578–1584. [DOI] [PubMed] [Google Scholar]
- 42. Louvi A., Artavanis-Tsakonas S.. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006; 7:93–102. [DOI] [PubMed] [Google Scholar]
- 43. Bray S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016; 17:722–735. [DOI] [PubMed] [Google Scholar]
- 44. Giniger E. Notch signaling and neural connectivity. Curr. Opin. Genet. Dev. 2012; 22:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khodosevich K., Monyer H.. Signaling in migrating neurons: from molecules to networks. Front Neurosci. 2011; 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ables J.L., Breunig J.J., Eisch A.J., Rakic P.. Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 2011; 12:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yugawa T., Handa K., Narisawa-Saito M., Ohno S., Fujita M., Kiyono T.. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 2007; 27:3732–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lagadec C., Vlashi E., Alhiyari Y., Phillips T.M., Bochkur Dratver M., Pajonk F.. Radiation-induced Notch signaling in breast cancer stem cells. Int. J. Radiat. Oncol. Biol. Phys. 2013; 87:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Santini S., Stagni V., Giambruno R., Fianco G., Di Benedetto A., Mottolese M., Pellegrini M., Barila D.. ATM kinase activity modulates ITCH E3-ubiquitin ligase activity. Oncogene. 2014; 33:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim M.Y., Ann E.J., Kim J.Y., Mo J.S., Park J.H., Kim S.Y., Seo M.S., Park H.S.. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol. Cell. Biol. 2007; 27:6506–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yashiro-Ohtani Y., He Y., Ohtani T., Jones M.E., Shestova O., Xu L., Fang T.C., Chiang M.Y., Intlekofer A.M., Blacklow S.C. et al.. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009; 23:1665–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nedjic J., Aifantis I.. RNA-binding proteins come out of the shadows. Nat. Immunol. 2010; 11:697–698. [DOI] [PubMed] [Google Scholar]
- 53. Kanner S., Goldin M., Galron R., Jacob Ben, Bonifazi E., Barzilai A.. Astrocytes restore connectivity and synchronization in dysfunctional cerebellar networks. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:8025–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou Z., Bruhn C., Wang Z.Q.. Differential function of NBS1 and ATR in neurogenesis. DNA Repair (Amst.). 2012; 11:210–221. [DOI] [PubMed] [Google Scholar]
- 56. Adelman C.A., De S., Petrini J.H.. Rad50 is dispensable for the maintenance and viability of postmitotic tissues. Mol. Cell. Biol. 2009; 29:483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li R., Yang Y.G., Gao Y., Wang Z.Q., Tong W.M.. A distinct response to endogenous DNA damage in the development of Nbs1-deficient cortical neurons. Cell Res. 2012; 22:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu Z., Brunskill E., Varnum-Finney B., Zhang C., Zhang A., Jay P.Y., Bernstein I., Morimoto M., Kopan R.. The intracellular domains of Notch1 and Notch2 are functionally equivalent during development and carcinogenesis. Development. 2015; 142:2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kopan R., Ilagan M.X.. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009; 137:216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoon K., Gaiano N.. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 2005; 8:709–715. [DOI] [PubMed] [Google Scholar]
- 61. Berezovska O., McLean P., Knowles R., Frosh M., Lu F.M., Lux S.E., Hyman B.T.. Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience. 1999; 93:433–439. [DOI] [PubMed] [Google Scholar]
- 62. Jang E.R., Choi J.D., Lee J.S.. Acetyltransferase p300 regulates NBS1-mediated DNA damage response. FEBS Lett. 2011; 585:47–52. [DOI] [PubMed] [Google Scholar]
- 63. Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knochel W., Borggrefe T.. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 2005; 25:10379–10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sato C., Zhao G., Ilagan M.X.. An overview of notch signaling in adult tissue renewal and maintenance. Curr Alzheimer Res. 2012; 9:227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barazzuol L., Ju L., Jeggo P.A.. A coordinated DNA damage response promotes adult quiescent neural stem cell activation. PLoS Biol. 2017; 15:e2001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Su Y., Ming G.L., Song H.. DNA damage and repair regulate neuronal gene expression. Cell Res. 2015; 25:993–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Arumugam T.V., Baik S.H., Balaganapathy P., Sobey C.G., Mattson M.P., Jo D.G.. Notch signaling and neuronal death in stroke. Prog. Neurobiol. 2018; 165–167:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vermezovic J., Adamowicz M., Santarpia L., Rustighi A., Forcato M., Lucano C., Massimiliano L., Costanzo V., Bicciato S., Del Sal G. et al.. Notch is a direct negative regulator of the DNA-damage response. Nat. Struct. Mol. Biol. 2015; 22:417–424. [DOI] [PubMed] [Google Scholar]
- 69. Adamowicz M., Vermezovic J., d’Adda di Fagagna F.. NOTCH1 inhibits activation of ATM by impairing the formation of an ATM-FOXO3a-KAT5/Tip60 complex. Cell Rep. 2016; 16:2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are deposited at NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE137506.