Abstract
Lung adenocarcinoma (LA) is a subtype of lung cancer that accounts for about 40% of all lung cancers. Analysis of molecular mechanisms controlling this cancer can help scientists to detect, control and treat LA. Here, a microarray dataset (GSE118370) containing six normal lung (NL) and six LA samples was screened using GEO2R to find differentially expressed genes (DEGs). Then, DAVID, KEGG and ChEA were used to analyze DEGs-related gene ontology, pathways and transcription factors (TFs), respectively. The Protein–protein interaction network for DEGs and TFs was constructed by STRING and Cytoscape. To find microRNAs and metabolites associated with DEGs, miRTarBase and HMDB were used, respectively. It was found that 350 genes were upregulated and 608 genes were downregulated in LA. The upregulated genes or LA-related gens were enriched in biological process and pathways such as extracellular matrix disassembly and p53 signaling pathway, whereas the downregulated genes or NL-related genes were enriched in cell adhesion and cell-surface receptor signaling pathway. ESR1, KIF18B, BIRC5, CHEK1, CCNB1 and AURKA were determined as hub genes for LA. FOXA1 and TFAP2A had the highest number of connectivity in LA-related TFs. hsa-miR-192-5p and hsa-miR-215-5p could target the highest number of LA-related genes. Metabolite analysis showed that Estrone and NADPH were among the top ten metabolites associated with LA-related genes. Taken together, LA-related genes, especially the hub genes, TFs, and metabolites might be used as novel markers for LA, as well as for diagnosis and guiding therapeutic strategies of LA.
Keywords: Lung adenocarcinoma, Protein–protein interaction, Bioinformatics analysis, Transcription factor, miRNA, Metabolite
Introduction
Lung cancer is a highly heterogeneous malignant epithelial tumor that is one of the most dangerous cancers that kill more than one million people of both sex each year (Chen et al. 2018). Approximately 50% of patients diagnosed with lung cancer die in the first year after diagnosis and the 5-year survival is < 20% (Howlader et al. 2015). Approximately 85% of lung cancer cases are diagnosed as non-small cell lung cancers (NSCLC), which are subdivided to squamous cell carcinoma (SCC), lung adenocarcinoma (LA) and large cell carcinoma (LCC) subtypes (Ettinger et al. 2018). Among them, LA accounts for 40% of all lung cancers and it is the most prevalence lung cancer in people who have never smoked (de Groot et al. 2018). Unfortunately, despite of different treatment options such as surgery, chemotherapy and radiotherapy for LA patients, but the five-year survival rate remains less than 20% (Howlader et al. 2015).
To this end, scientists and researchers studied many factors involved in lung cancer development. For example, tobacco smoke is the main reason for 85% of lung cancers (Weber et al. 2019). It seems that tobacco smoke lead to an increase at genetic and epigenetic abnormalities, which finally lead to invasive lesion and metastasis phenomenon of lung cancers (Saji et al. 2019). Several NSCLC inducing mutations have been found in some important factors including epidermal growth factor receptor (EGFR), parallel phosphatidylinositol 3-kinase (PI3K) pathway and mitogen-activated protein kinase kinase (MAP2K1) (Shtivelman et al. 2014; Nicetto and Zaret 2019). Moreover, the amplification of proto-oncogenes such as fibroblast growth factor receptor 1 (FGFR1) and discoidin domain receptor 2 (DDR2) is involved in NSCLC (Cooper et al. 2013). In addition, the role of tumor suppressor genes is increasingly recognized with aberrations reported in tumor protein P53 (TP53), phosphatase and tensin homolog (PTEN), retinoblastoma 1 (RB1) and liver kinase b1 (LKB11) (Brambilla and Gazdar 2009; Shtivelman et al. 2014). Despite of these important and interesting findings, as well as development of new therapeutic approaches, LA as the most important subtype of NSCLC, kills many people each year. To efficient treatment of LA, it seems necessary and useful to analyze this subtype precisely, especially at gene expression level. It is possible that other unknown molecular and cellular players have vital roles in LA development, progression and metastasis.
Comprehensive elucidation of the biological process, transcriptional regulation, and metabolic landscape in LA has the potential to find biomarkers for early detection and prediction of prognosis, as well as to guide the development of strategies that target key signaling and metabolic pathways and TFs supporting LA. Bioinformatics analysis of microarray data obtained from of LA and normal lung (NL) can be used for comprehensive understanding of molecular and cellular factors controlling LA behaviors and identity. Here, we obtained a microarray data of LA and NL. Then we determined differentially expressed genes (DEGs), and then analyzed the upregulated genes in LA (as LA-related gens) and downregulated genes (as NL-related genes) to find the key biological process, transcriptional regulatory networks and TFs, miRNAs and metabolites. Here we found some new key cellular and molecular factors driving LA. They can be considered as biomarkers for detection of LA, as well as molecular targets in “targeted therapy” using monoclonal antibodies and small molecules.
Materials and Methods
Gene profile extraction and identification of DEGs
A literature review for high-throughput gene expression studies was performed to find a suitable study having the gene expression profile for LA and NL samples. A study reported by Xu et al. was selected for a precise and comprehensive computational analysis (Xu et al. 2018). The microarray data was obtained from NCBI’s Gene Expression Omnibus database (http://www.ncbi.nlm. nih.gov/geo) with GEO accession number GSE118370, composed of overall 12 different samples including 6 LA and 6 NL. All GSE series matrix files set were downloaded and parsed by GEOquery package (Davis and Meltzer 2007) with R version 3.6.0 and the GSE series values normalized with R. Also the linear models for microarray data (Limma) package (Smyth 2004) was used to determined DEGs, which were significantly upregulated and downregulated in LA. DEGs were selected with univariate tests according to the p-value (p < 0.05) and log2 fold change (log2 FC ≥ 2).
Functional and Pathway Enrichment Analyses
To investigate the biological characteristics and functional enrichment of candidate DEGs, functional enrichment analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.ncifcrf.gov), which is a free and online web-tool for investigators to understand biological themes behind large list of genes. Here, DAVID was used for finding the DEGs-related biological process (BB), molecular function (Weber et al. 2019) and cellular component (CC) (Huang et al. 2007). To find DEGs-related pathways, DEGs were analyzed using Kyoto Encyclopedia of Genes and Genomes database (KEGG, www.genome.jp/kegg/), which is a database resource for understanding high-level functions and utilities of the biological system from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (Ogata et al. 1999).
Transcription Factor analysis and construction of protein-protein interaction network
To find the transcriptional regulators/transcription factors (TFs) that control the expression of LA- and NL-related genes, the ChIP enrichment analysis (ChEA) database was used. The information in ChEA database has been extracted manually from diverse ChIP-based experimental procedure including ChIP-chip, ChIP-seq, ChIP-PET, and DamID-seq techniques (Lachmann et al. 2010). Moreover, the TFs listed in the recent paper entitled: The Human Transcription Factors (Lambert et al. 2018), were used to select the TFs among LA- and NL-related genes.
In order to analyze protein-protein interaction between LA-related genes, NL-related genes or candidate TFs, STRING (https://string-db.org) was used. This online biological database is web resource of known and predicted PPI (Szklarczyk et al. 2018). Moreover, Cytoscape software (Version 3.4.0, http://www.cytoscape.org/) was also used to illustrate desired network for DEGs and TFs (Shannon et al. 2003).
Survival analysis for LA hub genes
To evaluate the prognostic value of the LA hub genes, the Kaplan–Meier plotter was applied to the patient data in the GEPIA database, which is a web-based tool to deliver fast and customizable functionalities based on TCGA and GTEx data (Tang et al. 2017). Using this analysis, the overall survival for the patients with lung cancer is obtained according to the low and high expression of the candidate genes. In addition, The Human Protein Atlas Database was used to illustrate the expression level of hub genes in NL and LA (Uhlén et al. 2005).
Analysis of DEGs-related Metabolite
In order to find DEGs-related metabolites, the Enrichr dataset-linked Human Metabolome Database (HMDB) was used (http://amp.pharm.mssm.edu). HMDB is a web-enabled metabolomics database containing comprehensive information about human metabolites along with their biological roles, physiological concentrations, disease associations, chemical reactions and metabolic pathways (Wishart et al. 2017). Top ten metabolites related to DEGs were selected and ranked based on p-value (P ≤ 0.05).
Perdition of miRNAs targeting LA-related genes
To find the top ten microRNAs that presumably target LA-related genes, Enrichr dataset-linked miRTarBase was used (http://amp.pharm.mssm.edu). miRTarBase provides information about experimentally validated miRNA-target interactions (MTIs) and its new update contains 422517 curated MTIs from 4076 miRNAs and 23054 target genes collected from over 8500 articles (Chou et al. 2017). Top ten miRNAs targeting LA-related genes were selected and ranked based on p-value (P ≤ 0.05).
Results
Identification of LA- and NL-related genes
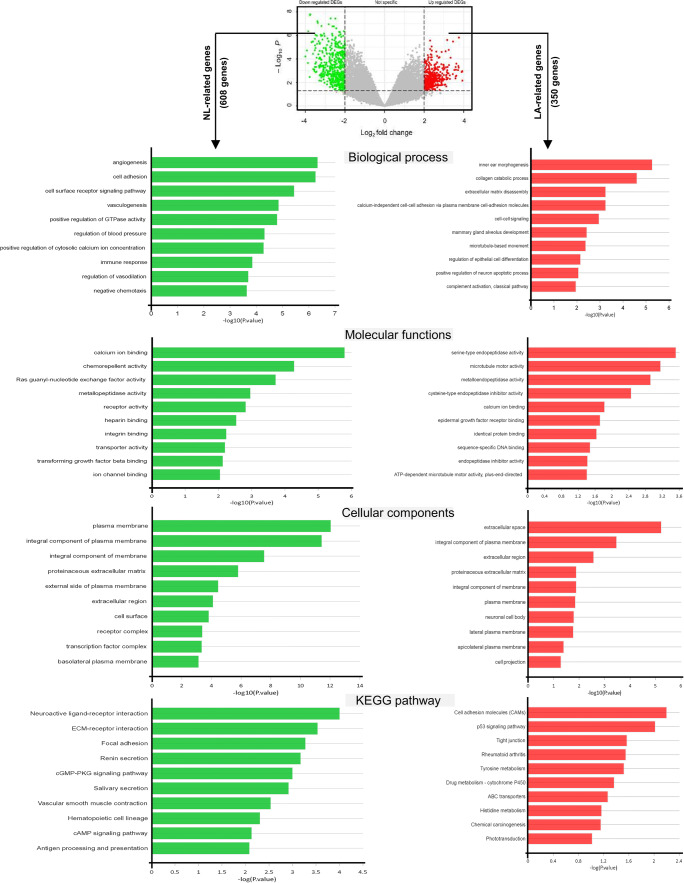
To find the DEGs in human invasive LA, the microarray data of GSE118370 containing six invasive LA tissues and six NL tissues was analyzed. The Limma package was applied to detect the DEGs, using adjust P value < 0.05 and |logFC| ≥ 2 as cut-off standard. A total of 958 DEGs were obtained, among them 350 genes were upregulated and 608 genes were downregulated in LA (Fig. 1, top). Here, we determined the upregulated and downregulated genes as LA- and NL-related genes, respectively.
Fig. 1.
GO functional annotation and pathway enrichment analysis of upregulated and downregulated genes in LA (here as LA- and NL-related genes). LA: lung adenocarcinoma, NL: normal lung
GO function and KEGG pathway enrichment analysis
For a more in-depth understanding of the candidate LA- and NL-related genes, GO function and pathway enrichment were screened using DAVID and KEGG database, respectively. It was found that LA- and NL-related genes were particularly enriched in BP, including collagen catabolic process, extracellular matrix disassembly, calcium-independent cell-cell adhesion, cell-adhesion molecules and cell-cell signaling for LA-related genes (Fig. 1, right). In contrast, NL-related genes were mainly involved in cell adhesion, cell surface receptor signaling pathway and angiogenesis and vasculogenesis (Fig. 1, left). The analysis of MF showed that the LA-related genes were significantly enriched in serine-type endopeptidase activity and microtubule motor activity (Fig. 1, right), whereas the NL-related genes were mainly involved in calcium ion binding, chemorepellent activity and Ras guanyl-nucleotide exchange factor activity (Fig. 1, left). Moreover, for LA-related genes, the most significantly enriched GO terms on CC were extracellular space, integral component of plasma membrane and extracellular region (Fig. 1, right), whereas the NL-related genes were significantly associated with plasma membrane and integral component of plasma membrane (Fig. 1c, right).
KEGG analysis results revealed that LA-related genes were particularly enriched in cell adhesion molecules (CAMs), p53 signaling pathway, and tight junction (Fig. 1, right), whereas the NL-related genes were significantly enriched in neuroactive ligand-receptor interaction, ECM-receptor interaction and focal adhesion (Fig. 1, left).
PPI network construction and hub genes selection
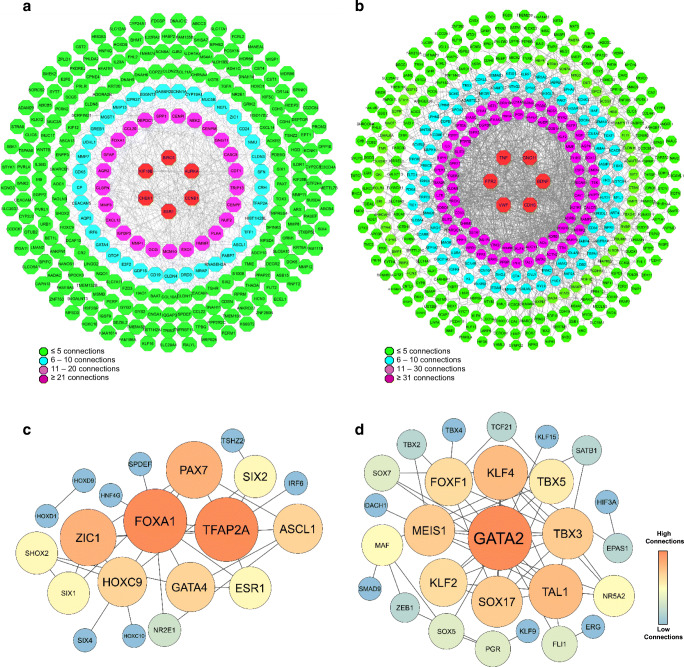
LA- and NL-related genes were analyzed using STRING database and illustrated using Cytoscape software. The PPI network constructed for LA-related genes showed that 31 genes could interact with more than 10 genes (Fig. 2a). Among them, Estrogen Receptor 1 (ESR1), Kinesin Family Member 18B (KIF18B), Baculoviral IAP Repeat Containing 5 (BIRC5), Checkpoint Kinase 1 (CHEK1), Cyclin B1 (CCNB1) and Aurora Kinase A (AURKA) have the highest connectivity with others (≥ 20 interactions), and they were identified as the hub genes for LA (Fig. 2a). The PPI network constructed for NL-related genes showed that 79 genes have more than 11 connections with others. Top six genes with highest degree of connectivity (≥ 31) were Tumor Necrosis Factor (TNF), G Protein Subunit Gamma 11 (GNG11), Brain Derived Neurotrophic Factor (BDNF), Cadherin 5 (CDH5), Von Willebrand Factor (VWF) and Formyl Peptide Receptor 2 (FPR2).
Fig. 2.
PPI network analysis. a PPI network of LA-related genes, b PPI network of NL-related genes, c PPI network of TFs existing in LA-related genes, d PPI network of TFs existing in NL-related genes
To extract TFs from the list of LA- and NL-related genes, the TFs listed in the recently published paper entitled “The Human Transcription Factors” was used (Lambert, Jolma et al. 2018). It was found that 33 and 35 TFs existed among LA- and NL-related genes, respectively. The PPI network constructed using the TFs existing in LA-related genes showed that Forkhead Box A1 (FOXA1), Transcription Factor AP-2 Alpha (TFAP2A), Paired Box 7 (PAX7), Zic Family Member 1 (ZIC1), Homeobox C9 (HOXC9), Achaete-Scute Complex-Like 1 (ASCL1), GATA Binding Protein 4 (GATA4), Estrogen Receptor 1 (ESR1) and SIX Homeobox 1 (SIX1) have more than three connectivities with each other or other TFs (Fig. 2c).Importantly, FOXA1 and TFAP2A had the highest connectivity with others, suggesting an important role for these TFs in LA. The PPI network constructed by TFs existing in NL-related genes illustrated that 11 TFs have ≥ 4 connectivities with other TFs in the network (Fig. 3d). In this network, GATA Binding Protein 2 (GATA2), Kruppel Like Factor 4 (KLF4), Meis Homeobox 1 (MEIS1), SRY-Box Transcription Factor 17 (SOX17), T-Cell Acute Lymphocytic Leukemia 1 (TAL1) and T-Box Transcription Factor 3 (TBX3) were selected as the top six TFs, which had the highest degree of connectivity with others. Notably, GATA2 had the highest number of connectivity with other TFs, suggesting its critical role in preserving NL identity (Fig. 2d).
Fig. 3.
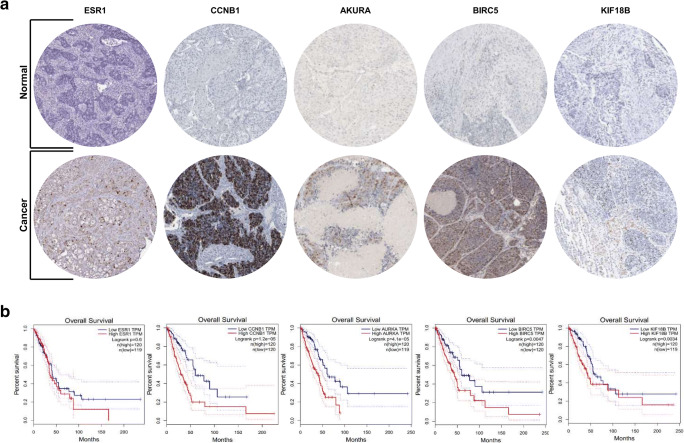
LA hub genes analysis. a Immunohistochemistry of the five hub genes in LA and NL samples (based on the Human Protein Atlas). b Overall survival analysis of the five hub genes based on Kaplan Meier-plotter. LA: Lung adenocarcinoma, NL: Normal lung
To find the upstream TFs that potentially regulate the expression of LA- and NL-related genes, ChEA database was used. The analyses revealed that 64 TFs could target the LA-related genes, whereas 90 TFs could potentially target NL-related genes (FDR < 0.05). The top ten TFs connected to LA-related genes were EHF, KLF5, ZBTB7C, GRHL2, ELF3, TFCP2L1, PAX9, E2F8, IRF6 and OVOL1 (Table1). They all had more than 20 targets among LA-related genes. In contrast, BCL6B, SOX18, MEOX1, SOX7, EPAS1, SOX17, MEOX2, TCF21, ERG and EBF1 were identified as the top ten TFs associated with NL-related genes. Each of them could target at least 40 NL-related genes.
Table 1.
Top ten TFs controlling the expression of LA- and NL-related genes
| TF | Description | Number of targets | FDR | |
|---|---|---|---|---|
| TFs regulating LA-related genes | EHF | ETS Homologous Factor | 32 | 8.18E-11 |
| KLF5 | Kruppel Like Factor 5 | 30 | 1.23E-9 | |
| ZBTB7C | Zinc Finger And BTB Domain Containing 7C | 28 | 2.16E-8 | |
| GRHL2 | Grainyhead Like Transcription Factor 2 | 27 | 7.86E-8 | |
| ELF3 | E74 Like ETS Transcription Factor 3 | 26 | 2.95E-7 | |
| TFCP2L1 | Transcription Factor CP2 Like 1 | 23 | 2.0E-5 | |
| PAX9 | Paired Box 9 | 22 | 5.33E-5 | |
| E2F8 | E2F Transcription Factor 8 | 22 | 5.33E-5 | |
| IRF6 | Interferon Regulatory Factor 6 | 22 | 5.33E-5 | |
| OVOL1 | Ovo Like Transcriptional Repressor 1 | 21 | 1.41E-4 | |
| TFs regulating NL-related genes | BCL6B | BCL6B Transcription Repressor | 58 | 5.88E-29 |
| SOX18 | SRY-Box Transcription Factor 18 | 58 | 5.88E-29 | |
| MEOX1 | Mesenchyme Homeobox 1 | 54 | 1.38E-25 | |
| SOX7 | SRY-Box Transcription Factor 7 | 51 | 7.09E-23 | |
| EPAS1 | Endothelial PAS Domain Protein 1 | 44 | 3.26E-17 | |
| SOX17 | SRY-Box Transcription Factor 17 | 42 | 8.58E-16 | |
| MEOX2 | Mesenchyme Homeobox 2 | 41 | 3.77E-15 | |
| TCF21 | Transcription Factor 21 | 41 | 4.2E-15 | |
| ERG | ETS Transcription Factor ERG | 40 | 1.86E-14 | |
| EBF1 | EBF Transcription Factor 1 | 36 | 1.58E-11 |
The prognostic value of hub genes
In order to confirm the expression hub genes of LA (ESR1, KLF18, BIRC5, CCNB1 and AURKA), human protein atlas database was used. Interestingly, all five hub genes had clearly higher level of protein expression in the LA samples compared to NL ones (Fig. 3a). Subsequently, a Kaplan‑Meier plotter was used to perform overall survival analysis (Fig. 3b). It’s important to note that the samples for overall survival analysis, derived from the Kaplan‑Meier plotter, were different from those used in the analysis of DEGs. It was found that the level of hub genes expression was associated with overall survival rate. Accordingly, patients with higher expression level of all hub genes exhibited poorer overall survival (Fig. 3b).
Top ten metabolites associated with LA- and NL-related genes
As we found using KEGG pathway enrichment, DEGs are involved in many metabolisms-related pathways and intermediate metabolites. The analysis of DEGs using HMDB showed that LA- and NL-related genes were linked to different metabolites. The top ten metabolites associated with LA-related genes were 3,4-dihydroxymandelaldehyde, acetaldehyde, estradiol, NADPH, melatonin, androstenedione, uridine diphosphate glucose, estrone, and 19S-hydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid (Table 2). Interestingly, three reproductive hormones/metabolites were among the top ten metabolites. Moreover, Cytochrome P450-related genes such as CYP2J2, CYP2C8 and CYP19A1 were most important LA-related genes that were associated with most of metabolites (Table 2). In contrast, the NL-related genes were associated with metabolism of serotonin, cyclic AMP, L-Arginine, imipramine, cyclic GMP, norepinephrine, epinephrine, hydrogen peroxide, sodium and zinc (Table 2). Each metabolite was linked, at least, to three NL-related genes.
Table 2.
Top ten Metabolites associated with LA- and NL-related genes
| Metabolite | P-value | Genes | |
|---|---|---|---|
|
Metabolites associated with LA-related genes |
3,4-Dihydroxymandelaldehyde | 0.001663 | ALDH1A3, ALDH3B2, ADH1C |
| Uridine diphosphate glucose | 0.005189 | GYS2, ENPP3, GYG2 | |
| Acetaldehyde | 0.005189 | ALDH1A3, ALDH3B2, ADH1C | |
| NAP | 0.005489 | CYP2J2, ALDH3B2, ALDH1A3, NQO1, CYP2C8, CYP24A1, ADH1C, HSD17B2, DECR2, CYP19A1 | |
| Estradiol | 0.005819 | CYP2J2, CYP2C8, HSD17B2, CYP19A1, ESR1 | |
| NADPH | 0.00644 | CYP2J2, ALDH3B2, ALDH1A3, NQO1, CYP2C8, CYP24A1, ADH1C, HSD17B2, DECR2, CYP19A1 | |
| Melatonin | 0.007036 | CYP2J2, CYP2C8, CYP19A1, ESR1 | |
| Androstenedione | 0.008626 | CYP2J2, CYP2C8, HSD17B2, CYP19A1 | |
| Estrone | 0.008633 | CYP2J2, CYP2C8, HSD17B2, CYP19A1, ESR1 | |
| 19S-hydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid | 0.0146 | CYP2J2, CYP2C8, CYP19A1 | |
|
Metabolites associated with NL-related genes |
Epinephrine | 2.14E-04 | MAOB, ADRB1, ADRB2, ADRA1A |
| Norepinephrine | 0.001681 | MAOB, ADRB1, ADRB2, ADRA1A | |
| Cyclic GMP | 0.001794 | GUCY1A2, PDE1C, NPR1, ADCY4, PDE3A, PDE5A | |
| Imipramine | 0.003582 | MAOB, FMO2, SLC6A4 | |
| L-Arginine | 0.00507 | ADPRH, PADI4, NOS1, ART4 | |
| Zinc | 0.010875 | ZEB1, ACE;BMP1, CA2, ADH1B, CA4, ALDOB | |
| Cyclic AMP | 0.019569 | RAP2A, MEIS1, PDE1C, TRPV2, PDE3A, ADCY4, PDE5A, PPBP, RXFP1, PTGDR | |
| Serotonin | 0.029554 | MAOB, INMT, SLC6A4 | |
| Sodium | 0.030329 | SCN10A, NFASC, KCNT2, CA4, ATP1A4, TNR, SCN7A, ATP1A2, SCN4B, SLC5A4, SLC6A4 | |
| Hydrogen peroxide | 0.035222 | AOC3, DUOX1, MAOB, GPX3 |
Top ten miRNAs targeting LA-related genes
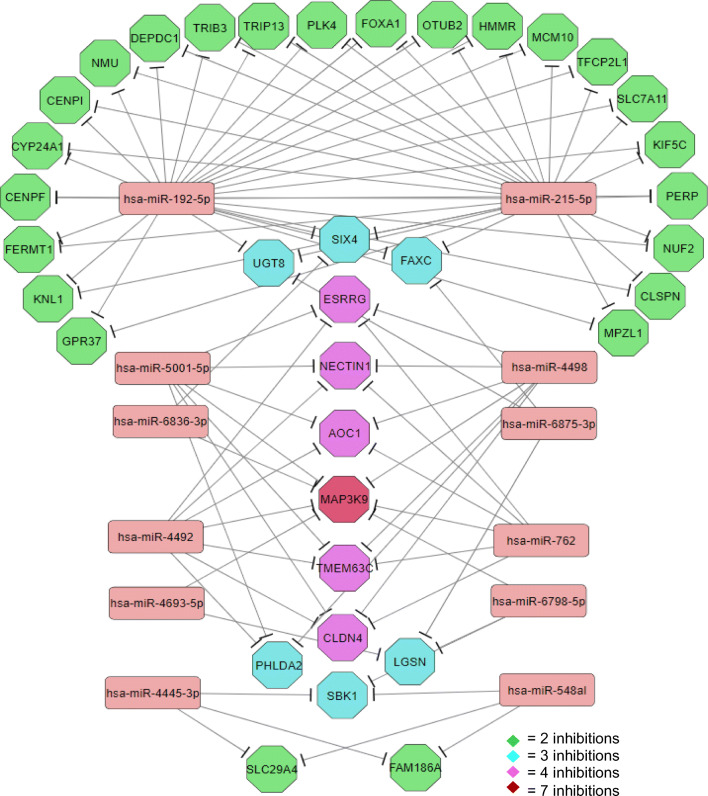
In order to find the candidate miRNAs that potentially could target LA-related genes, miRTarBase was used. It was found 1857 miRNAs could theoretically target at least one of the LA-related genes. Among them, those significantly targeted the highest number of LA-related genes were selected as top ten miRNAs (Table 3). Notably, hsa-miR-192-5p, hsa-miR-215-5p, hsa-miR-6875-3p and hsa-miR-6836-3p could target 32, 26, 11 and 10 mRNAs of LA-related genes, respectively. Interestingly, hsa-miR-192-5p and hsa-miR-215-5p had many common targets. Accordingly, hsa-miR-192-5p could target all targets of hsa-miR-215-5p, except SIX Homeobox 4 (SIX4) (Fig. 4). In addition, hsa-miR-762, hsa-miR-4492, hsa-miR-4498 and hsa-miR-5001-5p had the same targets between the LA-related genes (Fig. 4; Table 3). Among LA-related genes, MAP3K9 was the only one that could be targeted by seven miRNAs, suggesting its important role in LA development (Fig. 4; Table 3).
Table 3.
Top ten miRNAs targeting the LA-related genes
| miRNA | Number of targets | P-value | List of target genes |
|---|---|---|---|
| hsa-miR-548al | 3 | 0.005189 | SBK1, FAM186A, SLC29A4 |
| hsa-miR-4445-3p | 3 | 0.00854 | SBK1, FAM186A, SLC29A4 |
|
hsa-miR-4492, hsa-miR-4498, hsa-miR-5001-5p |
7 | 0.01872 | CLDN4, AOC1, TMEM63C, ESRRG, MAP3K9, PHLDA2, NECTIN1 |
| hsa-miR-6798-5p | 7 | 0.019506 | CCL22, SBK1, LGSN, DNLZ, BICDL1, MAP3K9, KLF16 |
| hsa-miR-6836-3p | 10 | 0.021782 | CDT1, RGS17, SIX4, ABCB5, CENPM, E2F2, MAP3K9, GYG2, KCNK1, AURKA |
| hsa-miR-762 | 7 | 0.022004 | CLDN4, AOC1, TMEM63C, ESRRG, MAP3K9, PHLDA2, NECTIN1 |
| hsa-miR-215-5p | 26 | 0.023077 | FOXA1, OTUB2, SMARCB1, FAXC, MCM10, TFCP2L1, HMMR, SLC7A11, UGT8, SIX4, KIF5C, PERP, NUF2, CLSPN, MPZL1, PLK4, GPR37, KNL1, FERMT1, CENPF, CYP24A1, CENPI, NMU, DEPDC1, TRIB3, TRIP13 |
| hsa-miR-4693-5p | 4 | 0.024688 | LGSN, TMEM200C, SHOX2, MAP3K9 |
| hsa-miR-192-5p | 32 | 0.029008 | FOXA1, OTUB2, HCN3, SMARCB1, FAXC, MCM10, TFCP2L1, HMMR, SLC7A11, C1ORF53, UGT8, SIX4, KIF5C, PERP, NUF2, CLSPN, E2F5, MPZL1, PLK4, ABCC3, GPR37, KNL1, ESR1, FAM111B, FERMT1, CENPF, CYP24A1, CENPI, NMU, DEPDC1, TRIB3, TRIP13 |
| hsa-miR-6875-3p | 11 | 0.036256 | GYS2, UGT8, LGSN, ZNF280B, FAXC, TSHZ2, C2CD4A, BIRC5, ASCL1, SYT7, ARFGEF3 |
Fig. 4.
miRNA-target gene network constructed for the miRNAs targeting the LA-related genes and their associated target genes (miRNA were selected based on the number of target genes and P < 0.05)
Discussion
It is estimated that cancer has existed for more than one million year in the human species, but the effective treatments targeting molecular and cellular mechanisms have not been achieved for most cancers (Patterson et al. 2018). Today, it is well accepted that multiple factors play role in the development of different cancers. LA as one of the important cancers still seriously threats many people’s health. To this end, many studies have been performed in the field of lung cancers, and they have found several molecular and cellular mechanisms that might be involved in LA formation, progression and metastasis. In the current study, we used different bioinformatic analyses to comprehensively determine the key factors involved in LA. We believe that bioinformatic analyses is very worthwhile, and even essential, for finding factors (e.g. pathways, TFs, miRNA and metabolite) and mechanisms supporting LA. The results of bioinformatic analysis are helpful for scientists to develop new strategies for diagnosis and treating this cancer, especially with focus on precision medicine and personalized medicine. Here, we analyzed the microarray data of NL and LA samples (Xu et al. 2018). We found that 350 and 608 genes were upregulated and downregulated in LA, respectively (Log FC ≥ 2). We determined the upregulated genes as LA-related genes, and the downregulated genes as NL-related genes. To comprehensively elucidate the molecular mechanisms supporting LA, we analyzed LA-related genes through 5 next steps: (1) Analysis of GO (BB, CC, MF) and pathways linked to LA- and NL-related genes, (2) Construction of PPI networks for LA- and NL-related genes and finding hub genes and key TFs, (3) Analysis of prognostic value of LA hub genes, (4) Prediction miRNAs targeting LA-related genes and construction of miRNA-targets network, (5) Finding metabolites associated to LA- and NL-related genes.
LA-related genes were involved in different biological processes such as cell-cell signaling, collage catabolic process, extracellular matrix disassembly and calcium-independent cell adhesion. Whereas, the NL-related genes were involved in cell adhesion, surface receptor signaling and angiogenesis. Our findings are supported by previous studies indicating that the extracellular factors in tumor microenvironment (TME) can regulate cancer development and metastasis (Allinen et al. 2004). It seems that the stromal and epithelial cells in cancer microenvironment jointly disrupt ECM components and their dynamics can be a hallmark of cancer (Finak et al. 2008). Moreover, abnormal ECM, such as disordered organization/assembly and changes in essential composition of the ECM associate to cancer initiation and metastasis by remodeling the behavior of stromal cells and promoting tumor-related angiogenesis and inflammation (Lu et al. 2012). In line with our findings, It has been reported that the expression of collagens (COL10A1, COL11A1), matrix metallopeptidases (MMP1, MMP12), secreted factors (S100A2), glycoproteins (CTHRC1, SPP1), is increased in LA (Lim et al. 2017). Whereas, the expression of surfactant proteins (SFTPC, SFTPA2, SFTPD), secreted proteins (CHRDL1, WIFI), ECM-regulated genes (CPB2, MAMDC2, HHIP, LPL, CD36, ADAMTS8), collagen (COL6A6), ECM-affiliated proteins (FCN3), ECM glycoproteins (TNNC1, ABI3BP), and proteoglycan (OGN) are decreased in LA (Lim et al. 2017). We also found calcium-independent cell adhesion as one of BBs enriched for LA-related genes. It has been shown that calcium-independent cell adhesion is involved in female lung cancer, even who never smoked (Shi et al. 2019a, b). Understanding how lung-derived ECM components change in the diseased state may help to identify new prognostic or therapeutic targets. In addition to GO analysis, KEGG analysis led to valuable information regarding pathways in LA. We found that LA-related genes were significantly enriched in P53 signaling, cell adhesion molecules, tyrosine and histidine metabolism and drug metabolism (cytochrome 450). The TP53 gene, first described in 1979, was the first tumor suppressor gene to be identified, and it has been shown that somatic mutations and upregulation of TP53 were frequently found in ∼23% and ∼65% of NSCLC, respectively (Mogi and Kuwano 2011). Moreover, intercellular cell adhesion molecule-1 (ICAM-1) has been identified in most cases of NSCLC cultured lines, and its expression levels seem to have a prognostic value for NSCLC (Kotteas et al. 2014). Notably, nutrient transporters and metabolic enzymes can be regulated by the oncogenic signals that drive cell division and many cancers show an increased demand for specific amino acids, and become dependent on either an exogenous supply or in vivo synthesis (Lukey et al. 2017). Depletion of glutamine, arginine, tryptophan, and Serine/glycine from serum or targeting their transporter or metabolic enzymes have been tested for cancer therapy (Lukey et al. 2017; Zhang et al. 2019). Here, we found that histidine and tyrosine metabolism are upregulated in LA and their depletion or targeting their regulation might help to treat LA. It has been shown that histidine catabolism coupled with the folate cycle contributes to methionine synthesis, which promotes protein methylation, which in turn induces cytidine triphosphate synthase (CTPS), an important metabolic enzyme in the de novo pyrimidine biosynthetic pathway (Lin et al. 2018). Therefore, inhibition of histidine-dependent protein transmethylation leads to impaired filament formation and destroys CTPS required for rapid use upon nutrient replenishment. On the other hand, tyrosine is one of the most important amino acids in receptor of tyrosine kinases (RTKs) such as epidermal growth factor receptor (EGFR) and vascular endothelial growth factor receptor (VEGFR). Inhibition of these RTKs using small molecules have been studied as promising strategy for cancer treatment (Poliaková et al. 2018; Li et al. 2019b). Our analysis suggests that tyrosine depletion from the serum or targeting its metabolism might help to treat LA.
Construction PPI network for LA-related genes revealed that ESR1, KIF18B, CCNB1, BIRC5, AURKA and CHEK1 have the highest connectivity with other upregulated genes. Accordingly, each of them interacted with more than 20 genes from the list of LA-related genes, suggesting that they can be a marker for LA. In this study, we selected these genes as the hub genes. Our finding was confirmed by the human protein atlas, showing that the identified hub genes have higher expression at protein level in LA samples compared to NL ones. The analysis of overall survival indicated that higher expression of ESR1, KIF18B, CCNB1, BIRC5 and AURKA is correlated with mortality rate. Accordingly, patients whose tumors expressed a high level of these genes had a significantly shorter survival time than patients whose tumors expressed a low level of these genes. The role of these hub genes have been investigated or confirmed in different cancers. ESR1 (also known as ERα) is also expressed in many cancers. For example, approximately 70% of breast cancers are ESR1 positive (Jeselsohn et al. 2015). Interestingly, several studies have reported the expressions of ESR1 as a prognosticator for NSCLC (Hsu et al. 2017). It has been proposed that the differences in NSCLC behavior, prognosis, and response to treatment are related to sex and hormonal status (Rodriguez-Lara et al. 2018). Accordingly, the premenopausal women present the worst prognosis compared to postmenopausal women and men. Additionally, it has been reported that the use of hormonal replacement therapy increases NSCLC mortality, supporting the role of estrogen signaling and ESR1 in the pathogenesis of lung cancer (Rodriguez-Lara et al. 2018). Therefore, the level of ESR1 expression can be a predictive marker for LA, as well as targeting this receptor or related hormone seems to be a promising opportunity in the development of novel therapeutics such as endocrine therapy. Very recently, it has been shown that KIF18B, a member of the kinesin family, is overexpressed in many cancers such as cervical cancer (Wu et al. 2018), hepatocellular carcinoma (Yang et al. 2020) and LA (Ji et al. 2019). These studies demonstrated that KIF18B overexpression promotes proliferation, migration, and invasion of these cancers. KIF18B exerts its role by activating Wnt/β-catenin signaling pathway in cervical cancer and hepatocellular carcinoma (Wu et al. 2018; Yang et al. 2020), while it activate Rac1 and mediating the AKT/mTOR signaling pathway in LA (Ji et al. 2019). Therefore, KIF18B can serve as a novel oncogene that promotes the tumorigenicity in LA. BIRC5 (also known as survivin) is a dual functional regulator that inhibits apoptosis and promotes proliferation in many embryonic tissues and cancers (Nitschkowski et al. 2019). Recent studies indicate that BIRC5 plays roles in the regulation of cytokinesis and cell cycle progression, as well as participates in signaling pathways such as the p53, Wnt, hypoxia, and Notch signaling pathways (Chen et al. 2016). Targeting BIRC5 for treatment of LA might be achievable via 5 strategies: (i) BIRC5-partner protein interaction inhibitors, (ii) BIRC5 homodimerization inhibitors, (iii) BIRC5 expression inhibitors, (iv) BIRC5 mRNA inhibitors and (v) BIRC5 immunotherapy (Li et al. 2019a). Today it is well accepted that CCNB1 is highly expressed in LA. CCNB1 is one of the hub genes in most studies in lung cancers (Shi et al. 2019a, b). Therefore, CCNB1 might be potential biomarker for diagnosis and prognosis targets for LA (Liu et al. 2019b). Interestingly, CCNB1 polymorphisms may also contribute to the clinical efficiency of platinum-based chemotherapy (one of the first-line chemotherapy regimen) in advanced NSCLC patients, and it is helpful for the personalized medicine (Di Liu et al. 2017). It was shown that downregulation of AURKA inhibits cell growth, cause cell cycle arrest and apoptosis in LA (Zhong et al. 2016). It seems that AURKA inhibition-induced cell cycle arrest and apoptosis are associated with downregulated RAF-1, CCND2, CCND3, CDK4, PAK4, EGFR and upregulated WEE1 expression (Zhong et al. 2016). Interestingly, it was shown that Inhibition of AURKA enhances radiosensitivity in lung cancer, and it might be a promising inhibition target, especially for P53-competent LA (Liu et al. 2019a). In addition to hub genes discussed above, PPI also showed that 25 genes that have 10–20 connectivity with other LA-related genes. MMP1, MMP3, FOXA1, NEK2, AGR2, EXO1 and SPP1 are among these 25 genes. Therefore, these 25 genes are also important for LA and they might be as markers or therapeutic target for LA. On the other hand, we found that VWF, CDH5, BDNF, GNG11, TNF and FPR2 as hub genes for NL. Therefore, it seems that the expression of these genes are required for sustaining normal identity and functions of lung cells. Expression level of these genes might be a predictor or marker for LA development or its stages.
Since TFs are one of the most important players controlling cellular behaviors, we identified TFs among LA- and NL-related genes. Constructing PPI network for the TFs existing in the list of LA-related genes showed that FOXA1, TFAP2A, PAX3, GATA2, ZIC1, HOXC9, ASCL1 and ESR1 have higher number of interactions with each other and other TFs in the list. Based on the number of interactions, it seems that FOXA1, TFAP2A, ZIC1 and PAX7 are the core TFs in LA. Targeting this key TFs or destruction their network can be an approach for treating LA. In contrast, GATA2, MEIS1, FOXF1, KLF4, KLF2, TBX3, TBX5 and SOX17 were the most important TFs inside the list of NL-related genes. It seems that they work together to preserve the identity of NL. Among them, GATA2 is the key TF with the highest number of interactivity with others. Therefore, it seems that sustaining their expression and maintaining their regulatory network via different mechanisms (e.g. epigenetic mechanisms and signaling pathways) can hinder LA development. Moreover, overexpression of these TFs might be useful for reprogramming/reversing LA to normal cells, as well as for generating lung cells via differentiation and transdifferentiation strategy. We also analyzed the upstream TFs that can bind the promotor of above-mentioned TFs and other LA-related genes. We found that EHF, KLF5, ZBTB7C, GRHL2, ELF3, PAX9, E2F8, IRF6 and OVOL1 are the most valuable upstream TFs that can regulate the expression of the LA-related genes. They may also exert their effects via regulating the core TFs. Interestingly, among these 10 upstream TFs, the roles of ZBTB7C, GRHL2, IRF6 and OVOL1 are not well-studied in LA, even in other cancers. It seems to be useful and interesting to investigate their roles in LA development, as well as their relations with other cancer-linked factors (epigenetic regulators, signaling pathways, TFs and metabolic factors). Moreover, our analysis identified SOX18, SOX7, SOX17, MEOX1, MEOX2, TCF21, ERG, EPAS1 and BCL6B the as most valuable upstream TFs regulating NL-related genes. All these factors are involved in normal development, function and homeostasis. They sustain the identity of lung cells by direct regulation of NL-related genes, as well as indirectly via controlling TFs existing inside the NL-related genes. Therefore, preserving their expression might prevent LA development, as well as be useful for reprogramming LA to normal cells.
Today, it is well understood that the metabolism of cancer cells differs markedly from that of normal cells. Cancer cells often relinquish the efficient energy-producing pathways used by normal cells and move to substitute mechanisms that yield less energy but generate more materials required for increasing cell number. Therefore, rapidly dividing cells become dependent on these substitute mechanisms and their metabolites and chemicals. Here, we found that LA-related genes are involved in generation some metabolites including NAP, NADPH, Estradiol, Estrone, Androstenedione, Melatonin, Acetaldehyde, Uridine diphosphate glucose, 3,4-Dihydroxymandelaldehyde and 19S-hydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid. Production of these metabolites in LA, or generation of higher quantity of them in LA suggests a critical role for them in LA. For example, LA generates high quantities of NADPH in the mitochondria and in the cytosol, in order to limit the accumulation of ROS (Ciccarese and Ciminale 2017). High expression of genes responsible for generation reproductive hormones- androstenedione, estradiol and estrone- in LA proposes a vital role for them, as well as increases the risk of estrogen therapy. It has been reported that estrogen is extensively metabolized in the lung to form different metabolites such as 4-OHEs that can bind and activate estrogen receptors and generate free radicals that damage DNA and cause oncogenic mutations (Peng et al. 2017). in addition, the use of hormone therapy (estrogen plus progestin) significantly increases the risk of death from lung cancer in postmenopausal women (Chlebowski et al. 2009). Interestingly, tamoxifen, an antiestrogen used for breast cancer patients, reduces mortality from lung cancer (Bouchardy et al. 2011). In addition, Acetaldehyde, a metabolite generated from ethanol, interferes with anti-oxidative defense systems and generates ROS. In overall, changes at the level of these metabolites can be a biomarker for LA, as well as controlling the level of these metabolites or targeting their metabolic pathways might be promising strategies for treating LA in the future.
miRNAs play critical roles in many biological processes by controlling gene expression at the post-transcriptional level. To this end, the role of several miRNAs have been reported in different cancers such as LA (Iqbal et al. 2018; Wu et al. 2019a). Targeting the genes involved in LA by synthetic miRNA mimics or miRNA expression plasmids seems to be a promising therapeutic strategy. Therefore, we analyzed the miRNAs targeting LA-related genes and we found that hsa-miR-192-5p, hsa-miR-215-5p, hsa-miR-6875-3p and hsa-miR-6836-3p target 32, 26, 11 and 10 genes, respectively. The roles of hsa-miR-192-5p and hsa-miR-215-5p have been already demonstrated in lung cancer development and metastasis (Wu et al. 2019b). Interestingly, both had many identical targets such as FOXA1, which is one of the key TFs of PPI network constructed for LA. The role of hsa-miR-6875-3p and hsa-miR-6836-3p is not well defined in LA, and still needs to be investigated. Interestingly, seven miRNAs including hsa-miR-4492, hsa-miR-4498, hsa-miR-5001-5p, hsa-miR-6798-5p, hsa-miR-6836-3p, hsa-miR-762, hsa-miR-4693-5p can target MAP3K9, suggesting its great importance in LA. The miRNAs found in our study or their mimics might be used alone or along with other therapeutic approaches to treat LA. It is important to note that single nucleotide polymorphisms in the targets of miRNAs and in the sequence of the miRNAs might increase the risk, susceptibility and progression of LA in different population.
Taken together, we found molecular pathways, hub genes, TFs and metabolites that might have vital role in LA development, progress or metastasis. These factors and related pathways may act as biomarkers or therapeutic targets for LA. Furthermore, we analyzed the molecular and cellular factors regulating the identity and function of normal lung. Supporting these factors also might be useful and necessary to prevent LA development. We also predicted a list of miRNAs that potentially target LA-related genes and might be helpful to treat LA via synthetic miRNA mimics or miRNA expression plasmids.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Bouchardy C, Benhamou S, Schaffar R, Verkooijen HM, Fioretta G, Schubert H, Vinh-Hung V, Soria JC, Vlastos G, Rapiti E. Lung cancer mortality risk among breast cancer patients treated with anti‐estrogens. Cancer. 2011;117(6):1288–1295. doi: 10.1002/cncr.25638. [DOI] [PubMed] [Google Scholar]
- Brambilla E, Gazdar A. Pathogenesis of lung cancer signalling pathways: roadmap for therapies. Eur Respir J. 2009;33(6):1485–1497. doi: 10.1183/09031936.00014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Duan N, Zhang C, Zhang W. Survivin and tumorigenesis: molecular mechanisms and therapeutic strategies. J Cancer. 2016;7(3):314. doi: 10.7150/jca.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zeng C, Ye Y, Wu D, Mu Z, Liu J, Xie Y, Wu H. Promoter methylation of TCF21 may repress autophagy in the progression of lung cancer. J Cell Commu Signal. 2018;12(2):423–432. doi: 10.1007/s12079-017-0418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT, Schwartz AG, Wakelee H, Anderson GL, Stefanick ML, Manson JE, Rodabough RJ, Chien JW, Wactawski-Wende J, Gass M. Oestrogen plus progestin and lung cancer in postmenopausal women (Women’s Health Initiative trial): a post-hoc analysis of a randomised controlled trial. Lancet. 2009;374(9697):1243–1251. doi: 10.1016/S0140-6736(09)61526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, Huang W-C, Sun T-H, Tu S-J, Lee W-H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucl Acids Res. 2017;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarese F, Ciminale V. Escaping death: mitochondrial redox homeostasis in cancer cells. Front Oncol. 2017;7:117. doi: 10.3389/fonc.2017.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WA, Lam DC, O’Toole SA, Minna JD. Molecular biology of lung cancer. J Thorac Dis. 2013;5(Suppl 5):S479. doi: 10.3978/j.issn.2072-1439.2013.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- de Groot P, Wu C, Carter B, Munden R. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018;7:220–233. doi: 10.21037/tlcr.2018.05.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Liu WX, Ding X, Yang Y, Su B, Fei K. Polymorphisms of CCNB1 associated with the clinical outcomes of platinum-based chemotherapy in Chinese NSCLC patients. J Cancer. 2017;8(18):3785. doi: 10.7150/jca.21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger DS, Aisner DL, Wood DE, Akerley W, Bauman J, Chang JY, Chirieac LR, D’Amico TA, Dilling TJ, Dobelbower M. NCCN guidelines insights: non–small cell lung cancer, version 5.2018. J Natl Compr Canc Netw. 2018;16(7):807–821. doi: 10.6004/jnccn.2018.0062. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Howlader N, Noone A, Krapcho M (2015) National Cancer Institute. SEER Cancer Statistics Review: 1975–2011
- Hsu L-H, Chu N-M, Kao S-H. Estrogen, estrogen receptor and lung cancer. Int J Mol Sci. 2017;18(8):1713. doi: 10.3390/ijms18081713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucl Acids Res. 2007;35(suppl_2):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA (2018) MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Molec Aspects Med [DOI] [PubMed]
- Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12(10):573. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Pan X, Shang Y, Ni D-T, Wu F-L. KIF18B as a regulator in microtubule movement accelerates tumor progression and triggers poor outcome in lung adenocarcinoma. Tissue Cell. 2019;61:44–50. doi: 10.1016/j.tice.2019.09.001. [DOI] [PubMed] [Google Scholar]
- Kotteas EA, Boulas P, Gkiozos I, Tsagkouli S, Tsoukalas G, Syrigos KN. The intercellular cell adhesion molecule-1 (icam-1) in lung cancer: implications for disease progression and prognosis. Anticancer Res. 2014;34(9):4665–4672. [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR and A. Ma’ayan (2010) ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26(19): 2438–2444 [DOI] [PMC free article] [PubMed]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. The human transcription factors. Cell. 2018;172(4):650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res. 2019;38(1):368. doi: 10.1186/s13046-019-1362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Halfter K, Zhang M, Saad C, Xu K, Bauer B, Huang Y, Shi L, Mansmann UR. Computational analysis of receptor tyrosine kinase inhibitors and cancer metabolism: implications for treatment and discovery of potential therapeutic signatures. BMC Cancer. 2019;19(1):600. doi: 10.1186/s12885-019-5804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SB, Tan SJ, Wan-Teck L, Lim CT. An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nature Commu. 2017;8(1):1–11. doi: 10.1038/s41467-017-01430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-C, Chakraborty A, Huang S-C, Wang P-Y, Hsieh Y-J, Chien K-Y, Lee Y-H, Chang C-C, Tang H-Y, Lin Y-T. Histidine-dependent protein methylation is required for compartmentalization of CTP synthase. Cell Rep. 2018;24(10):2733–2745 e2737. doi: 10.1016/j.celrep.2018.08.007. [DOI] [PubMed] [Google Scholar]
- Liu N, Wang YA, Sun Y, Ecsedy J, Sun J, Li X, Wang P. Inhibition of Aurora A enhances radiosensitivity in selected lung cancer cell lines. Respiratory Res. 2019;20(1):230. doi: 10.1186/s12931-019-1194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ouyang S, Zhou Z, Wang M, Wang T, Qi Y, Zhao C, Chen K, Dai L. Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: Analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. Mol Genet Genom Med. 2019;7(2):e00528. doi: 10.1002/mgg3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukey MJ, Katt WP, Cerione RA. Targeting amino acid metabolism for cancer therapy. Drug Discov Today. 2017;22(5):796–804. doi: 10.1016/j.drudis.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi A, Kuwano H (2011) TP53 mutations in nonsmall cell lung cancer. BioMed Research International 2011 [DOI] [PMC free article] [PubMed]
- Nicetto D, Zaret KS. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr Opin Genet Dev. 2019;55:1–10. doi: 10.1016/j.gde.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschkowski D, Marwitz S, Kotanidou SA, Reck M, Kugler C, Rabe KF, Ammerpohl O, Goldmann T. Live and let die: epigenetic modifications of Survivin and Regucalcin in non-small cell lung cancer tissues contribute to malignancy. Clin Epigen. 2019;11(1):1–4. doi: 10.1186/s13148-019-0770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AD, Gonzalez FJ, Perdew GH, Peters JM. Molecular Regulation of Carcinogenesis: Friend Foe. Toxicol Sci. 2018;165(2):277–283. doi: 10.1093/toxsci/kfy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Meireles SI, Xu X, Smith WE, Slifker MJ, Riel SL, Zhai S, Zhang G, Ma X, Kurzer MS. Estrogen metabolism in the human lung: impact of tumorigenesis, smoke sex race/ethnicity. Oncotarget. 2017;8(63):106778. doi: 10.18632/oncotarget.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliaková M, Aebersold DM, Zimmer Y, Medová M. The relevance of tyrosine kinase inhibitors for global metabolic pathways in cancer. Mol Cancer. 2018;17(1):27. doi: 10.1186/s12943-018-0798-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lara V, Hernandez-Martinez J-M, Arrieta O. Influence of estrogen in non-small cell lung cancer and its clinical implications. J Thorac Dis. 2018;10(1):482. doi: 10.21037/jtd.2017.12.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saji S, Patil SS, Alleyn M, Lockey R, Kolliputi N (2019) Nicotine in E-cigarette smoke: cancer culprit? J Cell Commu Signal: 1–2 [DOI] [PMC free article] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Li N, Yang M, Li W. Identification of key genes and pathways in female lung cancer patients who never smoked by a bioinformatics analysis. J Cancer. 2019;10(1):51. doi: 10.7150/jca.26908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Li Y, Yan C, Su H, Ying K. Identification of key genes and evaluation of clinical outcomes in lung squamous cell carcinoma using integrated bioinformatics analysis. Oncol Lett. 2019;18(6):5859–5870. doi: 10.3892/ol.2019.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E, Hensing T, Simon GR, Dennis PA, Otterson GA, Bueno R, Salgia R. Molecular pathways therapeutic targets in lung cancer. Oncotarget. 2014;5(6):1392. doi: 10.18632/oncotarget.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl Acids Res. 2018;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucl Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Björling E, Agaton C, Szigyarto CA-K, Amini B, Andersen E, Andersson A-C, Angelidou P, Asplund A, Asplund C. A human protein atlas for normal and cancer tissues based on antibody proteomics. Molec Cell Proteom. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- Weber M, McWilliams A, Canfell K (2019) Prospects for cost-effective lung cancer screening using individualised risk calculators [DOI] [PMC free article] [PubMed]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N. HMDB 4.0: the human metabolome database for 2018. Nucl Acids Res. 2017;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang A, Zhu B, Huang J, Lu E, Xu H, Xia W, Dong G, Jiang F, Xu L. KIF18B promotes tumor progression through activating the Wnt/β-catenin pathway in cervical cancer. Onco Targets Ther. 2018;11:1707. doi: 10.2147/OTT.S157440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K-L, Tsai Y-M, Lien C-T, Kuo P-L, Hung J-Y. The roles of MicroRNA in lung cancer. Int J Mol Sci. 2019;20(7):1611. doi: 10.3390/ijms20071611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-G, Chang T-H, Liu Y-N, Shih J-Y. MicroRNA in lung cancer metastasis. Cancers. 2019;11(2):265. doi: 10.3390/cancers11020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Lu C, Huang Y, Zhou J, Wang X, Liu C, Chen J. SPINK1 promotes cell growth and metastasis of lung adenocarcinoma and acts as a novel prognostic biomarker. BMB Rep. 2018;51(12):648. doi: 10.5483/BMBRep.2018.51.12.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wang S, Xie H, Wang C, Gao X, Rong Y, Liu Z, Lu Y (2020) KIF18B promotes hepatocellular carcinoma progression through activating Wnt/β-catenin‐signaling pathway. J Cell Physiol [DOI] [PubMed]
- Zhang H-l, Zhang A-h, Miao J-h, Sun H, G.-l. Yan, F.-f. Wu and X.-j. Wang (2019) Targeting regulation of tryptophan metabolism for colorectal cancer therapy: a systematic review. RSC Adv 9(6): 3072–3080 [DOI] [PMC free article] [PubMed]
- Zhong N, Shi S, Wang H, Wu G, Wang Y, Ma Q, Wang H, Liu Y, Wang J. Silencing Aurora-A with siRNA inhibits cell proliferation in human lung adenocarcinoma cells. Int J Oncol. 2016;49(3):1028–1038. doi: 10.3892/ijo.2016.3605. [DOI] [PubMed] [Google Scholar]