Abstract
Transgenic T-cell receptor (TCR) adoptive cell therapies recognizing tumor antigens are associated with robust initial response rates, but frequent disease relapse. This usually occurs in the setting of poor long-term persistence of cells expressing the transgenic TCR, generated using murine stem cell virus (MSCV) γ-retroviral vectors. Analysis of clinical transgenic adoptive cell therapy products in vivo revealed that despite strong persistence of the transgenic TCR DNA sequence over time, its expression was profoundly decreased over time at the RNA and protein levels. Patients with the greatest degrees of expression suppression displayed significant increases in DNA methylation over time within the MSCV promoter region, as well as progressive increases in DNA methylation within the entire MSCV vector over time. These increases in vector methylation occurred independently of its integration site within the host genomes. These results have significant implications for the design of future viral-vector gene engineered adoptive cell transfer therapies.
Keywords: adoptive cell therapy, epigenetics, DNA methylation, melanoma, sarcoma
INTRODUCTION
Genetically engineered adoptive cell therapy (ACT) is revolutionizing cancer treatment, with sustained clinical responses seen in a variety of malignancies. Current approaches utilize retroviral or lentiviral vectors for ex vivo transduction of a patient’s T-cells to express either a cancer antigen-specific T-cell receptor (TCR) or a chimeric antigen receptor (CAR). These reinfused cells then create a focused anti-tumor response in a variety of cancer subtypes (1, 2). However, while these treatments lead to durable clinical responses in many patients, a significant number of patients remain who do not respond, or who eventually relapse. Previous ACT clinical trials conducted by our group and others against the tumor antigens MART-1 (in melanoma) and NY-ESO-1 (in sarcoma and melanoma) have demonstrated that detectable surface expression of the transgenic TCR is rapidly lost in circulating T-cells following infusion (3-6). The transduction of these cells relies on retroviral vectors, most commonly the murine stem cell virus (MSCV), a γ-retrovirus which has been optimized for highly efficient transgene expression, and has been used for a variety of such applications in vivo (7). However, it has subsequently been shown to be vulnerable to epigenetic silencing via DNA methylation of CpG loci which are clustered within its 5’ long tandem repeat (LTR) promoter region (8, 9).
Given our observation of this phenomenon of rapid loss of surface expression of the transgenic TCR in circulating T-cells in vivo, along with the vulnerability of the MSCV vector to epigenetic silencing via DNA methylation, we hypothesized that acquisition of DNA methylation within the retroviral 5’LTR promoter was associated with loss of expression of the transgenic TCRs in these clinical samples. Herein we describe the analysis of clinical transgenic ACT samples for persistence of the transgenic TCR DNA sequence and accompanying expression of the TCR itself, as well as characterizing the DNA methylation status of the MSCV vector over time, and the relationship between vector methylation and suppression of transgenic TCR expression.
RESULTS
Trial conduct, patient characteristics, and outcomes
16 patients from our previous transgenic TCR ACT trials directed against MART-1 (4) and NY-ESO-1 (5) were selected for analysis. Patient demographics, clinical characteristics, and outcomes are summarized in Table 1. Following conditioning chemotherapy, patients were all treated with up to 1x109 autologous transgenic TCR T-cells, which were generated via ex vivo transduction using the MSCV γ-retrovirus encoding for the F5-MART-1 TCR or the NY-ESO-1 TCR (Figure S1). Of the 16 patients selected, seven out of eight patients treated with F-5 MART-1 TCR transgenic T-cells, and six out of eight patients treated with NY-ESO-1 TCR transgenic T-cells, demonstrated a transient objective response to therapy.
Table 1.
Patient Demographics and Outcomes
| Patient study number |
Sex (M/F) |
Ethnicity | Age | Type of Cancer |
Active Disease Sites |
Stage | Number of TCR transgenic cells |
IL-2 doses | DC doses | Evidence of transient tumor response |
Response at EOS (day 90) |
PFS (mo) |
OS (mo) |
Current Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F5-1 | M | Caucasian | 60 | Melanoma | Lung, Stomach, Liver, Pancreas, Peritoneum, Soft tissues | M1c | 1 x 109 | 12/14 | 3/3 | Yes by PET/CT | PD | 3 | 5 | Died of disease |
| F5-3 | M | Caucasian | 61 | Melanoma | Lung, Liver | M1c | 1 x 109 | 13/14 | 3/3 | Yes by PET/CT | SD | 7 | 86 | Died of disease |
| F5-6 | M | Caucasian | 59 | Melanoma | Lung, LN | M1b | 1 x 109 | 13/14 | 3/3 | Yes by PE | SD | 3 | 4 | Died of disease |
| F5-7 | M | Caucasian | 48 | Melanoma | SC, Bone | M1c | 1 x 109 | 9/14 | 3/3 | Yes by CT | SD | 4 | 11 | Died of disease |
| F5-8 | M | Caucasian | 44 | Melanoma | LN, Liver | M1c | 1 x 109 | 11/14 | 3/3 | Yes by PET/CT | SD | 4 | 11 | Died of disease |
| F5-9 | F | Caucasian | 46 | Melanoma | Skin, LN | M1a | 1 x 109 | 11/14 | 3/3 | No | PD | 3 | 20 | Died of disease |
| F5-12 | M | Caucasian | 40 | Melanoma | Lung, LN | MIVb | 3.9 x 109 | 6/9 | 3/3 | Yes by PET/CT | SD | 5 | 8 | Died of disease |
| F5-13 | M | Caucasian | 60 | Melanoma | Lung, Abdomen, SC | MIIIb | 4.41 x 109 | 4/9 | 3/3 | Yes by PET/CT | SD | 3 | 8 | Died of disease |
| ESO-1 | M | Hispanic | 47 | Liposarcoma | Right Renal Fossa; Liver left Lobe; Hepatic Segment; Peritoneal, Perihepatic Nodule | IV | 7.7 x 108 | 28/28 | 3/3 | No | PD | 2.6 | 16 | Died of disease |
| ESO-3 | F | Caucasian | 24 | Synovial Sarcoma | Right Lung; Multiple pulmonary Nodules | IV | 1 x 109 | 19/28 | 1/3 | Yes by PET/CT | PR | 67 | 67 | Alive with CR |
| ESO-4 | M | Caucasian | 41 | Synovial Sarcoma | Left Infraclavicular Mass; Left Pectoralis Mass | III | 1 x 109 | 18/28 | 3/3 | Yes by PET/CT | PD | 3 | 25 | Died of disease |
| ESO-5 | F | Caucasian | 43 | Synovial Sarcoma | Right popliteal fossa; Lung | IV | 1 x 109 | 14/14 | 3/3 | Yes by PET/CT | PR | 9 | 41.5 | Died of disease |
| ESO-6 | M | Caucasian | 26 | Osteosarcoma | Lung | IV | 1 x 109 | 14/14 | 3/3 | Yes by PET/CT | PD | 2.5 | 19 | Died of disease |
| INY-2 | M | Caucasian | 66 | Melanoma | LN, Liver | IV | 1 x 109 | 21/28 | 2/3 | Yes by PET/CT | PD | 3 | 6 | Died of disease |
| INY-3 | F | Caucasian | 44 | Synovial Sarcoma | Right popliteal fossa; Lung | IV | 1 x 109 | 14/14 | 3/3 | Yes by PET/CT | PD | 3 | 31 | Died of disease |
| INY-4 | M | Hispanic | 24 | Melanoma | Lung, LN, adrenal gland, liver, trachea, brain | IV | 1 x 109 | 10/14 | 3/3 | No | PD | NA | 3 | Died of disease |
Abbreviations: F: female; LN: lymph nodes; M: male; PFS: progression-free survival; OS: overall survival; EOS: end of study; DC: dendritic cells; TCR: T cell receptor; IL-2: interleukin-2; mo: months
Transgenic TCR-engineered T-cells display strong persistence of the transgene DNA sequence, but with greatly reduced expression of the RNA and surface protein over time
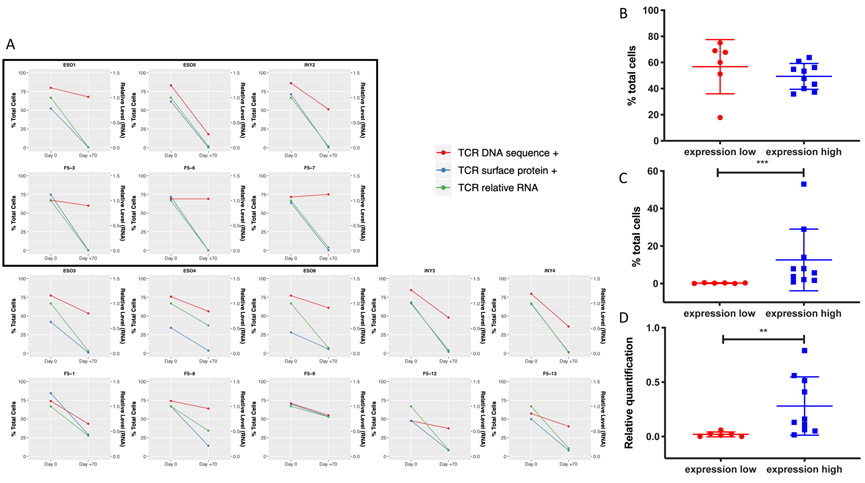
The above 16 patients had peripheral mononuclear blood cell (PBMC) samples from both their infusion (day 0) and 70 days after treatment (in peripheral circulation) analyzed for persistence of the transgenic TCR and RNA and surface TCR protein expression. All infusion products demonstrated robust presence of the transgenic TCR gene and its RNA and surface protein expression. However, we observed that despite largely decreased expression of the RNA transcript and the surface protein at day +70, persistence of the transgenic TCR still accounted for the vast majority of circulating TCR DNA clonotypes as measured by TCR sequencing using the ImmunoSEQ platform (Figure 1A, Figure S1, S2, S3). The lowest tertile of TCR surface protein expression (n=6) were noted to have <0.5% surface TCR protein expression of circulating CD3+ cells, the threshold previously established as a highly expanded clone (HEC) (10, 11). These six patients were designated as an “expression-low” cohort for further analyses, while the remaining ten patients were designated as “expression-high.” Although the expression-high cohort demonstrated correlation between transgenic TCR DNA and protein proportions when analyzed by linear regression, this correlation was not observed in the expression-low cohort (Figure S4A, B). While the degree of RNA and surface protein expression of the transgenic TCRs were significantly lower at day +70 in the expression-low group compared to the expression-high group, there were no statistically significant differences between the two cohorts’ proportion of transgenic TCR DNA in circulating PBMCs at day +70, which still accounted for the majority of circulating TCR clonotypes (Figure 1B-D, Figure S5, S6). There were also no statistically significant differences between the two groups’ transgenic TCR proportions and surface protein expression at day 0 (Figure S7A, B).
Figure 1. Persistence of transgenic TCR DNA and RNA/protein expression of TCR-engineered T-cells over time.
A) Comparison between infusion products (day 0) and post-infusion recovery products 70 days later from patients on NYESO-1 TCR-engineered cell therapy trials (ESO and INY) or F5-MART-1 TCR-engineered cell therapy trial (F5). Data displayed are percentage of cells containing the transgenic TCR DNA sequence via TCR sequencing (red, left axis), the percentage of cells expressing the TCR protein via MHC dextramer FACS (green, left axis), and the relative level of RNA transcript via qRT-PCR (blue, right axis). Inset boxed patients represent those with surface TCR expression <0.5% at day +70. B, C, D) Statistical comparisons between expression-high and expression-low patient cohorts demonstrating no significant differences between day +70 transgenic TCR DNA (B), while surface protein expression (C) and relative RNA level (D) at day +70 are significantly lower in the expression-low cohort (** p<0.01, ***p<0.001, Mann-Whitney U test).
Increased MSCV 5’LTR methylation is associated with decreased expression of the transgenic RNA and protein, despite persistence of the transgenic DNA
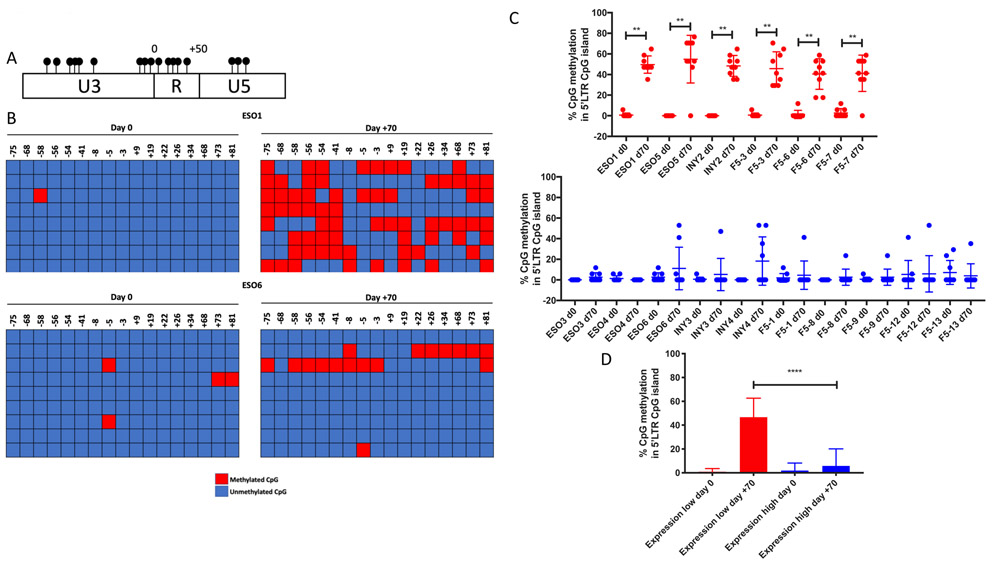
Given the overall strong predominance in transgenic TCR’s representation within the TCR repertoire of circulating T-cells despite profound loss of its expression, we explored the degree of CpG methylation within the MSCV 5’ LTR promoter region, which contains a CpG island characterized by a high concentration of clustered CpG loci over a relatively small region of DNA (Figure 2A). Genomic DNA was isolated from PBMCs of each patient sample at baseline (day 0) and 70 days post-infusion, and bisulfite converted. The area of DNA within the MSCV 5’LTR containing the CpG island was amplified by PCR, purified, and sequenced. We found that while all day 0 infusion products contained relatively little CpG methylation within the 5’LTR promoter region of the MSCV vector, the six patients in the expression-low cohort individually demonstrated significantly increased levels of CpG methylation at day +70 (Figure 2B-C, Figures S8, S9). The average proportion of promoter methylation was anticorrelated with transgenic TCR surface protein expression among all patients at day +70 (Figure S10). Furthermore, when the two cohorts were compared with one another, the expression-low cohort displayed significantly greater CpG methylation within the 5’LTR promoter region when compared to the expression-high cohort at day +70 (Figure 2D). While there were no differences in progression-free survival or overall survival between the two cohorts, the expression-low cohort displayed inferior decrease in tumor burden compared to the expression-high cohort, which nearly achieved statistical significance (p = 0.07, Figure S11A, B).
Figure 2. Increased MSCV 5’LTR methylation is associated with decreased expression of the transgenic RNA and protein, despite persistence of the transgenic DNA.
A) CpG island within the MSCV 5’ LTR, where each CpG loci is represented by a filled circle. Numbering is relative to the transcription start site. B) Bisulfite conversion was performed on genomic DNA from patient PBMCs at day 0 (infusion) and day +70, and the CpG island within the MSCV 5’LTR promoter region was PCR amplified and purified; two representative patients are shown. Each row represents sequencing of an individual experiment, with methylated cytosine loci indicated by red boxes and unmethylated loci by blue boxes. CpG loci positions are listed at the top of the graph relative to the TSS for the transgenic TCR. C) Percentage of CpG methylation within 5’LTR at day 0 and day +70 in individual expression-low (red) and expression-high (blue) patients (** p<0.01, Wilcoxon matched-pairs signed rank test); comparison between aggregate percent CpG methylation within the 5’LTR between all expression-high and expression-low patients at day 0 and day +70 is shown in D (**** p<0.0001, Mann-Whitney U test).
CpG methylation is increased across MSCV vector over time in all patients, and is significantly greater in those with decreased transgenic TCR expression over time
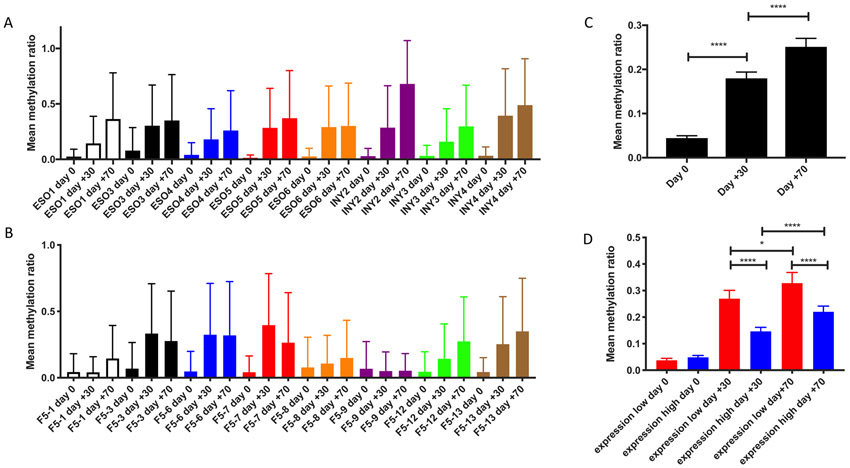
In order to expand our ability to characterize CpG methylation across the entire MSCV vector over time, we performed bisulfite conversion on genomic DNA library preparations isolated from all patients’ PBMC samples at day 0 (infusion), day +30, and day +70. We then carried out target enrichment using RNA probes to capture the MSCV vector. Bisulfite-converted libraries were then aligned against the human genome version 38 (hg38) with MSCV transgenic TCR vector reference sequences utilizing BSBolt, an integrated alignment and analysis platform for bisulfite-converted DNA. Each patient sample demonstrated overall progressive increases in CpG methylation across the transgenic TCR MSCV vector sequence over time (Figure 3A-B). When all patient sample data were aggregated, the increases in MCV vector CpG methylation were statistically significant at day +30 compared to day 0, and day +70 compared to day +30 (Figure 3C). When the data were further stratified to compare the expression-high and expression-low patient cohorts, we observed that while there were no significant differences between baseline levels of CpG methylation at day 0, the day +30 and day +70 methylation ratios, which progressively increased over time in both cohorts, were significantly higher in the expression-low cohort when compared to the expression-high cohort, consistent with what was observed in the targeted analysis of the 5’LTR promoter region (Figure 3D).
Figure 3. CpG methylation is increased across MSCV vector over time in all patients, and is significantly greater in those with decreased transgenic TCR expression over time.
Mean methylation ratio across MSCV vector at day 0, day +30, and day +70 for each patient treated with NY-ESO-1 TCR (A) and F5-MART-1 TCR (B) transgenic T-cells. C) Statistical comparisons between methylation values in all patients at day 0, day +30, and day +70; data are stratified to compare the increases in methylation values over time between the expression-high and expression-low patients in D (*, p<0.05, **** p<0.0001, Mann-Whitney U test).
MSCV-TCR vector integration occurs sporadically throughout the host genome relative to transcription start sites
While capturing the MSV vector fragments, we also obtained fragments that spanned the junction of the insertion site between the vector and the genome. These reads allowed us to explore the integration patterns of the MSCV vector within the patients’ genomes. Specifically, we utilized BSBolt to map discordant paired reads, where individual reads from the pair map to both the human genome and the MSCV transgenic TCR vector. Integration sites were characterized by their relative and absolute distance to transcription start sites (TSS). We observed that the MSCV vector integration sites occurred at sites generally distal to gene TSS (Figure S12A, B). Furthermore, when we compared the proportions of vector integration sites by their relative distance to TSS, we observed no significant differences between the expression-low cohort (i.e. those with high CpG methylation levels) and the expression-high cohort (i.e. those with low CpG methylation levels), suggesting that the integration site relative to TSS did not impact the degree of CpG methylation observed within a given vector read.
DISCUSSION
TCR transgenic ACT has established itself as a potent form of cancer immunotherapy for a wide variety of tumor subtypes. However, despite frequent early responses and reduction in tumor burden, the durability of these responses is often poor, and tumors often progress within several months. Our clinical experiences with transgenic TCR ACT generated with γ-retroviral vectors, as well as those of other groups, have consistently demonstrated that the presence of detectable circulating transgenic TCR surface expression rapidly diminishes within 1-2 months following cell transfer, in keeping with the timeline of disease progression after the initial transient response to therapy (3-6). This is in stark contrast to ACT using autologous cancer-antigen-specific TCR clones, which are isolated, expanded ex vivo, and reinfused to the patient without the aid of any viral vectors to transduce and generate these cells in large numbers. Previous ACT studies utilizing such endogenous TCR clones have shown remarkably strong persistence of cancer-antigen-specific TCR clones in circulation following cell transfer (12, 13). This discrepancy implies that genetically engineered ACT products have a fundamental vulnerability in the suppression of their transgenic TCR.
Given the known vulnerability of the MSCV vector to epigenetic silencing via CpG methylation, as well as the observed discordance between the strong persistence of the TCR transgenes and their poor expression in circulation, we hypothesized that increases in DNA methylation within the vector were associated with this phenomenon. While one small series of patients previously found no significant retroviral promoter methylation to cause suppression of transgenic TCR expression (14), that study only examined the methylation status of the MSCV vector promoter in a total of four patients. Our examination of 16 samples from patients receiving transgenic ACT demonstrated that samples from only six of these patients displayed significantly discordant, profound decreases in expression of the transgenic TCR which was associated with significant increases in MSCV vector methylation. This suggests that the phenomenon occurs in a minority of patients, and could be missed by sampling too small a cohort. Indeed, there may be cell phenotype-specific or patient-specific predispositions to rapid acquisition of CpG methylation of γ-retroviruses which would not be present in every subject studied. While further detailed studies of factors such as patient-specific polymorphisms in DNA methyltransferase enzymes would be needed to derive even speculative inferences into such predispositions, our studies did not demonstrate any significant association with the MSCV integration site’s distance to a given TSS at infusion and its propensity to acquire CpG methylation over time. Furthermore, our characterization of the MSCV vector integration sites was consistent with previously published studies dealing with the γ-retroviral vector murine leukemia virus (MLV), which showed that only ~25% of γ-retrovirus integration sites are within ±2.5-kb around the TSS (15) in human cells, consistent with our results. While MSCV has previously shown increased integration near TSS in murine bone marrow cells (16), it may be that there are species-specific factors which influence integration site, as our data are consistent with previously published data in humans.
The vulnerability of retroviral vectors to epigenetic suppression in transgenic TCR ACT products seen here raises an important question about how to overcome this potential weakness in clinical practice. One possibility would be to utilize dual therapy with systemic hypomethylating agents such as decitabine to prevent the acquisition of CpG methylation within the retroviral vector encoding the transgenic TCR. Such agents have also been shown in murine models to increase the expression of tumor antigens commonly targeted by transgenic TCR ACT, such as NY-ESO-1 (17, 18), theoretically further enhancing the immunotherapeutic effect of the transgenic T-cells. However, DNA methylation is often followed by histone recruitment and modification (de-acetylation and/or methylation), which further contribute to epigenetic suppression. Therefore, such pharmacologic interventions would potentially be insufficient to fully reverse epigenetic suppression, as it would not have an effect on the histone modifications and recruitment. Stimulation of T-cells with IL-2 and anti-CD3/CD28 beads has previously been shown to partially restore transgenic TCR expression in vitro in some patients, likely due to nonspecific modulation of these epigenetic factors (14). New non-viral approaches to generating transgenic TCR T-cells using CRISPR-Cas9 to deliver constructs under control of the native TCR promoter (rather than a viral promoter) would also potentially avoid this risk of epigenetic suppression of viral vector-encoded products (19). Furthermore, there may be utility in new modalities that provide a continuous supply of transgenic T-cells to the patient. Preclinical models have demonstrated that CD34+ hematopoietic stem cells encoding a transgenic TCR can endogenously differentiate into fully functional T-cells expressing the TCR (20, 21). We currently have recently an open phase I clinical trial which utilize this approach against NY-ESO-1 in solid tumors (NCT03240861), utilizing a lentiviral stem cell transduction for long-term expression.
One major limitation of our study is that we only examined transgenic TCR ACT products generated using the MSCV γ-retrovirus, which is known to be potentially vulnerable to epigenetic silencing via CpG methylation. Other types of cell therapy products, including the CD19 CAR-T product Kymriah™ (tisagenlecleucel), utilize lentiviral vectors, which have previously demonstrated remarkable persistence of transgene expression in vivo and do not appear vulnerable to CpG methylation (22), which may limit the broader applicability of our findings. Indeed, transgenic TCR ACT products manufactured using lentiviral vectors have been shown to have far superior persistence of detectable surface expression of the TCR (23). While the other commercially available CD19 CAR-T product Yescarta™ (axicabtagene ciloleucel) does utilize a γ-retroviral vector for its manufacture, its rates of durable complete and partial remission are far superior to any published transgenic TCR product (2). This is likely due to the rapid systemic clearance of lymphoma cells seen when using these ACT products, implying that long-term persistence of the transgenic T-cells in this setting is potentially less important than in treating refractory solid tumors with such therapeutics. Indeed, our examination of patient samples at day +70 was chosen due to this being the average point of circulating transgenic T-cell nadir in our previously published trials (4, 5). We were unable to determine any association of γ-retroviral vector methylation with patient survival, likely owing to the overall small number of long-term responders inherent to this therapy in solid tumors.
In summary, we have shown that progressive increases in CpG methylation within the MSCV γ-retroviral vector are associated with rapid suppression of transgenic TCR expression over time in clinical transgenic ACT, despite strong persistence of the transgene itself. This phenomenon did not appear to have any correlation with vector integration site within the host genome. These findings have significant implications in how the cellular therapeutics community should approach the design of future generations of these products.
MATERIALS AND METHODS
Clinical trial, patients, and manufacturing of MART-1 and NY-ESO-1 TCR engineered T-cells
For the F5-MART-1 transgenic TCR adoptive cell therapy clinical trial, patients positive for HLA-A*0201 with a MART-1-positive metastatic melanoma were enrolled under NCT00910650 (UCLA IRB #08-02020 and #10-001212) from April 2009 to September 2011, under investigational new drug (IND) #13859 (4). For the NYESO-1 transgenic TCR adoptive cell therapy clinical trials, patients positive for HLA-A*0201 with an NYESO-1-positive sarcoma or melanoma were enrolled under NCT02070406 or NCT01697527 (UCLA IRB #12-000153 and #13-001624, respectively) under IND#15167 (5). Written informed consent was obtained from all patients, and studies were conducted in accordance with local regulations, the guidelines for Good Clinical Practice, and the principles of the Declaration of Helsinki. All studies were approved by the UCLA institutional review board under the above approval numbers. Clinical trial design and manufacturing of the MART-1 and NYESO-1 TCR transgenic T-cells are previously described (4, 5). Briefly, non-mobilized autologous PBMCs were stimulated in culture with IL-2/OKT3 and transduced with clinical grade MSCV retrovirus vector expressing the MART-1 F5 TCR or the NYESO-1 TCR on two consecutive days, then continually expanded ex vivo for 6-7 days. Up to 1x109 transgenic TCR transgenic lymphocytes were administered to each patient following conditioning chemotherapy with cyclophosphamide and fludarabine, along with post-infusion systemic IL-2 for 7-14 days, and dendritic cell vaccine boosts, as previously described (4, 5).
Quantification of transgenic TCRβ genomic DNA persistence
Genomic DNA and RNA was isolated from patient-matched infusion products and post-infusion PBMCs recovered at day +70 (+/− 10 days), with an AllPrep DNA/RNA isolation kit according to the manufacturer’s instructions (Qiagen). TCRβ alleles were sequenced at 100,000 reads by Adaptive Biotechnologies (Seattle, WA). Bulk TCR sequencing was utilized due to its superior sampling depth (previously characterized as 20-fold greater) and sensitivity over scRNAseq-based approaches in detection of non-transgenic TCR clonotypes (24). Briefly, this process utilizes a synthetic immune repertoire, corresponding to every possible biological combination of Variable (V) and Joining (J) gene segments for each T-cell receptor locus, spiked into every sample at a known concentration. These inline controls enable rigorous quality assurance for every sample assayed and allow for correction of multiplex PCR amplification bias, providing an absolute quantitative measure of T-cells containing the transgenic TCR relative to the other endogenous TCR clonotypes, with no difference in amplification efficiency (25). Productive TCRβ sequences, i.e. those that could be translated into open reading frames and did not contain a stop codon, were reported. The transgenic F5-MART-1 and NY-ESO-1 TCR sequences’ persistence were identified based on comparison of reads with the known TCRβ sequence for the transgenic product, and expressed as a percentage of total productive TCRβ sequences present within a given sample/timepoint.
qRT-PCR
Total RNA isolated from patient samples (as described above) was used for analysis of relative abundance of the transgenic F5 MART-1 TCR or the NYESO-1 TCR. Samples were converted to cDNA using iScript TM Reverse Transcription Supermix for RT-PCR (Bio-Rad), then cDNA was amplified and quantified using iTaq TM Universal SYBR Green Supermix (Bio-Rad) on an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems). PCR conditions were 1 cycle of 1 min at 95°C, 35 cycles of 15 sec at 95°C and 60 sec at 60°C, and 5 min incubation at 72°C. Replicate samples were run with test primer sets for the F5-MART-1 TCR, the NYESO-1 TCR, or the endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH); primer sequences are available upon request. Data were analyzed according to the comparative Ct method.
MHC dextramer immunologic monitoring for surface expression of transgenic TCRs
Detection and quantification of F5 MART-1 TCR or NY-ESO-1 TCR expression using fluorescent MHC dextramer analysis for MART-1 or NY-ESO-1 (Immudex) was performed on patient-matched infusion products and post-infusion PBMCs recovered at day +70, as previously described (4, 5, 26). Our definitions for a positive or negative immunologic response using standardized MHC multimer assays were used, which are based on assay performance specifications by defining changes beyond the assay variability with a 95% confidence level (26).
Bisulfite sequencing of MSCV retroviral promoter in patient samples
Genomic DNA isolated from patient PBMC samples was bisulfite converted using the EZ DNA Methylation-Gold Kit (Zymo Research) according to the manufacturer’s instructions. A CpG island within the MSCV 5’LTR promoter U3/R/U5 region, defined as an area >100bp, with a GC content of >50%, and possessing an observed:expected CpG ratio of >0.6, was determined using MethPrimer software (27), which also designed PCR primers capable of amplifying the methylated and non-methylated bisulfite-converted DNA sequence of interest. CpG islands were PCR amplified, purified, and subjected to DNA Sanger sequencing (Laragen). The methylation status of each CpG locus within the individual amplicons was determined using QUMA software (28).
Targeted bisulfite sequencing library preparation and sequencing
Purified genomic DNA from patient samples (isolated as described above) was quantified using the Qubit dsDNA BR Assay (Thermo Fisher Scientific). For each sample, 250ng of DNA was sonicated using a Bioruptor Pico (Diagenode) for 15 cycles (30 sec ON; 60 sec OFF). Libraries were prepared using the NEBNext Ultra II DNA kit (NEB) according to manufacturer instructions with few modifications. Briefly, sonicated DNA was subjected to EndPrep (End Repair and A-tailing), followed by Adapter Ligation using 2.5μL of Illumina TruSeq pre-methylated Adapters (Illumina). Samples were purified using 0.85x NEB Purification Beads and eluted in 15μL of 10mM Tris-HCl, pH 8. Samples were mixed in 16-sample pools and column purified using a DCC-5 (Zymo Research). Elution was performed with 10μL of 60℃ 10mM Tris-HCl, pH 8. Each sample pool was subject to hybrid capture with custom biotinylated RNA probes designed to tile the MSCV vector (MyBaits Arbor Bioscience - Human_6K and Human patch2) according to the manufacturer’s protocol. The hybridization was carried out for 20 hours overnight at 65℃. Captured DNA was eluted by heating in 20μL of 10mM Tris-HCl pH 8+0.05% Tween-20. The eluted DNA was then subject to bisulfite conversion using the DNA Methylation Lightning kit (Zymo Research). Converted DNA was then amplified using xGen Library Amplification Primer Mix (IDT) and Kapa Uracil+ Ready Mix using the following conditions: 98℃ for 2 min; 20 cycles of 98℃−20 sec, 60℃−30 sec, 72℃−30 sec; Final Extension 72℃−5 min; hold 4℃. PCR products were purified using 0.9 volumes of NEBNext Purification Beads and eluted in 15μL of 10mM Tris-HCl, pH 8. Final libraries were then quantified using the Qubit dsDNA BR Assay (Thermo Fisher Scientific) and visualized using a D1000 ScreenTape (TapeStation 2200 system - Agilent Technologies). Each pool was then sequenced at 150bp PE on a HiSeq3000 instrument (Illumina).
Targeted bisulfite sequencing alignment and methylation calling
Paired end, 150bp targeted bisulfite sequencing reads were aligned to the combined hg38 and MSCV transgenic TCR vector sequence bisulfite converted references using BSBolt v0.1.2 (https://github.com/NuttyLogic/BSBolt) local alignment. Alignments with <5 mismatches and an alignment score >160 were considered valid, up to 10 alignments per read pair were considered. Alignments where read pairs did not meet expected paired end constraints, an insert size >500bp or alignments on separate chromosomes, were reported as discordant. Reads pairs with only one valid alignment were reported as mixed. Duplicate reads were removed using SAMtools v1.9 (29). Following duplicate removal methylation values were called for all observed cytosines with ≥5 reads with a base call quality above 25 using BSBolt v0.1.2 (https://github.com/NuttyLogic/BSBolt). Sequencing data available in NCBI’s Gene Expression Omnibus (accession number GSE153820).
Vector integration site detection
Discordant reads pairs with a vector alignment and a genome alignment were evaluated as potential integration sites. Discordant read pairs were further filtered by removing reads that aligned to genomic regions homologous with the vector sequence or aligned outside the expected integration region within the vector sequence. Alignments with an alignment score greater than 160 and with secondary alignments that repeated no more than 10% of the primary alignment sequence were considered integration site supporting alignments. Integration sites were reported as the closest genomic base to vector alignment or the average integration site position for sites with multiple supporting reads. The integration site selection pipeline was implemented using custom python code (https://github.com/NuttyLogic/Epigenetic-suppression-of-transgenic-TCR-expression-in-ACT). The vector integration detection pipeline was validated against simulated 150bp, paired end bisulfite converted vector integration libraries; see Supplemental Methods for further details.
Statistical analysis
Graphing and descriptive statistical analyses were carried out with GraphPad Prism version 7.0 (GraphPad). Where indicated, Mann-Whitney U test or Wilcoxon matched-pairs signed rank test were used for comparison of two groups. Linear regression was performed comparing the correlation of transgenic TCR DNA and surface protein expression in the expression-high and expression-low cohorts (as the independent and dependent variables, respectively), as well as comparing CpG promoter methylation and transgenic TCR expression in all patient samples (as independent and dependent variables, respectively). P values of <0.05 were considered statistically significant.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Cellular immunotherapies’ reliance on retroviral vectors encoding foreign genetic material can be vulnerable to progressive acquisition of DNA methylation and subsequent epigenetic suppression of the transgenic product in TCR ACT. This must be considered in the design of future generations of cellular immunotherapies for cancer.
ACKNOWLEDGEMENTS
Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by NIH awards P30 CA016042 and 5P30 AI028697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
GRANT SUPPORT: This study was funded in part by NIH grants R35 CA197633 and P01 CA168585, the Parker Institute for Cancer Immunotherapy, and the Ressler Family Fund (to A.R.). T.S.N. is supported by the NIH/NICHD grant K12-HD000850 (Pediatric Scientist Development Program), the Tower Cancer Research Foundation Career Development Award, and the Hyundai Hope on Wheels Young Investigator Award. K.M.C. is supported by the UCLA Tumor Immunology Training Grant (NIH T32CA009120) and the Cancer Research Institute Irvington Postdoctoral Fellowship Program.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST: T.S.N. has received honoraria from consulting with Allogene Therapeutics. A.R. has received honoraria from consulting with Amgen, Bristol-Myers Squibb, Chugai, Genentech, Merck, Novartis, Roche and Sanofi, is or has been a member of the scientific advisory board and holds stock in Advaxis, Arcus Biosciences, Bioncotech Therapeutics, Compugen, CytomX, Five Prime, FLX-Bio, ImaginAb, Isoplexis, Kite-Gilead, Lutris Pharma, Merus, PACT Pharma, Rgenix and Tango Therapeutics, and has received research funding from Agilent and Bristol-Myers Squibb through Stand Up to Cancer (SU2C). B.C-A. has received honoraria from consulting with Advarra IBC. K.M.C. is a shareholder in Geneoscopy LLC. The rest of the authors declare no potential conflicts of interest.
DISCLAIMER: The contents of this article are solely the responsibility of the authors and do not necessarily represent the official view of the NIH, the NCI, the NICHD, the Parker Institute for Cancer Immunotherapy, or the Ressler Family Foundation.
REFERENCES
- 1.Yang JC, Rosenberg SA. Adoptive T-Cell Therapy for Cancer. Advances in immunology 2016, 130:279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June CH, Sadelain M. Chimeric Antigen Receptor Therapy. N Engl J Med 2018, 379:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006, 314:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chodon T, Comin-Anduix B, Chmielowski B, et al. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin Cancer Res 2014, 20:2457–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowicki TS, Berent-Maoz B, Cheung-Lau G, et al. A Pilot Trial of the Combination of Transgenic NY-ESO-1-reactive Adoptive Cellular Therapy with Dendritic Cell Vaccination with or without Ipilimumab. Clin Cancer Res 2019, 25:2096–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins PF, Kassim SH, Tran TL, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res 2015, 21:1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawley RG, Lieu FH, Fong AZ, et al. Versatile retroviral vectors for potential use in gene therapy. Gene Ther 1994, 1:136–138. [PubMed] [Google Scholar]
- 8.Swindle CS, Kim HG, Klug CA. Mutation of CpGs in the murine stem cell virus retroviral vector long terminal repeat represses silencing in embryonic stem cells. J Biol Chem 2004, 279:34–41. [DOI] [PubMed] [Google Scholar]
- 9.Yao S, Sukonnik T, Kean T, et al. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther 2004, 10:27–36. [DOI] [PubMed] [Google Scholar]
- 10.Klarenbeek PL, de Hair MJ, Doorenspleet ME, et al. Inflamed target tissue provides a specific niche for highly expanded T-cell clones in early human autoimmune disease. Ann Rheum Dis 2012, 71:1088–1093. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y, Li B, Li Y, et al. A comprehensive immune repertoire study for patients with pulmonary tuberculosis. Mol Genet Genomic Med 2019, 7:e00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudley ME, Wunderlich JR, Robbins PF, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 2002, 298:850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunder NN, Wallen H, Cao J, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med 2008, 358:2698–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns WR, Zheng Z, Rosenberg SA, et al. Lack of specific gamma-retroviral vector long terminal repeat promoter silencing in patients receiving genetically engineered lymphocytes and activation upon lymphocyte restimulation. Blood 2009, 114:2888–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Ravin SS, Su L, Theobald N, et al. Enhancers are major targets for murine leukemia virus vector integration. J Virol 2014, 88:4504–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aker M, Tubb J, Miller DG, et al. Integration bias of gammaretrovirus vectors following transduction and growth of primary mouse hematopoietic progenitor cells with and without selection. Mol Ther 2006, 14:226–235. [DOI] [PubMed] [Google Scholar]
- 17.Chou J, Voong LN, Mortales CL, et al. Epigenetic modulation to enable antigen-specific T-cell therapy of colorectal cancer. J Immunother 2012, 35:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollack SM, Li Y, Blaisdell MJ, et al. NYESO-1/LAGE-1s and PRAME are targets for antigen specific T cells in chondrosarcoma following treatment with 5-Aza-2-deoxycitabine. PLoS One 2012, 7:e32165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth TL, Puig-Saus C, Yu R, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 2018, 559:405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Baltimore D. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc Natl Acad Sci U S A 2005, 102:4518–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vatakis DN, Koya RC, Nixon CC, et al. Antitumor activity from antigen-specific CD8 T cells generated in vivo from genetically engineered human hematopoietic stem cells. Proc Natl Acad Sci U S A 2011, 108:E1408–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeifer A, Ikawa M, Dayn Y, et al. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc Natl Acad Sci U S A 2002, 99:2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Angelo SP, Melchiori L, Merchant MS, et al. Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov 2018, 8:944–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yost KE, Satpathy AT, Wells DK, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019, 25:1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson CS, Emerson RO, Sherwood AM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nature communications 2013, 4:2680. [DOI] [PubMed] [Google Scholar]
- 26.Comin-Anduix B, Gualberto A, Glaspy JA, et al. Definition of an immunologic response using the major histocompatibility complex tetramer and enzyme-linked immunospot assays. Clin Cancer Res 2006, 12:107–116. [DOI] [PubMed] [Google Scholar]
- 27.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002, 18:1427–1431. [DOI] [PubMed] [Google Scholar]
- 28.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 2008, 36:W170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.