Abstract
Groundbreaking structural and spectroscopic studies of class A G protein-coupled receptors (GPCRs), such as rhodopsin and the β2 adrenergic receptor, have provided a picture of how structural rearrangements between transmembrane helices control ligand binding, receptor activation and effector coupling. However, the activation mechanism of other GPCR classes remains more elusive in large part due to complexity in their domain assembly and quaternary structure. In this review, we focus on the class C GPCRs, which includes metabotropic glutamate receptors (mGluRs) and gamma-aminobutyric acid B (GABAB) receptors most prominently. We discuss the unique biophysical questions raised by the presence of large extracellular ligand binding domains (LBD) and constitutive homo/hetero-dimerization. Furthermore, we discuss how recent studies have begun to unravel how these fundamental class C GPCR features impact the processes of ligand binding, receptor activation, signal transduction, regulation by accessory proteins and crosstalk with other GPCRs.
Keywords: G protein-coupled receptors, metabotropic glutamate receptor, GABAB receptor, calcium-sensing receptor
G Protein-Coupled Receptors
G protein-coupled receptors (GPCRs) serve as signal transducers which convert extracellular signals into intracellular signaling events via heterotrimeric G proteins, and serve as major drug targets for human disorders [1]. All GPCRs share the same general structure: an amino-terminal extracellular domain (ECD), a seven-helix transmembrane domain (TMD), and an intracellular carboxy-terminal domain (CTD). Variability in the ECD and the location of ligand binding provides a basis for grouping GPCRs into major classes. Thus, ligands bind within the TMD for class A GPCRs which typically contain short (<50 residues) ECDs, while peptides that bridge the ECD (~120–160 residues) and the TMD activate class B GPCRs [2]. Class C GPCRs, on the other hand, bind ligands within a large (400–600 amino acid) N-terminal ECD [3]. Class F GPCRs (i.e. smoothened, frizzled) [4, 5] and adhesion GPCRs [6] also contain large ECDs and are activated via complex mechanisms that are relatively poorly understood.
Our base of understanding of TMD activation comes primarily from extensive structural studies of class A GPCRs [7, 8]. While most structures represent inactive states, an increasing number are considered to capture active states with such structures typically captured bound to G proteins or G protein-mimetic nanobodies (see Glossary). Comparison between structures in inactive and active states have revealed conserved “molecular switches” that facilitate allosteric communication between the ligand binding and intracellular transducer sites [9]. For example, the so-called “ionic lock” describes a salt bridge between Arg and Glu residues at the intracellular ends of TM3 and TM6, respectively, which stabilizes the inactive state and, upon breaking, allows the outward movement of the intracellular end of TM6 to create a cavity on the cytoplasmic face of the receptor that accommodates the Gα C-terminus to initiate signaling. The same cleft appears to also be engaged by G protein-coupled receptor kinases (GRKs) and arrestins [7, 8], which serve to both desensitize the receptor and initiate signaling cascades. Spectroscopic and computational methods have revealed that GPCRs exist in a dynamic ensemble of conformations that are selectively stabilized by ligands and, especially, by G proteins [10–12]. This conformational heterogeneity underlies the range of signaling dynamics initiated or inhibited by the many types of orthosteric and allosteric ligands that exist for GPCRs [13].
In this review, we analyze the assembly and ligand-regulated structural properties of class C GPCRs as revealed by a number of recent breakthroughs. Critically, a deeper understanding of the basic biophysical properties of GPCRs should also lead to improvements in the repertoire of pharmacology available for both basic science and clinical use (see [14]).
Class C GPCRs: Assembly Mechanism and Overall Architecture
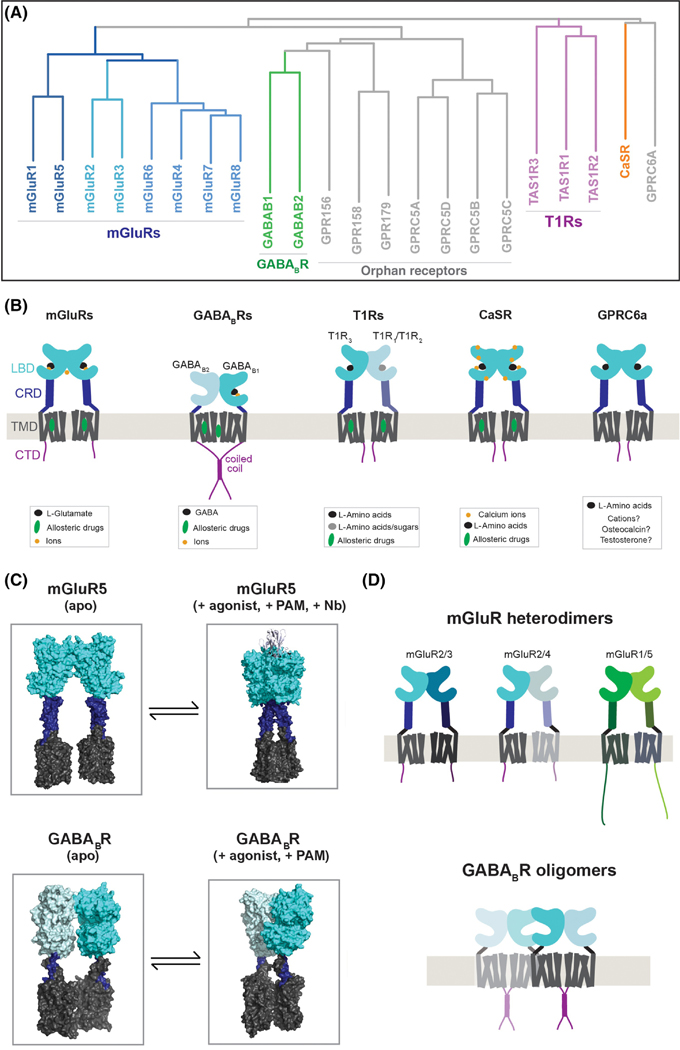
Class C GPCRs form a diverse, physiologically-important family (Fig 1A), including eight metabotropic glutamate receptor (mGluRs or mGlu Receptors) [15], two GABAB receptor subunits (GABABRs) [16], the calcium-sensing receptor (CaSR) [17], three T1R taste receptor subunits (T1Rs) [18], GPRC6a [19], a promiscuous L-amino acid receptor, and several orphan receptors. This family is distinguished from other GPCRs by two features: large ECDs and constitutive dimerization (Fig. 1B) which together raise many biophysical questions that make up the focus of this review. In all well-characterized class C GPCRs, the ECD contains a bi-lobed “clamshell” ligand binding domain (LBD), sometimes referred to as a venus flytrap motif, which is homologous to bacterial periplasmic amino-acid binding proteins and contains the orthosteric binding site for native ligands. Except for GABABRs, all non-orphan class C GPCRs contain an intermediate cysteine-rich domain (CRD) between the LBD and TMD (Fig. 1B). Class C orphan receptors (GPR156, GPR158, GPR179, GPRC5A, GPRC5B, GPRC5C, and GPRC5D) either have no ECD or an unrelated ~400 amino acid ECD. In 2019 the first snapshots of a dimeric, full-length class C GPCR, mGluR5, were captured using cryogenic electron microscopy (cryo-EM) [20] and more recently a series of papers reported full-length structures of the GABABR [21–24], providing a framework for our discussion and analysis (Fig. 1C).
Figure 1, Molecular diversity of class C GPCRs.
A, Phylogenetic tree showing all class C GPCRs grouped into the major subfamilies. B, Summary of domain structure, homo- and hetero-dimerization and ligand binding properties of well-characterized, non-orphan class C GPCRs. T1R1/T1R3 heterodimers form the umami receptor and T1R2/T1R3 heterodimers form the sweet receptor. While T1R2, CaSR and GPRC6a show promiscuity with regard to L-amino acids, they prefer glutamate, tryptophan and basic amino acids, respectively. In the case of GPRC6a, controversy exists over whether osteocalcin and testosterone can bind and where their binding sites are and a defined cation binding site has not been proposed. C, Summary of cryo-EM structures of mGluR5 [20] and the GABABR [24] showing the apo-state (left) and an agonist and PAM-bound state (right). Note: an agonistic nanobody is shown in the agonist-bound mGluR5 structure. D, Schematic of further complexity in the assembly of class C GPCRs. Various heterodimeric mGluR combinations have been identified (top) and evidence for higher order assembly (bottom) exists, with the strongest data obtained for GABABRs. Oligomeric inter-LBD and inter-TMD interfaces involving GABAB1 subunits have been proposed for tetrameric or higher order GABAB complexes.
mGluRs were first found to dimerize using biochemical assays, which identified an inter-subunit disulfide bridge formed via a conserved cysteine within the LBD, allowing for dimers to be observed in a denaturing gel [25]. Similar disulfide-mediated dimerization has been observed in denaturing gels for both the CaSR [26] and GPRC6a [27]. In recent years, fluorescence methods have confirmed the dimerization of all mGluRs, including the demonstration of strict dimerization using single molecule photobleaching analysis of GFP-tagged mGluRs in the plasma membrane of live cells [28]. Supporting the fundamental role of dimerization in mGluR activation, a nanodisc reconstitution study showed that mGluR2 dimerization is required for glutamate-driven G protein activation [29].
In the case of both GABABRs and T1Rs, the requirement of co-expression of two receptor subtypes for function enabled the demonstration of requisite heterodimerization. GABABRs employ a trafficking-based mechanism where GABAB1 and GABAB2 subunits form a coiled-coil between the CTDs, which masks an endoplasmic reticulum (ER) retention motif in GABAB1 to enable surface targeting of heterodimers [30]. Furthermore, within this complex only the GABAB1 subunit binds GABA and only the GABAB2 subunit binds G proteins[30]. Interestingly, GABAB2 can traffic in the absence of GABAB1 [31], GABAB responses have been observed in GABAB2 KO mice [32] and a cryo-EM structure of a GABAB1 homodimer in an active-like state was recently solved [21], suggesting that other biologically-relevant GABABR complexes may exist. In the case of T1Rs, it is unclear how specific heterodimer assembly and trafficking is controlled and whether monomeric or homodimeric species can form. mGluRs have also been shown to form a number of different hetero-dimer combinations largely through heterologous studies in cultured cells [28, 33, 34] and co-immunoprecipitation from brain lysates [35] (Fig. 1D). A consensus finding is that heterodimers can form either between group II and III mGluRs or between group I mGluRs. This assembly pattern seems to ensure the maintenance of a strictly Gq/11 (Group I) or Gi/o-coupled (Group II/III) receptor dimer. While biophysical studies have dissected the myriad effects of mGluR heterodimerization, the biological prominence and functional roles of such complexes remain poorly understood. Supporting a prominent role for heterodimerization in mGluR biology, a recent study used single cell RNA sequencing analysis to show a high degree of mGluR co-expression in cortical pyramidal cells and quantitative fluorescence-based analysis of dimerization propensities found many instances where heterodimerization is preferred over homodimerization [36].
Based on truncation studies of mGluRs [28, 29], class C GPCR dimers are known to form primarily via an inter-LBD interface with secondary contributions from TMDs. Crystal structures of isolated LBD dimers [37–40], as well as full-length mGluR5 structures [20], have revealed a core hydrophobic interface between the upper lobes (LB1) containing highly-conserved residues within helix B and C. Mutations within this hydrophobic core substantially reduce mGluR dimerization [28]. Further work is needed to dissect these interfaces in detail and decipher how they contribute to the specificity of homo- and hetero-dimeric interactions within the T1R and mGluR subfamilies. Interestingly, mutation of the cysteine involved in the inter-LBD disulfide bridge produces only subtle effects on mGluR dimerization, and appears to impair the activation process [28]. This cysteine is found within an unstructured, variable loop that is ~20 residues long and remains poorly understood. In the T1R2/R3 LBD structure [40] an asymmetric inter-subunit disulfide is formed between cysteine residues in different loops, likely providing stability and specificity to the heterodimer.
While dimers are widely accepted as the minimal functional unit of class C GPCRs, the formation of higher order oligomers has also been proposed (Fig. 1D). A live-cell single molecule imaging study used fluorescence intensity analysis to infer tetrameric and higher order GABABR complexes that formed in a density-dependent manner [31], consistent with prior FRET studies [41, 42]. The structural basis of such complexes remains unclear although inter-LBD[43] and inter-TMD[44] interactions have been proposed to mediate higher order oligomerization. Recently, brightness analysis in cultured neurons provided evidence for ligand-induced higher order oligomers of heterologously-expressed mGluR2 [45] while single molecule imaging of endogenous mGluR4 in cerebellar parallel fibers revealed primarily dimers [46], suggesting that higher-order complexes may exist for other class C GPCRs depending on receptor subtype and biological context. Further investigation of class C GPCR oligomerization and spatial distribution in live cells, as well as biochemical isolation and structural characterization of such complexes, will be required to clarify this issue.
Ligand Binding Domains: Intra- and Inter-subunit Dynamics
The ligand binding and conformational properties of class C GPCR LBDs (Fig. 2A) have been studied extensively. All crystal structures of isolated LBDs have confirmed a similar bi-lobed structure where orthosteric ligands bind in the cleft between lobes [37–40, 47]. LBDs have been observed in open (interlobe angle ~40–50°) and closed (interlobe angle ~25°) conformations in both the presence and absence of ligands, but it is clear that agonists stabilize the closed state through specific interactions with residues in both the upper (LB1) and lower (LB2) lobes [37–40] (Fig. 2B). In contrast, orthosteric antagonists primarily interact with LB1 residues with minimal LB2 contacts in mGluR and GABABR structures [37, 38]. Interestingly, at the individual LBD level, closed state mGluR structures with agonists of variable efficacy show a very similar degree of LBD closure [48].
Figure 2, Structural dynamics of class C GPCR ligand binding domains (LBDs).
A, LBD dimers (dashed box) initiate activation following binding to orthosteric ligands, as shown in the full-length mGluR5 structure. B, Summary of mGluR LBD conformational changes. In an apo structure of mGluR1 (left) LBDs are found in the “open” state with upper (LB1) and lower (LB2) lobes far apart. At the inter-subunit level this structure is characterized as “relaxed” due to the lack of an inter-LBD interface. In a glutamate-bound structure of mGluR2 (right) both LBDs are in the closed state and dimer reorientation has allowed for the formation of an electrostatic LBD interface to form the “active state”. Note that the O-O/R and C-C/A are thought of as extreme conformations, that many intermediates exist and that the correlation between ligand occupancy and conformation is complex. C, 6-state model of mGluR LBD activation incorporating intra-subunit and inter-subunit conformational changes. * indicates states that have been captured in crystal structures. A 3-state model based on smFRET studies is highlighted with donor and acceptor fluorophore positions shown as green and red ovals. Right, representative smFRET trace showing the transition of mGluR2 between three states (dotted lines) on tens of ms time scale at approximately EC 50 glutamate levels. Inset shows the relative occupancy of the C-C/A state observed for mGluR2 under different conditions. D, Free energy diagrams summarizing differences in relative stability of relaxed and active states for mGluR2, 3 and 7. mGluR3 shows basal occupancy of the active state while mGluR7 shows minimal occupancy of the active state, even under saturating agonist conditions. E, GABABR LBD dimer structures showing a comparatively subtle dimer reorientation associated with activation.
A major means of regulating class C GPCR LBDs appears to be through the binding of ions. This is most clear in the case of the CaSR which responds to millimolar extracellular calcium with high cooperativity [49]. CaSR LBD crystal structures showed four calcium binding sites and a binding site for L-tryptophan (L-Trp) in a site homologous to the orthosteric site for mGluRs and the GABABR [39, 47]. Functional studies confirm that L-Trp and calcium function as co-agonists to stabilize LBD closure and induce downstream signaling [39, 50]. Similarly, ions appear to serve as co-agonists of mGluRs, GPRC6a and the GABABR, although controversy exists over whether divalent cations (i.e. 1–10 mM Ca2+) [19, 51–53] or chloride ions (50–100 mM) [54] mediate such effects and the location of the associated binding sites. Notably, an inactive cryo-EM structure of the GABABR revealed a Ca2+ binding site on the LB2 surface of the GABAB1 subunit, distinct from the four sites identified in the CaSR but adjacent to the orthosteric site, provides a means of modulating agonist affinity [23]. Supporting the functional relevance of this site, either mutation of coordinating residues (E309K, E423R) or chelation of Ca2+ decreased basal activity of the GABABR [23].
While a 2-state open-closed model likely captures the dynamics of a single LBD, complexity arises at the level of inter-subunit rearrangement. Class C GPCR LBD structures have been solved as dimers which interact in a shoulder-to-shoulder fashion while facing opposite directions (Fig. 2B). Such structures can be sorted into two broadly-defined forms: “relaxed”, with LB2 domains far apart, and “active”, with an inter-LB2 interface engaged (Fig. 2B). Mutations at the electrostatic LB2 interface modify the activation of mGluRs [28, 52]. Combining intra-subunit (closed/open, C/O) and inter-subunit (active/relaxed, A/R) states provides a framework for modeling the conformational dynamics of LBD dimers (Fig. 2C). Consistent with the diversity of conformations that have been crystallized (Fig. 2C), substantial dynamics between LBDs have been measured in FRET studies using N-terminally SNAP-tagged mGluRs [28, 52, 55–57]. Vafabakhsh et al [52] used smFRET on full-length mGluR2 and mGluR3 to observe inter-LBD conformational changes, revealing the presence of at least 3 inter-convertible states populated on the 30–100 ms time scale (Fig. 2C). Agonist efficacy was closely correlated with the relative stabilization of the low FRET state which likely represents the C-C/A conformation. Based on mutagenesis, structural and kinetic analyses, the transient intermediate state was proposed to be an inactive O-C/R state, allowing for the proposal of a 3-state model of LBD activation (Fig. 2C). To assess the role of ligand occupancy in mGluR2, Levitz et al [28] used tethered photoswitchable ligands attached to either one or both subunits to find that binding of a single agonist leads to weak (~20%) activation, suggesting that transient closure of the un-liganded subunit produces active state occupancy. This is consistent with smFRET [52] and functional [58] studies of mGluR dimers where one subunit is mutated to decrease glutamate affinity. Together this work motivates further studies using high spatial and temporal resolution spectrosocopic and computational methods to develop a complete description of the ligand-induced conformational trajectories of mGluR2 as a model class C GPCR.
Other mGluR subtypes have been shown using smFRET to populate the same states, but with different kinetics and occupancies (Fig. 2D). Strikingly, even in the presence of saturating glutamate mGluR7 homodimers show rare, brief visits to the active state, but heterodimerization with mGluR2 produces a receptor with full active-state occupancy even in response to ligand binding in a single subunit [57]. Future work will be needed to investigate what determines the differences in conformational dynamics between closely-related subtypes and how this controls their distinct roles in sensing synaptic glutamate dynamics. Mutations to the LB1 and LB2 inter-LBD dimer interfaces can have large effects on conformational dynamics [28], suggesting a critical role for inter-subunit interactions in tuning receptor activation. Limited work has addressed the dynamics of inter-subunit conformational changes in other class C GPCRs, but crosslinking of LB2 residues stabilizes the active state of the GABABR supporting a conserved role of a “relaxed” to “active” inter-subunit transition [38]. However, compared to mGluRs, GABABR and CaSR structures show a much subtler dimer reorientation (Fig. 2E), which may accommodate the need to activate following closure of a single LBD. Ensemble FRET measurements of GABABRs tagged at the N-terminus or within the LBD suggest subtle and distinct inter-subunit conformational changes compared to mGluRs [59], motivating further analysis to dissect the distinct conformational pathways employed by different class C GPCRs.
Transmembrane Domain Activation and G Protein-Coupling
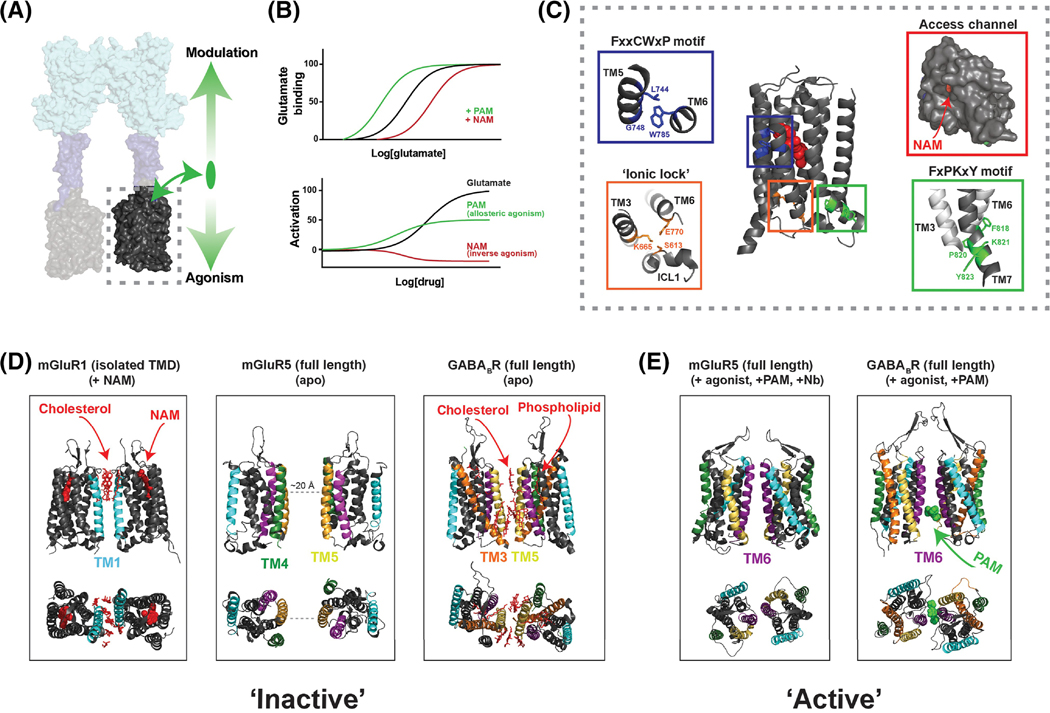
While orthosteric ligand binding and the associated conformational changes of the LBDs are beginning to be well understood, the mechanistic properties of class C GPCR TMDs remain unclear. Whereas the TMDs do not contain binding sites for known native ligands, they do bind synthetic positive (PAMs) and negative (NAMs) allosteric modulators making them major targets for subtype-selective drug development for clinical applications [60, 61]. By binding to these sites, an allosteric ligand can modulate the action of the orthosteric ligand by altering its affinity and/or efficacy (Fig. 3A-B). Previously it was thought that allosteric ligands lack intrinsic efficacy on full-length receptors, and that only upon deletion of the ECD can PAMs act as agonists on both mGluRs [29, 62] and GABABRs [63]. However, it has become clear that many mGluR PAMs act as agonists, even when the LBD is bound to an orthosteric antagonist [64–67]. Allosteric ligands with intrinsic efficacy have also been reported for GABABRs, CaSR, and T1Rs [59, 68, 69]. Functional and conformational readouts have shown that mGluR PAMs possess a range of efficacies and kinetics of action, which may be modulated by drug-membrane interactions [67]. Similarly, while some NAMs serve merely as neutral antagonists that inhibit agonist-mediated activation, others can serve as inverse agonists to decrease basal activity (Fig. 3B) [67, 70].
Figure 3, Allosteric modulation and conformational dynamics of class C GPCR transmembrane domains.
A, TMDs (dashed box) respond to agonist-binding in the LBD and mediate G protein activation. TMDs also bind allosteric ligands which can directly alter receptor activation (downward arrow) and modulate the response to orthosteric ligands (upward arrow). B, Theoretical dose-response curves showing the classical effects of allosteric modulators on orthosteric agonist binding (top) and on receptor activation (bottom). Many ligands show properties of both allosteric modulation and agonism or inverse agonism. C, NAM-bound mGluR5 TMD structure (PDB: 4OO9) reveals microswitches similar to those seen in class A structures, including the ionic lock interaction between Lys665 and Glu770 (orange box), the FxxCWxP motif (blue box) which is thought to serve as a “trigger switch” to couple ligand binding to TM6 rearrangement, and the FxPKxY motif (green box) at the intracellular end of TM7 which is thought to stabilize the active conformation. Entrance to the allosteric pocket is restricted by a narrow access channel formed by the helical bundle and extracellular loop 2 (red box). D-E, Structural data showing various inter-TMD dimer interfaces. NAM-bound mGluR1 TMD structures (D, left) revealed an inter-TM1 interface mediated, in part, by cholesterol molecules (red). The cryo-EM structure of mGluR5 showed no direct interface in the apo-state (D, center), but inactive GABABR cryo-EM structures show an interface consisting primarily of TM5 and the cytosolic end of TM3 as well as bound phospholipids and cholesterol (D, right)[23]. The agonist and PAM-bound cryo-EM structure of mGluR5 show an inter-TM6 interface (E, left) [24] that is similar to that seen in agonist and PAM-bound GABABR structures (E, right).
The highest resolution (<3.0 Å) structural data that exist for class C GPCR TMDs come from NAM-bound crystal structures of the isolated TMDs of mGluR1 and mGluR5 [71–74], which show structural homology with class A and B GPCRs. As expected, the NAM binding pockets overlap with those for orthosteric class A GPCR ligands, and both structures reveal a narrow opening that restricts entrance to the allosteric pocket (Fig. 3C), potentially explaining the slow binding kinetics of allosteric drugs [67]. Several molecular switches characterized in class A GPCRs have been identified in these structures, with some notable differences (see Fig. 3C). For instance, in group I mGluRs an “ionic lock” forms between intracellular ends of TM3 and TM6 although the positive charge on TM3 is provided by K3.46 rather than R3.50 of the (D/E)RY motif as seen in class A GPCRs. Furthermore, a conserved serine in ICL1 is also positioned to interact with the ionic lock (Fig. 3C). Consistent with the classical role of the ionic lock, mutations to these residues increase constitutive activity in mGluR5 [72] and enhance agonist affinity in the GABABR [63]. The lack of active mGluR TMD structures has motivated computational work to try to assess the structural changes that drive PAM- or agonist-induced activation [75, 76]. Perez-Benito et al [75] used MD simulations and mutagenesis experiments on mGluR2 to propose a role for a “trigger” switch involving allosteric ligand-induced rearrangement of W6.48 and a “transmission” switch involved in relaying conformational changes to the intracellular face of the TMD (Fig. 3C), which is analogous to models of class A GPCR activation [9]. While the mGluR5 TMD did not show clear structural differences in apo and agonist-bound states in cryo-EM structures[20], full-length GABABR structures bound to an agonist and a PAM revealed subtle movements of TM3, TM4 and TM5 relative to the apo state within the GABAB2 subunit [22, 24]. The lack of major TM6 motions and the potential constraints provided by a TM6-TM6 dimer interface (see below) suggests that class C GPCRs use different structural rearrangement compared to class A GPCRs to enable G protein coupling. Intriguingly, phospholipids were found in cryo-EM structures bound within the core of GABABR TMDs [21, 23], suggesting that lipid composition is also a critical regulator of TMD conformation. Ultimately, further work is needed to define the TMD molecular switches across class C GPCRs, to characterize the intra-helical rearrangements that they regulate and to decipher how different PAMs can bind to and stabilize active states.
Class C GPCR TMDs exist within dimeric or oligomeric contexts raising the question of how they sense allosteric input from the dimeric LBDs and rearrange during the activation process. Single molecule imaging of detergent-solubilized TMDs revealed that inter-TMD interactions contribute to dimerization in mGluRs [28], although to a different degree for different subtypes [67]. While the NAM-bound mGluR1 TMD structure [71] showed a cholesterol-rich TM1 dimer interface (Fig. 3D), the mGluR5 TMD structure did not appear dimeric [72]. An inter-subunit crosslinking study on mGluR2 proposed a dimer reorientation model where a TM4/TM5 interface in the inactive state rotates to form a TM6 interface upon activation [77]. Similarly, GABABR crosslinking suggested a rearrangement from a TM5 interface to a TM6 interface upon activation [44]. Such a TM6 interface has now been confirmed in the full-length glutamate- and PAM-bound mGluR5 cryo-EM structure in a detergent micelle [20] as well as in full-length, detergent-solubilized GABABR agonist and PAM-bound structures [21–24] (Fig. 3E). In contrast, no TMD interface was seen in the nanodisc-reconstituted mGluR5 apo state with TM5 of each subunit nearly 20 Å way from each other [20], while GABABR structures show an inactive interface formed via TM5, the intracellular end of TM3 and either cholesterol or detergent molecules [21–23]. Further work is needed to assess the inter-TMD interactions across receptor subtypes in different conditions, but a number of mechanistically-relevant observations have emerged from the aforementioned structural studies. Supporting the importance of inter-TMD interactions, Park et al [23] showed that the inactive GABABR TM3/TM5 dimer interface is secured by a network of salt bridges referred to as an “intersubunit latch”, the disruption of which enhances the receptor’s constitutive activity. This observation further supports a model where inactive TMD interfaces transition to a TM6 interface to initiate activation. Consistent with the critical role of the TM6 interface, GABABR PAMs were found bound between TM6 residues in adjacent subunits [22, 24] (Fig. 3E), presumably stabilizing the active intersubunit orientation. Future work will be needed to determine if such an intersubunit allosteric binding site is seen in other class C GPCRs.
While extremely informative, the static nature of structural approaches provides limited information about the dynamics that underlie receptor activation, motivating the use of spectroscopic methods to assess receptor conformation in the plasma membrane of living cells. Extensive intra- and inter-subunit FRET studies using group I mGluR constructs tagged with fluorescent proteins (FPs) have been performed. Introduction of FPs into intracellular loop 2 (IL2) of each subunit leads to a FRET increase upon glutamate application, while simultaneous introduction of FPs into IL2 and the CTD of the same subunit produces a glutamate-induced FRET decrease [78, 79]. Such probes have been used for high resolution kinetic studies that revealed that glutamate-induced activation involves sequential inter- (~1 ms) and intra-subunit (~20 ms) conformational changes [80]. This is consistent with a model where TMD dimer re-arrangement precedes the intrasubunit conformational changes that enable G protein-coupling. While most inter-TMD FRET studies have focused on orthosteric agonists, Gutzeit et al [67] showed that PAMs can induce inter-subunit FRET increases in isolated mGluR TMD constructs, suggesting that TMD dimer reorientation can proceed without direct allosteric input from the LBDs. Though extreme caution is warranted when interpreting such FRET results structurally, these studies complement structural analysis to show that dynamic inter-TMD interactions are central to class C GPCR activation.
A major remaining question regarding class C GPCRs is how agonist binding within the LBD is allosterically coupled to the TMD. In mGluRs, mutational and cross-linking studies have shown that LBD-level conformational changes are relayed to the CRD via an inter-domain disulfide bond [81] and that inter-CRD interactions within a dimer stabilize the active state [82]. Based on cryo-EM structures, it was proposed that interactions between the CRD and extracellular loop 2 in mGluRs [20], or between the LBD-TMD linker and EL2 in the GABABR [21–24], initiate structural rearrangements at the level of the TMD. While attractive, experimental work is needed to test this model and extend this analysis across the class C GPCR family.
Lastly, little is known about the coupling mechanism between class C GPCR TMDs and G proteins. In class A GPCRs activation-associated structural changes are driven by both ligand and G protein binding [11], which may explain why no major intra-subunit rearrangements are seen in the full-length glutamate-bound mGluR5 structure that lacks G protein [20]. Mao et al [22] report GABABR structures in the presence of Gαi1 although the low resolution of the G proteins prevented modeling of the interaction interface. The lack of canonical outward TM6 motions in these structures suggests either that alternative means of controlling G protein access are used by class C GPCRs or such structures may have captured an intermediate state in the G protein coupling process. The stoichiometry of G protein coupling in GPCR dimers is not clear, except for the well-characterized GABABRs in which only the GABAB2 subunit is capable of coupling to G proteins [83]. Consistent with this, analysis of full-length cryo-EM structures suggests that the dimeric arrangement of TMDs precludes the binding of two G protein heterotrimers simultaneously. Evidence for asymmetric signal transduction, where only one TMD can adopt the active-state at a time exists for mGluR homodimers [84, 85] and, similarly, in mGluR2/4 heterodimers, the mGluR4 subunit is thought to couple to G protein unless the asymmetry is reversed via mGluR2 PAMs or mGluR4 NAMs [86]. In addition, the mechanisms by which class C GPCRs, recognize specific Gα protein families remains unclear. While it has been widely accepted that the C-terminus of Gα is the primary G protein mediator of selectivity [87], variable intracellular regions of GPCRs contribute to selectivity [88, 89]. Chimera studies of mGluRs have found that G protein coupling selectivity is determined primarily by IL2 [90], but further work is needed to decipher the mechanism. Finally, relatively little is known about coupling between class C GPCR TMDs and β-arrestins with conflicting studies and compelling evidence for β-arrestin-independent desensitization and internalization of some subtypes [91–93]. However, recent studies have demonstrated β-arrestin interactions for both mGluR7 [94] and the CaSR [95], motivating detailed study of class C GPCR/β-arrestin complexes. In addition to interactions with transducers, class C GPCRs also interact with an ever-increasing amount of accessory and regulator proteins (box 1), that tune the activation, signaling and localization of these receptors.
Box 1: A Diverse Array of Interaction Partners
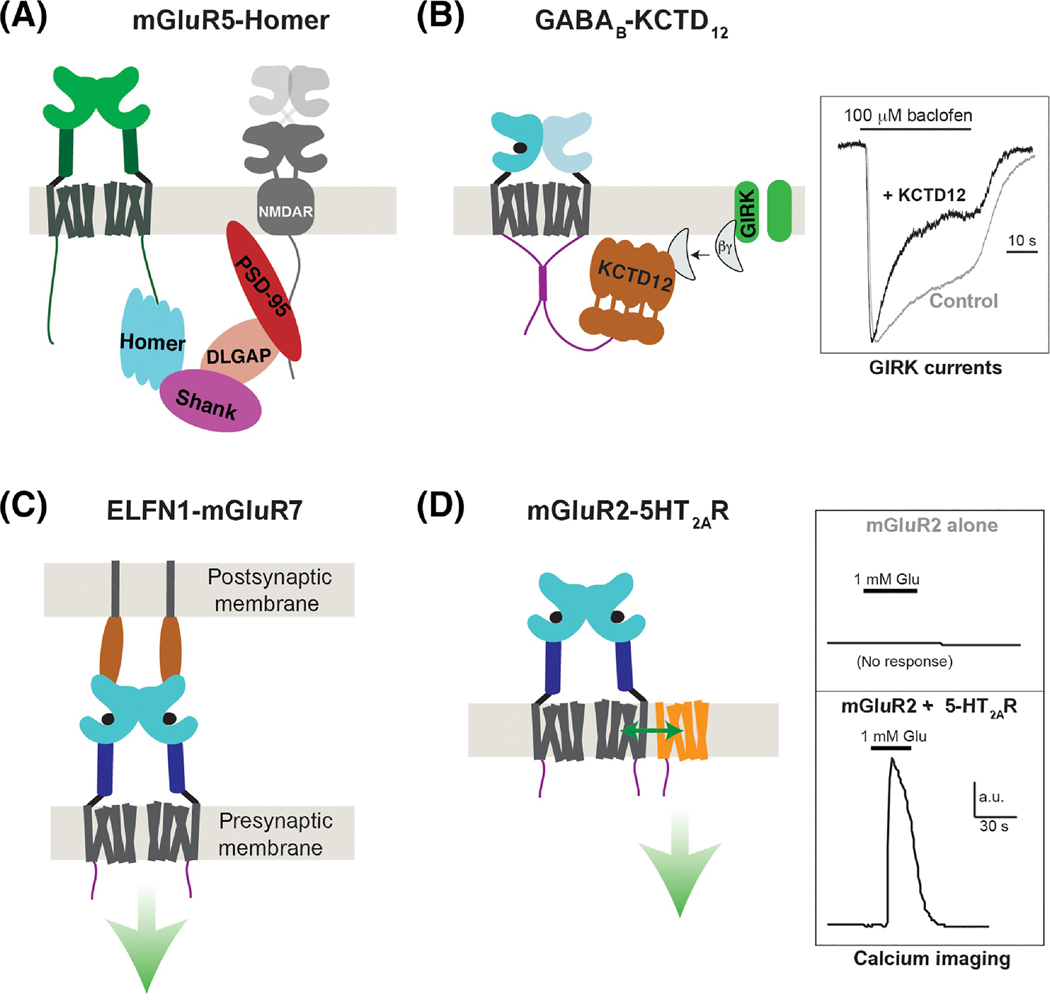
An increasingly appreciated aspect of class C GPCR function is the co-assembly with accessory proteins. While little structural information exists to understand such interactions at high resolution, biochemical and functional studies have provided a wealth of information. The CTDs of different mGluR subtypes are the targets of synaptic scaffold proteins (Fig. 4A) and a plethora of kinases and phosphatases target residues on the CTD to control trafficking and activation [3], although it is unknown how regulation is enacted through this unstructured domain. The precise localization of class C GPCRs likely determines their network of interacting proteins under particular physiological conditions. This was demonstrated in a recent super-resolution imaging study which showed striking co-localization of native mGluR4 and Munc-18 in presynaptic terminals[46], consistent with a previous functional and biochemical study that revealed mGluR4/Munc-18 interactions[114]. One of the best-established intracellular accessory subunits of a class C GPCR are the KCTD proteins (KCTD8, 12, 16) which bind tightly to the GABAB2 CTD and control receptor trafficking, activation and desensitization via direct interaction with G proteins [115, 116]. Recent structural snapshots of GABAB/KCTD and KCTD/Gβγ complexes revealed unique pentameric ring structures with highly cooperative G protein binding [110, 117] that enables efficient scavenging of G proteins from GIRK channels to induce rapid desensitization (Fig. 4B). KCTD-GABABR interactions were discovered via immunoprecipitation-based mass spectrometry and this same approach has revealed many other GABABR interacting proteins [111], motivating further application of this approach to identify class C GPCR interacting proteins.
It has become clear that class C ECDs are also targets for protein-protein interactions. Among these interactions, the most well-documented is the physiologically critical interaction between the ELFN proteins and group III mGluRs [118, 119]. However, the nature of the trans-synaptic ELFN/mGluR interaction in terms of precise binding sites and the allosteric effects on receptor signaling [120–122] remain to be resolved (Fig. 4C). Trans-synaptic interactions have also been identified for the orphan class C GPCRs, GPR158 [123] and GPR179 [124]. Clinically-relevant extracellular interactions between class C GPCRs and soluble proteins have also been reported, including the recent discovery that the amyloid-β precursor protein (APP) directly binds to the N-terminal sushi domain of the GABAB1a splice variant to control pre-synaptic targeting and function and amyloid formation [111, 125, 126]. The sushi domains of GABAB1a also bind AJAP-1 and PIANP proteins, which can also influence the pre-synaptic targeting of the receptor [126]. mGluR5 has also been proposed to play a role in Alzheimer’s pathophysiology by serving as a receptor for amyloid-β oligomers via interaction with cellular prion protein [127, 128]. Finally, an intriguing recent study provided evidence that the extracellular domain of mGluR2 serves as a receptor for the rabies virus glycoprotein to facilitate rabies infection [129]. Overall, extensive structural and biophysical work will be needed to understand the aforementioned complexes and may, in turn, lead to novel approaches for targeting specific complexes in the treatment of neurological disorders.
Oligomers between Class C and Class A GPCRs
While obligate dimerization of class C GPCRs is widely accepted, the significance of class A GPCR oligomerization in living mammalian cells remains controversial. Many studies, using a wide-range of experimental techniques, have suggested that class A GPCR homo- and heteromerization alters receptor pharmacology, trafficking and function [96, 97]. However, reconstituted monomeric class A GPCRs effectively couple to G proteins and recent cellular studies have suggested that homodimers for various class A GPCRs exist only transiently or at high expression levels [96, 97]. Despite their divergent modes of assembly and gating, a number of class A and class C GPCR have been shown to form complexes based on either the use of co-IP, the design and application of bivalent ligands and resonance-based proximity assays. This includes interactions between mGluR5 and the adenosine A2A receptor [98], the μ-opioid receptor [99] and the D1 dopamine receptor [100]. These inter-family GPCR complexes provide a means of cross-talk between neuromodulatory systems, and have been implicated in mechanisms underlying Parkinson’s disease [98, 99] and inflammatory pain [99].
The most extensively characterized class A-class C complex is formed between mGluR2 and the serotonin 2A receptor (5-HT2AR). Strong evidence exists for the formation of this heteromer in vitro, as well as in rodent and human frontal cortex, and it is thought to be involved in the signaling and behavioral responses induced by psychedelic 5-HT2AR agonists and atypical antipsychotics [101–105]. Initial cultured cell studies showed that activation of the Gi/o-coupled mGluR2, but not mGluR3, leads to Gq/11-dependent intracellular Ca2+ release in cells co-expressing 5-HT2AR (Fig. 4D). However, an important factor to consider with GPCR heteromers is how absolute and relative levels of expression of the individual components define the nature of the crosstalk. For example, agonist-induced trans-activation between mGluR2 and 5-HT2AR has been validated in some, but not all recent studies [106–109], likely due to differences in expression and cellular conditions. Under some conditions inverse conformational coupling, where the activated state of one receptor favors the inactive state of the partner receptor, has been observed [102]. This inverse coupling likely underscores the synergistic antipsychotic effects of inverse agonists of 5-HT2AR and agonists of mGluR2 [102].
Figure 4, Modulation of class C GPCRs via accessory proteins.
A, Schematic showing group I mGluR (mGluR1/5) coupling to a network of intracellular scaffold proteins which control signaling within the post-synaptic density. In brief, mGluR1 or mGluR5 CTDs bind directly to Homer proteins which, via SHANK, DLGAP and PSD-95, facilitate cross-talk with NMDA-type ionotropic glutamate receptors. Homer has also been shown to facilitate coupling to IP3 receptors and other elements of the scaffold and trafficking machinery. B, Interaction between the GABAB2 CTD and KCTD12 enhances the activation kinetics and desensitization of agonist-induced GIRK potassium channel currents by binding Gβγ subunits (right). Note: crystal structures revealed that the KCTD12 BTB domain forms a ring structure that binds one CTD and the H1 domain forms a 5:5 pentameric complex with Gβγ to effectively scavenge the G proteins away from the channel. C, Trans-synaptic interactions between group III mGluRs and ELFN proteins control the synaptic localization of mGluRs and have been shown to allosterically modulate receptor activation properties (green arrow), although the effects on signaling are unclear. D, Heteromerization between mGluR2 and class A 5-HT2ARs enables various forms of functional crosstalk, including trans-activation of 5-HT2ARs to produce Gq signaling (i.e. calcium elevation) following mGluR2 agonism (right). The stability and stoichiometry of such heteromers are not clear but an interface involving the cytoplasmic end of TM4 has been demonstrated.
Based on biophysical studies using chimeric constructs between mGluR2 and mGluR3, it has been suggested that crosstalk requires direct heteromerization between 5-HT2AR and mGluR2 that is dependent on three Ala residues within the intracellular half of TM4 of mGluR2 [103]. Furthermore, viral overexpression of wild-type, but not mGluR2ΔTM4 (an mGluR2/mGluR3 chimeric construct that is not able to interact with 5-HT2AR), was able to rescue a deficit in head-twitch behavior in mGluR2-KO mice [103]. Interestingly, residues at the extracellular end of TM4 have been proposed to contribute to mGluR2 homodimerization [77], raising the question of how this heteromeric complex assembles and what determines the mode of crosstalk (trans-activation versus inverse coupling). Overall, further work is needed to define how TM4 Ala residues contribute to the inter-family GPCR interface and to define the stability and stoichiometry of this biologically and clinically-relevant heteromer.
Together, the existence of class A-class C complexes indicates that class C GPCRs can be directly influenced by trans-interactions at the level of the TMD and can, in turn, influence other GPCRs this way. Further work is necessary to determine the stoichiometry, stability, inter-subunit interfaces, conformational dynamics, pharmacology, signaling and biological significance of class A-class C heteromers.
Concluding Remarks
Over 30 years of research has established that class C GPCRs form a physiologically-important family of complex signaling machines. Progress has accelerated in recent years in large part due to technological breakthroughs that opened up new ways to observe or perturb GPCRs. Improved methods for x-ray crystallography and cryo-EM have produced insightful, atomic-level structures of class C GPCRs and their interacting proteins [20, 21–24, 110]. In parallel, the application of spectroscopic methods, especially at the single molecule level, have begun to reveal the dynamic nature of the class C GPCR activation process [28, 31, 52]. Proteomic methods continue to identify new receptor-specific accessory and effector proteins [111] and photopharmacology has enabled studies of receptor signaling with high spatiotemporal precision in native systems, including in vivo [112, 113]. In addition, the oligomeric nature of class C GPCRs has begun to become appreciated and examples of pharmacological treatments to treat disease based on receptor heteromers are emerging [102]. Future work will further develop and apply biophysical approaches coupled to genetic manipulation to enable an integration of our understanding of the molecular biophysical properties of class C GPCRs with a description of their cellular signaling dynamics in health and disease (see Outstanding Questions Box).
Outstanding questions:
How are specific homo- and hetero-dimeric class C GPCR combinations formed and what inter-subunit interactions mediate specific assembly? How do these different inter-subunit interactions control receptor activation?
What conformational dynamics mediate orthosteric versus allosteric activation and modulation of different receptor subtypes? How does occupancy within a dimer for each class of ligand drive conformational and functional responses?
How do intra- and inter-subunit conformational changes at the LBD correspond to intra- and inter-subunit conformational changes at the TMD level and how are the LBDs allosterically coupled to TMDs? Why is dimerization requirement for allosteric coupling from the LBD to the TMD?
Do class C GPCR TMDs activate with analogous TM6 motion and G protein interactions compared to class A GPCRs? Are similar interfaces engaged by GRKs and arrestins?
How do specific inter-family GPCR heteromers assemble and exert functional crosstalk?
How are class C GPCR signaling units (i.e. receptors + accessory proteins + transducers + effectors) arranged in space and time under physiological conditions? Are large, stable complexes employed or do transient interactions mediate signaling?
Highlights:
Class C GPCRs show a unique multi-domain structure and dimeric assembly that allows for a complex interplay between orthosteric LBD-targeting and allosteric TMD-targeting ligands
Rapid inter-LBD re-arrangement on the milliseconds time scale drives receptor activation, in part, by repositioning the TMDs relative to each other.
Class C GPCRs interact with a plethora of accessory proteins, including intracellular scaffold proteins, secreted proteins, inter-synaptic scaffolds and class A GPCRs, which tune their biophysical and signaling properties.
Acknowledgements
The authors thank David Eliezer for helpful comments on the manuscript. JGM is supported by grants from NIMH (R01 MH084894 and R01 MH111940), DEL is supported by a grant from NIHL (R01 HL059949–23) and JL is supported by a grant from NIGMS (R35 GM124731) and the Rohr Family Research Scholar Award.
GLOSSARY:
- Allosteric Ligand:
a ligand that binds to an allosteric site that is spatially-distinct from, but conformationally-linked to, the orthosteric binding site.
- Efficacy:
the maximum magnitude of response that a drug can produce regardless of the dose. Partial agonists have less efficacy (i.e. maximum activation level) compared to full agonists.
- ELFN:
Extracellular-leucine-rich repeat (LRR) fibronectin domain protein. ELFN1 and 2 are pre- or post-synaptic adhesion proteins that interact across the synapse with group III mGluRs throughout the nervous system.
- FRET:
Förster resonance energy transfer; a method for measuring the distance, in the 20–100 Å range, between two fluorescent probes based on the distance-dependent energy transfer between donor and acceptor probes. Ideal for detecting conformational changes or dynamic assembly of proteins, as is widely used in studies of class C GPCRs.
- KCTD:
Potassium channel tetramerization domain protein family, so called due to the sequence similarity between their conserved N-terminal region and the tetramerization domain in some voltage-gated potassium channels. Some family members (KCTD8, 12, 16) associate with the C-terminus of GABAB2 and function to regulate the kinetics of potassium and calcium currents activated by GABAB receptors by reversibly binding Gβγ subunits.
- Inverse agonist:
A ligand that decreases constitutive/basal receptor activity and thus produces a response opposite in direction to that of an agonist, despite binding to the same receptor binding site.
- Molecular switches:
Non-covalent intramolecular interactions that exist in a given state for a GPCR and undergo breakage/formation during the receptor’s transition to a functionally distinct state. The canonical example is the “ionic lock” that forms between charged residues at the intracellular end of TM3 and TM6.
- Nanobody (Nb):
A recombinant single domain antibody fragment that contains the unique structural and functional properties of naturally occurring heavy-chain-only antibodies in camelids, yet in contrast to conventional antibodies, their compact shape facilitates their access to the transducer-binding cleft of a receptor to induce structural rearrangements similar to those induced by functional transducers such as G proteins.
- Neutral antagonist:
also known as silent antagonist, a drug that has no intrinsic activity itself, but by binding to a receptor, attenuates the responses to agonists or inverse agonists.
- Orthosteric Ligand:
A ligand (agonist or antagonist) that binds to the same receptor binding site recognized by the endogenous ligand for that receptor.
- Photopharmacology:
The manipulation of a given biological process using a synthetic photoswitch; a chemical moiety that undergoes a change in its structure upon irradiation with light, which can be incorporated into the structure of a given chemical compound. Most photopharmacological ligands incorporate an azobenzene photoswitch that toggles between cis and trans states in response to different wavelengths of light. Orthosteric and allosteric mGluR-targeting photopharmacological ligands have been developed.
- Photoswitchable Tethered Ligand (PTL):
A photopharmacological ligand that is covalently attached to a receptor target through genetic engineering of the receptor itself, allowing a high degree of subtype-specificity and genetic targeting with optimal spatial and temporal precision. Agonistic PTLs have been attached to mGluR LBDs via either cysteine-chemistry, through attachment to N-terminal SNAP-, CLIP- or Halo-tags or via nanobodies.
- Proteomics:
the study of large sets of proteins. Various forms of mass spectrometry are typically used for such studies, including for the analysis of co-immunoprecipitated proteins, as has been done to identify accessory proteins of the GABABR.
- smFRET:
Single molecule Förster resonance energy transfer; a method for measuring FRET from individual molecules, thus gaining the ability to resolve conformational dynamics with high precision on the millisecond time scale in order to develop state models and resolve microscopic kinetics.
- Sushi domain:
Short protein domains (~70 aa) involved in extracellular protein recognition in a variety of contexts; also known as complement control protein (CCP) modules or short consensus repeats (SCR); two sushi domains are present in the GABAB1a splice variant and are involved in plasma membrane and axonal targeting of the receptor..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Sriram K and Insel PA (2018) G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol 93 (4), 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Graaf C. et al. (2017) Extending the Structural View of Class B GPCRs. Trends Biochem Sci 42 (12), 946–960. [DOI] [PubMed] [Google Scholar]
- 3.Pin JP and Bettler B. (2016) Organization and functions of mGlu and GABAB receptor complexes. Nature 540 (7631), 60–68. [DOI] [PubMed] [Google Scholar]
- 4.Kowatsch C. et al. (2019) Structures of vertebrate Patched and Smoothened reveal intimate links between cholesterol and Hedgehog signalling. Curr Opin Struct Biol 57, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X. et al. (2018) Structural and Druggability Landscape of Frizzled G Protein-Coupled Receptors. Trends Biochem Sci 43 (12), 1033–1046. [DOI] [PubMed] [Google Scholar]
- 6.Bassilana F. et al. (2019) Adhesion G protein-coupled receptors: opportunities for drug discovery. Nat Rev Drug Discov 18 (11), 869–884. [DOI] [PubMed] [Google Scholar]
- 7.Erlandson SC et al. (2018) Structural Basis for G Protein-Coupled Receptor Signaling. Annu Rev Biophys. [DOI] [PubMed] [Google Scholar]
- 8.Weis WI and Kobilka BK (2018) The Molecular Basis of G Protein-Coupled Receptor Activation. Annu Rev Biochem 87, 897–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thal DM et al. (2018) Structural insights into G-protein-coupled receptor allostery. Nature 559 (7712), 45–53. [DOI] [PubMed] [Google Scholar]
- 10.Latorraca NR et al. (2017) GPCR Dynamics: Structures in Motion. Chem Rev 117 (1), 139–155. [DOI] [PubMed] [Google Scholar]
- 11.Manglik A. et al. (2015) Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell 161 (5), 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregorio GG et al. (2017) Single-molecule analysis of ligand efficacy in beta2AR-G-protein activation. Nature 547 (7661), 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wacker D. et al. (2017) How Ligands Illuminate GPCR Molecular Pharmacology. Cell 170 (3), 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Congreve M. et al. (2020) Impact of GPCR Structures on Drug Discovery. Cell 181 (1), 81–91. [DOI] [PubMed] [Google Scholar]
- 15.Reiner A. and Levitz J. (2018) Glutamatergic Signaling in the Central Nervous System: Ionotropic and Metabotropic Receptors in Concert. Neuron 98 (6), 1080–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heaney CF and Kinney JW (2016) Role of GABA(B) receptors in learning and memory and neurological disorders. Neurosci Biobehav Rev 63, 1–28. [DOI] [PubMed] [Google Scholar]
- 17.Hannan FM et al. (2018) The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases. Nat Rev Endocrinol 15 (1), 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer RK (2019) A Pharmacological Perspective on the Study of Taste. Pharmacol Rev 71 (1), 20–48. [DOI] [PubMed] [Google Scholar]
- 19.Clemmensen C. et al. (2014) The GPCR, class C, group 6, subtype A (GPRC6A) receptor: from cloning to physiological function. Br J Pharmacol 171 (5), 1129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koehl A. et al. (2019) Structural insights into the activation of metabotropic glutamate receptors. Nature 566 (7742), 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papasergi-Scott MM et al. (2020) Structures of metabotropic GABAB receptor. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao C. et al. (2020) Cryo-EM structures of inactive and active GABAB receptor. Cell Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J. et al. (2020) Structure of human GABAB receptor in an inactive state. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaye H. et al. (2020) Structural basis of the activation of a metabotropic GABA receptor. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romano C. et al. (1996) Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J Biol Chem 271 (45), 28612–6. [DOI] [PubMed] [Google Scholar]
- 26.Bai M. et al. (1998) Dimerization of the extracellular calcium-sensing receptor (CaR) on the cell surface of CaR-transfected HEK293 cells. J Biol Chem 273 (36), 23605–10. [DOI] [PubMed] [Google Scholar]
- 27.Norskov-Lauritsen L. et al. (2015) N-glycosylation and disulfide bonding affects GPRC6A receptor expression, function, and dimerization. FEBS Lett 589 (5), 588–97. [DOI] [PubMed] [Google Scholar]
- 28.Levitz J. et al. (2016) Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors. Neuron 92 (1), 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Moustaine D. et al. (2012) Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc Natl Acad Sci U S A 109 (40), 16342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bettler B. et al. (2004) Molecular structure and physiological functions of GABA(B) receptors. Physiol Rev 84 (3), 835–67. [DOI] [PubMed] [Google Scholar]
- 31.Calebiro D. et al. (2013) Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci U S A 110 (2), 743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gassmann M. et al. (2004) Redistribution of GABAB(1) protein and atypical GABAB responses in GABAB(2)-deficient mice. J Neurosci 24 (27), 6086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doumazane E. et al. (2011) A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J 25 (1), 66–77. [DOI] [PubMed] [Google Scholar]
- 34.Moreno Delgado D. et al. (2017) Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin S. et al. (2014) Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J Neurosci 34 (1), 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J. et al. (2020) Defining the Homo- and Heterodimerization Propensities of Metabotropic Glutamate Receptors. Cell Rep 31 (5), 107605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunishima N. et al. (2000) Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407 (6807), 971–7. [DOI] [PubMed] [Google Scholar]
- 38.Geng Y. et al. (2013) Structural mechanism of ligand activation in human GABA(B) receptor. Nature 504 (7479), 254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geng Y. et al. (2016) Structural mechanism of ligand activation in human calcium-sensing receptor. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuemket N. et al. (2017) Structural basis for perception of diverse chemical substances by T1r taste receptors. Nat Commun 8, 15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurel D. et al. (2008) Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat Methods 5 (6), 561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comps-Agrar L. et al. (2011) The oligomeric state sets GABA(B) receptor signalling efficacy. EMBO J 30 (12), 2336–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart GD et al. (2018) Allosteric interactions between GABAB1 subunits control orthosteric binding sites occupancy within GABAB oligomers. Neuropharmacology 136 (Pt A), 92–101. [DOI] [PubMed] [Google Scholar]
- 44.Xue L. et al. (2019) Rearrangement of the transmembrane domain interfaces associated with the activation of a GPCR hetero-oligomer. Nat Commun 10 (1), 2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moller TC et al. (2018) Oligomerization of a G protein-coupled receptor in neurons controlled by its structural dynamics. Sci Rep 8 (1), 10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siddig S. et al. (2020) Super-resolution imaging reveals the nanoscale organization of metabotropic glutamate receptors at presynaptic active zones. Sci Adv 6 (16), eaay7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C. et al. (2016) Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci Adv 2 (5), e1600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muto T. et al. (2007) Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A 104 (10), 3759–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C. et al. (2016) Molecular Basis of the Extracellular Ligands Mediated Signaling by the Calcium Sensing Receptor. Front Physiol 7, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conigrave AD et al. (2000) L-amino acid sensing by the extracellular Ca2+-sensing receptor. Proc Natl Acad Sci U S A 97 (9), 4814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubo Y. et al. (1998) Structural basis for a Ca2+-sensing function of the metabotropic glutamate receptors. Science 279 (5357), 1722–5. [DOI] [PubMed] [Google Scholar]
- 52.Vafabakhsh R. et al. (2015) Conformational dynamics of a class C G-protein-coupled receptor. Nature 524 (7566), 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galvez T. et al. (2000) Ca(2+) requirement for high-affinity gamma-aminobutyric acid (GABA) binding at GABA(B) receptors: involvement of serine 269 of the GABA(B)R1 subunit. Mol Pharmacol 57 (3), 419–26. [DOI] [PubMed] [Google Scholar]
- 54.Tora AS et al. (2015) Allosteric modulation of metabotropic glutamate receptors by chloride ions. FASEB J 29 (10), 4174–88. [DOI] [PubMed] [Google Scholar]
- 55.Doumazane E. et al. (2013) Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc Natl Acad Sci U S A 110 (15), E1416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olofsson L. et al. (2014) Fine tuning of sub-millisecond conformational dynamics controls metabotropicglutamate receptors agonist efficacy. Nat Commun 5, 5206. [DOI] [PubMed] [Google Scholar]
- 57.Habrian CH et al. (2019) Conformational pathway provides unique sensitivity to a synaptic mGluR. Nat Commun 10 (1), 5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kniazeff J. et al. (2004) Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol 11 (8), 706–13. [DOI] [PubMed] [Google Scholar]
- 59.Lecat-Guillet N. et al. (2017) FRET-Based Sensors Unravel Activation and Allosteric Modulation of the GABAB Receptor. Cell Chem Biol 24 (3), 360–370. [DOI] [PubMed] [Google Scholar]
- 60.Leach K. and Gregory KJ (2017) Molecular insights into allosteric modulation of Class C G protein-coupled receptors. Pharmacol Res 116, 105–118. [DOI] [PubMed] [Google Scholar]
- 61.Lindsley CW et al. (2016) Practical Strategies and Concepts in GPCR Allosteric Modulator Discovery: Recent Advances with Metabotropic Glutamate Receptors. Chem Rev 116 (11), 6707–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goudet C. et al. (2004) Heptahelical domain of metabotropic glutamate receptor 5 behaves like rhodopsin-like receptors. Proc Natl Acad Sci U S A 101 (1), 378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Binet V. et al. (2004) The heptahelical domain of GABA(B2) is activated directly by CGP7930, a positive allosteric modulator of the GABA(B) receptor. J Biol Chem 279 (28), 29085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitsukawa K. et al. (2005) A selective metabotropic glutamate receptor 7 agonist: activation of receptor signaling via an allosteric site modulates stress parameters in vivo. Proc Natl Acad Sci U S A 102 (51), 18712–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noetzel MJ et al. (2012) Functional impact of allosteric agonist activity of selective positive allosteric modulators of metabotropic glutamate receptor subtype 5 in regulating central nervous system function. Mol Pharmacol 81 (2), 120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rovira X. et al. (2015) Overlapping binding sites drive allosteric agonism and positive cooperativity in type 4 metabotropic glutamate receptors. FASEB J 29 (1), 116–30. [DOI] [PubMed] [Google Scholar]
- 67.Gutzeit VA et al. (2019) Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keller AN et al. (2018) Identification of Global and Ligand-Specific Calcium Sensing Receptor Activation Mechanisms. Mol Pharmacol 93 (6), 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toda Y. et al. (2018) Positive/Negative Allosteric Modulation Switching in an Umami Taste Receptor (T1R1/T1R3) by a Natural Flavor Compound, Methional. Sci Rep 8 (1), 11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porter RH et al. (2005) Fenobam: a clinically validated nonbenzodiazepine anxiolytic is a potent, selective, and noncompetitive mGlu5 receptor antagonist with inverse agonist activity. J Pharmacol Exp Ther 315 (2), 711–21. [DOI] [PubMed] [Google Scholar]
- 71.Wu H. et al. (2014) Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344 (6179), 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dore AS et al. (2014) Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511 (7511), 557–62. [DOI] [PubMed] [Google Scholar]
- 73.Christopher JA et al. (2015) Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J Med Chem 58 (16), 6653–64. [DOI] [PubMed] [Google Scholar]
- 74.Christopher JA et al. (2019) Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures. J Med Chem 62 (1), 207–222. [DOI] [PubMed] [Google Scholar]
- 75.Perez-Benito L. et al. (2017) Molecular Switches of Allosteric Modulation of the Metabotropic Glutamate 2 Receptor. Structure 25 (7), 1153–1162 e4. [DOI] [PubMed] [Google Scholar]
- 76.Lans I. et al. (2020) Exploring the Activation Mechanism of the mGlu5 Transmembrane Domain. Front Mol Biosci 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xue L. et al. (2015) Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat Chem Biol 11 (2), 134–40. [DOI] [PubMed] [Google Scholar]
- 78.Tateyama M. et al. (2004) Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1alpha. Nat Struct Mol Biol 11 (7), 637–42. [DOI] [PubMed] [Google Scholar]
- 79.Marcaggi P. et al. (2009) Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. Proc Natl Acad Sci U S A 106 (27), 11388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grushevskyi EO et al. (2019) Stepwise activation of a class C GPCR begins with millisecond dimer rearrangement. Proc Natl Acad Sci U S A 116 (20), 10150–10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rondard P. et al. (2006) Coupling of agonist binding to effector domain activation in metabotropic glutamate-like receptors. J Biol Chem 281 (34), 24653–61. [DOI] [PubMed] [Google Scholar]
- 82.Huang S. et al. (2011) Interdomain movements in metabotropic glutamate receptor activation. Proc Natl Acad Sci U S A 108 (37), 15480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galvez T. et al. (2001) Allosteric interactions between GB1 and GB2 subunits are required for optimal GABA(B) receptor function. EMBO J 20 (9), 2152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goudet C. et al. (2005) Asymmetric functioning of dimeric metabotropic glutamate receptors disclosed by positive allosteric modulators. J Biol Chem 280 (26), 24380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hlavackova V. et al. (2005) Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J 24 (3), 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J. et al. (2017) Allosteric control of an asymmetric transduction in a G protein-coupled receptor heterodimer. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Conklin BR et al. (1993) Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature 363 (6426), 274–6. [DOI] [PubMed] [Google Scholar]
- 88.Flock T. et al. (2017) Selectivity determinants of GPCR-G-protein binding. Nature 545 (7654), 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inoue A. et al. (2019) Illuminating G-Protein-Coupling Selectivity of GPCRs. Cell 177 (7), 1933–1947 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Havlickova M. et al. (2003) The second intracellular loop of metabotropic glutamate receptors recognizes C termini of G-protein alpha-subunits. J Biol Chem 278 (37), 35063–70. [DOI] [PubMed] [Google Scholar]
- 91.Fourgeaud L. et al. (2003) The metabotropic glutamate receptor mGluR5 is endocytosed by a clathrin-independent pathway. J Biol Chem 278 (14), 12222–30. [DOI] [PubMed] [Google Scholar]
- 92.Perroy J. et al. (2003) Phosphorylation-independent desensitization of GABA(B) receptor by GRK4. EMBO J 22 (15), 3816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suh YH et al. (2018) Metabotropic glutamate receptor trafficking. Mol Cell Neurosci 91, 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee S. et al. (2019) Nedd4 E3 ligase and beta-arrestins regulate ubiquitination, trafficking, and stability of the mGlu7 receptor. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gorvin CM et al. (2018) A calcium-sensing receptor mutation causing hypocalcemia disrupts a transmembrane salt bridge to activate beta-arrestin-biased signaling. Sci Signal 11 (518). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ferre S. et al. (2014) G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev 66 (2), 413–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sleno R. and Hebert TE (2018) The Dynamics of GPCR Oligomerization and Their Functional Consequences. Int Rev Cell Mol Biol 338, 141–171. [DOI] [PubMed] [Google Scholar]
- 98.Ferre S. et al. (2002) Synergistic interaction between adenosine A2A and glutamate mGlu5 receptors: implications for striatal neuronal function. Proc Natl Acad Sci U S A 99 (18), 11940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Akgun E. et al. (2013) Ligands that interact with putative MOR-mGluR5 heteromer in mice with inflammatory pain produce potent antinociception. Proc Natl Acad Sci U S A 110 (28), 11595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sebastianutto I. et al. (2020) D1-mGlu5 heteromers mediate noncanonical dopamine signaling in Parkinson’s disease. J Clin Invest 130 (3), 1168–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gonzalez-Maeso J. et al. (2008) Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature 452 (7183), 93–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fribourg M. et al. (2011) Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell 147 (5), 1011–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moreno JL et al. (2012) Identification of three residues essential for 5-hydroxytryptamine 2A-metabotropic glutamate 2 (5-HT2A.mGlu2) receptor heteromerization and its psychoactive behavioral function. J Biol Chem 287 (53), 44301–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moreno JL et al. (2016) Allosteric signaling through an mGlu2 and 5-HT2A heteromeric receptor complex and its potential contribution to schizophrenia. Sci Signal 9 (410), ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hideshima KS et al. (2018) Role of mGlu2 in the 5-HT2A receptor-dependent antipsychotic activity of clozapine in mice. Psychopharmacology (Berl) 235 (11), 3149–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murat S. et al. (2019) 5-HT2A receptor-dependent phosphorylation of mGlu2 receptor at Serine 843 promotes mGlu2 receptor-operated Gi/o signaling. Mol Psychiatry 24 (11), 1610–1626. [DOI] [PubMed] [Google Scholar]
- 107.Olivero G. et al. (2018) 5-HT2A-mGlu2/3 receptor complex in rat spinal cord glutamatergic nerve endings: A 5-HT2A to mGlu2/3 signalling to amplify presynaptic mechanism of auto-control of glutamate exocytosis. Neuropharmacology 133, 429–439. [DOI] [PubMed] [Google Scholar]
- 108.Hamor PU et al. (2018) Chronic methamphetamine self-administration dysregulates 5-HT2A and mGlu2 receptor expression in the rat prefrontal and perirhinal cortex: Comparison to chronic phencyclidine and MK-801. Pharmacol Biochem Behav 175, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Delille HK et al. (2012) Heterocomplex formation of 5-HT2A-mGlu2 and its relevance for cellular signaling cascades. Neuropharmacology 62 (7), 2184–91. [DOI] [PubMed] [Google Scholar]
- 110.Zheng S. et al. (2019) Structural basis for KCTD-mediated rapid desensitization of GABAB signalling. Nature 567 (7746), 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwenk J. et al. (2016) Modular composition and dynamics of native GABAB receptors identified by high-resolution proteomics. Nat Neurosci 19 (2), 233–42. [DOI] [PubMed] [Google Scholar]
- 112.Zussy C. et al. (2018) Dynamic modulation of inflammatory pain-related affective and sensory symptoms by optical control of amygdala metabotropic glutamate receptor 4. Mol Psychiatry 23 (3), 509–520. [DOI] [PubMed] [Google Scholar]
- 113.Acosta-Ruiz A. et al. (2020) Branched Photoswitchable Tethered Ligands Enable Ultra-efficient Optical Control and Detection of G Protein-Coupled Receptors In Vivo. Neuron 105 (3), 446–463 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nakajima Y. et al. (2009) Ca2+-dependent release of Munc18–1 from presynaptic mGluRs in short-term facilitation. Proc Natl Acad Sci U S A 106 (43), 18385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schwenk J. et al. (2010) Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 465 (7295), 231–5. [DOI] [PubMed] [Google Scholar]
- 116.Turecek R. et al. (2014) Auxiliary GABAB receptor subunits uncouple G protein betagamma subunits from effector channels to induce desensitization. Neuron 82 (5), 1032–44. [DOI] [PubMed] [Google Scholar]
- 117.Zuo H. et al. (2019) Structural basis for auxiliary subunit KCTD16 regulation of the GABAB receptor. Proc Natl Acad Sci U S A 116 (17), 8370–8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tomioka NH et al. (2014) Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun 5, 4501. [DOI] [PubMed] [Google Scholar]
- 119.Cao Y. et al. (2015) Mechanism for Selective Synaptic Wiring of Rod Photoreceptors into the Retinal Circuitry and Its Role in Vision. Neuron 87 (6), 1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dunn HA et al. (2018) Synaptic adhesion protein ELFN1 is a selective allosteric modulator of group III metabotropic glutamate receptors in trans. Proc Natl Acad Sci U S A 115 (19), 5022–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dunn HA et al. (2019) ELFN2 is a postsynaptic cell adhesion molecule with essential roles in controlling group III mGluRs in the brain and neuropsychiatric behavior. Mol Psychiatry 24 (12), 1902–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stachniak TJ et al. (2019) Elfn1-Induced Constitutive Activation of mGluR7 Determines Frequency-Dependent Recruitment of Somatostatin Interneurons. J Neurosci 39 (23), 4461–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Condomitti G. et al. (2018) An Input-Specific Orphan Receptor GPR158-HSPG Interaction Organizes Hippocampal Mossy Fiber-CA3 Synapses. Neuron 100 (1), 201–215 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Orlandi C. et al. (2018) Transsynaptic Binding of Orphan Receptor GPR179 to Dystroglycan-Pikachurin Complex Is Essential for the Synaptic Organization of Photoreceptors. Cell Rep 25 (1), 130–145 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rice HC et al. (2019) Secreted amyloid-beta precursor protein functions as a GABABR1a ligand to modulate synaptic transmission. Science 363 (6423). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dinamarca MC et al. (2019) Complex formation of APP with GABAB receptors links axonal trafficking to amyloidogenic processing. Nat Commun 10 (1), 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Um JW et al. (2013) Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron 79 (5), 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brody AH and Strittmatter SM (2018) Synaptotoxic Signaling by Amyloid Beta Oligomers in Alzheimer’s Disease Through Prion Protein and mGluR5. Adv Pharmacol 82, 293–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang J. et al. (2018) Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog 14 (7), e1007189. [DOI] [PMC free article] [PubMed] [Google Scholar]