Abstract
Collective invasion can be led by breast cancer cells expressing basal epithelial markers, typified by keratin-14 (KRT14). We analyzed gene expression data from The Cancer Genome Atlas and demonstrated a significant correlation between a KRT14+ invasion signature and a stromal mediated extracellular matrix (ECM) organization module. We then developed a novel co-culture model of tumor organoids with autologous stromal cells. Co-culture significantly increased KRT14 expression and invasion of organoids from both luminal and basal murine breast cancer models. However, stromal cell conditioned medium induced invasion but not KRT14 expression. Cancer cells released TGF-β and that signaling pathway was required for stromal cell-induced invasion and KRT14 expression. Mechanistically, TGF-β induced NOX4 expression in stromal cells and NOX4 inhibition reduced invasion and KRT14 expression. In summary, we developed a novel co-culture model and revealed dynamic molecular interactions between stromal cells and cancer cells that regulate both basal gene expression and invasive behavior.
Introduction
Invasion is a fundamental early step in metastasis, which is the dominant cause of cancer mortality. Cancer cells can invade surrounding tissues through a continuum of single amoeboid, single cell mesenchymal, or collective invasion strategies (1). Recent studies across multiple tumor types suggest that collective invasion is more frequently observed than single cell invasion in patient tumor samples (2). Collective invasion has also been implicated as the source of circulating tumor cell (CTC) clusters, which have been demonstrated to be more efficient at seeding metastases than single cells (3,4). Detection of CTC clusters also correlates with poor outcomes in patients (3). Therefore, understanding the molecular mechanisms that regulate collective invasion may help develop therapeutic strategies to limit or prevent invasion and metastases.
Collective invasion in invasive ductal breast cancer (IDC) can be led by specialized cancer cells that retain a basal epithelial phenotype (5). These leader cells are identified by expression of basal epithelial genes (e.g. KRT14 and p63) and the absence of myoepithelial markers (e.g. α-SMA, SMM) (5). KRT14-positive cells are detected in tumors from diverse breast cancer subtypes and KRT14 expression correlates with poor survival independent of tumor size or hormone receptor status (6). Furthermore, KRT14 expression has been shown in mouse models to be required for both invasion and distant metastasis (4,5,7). However, it is increasingly clear that tumor-level phenotypes typically arise through reciprocal interactions among the cancer cells, the extracellular matrix (ECM), and various stromal cell populations (8). Indeed, multiple groups have reported guidance of cancer cell invasion by macrophages (9) and fibroblasts (10–12).
The aim of this study was to identify mechanisms regulating the activation of the KRT14+ basal invasion program. We utilized a systems biology approach and analyzed existing transcriptomic data from human tumors. This analysis identified a group of ECM related genes that were highly correlated with a breast cancer invasion gene expression signature defined by KRT14+, suggesting that stromal cells could regulate the KRT14+ basal epithelial invasion phenotype. We then developed a novel 3D culture model to test the effects of autologous stromal cells on both invasion and KRT14 expression.
Materials and Methods
Bioinformatics analysis
RSEM normalized RNA-Sequencing (V2) data with accompanying clinical traits, ABSOLUTE tumor purity measurements, and reverse phase protein microarray signature scores were downloaded from the Ciriello et al. study (13). Gene expression data were log2(normalized counts + 1) transformed, normalized for tumor purity, and z-score scaled. Weighted gene correlation network analysis (WGCNA) was performed using the wgcna R package as described previously (14). The KRT14+ gene signature was defined using the sum of z-scores for genes significantly up-regulated (FDR Adj.P < 1e-6 & log2 (Fold Change) > 3) in KRT14+ vs KRT14- tumor cells, as detected by RNA-Sequencing in a previous study (4). Full details of all bioinformatics analyses are provided in the Supplementary Materials and Methods section.
Mouse Lines and Breeding
Animal use was conducted in accordance with protocols approved by the Johns Hopkins Institutional Animal Care and Use Committee. The FVB-Tg(C3–1-Tag)cJeg/JegJ (C31-Tag), FVB/N-Tg(MMTV-PyVT)634Mul/J (MMTV-PyMT), and B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) mouse lines were acquired from the Jackson Labs. The TgN(ActbECFP)1Nagy(CK6/ECFP) (β-actin::CFP) transgenic line (15) was acquired from the Hadjantonakis Lab (MSKCC). All lines were backcrossed and maintained on the FVB/NCrl (Charles River) background.
Tumor organoid and stromal cell isolation
Tumor organoids were isolated from primary tumors as described previously (5), with the ECM prepared as in (16). Stromal cells were isolated during the differential centrifugations that are used to separate organoids from the single cells. The supernatant from these centrifugation steps was collected and centrifuged at 1500 rpm for 5 minutes. The cell pellet was re-suspended in cell depletion buffer (PBS + 2% FCS + 1 mM EDTA), counted and diluted to 1×108 cells/ml. Biotin conjugated antibodies targeting CD326 (EpCAM; G8.8; 2 μg/ml; Biolegend), CD45 (30-F11; 2 μg/ml; Biolegend) and erythroid cells (TER-119; 1 μg/ml; STEMCELL Technologies) were used to deplete epithelial/immune/erythroid cells using the EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit (STEMCELL Technologies), per the manufacturer’s instructions. The efficiency of cell depletion was assessed by flow cytometry using standard protocols. Details of this analysis are provided in Supplementary Materials and Methods.
Cell culture
All cell cultures were maintained in 37°C humidified incubators at 5% CO2. DMEM/F12 supplemented with Gluta-MAX, insulin/transferrin/selenium, and penicillin/streptomycin was used as base media for all cultures. Organotypic cultures were carried out as previously described (5) with some alterations; full details are provided in Supplementary Materials and Methods. For contractility assays, 150,000 stromal cells were embedded in 350 μl 1.5 mg/ml collagen gels, cast in 24 well plates. The gels were incubated for 1 hour to set, 1 ml of base media was added, they were incubated overnight incubation, then the gels were detached from the well. After 72 hours (C31-Tag) or 110 hours (MMTV-PyMT) gels were collected and weighed.
Organotypic culture staining and analysis
For end-point analysis at 72 hours, organotypic cultures were fixed, stained, and imaged using a confocal microscope as described previously (5), please see Supplementary Materials and Methods for full details.
Image analysis was carried out in an automated manner using custom image processing macros. Full details are provided in the Supplementary Materials and Methods.
Results
A KRT14+ gene expression signature correlates with genes expressed by stromal cells involved in ECM organization
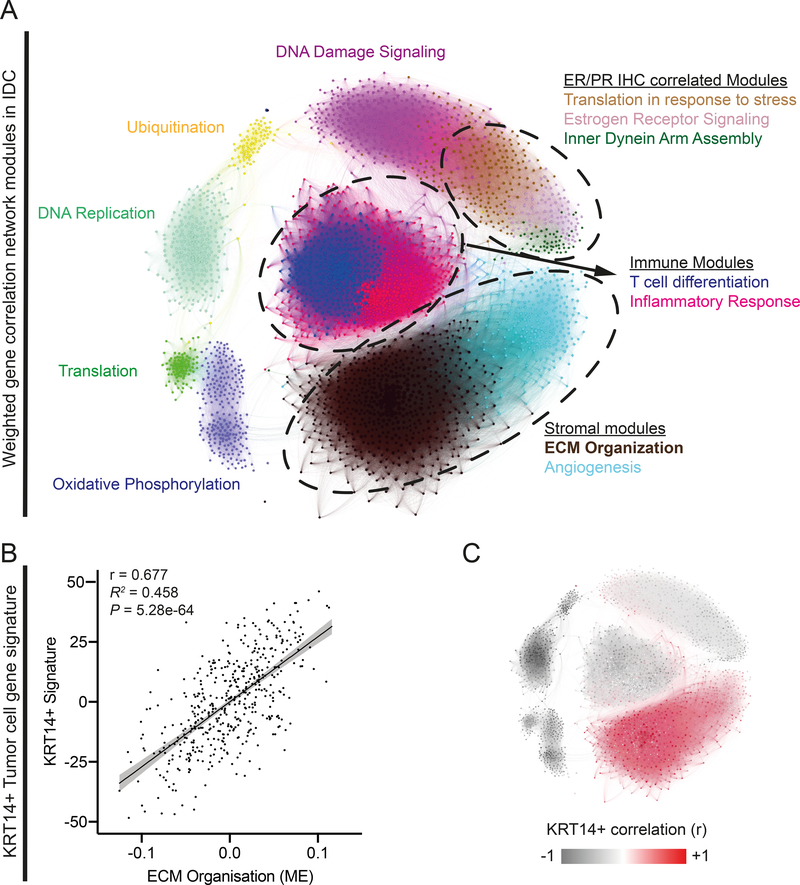
We previously identified conserved basal protein expression in cells leading collective breast cancer invasion (7). The most conserved marker for invasive leader cells was KRT14 expression and we used RNA-seq to define the broader gene expression differences between KRT14+ and KRT14- cancer cells (6). We now apply this KRT14+ signature to examine RNAseq data from human invasive ductal carcinoma (IDC) samples (Cancer Genome Atlas; n=490) using weighted gene correlation network analysis (WGCNA) (13). WGCNA describes patterns in gene expression, identifying groups of genes (modules) that are highly correlated, to suggest genes that could be functionally related.
Our analysis identified 12 modules corresponding to critical biological processes within these tumors (Figure 1A, Supplementary Table 1), including previously described features of the IDC intrinsic molecular subtypes (17). For example, a “DNA replication” module was identified, which correlated with reverse-phase protein microarray (RPPA) profile scores for proliferation (r = 0.85, p = 3.1e-131) and cell-cycle (r = 0.80, p = 1.1e-107) signatures (Supplementary Table 1). This module was most highly expressed in the basal PAM50 subtype. Increased expression was also seen in Luminal B compared to Luminal A subtypes, consistent with the well-documented variation in proliferative capacity between these subtypes (17) (Figure S1A). Three modules were significantly correlated with estrogen receptor IHC status (r > 0.59, p < 7.0e-47; Supplementary Table 1), including an ‘Estrogen receptor signaling’ module. This module was up-regulated in luminal breast cancer subtypes and contained numerous previously described markers of luminal breast cancer (XBP1, GATA3, BCL2, ERB4 and SLC39A6 (17); Figure S1A).
Figure 1. Weighted gene correlation network analysis (WGCNA) of primary tumor samples from human invasive ductal breast cancer reveals a highly significant correlation between a Keratin 14 positive tumor cell gene signature and the expression of genes involved in extra-cellular matrix (ECM) organization.
A) Graphical representation of the topological overlap between genes WGCNA where each node (gene) is colored by their module assignment within the network and sized by their connectivity within the assigned module. Modules are labeled by the most prominent gene ontology term enrichment (see Table S1 for details). B) Linear Regression analysis comparing the ECM organization module eigengene to KRT14+ signature score. C) Network Map (equivalent to that shown in A) where each node is colored to represent correlation (Pearson’s r) to the KRT14+ gene signature.
We next tested for correlations between our KRT14+ defined basal signature and the WGCNA gene expression modules. This identified a highly significant correlation with the “ECM Organization” module (module eigengene correlation: r = 0.68, p = 5.3e-64; Figure 1 B–C and Supplementary Table 2). This correlation was consistently observed across PAM50 subtypes (Figure S1B). Moreover, the “ECM organization” module correlates with Luminal B, Her2 and Basal PAM 50 subtypes (Figure S1C). Expression of genes in the “ECM Organization” module suggests the intratumoral accumulation of cancer associated fibroblasts (CAFs), as these cells are a prominent stromal component of IDC tumors that both secrete and remodel ECM (18). Furthermore, the module contained numerous previously described CAF markers (ACTA2 (19), FAP (20), NOX4 (21), LOX (22)). Gene Set Enrichment Analysis (GSEA) also showed significant enrichment of genes associated with invasion-associated desmoplasia, regulated by CAFs (23) (Figure S1D, NES = 2.9, Adj.P = 1.2e-3). Taken together, our analysis reveals a correlation between the basal epithelial transcriptional program expressed in KRT14+ breast cancer cells and gene expression markers typical of CAF associated ECM remodeling, across molecular subtypes of breast cancer.
Stromal cells directly enhance collective invasion and KRT14 expression in primary tumor organoids
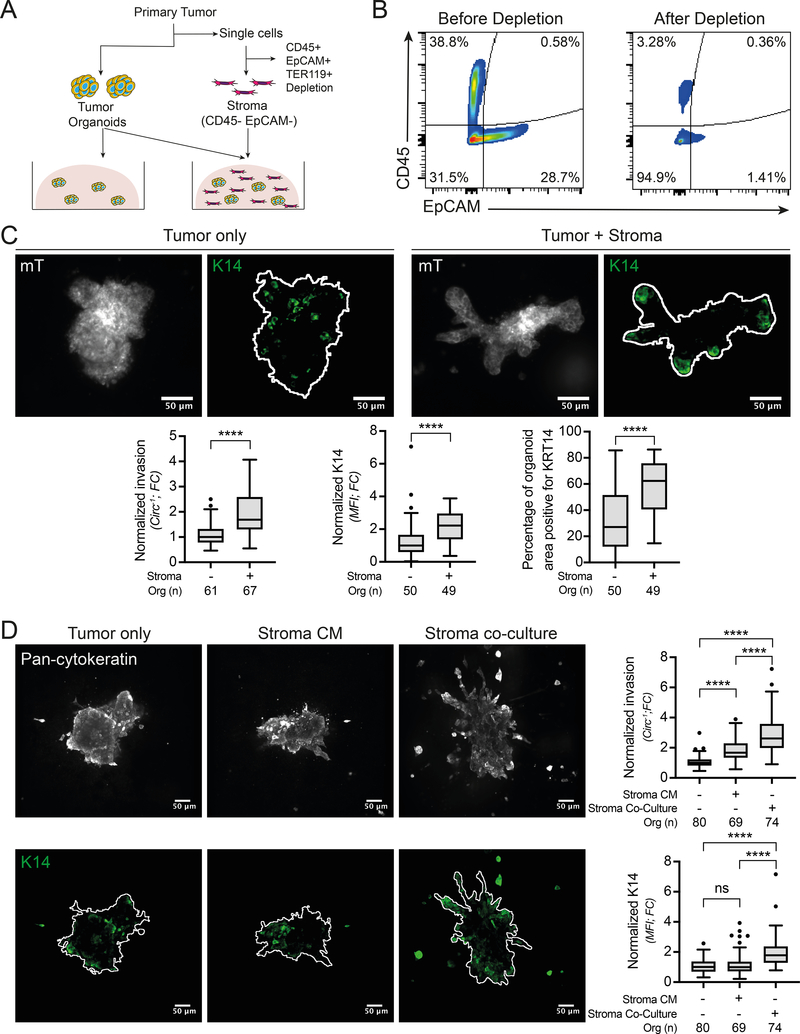
The correlation between basal gene expression and CAF markers led us to hypothesize that stromal cells could regulate the KRT14+ basal phenotype and thereby enhance cancer invasion. To test this hypothesis, we developed a novel protocol to isolate autologous tumor organoids and stromal cells from primary tumors and combine them in 3D organotypic cultures (Figure 2A). We note that the fixation and quantification of invasion was done at an earlier time point compared to our past publications, resulting in a relatively lower amount of invasion in the monocultures. This gave us more sensitivity to detect stromal mediated changes in invasive behavior. Tumor organoids were isolated as previously (5), and concurrently stromal cells were isolated by depleting immune (CD45+), epithelial (EpCAM+) and erythroid (TER119+) cells from the single cell fraction that is normally discarded during organoid isolation (Figure 2A). This procedure resulted in ~95% stromal cell enrichment, as validated by flow cytometry (Figure 2B).
Figure 2: Direct tumor-stroma co-culture increases tumor cell invasion and KRT14 expression.
A) Schematic describing the experimental set-up. B) Flow cytometry validation of epithelial (EpCAM+) and immune cells (CD45+) depletion from stromal cells. C) Representative confocal images of MMTV-PyMT primary tumor organoids, expressing a membrane Tomato (mT) reporter and stained for KRT14, cultured in collagen gels for 72h either in mono-culture or in co-culture with stromal cells (the segmented organoid outline is shown in white). Scale bar, 50μm. The associated quantifications of organoid invasion and KRT14 expression are shown below. Statistical testing was performed using a Mann-Whitney test comparing organoids isolated from 3 tumors, ****p<0.0001. D) MMTV-PyMT organoids were cultured either in mono-culture, in mono-culture with stromal cell conditioned media or in co-culture. The left panel shows representative confocal images of organoids stained for pan-cytokeratin and KRT14. Scale bar, 50μm. Quantification of invasion and KRT14 expression from 3 different tumors is shown on the right. ****p<0.0001, ns = not-significant (p>0.05) (ANOVA test). Micrographs are displayed as maximum intensity Z-projections of 45μm in Z. Circ−1 = inverse circularity; FC = Fold Change; MFI = Mean Fluorescence Intensity.
Following isolation, tumor organoids were cultured in collagen gels +/− stromal cells for 72 hours (Supplementary Video 1). Individual organoids were imaged by confocal microscopy, then segmented and quantitatively analyzed using a custom image processing macro. Invasion was measured as 1/circularity to reflect the transition from a circular shape in non-invasive organoids to a progressively less circular shape following invasion. KRT14 protein levels within each organoid were quantified using immunofluorescence as a marker for the broader basal epithelium program (4,5). Co-culture with stromal cells significantly increased invasion, KRT14 expression and the percentage of organoid area positive for KRT14 in MMTV-PyMT organoids (Figure 2C).
To identify the type of interaction involved in stromal cell mediated increases in tumor organoid invasion and KRT14 expression, we compared tumor organoids co-cultured with stromal cells and organoids in monoculture supplemented with stromal cell-conditioned media. Conditioned media supplementation induced a significant increase in invasion compared to monoculture controls but did not induce a significant increase in KRT14 levels (Figure 2D). Moreover, the magnitude of the increase in invasion induced by co-culture was significantly greater than that induced by conditioned media addition (Figure 2D). These data suggest that stromal cell induction of KRT14 expression involves juxtacrine signaling, while invasion can be induced by juxtacrine signaling and soluble factors.
Stromal cells are in close physical contact with invasive organoids
Previous research has demonstrated diverse mechanisms by which stromal cells can increase cancer invasion, including paracrine signaling (19), ECM remodeling (22) and directly leading collective invasion (10). To investigate the function of stromal cells within our 3D co-cultures, we performed time-lapse differential interference contrast (DIC) microscopy. Visual inspection revealed stromal cells to be highly motile within the collagen I matrix (Supplementary Video 1). They also directly interacted with and remodeled collagen I fibers (Supplementary Video 2). DIC images allowed real-time analysis of stromal cell dynamics; however, without cell type-specific labels it was difficult to unambiguously determine the location and type of cancer cell-stroma interactions. We therefore isolated organoids from MMTV-PyMT (β-actin::CFP; cyan fluorescent label) tumors and co-cultured them with stromal cells from MMTV-PyMT (mT/mG; red fluorescent), using mice of comparable age and tumors of comparable size. KRT14 immunofluorescence was imaged by confocal microscopy to quantify the location of stromal cells in relation to the organoids. Stromal cells localized to the bifurcation point between an invasion strand and the body of the organoid (~80%), adjacent to invading epithelial cells (~50%) and at the leading edge (~30%) (Figure S2A–B). The numbers add to >100% due to the presence of stromal cells in more than one location in a typical organoid. Taken together, the location and behavior of stromal cells in these co-cultures suggests that their regulation of invasion may be mediated by cell-cell contacts, in addition to matrix remodeling.
Their close physical proximity led us to consider the possibility of contact-mediated juxtracrine signaling between stromal cells and invasive organoids. The Notch pathway was an attractive candidate pathway both based on its general role in regulating cell fate decisions among adherent cells and the fact that we previously showed that Jag1 (coding NOTCH receptor ligand Jagged-1) was significantly up-regulated in KRT14+ cells compared to KRT14- cells (4). The role of NOTCH signaling in the tumor microenvironment is not fully understood but Jag1 has been shown to promote fibroblast activation in prostate cancer (24). Therefore, we hypothesized that NOTCH signaling may be involved in the stromal cell mediated invasion and KRT14 expression observed in our model system. To test this, we treated organoid monocultures and organoid-stromal cell co-cultures with the γ-secretase inhibitor DAPT. DAPT had no significant effect on stromal cell-mediated increases in organoid invasion or KRT14 expression. However, DAPT significantly increased KRT14 expression when organoids were grown in monoculture (Figure S2C). These results suggest that the Notch pathway is not involved in stromal cell mediated KRT14 expression and invasion but may limit KRT14 expression in the absence of stromal cells. We were therefore motivated to consider the role of other signaling pathways in regulating invasion.
Inhibition of TGFβ-induced NOX1/4 signaling blocks ECM remodeling and prevents the stromal induction of invasion and KRT14 expression
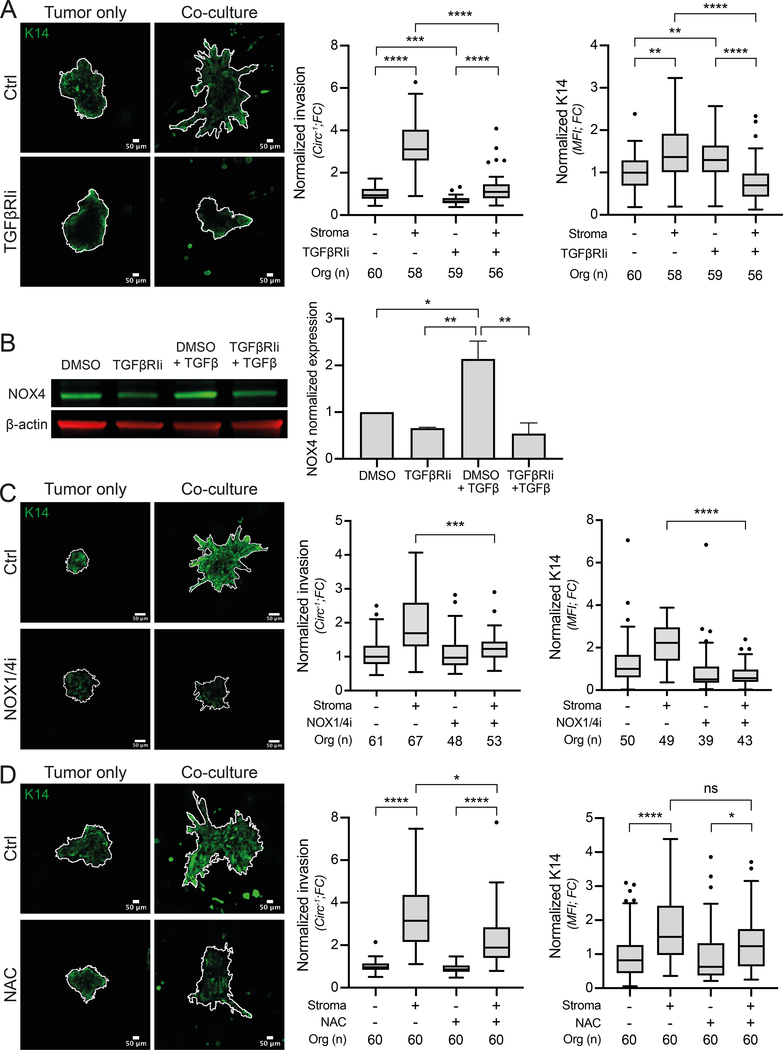
To explore alternative signaling mechanisms, we examined the WGCNA ECM Organization module. GSEA identified significant enrichment of genes up-regulated in TGF-β-treated fibroblasts (Figure S3A). Therefore, we examined the role of TGF-β signaling in our co-culture model. Inhibition of TGF-β receptor I (TGFβRIi), resulted in a significant decrease in organoid invasion but an increase in KRT14 expression in monoculture (Figure 3A, consistent with (25)). In contrast, TGFβRIi induced a significant decrease in both stromal cell-induced invasion and KRT14 expression (Figure 3A). These results reveal that TGF-β signaling regulates the ability of stromal cells role to induce invasion and KRT14 expression. Further studies will be required to disentangle the distinct contributions of stromal vs. epithelial TGF-β signaling on KRT14 expression.
Figure 3: TGFβ-NOX4-ROS pathway regulates organoid invasion and KRT14 expression.
A) MMTV-PyMT organoids, cultured either alone or in co-culture with stromal cells, were treated with TGFβ receptor I inhibitor (TGFβRIi) at 1μM or with a vehicle (DMSO) as control. The left panel shows representative confocal images of organoids invasion (shown by the organoid outline in white) and KRT14 expression. Scale bar, 50 μm. The panel on the right shows the associated quantifications of invasion and KRT14 intensity from 3 different tumors. *p<0.0332, **p<0.0021, ****p<0.0001 (ANOVA test). B) Stromal cells were treated with either DMSO as control, TGFβRIi (1μM), TGFβ1 (2nM) or the combination of both TGFβRIi (1μM) and TGFβ1 (2nM). NOX4 expression was analyzed by immunoblot and β-actin was used as a loading control. The bar graph represents the quantification of NOX4 expression from 3 independent experiments. *p= 0.0332, **p=0.0021 (ANOVA test). C) MMTV-PyMT organoids, either in mono-culture or co-culture and treated or not with 40 μM NOX1/4 inhibitor (GKT831). Left panel shows representative confocal images of organoid invasion (organoid outline in white) and KRT14 expression. Scale bar, 50 μm. Right panel shows quantification of invasion and KRT14 intensity from 3 different tumors. ***p<0.0002, ****p<0.0001 (ANOVA test). D) MMTV-PyMT organoids, either in mono-culture or in co-culture, were treated with ROS inhibitor (NAC) at 1mM or with a vehicle (DMSO) as control. Left panel shows representative confocal images of organoid invasion (shown by the organoid outline in white) and KRT14 expression. Scale bar, 50 μm. Right panel shows quantification of invasion and KRT14 intensity from 3 different tumors. ns = not-significant (p>0.05), *p<0.0332, ****p<0.0001 (ANOVA test). Micrographs are displayed as maximum intensity Z-projections of 45μm in Z. Circ−1 = inverse circularity; FC = Fold Change; MFI = Mean Fluorescence Intensity.
NADPH oxidase 4 (NOX4) was identified among the WGCNA ECM organization module hub genes and is known to mediate TGF-β’s role in converting fibroblasts to tumor-promoting myofibroblasts (21,26,27). Therefore, we hypothesized that NOX4 may be involved in the TGF-β dependent effects on stromal-cell mediated organoid invasion and KRT14 expression. We selected NOX4 for further study based on its significance in this computational analysis, its mechanistic plausibility in regulating key aspects of invasion in this system (25), and the ready availability of antibodies and a potent inhibitor. In support of this hypothesis, we demonstrated that tumor organoids secrete TGF-β (Figure S3B) and that stromal cell expression of NOX4 is TGFβRI dependent, when treated with either recombinant TGF-β or organoid conditioned media (Figure 3B and S3C). To test the role of NOX4 in stromal-cell mediated invasion and KRT14 expression we treated mono- and co-cultures with a NOX1/4 inhibitor (GKT831). This treatment did not affect monocultures, but significantly reduced the ability of stromal cells to promote invasion and KRT14 expression (Figure 3C). We repeated this experiment using siRNA targeting NOX4, which produced ~55% knockdown of NOX4 expression and significantly reduced organoid invasion (Figure S3D–F). CAF/myofibroblast mediated ECM remodeling is known to facilitate tumor cell invasion (22). Therefore, we analyzed whether NOX4 was responsible for stromal cell mediated collagen remodeling, using a gel contraction assay, which showed that NOX4 inhibition reduced stromal cell contractility (Figure S3G). Those results were also validated in the murine C31-Tag basal breast cancer model (Figure S3G–H). NOX4’s primary function is reactive oxygen species (ROS) production (28). Therefore, we treated mono- and co-cultures with the antioxidant N-acetylcysteine (NAC) to determine whether non-specific ROS inhibition would phenocopy NOX4 inhibition. NAC significantly reduced stromal cell induced organoid invasion. However, at 1 mM, it had no significant effect on KRT14 expression (Figure 3D).
Discussion
In this study we used WGCNA as a hypothesis-generating platform to suggest mechanisms regulating the KRT14+ basal invasive phenotype. These efforts suggested a role for CAFs, based on correlation between a Krt14+ gene signature and gene expression modules identified in TCGA data. We then used primary 3D organotypic culture to test this hypothesis and to reveal a novel mechanism for stromal regulation of cancer cell invasion. Juxtacrine signaling mediated by stromal cells induces tumor cells to increase activation of a basal invasive program. These observations were facilitated by development of a novel method to co-culture tumor organoids and autologous stromal cells in 3D, which enabled tumor-stromal interactions to be analyzed in a physiologically relevant setting. Furthermore, we demonstrated that these pro-tumorigenic effects of stromal cells are sensitive to inhibition of NOX1/4 signaling.
Paracrine signaling from stromal cells has been shown previously to increase tumor cell invasion, with multiple demonstrated molecular mediators of these signaling interactions (e.g. HGF) (19). Prior studies have predominantly utilized transwell assays to monitor tumor cell invasion, which precludes investigation of collective invasion. This technical difference in assays may account for why we observed a more prominent role for juxtacrine signaling and suggests that different mechanisms of tumor-stromal interaction may drive different modes of tumor cell invasion.
Fibroblasts have been shown to lead collective invasion of squamous cell carcinoma cells, by creating tracks through the ECM that cancer cells could not efficiently create on their own (10). Similar fibroblast leader cells have not been described in IDC; instead CAFs have been shown to regulate breast cancer invasion at a distance, through Hippo signaling-dependent remodeling of ECM (12). Our analysis of CAF localization revealed stromal cells at both the leading edge of invasion strands and at the junction connecting the strand to the organoid body, suggesting multiple roles beyond generating tunnels. Our study is the first to demonstrate regulation of KRT14+ basal phenotype by the stromal microenvironment. However, KRT14+ epithelial cells are capable of leading collective invasion in the absence of stromal cells in organoids from both mouse models, suggesting both cancer cell-intrinsic and stromal-dependent mechanisms of induction of the KRT14 basal phenotype and collective invasion (5). We also acknowledge that our isolation method cannot distinguish the relative contributions of normal vs. cancer associated fibroblasts to this induction of invasive phenotype and behavior. The autologous co-culture model system developed here should prove broadly useful to investigate this signaling interaction further in breast cancer and also to extend these analyses to other solid tumors.
Stromal cells may also regulate the KRT14+ basal phenotype and invasion indirectly, through ECM remodeling (12). Time-lapse imaging revealed stromal cell remodeling of collagen I fibers in co-cultures. Invasive KRT14+ tumor islands are also more frequently found in regions with aligned and elongated fibrillar collagen in vivo in mouse models (5). The CAFs most representative of the ECM Organization module, those activated by TGF-β, have also been shown to regulate collagen structure in the stroma of multiple solid tumors (29). Furthermore, collagen cross-linking and alignment correlates with poor prognosis in breast cancer, regulating invasion and metastases (22).
Strategies to therapeutically target cancer invasion are critically needed in multiple cancers. Our study suggests that targeting the fibroblastic stroma through NOX4 inhibition could reduce invasion and therefore metastasis, though further work in animal models is needed. Further analysis of the ECM organization module hub genes (Supplementary Table 2) could also identify alternative candidate targets.
Supplementary Material
Implications.
Fibroblasts within mammary tumors can regulate the molecular phenotype and invasive behavior of breast cancer cells.
Acknowledgements
The authors thank Joel Bader for helpful comments on the manuscript. C.J. Hanley and G.J. Thomas received support for this project through grants from Cancer Research UK. A.J. Ewald received support for this project through grants from: the Breast Cancer Research Foundation/Pink Agenda (BCRF-19-048), the Commonwealth Foundation, and the National Institutes of Health / National Cancer Institute (U01CA217846, U54CA2101732, 3P30CA006973). The results from Figure 1 are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. The GKT831 compound was kindly provided by Genkyotex.
Footnotes
Conflict of Interest Statement: AJE is an inventor on patents related to the use of keratin-14 as a biomarker for invasive cancer cells and on the use of antibodies to treat cancer. GJH and CJH are inventors on a patent on the use of NOX4 inhibitors to treat cancer
References
- 1.Te Boekhorst V, Preziosi L, Friedl P. Plasticity of Cell Migration In Vivo and In Silico. Annu Rev Cell Dev Biol 2016;32:491–526 [DOI] [PubMed] [Google Scholar]
- 2.Bronsert P, Enderle-Ammour K, Bader M, Timme S, Kuehs M, Csanadi A, et al. Cancer cell invasion and EMT marker expression: a three-dimensional study of the human cancer-host interface. The Journal of pathology 2014;234:410–22 [DOI] [PubMed] [Google Scholar]
- 3.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci U S A 2016;113:E854–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013;155:1639–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Silva Rudland S, Platt-Higgins A, Winstanley JH, Jones NJ, Barraclough R, West C, et al. Statistical association of basal cell keratins with metastasis-inducing proteins in a prognostically unfavorable group of sporadic breast cancers. Am J Pathol 2011;179:1061–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung KJ, Ewald AJ. A collective route to metastasis: Seeding by tumor cell clusters. Science 2016;352:167–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nat Rev Cancer 2015;15:473–83 [DOI] [PubMed] [Google Scholar]
- 9.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer 2011;11:573–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labernadie A, Kato T, Brugues A, Serra-Picamal X, Derzsi S, Arwert E, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nature cell biology 2017;19:224–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 2007;9:1392–400 [DOI] [PubMed] [Google Scholar]
- 12.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 2013;15:637–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163:506–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol 2002;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen-Ngoc KV, Shamir ER, Huebner RJ, Beck JN, Cheung KJ, Ewald AJ. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol Biol 2015;1189:135–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yersal O, Barutca S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World journal of clinical oncology 2014;5:412–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005;121:335–48 [DOI] [PubMed] [Google Scholar]
- 19.Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, et al. Stromal features are predictive of disease mortality in oral cancer patients. The Journal of pathology 2011;223:470–81 [DOI] [PubMed] [Google Scholar]
- 20.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 2010;330:827–30 [DOI] [PubMed] [Google Scholar]
- 21.Hanley CJ, Mellone M, Ford K, Thirdborough SM, Mellows T, Frampton SJ, et al. Targeting the Myofibroblastic Cancer-Associated Fibroblast Phenotype Through Inhibition of NOX4. Journal of the National Cancer Institute 2018;110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009;139:891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC medical genomics 2010;3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Q, Zhang B, Zhang L, Dang T, Rowley D, Ittmann M, et al. Jagged1 upregulation in prostate epithelial cells promotes formation of reactive stroma in the Pten null mouse model for prostate cancer. Oncogene 2017;36:618–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019;573:439–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, Dikalov S, et al. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ Res 2005;97:900–7 [DOI] [PubMed] [Google Scholar]
- 27.Hecker L, Vittal R, Jones T, Jagirdar R, Luckhardt TR, Horowitz JC, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med 2009;15:1077–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiose A, Kuroda J, Tsuruya K, Hirai M, Hirakata H, Naito S, et al. A novel superoxide-producing NAD(P)H oxidase in kidney. J Biol Chem 2001;276:1417–23 [DOI] [PubMed] [Google Scholar]
- 29.Mellone M, Hanley CJ, Thirdborough S, Mellows T, Garcia E, Woo J, et al. Induction of fibroblast senescence generates a non-fibrogenic myofibroblast phenotype that differentially impacts on cancer prognosis. Aging 2016;9:114–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.