Abstract
Extreme environments often result in the evolution of dramatic adaptive features. The Mexican tetra, Astyanax mexicanus, includes 30 different populations of cave-dwelling forms that live in perpetual darkness. As a consequence, many populations have evolved eye loss, reduced pigmentation, and amplification of non-visual sensory systems. Closely-related surface-dwelling morphs demonstrate typical vision, pigmentation, and sensation. Transcriptomic assessments in this system have revealed important developmental changes associated with the cave morph, however they have not accounted for photic rearing conditions. Prior studies reared individuals under a 12:12 hr light/dark (LD) cycle. Here, we reared cavefish under constant darkness (DD) for 5+ years. From these experimental individuals, we performed mRNA sequencing and compared gene expression of surface fish reared under LD conditions to cavefish reared under DD conditions to identify photic-dependent gene expression differences. GO enrichment analyses revealed a number of previously underappreciated cave-associated changes impacting blood physiology and olfaction. We further evaluated the position of differentially expressed genes relative to QTL positions from prior studies, and found several candidate genes associated with these ecologically relevant lighting conditions. In sum, this work highlights photic condition as a key environmental factor impacting gene expression patterns in blind cave-dwelling fish.
Keywords: RNA-seq, Mexican tetra, troglomorphy, regressive evolution
Graphical Abstract
INTRODUCTION
Dramatic environmental shifts can have equally dramatic impacts on an organism’s phenotype. These extreme changes can impact behavioral (Dall et al. 2012), morphological (Temerin and Cant 1983) or pigmentation phenotypes (Greenwood et al. 2011). However, these divergent phenotypes are often accompanied by changes at the molecular level, i.e. alterations to genetic regulation or structure (Carroll 2008, West-Eberhard 2003, Hoekstra et al. 2006), effectively linking the environment with the genomic and transcriptomic architectures. While taking many forms, deciphering this connection has been a key challenge in contemporary biological research. One increasingly utilized model to examine this link is the blind Mexican cavefish, Astyanax mexicanus. This species exists in two distinct morphotypes – a surface morph and cave-adapted morphs, found naturally in 30 distinct populations. Despite substantial similarities in their genomes, embryonic development, and geographic origin, these animals demonstrate very different phenotypes. The extant surface morphotype inhabits rivers and streams surrounding the Sierra de El Abra cave system of northeastern Mexico. Cavefish reside amidst a complex series of limestone caves in this region, and have evolved a number of “regressive” traits (Protas et al. 2006; Jeffery 2009a; Duboué et al. 2011), including the loss of eyes and a reduction of melanic pigmentation (Şadoğlu 1957; 1967; Jeffery 2001; 2008; Yamamoto et al. 2003; Wilkens 2007), as well as “constructive” traits such as the expansion of non-visual sensory systems (Hassan 1989; Yoshizawa et al. 2014).
Despite these phenotypic differences, cave and surface morphs are members of the same species owing to their high genetic similarity and ability to interbreed to produce viable offspring (Şadoğlu 1957; Avise and Selander 1972). Robust differences in gene expression (e.g., shh, twhh, and pax6) accompany many of the well-known phenotypic differences between the two morphotypes (Strickler et al. 2001; Yamamoto et al. 2003; Retaux et al. 2008; Pottin et al. 2011). Previous transcriptomic studies in this system capitalized on the genetic similarity between cave and surface fish to characterize genome-wide differential gene expression (Gross et al. 2013), eye loss (Hinaux et al. 2013) and convergent embryonic expression patterns (Stahl and Gross 2017).
Importantly, caves are characterized by a number of selective forces, including low food resources and absence of light (Elliott 2015; Jeffery 2015). Despite this absence of light, prior transcriptomic studies were carried out on cavefish reared under a 12:12 LD schedule. However, darkness is known to influence a number of regressive characters (Gonzalez and Aston-Jones 2008; Emerling and Springer 2014), such as downregulation of phototransduction genes in the Chinese cavefish, Sinocyclocheilus anophthalmus (Meng et al. 2013).
Mexican cavefish respond to light, despite the fact that most populations live in complete darkness and lose eyes through development. This light-sensing ability occurs through the pineal gland (Axelrod 1974; Yoshizawa and Jeffery 2008), and, bright light causes structural changes to photoreceptor cells of the pineal gland (Omura 1975). Conversely, surface fish raised in total darkness have smaller retinas, and their eyes appear darker owing to less guanine deposition (Wilkens 1988; Gross et al. 2016), and they lose normal shoaling behaviors (Gregson and Burt de Perera 2007). When cavefish are reared in their natural photic conditions (constant darkness), they show very different motor activity and tank usage compared to those reared in light-dark conditions (Carlson and Gross 2018). Therefore, despite its effect on diverse phenotypes in Astyanax, the effect of light on gene expression has not yet been characterized in this system.
Here we present a transcriptomic analysis of cavefish and surface fish reared under two photic conditions each, constant darkness (DD) and a diurnal light/dark photoperiod (LD). Our interests centered mainly on the contrasts in gene expression under “natural” photic conditions, i.e. between cavefish reared in constant darkness and surface fish reared under light/dark conditions. Cavefish and surface fish demonstrate measurable behavioral differences under different lighting conditions (Carlson et al., 2018). Therefore, we predicted that this analysis would uncover numerous ecologically-relevant gene expression changes that would otherwise go unnoticed in studies that do not account for lighting condition. We found that gene expression differences did not simply reflect an expansion of previously established differences, but rather we found novel patterns associated with these normative lighting regimes. This work, which examines lighting as an essential regulator of gene expression, enables a clearer understanding of the interaction between genotype, phenotype, and environment in this fascinating natural model system.
METHODS
Animal Rearing
All experimental animals were maintained under identical light-dark (LD) or constant darkness (DD) rearing conditions. Our approach was to perform comprehensive RNA-sequencing of four conditions (two morphotypes under two lighting regimes) and perform pair-wise comparisons to examine how light impacts gene expression in cave and surface fish. Under the LD routine, animals experienced light within the room (494.62 ± 28.43 Lux) for 12 hours between 8:00am to 8:00pm (EST), followed by darkness (0.034 ± 0.007 Lux) for 12 hours. All light measurements were performed using an LT300 light meter (Extech Instruments, Nashua, NH). All fish were maintained in 5-gallon glass tanks with continuous flow of conditioned water (pH = 7.4 ±0.2, conductivity = ~800 μS/cm; Aquaneering, San Diego, CA). All fish were maintained on a diet of flake food (TetraMin Pro). A subset of Pachón cavefish and surface fish embryos (>12 hours post-fertilization) were reared under dark-dark (DD) conditions for over five years. Animals were dark-reared in 5-gallon glass tanks wrapped in impermeable black construction paper. While the light level in LD tanks averaged 48.59 ± 6.32 lux, the DD tanks measured an average of only 0.27 ± 0.06 lux. This difference in illumination inside tanks is likely due to presence of shadows, which are a function of the physical placement of the tanks within our husbandry facility. We still observed a ~180-fold difference in light intensity between the LD and DD conditions, irrespective of tank placement within the husbandry facility. All other variables were held constant between LD and DD tanks and individuals. These experiments were conducted in compliance with the University of Cincinnati Institutional Animal Care and Use Committee (Protocol # 10-01-21-01).
RNA Isolation, Sequencing and Processing
RNA-sequencing was conducted on (n=4) adult individuals from each of the following groups: LD-reared surface fish (SLD), DD-reared surface fish (SDD), LD-reared cavefish (CLD) and DD-reared cavefish (CDD), for a total of 16 biological samples. All individuals were ~ five-years-old at the time of RNA extraction. We evaluated two lab-reared populations of Astyanax mexicanus originating from the Sierra de El Abra region of Mexico. These included individuals from the Pachón cave (pedigree 138) in Tamaulipas, Mexico; and surface-dwelling fish (pedigree 155) from the Río Sabinas and Río Valles drainages near Ciudad Valles, Mexico. Fish were originally obtained and generously provided to us by Dr. Richard Borowsky (NYU). Approximately four clutches of each morphotype were divided in half and randomly assigned to each lighting condition. RNA extractions were performed between 11am and 2pm (EST) to minimize variation associated with circadian patterns of gene expression (Idda et al. 2012). RNA extractions were performed under red-light illumination for DD individuals. Whole RNA isolation was performed separately for head and flank tissues, processed immediately using liquid nitrogen for tissue homogenization, and carried out using the RNeasy Universal Mini Kit according to the manufacturer instructions (Qiagen, Germantown, MD). Each sample was quality-assessed using a spectrophotometer, and samples were then pooled together and submitted to a sequencing core. Owing to our approach of pooling samples, we acknowledge the possibility that certain gene expression differences between cave and surface fish may arise as a simple consequence of presence/absence of certain structures (e.g., such as the eye). Having said this, the absence of certain vision-related genes can also serve as an important control for gene expression and GO enrichment analyses given our prior demonstration of reduced expression of vision-related genes in cave morphs (Gross et al., 2013). Samples were stored at −80°C prior to library synthesis and sequencing.
All RNA samples were then subjected to QC analysis (RQN>7) at the Cincinnati Childrens DNA Sequencing Core. Libraries were generated using only the pooled RNA extracts of head tissue, with the Illumina TruSeq 2 Poly-A stranded tail kit and sequenced as paired-end, 75bp reads to a depth of 10 million reads per sample (Illumina HiSeq 2500). Raw fastq-formatted reads were assessed using FastQC (v0.11.5) and filtered to remove TruSeq3-PE adapter sequences, trimmed for low quality leading- and trailing-bases below a quality measure of 3 (sliding window 4–15), and read lengths less than 36 bp (Trimmomatic software; Bolger et al. 2014). Following trimming, FastQC analyses were performed again to confirm removal of adapter and poor-quality sequences.
Differential Expression Analyses
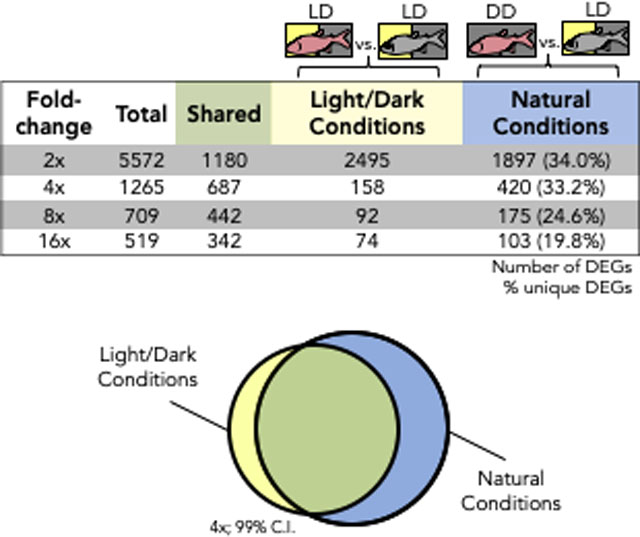
Reads were aligned to the draft cavefish genome (v.93, ensembl.org) using ArrayStar (v15.0, DNAStar, Madison, WI). Each of our pooled samples was subjected to high-throughput sequencing as technical triplicates. Following alignment, we grouped all sequencing runs according to triplicate group and photic condition in order to evaluate within-sample variations across different sequencing runs. Normalized expression (RPKM; Mortazavi et al. 2008) was calculated for each experimental group: CLD, SLD, CDD and SDD. Expression similarity was quantified based on a global correlative value between experimental groups (Fig. 1). “Natural Conditions” refers to the comparison of CDD and SLD (Fig. 1C), and “Light/Dark Conditions” refers to comparisons between CLD and SLD (Fig. 1B). The number of differentially expressed genes (DEGs) were determined by fold-change and confidence interval thresholds, and all DEG numbers were reported at a 99% confidence interval (Fig. 2; 3A). Genes that were deemed sensitive to lighting condition were based on differences in fold-change between natural conditions and light/dark conditions. The fold-change cutoff reported in Table 1 was generated based on a 2.5x threshold in CDD and SLD.
Figure 1. Correlations between morphotypes are least similar under natural conditions.
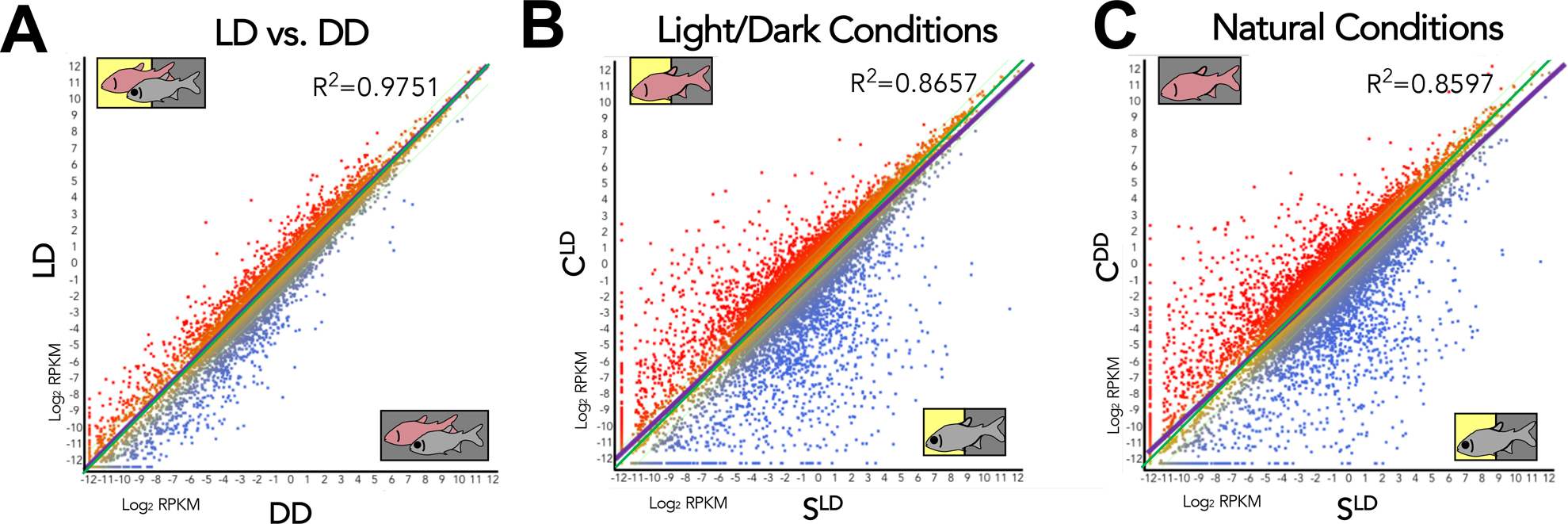
Each point on this scatterplot represents an individual gene. Some genes (red) have a higher expression in the condition on the y-axis. Other genes (blue) have a higher expression in the condition on the x-axis. LD vs. DD (A) is more similar than light/dark conditions (B), while natural conditions (C) are the least related based on the R2 and spread of points about the line of regression (purple). As each of these scatterplots encapsulates all 25,271 genes, any visible changes correlate to large scale changes in global gene expression. The increasing distance between the line of regression (purple) and the zero-fold line (green) viewed between panels A to C yields an approximation of the increase in difference between the two conditions.
Figure 2. More genes are under-expressed in cavefish, regardless of rearing conditions.
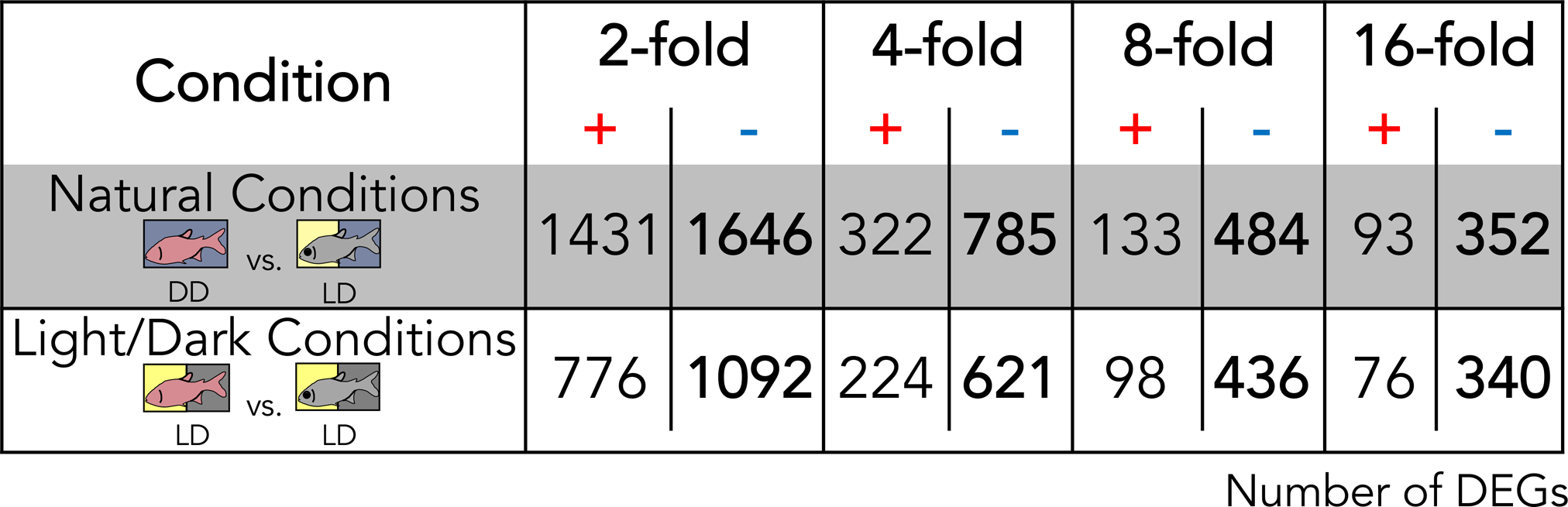
Genes with a higher (plus) or lower (minus) expression in cavefish in comparison to surface fish under both natural and light/dark conditions reveal a higher number of genes with a lower expression in cavefish regardless of the lighting condition. This pattern may indicate that the function of many genes is reduced in the cavefish compared to the surface fish. The replication of this pattern in the dark indicates that this pattern of under-expression in cavefish is conserved regardless of lighting condition.
Figure 3. Rearing under natural photic conditions reveals novel changes to gene expression.
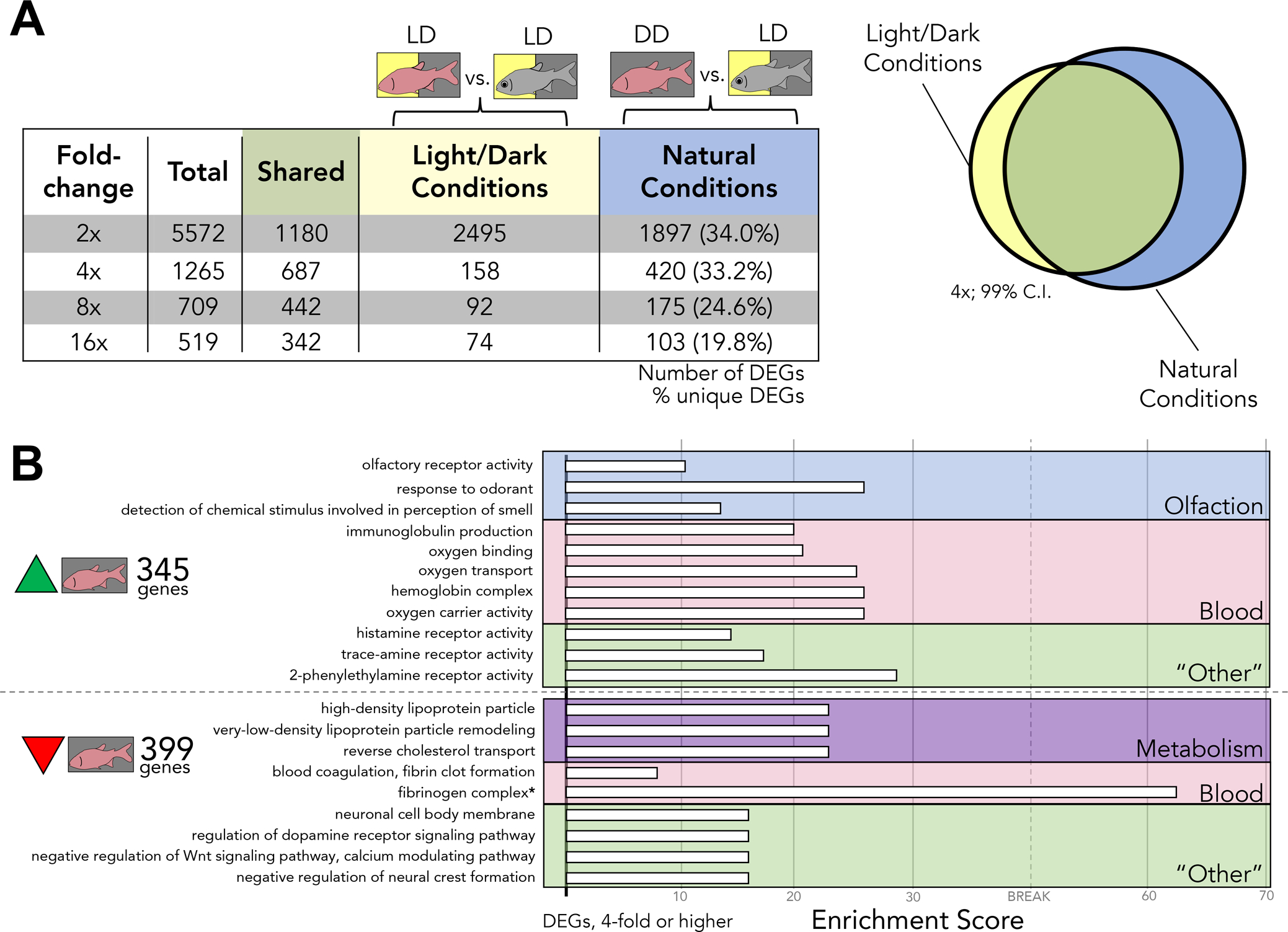
A) The number of differentially expressed genes unique to either light/dark conditions or natural conditions is represented in the 4th and 5th columns of the table, respectively. These DEGs are inclusive of genes with a higher expression in either cave or surface fish on their respective lighting conditions, represented diagrammatically to the right of the table (Venn diagram). There are a large number of differentially expressed genes that are revealed under natural conditions alone. This large number of DEGs represent a set of genes that have not been previously explored B) Test sets of genes were selected based on their expression at a 4-fold threshold. DEGs unique to natural conditions were assessed. Under light/dark conditions, 345 genes have a higher expression in CDD compared to SLD and 399 have a lower expression in CDD compared to SLD. The proportion of GO terms in these test sets was compared to the proportion of GO terms found in the whole transcriptome as a reference. The observed occurrence of each term is taken over the expected occurrence of each term based on is proportion in the reference. Each white bar to the right of a term represents an “Enrichment Score” (observed/expected). This indicates that dark-rearing may potentially play a role in the development and function of the circulatory, olfactory, and metabolic systems in A. mexicanus cavefish.
Table 1.
Genes identified with the greatest fold change difference when comparing surface fish and cavefish reared under their natural photic conditions (SLD vs CDD).
| Gene | Gene Description | SLD RPKM |
CLD RPKM |
CDD RPKM |
LD Conditions Fold Change | Natural Conditions Fold Change |
|---|---|---|---|---|---|---|
| hbaa1 | Hemoglobin subunit alpha 1 | 63.3 | 98.3 | 1337.4 | 1.6 up | 21.1 up |
| adgre6 | Adhesion G protein-coupled receptor E6 (Fragment) | 0.2 | 1.7 | 2.3 | 9.9 up | 13.6 up |
| hbae1.3 | Hemoglobin, alpha embryonic 1.3 (Fragment) | 0.2 | 1.1 | 2.5 | 5.4 up | 11.8 up |
| ba1 | Hemoglobin subunit beta-1 | 399.3 | 530.4 | 3996.7 | 1.3 up | 10.0 up |
| faua | 40S ribosomal protein S30 | 12.4 | 17.2 | 116.5 | 1.4 up | 9.4 up |
| hbaa1 | Hemoglobin subunit alpha 1 | 337.6 | 527.1 | 2828.7 | 1.6 up | 8.4 up |
| ba1 | Hemoglobin subunit beta-1 | 32.3 | 43.1 | 151.1 | 1.3 up | 4.7 up |
| tnni2a.4 | Fast muscle troponin I | 26.9 | 70.7 | 117.6 | 2.6 up | 4.4 up |
| gja5b | Gap junction protein 5b | 0.6 | 0.7 | 1.8 | 1.3 up | 3.4 up |
| hbba2 | Hemoglobin, beta adult 2 | 585.5 | 1111.1 | 1527.1 | 1.9 up | 2.6 up |
| ckbb | Brain-subtype creatine kinase | 134.0 | 68.0 | 41.8 | 2.0 down | 3.2 down |
| hpx | Hemopexin | 530.1 | 329.8 | 151.2 | 1.6 down | 3.5 down |
| apoa1 | Apolipoprotein A-I | 3891.5 | 2905.4 | 1057.4 | 1.3 down | 3.7 down |
| slc25a48 | Solute carrier family 25 member 48 | 2.1 | 1.8 | 0.5 | 1.2 down | 4.0 down |
| serpina1 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 730.8 | 282.2 | 168.6 | 2.6 down | 4.3 down |
| fgb | Fibrinogen beta chain | 172.6 | 105.3 | 38.9 | 1.6 down | 4.4 down |
| fgg | Fgg protein | 177.3 | 91.3 | 35.1 | 1.9 down | 5.1 down |
| abl2 | Tyrosine-protein kinase | 1.8 | 1.4 | 0.2 | 1.3 down | 7.6 down |
| cebpb | CCAAT enhancer-binding protein beta | 224.1 | 123.1 | 22.2 | 1.8 down | 10.1 down |
| cebpd | CCAAT enhancer-binding protein delta | 323.7 | 200.5 | 30.1 | 1.6 down | 10.7 down |
qPCR Validation
We assessed the expression of 16 genes demonstrating diverse patterns of expression by morphotype and lighting conditions, based on RNA-sequencing results (Figure S2). qPCR validation was carried out using a BioRad CFX96 real time PCR instrument (Hercules, California). RNA isolates from each treatment group (SLD, SDD, CLD and CDD) were used to generate cDNA. 1μg of RNA was thawed and annealed with 0.25μg of Oligo(dT)12–18 Primer (Invitrogen) at 65°C for 10 minutes (C1000 Thermocycler, BioRad). To this preparation, we added 4μL of 5X Transcriptor RT reaction buffer (Millipore-Sigma), 0.5μL Protector RNase Inhibitor (Millipore-Sigma), 2μL dNTP mix (10 mM; Roche), and 0.5μL Transcriptor Reverse Transcriptase (Millipore-Sigma), and reactions were incubated at 50°C for 60 min, followed by inactivation of RT enzyme at 85°C for 5 minutes. cDNA preps were then stored at −20°C until use.
Amplifications were carried out using SsoFast EvaGreen Supermix (BioRad, Hercules, CA). Cycling conditions were: 1) activation: 95°C for 30s, 2) denaturation: 95°C for 5s, 3) annealing: 60°C for 5s, for 35 cycles, with 4) the addition of a melt curve: 65–95°C in 0.5°C increments of 5s/step. Normalized expression was calculated using the housekeeping gene bactin1 (actb1; ENSAMXG00000004264.1; McCurley and Callard 2008) using CFX Maestro software (v.4 BioRad, Hercules CA). Primers used for qPCR were blasted against the Astyanax reference genome, and confirmed to harbor a single match to the gene of interest using Primer-BLAST (NCBI, Primer3, Ye et al. 2012). Further, all amplicons demonstrated a single melt temperature, indicating the absence of multiple amplicons via qPCR analyses. Correlations between RNA-seq (RPKM) and qPCR expression values (ΔΔCt) were determined using Pearson’s correlation test (MS Excel). The following primers were used for qPCR validation: atp5pd forward 5′-CGGTCCGCTAGGTGTCAGTA-3′, reverse 5′-TACCAAACCGCTGCAGAGAC-3′; bactin1 forward 5′- ACACTGACATGTTGAACCCAA-3′, reverse 5′- ACCATGCAGCAGGATAAACATTG-3′; cebpb forward 5′- CAAAGGCAAGAAGCGCCTG-3′, reverse 5′- CATCTTGGCTTTATCGCGGC-3′; col10a1b forward 5′- TGCACAAGGTGTCTCATGCTA-3′, reverse 5′- CAGGCCTGGAGACCAAAGAA-3′; ddit4 forward 5′- GACAGATGCTATTCTGTCAACTGC-3′, reverse 5′- AGCTGCCACAGCAAGTTTC-3′; f8 forward 5′- GCAGTGAAGCCGCCTACAAA-3′, reverse 5′- GATGAATCGGGTGAATGCGG-3′; hbz forward 5′- TGGGACCTCTTCTGGGGTTA-3′, reverse 5′- TCTGCGTCTTCTCAACTCAGG-3′; nfil3–5 forward 5′- GTCCAACAGTGGTACCCAGG-3′, reverse 5′- CTGTGTGACAGCCAGGTAGG-3′; nfil3–6 forward 5′- GGAAATTCCTGAAGGCATGA-3′, reverse 5′- ACCGATAGGCTGCAGTTGAC-3′; olfml3a forward 5′- CGAGCATCCGAGCCATGAAG-3′, reverse 5′- TACAGGGTGTCCTCGGACGTT-3′; opn1w1 forward 5′- GTCTAATCCAGCCCAGGCAT-3′, reverse 5′- CTGCCTGTCGTCAAAGGAGC-3′; rgs2 forward 5′- GACCTGCGAAGAGTTTCGAC-3′, reverse 5′- TCGGCGACTGACTCTTGATG-3′; rho forward 5′- TCACCATCGAGCACAAGAAG-3′, reverse 5′- CGAAGACGAAGTAGCCGTTC-3′; rps18 forward 5′- ACACGAACATCGATGGTAGGAG-3′, reverse 5′- TTGTTGAGGTCGATGTCTGC-3′; serpinh1b forward 5′- CTCGGAGGAAAGTCGTCCAC-3′, reverse 5′- TCAGAAGCTCGGACAAACCC-3′; smtlb forward 5′- TGCACCTCCAACATCTTCCC-3′, reverse 5′- CCACCAGGGGATCAATCCAG-3′.
Gene Ontology Enrichment Analyses
Gene ontology (GO) enrichment analyses were performed using Blast2GO v.5 (Ashburner et al. 2000; Harris et al. 2004). The complete transcriptome predicted from the Astyanax genome (25,271 genes) was used as the GO terminology reference set. We generated a local database from the Danio rerio proteome using Blast2GO, which was obtained from the UniProt-Proteomes Database (UniProt ID: UP000000437). Pairwise GO enrichment analyses were performed for genes expressed at a 4-fold or higher difference.
QTL Comparison
QTL identified from prior studies (Borowsky and Wilkens 2002; Protas et al. 2007; Protas et al. 2008; Yoshizawa et al. 2012; O’Quin et al. 2013; Kowalko et al. 2013a; b; Gross et al. 2014) were localized to the current draft of the Astyanax genome (NCBI; McGaugh et al. 2014) through routine BLAST searches of the closest linked marker to each locus. Blast2GO was used to compare Ensembl and NCBI databases to the Danio rerio UniProt Database (UniProt ID: UP000000437). We used this resource to determine the chromosomal position of the Astyanax Ensembl database relative to the estimated chromosomal positions in the NCBI draft Astyanax genome. Prior analyses identified the genes pmela and tyrp1b as mediators of melanophore number in Astyanax cavefish (Stahl et al. 2018). As a proof-of-concept, we successfully retrieved the location of these genes in the current draft of the Astyanax genome (NCBI, Astyanax 2.0, v.102), using their associated QTL markers.
To identify candidate genes for eye loss, all genes with a known chromosomal position, a known QTL, and an expression difference of 2-fold or higher between cave and surface (under any lighting regime) were considered. 268 vision-related genes were selected with GO terms inclusive of ‘eye’, ‘lens’, ‘visual’, and ‘retina’ were included. 61 pigmentation-related genes with GO terms including the words ‘chromophore’, ‘melanocyte’, ‘melanosome’, ‘melanin’, or ‘pigmentation’ were included. Positional information for QTL (relative to selected candidate genes) was visualized using Circos (Fig. 4; Krzywinski, et al. 2009).
Figure 4. Vision and pigmentation genes localized to previously identified QTL providing novel candidate genes.
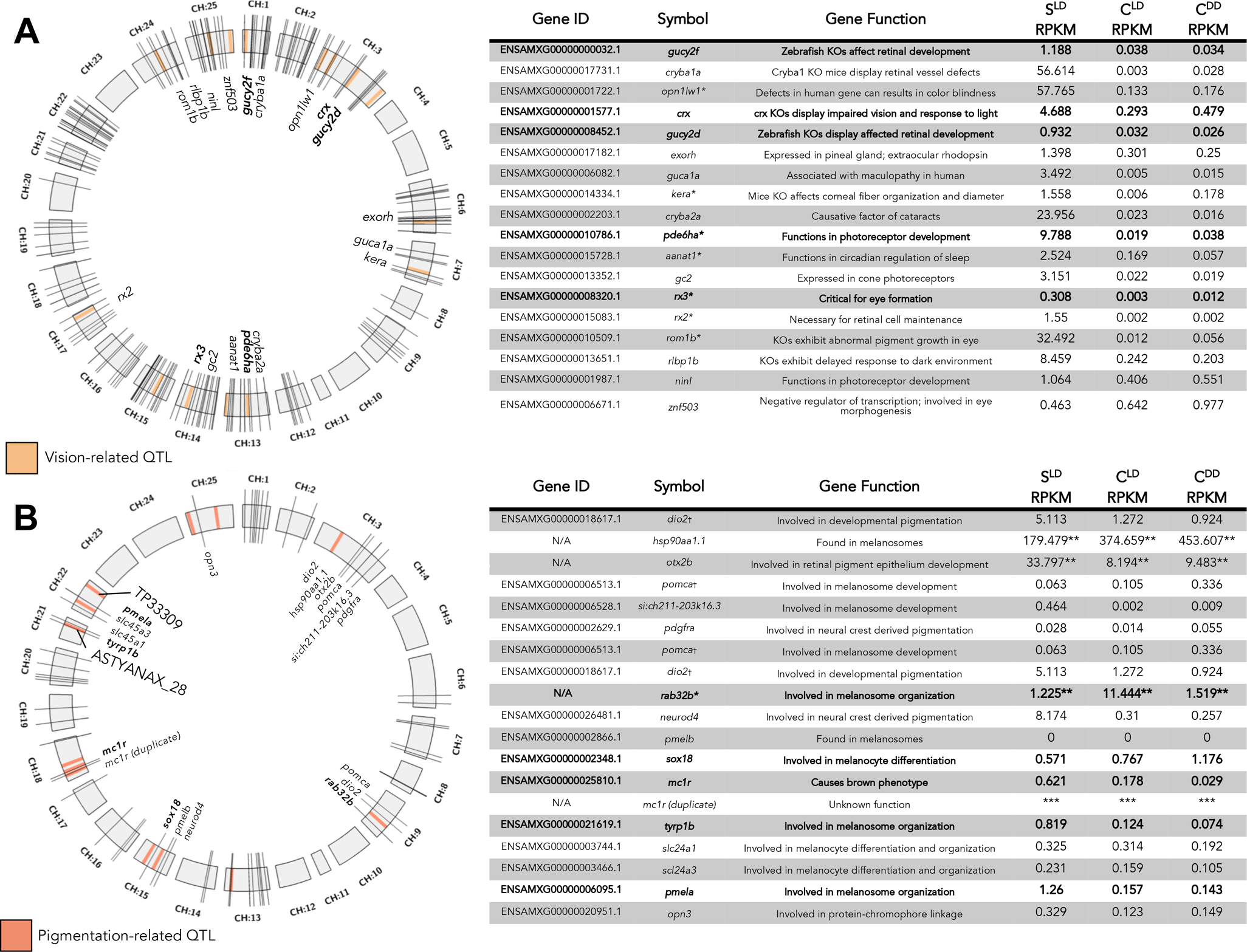
The distribution of vision-related (A) and pigmentation-related (B) QTL (light orange or light pink boxes) across the 25 chromosomes of the draft Astyanax genome (gray boxes, CH:1–25). Gene identity and function are represented in the tables to the right. Values are given in RPKM (normalized expression). Some genes fall remarkably close to QTL (represented as bold hash marks and text) and demonstrate relevant alterations in development when function is disrupted in other systems. * indicates gene name was derived from Danio rerio based on sequence similarity. ** indicates RPKM was obtained via RNA-seq alignment to Astyanax NCBI draft genome by chromosome, and cannot be directly compared to other values, although intra-gene comparisons of expression based on morphotype and lighting condition are valid. † indicates an A. mexicanus gene that aligns to the same protein in D. rerio as another A. mexicanus gene by BLAST. *** is used in place of expression values for the duplicate mc1r gene due to incomplete functional and coding information for this copy number variant.
RESULTS
Alternate photic conditions reveal light-sensitive gene expression in Astyanax
We compared gene expression of all fish (i.e., both cave and surface) reared under LD-conditions to those reared under DD conditions, and observed a high correlation (R2=0.9751; Figure 1A; Table S1). We then evaluated expression of all grouped cavefish data (irrespective of lighting condition) to all surface fish individuals and observed a slightly lower correlation (R2=0.8873; Table S1). When examining expression patterns between cave and surface fish reared under light/dark conditions (CLD × SLD), the correlative value dropped further (R2=0.8657; Figure 1B; Table S1). The lowest correlation of global expression, however, was observed when we compared “natural conditions”, i.e., gene expression in cavefish reared in darkness and surface fish reared under an LD photic regime (CDD × SLD; R2=0.8597; Figure 1C; Table S1). This suggests that global expression is least similar under photic regimes mimicking the natural conditions in which cave and surface morphs reside.
We next sought to understand the polarity of differential gene expression, to determine if a pattern exists with respect to over- and under-expressed genes in cavefish compared to surface fish. We found that at several fold-change thresholds (2-, 4-, 8-, or 16-fold), more genes are overexpressed in surface fish compared to cavefish (Fig. 2). For instance, considering genes displaying 4x fold change under LD photic conditions, 621 genes demonstrate lower expression values in CLD compared to 224 genes with lower expression in SLD. When comparing “natural” photic conditions, 785 genes are expressed at lower levels in CDD compared to 322 genes demonstrating lower expression in SLD (Fig. 2). As reported in prior studies (Stahl and Gross, 2017) the transcriptome reflects the pattern of more genes with lower expression in cavefish compared to surface fish, under light/dark and natural photic conditions.
These expression results are bolstered by a strong correlation (mean Pearson’s coefficient = 0.868) between RNA-seq expression and qPCR analysis (Table S2; Fig. S2). Within our test group, five genes exhibited a correlative value > 0.95.
“Natural” photic rearing reveals novel differences in gene expression
We reasoned that rearing cave and surface fish under photic conditions that more closely mimic their natural photic environments would reveal gene expression differences not captured in prior studies. We began by analyzing expression differences at multiple fold change thresholds (2-, 4-, 8- and 16-fold) to compare transcriptomic responses under different lighting conditions (Fig. 3A). Interestingly, a significant number of the same genes were differentially expressed in both lighting conditions (“shared”; Fig. 3A). At each threshold, we discovered a number of differentially expressed genes that were only present when evaluating natural photic conditions. At a 2-fold threshold, 1897 genes were differentially expressed uniquely under natural photic conditions. This represents 34% of the DEGs at this threshold. At thresholds of 4-, 8- and 16-fold, at least 20% of genes were differentially expressed only in natural conditions. In total, 8065 genes were differentially expressed at 2-fold or higher, of which 2595 genes were uniquely identified under natural photic conditions (> 32%). In sum, rearing Astyanax under natural photic conditions uncovered a considerable number of DEGs not observed in LD transcriptomic analyses.
To understand the novel changes to the transcriptome under natural photic conditions, we selected the most differentially-expressed genes in CDD compared to SLD (Table 1). Interestingly, we found that fold-change differences always increased in magnitude under “natural” conditions compared to light/dark conditions (Table 1), underscoring the importance of light in certain gene expression patterns.
Of the genes demonstrating the largest expression differences, several were associated with commonly-studied traits of cave animals including pigmentation and metabolism. One gene known to affect pigmentation is gja5, which encodes a gap junction protein belonging to the connexin family. Djurdjebič et al. 2019 hypothesized that gja5 could be influential to forming a spotted pattern in brown trout by facilitating communication between multiple erythrophores, or between erythrophores and melanophores. A second gene implicated metabolism, apoa1, is linked to multiple human diseases, such as cardiovascular and early-onset nonfamiliar Alzheimer’s disease, owing to its role in lipid metabolism (Lai et al. 2005; Vollbach et al. 2005).
At least eight of these genes influence blood physiology – an understudied system within cave animals. These genes can be categorized into two groups: hemoglobin production and detoxification/immunological function. Genes involved in detoxification and immunology displayed elevated expression in SLD compared to CLD, and even higher expression compared to CDD. Two genes, fgb and fgg, are well known for their contributions to specific subunits of fibrinogen proteins. This protein can be converted to fibrin – which plays a key role in blood clotting. Additionally, serpina1 and hpx maintain the highest patterns of expression in SLD followed by CLD, and then CDD. Serpina1 encodes serpin proteins, which inhibit enzymes playing a key role in inflammation, thrombosis and the complement cascade (Gooptu and Lomas 2008). Hpx encodes hemopexin, a glycoprotein with a high affinity for free hemoglobin, acting as an antioxidant following blood heme overload. Interestingly, all four of these genes were identified by Jima et al. (2009) as upregulated in mutant zebrafish that show signs of an “enhanced” innate immune response, to compensate for a lack of an effective adaptive immune system.
Four additional hemoglobin (hbaa1, hbae1, ba1 and hbaa2) genes were highly differentially expressed in this analysis. Unlike the detoxification/immunological genes, these genes were expressed highest in CDD, followed by CLD and SLD. This result was surprising in light of historic reports suggesting photic condition does not contribute to hemoglobin production or concentration. Hollwich (1979) and Robscheit-Robbins (1929) described experiments involving a range of taxa (horses, mules, humans, dogs, rabbits, roosters, frogs, fish) that displayed no long-term hematological changes in response to dark rearing. However, these studies were limited to animals with lineages of constant exposure to light, and were not introduced to darkness immediately following birth or fertilization.
To determine if the photic conditions we assessed were associated with functional shifts in gene expression, we performed a series of Gene Ontology (GO) enrichment analyses. We began with an analysis of genes demonstrating higher expression in cavefish under LD conditions (compared to cavefish under DD conditions), and found enrichment of terms associated with olfaction (Figure S1). Unsurprisingly, genes expressed higher in SLD compared to CLD were associated with pigmentation, metabolism, and vision (Fig. S1). When comparing SLD to CDD, an unexpected category of terms was identified, blood physiology (Fig 3B). Genes with higher expression levels in cavefish reared in darkness were associated with pathways such as “oxygen transport”, “oxygen binding” and “hemoglobin complex.” Further, genes with reduced expression levels in CDD were similarly associated with blood physiology, but in pathways associated primarily with blood coagulation. In sum, cave and surface fish reared under their natural photic regimes revealed a functional enrichment for genes involved in blood physiology (specifically associated with hemoglobin genes).
Olfaction also appears to be impacted by darkness, based on the enrichment of GO terms ‘olfactory receptor activity’ and ‘response to odorant’ in overexpressed genes in CDD compared to CLD (Figure 3B). Although these terms are also enriched in CLD individuals, additional olfaction-related genes have a higher expression in CDD relative to their light-dark reared counterparts. This suggests cavefish olfaction is sensitive to lighting condition, including particular odorant receptor genes expressed in olfactory neurons (Vogt et al. 1997; Alioto and Ngai 2005; Figure 3B). Olfactory receptor genes demonstrating a higher expression in cavefish include or101–1, or118–2 (higher expression in CLD compared to SLD; data not shown). This pattern is repeated in CDD compared to SLD fish (e.g. or126–1, or125–5; higher expression in CDD compared to SLD).
Novel DEGs map near previously discovered QTL associated with regressive loss
To determine if any of the DEGs we identified map near the position of QTL discovered in prior studies, we assessed the positions of the most differentially expressed genes in our datasets. We began with 15 markers from prior QTL studies associated with vision loss (Figure 4A, light orange boxes), and surveyed across our transcriptomic analysis. In total, we evaluated 268 vision-related genes (Figure 4A, black hash marks) with differential expression between cave and surface fish, and identified 18 genes as potential candidates for eye loss (Figure 4A, bold hash marks and text). Two of these genes are gucy family members, gucy2d, gucy2f. A third candidate, pde6ha, previously identified in a microarray analysis (Strickler and Jeffery, 2009), was localized to chromosome 13 of the Astyanax genome, residing ~1.6Mb from eye loss QTL marker 30C (Fig. 4A). We also found the gene rx3, which lies ~5 Mb from marker 55A on chromosome 14 (Fig. 4A), which was originally proposed as a candidate for eye loss following a genomic analysis (McGaugh et al. 2014). We propose that these three genes, identified based on differential response to lighting condition, represent promising candidates for a role in the genetic basis for vision loss in Astyanax.
We then mapped the genomic position for 61 pigmentation genes. We identified candidates associated with the position of 11 pigmentation-associated QTL (Fig. 4B), and as a proof-of-concept, co-analyzed the position and expression of two genes (pmela and tyrp1b) previously implicated in pigmentation loss in cavefish (Stahl et al. 2018). Interestingly, the expression of these genes decreases slightly in cavefish under dark rearing (Figure 4B). Included among our findings were the genes sox18, gja5, and rab32b. Located ~5Mb upstream of marker 206A (Figure 4B), rab32b is expressed 9.34-fold higher in CLD fish compared to SLD fish, and encodes a GTPase enzyme involved in membrane trafficking of tyrosinase and tyrosinase-related-protein between organelles in eukaryotic cells (Diekmann et al. 2011; Coppola et al. 2016; Braasch et al. 2009; Wasmeier et al. 2006). Although this gene is overexpressed in cavefish, downstream targets (tryp1b) are expressed at much lower levels (Stahl et al. 2018). However, we found a modest increase (1.24-fold) in rab32b expression in CDD compared to SLD fish (Figure 4B), suggesting this gene is mildly sensitive to light-dark conditions. The absence of light also affected the pigmentation-related gene, gap junction protein 5b (gja5b), which exhibits a 3.35-fold increase in expression in CDD compared to SLD (Table 1). Interestingly, this gene is expressed at similar levels in cave and surface fish reared on light/dark conditions (0.73 and 0.55 RPKM, respectively; Table 1). Based on their established roles in other systems, these three light-sensitive genes represent promising candidates for the complex genetic basis for pigmentation regression.
DISCUSSION
Dark-rearing reveals novel patterns of differential expression in the Mexican tetra
Previous studies in Astyanax characterized substantial variability in gene expression between cave and surface morphotypes (Gross et al. 2013; Hinaux et al. 2013). Many expression differences are rooted early in development, and likely contribute to phenotypic differences between cave and surface fish adults (Stahl and Gross 2017). Although these studies found several DEGs between cave and surface forms, the work we present here reveals that additional differences emerge when evaluating cavefish reared under their natural photic condition of complete darkness. We found that differential expression is mostly marked by under-expression in cavefish (Figure 2). Interestingly, this pattern of differential gene expression is not observed in transcriptomic analyses of development in cavefish compared to surface fish (Stahl and Gross 2017). In fact, the inverse appears to be true, with cavefish demonstrating a higher expression in more genes than surface fish; although, this pattern reverses as the embryos develop (Stahl and Gross 2017). The under-expression of certain genes in cavefish may indicate lower expression of certain genes contributes to trait regression.
Natural photic conditions reveal an effect of darkness on blood physiology and olfaction
We discovered that the transcription of several blood-related genes increases in darkness, including a number of hemoglobin genes (Table 1). Although the functional relevance and mechanism that underlies dark-dependent regulation of blood-related genes is unclear, one of the principle abiotic factors characterizing life in caves is reduced oxygen (Coineau 2000). The up-regulation of hemoglobin genes in CDD likely evolved as an adaptation to reduced oxygen across the El Abra cave network (Ornelas-Garcia et al. 2018).
We noted a dramatic increase in expression of several hemoglobin subunits (ba1, hbaa1, hbba2, hbae1.3) exclusively in dark-reared cavefish (Table 1). A GO enrichment analysis revealed the terms “blood coagulation” and “fibrinogen” are enriched in down-regulated genes of CDD (Figure 3B). This is interesting since we observed a decrease in expression of two fibrinogen genes (fgg and fgb), which may mitigate blood clotting in light of increased hemoglobin concentration of blood (Table 1). This result contradicts historic literature. Sonnenlichtes (1919) suggested, “the belief that sunlight is indispensable to human welfare has become strongly entrenched in modern hygienic propaganda…it is therefore improbable that sunlight is in any sense directly concerned with the hematopoietic functions.”
This discrepancy may also reflect an evolutionary adaptation evolving under intense environmental pressure. For instance, cavefish have lived in total darkness for thousands of generations, while animals in classic studies spent no more than a single generation in darkness. An alternative explanation could be phenotypic plasticity of Astyanax. Bilandžija et al. (2019) utilized dark-rearing, and found substantial plastic responses in this species, suggesting it may have facilitated successful recurrent colonization of epigean forms to the subterranean environment.
The enhancement to olfaction is less surprising. For instance, cavefish have an expanded telencephalon (wherein the olfactory bulbs are situated) compared to surface fish (Rétaux et al. 2008). It is unclear, however, why olfactory gene expression would be increased as a consequence of photic condition, but darkness may provide an environmental cue to increase the sensitivity of non-visual sensory systems (Yoshizawa et al. 2014).
Expression analyses reveal candidate genes for vision and pigmentation loss
Prior studies in Astyanax created hybrid pedigrees (Şadoğlu 1957) permitting genetic studies of various cave-associated traits (Borowsky and Wilkins 2002) including pigmentation and eye loss (Gross et al. 2009; McGaugh et al. 2014; Stahl et al. 2018). We reasoned that our rearing paradigm could provide a source of additional candidate genes through co-analysis of expression data and positions of previously established QTL.
After reviewing several QTL studies (Borowsky and Wilkins 2002; Protas et al. 2007; Protas et al. 2008; Yoshizawa et al. 2012; O’Quin et al. 2013; Kowalko et al. 2013a; b; Gross et al. 2014), we compared positional information for 15 vision-related QTL (Fig. 4A). A putative role for certain candidate genes is illustrated by structural impacts in other species. For instance, defects in human gucy2d are linked to Leber’s congenital amaurosis, a disorder of severe visual impairment caused by structural defects in the retina (Milam et al. 2003). Knockdown of gucy2f in zebrafish also causes visual impairment, accompanied by histological changes to the eye in Danio (Stiebel-Kalish et al. 2012). Further, both gucy2d and gucy2f are inactivated in subterranean-dwelling moles (Heterocephalus glaber and Chrysochloris asiatica; Emerling and Springer, 2014).
Another candidate gene, pde6ha, encodes a component of cone photoreceptors essential for phototransduction (Ionita and Pittler 2007; Collery and Kennedy 2010). Structural alterations to this gene in humans are associated with achromatopsia, an abnormality causing reduced visual acuity (Kohl et al. 2012). Although functional analyses of pde6h in mice did not cause noticeable changes in phototransduction (Brennenstuhl et al. 2015), in Danio this gene is upregulated in response to light (Weger et al. 2011).
We then mapped the genomic positions for 61 pigmentation genes, including rab32b (Figure 4B). Interestingly, in zebrafish embryos, rab32 paralogs are expressed in the retinal pigment epithelium and developing melanoblasts (Coppola et al. 2016). Additionally, Sox-family transcription factors, such as sox18, regulate diverse events throughout early development, including neural crest migration (Kiefer 2007). Sox18-deficient mice have alterations to pheomelanin content in hair shafts, resulting in a darker appearance (Pennisi et al. 2000).
Another candidate gene we identified, gja5, is associated with human idiopathic atrial fibrillation, due to its role in activation of the atria (Gollob et al. 2006). In zebrafish, a mutation in gja5b is associated with the leopard mutant, which reorganizes stripe pigmentation into leopard-like spots (Irion et al. 2014). Taken together, the approach we use here illustrates the relevance of light as an environmental factor that likely impacts on quantitative changes in phenotype. This environmental factor is perhaps especially relevant for pigmentation, since cavefish harbor a number of coloration defects (Jeffery 2009b). Although neutral mutations may be an explanation for pigmentation loss, it is possible that secondary benefits arise with these specific mutations (Jeffery 2009b). For instance, genetic changes to key pigmentation genes may cause L-tyrosine (a melanin intermediate) to be blocked and shunted from its role in coloration, and utilized for L-dopamine production which in turn influence metabolism and foraging (Bilandžija et al., 2013). Future analyses of key environmental factors, such as photic condition, may further inform the adaptive nature of these complex interactions.
CONCLUSIONS
The blind Mexican cavefish has evolved under the pressure of constant darkness. Both genetic variation and plasticity play key roles in ecologically relevant traits. Although a captive laboratory environment provides exceptional control over numerous confounding variables, this work reveals that inclusion of key environmental variables (e.g., lighting condition) is essential for understanding the complex processes by which animals evolve in the wild. This study revealed that rearing under ‘natural photic conditions’ (darkness) yields substantially different gene expression patterns compared to light/dark rearing. Here, we report that dark-rearing impacts expression of suites of genes associated with well-known troglomorphic traits such as pigmentation reduction and vision loss. This work also revealed expression differences associated with less understood traits evolving in cavefish, such as blood physiology. Our expression analyses demonstrating labile expression associated with photic condition, alongside prior QTL studies, revealed additional candidate genes for eye or pigmentation loss. Moreover, this study indicates the importance of photic condition as an environmental pressure in the blind Mexican cavefish, and underscores the importance of accounting for natural lighting conditions when examining genetic and phenotypic expression in this system.
Supplementary Material
Figure S1. GO enrichment analysis under light/dark conditions reveal enrichment of vision, pigmentation, and metabolism GO terms among other “cave adaptive” traits. Test sets of genes were selected based on their expression at a 4-fold threshold. Under light/dark conditions, 673 genes have a higher expression in cavefish and 1197 have a higher expression in surface fish. The proportion of GO terms in these test sets was compared to the proportion of GO terms found in the whole transcriptome as a reference. The observed occurrence of each term is taken over the expected occurrence of each term based on is proportion in the reference. Each white bar to the right of a term represents an “Enrichment Score” (observed/expected). Under light/dark conditions, enriched GO terms reveal patterns of cave evolution (as demonstrated by the enrichment of these terms in surface fish) such as a loss of melanic pigmentation, and a reduction of eye development and phototransduction in cavefish. The enrichment of these terms in either cave or surface fish confirms that analyses of enriched GO terms support the underlying biology of the organism.
Figure S2. Gene expression determined by qPCR validates RNA-seq expression. Qualitative (gel, inset, A; B) expression and quantitative RNA-seq (in RPKM) expression is highly similar in both genes with differential expression such as rhodopsin (A; C) and in genes with more similar expression between conditions such as the housekeeping/reference gene bactin1 (B). Comparisons of RNA-seq (RPKM, x-axes) and qPCR-derived (ΔΔc(t), y-axes) quantitative expression indicate a high degree of similarity in expression for many genes as indicated by Pearson’s correlation. This is exemplified for the genes rho (C), nfil3–5 (D), f8 (E) and cepbp (F). This high degree of correlation is recapitulated across 16 tested genes, with a mean correlative value of 86.8%.
Table S1. Correlative expression value for eight comparisons evaluating variable lighting on global gene expression.
Table S2. Similarity in gene expression derived by RNA-seq and qPCR methodologies assessed by Pearson’s correlation.
ACKNOWLEDGEMENTS
The authors wish to thank members of the Gross lab for help with this project. This work was supported in part by a Wieman-Benedict award to CRS. Special thanks to Daniel Berning for assistance with fish husbandry, and Brian Carlson for guidance on transcriptomic data analyses. JBG is supported by the National Science Foundation (DEB-1457630), and the National Institutes of Health (NIDCR-DE025033). The data from this report will be available in the NCBI Sequencing Read Archive under SRA accession PRJNA605208 (https://www.ncbi.nlm.nih.gov/sra/PRJNA605208). The authors declare no conflicts of interest.
REFERENCES
- Alioto TS, and Ngai J, 2005. The odorant receptor repertoire of teleost fish. BMC Genomics. 6: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. , 2000. Gene Ontology: Tool for the unification of biology. Nature Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC and Selander RK, 1972. Evolutionary Genetics of Cave-Dwelling Fishes of the Genus Astyanax. Evolution. 26: 1–19. [DOI] [PubMed] [Google Scholar]
- Axelrod J, 1974. The pineal gland: A neurochemical transducer. Science, 184: 1341–1348. [DOI] [PubMed] [Google Scholar]
- Bilandžija H, Ma L, Parkhurst A and Jeffery WR. 2013. A potential benefit of albinism in Astyanax cavefish: Downregulation of the oca2 gene increases tyrosine and catecholamine levels as an alternative to melanin synthesis. PLoS One, 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilandžija H, Hollifield B, Steck M, Meng G, Ng M, et al. , 2019. Phenotypic plasticity as an important mechanism of cave colonization and adaption in Astyanax cavefish. bioRxiv 657460; doi: 10.1101/657460. [DOI] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B, 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky R, and Wilkens H, 2002. Mapping a cave fish genome: Polygenic systems and regressive evolution. J. Hered 93: 19–21. [DOI] [PubMed] [Google Scholar]
- Braasch I, Brunet F, Volff JN, and Schartl M, 2009. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol. Evol 1: 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennenstuhl C, Tanimoto N, Burkard M, Wagner R, Bolz S, et al. , 2015. Targeted ablation of Pde6h in mice reveals cross-species differences in cone and rod phototransduction protein inventory. J. Biol. Chem 290: 10242–10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM, and Gross JB, 2018. Characterization and comparison of activity profiles exhibited by the cave and surface morphotypes of the blind Mexican tetra, Astyanax mexicanus. Comp. Biochem. Physiol. C Toxicol. Pharmacol 208: 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM, Klingler IB, Meyer BJ and Gross JB, 2018. Genetic analysis reveals candidate genes for activity QTL in the blind Mexican tetra, Astyanax mexicanus. PeerJ. 6:e5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, 2008. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Coineau N, 2000. Adaptations to interstitial groundwater life, pp. 189–210 in Ecosystems of the World: Subterranean Ecosystems, edited by Wilkens H, Culver DC, and Humphreys WF. Elsevier Science B. V., Amsterdam. [Google Scholar]
- Collery RF, and Kennedy BN, 2010. Photoreceptor guanylate cyclases and cGMP phosphodiesterases in zebrafish, pp. 55–61 in Retinal Degenerative Diseases. Advances in Experimental Medicine and Biology, vol. 664, edited by Anderson R, Hollyfield J, and LaVail M. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Coppola U, Annona G, D’Aniello S, and Ristoratore F, 2016. Rab32 and Rab38 genes in chordate pigmentation: An evolutionary perspective. BMC Evol. Biol 16: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall ARX, Bell AM, Bolnickand DI and Ratnieks FLW, 2012. An evolutionary ecology of individual differences. Ecol. Lett 15: 1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann Y, Seixas E, Gouw M, Tavares-Cadete F, Seabra MC, et al. , 2011. Thousands of rab GTPases for the cell biologist. PLoS Comput. Biol 7: e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurdjevič I, Furmanek T, Miyazawa S, and Bajec SS, 2019. Comparative transcriptome analysis of trout skin pigment cells. BMC Genomics. 20:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboué ER, Keene AC and Borowsky RL, 2011. Evolutionary Convergence on Sleep Loss in Cavefish Populations. Curr. Biol 8: 671–676. [DOI] [PubMed] [Google Scholar]
- Elliott WR, 2015. Cave biodiversity and ecology of the Sierra de El Abra region, pp. 59–75 in Biology and Evolution of the Mexican Cavefish, edited by Keene AC, Yoshizawa M, and McGaugh SE. Elsevier, San Diego. [Google Scholar]
- Emerling CA, and Springer MS, 2014. Eyes underground: Regression of visual protein networks in subterranean mammals. Mol. Phylogenetics Evol 78: 260–270. [DOI] [PubMed] [Google Scholar]
- Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, et al. , 2006. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N. Engl. J. Med 354: 2677–2688. [DOI] [PubMed] [Google Scholar]
- Gonzalez MMC, and Aston-Jones G, 2008. Light deprivation damages monoamine neurons and produces a depressive behavioral phenotype in rats. PNAS. 105: 4898–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooptu B, and Lomas DA, 2008. Polymers and inflammation: disease mechanisms of the serpinopathies. J. Exp. Med 7: 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson JNS and Burt de Perera T, 2007. Shoaling in eyed and blind morphs of the characin Astyanax fasciatus under light and dark conditions. J. Fish Biol 70: 1615–1619. [Google Scholar]
- Greenwood AK, Jones FC, Chan YF, Brady SD, Absher DM, et al. , 2011. The genetic basis of divergent pigment patters in juvenile threespine sticklebacks. Heredity. 107: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Borowsky R, and Tabin CJ, 2009. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 5: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Furterer A, Carlson BM, and Stahl BA, 2013. An integrated transcriptome-wide analysis of cave and surface dwelling Astyanax mexicanus. PLoS ONE. 8: e55659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Krutzler AJ, and Carlson BM, 2014. Complex craniofacial changes in blind cave-dwelling fish are mediated by genetically symmetric and asymmetric loci. Genetics. 196: 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Powers AK, Davis EM, and Kaplan SA, 2016. A pleiotropic interaction between vision loss and hypermelanism in Astyanax mexicanus cave x surface hybrids. BMC Evol. Biol 16: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, et al. , 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32: D258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan ES, 1989. Hydrodynamic imaging of the surroundings by the lateral line of the blind cave fish Anoptichthys jordani, pp. 217–227 in The Mechanosensory Lateral Line, edited by Coombs S, Görner P, and Münz H. Springer, New York. [Google Scholar]
- Hinaux H, Poulain J, Da Silva C, Noirot C, Jeffery WR, et al. , 2013. De novo sequencing of Astyanax mexicanus surface fish and Pachón cavefish transcriptomes reveals enrichment of mutations in cavefish putative eye genes. PloS ONE. 8: e53553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, 2006. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 97: 224–234. [DOI] [PubMed] [Google Scholar]
- Hollwich F, 1979. Light and Blood Count, pp 39–42 in The influence of ocular light perception on metabolism in man and in animal. Springer, New York. [Google Scholar]
- Idda ML, Bertolucci C, Vallone D, Gothilf Y, Sánchez-Vázquez FJ, and Foulkes NS, 2012. Circadian clocks: Lessons from fish, pp. 41–57 in Progress in Brain Research. Elsevier, San Diego. [DOI] [PubMed] [Google Scholar]
- Ionita MA and Pittler SJ, 2007. Focus on molecules: rod cGMP phosphodiesterase type 6. Exp. Eye Res 84: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Frohnhöfer HG, Krauss J, Champollion TC, Maischein HM, et al. , 2014. Gap junctions composed of connexins 41.8 and 39.4 are essential for colour pattern formation in zebrafish. eLife. 3: e05125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR 2001. Cavefish as a model system in evolutionary developmental biology. Dev. Biol 231: 1–12. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, 2008. Emerging model systems in evo‐devo: Cavefish and microevolution of development. Evol. Dev 10: 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, 2009a. Evolution and development in the cavefish Astyanax. Curr. Top. Dev. Biol 86: 191–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR 2009b. Regressive evolution in Astyanax cavefish. Annu. Rev. Genet 43: 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, 2015. Pigment regression and albinism in Astyanax cavefish, pp. 155–174 in Biology and Evolution of the Mexican Cavefish, edited by Keene AC, Yoshizawa M, and McGaugh SE. Elsevier, San Diego. [Google Scholar]
- Jima DD, Shah RN, Orcutt TM, Joshi D, McHugh Law J, et al. , 2009. Enhanced transcription of complement and coagulation genes in the absence of adaptive immunity. Mol. Immunol 7: 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JC, 2007. Back to basics: Sox genes. Dev. Dyn 236: 2356–2366. [DOI] [PubMed] [Google Scholar]
- Kohl S, Coppieters F, Meire F, Schaich S, Roosing S, et al. , 2012. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am. J. Hum. Genet 91: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko JE, Rohner N, Linden TA, Rompani SB, Warren WC, et al. , 2013a. Convergence in feeding posture occurs through different genetic loci in independently evolved cave populations of Astyanax mexicanus. PNAS. 110: 16933–16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CQ, Parnell LD and Jose M, 2005. The APOA1/C3/A4/A5 gene cluster, lipid metabolism and cardiovascular disease risk. Curr. Opin. Lipidol 16: 153–166. [DOI] [PubMed] [Google Scholar]
- McCurley AT and Callard GV, 2008. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC. MOL. BIOL 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, Gross JB, Aken B, Blin M, Borowsky R, et al. , 2014. The cavefish genome reveals candidate genes for eye loss. Nat. Commun 5: 5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Braasch I, Phillips JB, Lin X, Titus T, et al. , 2013. Evolution of the eye transcriptome under constant darkness in Sinocyclocheilus cavefish. Mol. Biol. Evol 30: 1527–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milam AH, Barakat MR, Gupta N, Rose L, Aleman TS, et al. , 2003. Clinicopathologic effects of mutant GUCY2D in Leber congenital amaurosis. Ophthalmology, 110: 549–558. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, and Wold B, 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5: 621. [DOI] [PubMed] [Google Scholar]
- O’Quin KE, Yoshizawa M, Doshi P, and Jeffery WR, 2013. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PloS ONE, 8: e57281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura Y, 1975. Influence of light and darkness on the ultrastructure of the pineal organ in the blind cave fish, Astyanax mexicanus. Cell Tissue Res, 160: 99–112. [DOI] [PubMed] [Google Scholar]
- Ornelas-García P, Pajares S, Sosa-Jiménez VM, Rétaux S, and Miranda-Gamboa RA, 2018 Microbiome differences between river-dwelling and cave-adapted populations of the fish Astyanax mexicanus (De Filippi, 1853). PeerJ, 6: e5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, et al. , 2000. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nature Genet. 24: 434–437. [DOI] [PubMed] [Google Scholar]
- Pottin K, Hinaux H, and Rétaux S, 2011. Restoring eye size in Astyanax mexicanus blind cavefish embryos through modulation of the Shh and Fgf8 forebrain organising centres. Development, 138: 2467–2476. [DOI] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, et al. , 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nature Genet. 38: 107. [DOI] [PubMed] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, and Borowsky R, 2007. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr. Biol 17: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas M, Tabansky I, Conrad M, Gross JB, Vidal O, et al. , 2008. Multi‐trait evolution in a cave fish, Astyanax mexicanus. Evol. Dev 10: 196–209. [DOI] [PubMed] [Google Scholar]
- Retaux S, Pottin K, and Alunni A, 2008. Shh and forebrain evolution in the blind cavefish Astyanax mexicanus. Biol. Cell 100: 139–147. [DOI] [PubMed] [Google Scholar]
- Robscheit-Robbins FS, 1929. The regeneration of hemoglobin and erythrocytes. Physiol. Rev 4: 666–709. [Google Scholar]
- Şadoğlu P, 1957. A Mendelian gene for albinism in natural cave fish. Cell. Mol. Life Sci 13: 394–394. [Google Scholar]
- Şadoğlu P, 1967. The selective value of eye and pigment loss in Mexican cave fish. Evolution. 21: 541–549. [DOI] [PubMed] [Google Scholar]
- Sonnenlichtes E, 1919. The composition of the blood in sunlight and darkness. Jama-J. Am. Med. Assoc 73:1446. [Google Scholar]
- Stahl BA and Gross JB, 2017. A comparative transcriptomic analysis of development in two Astyanax cavefish populations. J. Exp. Zool. Part B 328: 515–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl BA, Sears CR, Ma L, Perkins M, and Gross JB, 2018. Pmela and Tyrp1b contribute to melanophore variation in Mexican cavefish, pp. 3–22 in Origin and Evolution of Biodiversity, edited by Pontarotti P. Springer, Cham. [Google Scholar]
- Stiebel-Kalish H, Reich E, Rainy N, Vatine G, Nisgav Y, et al. , 2012. Gucy2f zebrafish knockdown–a model for Gucy2d-related leber congenital amaurosis. Eur. J. Human Genet 20: 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler AG, Yamamoto Y, and Jeffery WR, 2001. Early and late changes in Pax6 expression accompany eye degeneration during cavefish development. Dev. Genes Evol 211: 138–144. [DOI] [PubMed] [Google Scholar]
- Strickler AG, and Jeffery WR, 2009. Differentially expressed genes identified by cross‐species microarray in the blind cavefish Astyanax. Integr. Zool 4:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temerin LA and Cant JGH, 1983. Evolutionary divergence of old world monkeys and apes. Am. Nat 122: 335–351. [Google Scholar]
- Vogt RG, Lindsay SM, Byrd CA, and Sun M, 1997. Spatial patterns of olfactory neurons expressing specific odor receptor genes in 48-hour-old embryos of zebrafish Danio rerio. J. Exp. Biol 200: 433–443. [DOI] [PubMed] [Google Scholar]
- Vollbach H, Heun R, Morris CM, Edwardson JA, McKeith IG, et al. , 2005. APOA1 polymorphism influences risk for early-onset nonfamiliar AD. Ann. Neurol 58: 436–441. [DOI] [PubMed] [Google Scholar]
- Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, et al. , 2006. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol 175: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger BD, Sahinbas M, Otto GW, Mracek P, Armant O, et al. , 2011. The light responsive transcriptome of the zebrafish: Function and regulation. PLoS ONE. 6: e17080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ (2003) Developmental Plasticity and Evolution. New York, New York: Oxford University Press. [Google Scholar]
- Wilkens H, 1988. Evolution and Genetics of Epigean and Cave Astyanax fasciatus (Characidae, Pisces). Evol. Biol 23: 271–367. [Google Scholar]
- Wilkens H, 2007. Regressive evolution: ontogeny and genetics of cavefish eye rudimentation. Biol. J. Linnean Soc 92: 287–296. [Google Scholar]
- Yamamoto Y, Espinasa L, Stock DW, and Jeffery WR, 2003. Development and evolution of craniofacial patterning is mediated by eye‐dependent and‐independent processes in the cavefish Astyanax. Evol. Dev 5: 435–446. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, et al. , 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, and Jeffery WR, 2008. Shadow response in the blind cavefish Astyanax reveals conservation of a functional pineal eye. J. Exp. Biol 211: 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Yamamoto Y, O’Quin KE, and Jeffery WR, 2012. Evolution of an adaptive behavior and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Jeffery WR, van Netten SM, and McHenry MJ, 2014. The sensitivity of lateral line receptors and their role in the behavior of Mexican blind cavefish (Astyanax mexicanus). J. Exp. Biol 217: 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. GO enrichment analysis under light/dark conditions reveal enrichment of vision, pigmentation, and metabolism GO terms among other “cave adaptive” traits. Test sets of genes were selected based on their expression at a 4-fold threshold. Under light/dark conditions, 673 genes have a higher expression in cavefish and 1197 have a higher expression in surface fish. The proportion of GO terms in these test sets was compared to the proportion of GO terms found in the whole transcriptome as a reference. The observed occurrence of each term is taken over the expected occurrence of each term based on is proportion in the reference. Each white bar to the right of a term represents an “Enrichment Score” (observed/expected). Under light/dark conditions, enriched GO terms reveal patterns of cave evolution (as demonstrated by the enrichment of these terms in surface fish) such as a loss of melanic pigmentation, and a reduction of eye development and phototransduction in cavefish. The enrichment of these terms in either cave or surface fish confirms that analyses of enriched GO terms support the underlying biology of the organism.
Figure S2. Gene expression determined by qPCR validates RNA-seq expression. Qualitative (gel, inset, A; B) expression and quantitative RNA-seq (in RPKM) expression is highly similar in both genes with differential expression such as rhodopsin (A; C) and in genes with more similar expression between conditions such as the housekeeping/reference gene bactin1 (B). Comparisons of RNA-seq (RPKM, x-axes) and qPCR-derived (ΔΔc(t), y-axes) quantitative expression indicate a high degree of similarity in expression for many genes as indicated by Pearson’s correlation. This is exemplified for the genes rho (C), nfil3–5 (D), f8 (E) and cepbp (F). This high degree of correlation is recapitulated across 16 tested genes, with a mean correlative value of 86.8%.
Table S1. Correlative expression value for eight comparisons evaluating variable lighting on global gene expression.
Table S2. Similarity in gene expression derived by RNA-seq and qPCR methodologies assessed by Pearson’s correlation.


