Abstract
T cell costimulation is mediated by the interaction of a number of receptors and ligands present on the surface of the T cell and antigen-presenting cell, respectively. Stimulatory or inhibitory signals from these receptor-ligand interactions work in tandem to preserve immune homeostasis. BTNL2 is a type-1 membrane protein that provides inhibitory signal to T cells and plays an important role in several inflammatory and autoimmune diseases. Therefore, manipulation of the molecular interaction of BTNL2 with its putative receptor could provide strategies to restore immune homeostasis in these diseases. Hence, it is imperative to study the structural characteristics of this molecule which will provide important insights into its function as well. In this study, the membrane-distal ectodomain of murine BTNL2 was expressed in bacteria as inclusion bodies, refolded in vitro and purified for functional and structural characterization. The domain is monomeric in solution as demonstrated by size-exclusion chromatography and analytical ultracentrifugation, and also binds to its putative receptor on naïve B cells and activated T cell subsets. Importantly, for the first time, we report the structure of BTNL2 as determined by solution NMR spectroscopy and also the picosecond-nanosecond timescale backbone dynamics of this domain. The N-terminal ectodomain of BTNL2, which was able to inhibit T cell function as well, exhibits distinctive structural features. The N-terminal ectodomain of BTNL2 has a significantly reduced surface area in the front sheet due to the non-canonical conformation of the CC’ loop, which provides important insights into the recognition of its presently unknown binding partner.
Keywords: Determination of protein structure by NMR spectroscopy, immunoglobulin fold, immunoglobulin-variable domain, Butyrophilins, T cell costimulation
Introduction
Naïve T cells, which have undergone maturation in the thymus but are yet to encounter any antigen, get activated following multi-step signaling cascades to generate effector T cells. The initial step is the recognition of the peptide-loaded major histocompatibility complex (pMHC), which is expressed on the surface of an antigen presenting cell (APC), by the T cell receptor (TCR). While the antigen-dependent pMHC-TCR interaction initiates the process of T cell activation, a number of antigen-independent signals are essential for optimal T cell function. These antigen-independent signals are provided by interactions between costimulatory or coinhibitory receptors, expressed on the surface of T cells, with their specific ligands present on the surface of APC. Naïve T cells that only encounter the peptide-antigen but do not undergo costimulation become non-responsive and may even perish (1–3).
While costimulatory signaling (derived from receptor-ligand pairs present on the T cell and the APC respectively) leads to the activation of the T cell, to re-establish homeostasis, the activated T cell must employ inhibitory receptors to regulate and suppress any unwanted and prolonged immune response. This phenomenon is well illustrated by the CD28/CTLA-4: B7 ligands axis. Post TCR-pMHC engagement, the interaction of the B7 ligands with the constitutively expressed costimulatory molecule, CD28 leads to T cell activation. Interestingly, the engagement of those same set of ligands with CTLA-4, a T cell coinhibitory receptor expressed on activated T cells, results in restoration of homeostasis by dampening the response of the activated T cell (1, 4, 5). Similarly, the interaction of PD-1, an inhibitory receptor on the T cells, with its ligands PD-L1 (B7-H1, expressed on many peripheral tissues) and PD-L2 (B7-DC, expressed mainly on APCs) leads to immune suppression and is one of the most important peripheral tolerance mechanisms (6). Additional signaling from several other immune-modulatory molecules with similar activating or inhibitory functions helps shape the final outcome of an adaptive immune response as well as preserve tolerance to self-antigens in the periphery.
Butyrophilin-like 2 (BTNL2), a protein belonging to the butyrophilin-like (BTNL) protein family, has been implicated in regulating immune responses (7, 8) in several inflammatory conditions such as intestinal inflammation (9), graft-versus-host disease (10), type 1 diabetes (11) and infections like cerebral malaria (12), in both mice and humans. Importantly, BTNL2 polymorphisms have been found to have genetic association with different diseases like sarcoidosis (13–15), osteoarthritis (16), cardiomyopathy (17) and ulcerative colitis (18). BTNL2 acts as T cell inhibitory molecule which can induce FoxP3+ Treg differentiation, thus effectively suppressing effector T cells during inflammation (8). Such an action by BTNL2 has the potential to be manipulated to either induce immune-suppression during unwanted autoreactive responses or to trigger stronger immune responses in the tumor microenvironment. Indeed, missense mutations in the BTNL2 gene have been found to play an important role in susceptibility to familial and sporadic prostate cancer development (19), thus underlining the impact of this molecule in tumor development and progression.
Immunotherapeutic applications of BTNL2 require its structural description as well as the identification of its binding partner but these are yet to be determined. The extracellular region of BTNL2 comprises of multiple immunoglobulin variable (IgV) and immunoglobulin constant (IgC) domains, followed by a single-pass transmembrane region and a short cytoplasmic region (7). Unlike most of the other butyrophilin and butyrophilin-like members, BTNL2 does not contain the cytoplasmic B30.2 signaling domain (20). Although its binding partner remains unidentified, in vitro studies have shown that B cells express receptors for BTNL2 on their surface, while CD4+ and CD8+ T cells do so upon activation (7, 9). The use of BTNL2 as a potential therapeutic agent for restoring immune-tolerance under inflammatory conditions, as well as manipulation of BTNL2-mediated immune response for the development of novel therapeutics can be fully realized if its structure-function relationship can be elucidated at a molecular level. Here, we report the solution NMR structure of the N-terminal IgV domain of mouse BTNL2, which shares 65% sequence identity with its human counterpart. We also demonstrate that the N-terminal IgV domain of mouse BTNL2 is able to recognize its putative binding partner present on naïve B cells and also on phorbol myristate acetate (PMA)/ionomycin activated CD4+ and CD8+ T cells, which suggest that the purified IgV domain is functionally active. Additionally, this IgV domain alone is shown to inhibit IL-2 secretion by activated CD4+ T cells, indicative of the ability of the IgV domain alone to execute the T cell inhibitory function of BTNL2 unlike previous studies where full-length ectodomains of BTNL2 or Fc-fused BTNL2 were used for functional studies (7, 9). Being the first atomic level structure from the BTNL family across all species, the current study also provides insights into how BTNL2 recognizes its binding partner in a manner distinct from the classical B7 family of immune receptors.
Results
BTNL2 exists as monomer in solution
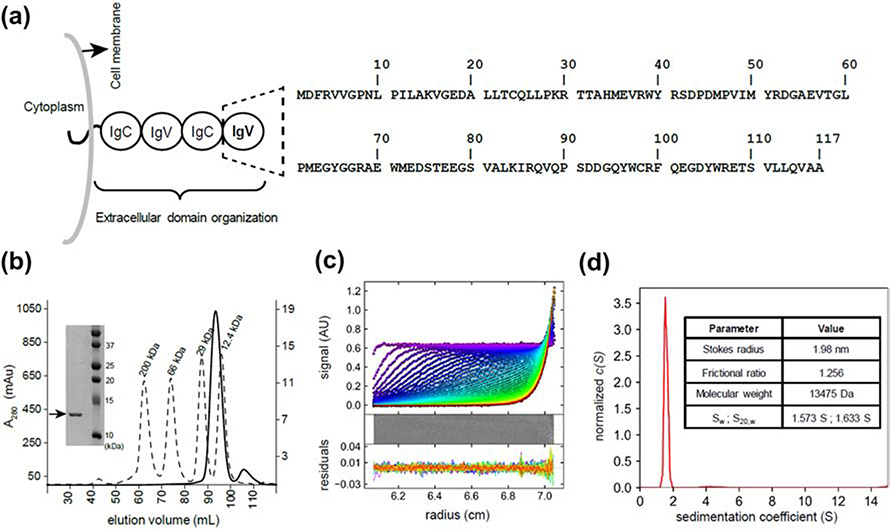
We have characterized the N-terminal IgV domain of mouse BTNL2 containing residues D28 to A143 (BTNL2D28-A143), which are renumbered as D2 to A117 (Fig 1A). The first 27 residues of BTNL2 form the signaling sequence and hence are not considered. In size exclusion chromatography (SEC), refolded BTNL2 eluted as a single sharp peak indicating a monodispersed and homogeneous protein sample (Fig. 1B). BTNL2 eluted at 93.7 ml, which corresponds to an estimated molecular mass of 15.4kDa (Fig. S1). This is in good agreement with its theoretically calculated molecular mass of 13.3kDa and indicates a monomeric protein. The eluted protein from SEC was highly pure as determined by SDS-PAGE (Fig. 1B).
Figure 1: BTNL2 IgV domain exists as a monomer in solution.
A. Domain architecture of full-length mouse BTNL2 showing four extracellular domains, a membrane-spanning region followed by a short cytoplasmic stretch. The N-terminal IgV domain is highlighted (dashed lines) and the corresponding sequence is indicated. B. BTNL2 elutes at 93.5 ml (solid line) in HiLoad™ 16/600 Superdex™ 200 pg SEC column. Molecular weight standards are shown as broken lines, and their absorbance is plotted as a secondary Y-axis. Inset shows SDS PAGE gel of purified BTNL2, indicated by an arrow. C. Sedimentation profile of absorbance at 280nm for BTNL2. Residuals indicate the goodness of fit. D. Continuous sedimentation coefficient distribution, c(S) curve, for BTNL2, shows the presence of a single dominant population in solution. The table lists biophysical parameters determined by sedimentation velocity analytical ultracentrifugation.
BTNL2 was further characterized by analytical ultracentrifugation (AUC), which provides insight into biophysical parameters such as the hydrodynamic size, shape and mass of proteins in bulk solution. From sedimentation velocity experiment, the Stokes radius of 1.98 nm, sedimentation coefficient (Sw) of 1.57 S and frictional ratio of 1.256 was determined for BTNL2 (Fig. 1C, D). The Sw value correlates to a molecular weight of 13.5 kDa, which closely matches with the expected value. Additionally, the frictional ratio of 1.256 for BTNL2 strongly suggests that it is a globular protein (21, 22). The C(s) distribution model for fitted data clearly indicates the presence of a single dominant species in solution (Fig. 1D) which reiterates that BTNL2 exists as a homogeneous monomeric protein in solution.
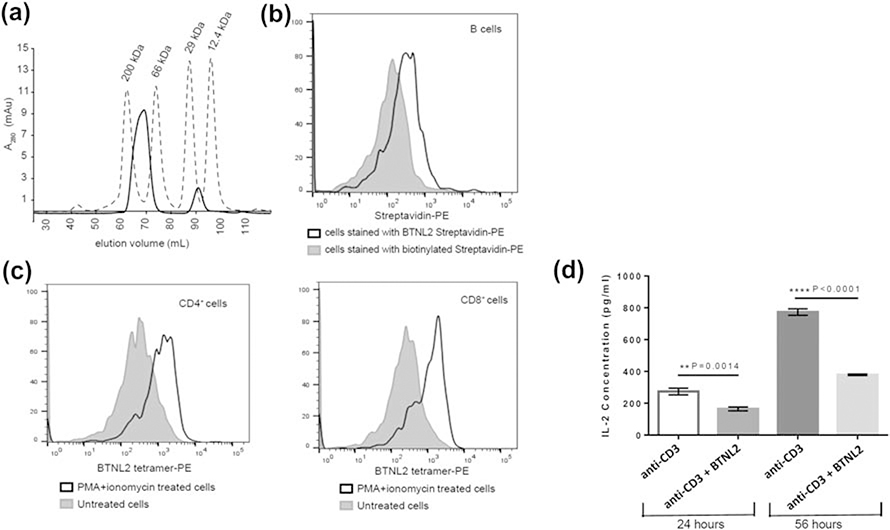
For functional assays of BTNL2, a tetrameric protein was generated by binding biotinylated BTNL2 to streptavidin in a 4:1 stoichiometric ratio. This protein complex eluted out at 69.2 ml in SEC column (Fig. 2A). This corresponds to 109 kDa (Fig. S1), which is in good agreement with the expected molecular weight of 112 kDa for the complex.
Figure 2: BTNL2 IgV domain binds to its putative receptor expressed on naïve B cells and in vitro activated CD4+ and CD8+ T cells.
A. Biotinylated BTNL2 was tetramerized using streptavidin, which elutes at 69.2 ml (solid line) in HiLoad™ 16/600 Superdex™ 200 pg SEC column. Molecular weight standards are also indicated B. BTNL2 was tetramerized with PE-conjugated streptavidin. Binding of BTNL2 tetramer was analyzed by gating on CD19+ B cells using biotinylated streptavidin-PE as a control to account for any non-specific binding of the tetramer. C. Untreated as well as PMA/ionomycin activated splenocytes were stained with fluorochrome-conjugated anti-CD3, and anti CD4 (left panel) or anti-CD8 (right panel) antibodies followed by incubation with PE-conjugated BTNL2 tetramer. The solid gray histogram represents BTNL2 tetramer binding to untreated cells, while the white histogram represents binding of the tetramer to PMA/ionomycin treated cells. D. Sorted CD4+ T cells were stimulated with plate-bound anti CD3 antibody with or without immobilized BTNL2 IgV monomer for 24 hours (p value = 0.0014) and 56 hours (p value ˂ 0.0001). The supernatant were collected and analyzed for IL-2 secretion. Data represents mean with SEM of 3 biological replicates for each set. p value was calculated using unpaired T test.
Refolded BTNL2 recognizes its putative binding partner present on naïve B cells, and activated T cells and is functionally active
In order to ascertain whether refolded BTNL2 is able to bind to its putative receptor present on naïve B cell or in vitro activated T cell subsets, the monomeric BTNL2 membrane-distal IgV domain was tetramerized using PE-conjugated streptavidin for flow cytometry-based cell binding assays. The data clearly demonstrated the binding of the BTNL2 IgV domain to CD19+ B cells (Fig. 2B). This is in good agreement with the previous report that naïve B cells express binding partner for BTNL2 on their surface (7). Additionally, it was also found that BTNL2 tetramer was able to bind to in vitro activated CD4+ and CD8+ T cells (Fig. 2C). Both of these findings unequivocally demonstrated that the N-terminal IgV domain of BTNL2, expressed in E.coli and refolded in vitro is able to bind to its putative receptors present on naïve B cells and activated CD4 and CD8 T cells.
Furthermore, when CD4+ T cells were cultured in presence of plate bound anti-CD3 and BTNL2 IgV monomer, there was significant inhibition of IL-2 secretion by the T cells in 24 hours, which was further down-regulated after 56 hours of culture (Fig. 2D). This observation is strongly indicative of the ability of the IgV domain of BTNL2 to execute its previously reported T cell inhibitory function by down-regulating IL-2 secretion.
Backbone assignment reveals well-folded BTNL2 comprising mostly of beta strands
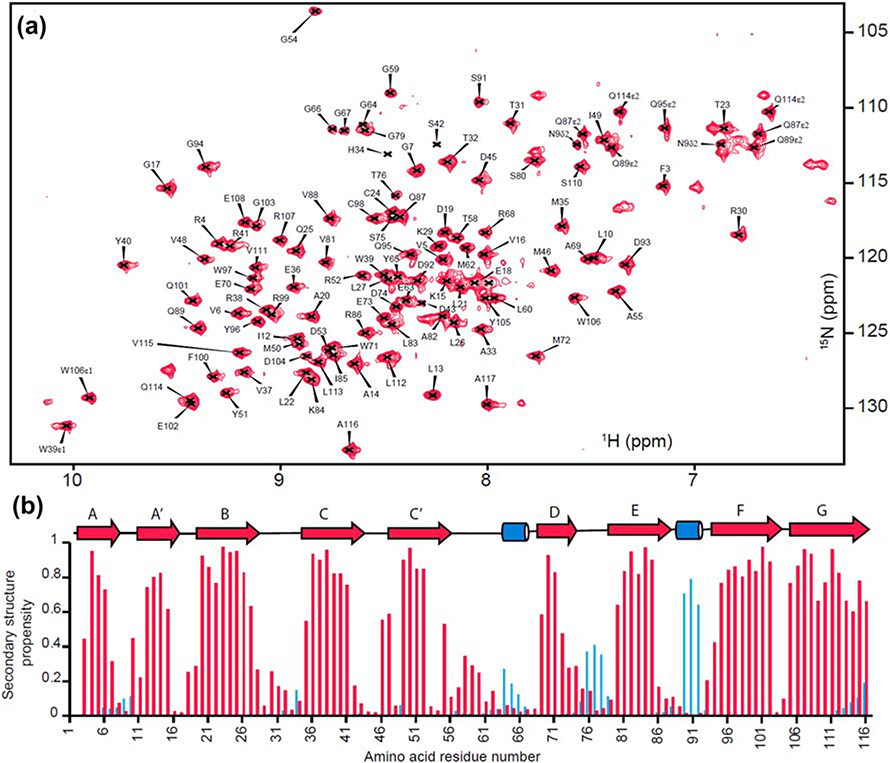
The refolded BTNL2 was structurally characterized by solution NMR spectroscopy. The 15N-HSQC spectrum of BTNL2 has well-dispersed peaks, thereby indicating a properly folded domain (Fig. 3A). Apart from the N-terminal non-native methionine, out of 110 non-proline residues 107 N-H peaks from the protein backbone have been assigned, which corresponds to 97.3% completion of the backbone assignment. The backbone chemical shifts of 1Hɑ, 1HN, 15N, 13Cɑ, 13Cβ and 13CO were used to compute the secondary structure propensity of BTNL2 using MICS (23). This indicates the presence of nine beta strands and two short helical turns, which is in good agreement with many proteins of the immunoglobulin superfamily (Fig. 3B).
Figure 3: Backbone assignment and secondary structure propensity of BTNL2 IgV domain.
A. Assigned 15N-1H HSQC spectra of mouse BTNL2 IgV domain. B. Secondary structure propensity calculated from backbone chemical shifts using MICS. β-strands and α-helices are colored in red and blue, respectively, and also shown as arrows and cylinders, respectively.
BTNL2 has two cysteine residues, Cys24 and Cys98 (Fig. 1A), which are typically involved in the formation of a disulfide bond in IgV folds (24). The Cβ chemical shifts of cysteines are very sensitive to the oxidized state of the adjacent sulphur atom. For oxidized and reduced states the chemical shift value is greater than 40 ppm and less than 35 ppm, respectively (25). The Cβ chemical shifts of Cys24 and Cys98 of BTNL2 are 49 ppm and 41.6 ppm, respectively. This strongly suggests that Cys24 and Cys98 form a disulfide bond.
The IgV domain of BTNL2 adopts an immunoglobulin (Ig) fold
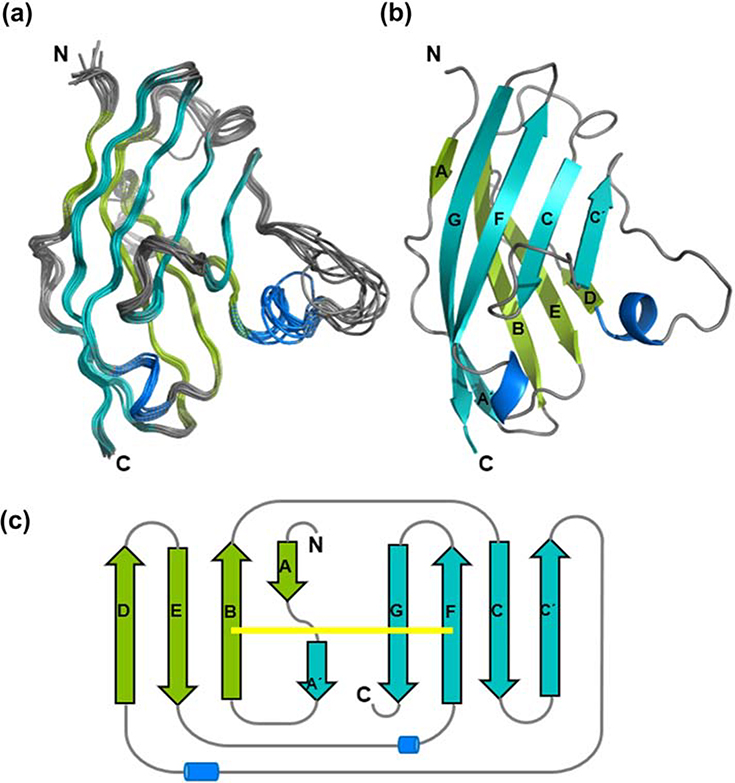
The structural ensemble of BTNL2 was calculated using PONDEROSA-C/S coupled with Xplor-NIH (26, 27). Nuclear Overhauser enhancement (NOE)-derived distance and backbone chemical-shift-derived dihedral angle restraints were used for structure calculation (Table 1). An ensemble of 10 best energy minimized models has a backbone atom RMSD of 0.56Å and heavy atom RMSD of 0.76Å across all rigid residues which form regular secondary structural elements (Fig. 4A).
TABLE 1.
Statistical parameters of solution NMR structure of membrane-distal ectodomain of mouse BTNL2.
| Parameter | Value |
|---|---|
| NOE derived distance constraints | |
| Short range (|i − j| ≤ 1) | 814 |
| Medium range (1 < |i − j| < 5) | 79 |
| Long range (|i − j| ≥ 5) | 650 |
| Total | 1543 |
| Hydrogen bond constraints | 40 |
| Dihedral angle constraints | |
| Φ | 92 |
| Ψ | 97 |
| Total | 189 |
| Average root mean square deviations against lowest energy model coordinates (Å) for well-defined residues† | |
| Backbone atoms (N, Cα, C, O) | 0.56±0.08 |
| All heavy atoms | 0.76±0.13 |
| Ramachandran plot summary for well-defined residues† averaged over the NMR ensemble | |
| Favoured | 95.1% |
| Allowed | 4.6% |
| Outliers | 0.3% |
| All atom Clashscore | 6 |
| Side chain outliers | 3% |
| Accession numbers associated with this structure | |
| BMRB ID | 28032 |
| PDB ID | 6L7Z |
Well-defined core protein residues: 3–27, 32–57, 68–117
Structural quality was evaluated by validation pipeline (wwPDB-VP:2.13)
Figure 4: Solution NMR structure of BTNL2 IgV domain.
A. Overlay of 10 lowest energy structures of BTNL2 determined by solution NMR spectroscopy. Loops are colored in grey; the beta strands are colored in teal and green, respectively. The helical turns are in blue. B. Cartoon representation of the lowest energy structure of BTNL2. The strands are labelled following standard nomenclature. C. Topology diagram of BTNL2. The front face comprising of A’, G, F, C and C’ strands is colored in teal, while the back face, comprising of A, B, E and D strands is colored in green. The canonical disulfide bond bridging the B and F strands is in yellow.
The mouse IgV domain of BTNL2 adopts a beta sandwich fold consisting of nine beta strands and two short helical turns (Fig. 4B). The canonical beta sandwich topology of the immunoglobulin-fold is formed by A, B, E and D strands present on the back sheet and A′, C, C′, F and G strands present on the front sheet. In case of BTNL2, the A strand splits into A and A′ strands; while the A strand remains with the back sheet, the A’ strand get included in the front sheet of BTNL2. The two beta-sheets are bridged by a canonical disulfide bond formed between Cys24 on the B strand and Cys98 on the F strand (Fig. 4C).
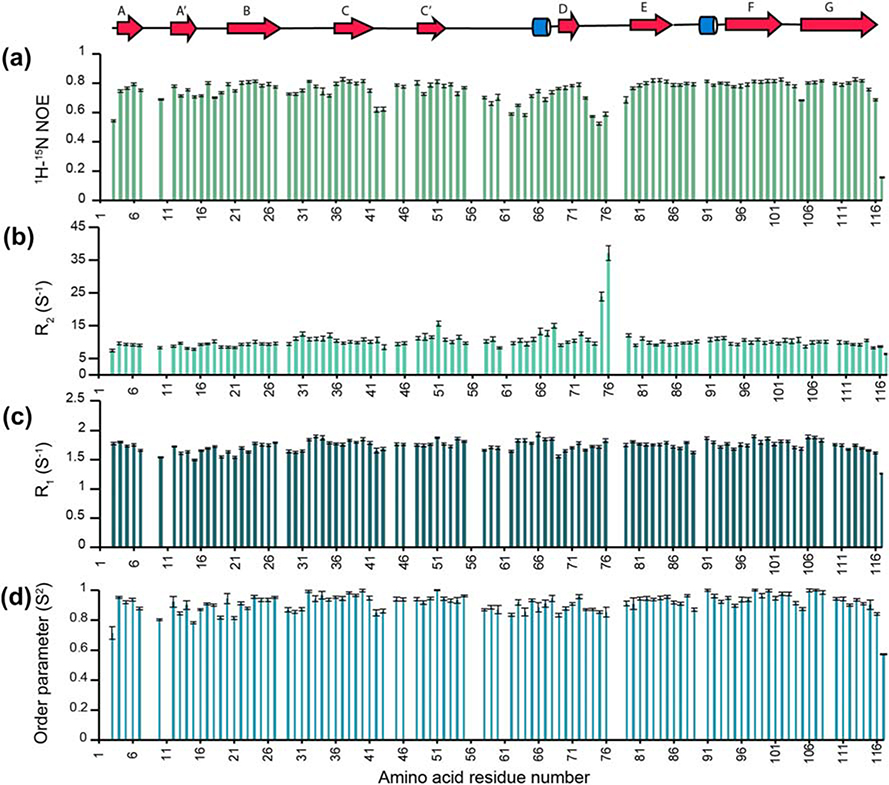
15N backbone dynamic studies reveal that BTNL2 is a rigid domain with a few flexible loops
The global hydrodynamic and local dynamic properties of BTNL2 were determined using the amide 15N R1, R2, and heteronuclear NOE relaxation experiments at 25°C (Fig. 5). Heteronuclear {1H}–15N NOE values are very sensitive to the fast-internal backbone motions in the picosecond to nanosecond timescale. These values are approximately + 0.8 for rigid 15N–1HN bond vectors in a folded domain and rapidly decrease to – 4.0 for disordered regions with high mobility (28). The heteronuclear NOE values of residues in the beta strands are ~ 0.78 ± 0.05, indicating that they are well ordered on this fast timescale (Fig. 5A). In contrast, loop regions show NOE values as low as ~ 0.5, indicative of enhanced fast backbone dynamics. These observations are consistent with increased RMSDs of the loop regions in the structural ensembles indicating increased flexibility of these regions (Fig. 4A).
Figure 5: Backbone dynamics of BTNL2 IgV domain.
Heteronuclear {1H}−15N NOE (A), amide 15N R2 (B) and amide 15N R1 (C) for BTNL2 indicate that the protein has an overall rigid domain. Missing data correspond to prolines, residues with overlapping signals, unassigned residues or residues not adequately fit during data analysis. D. Order parameter from the above data has been determined using TENSOR2.
Using the R2/R1 ratios for rigid residues, the isotropic tumbling correlation time τc for BTNL2 was calculated to be 6.79 ± 0.02 ns. This global τc value is consistent with the molecular mass of the protein and confirms that it is indeed monomeric under these experimental conditions, which is also consistent with our SEC and AUC studies. The 15N relaxation data were fit according to isotropic model-free formalism (29). The amides in β-strands are well-ordered, whereas those at the termini and in loop regions are more flexible. Overall, the dynamics studies show that BTNL2 is a monomer in solution and has a rigid structure.
Discussion
Butyrophilins, that include both butyrophilin (BTN) and butyrophilin-like (BTNL) families, are single-pass type-1 membrane glycoproteins and are considered to be related to the B7 family of costimulatory receptors and ligands (20). Especially, members of BTNL family cluster well with B7 family of ligands such as B7–1, B7–2, B7-H3, B7-H4, as well as PD-L1 and PD-L2 (Fig. S2). While the majority of B7 ligands have been crystallized, either alone or in complex with their respective binding partner, and their structure-function relations are well established, our solution NMR structure of BTNL2 is the first representative structure of BTNL family.
The membrane distal IgV domain of BTNL2 was expressed as inclusion bodies, refolded and purified. Both SEC and AUC studies suggest that BTNL2 exists as stable monomeric population in solution even in a very high concentration, which makes it an ideal candidate for NMR studies. Since this protein has been refolded in vitro, a series of cell binding experiments have been performed to assess functionality. Splenic B cells, and in vitro activated CD4+ and CD8+ T cells are used to demonstrate the binding of BTNL2 tetramer with its putative binding partner that has been reported to be present on the surface of these cells. Apart from showing that the N-terminal ectodomain of BTNL2 is functional, the results of these cell binding assays indicate that BTNL2 tetramer, produced inexpensively from bacterial over-expression system, can be exploited for large scale experiments to identify binding partner(s) of BTNL2. Moreover, these results confirm that BTNL2 receptors are expressed in all of these three cell types. Furthermore, the ability of the monomeric IgV domain of BTNL2 to inhibit IL-2 secretion from in vitro activated CD4+ T cells is indicative of the importance of this domain in the interaction of the BTNL2 protein with its putative receptor. Such data, along with our structural studies of the IgV domain of BTNL2 can provide important insights on its binding partner as well.
Our solution NMR studies reveal that the membrane-distal N-terminal domain of BTNL2 adopts a canonical beta sandwich topology of the immunoglobulin folds where the G, F, C, C′ and A′ strands constitute the front sheet and the A, B, E and D strands constitute the back sheet of the IgV domain (Fig. 4B). Cys24 present on B strand and Cys98 on F strand are engaged in the formation of a disulfide bond, a characteristic feature of an Ig fold which helps to hold these two beta sheets together. It is interesting to observe that positively and negatively charged residues cluster on the protein surface. The front face contains largely positively charged residues whereas the back face has largely negative charged residues. A stretch of arginine residues is present on the front face of the IgV domain. Arg4 on A strand, Arg38 on C strand, Arg52 on C′ strand, Arg99 on G strand and Arg107 on the F strand form this unique stretch of positively charged residues (Fig. S3B). Interestingly, Arg4, Arg38 and Arg52 in this stretch of arginine residues are conserved across species (Fig. S3A).
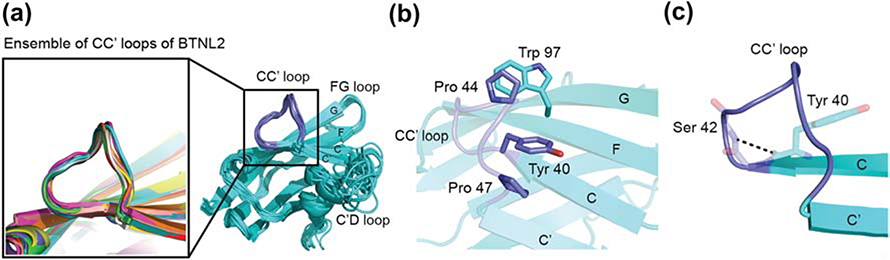
Another intriguing feature of the BTNL2 structure is the presence of a non-canonical conformation of the CC′ loop. The CC′ loop is partially peeled off, away from the hydrophobic core of the IgV domain and is closer to the FG loop. In contrast, in a large number of IgV structures in related families, the CC′ loop packs with the hydrophobic core of the domain. In all 10 models of the BTNL2 structure, the CC′ loop adopts this rigid non-canonical conformation (Fig. 6A). This relative rigidity of the CC’ loop is also evident from the 15N-NOE data (Fig. 5A), which shows the central loop residues D45 and M46 with NOE values of ~0.8, similar to the rigid residues in the protein. In comparison, multiple residues in the C’D and DE loops have NOE values as low as 0.6, thereby indicating their flexible nature (Fig 5). The CC’ loop is rich in prolines and its conformation appears to be stabilized by CH/π interactions between Pro44 (CC′ loop) and Trp97 (F strand) and also between Tyr40 (C strand) and Pro47 (CC′ loop) (30, 31) (Fig. 6B). All these residues are completely conserved in BTNL2 (Fig. S3A). The four hydrophobic residues Tyr40, Pro44, Pro47 and Trp97 on the surface exposed region of the CC′ loop form a small hydrophobic patch, which might also provide additional stability for this non-canonical CC′ loop conformation. The loop is further stabilized by a potential hydrogen bond between Tyr40 and Ser42 (Fig. 6C), which are also conserved residues (Fig. S3A). Taken together, all of these interactions most likely stabilize the rigid non-canonical conformation of the CC′ loop in BTNL2. However, it should be noted that at this point we do not have functional data to explain the biological significance of this unique CC’ loop conformation.
Figure 6: Non-canonical conformation of the CC′ loop of BTNL2.
A. Lowest energy 10 structures of BTNL2 are shown in cartoon (cyan). Inset illustrates the rigid conformation of the CC′ loop observed in all the structures. For comparison, the variability in conformation for the C′D loop is shown. B. Pro44 (sticks, blue) on the CC′ loop potentially forms CH-π interaction with Trp97 (sticks, cyan) present on the F strand. Similarly, Tyr40 and Pro47 on the CC′ loop also interact in a similar fashion to stabilize and maintain the rigid non-canonical conformation of the CC′ loop. C. Potential hydrogen bond between Tyr40 and Ser42 is shown. This main-chain to main-chain H bond helps supplement the non-canonical CC′ loop conformation by lifting up the loop and causing it to peel away from the sheet surface.
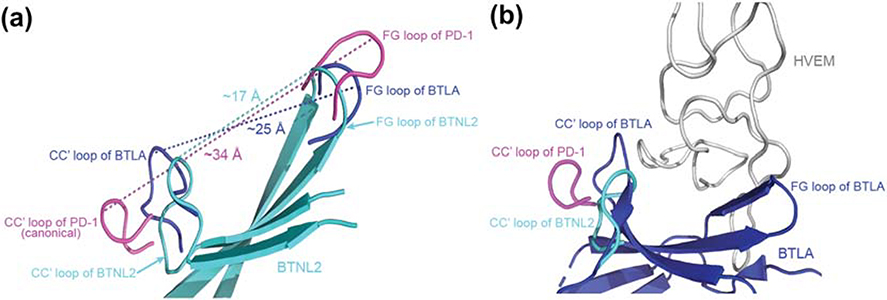
Most IgV domains bind their receptors via the front-sheet (32, 33). In mBTNL2, this putative interaction interface has a continuous stretch of positively charged arginine residues and also has significantly reduced surface area for binding to its presently unknown receptor, due to the non-canonical conformation of the CC′ loop. The distance between the CC′ loop and the FG loop in the canonical IgV domain of the immune checkpoint receptor PD-1 is ~34Å, whereas in mBTNL2 these loops are separated by a distance of ~ 17Å, thereby significantly reducing the available surface area of the front-sheet of mBTNL2 (Fig. 7A). This non-canonical conformation of the CC′ loop of BTNL2 is similar to that observed in immune checkpoint receptors like CD160 in the CD160:HVEM complex (PDB: 6NG3) and BTLA in the BTLA:HVEM complex (PDB: 2AW2), where a similar non-canonical conformation of CC′ loop is stabilized by a non-canonical disulfide linkage (34, 35). The binding partner, HVEM, is a TNF superfamily member and is significantly narrower than a typical IgV domain, thus being able to fit the smaller binding surface of the partner proteins. The distance between the CC′ loop and FG loop of BTLA is ~25Å, similar to that of mBTNL2 (Fig. 7A). The CC′ loop of BTLA and CD160 restricts the interaction of these proteins with partner molecules whose interacting domains are significantly narrower than IgV domain (Fig. 7B). Moreover, mutations in the CC′ loop of CD160 have been shown to substantially reduce the binding affinity with HVEM (34). Hence, it can be postulated that the non-canonical conformation of the CC′ loop of mBTNL2 could prevent its interaction with other IgV domains. This also indicates that the binding partner of mBTNL2, if it interacts with the front face of BTNL2, would possess a structural domain narrower than a canonical IgV domain. It should be noted here that this hypothesis is based on the structural data and needs to be explored further through functional assays.
Figure 7: Non-canonical CC′ loop reduces the surface area of the front face of BTNL2.
A. Superimposed structures of BTNL2 (cyan), BTLA (blue) and PD-1 (magenta) depicts distances between their respective CC′ and FG loops. The CC′ and the FG loops (magenta) of PD-1 (a canonical IgV domain) flanks the front face binding interface and is separated by a distance of ~34Å. Contrastingly, the CC′ and FG loops (blue) of BTLA which binds to HVEM (a TNF receptor superfamily member) is separated by a distance of ~25Å. The CC′ and FG loops (cyan) of BTLN2 is even closer and is separated only by ~17Å. B. The CC′ loop of PD-1 flanks the interaction interface area of PD-1 which interacts with a wider IgV domain, whereas the CC′ loop of BTLA (blue) flanks the front face of BTLA, which in turn contributes to the binding of relatively narrow HVEM (grey). The CC′ loop of BTNL2 (cyan) appears to assume a similar conformation to that of BTLA, which could hint at a non IgSF binding partner whose interacting domain is of a similar size or even lower than HVEM. PDB files used are 2AW2 for BTLA and 3RRQ for PD-1.
Although this is the first structural report of any of the BTNL family members, the structure of BTN3A1, a member of BTN family that modulates γδ T cell function was reported earlier (36). Interestingly, it has been suggested that a positively charged pocket present on the IgV domain of BTN3A1 can bind to phosphorylated antigens (36), although later studies show that instead, it is the B30.2 domain located in the cytoplasmic region of BTN3A1 which interacts with the phosphoantigens (37, 38). Notably, a positively charged stretch also exists on the front sheet of the N-terminal IgV domain of BTNL2, which incidentally, lacks the B30.2 domain. Although, a number of studies highlight the functional relevance of BTNL family members especially for BTNL2, the binding partner of BTNL2 remains to be identified. The current study provides thorough structural insights of a functionally active BTNL2 through solution NMR studies that could help us to identify its presently unknown binding partner.
Materials and methods
Cloning, expression, refolding and purification of BTNL2
A gene construct corresponding to the extracellular N-terminal IgV domain (residues Asp28 to Ala143) of mouse BTNL2 (Uniprot ID: O70355) with an additional N-terminal methionine was cloned into pET3a vector and transformed into E. Coli BL21 (λDE3) strain. Transformed bacteria were grown in Luria Bertani broth (Miller) at 37°C till OD600 of 1.0, induced with 1mM IPTG and harvested after five hours. The cell pellet was resuspended in lysis buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 20% sucrose and freshly prepared 10 mM dithiotreitol (DTT), sonicated and centrifuged at 16000×g for 15 minutes. Since BTNL2 is over-expressed as inclusion bodies, the pellet was collected and washed vigorously with buffer containing 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and 10 mM DTT. It was then centrifuged at 16000×g for 15 minutes at 25° C. The supernatant was discarded and washed two more times. The final pellet was homogenized with wash buffer devoid of Triton X-100 and centrifuged as before. The resulting washed inclusion bodies were dissolved in buffer containing 6 M guanidine hydrochloride, 10 mM sodium acetate, 5 mM EDTA and 1 mM DTT. This was then centrifuged at 16000×g for 15 minutes at 4° C and the supernatant containing solubilized BTNL2 was collected.
The solubilized protein was refolded by fast dilution method as described earlier (39, 40). Briefly, solubilized protein was injected in refolding buffer containing 400 mM arginine monohydrochloride, 100 mM Tris-HCl (pH 8.0), 1 mM EDTA, 5 mM reduced glutathione and 0.5 mM oxidized glutathione at 4 °C with constant stirring. The refolded protein was concentrated and exchanged to final working buffer comprising of 10 mM Tris-HCl (pH 7.4) and 150 mM NaCl using Amicon™ stirred cell. The resulting solution was then injected into a HiLoad™ 16/600 Superdex™ 200 size exclusion chromatography (SEC) column (GE Healthcare) pre-equilibrated with buffer containing 10 mM Tris-HCl (pH 7.4) and 150 mM NaCl. The fractions were collected and analyzed for purity by SDS-PAGE.
Analytical ultracentrifugation sedimentation velocity studies
Sedimentation velocity studies of BTNL2 were carried out at 20° C in a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge. The epoxy double-sector centerpiece was filled with 400μl of sample buffer and 380 μl of BTNL2 having an A280 value of 0.7, respectively. The protein was centrifuged at 40,000 rpm and A280 scans were acquired at an interval of three minutes till sedimentation was completed (~28 hours). The viscosity and density of the sample buffer, as well as the partial specific volume of the protein were determined using SEDNTERP and the sedimentation data was analyzed using SEDFIT (41). Final figures were made using GUSSI (42).
Sample preparation for NMR studies
For NMR studies, E.Coli BL21(λDE3) transformed with pET3a that contains BTNL2D28-A143 gene was grown in M9-minimal media containing 3g/L 13C glucose and 1g/L 15N ammonium chloride, as sole carbon and nitrogen source, respectively. The expression and purification steps were similar to unlabeled samples as described earlier. Final samples had 0.6 mM protein in 500μL of 20 mM sodium phosphate at pH 6.5, 50 mM NaCl and 7% D2O for spin lock. For the long-term stability of the samples, 0.4 mM phenylmethylsulfonyl fluoride (PMSF) and 0.04% NaN3 were also added.
Cloning, expression and purification of BTNL2 with a C-terminal AviTag™
The gene corresponding to BTNL2D28-A143 was cloned into pNIC28-Bsa4 vector with appropriate primers in order to code for an additional C-terminal AviTag™. The AviTag™ comprises of a 15 amino acid sequence GLNDIFEAQKIEWHE to which biotin can be conjugated in an ATP dependent manner by biotin ligase enzyme BirA (43, 44). This BTNL2-AviTag construct was over-expressed in E. Coli BL21 (λDE3) as inclusion bodies, refolded in vitro and purified using the same protocol as described.
Preparation of BTNL2 tetramer for functional studies
BTNL2-AviTag was incubated with biotin ligase BirA at 25°C for 3 hours in buffer containing 20mM Tris-HCl (pH 7.4), 150 mM NaCl, 3mg/ml ATP, 7.5 mM MgCl2 and 50μM D-biotin. The biotinylated protein was purified by SEC and added to streptavidin in a 4:1 stoichiometric ratio. This was incubated at room temperature for 15 minutes and the efficacy of tetramer formation was assessed by SEC. For flow cytometry-based experiments, biotinylated BTNL2 was incubated in a 4:1 stoichiometric ratio with streptavidin conjugated with phycoerythrin (PE-streptavidin).
Animals
BALB/c mice were procured from National Institute of Nutrition, Hyderabad, India and were maintained in the central animal research and housing facility, IIT Kharagpur. All experiments involving mice were carried out in compliance with guidelines established by the Institutional Animal Ethical Committee, IIT Kharagpur.
Flow cytometry-based cell binding assay
All flow cytometric experiments were carried out in a BD LSR Fortessa™ instrument and analyses were done using FlowJo™ software (for Windows), version 10.6.0. Ashland, OR: Becton, Dickinson and Company; 2019. In order to establish the functionality of refolded and purified IgV domain BTNL2, splenocytes were isolated from healthy BALB/c mice and stained with B cell specific anti-CD19 antibody (BD Biosciences™) prior to incubation with PE-conjugated BTNL2 tetramer or biotinylated streptavidin-PE to account for non-specific binding. To further check the ability of the BTNL2 tetramer to bind to its putative receptor on in vitro activated CD4+ and CD8+ cells, splenocytes were isolated from BALB/c mice and cultured with or without 10 ng/ml PMA and 500 ng/ml ionomycin for 24 hours. The cells were collected and stained with fluorophore tagged anti-CD3, anti-CD4 and anti-CD8 antibodies (BD Biosciences™), followed by incubation with PE-conjugated BTNL2 tetramer (5μg/ml) for 20 minutes. Cells were then washed, fixed and analyzed by flow cytometry for binding of PE-conjugated BTNL2 tetramer on CD4+ or CD8+ cells.
IL-2 ELISA from in-vitro activated T cells
CD4+ T cells were isolated by magnetic separation using MACS CD4+ T cell Isolation kit, (Miltenyi Biotec Inc.). The isolated CD4+ T cells were stimulated with 2μg/ml plate-bound purified anti-mouse CD3e antibody (clone: 145–2C11; BD Pharmingen) antibody and cultured in presence or absence of immobilized BTNL2 IgV monomer (20μg/ml) for 24 hours and 56 hours. The supernatant were collected and analyzed for IL-2 secretion by ELISA using manufacturer’s instructions (mouse IL-2 Duoset ELISA kit, R & D Systems).
Chemical shift assignment and structure calculation
All NMR experiments were carried out at 25° C in a cryoprobe equipped Bruker Avance III 600 MHz spectrometer. Spectra were processed using NMRPipe (45) and analyzed by NMRFAM-Sparky (46). Signals from the backbone 1H, 15N and 13C nuclei of BTNL2 were sequentially assigned by analyzing a set of triple resonance experiments CBCA(CO)NH, HNCACB, HN(CA)CO, HNCO and HN(CA)NNH. Aliphatic side chain 1H and 13C nuclei chemical shifts were assigned using HCCH-TOCSY, HBHA(CO)NH, H(CC)(CO)NH-TOCSY, (H)CC(CO)NH-TOCSY and 15N-HSQC-TOCSY experiments (47). For distance restraints, 15N and 13C edited NOESY experiments with a mixing time of 110ms were collected. The chemical shifts of the backbone nuclei were used to determine the secondary structure propensity using MICS (23).
The solution structure of BTNL2 was determined using PONDEROSA-C/S suite (26) in conjunction with Xplor-NIH (27). Well-resolved NOE crosspeaks were assigned manually, followed by automated assignment using AUDANA (48). Structure calculations were performed in an iterative manner. At the initial stages, each cycle yielded 40 structures of which the 20 lowest energy structures were used as the basis for further spectral analysis and structure calculations. Once sufficient number of distance restraints were acquired, explicit water refinement was done to determine the final 100 structures. The best 10 structures having lowest energy were deposited in RCSB with PDB Id 6L7Z. The associated chemical shifts data have been deposited in BioMagResBank with accession number 28032.
Backbone amide 15N relaxation
Backbone amide 15N relaxation data i.e. R1, R2 and heteronuclear NOE, were acquired at 25 °C. Spectra for R1 (50, 100, 150, 200, 400, 600, 900, 1200 ms) and R2 (25, 50, 75, 100, 125, 150, 200, 250, 300, 400 ms) time series were collected in random sequence for each set. The relaxation rate constants for each residue were obtained by fitting peak heights to an exponential decay using in-house scripts. The residue wise heteronuclear {1H}–15N NOE values was determined from the ratio of peak heights versus a reference spectrum devoid of proton saturation. The backbone relaxation data and structural coordinates of BTNL2 were used as input in the program TENSOR2 (49) in order to determine the order parameter (S2) and global rotational correlation time (τC) by Lipari Szabo model free formalism.
Accession Numbers
The solution structure ensemble of BTNL2 comprising of best 10 structures having lowest energy has been deposited in RCSB with PDB Id 6L7Z. The associated chemical shifts data have been deposited in BioMagResBank with accession number 28032.
Supplementary Material
Research highlights.
BTNL2, a T cell inhibitory molecule regulates immune function in several diseases
This study shows BTNL2 N-terminal IgV domain binds to its putative receptor
This domain reduces IL2 production from in vitro activated CD4 T cells
The 3-D structure of this domain is solved here by solution NMR spectroscopy
This study provides important insights into the inhibitory function of BTNL2
Acknowledgements
The authors wish to acknowledge Dr. Udupi A. Ramagopal, Veda Sheersh Boorla, Bidisha Acharya and Saptarshi Banerjee for helpful discussions throughout the course of this work. The authors would like to thank the Central Research Facility (CRF) at IIT Kharagpur for the use of NMR and AUC facilities.
Funding and Additional information
This work has been supported by Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India: ECR/2016/000847 to SD and ECR/2016/000923 to DS; and Department of Biotechnology, Government of India: BT/PR26821/MED/30/1938/2017 to GM and DS. WL and computational resources at NMRFAM have been supported by the National Institutes of Health (NIH) grant: P41GM103399 and the National Science Foundation: DBI 1902076. AJB, SM, DM and KD acknowledge IIT Kharagpur for fellowship. AH acknowledges CSIR, Government of India, for fellowship: 09/081(1323)/2018-EMR-I. SVK acknowledges KSTePS, DST, Govt. of Karnataka, India for fellowship. All animal experiments were performed according to the Institutional Animal Ethical Committee guidelines and approval by CPCSEA (IE-3/GM-SMST/3.18).
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharpe AH & Abbas AK (2006). T-cell costimulation — biology, therapeutic potential, and challenges. N. Engl. J. Med 355, 973–975. [DOI] [PubMed] [Google Scholar]
- 2.Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB & Bluestone JA (1994). CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1, 406–413. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz JCD, Zhang X, Nathenson SG & Almo SC (2002). Structural mechanisms of costimulation. Nat. Immunol 3, 427–434. [DOI] [PubMed] [Google Scholar]
- 4.Esensten JH, Helou YA, Chopra G, Weiss A & Bluestone JA (2016). CD28 costimulation: from mechanism to therapy. Immunity. 44, 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krummel MF & Allison JP (1995). CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med 182, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharpe AH & Pauken KE (2018). The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol 18, 153–167. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen T, Liu XK, Zhang Y & Dong C (2006). BTNL2, a Butyrophilin-like molecule that functions to inhibit T cell activation. J. Immunol 176, 7354–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson RM, Gavin MA, Escobar SS, Rottman JB, Lipsky BP, Dube S, Li L, Bigler J, Wolfson M, Arnett HA & Viney JL (2013). Butyrophilin-like 2 modulates B7 costimulation to induce Foxp3 expression and regulatory T cell development in mature T cells. J. Immunol 190, 2027–2035. [DOI] [PubMed] [Google Scholar]
- 9.Arnett HA, Escobar SS, Gonzalez-Suarez E, Budelsky AL, Steffen LA, Boiani N, Zhang M, Siu G, Brewer AW & Viney JL (2007). BTNL2, a butyrophilin/B7-like molecule, is a negative costimulatory molecule modulated in intestinal inflammation. J. Immunol 178, 1523–1533. [DOI] [PubMed] [Google Scholar]
- 10.Cui C, Tian X, Lin Y, Su M, Chen Q, Wang SY & Lai L (2019). In vivo administration of recombinant BTNL2-Fc fusion protein ameliorates graft-versus-host disease in mice. Cell. Immunol 335, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian X, Lin Y, Cui C, Su M & Lai L (2019). BTNL2-Ig protein attenuates type 1 diabetes in non-obese diabetic (NOD) mice. Adv. Healthc. Mater 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramaniam KS, Spaulding E, Ivan E, Mutimura E, Kim RS, Liu X, Dong C, Feintuch CM, Zhang X, Anastos K, Lauvau G & Daily JP (2015). The T-cell inhibitory molecule Butyrophilin- like 2 is up-regulated in mild Plasmodium falciparum infection and is protective during experimental cerebral malaria. J. Infect. Dis 212, 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, Koch A, Lengauer T, Seegert D, Reiling N, Ehlers S, Schwinger E, Platzer M, Krawczak M, Muller-Quernheim J, Schurmann M & Schreiber S (2005). Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat. Genet 37, 357–364. [DOI] [PubMed] [Google Scholar]
- 14.Delaveri A, Rapti A, Poulou M, Fylaktou E, Tsipi M, Roussos C, Makrythanasis P, Kanavakis E & Tzetis M (2014). BTNL2 gene SNPs as a contributing factor to sarcoidosis pathogenesis in a cohort of Greek patients. Meta Gene. 2, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Wei J, Fan L & Cheng D (2015). BTNL2 gene polymorphism and sarcoidosis susceptibility: A meta-analysis. PLoS One. 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun W, Min H & Zhao L (2019). Association of BTNL2 single nucleotide polymorphisms with knee osteoarthritis susceptibility. Int. J. Clin. Exp. Pathol 12, 3921–3927. [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng L, Zhao R, Jin Z, Ren K, Deng C & Yu S (2015). Association of genetic polymorphisms on BTNL2 with susceptibility to and prognosis of dilated cardiomyopathy in a Chinese population. Int. J. Clin. Exp. Pathol 8, 10488–10499. [PMC free article] [PubMed] [Google Scholar]
- 18.Pathan S, Gowdy RE, Cooney R, Beckly JB, Hancock L, Guo C, Barrett JC, Morris A & Jewell DP (2009). Confirmation of the novel association at the BTNL2 locus with ulcerative colitis. Tissue Antigens. 74, 322–329. [DOI] [PubMed] [Google Scholar]
- 19.FitzGerald LM, Kumar A, Boyle EA, Zhang Y, McIntosh LM, Kolb S, Stott-miller M, Smith T, Karyadi DM, Ostrander EA, Hsu L, Shendure J & Stanford JL (2013). Germline missense variants in the BTNL2 gene are associated with prostate cancer susceptibility. Cancer. Epidemiol. Biomarkers. Prev 22, 1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes DA, Reith W & Trowsdale J (2016). Regulation of immunity by butyrophilins. Annu. Rev. Immunol 34, 151–172. [DOI] [PubMed] [Google Scholar]
- 21.Erickson HP (2009). Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biological Procedures Online. 11, 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekdemir A & Stellacci F (2016). A centrifugation-based physicochemical characterization method for the interaction between proteins and nanoparticles. Nat. Commun 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y & Bax A (2012). Identification of helix capping and β-turn motifs from NMR chemical shifts. J. Biomol. NMR 52, 211–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bork P, Holm L & Sander C (1994). The immunoglobulin fold structural classification, sequence patterns and common core. J. Mol. Biol 242, 309–320. [DOI] [PubMed] [Google Scholar]
- 25.Sharma D & Rajarathnam K (2000). 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR 18, 165–171. [DOI] [PubMed] [Google Scholar]
- 26.Lee W, Stark JL & Markley JL (2014). PONDEROSA-C / S: client – server based software package for automated protein 3D structure determination. J. Biomol. NMR 60, 73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwieters CD, Kuszewski JJ, Tjandra N & Clore GM (2003). The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson 160, 65–73. [DOI] [PubMed] [Google Scholar]
- 28.Maiti S, Acharya B, Boorla VS, Manna B, Ghosh A & De S (2019). Dynamic studies on intrinsically disordered regions of two paralogous transcription factors reveal rigid segments with important biological functions. J. Mol. Biol 431, 1353–1369. [DOI] [PubMed] [Google Scholar]
- 29.Lipari G & Szabo A (1982). Model-Free approach to the interpretation of nuclear magnetic Resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc 104, 4546–4559. [Google Scholar]
- 30.Brandl M, Weiss MS, Jabs A, Sühnel J & Hilgenfeld R (2001). C-H···π-interactions in proteins. J. Mol. Biol 307, 357–377. [DOI] [PubMed] [Google Scholar]
- 31.Plevin MJ, Bryce DL & Boisbouvier J (2010). Direct detection of CH / π interactions in proteins. Nat. Chem 2, 466–471. [DOI] [PubMed] [Google Scholar]
- 32.Chattopadhyay K, Lazar-Molnar E, Yan Q, Rubinstein R, Zhan C, Vigdorovich V, Ramagopal UA, Bonanno J, Nathenson SG &, Almo SC (2009). Sequence, structure, function, immunity: structural genomics of costimulation. Immunol. Rev 229, 356–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundapura SV & Ramagopal UA (2019). The CC ′ loop of IgV domains of the immune checkpoint receptors, plays a key role in receptor : ligand affinity modulation. Sci. Rep 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Garrett SC, Fedorov EV, Ramagopal UA, Garforth SJ, Bonanno JB & Almo SC (2019). Structural Basis of CD160 : HVEM Recognition. Structure. 27, 1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Compaan DM, Gonzalez LC, Tom I, Loyet KM, Eaton D, and Hymowitz SG (2005). Attenuating lymphocyte activity: The crystal structure of the BTLA-HVEM complex. J. Biol. Chem 280, 39553–39561. [DOI] [PubMed] [Google Scholar]
- 36.Vavassori S, Kumar A, Wan GS, Ramanjaneyulu GS, Cavallari M, Daker SE, Beddoe T, Theodossis A, Williams NK, Gostick E, Price DA, Soudamini DU, Voon KK, Olivo M, Rossjohn J, Mori L & Libero GD (2013). Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nat. Immunol 14, 908–918. [DOI] [PubMed] [Google Scholar]
- 37.Salim M, Knowles TJ, Baker AT, Davey MS, Jeeves M, Sridhar P, Wilkie J, Willcox CR, Kadri H, Taher TE, Vantourout P, Hayday A, Mehellou Y, Mohammed F & Willcox BE (2017). BTN3A1 discriminates γδ T cell phosphoantigens from nonantigenic small molecules via a conformational sensor in its B30.2 domain. ACS Chem. Biol 12, 2631–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandstrom A, Peigné CM, Léger A, Crooks JE, Konczak F, Gesnel MC, Breathnach R, Bonneville M, Scotet E & Adams EJ (2014). The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vγ9Vδ2 T cells. Immunity. 40, 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Schwartz JCD, Almo SC & Nathenson SG (2002). Expression, refolding, purification, molecular characterization, crystallization, and preliminary X-ray analysis of the receptor binding domain of human B7–2. Protein Expr. Purif 25, 105–113. [DOI] [PubMed] [Google Scholar]
- 40.Samanta D, Ramagopal UA, Rubinstein R, Vigdorovich V & Nathenson SG (2012). Structure of Nectin-2 reveals determinants of homophilic and heterophilic interactions that control cell – cell adhesion. Proc. Natl. Acad. Sci. USA 109, 14836–14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophysical. J. 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brautigam CA (2015). Calculations and publication-quality illustrations for analytical ultracentrifugation data. Method. Enzymol 562, 109–133. [DOI] [PubMed] [Google Scholar]
- 43.Cull MG & Schatz PJ (2000). Biotinylation of proteins in vivo and in vitro using small peptide tags. Method. Enzymol 326, 430–440. [DOI] [PubMed] [Google Scholar]
- 44.Samanta D, Mukherjee G, Ramagopal UA, Chaparro RJ, Nathenson SG, DiLorenzo TP & Almo SC (2011). Structural and functional characterization of a single-chain peptide – MHC molecule that modulates both naive and activated CD8+ T cells. Proc. Natl. Acad. Sci. USA. 108, 13682–13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J & Bax A (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 46.Lee W, Tonelli M & Markley JL (2015). Structural bioinformatics NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sattler M, Schleucher J & Griesinger C (1999). Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. nucl. mag. res. sp 34, 93–158. [Google Scholar]
- 48.Lee W, Petit CM, Cornilescu G, Stark JL & Markley JL (2016). The AUDANA algorithm for automated protein 3D structure determination from NMR NOE data. J. Biomol. NMR 65, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dosset P, Hus JC, Blackledge M & Marion D (2000). Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.