Abstract
Background
Male infertility is currently one of the most common problems faced by couples worldwide. We performed a GWAS on Greek population and gathered statistically significant SNPs in order to investigate whether they lie within or near lncRNA regions.
Objectives
The aim of this study was to investigate whether polymorphisms on or near lncRNAs affect interactions with miRNAs and can cause male infertility.
Materials and methods
In the present study, a GWAS was conducted, using samples from 159 individuals (83 normozoospermic individuals and 76 patients of known fertility issues). Standard procedures for quality controls and association testing were followed, based on case-control testing.
Results
We detected six lncRNAs (LINC02231, LINC00347, LINC02134, NCRNA00157, LINC02493, Lnc-CASK-1) that are associated with male infertility through their interaction with miRNAs. Furthermore, we identified the genes targeted by those miRNAs and highlighted their functions in spermatogenesis and the fertilization process.
Discussion and conclusion
lncRNAs are involved in spermatogenesis through their interaction with miRNAs. Thus, their study is very important, and it may contribute to the understanding of the molecular mechanisms underlying male infertility.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01937-w) contains supplementary material, which is available to authorized users.
Keywords: GWAS, Male infertility, Greek population, lincRNA
Introduction
Infertility is “a disease of the reproductive system defined by the failure to achieve a clinical pregnancy after 12 months or more of regular unprotected sexual intercourse” according to the World Health Organization [1] and a worldwide problem. Approximately one in six couples in the Western world are affected by infertility [2], which results in social and psychological impacts [3]. It is estimated that male factor is solely responsible for 30% of cases and co-contributes with female factor to 20%, resulting in a presence up to 50% of the total cases [4]. It is also estimated that 7% of the male population deals with infertility [5]; however, the statistics of male infertility do not reflect accurate rates in all countries due to cultural or religious peculiarities in some regions [6]. Male infertility is a complex disorder, and except for the genetic basis, including single-gene mutations and chromosomal abnormalities [5], environmental factors also contribute to the phenotype [7]. Many cases are also characterized as idiopathic, as no cause is identified [8]. Therefore, the identification of genetic factors, gene mutations, or polymorphisms, which are associated with idiopathic infertility, is of great importance for clinical application and for the discovery of new methods for diagnosis and treatment.
Transcriptomics analyses have identified, except for protein-coding, many non-coding transcripts [9–11]. Non-coding RNAs (ncRNAs) can be classified into two main categories according to their size: short non-coding RNAs (small ncRNAs), with a length shorter than 200 nucleotides, and long non-coding RNAs (lncRNAs), which are larger than 200 nucleotides but it is also possible to have a size of several kilobases [12]. MicroRNAs (miRNAs) are the most studied small non-coding RNAs. Non-coding RNAs are abundant in germ cells and play a critical role in the complex developmental process of spermatogenesis, which affects male infertility [13]. More specifically, stage-specific Dicer knock-out (KO) in mice led to infertility/subfertility in three different studies [14–16], indicating the important role of miRNA pathway in male fertility. Several studies that followed confirmed the specific role of miRNAs in mammalian meiosis, e.g., miR-21, miR-18a [17, 18]. PIWI-interacting RNAs (piRNAs) are another category of ncRNAs that are associated with PIWI proteins and they are expressed in germline cells, contributing to genome integrity [13].
However, lncRNAs constitute the largest proportion of ncRNAs [19]. Long intergenic non-coding RNAs (lincRNAs) are long RNAs of more than 200 nucleotides that lie within intergenic regions of the genome and do not overlap with any protein coding genes. LincRNAs take part in chromatin remodeling, gene regulation, and alteration of expression [20].
Moreover, lncRNAs regulate gene expression in several biological processes. Therefore, their dysregulation contributes to the development of several diseases [21]. The highest number of lncRNAs is expressed at brain and testis and many of them show testis-specific expression [22]. A recent study also suggests a key role of lncRNAs in reproductive capacity as lncRNAs and miRNAs escape meiotic sex chromosome inactivation (MSCI), an epigenetic process required for the transcriptional silencing of X- and Y-chromosomes during pachytene stage. The same study revealed a dynamic expression pattern of lncRNAs during spermatogenesis in mouse, too [23]. These studies have shed new light on the issue but the role of lncRNAs in male gametogenesis is still largely unexplored.
LncRNAs have been recognized to contribute to the development of several diseases [21, 24] through a variety of mechanisms, but an emerging field is the investigation of lncRNAs-miRNA interactions [25–27]. Recent reports suggest that lncRNAs may interact with other RNA classes, including miRNAs [28]. Male infertility might be also affected by miRNAs-lncRNA interactions as Lü et al. [29] showed that human spermatogenesis is regulated by a mechanism in which a lncRNA, NLC1-C, modulates miRNA expression at the transcriptional level. More specifically, there are several forms of interplay among lncRNAs and miRNAs. Mainly, miRNAs regulate the abundance of lncRNAs as they can decrease their stability and promote degradation. A similar effect on the levels of miRNAs is caused by the action of lncRNAs, as well as they exert a ‘sponge effect’ on miRNAs, control their concentration on cytoplasm, and sequester them away from their targets [25, 27].
LncRNAs can compete with miRNAs for binding to mRNAs or they can produce small RNAs and miRNAs, too [25]. However, our understanding of the impact of lncRNAs-miRNA networks on disease development is limited [24]. The mechanisms by which miRNAs and lncRNAs mediate complex processes like spermatogenesis and affect the occurrence and development of diseases like male infertility still need further exploration in order to provide new breakthroughs for clinical applications.
The aim of this study is to perform a genome-wide association analysis to identify genetic variants (single-nucleotide polymorphisms, SNPs) significantly related to male infertility, within or near lncRNA regions, by comparing normozoospermic (control group) and non-normozoospermic individuals (case group). The purposes were (a) to study and list SNPs associated with male infertility within or near lncRNAs and (b) to investigate the biological mechanism through which these SNPs act by understanding the role of these lncRNAs in spermatogenesis process and thus, in reproductive capacity and sperm quality, through their interactions with miRNAs using bioinformatics analysis. Identifying genomic regions, and preferably lncRNAs regions, responsible for genetic variation in male infertility will enhance the understanding of biological pathways involving this trait and contribute to a better understanding of the patient’s profile. This may point to opportunities for improving the prognosis and diagnosis of male infertility and even treatment.
Materials and methods
Patient selection and biological material
All semen samples used in this study were derived from individuals after they had given approved informed consent. The samples were collected in cooperation with the ‘Embryolab Fertility Clinic’ (Thessaloniki, Greece), after 3 to 4 days of abstinence, via masturbation. Individuals primarily from the Greek population were used in the study to ensure genetic homogeneity. According to Panoutsopoulou et al. [30], the Greek population is genetically homogenous, except for some minorities that are characterized as genetic isolates. None of the samples were collected from individuals from these isolates, since place of birth and relevant data are collected from individuals through the questionnaire which are required to fill in along with the consent form. All the individuals were also aged between 18 and 40 years, in order to ensure that increasing age does not contribute to reduced semen quality. Semen analysis was performed for all individuals enlisted in the study. The subjects were divided into two groups. The control group was defined as normozoospermic individuals (n = 92; volume > 1.5 mL, sperm concentration > 39 million/mL, total sperm number > 15 million, total motility > 40%, progressive motility > 32%, and viability > 58%) according to inclusion criteria of World Health Organization (2010). No samples exhibited normal sperm concentration but low total motile or low total count of spermatozoa. The case group consisted of non-normozoospermic individuals (n = 100). These were either oligozoospermic individuals (n = 18), with a total sperm number of less than 15 million/mL, or asthenozoospermic individuals with motility less than 40% (n = 17), teratozoospermic individuals with less than 4% normal morphology (n = 14), or combinations of the three categories (n = 51). None of the patients exhibited any fertility-related issues regarding varicocele or Y-chromosome microdeletions, in order to investigate for the existence of novel genetic markers beyond microdeletions, for the trait of male infertility.
DNA extraction
DNA was extracted from semen samples according to the protocol developed by Weyrich A. [31]. Briefly, semen has two basic components: spermatozoa and seminal fluid. Seminal fluid can affect the DNA quality due to its high concentration of fructose and proteins. Thus, ethanol was used to remove the seminal fluid and after that, cell lysis followed. Somatic cells were removed after incubation of the cell pellet with somatic cell lysis buffer SCLB (0.05% Triton and 0.05% SDS) for 30 min on ice. This detergent-based buffer ensures the rupture of somatic cells. The incubation was followed by centrifugation at 5000g for 8 min and subsequent removal of supernatant was performed. PBS was used to wash the pellet for any residual somatic cell contaminants. The pellet was then dissolved in lysis buffer (10 Mm Tris-HCl pH 8.0, 100 Mm NaCl, 10 Mm EDTA, 0.5% SDS Triton-X100 0.5%) that also contained DDT (0.1 M) to break the disulfide bonds of the lipidic membrane that protects spermatozoa, as well as proteinase K (100 mg/mL) and SDS to ensure cell lysis and protein destruction. Cell debris was removed after centrifugation and the supernatant was carefully collected. In this point, phenol-chloroform extraction was performed as an extra step, to fully separate nucleic acid from proteins and lipids and to obtain DNA of high quality and purity. The mixture of phenol-chloroform is immiscible with water, and as a result, two phases are created. The hydrophilic lipids are partitioned into the lower organic phase, the proteins remain at the interphase, and the nucleic acids are in the aqueous phase. Thus, the aqueous phase was collected, and the DNA was then precipitated with the use of ethanol, washed, and resuspended in ddH2O. Agarose gel electrophoresis was used to check the integrity of the extracted DNA and DNA concentration was assessed spectrophotometrically with Qubit 2.0 and subsequently adjusted to approximately 200 ng/mL. Purified DNA was stored at − 20 °C before use.
Genotyping
All the DNA samples were genotyped at the Human Genomics Facility (HuGe-F) of Erasmus MC (University Medical Centre Rotterdam, Netherlands) using the Illumina Infinium® Global Screening Array. This chip contains 642.824 single-nucleotide polymorphisms (SNPs) across the human genome. Moreover, 256.673 of these SNPs are found in intronic regions, thus providing a good representation of polymorphisms that are useful in order to study lncRNAs.
Quality control
The genotype data that were obtained, in .ped and .map files, were analyzed using PLINK [32], a software package with a wide range of functions, that is widely used for the analysis of GWAS data. All SNPs analyzed met the following quality control criteria: SNPs with a MAF ≤ 0.05 were excluded, individuals with more than 10% missing genotypes were excluded, and only SNPs with 90% genotyping rate were included in the study.
Association analysis
SNP association analysis was performed using Pearson’s Chi-square test. Chi-square test compares allele frequencies between case controls to test the null hypothesis that there is no association between each SNP and the phenotype of male infertility. A common strategy for the evaluation of individual SNPs involves the use of a genome-wide significance p value threshold of 5 × 10−8 under the assumption of independence among markers. In a GWAS, p -value provides statistical significance and is used as a safeguard against false positives. However, this threshold requires sample sizes of thousands and a sizable proportion of risk alleles are still being missed by this strategy. According to previous studies, it has been proved that the analysis of SNPs with a much higher significance threshold can provide important information if the GWAS is well-powered [33]. For this reason, odds ratios (ORs) were used in combination with p values to identify SNPs with the strongest effect on disease risk. Odds ratio measures the strength of association and is important in GWAS studies as many times despite their statistical significance, associations involving very low ORs that explain little about the genetic contribution to the disease. Taking all these into account, for the selection of SNPs, these thresholds were used: p value ≤ 0.05 and odds ratio > 1. After the odds ratio evaluation, all the SNPs that were selected had p -value ≤ 10−4.
Bioinformatics analyses
This study aimed at the correlation of lncRNAs with male infertility. Thus, all the SNPs were examined separately for genomic location (e.g., exonic, intronic, within a promoter or an enhancer, etc.) to identify SNPs that are located in regions that are transcribed into long non-coding RNAs via annotation using SNPnexus [34]. To assess the potential role of these lncRNAs to male infertility, the LNCipedia version 5.2 [35] was used. For each lncRNA, literature about the process of spermatogenesis or the general term of infertility was gathered. After that, we attempted to prove that some of these lncRNAs may have an impact on male infertility through their interactions with miRNAs. Thus, for a better understanding of the biological mechanism through which these SNPs act and for evaluating their functional impact, lncRNAs-miRNA interactions were also investigated using DIANA tools-LncBase Predicted v.2. [36], a database that provides information about in silico predicted miRNA targets on lncRNAs based on an algorithm. The threshold used for the search was 0.9, which is stringent. Then, the predicted miRNA targets were further studied to identify a potential role in male infertility or spermatogenesis process using miRBase [37]. miRNAs associated with the phenotype of interest, based on miRBase search results, were then investigated for their interaction networks with human mRNAs using miRTargetLinkHuman [38] to identify strong interactions and potential regulatory function of miRNAs with genes that have a role in the complex process of spermatogenesis and its regulation (GeneCards®: The Human Gene Database). Only experimentally validated networks were used.
Ethics statement
All the participants were informed about the study and they gave their consent in order to participate by filling out a questionnaire along with the consent form. Both the study and the consent procedure were approved by the ethics committee of the Medical Faculty of the University of Thessaly.
Results
GWAS: identification of SNPs within or near lncRNAs associated with male infertility
The role of lncRNAs in spermatogenesis process and male infertility is still largely unexplored. Thus, the purposes of this study were to identify and list SNPs significantly associated with male infertility within or near lncRNAs regions. For this reason, as described above, a total of 192 individuals were selected for genotyping using the Illumina Infinium® Global Screening Array, including 92 normozoospermic individuals (control group) and 100 non-normozoospermic individuals (case group). Genotyping revealed variance among 642,824 single-nucleotide polymorphisms (SNPs) across the human genome. Meanwhile, it was observed that some SNPs failed to be scored on at least 99% of all the individuals and had a MAF ≤ 0.05 in the whole dataset. Some individuals had also more than 1% missing genotypes. By excluding the SNPs and individuals described above, the remaining 308,495 SNPs and 159 individuals (control group, n = 76; case group, n = 83) were used for the GWAS analysis. The case group consisted of oligozoospermic, asthenozoospermic, teratozoospermic, and their combination individuals. A complete list of each individual’s phenotype is listed on Supplementary Table S3.
After association analysis was performed, 860 SNPs (Supplementary Table S1) were selected as significantly associated with male infertility because they satisfied our criteria for genome-wide significance (p value ≤ 0.05 and odds ratio > 1, “Materials and methods”). We should also note that after odds ratio evaluation, all the SNPs that were selected for further analysis had p value ≤ 10−4. The aim was to study SNPs associated with male infertility within or near lncRNAs; for this reason, the 860 SNPs associated with male infertility were examined via annotation for their genomic location using SNPnexus [34]. It was found that 92 SNPs were located in regions that lie in close proximity to lincRNA regions, as shown in Table 1.
Table 1.
SNPs associated with male infertility and located nearby lincRNA regions
| SNP | Chromosome | Position | Type of nearest upstream gene |
|---|---|---|---|
| rs7529824 | chr1 | 168500458 | lincRNA |
| rs1890734 | chr1 | 181108784 | lincRNA |
| rs76237371 | chr1 | 188773803 | lincRNA |
| rs12732389 | chr1 | 220563560 | lincRNA |
| rs10779404 | chr1 | 220568157 | lincRNA |
| rs6663920 | chr1 | 63554295 | lincRNA |
| rs72672087 | chr1 | 76531943 | lincRNA |
| rs17105542 | chr1 | 81336283 | lincRNA |
| rs6047591 | chr20 | 2223481 | lincRNA |
| rs6027995 | chr20 | 59623745 | lincRNA |
| rs2824534 | chr21 | 19259311 | lincRNA |
| rs16825349 | chr2 | 146425531 | lincRNA |
| rs816884 | chr2 | 151319753 | lincRNA |
| rs13405989 | chr2 | 15962207 | lincRNA |
| rs10495699 | chr2 | 19997343 | lincRNA |
| rs17574702 | chr2 | 220547930 | lincRNA |
| rs4674431 | chr2 | 220632013 | lincRNA |
| rs1424341 | chr2 | 222683948 | lincRNA |
| rs11892518 | chr2 | 5901174 | lincRNA |
| rs1517240 | chr3 | 152382636 | lincRNA |
| rs2729324 | chr3 | 2071931 | lincRNA |
| rs7648418 | chr3 | 30591844 | lincRNA |
| rs369145 | chr3 | 41174299 | lincRNA |
| rs115913570 | chr4 | 11139391 | lincRNA |
| rs13152779 | chr4 | 11142798 | lincRNA |
| rs116669694 | chr4 | 131208181 | lincRNA |
| rs11099266 | chr4 | 134964519 | lincRNA |
| rs295240 | chr4 | 161179694 | lincRNA |
| rs73871504 | chr4 | 165754338 | lincRNA |
| rs12507442 | chr4 | 17276304 | lincRNA |
| rs7690576 | chr4 | 31930676 | lincRNA |
| rs6825814 | chr4 | 37067592 | lincRNA |
| rs13127970 | chr4 | 80502390 | lincRNA |
| rs3109166 | chr4 | 80552199 | lincRNA |
| rs59714449 | chr5 | 10036524 | lincRNA |
| rs62390587 | chr5 | 174686982 | lincRNA |
| rs77726272 | chr5 | 21174806 | lincRNA |
| rs10057544 | chr5 | 98554365 | lincRNA |
| rs58485094 | chr6 | 132538110 | lincRNA |
| rs17165277 | chr6 | 157775271 | lincRNA |
| rs6931622 | chr6 | 169381577 | lincRNA |
| rs1247000 | chr6 | 5061594 | lincRNA |
| rs17460313 | chr7 | 114925322 | lincRNA |
| rs41502546 | chr7 | 114928699 | lincRNA |
| rs4566937 | chr7 | 15230201 | lincRNA |
| rs11765437 | chr7 | 41295987 | lincRNA |
| rs77064312 | chr8 | 122149175 | lincRNA |
| rs1354365 | chr8 | 134969520 | lincRNA |
| rs2565116 | chr8 | 40267939 | lincRNA |
| rs10982910 | chr9 | 118534500 | lincRNA |
| rs10756497 | chr9 | 13763734 | lincRNA |
| rs1888109 | chr9 | 24539632 | lincRNA |
| rs11793053 | chr9 | 37413413 | lincRNA |
| rs11016279 | chr10 | 130297630 | lincRNA |
| rs6481863 | chr10 | 33911780 | lincRNA |
| rs7914587 | chr10 | 3597405 | lincRNA |
| rs11812097 | chr10 | 3620969 | lincRNA |
| rs4765481 | chr12 | 127396035 | lincRNA |
| rs12425149 | chr12 | 65384706 | lincRNA |
| rs10878327 | chr12 | 66143939 | lincRNA |
| rs12309527 | chr12 | 84452099 | lincRNA |
| rs7960972 | chr12 | 84474458 | lincRNA |
| rs331984 | chr13 | 110340104 | lincRNA |
| rs9315332 | chr13 | 20686602 | lincRNA |
| rs1458276 | chr13 | 54470914 | lincRNA |
| rs9572694 | chr13 | 71803040 | lincRNA |
| rs960621 | chr13 | 75225241 | lincRNA |
| rs75561824 | chr13 | 75242252 | lincRNA |
| rs7334899 | chr13 | 75264572 | lincRNA |
| rs538034 | chr13 | 75842148 | lincRNA |
| rs9544122 | chr13 | 76591775 | lincRNA |
| rs17529436 | chr14 | 27704132 | lincRNA |
| rs10141127 | chr14 | 87773053 | lincRNA |
| rs11848918 | chr14 | 90950492 | lincRNA |
| rs74078723 | chr14 | 95456421 | lincRNA |
| rs234604 | chr14 | 97072324 | lincRNA |
| rs74377316 | chr15 | 27988150 | lincRNA |
| rs17705275 | chr15 | 36829838 | lincRNA |
| rs34387846 | chr15 | 79717219 | lincRNA |
| rs4307931 | chr15 | 98865620 | lincRNA |
| rs80112443 | chr16 | 48031408 | lincRNA |
| rs9925381 | chr16 | 51959612 | lincRNA |
| rs883794 | chr16 | 86027818 | lincRNA |
| rs3924409 | chr16 | 86028237 | lincRNA |
| rs237291 | chr17 | 12403835 | lincRNA |
| rs9890751 | chr17 | 35020814 | lincRNA |
| rs11081188 | chr18 | 5371579 | lincRNA |
| rs1012680 | chr18 | 63642446 | lincRNA |
| rs423826 | chrX | 139299448 | lincRNA |
| rs4933145 | chrX | 1986252 | lincRNA |
| rs12843591 | chrX | 41942431 | lincRNA |
| rs7876156 | chrX | 47181569 | lincRNA |
Bioinformatics analysis: investigation of the potential role of lncRNAs
Identification of lncRNAs that are involved in spermatogenesis could provide useful information in order to better understand the molecular mechanisms underlying male infertility. We identified SNPs that are located in close proximity to regions coding for lincRNAs, and in order to investigate the potential role of these lncRNAs, a literature search was performed using LNCipedia version 5.2. However, for each of the 92 lincRNAs that were previously detected, no result associated with male infertility or spermatogenesis process was found.
Bioinformatics analysis: interaction between lncRNAs-miRNAs
It is becoming increasingly evident that lncRNAs regulate gene expression through their interactions with miRNAs. For example, a general phenomenon is their competitive action for miRNA binding. Therefore, we hypothesized that lncRNAs may affect fertilization capacity through their interactions with miRNAs. To understand the role of lncRNAs in spermatogenesis process, a lncRNAs-miRNA interaction analysis was carried out using DIANA tools-LncBase Predicted v.2. This database performs in silico prediction of lncRNA-miRNA interactions using an appropriately adjusted DIANA-microT algorithm [36]. This microT-CDS algorithm can identify miRNA targets in 3′UTR, as well as in CDS regions [39]. More specifically, miRNA recognition elements (MREs) on lncRNAs are scored separately and each pair of lncRNA-miRNA interaction is characterized by a score that signifies the interaction strength [36]. The threshold that it was used for this score was quite stringent (0.9) in order to identify only interactions of high strength. The highly reliable miRNA-lncRNA interaction pairs are provided in Supplementary Material, Table S2. Approximately 1000 miRNAs were predicted to interact with the 92 lncRNAs; however, some of the lncRNAs were found to have no interaction with miRNAs or they were not found on the database at all.
Bioinformatics analysis: investigation of the role of miRNAs on male infertility
The predicted target miRNAs mentioned above (~ 1000) were input in miRBase in order to identify a potential role in meiosis or an effect on sperm quality. As shown in Table 2, approximately 90 of them were studied in seven different research papers and they were associated with the fertility process in males.
Table 2.
miRNAs associated with male infertility according to research papers
| miRNAs | Authors publication | Research paper |
|---|---|---|
|
hsa-miR-5699-5p, hsa-miR-653-3p, hsa-miR-410-3p, hsa-miR-6720-5p, hsa-miR-3192-3p, hsa-miR-7154-5p, hsa-miR-579-3p, hsa-miR-190a-5p, hsa-miR-579-3p, hsa-miR-7161-3p, hsa-miR-520g-5p, hsa-miR-7156-5p, hsa-miR-208a-5p, hsa-miR-656-3p, hsa-miR-670-3p, hsa-miR-7160-3p, hsa-miR-1296-5p, hsa-miR-181d-5p, hsa-miR-1288-3p, hsa-miR-670-3p, hsa-miR-7161-5p, hsa-miR-653-3p, hsa-miR-653-3p, hsa-miR-1180-5p, hsa-miR-7156-3p, hsa-miR-1288-3p, hsa-miR-370-3p, hsa-miR-891a-3p, hsa-miR-7152-5p, hsa-miR-7152-5p, hsa-miR-1288-3p, hsa-miR-942-5p, hsa-miR-597-3p, hsa-miR-7160-5p, hsa-miR-1301-3p, hsa-miR-1301-3p, hsa-miR-651-3p, hsa-miR-651-3p, hsa-miR-520f-5p, hsa-miR-7160-5p, hsa-miR-597-5p, hsa-miR-889-3p, hsa-miR-548f-3p, hsa-miR-7153-5p, hsa-miR-670-3p, hsa-miR-7156-5p, hsa-miR-1288-3p, hsa-miR-7160-5p, hsa-miR-1298-5p, hsa-miR-7160-3p, hsa-miR-597-3p, hsa-miR-889-3p, hsa-miR-7152-5p, hsa-miR-7152-5p, hsa-miR-7159-5p, hsa-miR-7156-5p, hsa-miR-3192-5p, hsa-miR-5699-3p, hsa-miR-942-5p, hsa-miR-580-5p, hsa-miR-1252-3p, hsa-miR-370-3p, hsa-miR-7161-5p |
Birth and expression evolution of mammalian microRNA genes [40] | Meunier et al. (2013) |
|
hsa-miR-513c-5p, hsa-miR-513b-5p, hsa-miR-513c-3p, hsa-miR-513b-5p, hsa-miR-513c-5p, hsa-miR-513c-3p, hsa-miR-513c-5p, hsa-miR-513c-5p, hsa-miR-513c-3p |
Rapid evolution of an X-linked microRNA cluster in primates [41] | Zhang et al. (2007) |
|
hsa-miR-6720-5p, hsa-miR-6509-3p, hsa-miR-6719-3p, hsa-miR-6718-5p, hsa-miR-6515-3p, hsa-miR-6515-5p, hsa-miR-6715a-3p, hsa-miR-6716-5p, hsa-miR-6515-5p |
Deep sequencing analysis of small non-coding RNAs reveals the diversity of miRNAs and piRNAs in the human epididymis [42] | Li et al. (2012) |
|
hsa-miR-429, hsa-miR-181a-5p |
Spermatogenesis in humans and its affecting factors [43] | Neto et al. (2016) |
|
hsa-miR-429, hsa-miR-34b-3p |
Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of miR-141, miR-429 and miR-7-1-3p [44] | Wu et al., 2013 |
|
hsa-miR-181a-5p, hsa-miR-513a-5p |
Altered profile of Seminal Plasma MicroRNAs in the Molecular Diagnosis of Male Infertility [45] | Wang et al. (2011) |
|
hsa-miR-429, hsa-miR-34b-3p |
Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility [46] | Abu-Halima et al. (2014) |
To better understand the potential mechanism of action of lncRNAs through their interaction with miRNAs on reproductive capacity of spermatozoa, these 90 miRNAs found to be associated with the phenotype of interest according to previous research were further studied. miRNAs regulate gene expression usually by base-pairing to the mRNA 3′-untranslated regions (UTRs) to repress protein synthesis. Other mechanisms of action include mRNA degradation and transcriptional inhibition. Therefore, miRNA-mRNA interactions are extremely important for regulation of various processes [47, 48]. Thus, miRNAs were then investigated for their interaction networks with human mRNAs using miRTargetLinkHuman [38]. Of the 90 miRNAs studied, 21 exhibited experimentally validated interaction networks, as shown in Fig. S1. Only networks with strong interactions were selected. To investigate the potential regulatory function of miRNAs over genes that have a role in the complex process of spermatogenesis, the target mRNAs were searched in GeneCards®: The Human Gene Database. It was observed that 18 genes were associated with male infertility or gametogenesis (Table 3). Information about the association of these genes with male infertility is provided on Supplementary Material, Table S4.
Table 3.
List of genes whose mRNAs are targeted by miRNAs and they are associated with male infertility
| Genes | ||
|---|---|---|
| ATM serine/threonine kinase (ATM) | Sirtuin 1 (SIRT1) | Cystic fibrosis transmembrane conductance regulator (CFTR) |
| MDM2 oncogene, E3 ubiquitin protein ligase (MDM2) | Proline-rich acidic protein 1 (PRAP1) | Vascular endothelial growth factor A (VEGF) |
| B cell CLL/lymphoma 2 (BCL2) | Progesterone receptor (PGR) | Nanog homeobox (NANOG) |
| Septin 7 (SEPT7) | Telomerase reverse transcriptase (TERT) | cAMP responsive element binding protein 1 (CREB1) |
| DNA methyltransferase 1 (DNMT1) | Interferon gamma (IFNG) | Signal transducer and activator of transcription 3 (STAT3) |
| TIMP metallopeptidase inhibitor 2 (TIMP2) | Αryl hydrocarbon receptor (AHR) | Insulin-like growth factor 1 (IGF1) |
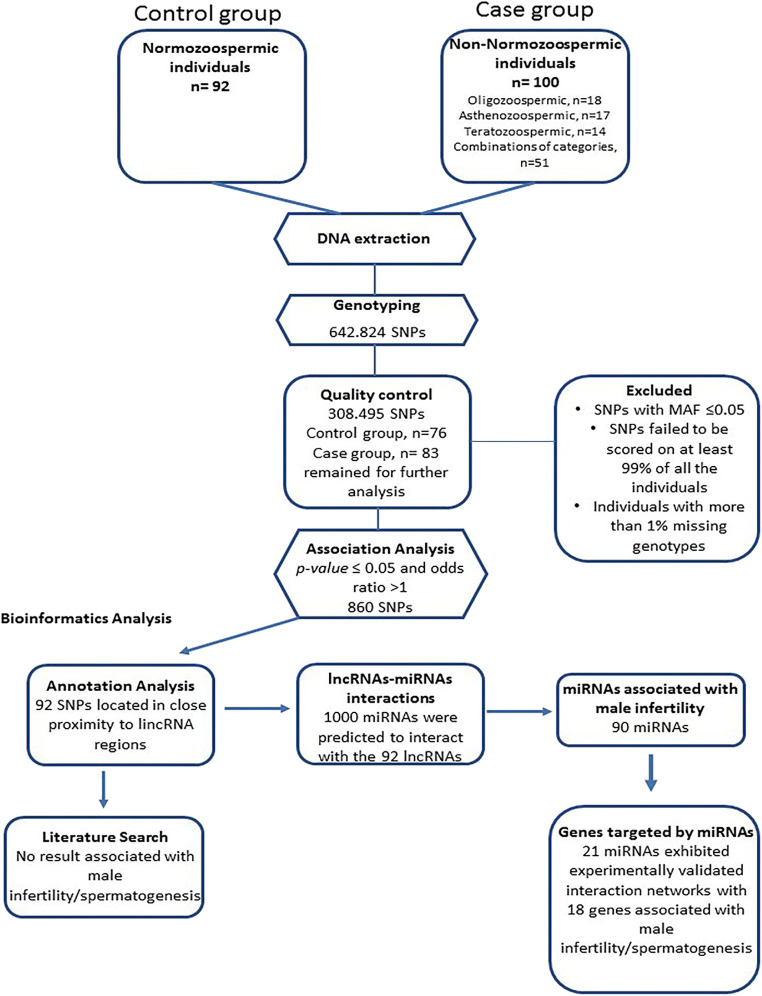
Finally, after genome-wide association study, we managed to identify genetic variants (SNPs) significantly associated with male infertility in close proximity to lncRNAs regions in the Greek population (Table 1). To better understand the molecular mechanisms underlying male infertility, a pipeline of analysis was constructed and it comprised identification of statistically significant SNPs, annotation of each SNP to its target (lncRNAs), analysis of lncRNAs-miRNAs interactions, and highlighting of interactions that can affect male fertility and its physiology (Fig. 1). This analysis has brought up six lncRNAs that are analyzed on Table 4.
Fig. 1.
The constructed pipeline used for analysis, which comprised identification of statistically significant SNPs, annotation of each SNP to its target (lncRNAs), analysis oflncRNAs-miRNA interactions, and highlighting of interactions that can affect male infertility. The results that we obtained from each step are presented here
Table 4.
List of lncRNAs with a potential role on male infertility through interaction with miRNAs
| lncRNAS | miRNAs | Gene targets |
|---|---|---|
| ENSG00000248995 | hsa-miR-410-3p | MDM2 |
| ENSG00000236678 | hsa-miR-429 | BCL2 |
| SEPT7 | ||
| DNMT1 | ||
| TIMP2 | ||
| hsa-miR-190a-5p | IGF1 | |
| ENSG00000260450 | hsa-miR-181d-5p | BCL2 |
| hsa-miR-181a-5p | BCL2 | |
| ATM | ||
| SIRT1 | ||
| PRAP1 | ||
| PGR | ||
| TERT | ||
| IFNG | ||
| AHR | ||
| STAT3 | ||
| ENSG00000231755 | hsa-miR-509-3p | CFTR |
| ENSG00000250819 | hsa-miR-34b-3p | BCL2 |
| VEGFA | ||
| NANOG | ||
| CREB1 | ||
| ENSG00000233103 | hsa-miR-509-3p | CFTR |
Discussion
Previous studies have shown that spermatogenesis is a complex process that requires a dynamic gene expression pattern of coding and non-coding elements like piRNAs, miRNAs, and lncRNAs [49]; thus, their dysregulation could affect male fertility. In this study, semen from normozoospermic and non-normozoospermic men of the Greek population was used for DNA genotyping and bioinformatics analysis in order to identify SNPs significantly associated with male infertility in close proximity to lncRNA regions. Based on our analysis, we have now demonstrated that six of these lncRNAs exert a potential role in male infertility through their interactions with miRNAs.
Semen samples were collected from control and case groups and DNA genotyping followed in order to identify SNPs associated with male infertility. GWAS has changed how research is conducted as it allows the identification of the genetic basis of complex traits and diseases like male infertility which are caused by a combination of environmental factors and gene variants with subtle effects [50]. In this particular study, we identified variants in close proximity to lncRNA regions. lncRNAs have been shown to have a regulatory function in spermatogenesis process but they are the least characterized ncRNAs [13].
Investigation of the potential role of lncRNAs in male infertility led to very few results as it is an emerging field of research and it must be analyzed further with the addition of studies with more samples as well as studies incorporating functional analysis. This was expected as there are a large number of data sets available, particularly of lncRNAs, due to RNA-seq and advances on transcriptomics but the mechanistic characterization of lncRNAs is rather poor. It is known that they regulate various cellular and molecular processes but the exact mechanism of function of many of them is still largely unexplored [51]. Even though they can be found in a broad range of species, lncRNAs are relatively poorly conserved in terms of nucleotide sequence. Thus, it is difficult to investigate their functions using the right animal models [19, 52]. However, some studies prove the association of lncRNAs with male infertility [23, 53]. More specifically, lncRNA Gm2044 plays an important role in the regulation of spermatogenic cell development and its expression is increased in non-obstructive azoospermia [53].
Several studies demonstrate that miRNAs are essential for gametogenesis [13]. As the molecular function of lncRNAs is mediated through interactions with other RNA species and proteins, it was important to also investigate lncRNA-miRNA interactions. A large number of miRNAs were found to interact with the lncRNAs that we studied (Supplementary Material, Table S2). Many of them belong to X-linked miRNA families and their evolution study revealed peculiar patterns. They are predominantly expressed in testis and these families were expanded through tandem gene duplication. The evolutionary force driving the duplication of X-linked miRNAs was MSCI as many miRNA gene copies allowed them to escape MSCI and to express in spermatocytes and spermatids. Moreover, amplified X-linked miRNA families show rapid sequence evolution, potentially driven by positive selection. Thus, X-linked miRNAs evolved higher expression levels and diverse functions during gametogenesis through selectively driven duplication divergence processes [40]. Consistent with these results, some other miRNAs that interact with the lncRNAs studied were also investigated from an evolutionary perspective. Conserved miRNAs are considered essential for spermatogenesis due to their regulatory roles. However, non-conserved miRNAs exhibit also an interesting evolutionary role as they contribute to functional novelties. Such an X-linked miRNA cluster evolved through changes in copy number and sequence substitutions leading to changes in male sexual maturation and spermatogenesis process [41].
Other miRNAs were found to be highly expressed on the epididymis. Epididymis plays an important role in sperm maturation and motility; thus, these miRNAs may be involved in physiological pathways affecting male infertility [42]. Some of the miRNAs interacting with lncRNAs, e.g., miR-429, miR-181a-5p, were also the most studied in infertile men but their function remains unknown [43]. Finally, another extremely important observation is the fact that many of these miRNAs could be potentially used as biomarkers for diagnosis of male infertility. miRNAs are expressed in various tissues and their dysregulation affects many cellular and molecular mechanisms and gene networks. That means that their detection can provide useful information for several diseases or pathophysiological processes. It has been already proposed that miRNAs could serve as biomarkers for cancer, nervous system disorders and diabetes [54]. Many of the miRNAs identified could be used as noninvasive biomarkers for non-obstructive azoospermia (NOA), one of the different types of spermatogenic impairments [44, 46]. Other miRNAs found to interact with the lncRNAs also exhibit a different expression profile in seminal plasma of infertile men and they could be used as a noninvasive approach for diagnosing male infertility [45].
miRNAs exert their function through interactions with mRNAs, which they can degrade. For many of the miRNAs mentioned above, their role on male infertility was confirmed by the fact that their target mRNAs were associated with the meiosis process on mammals (Supplementary Material, Table S4). Among the most important targets, BCL2 may regulate apoptosis of sperm cells during gametogenesis and it has been demonstrated to be associated with non-obstructive azoospermia (NOA) [55]. A promising field is also the investigation of epigenetics on male infertility, as DNMT1 plays a key role in DNA methylation and it could be associated with oligozoospermia [56]. Another gene usually studied is PGR (progesterone receptor) as hormones regulate different aspects of spermatogenesis process [57, 58]. Moreover, CFTR has been associated with male infertility as it affects sperm capacitation [59, 60], and STAT3 has been observed to affect human spermatozoa through regulation of mitochondrial activity [61].
It should also be noted that one strength of this study is the in-depth bioinformatics analysis of the lincRNAs that may play an important role in spermatogenesis, as SNPs significantly associated with male infertility were found in regions in close proximity with them. Several different databases were used (LNCipedia version 5.2., DIANA tools-LncBase Predicted v.2., miRBase) to ensure that we obtain all the information that is available about these lincRNAs and the miRNAs interacting with them. Moreover, rigorous quality control and the exclusion SNPs and samples that could affect the quality of our results and lead to false-positive or false-negative results were performed. However, we should keep in mind that this study is limited in terms of the number of samples. We began this study with 192 samples, but after quality control, the final number of samples that were analyzed was reduced to 159. It should also be noted that the generalizability of the study is also limited by the genetic homogeneity of the Greek population. Another limitation is that the interactions between lncRNAs and miRNAs were predicted using a database. As the interactions were not experimentally verified, maybe some of the miRNAs that we studied do not truly interact with the lncRNAs, though we used quite stringent criteria (threshold of 0.9) in order to obtain more reliable interactions and results.
In conclusion, the present study is significant as, for the first time, it explores the existence of genetic variants within or near lncRNAs and the subsequent association with male infertility in the Greek population. Moreover, we analyzed all these data collected from Greek individuals to investigate the potential role of lncRNAs in male infertility, which are the least characterized ncRNAs. The findings indicate a regulatory role of lncRNAs in mammalian meiosis through their interactions with miRNAs. Although little is currently known about the functions of lncRNAs in spermatogenesis process, identification of such lncRNAs provides a shortlist of key candidates for functional studies. The impact of the SNPs identified as significant with male infertility on the way lncRNAs function and regulate gene expression should be also investigated in future studies, for a larger number of individuals in order to surpass the limited generalizability of such studies. For the interpretation of our data, it is important also to bear in mind that we used a limited number of samples; thus, our data support the need for further studies of lncRNAs that we and others have identified on large scale in order to fully understand the molecular mechanisms underlying male infertility. Given the growing cases of male infertility worldwide, the study of spermatogenesis is extremely important as there is a growing need to obtain useful applications in clinical practice like diagnostic accuracy and successful treatment.
Electronic supplementary material
(DOCX 1199 kb)
Author’s contributions
Maria-Anna Kyrgiafini was responsible for all data analysis and composing the manuscript along with participating in reviewing important details. Maria Markantoni participated in the creation of the study concept and in data interpretation. Theologia Sarafidou was responsible for article significant revisions. Alexia Chatziparasidou and Nicolas Christoforidis were responsible for all samples acquisition along with consent forms. Zissis Mamuris was responsible for the study concept and funding acquisition, along with article revisions.
Funding
This work is supported by the Spermogene project which is co-financed by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship, and Innovation, under the call RESEARCH–CREATE–INNOVATE (Grant number Τ1ΕΔΚ-02787).
Data availability
All data are openly available in the form of supplementary materials.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All the participants were informed about the study and they gave their consent in order to participate by filling out a questionnaire along with the consent form. Both the study and the consent procedure were approved by the ethics committee of the Medical Faculty of the University of Thessaly.
Footnotes
Maria-Anna Kyrgiafini and Maria Markantoni are joint first authors
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K, et al. International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009∗. Fertil Steril. 2009, 2009;92:1520–4. [DOI] [PubMed]
- 2.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl. 2012;14:40–48. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith JF, Walsh TJ, Shindel AW, Turek PJ, Wing H, Pasch L, et al. Sexual, marital, and social impact of a man's perceived infertility diagnosis. J Sex Med. 2009;6:2505–15. [DOI] [PMC free article] [PubMed]
- 4.Winters BR, Walsh TJ. The epidemiology of male infertility. Urol Clin N Am. 2014;41:195–204. doi: 10.1016/j.ucl.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15:369–384. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabrielsen JS, Tanrikut C. Chronic exposures and male fertility: the impacts of environment, diet, and drug use on spermatogenesis. Andrology. 2016;4:648–661. doi: 10.1111/andr.12198. [DOI] [PubMed] [Google Scholar]
- 8.Jungwirth A, Diemer T, Kopa Z, Krausz C, Tournaye H. European Association of Urology (EAU) guidelines on male infertility. Arnhem, The Netherlands: European Association of Urology; 2015. [DOI] [PubMed] [Google Scholar]
- 9.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. [DOI] [PMC free article] [PubMed]
- 10.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009, 2009;458:223–7. [DOI] [PMC free article] [PubMed]
- 11.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee A, Koli S, Reddy KVR. Regulatory non-coding transcripts in spermatogenesis: shedding light on “dark matter.”. Andrology. 2014;2:360–369. doi: 10.1111/j.2047-2927.2014.00183.x. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3:e1738. [DOI] [PMC free article] [PubMed]
- 15.Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse germ line. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- 16.Korhonen HM, Meikar O, Yadav RP, Papaioannou MD, Romero Y, Da Ros M, et al. Dicer is required for haploid male germ cell differentiation in mice. PLoS One. 2011;6:e24821. [DOI] [PMC free article] [PubMed]
- 17.Kostereva N, Hofmann MC. Regulation of the spermatogonial stem cell niche. Reprod Domest Anim. 2008;43:386–392. doi: 10.1111/j.1439-0531.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Björk JK, Sandqvist A, Elsing AN, Kotaja N, Sistonen L. miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development. 2010;137:3177–3184. doi: 10.1242/dev.050955. [DOI] [PubMed] [Google Scholar]
- 19.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. [DOI] [PMC free article] [PubMed]
- 20.Ransohoff J, Wei Y, Khavari P. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. [DOI] [PMC free article] [PubMed]
- 23.Wichman L, Somasundaram S, Breindel C, Valerio DM, McCarrey JR, Hodges CA, et al. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility. Biol Reprod. 2017;97:313–23. [DOI] [PubMed]
- 24.Xue M, Zhuo Y, Shan B. MicroRNAs, long noncoding RNAs, and their functions in human disease. Methods Mol Biol. 1617;2017:1–25. doi: 10.1007/978-1-4939-7046-9_1. [DOI] [PubMed] [Google Scholar]
- 25.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA–LncRNA interactions. Methods Mol Biol. 2016;140:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 27.Sun B, Liu C, Li H, Zhang L, Luo G, Liang S, et al. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol Lett. 2020;19:595–605. [DOI] [PMC free article] [PubMed]
- 28.Dong Z, Zhang A, Liu S, Lu F, Guo Y, Zhang G, et al. Aberrant methylation-mediated silencing of lncRNA MEG3 functions as a ceRNA in esophageal cancer. Mol Cancer Res. 2017;15:800–10. [DOI] [PubMed]
- 29.Lü M, Tian H, Cao YX, He X, Chen L, Song X, et al. Downregulation of miR-320a/383-sponge-like long non-coding RNA NLC1-C (narcolepsy candidate-region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death Dis. 2015;6:e1960. [DOI] [PMC free article] [PubMed]
- 30.Panoutsopoulou K, Hatzikotoulas K, Xifara DK, Colonna V, Farmaki AE, Ritchie GR, et al. Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants. Nat Commun. 2014;5:5345. [DOI] [PMC free article] [PubMed]
- 31.Weyrich A. Preparation of genomic DNA from mammalian sperm. Curr Protoc Mol Biol. 2012;98:2.13.1–2.13.3. doi: 10.1002/0471142727.mb0213s98. [DOI] [PubMed] [Google Scholar]
- 32.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed]
- 33.International Schizophrenia Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. [DOI] [PMC free article] [PubMed]
- 34.Dayem Ullah AZ, Oscanoa J, Wang J, Nagano A, Lemoine NR, Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res. 2018;46:W109–W113. doi: 10.1093/nar/gky399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47:D135–9. [DOI] [PMC free article] [PubMed]
- 36.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–8. [DOI] [PMC free article] [PubMed]
- 37.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamberg M, Backes C, Fehlmann T, Hart M, Meder B, Meese E, et al. MiRTargetLink--miRNAs, genes and interaction networks. Int J Mol Sci. 2016;17:564. [DOI] [PMC free article] [PubMed]
- 39.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–73. [DOI] [PMC free article] [PubMed]
- 40.Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, et al. Birth and expression evolution of mammalian microRNA genes. Genome Res. 2013;23:34–45. [DOI] [PMC free article] [PubMed]
- 41.Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Wang H, Wan F, Liu F, Liu J, Zhang N, et al. Deep sequencing analysis of small non-coding RNAs reveals the diversity of microRNAs and piRNAs in the human epididymis. Gene. 2012;497:330–5. [DOI] [PubMed]
- 43.Neto L, Bach PV, Najari BB, Li PS, Goldstein M. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol. 2016;59:10–26. doi: 10.1016/j.semcdb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Wu W, Qin Y, Li Z, Dong J, Dai J, Lu C, et al. Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum Reprod. 2013;28:1827–36. [DOI] [PubMed]
- 45.Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, Li L. Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem. 2011;57:1722–1731. doi: 10.1373/clinchem.2011.169714. [DOI] [PubMed] [Google Scholar]
- 46.Abu-Halima M, Hammadeh M, Backes C, Fischer U, Leidinger P, Lubbad AM, et al. Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril. 2012;102:989–97. [DOI] [PubMed]
- 47.Andrés-León E, Gómez-López G, Pisano DG. Prediction of miRNA–mRNA interactions using miRGate. Methods Mol Biol. 2017;1580:225–237. doi: 10.1007/978-1-4939-6866-4_15. [DOI] [PubMed] [Google Scholar]
- 48.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 49.Lesch BJ, Page DC. Genetics of germ cell development. Nat Rev Genet. 2012;13:781–794. doi: 10.1038/nrg3294. [DOI] [PubMed] [Google Scholar]
- 50.Uitterlinden A. An introduction to genome-wide association studies: GWAS for dummies. Semin Reprod Med. 2016;34:196–204. doi: 10.1055/s-0036-1585406. [DOI] [PubMed] [Google Scholar]
- 51.Fritah S, Niclou SP, Azuaje F. Databases for lncRNAs: a comparative evaluation of emerging tools. RNA. 2014;20:1655–1665. doi: 10.1261/rna.044040.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Liang M, Hu K, He C, Zhou J, Liao Y. Upregulated lncRNA Gm2044 inhibits male germ cell development by acting as miR-202 host gene. Anim Cells Syst. 2019;23:128–134. doi: 10.1080/19768354.2019.1591506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2015;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Y, Ji J, Du G, Wu W, Dai J, Hu Z, et al. Comprehensive pathway-based analysis identifies associations of BCL2, GNAO1 and CHD2 with non-obstructive azoospermia risk. Hum Reprod. 2014;29:860–866. doi: 10.1093/humrep/deu013. [DOI] [PubMed] [Google Scholar]
- 56.Cheng P, Chen H, Zhang RP, Liu S, Zhou-Cun A. Polymorphism in DNMT1 may modify the susceptibility to oligospermia. Reprod BioMed Online. 2014;28:644–649. doi: 10.1016/j.rbmo.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Abid S, Gokral J, Maitra A, Meherji P, Kadam S, Pires E, et al. Altered expression of progesterone receptors in testis of infertile men. Reprod BioMed Online. 2008;17:175–84. [DOI] [PubMed]
- 58.Sen S, Dixit A, Thakur C, Gokral J, Hinduja I, Zaveri K, et al. Association of progesterone receptor gene polymorphism with male infertility and clinical outcome of ICSI. J Assist Reprod Genet. 2013;30:1133–9. [DOI] [PMC free article] [PubMed]
- 59.Puga Molina LC, Pinto NA, Torres Rodríguez P, Romarowski A, Vicens Sanchez A, Visconti PE, et al. Essential role of CFTR in PKA-dependent phosphorylation, alkalinization, and hyperpolarization during human sperm capacitation. J Cell Physiol. 2017;232:1404–14. [DOI] [PMC free article] [PubMed]
- 60.Tamburino L, Guglielmino A, Venti E, Chamayou S. Molecular analysis of mutations and polymorphisms in the CFTR gene in male infertility. Reprod BioM Online. 2008;17:27–35. doi: 10.1016/S1472-6483(10)60289-1. [DOI] [PubMed] [Google Scholar]
- 61.Lachance C, Goupil S, Leclerc P. Stattic V, a STAT3 inhibitor, affects human spermatozoa through regulation of mitochondrial activity. J Cell Physiol. 2012;228:704–713. doi: 10.1002/jcp.24215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1199 kb)
Data Availability Statement
All data are openly available in the form of supplementary materials.