Abstract
NKX3.1 is the most commonly deleted gene in prostate cancer and is a gatekeeper suppressor. NKX3.1 is haploinsufficient, and pathogenic reduction in protein levels may result from genetic loss, decreased transcription, and increased protein degradation caused by inflammation or PTEN loss. NKX3.1 acts by retarding proliferation, activating antioxidants, and enhancing DNA repair. DYRK1B-mediated phosphorylation at serine 185 of NKX3.1 leads to its polyubiquitination and proteasomal degradation. Because NKX3.1 protein levels are reduced but never entirely lost in prostate adenocarcinoma, enhancement of NKX3.1 protein levels represents a potential therapeutic strategy. As a proof of principle, we used CRISPR/Cas9-mediated editing to engineer in vivo a point mutation in murine Nkx3.1 to code for a serine to alanine missense at amino acid 186, the target for Dyrk1b phosphorylation. Nkx3.1S186A/-, Nkx3.1+/−, and Nkx3.1+/+ mice were analyzed over one year to determine the levels of Nkx3.1 expression and effects of the mutant protein on the prostate. Allelic loss of Nkx3.1 caused reduced levels of Nkx3.1 protein, increased proliferation, and prostate hyperplasia and dysplasia, whereas Nkx3.1S186A/- mouse prostates had increased levels of Nkx3.1 protein, reduced prostate size, normal histology, reduced proliferation, and increased DNA end-labeling. At 2 months of age, when all mice had normal prostate histology, Nkx3.1+/− mice demonstrated indices of metabolic activation, DNA damage response, and stress response. These data suggest that modulation of Nkx3.1 levels alone can exert long-term control over premalignant changes and susceptibility to DNA damage in the prostate.
Keywords: NKX3.1, prostate cancer, DYRK1B, tumor suppressor
Introduction
Genetic studies over decades have consistently implicated 8p21, the locus of NKX3.1, as the most commonly deleted chromosomal locus of prostate cancer (1–3). NKX3.1, is known to be the most commonly deleted gene in human prostate cancer (4). NKX3.1 is a prostate-specific homeobox gene that controls cell proliferation (5), epithelial differentiation (6), and stem cell maintenance (7). In preinvasive and primary prostate cancer reduced levels of the haploinsufficient NKX3.1 protein are seen in the majority of prostate cancers (8). During the advanced phases of prostate cancer there is ongoing selective pressure to decrease NKX3.1 expression (9). NKX3.1 protein loss may result from genetic loss, decreased mRNA, or protein degradation, all effecting reduced NKX3.1 protein levels early in the evolution of most prostate cancers (8,10). One mechanism by which NKX3.1 exerts tumor suppression is enhancement of the DNA repair response in the prostate and acceleration of ATM activation after DNA damage (11,12). Importantly, the early reduction of NKX3.1 in initiating prostate carcinogenesis predisposes to the pathognomonic translocation between TMPRSS2 and ERG that characterizes more than half of all prostate cancers (10,13).
NKX3.1 is under tight control by numerous post translational modifications, most notably phosphorylation (14). Steady state levels of NKX3.1 protein are tightly controlled by phosphorylation that triggers polyubiquitination and protein destruction with a half-life in vitro of less than 30 minutes (15). The primary mediator of NKX3.1 turnover is the kinase DYRK1B that phosphorylates NKX3.1 at serine 185 (16). Missense mutation of serine 185 to alanine substantially prolongs NKX3.1 half-life (15). We have shown that DYRK inhibitors prolong NKX3.1 half-life in vitro (16) and have hypothesized that in vivo DYRK inhibitors may prolong NKX3.1 half-life and thereby have a role in prostate cancer treatment. NKX3.1 expression is preserved even in advanced prostate cancer and therefore may be a target for therapeutic manipulation (17). Increased expression of NKX3.1 has the potential to arrest growth of even advanced stage prostate cancer cells. NKX3.1 is a profound growth suppressor and only very few prostate cancer cell lines, LNCaP, LAPC4, VCaP, and 22Rv1 have adapted to grow despite its expression. In fact, adaptation of the aggressive PC-3 cells to growth despite NKX3.1 expression is a rare event that requires constant selective pressure to sustain expression in vitro that is rapidly lost in vivo when the selective pressure cannot be maintained (18). For this reason, we developed a genetic model for inhibition DYRK1B phosphorylation of NKX3.1 by engineering the Nkx3.1(S186A) gene in mice.
NKX3.1 protein levels are tightly controlled and protein half-life is 20–30 minutes (15). Steady state protein turnover is controlled by phosphorylation at serine 185 that is targeted by the kinase DYRK1B (16). Nevertheless, NKX3.1 expression, although decreased, is never completely lost in prostate adenocarcinoma (17). For that reason, it may be beneficial to target increased NKX3.1 as a therapeutic mechanism in advanced prostate cancer. NKX3.1 is profoundly growth suppressive and very few cells have adapted in culture to tolerate its expression. In fact, cells that were selected to tolerate NKX3.1 expression in vitro rapidly lose expression upon growth as xenografts (18). Ultimately our goal is to mediate NKX3.1 levels by blocking phosphorylation to increase protein levels. As a proof-of-principle, we have engineered mice by CRISPR/Cas9-mediated editing to express Nkx3.1(S186A) containing a single mutation that abrogated the serine target of phosphorylation that triggers ubiquitination and degradation. We show here that a single copy of the mutant Nkx3.1S186A gene eliminates proneoplastic effects of Nkx3.1 heterozygosity.
Material and Methods
Mouse strains and generation of Nkx3.1S186A mutant mice
Wild type C57BL6 mice were purchased from Taconic. Swiss-Webster mice were purchased from Taconic, and B6D2F1 (C57BL/6 × DBA/2) mice were purchased from Jackson Laboratory. The Nkx3.1 null mutation has previously been described (6). Animal studies were conducted according to federal guidelines and approved by both the Columbia University Irving Medical Center and University of Arizona Institutional Animal Care and Use Committees.
Mice with the Nkx3.1(S186A) mutation were generated using CRISPR/Cas-9, as described previously (19). Cas9 mRNA was ordered from TriLink Biotech (L-6129). For gNkx3.1 mRNA preparation, T7 promoter sequence was appended to the sgRNA template by PCR amplification using the appropriate primer pair listed in Supplementary Table 1. The PCR reaction was performed with 30 ng of px330-Cas9 plasmid, Phusion high-fidelity DNA polymerase (NEB) and HF buffer per instruction of the vendor, with Tm=40°C, for 30 sec extension time at 72°C for 40 cycles. PCR products were purified by gel purification and ethanol precipitation. In vitro transcription of sgRNA was performed using 50 nM of the T7-sgRNA PCR product in a 20 μl reaction as the template with the MEGAshortscript T7 kit (Life Technologies) according to the kit protocol. Incubation time was 6 hours. The sgRNA was purified using the RNeasy kit and eluted with elution buffer according to the kit protocol. 200 ng of gRNA was heated at 70°C for 2 min, resolved on a 1% agarose gel in TAE buffer to confirm integrity before injection.
Single cell zygotes from superovulated 3–4 week-old B6D2F1 females mated to B6D2F1 males were injected with Cas9 mRNA (TriLink Biotech L-6129), sgRNA and ssDNA, and cultured overnight. Two cell embryos were transferred to oviducts of pseudopregnant Swiss Webster surrogate females. Resulting pups were genotyped for the Nkx3.1(S186A) mutation and a single founder animal was selected from among two for backcross mating to C57BL6 mice. The embryos we injected came from a mating of B6D2F1/J females (F1 from C57BL/6 and DBA/2 strain mating) crossed to B6D2F1/J males from Jackson labs. The pups are therefore B6D2 F2. ordered from Taconic. The S186A mutant founders were back crossed with C57BL/6 mice. The experiment used mice backcrossed at least 7 and up to 10 generations.
Genotyping
Mouse tail DNA was isolated with protocol from the Tsai laboratory (http://tsailaboratory.mit.edu/wp-content/uploads/2014/01/protocol-for-preparation-of-genomic-dna-for-genotyping.pdf). For analysis of the Nkx3.1(S186A) mutations we employed a positive control MYC-Nkx3.1(S186A) plasmid that had been validated by sequencing. For the plasmid template we used the oligonucleotide listed in Supplementary Table 2 (ssODN1) as forward primer, and 5’-CTT CCG ACT CCT TGA CAT C-3’ as a reverse primer. These primers generated a PCR product of 301 bp. PCR was performed with 500 ng of DNA in 50 μl at Tm=50°C for 40 cycles. PCR products were purified with the QIAGEN PCR purification kit. About 20 ng/μl DNA was eluted with 30 μl H2O. One μl of PCR product was used for TA-cloning with TOPO-TA cloning kit (ThermoFisher Scientific) that was then used for sequencing. 300–400 ng of PCR products was incubated with 0.5 unit of EciI at 37°C for 1 hour. The reaction was stopped with 0.2% SDS in loading buffer. The input and cleaved samples were resolved on a 2% agarose gel. For genotyping of Nkx3.1S186A/- mice, PCR was performed with primers listed in Supplementary Table 3. PCR products were purified and digested as described above. Nkx3.1S186A/- mutant mice identified by genotyping was further confirmed by sequencing through TA-cloning.
Histologic analysis
Anterior, ventral, and dorsolateral prostate lobes were fixed in 10% formalin and embedded in paraffin. Immunofluorescence staining was done using 5 μm paraffin sections and analyzed with a Leica TCS SP5 confocal microscope. Details of all the primary and secondary antibodies used in this study are provided in Supplementary Table 4. Deparaffinized slides were treated with 10 mM citrate buffer pH 6.0 in steamer for 40 minutes. Slides were permeabilized with 0.5% Triton X-100 in PBS for 30 minutes followed by incubation in 3% H2O2 for 30 minutes. All the primary antibodies were incubated at 4°C overnight, and secondary antibodies at room temperature for 30 minutes. Detection of mouse Nkx3.1 was enhanced using biotinylated anti-rabbit IgG (Vector Labs) and then incubating with Texas Red avidin DCS (Vector Labs). Signals for γH2AX, 8-oxoG, pATM, 53BP1, and pS6 were enhanced by tyramide amplification (Fisher Scientific) by incubating with horseradish peroxidase (HRP)-conjugated secondary antibody (Invitrogen), followed by incubation with tyramide 568 for 8 minutes. β-catenin, Ki67, CK5, CK8, nitrotyrosine, and cleaved caspase 8were revealed by Alexa fluor 488 or 568 antibodies. Supplementary Table 4 shows the dilution of each antibody used for immunofluorescence staining. Slides were mounted with anti-fade mounting medium with DAPI (Vector Labs).
For 8-oxoG staining mouse prostate tissue sections were deparaffinized by passing through 100% xylene, sequential 10-minute washes with 100%, 90% and 70% ethanol followed by 5 minutes in PBS. The slides were treated with RNase A (200 μg/mL in PBS) for 1 hour at 37°C. After washing with PBS, DNA was denatured by incubation in 2N HCl for 10 minutes. For proteolysis, slides were incubated with 20 μg/mL proteinase K in 10 mM Tris/HCl pH7.5 and 5 mM EDTA for 30 minutes at 37°C. Slides were rinsed with PBS twice, then incubated with 8-oxoguanine antibody (1:250 Millipore) at 4°C overnight. The fluorescence signal was amplified with HRP-conjugated anti-mouse IgG (1:100) for 30 min, and then incubated with tyramide for 8 minutes. Slides were mounted with anti-fade mounting medium with DAPI.
In situ TUNEL staining of murine prostate tissues
For TUNEL assay (20), slides of prostate tissue were fixed with 10% formalin for 24–48 hours. Deparaffinized slides were permeabilized with 20 μg/ml proteinase K in 10 mM Tris 7.5 and 5 mM EDTA at room temperature for 30 minutes. Slides were rinsed three times in PBS for 5 minutes each then incubated with TUNEL assay reagent (In Situ Cell Death Detection Kit, Fluorescein, Roche) at 37°C for 60 minutes. Slides were again rinsed three times with PBS with 0.1% Tween 20 for 5 minutes each and then mounted with anti-fade mounting medium with DAPI (Vector Labs).
Irradiation of mice used as positive controls for stress response and DNA damage were done at 2 months of age using a single dose of 15Gy from a Gammacell 40 Exactor (Best Theratronics, Ottawa, Canada). Mice were sacrificed 1.5–2 hours after irradiation.
Analysis of prostate secretory proteins
Prostate secretory proteins were prepared as reported by Fujimoto et al. (21). Each dissected prostate lobe from 12-week-old mice was rinsed well in saline and placed in an Eppendorf tube with 100 μl saline containing protease inhibitors (Roche). Each lobe was cut into four or five pieces and left to stand for 5 minutes on ice. After centrifugation at 10,000g for 5 minutes at room temperature, the supernatant was collected and assayed for protein concentration with the Protein Assay Kit (Bio-Rad Laboratories). Fifteen μg of sample protein was resolved by SDS-PAGE and stained with Coomassie brilliant blue.
Statistical analysis
Immunohistochemical staining was analyzed with Image J software. The intensity of Nkx3.1 was normalized to that of wild type mice at the same time points. Intensities of 53BP1, pATM and 8-oxoG were normalized to that of 2 month-old wild type mice. Nkx3.1+/+ and Nkx3.1S186A/- mice were compared with that of Nkx3.1+/− mice with t-test. A p-value of <0.05 was considered to be statistically significant. t-Test of a group was applied to all the mice of same genotype analyzed as one group and comparison was made between groups. In analysis of slopes, t-test comparison was performed between slopes of regression lines.
Results
Engineering Mice with Nkx3.1(S186A)
NKX3.1(S185A) is the target for DYRK1B phosphorylation that controls steady state turnover of the protein and triggers polyubiquitination and protein turnover (15,16). Comparison the human and murine NKX3.1 primary protein structure suggested that Nkx3.1 serine 186 was the analogous residue to serine 185 in the human protein (Supplementary Figure 1A). We introduced a serine to alanine missense mutation at Nkx3.1 serine 186 and observed that the protein half-life was prolonged similar to the effect of the serine 185 to alanine replacement in NKX3.1 (Supplementary Figure 1B). Based on this result, we used CRISPR-Cas9 to engineer the identical alteration in mice (Supplementary Figures 2A and B), generating two founder mice carrying the desired mutation. During the entire breeding process and subsequently no mice with the Nkx3.1S186A/S186A genotype were ever identified leading us to assume that this genotype conferred embryonic lethality. One of these founders established a line that was back-crossed to wild-type C57BL/6 for 7–10 generations to ensure that the Nkx3.1S186A point mutation was the only genetic alteration we were testing. Over one year, we followed Nkx3.1 wild type mice, Nkx3.1+/− heterozygous mice (heterozygotes for the null allele), and Nkx3.1S186A/- mice (point mutation in trans to the null allele), that were all maintained in a congenic C57BL/6 strain background.
Prostate Weight, Histology, and Nkx3.1 Expression
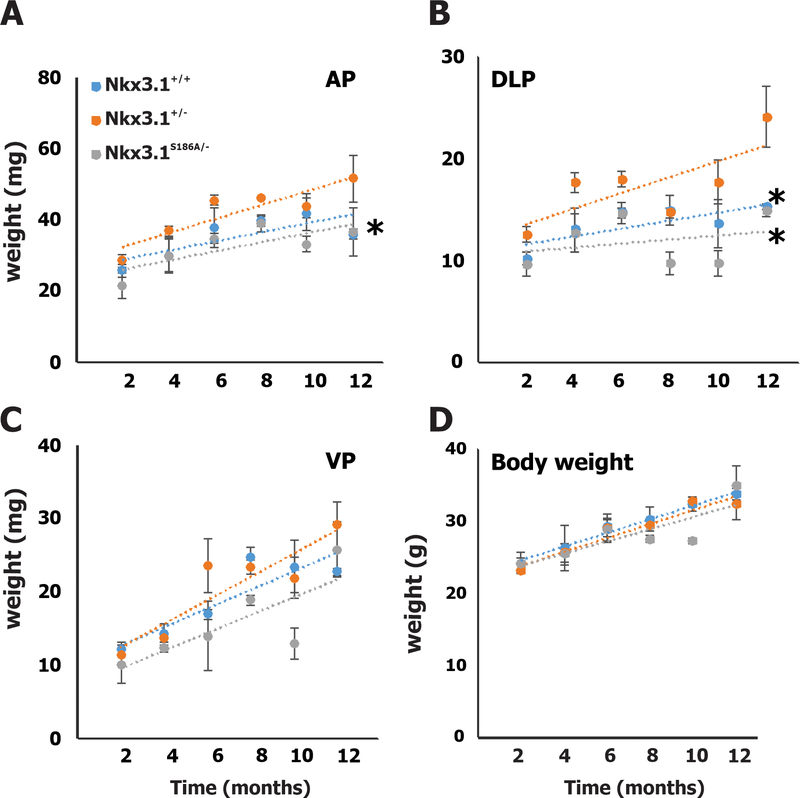
At two-month intervals prostate glands from each of three mice of each genotype were dissected and anterior, ventral, and dorsolateral lobes were analyzed separately. Anterior (Figure 1A) and dorsolateral lobes (Figure 1B) of the prostate glands were largest in the Nkx3.1+/− mice and smallest in the Nkx3.1S186A/- mice. The ventral prostates from Nkx3.1S186A/- mice were smaller than from the other two genotypes, but not to a degree of statistical significance (Figure 1C). Also, Nkx3.1 genotype had no effect on body weight over the year of experimental observation (Figure 1D).
Figure 1.
Prostate lobe weights at different time points. Statistical differences between Nkx3.1+/− mice with Nkx3.1+/+ and Nkx3.1S186A/- mutant mice was analyzed by t-test. * p<0.05.
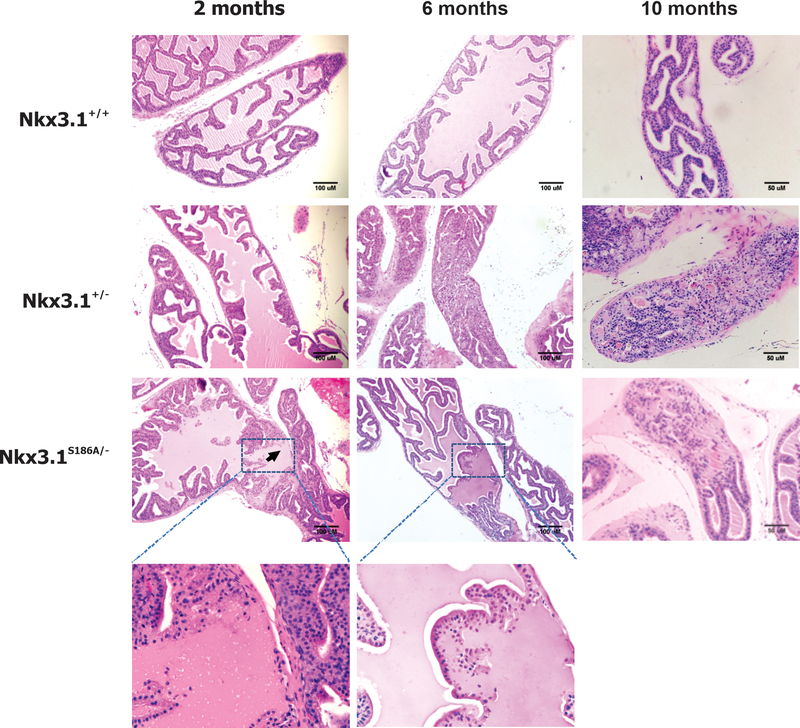
Histologic examination of the anterior prostate lobe shows the expected hyperplasia and dysplasia in Nkx3.1+/− mice that progressed as the mice aged (6). Compared to Nkx3.1+/+ the excess number of cells and distorted glandular architecture can be easily seen. In contrast, Nkx3.1S186A/- mice showed no evidence of hyperplasia but did have glands with areas of disrupted architecture as shown by the higher magnification panels in Figure 2. Similar histologic findings at the specified time points were seen in the dorsolateral prostatic lobes (Supplementary Figure 3A). The cellular hyperplasia and changes in nuclear morphology in Nkx3.1+/− mice can be seen at higher magnification, confirming previous observations about the effect of Nkx3.1 allelic loss (Supplementary Figures 3B and 3C). In contrast, despite effects on prostate weight, few effects of either Nkx3.1+/− or Nkx3.1S186A/- genotypes were seen in the ventral prostatic lobes during the course of the experiment.
Figure 2.
Histologic sections of anterior prostate lobe at different time points. H&E staining of anterior prostate sections are shown for each genotype at 2, 6, and 10 months of age. The higher magnifications show paucicellular areas seen in glands of Nkx3.1S186A/- mice but not in other genotypes.
At two months of age the histologic sections were nearly indistinguishable except for early hyperplasia in Nkx3.1+/− mice and the presence of paucicellular areas in prostates of Nkx3.1S186A/- mice. The mutations did not disrupt the normal architectural organization of the glands as further confirmed by immunohistochemical staining for cytokeratins 5 and 8 (Supplementary Figure 4A). To demonstrate further that prostate function was minimally affected in the different genotypes, we performed electrophoretic separation of prostate secretory proteins from the different prostate lobes and included seminal vesicle secretory proteins for comparison with an organ in which Nkx3.1 is not expressed. Similar to results previously published, we saw very few differences in secretions affected by genotype (6) (Supplementary Figure 4B).
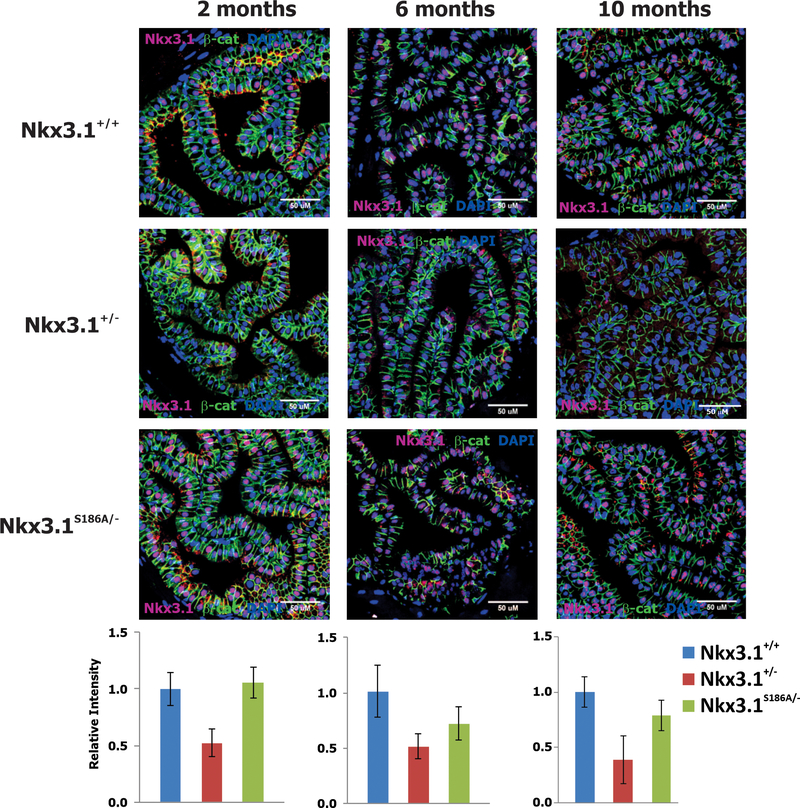
We predicted that the Nkx3.1S186A/- mice would have higher expression of Nkx3.1 compared to the congenic Nkx3.1+/− mice. We utilized a quantitative confocal assay for Nkx3.1 levels previously validated to compare Nkx3.1 protein expression across the three genotypes (8,10). In fact, Nkx3.1 levels were reduced in Nkx3.1+/− mice compared both to Nkx3.1+/+ and Nkx3.1S186A/- mice (p<0.01), but the protein expression in the latter two genotypes were not different statistically (Figure 3). Importantly, Nkx3.1(S186A) levels in the anterior prostate lobes was persistent throughout the duration of the observation period. This was not as true for the dorsolateral prostatic lobes where Nkx3.1(S186A) expression was sustained through six months, but was reduced at 10 months of age (Supplementary Figure 5).
Figure 3.
Nkx3.1 expression. Nkx3.1 was detected in paraffin sections from anterior prostate lobes of Nkx3.1+/+, Nkx3.1+/−, and Nkx3.1S186A/- mice at 2, 6, and 10 months of age. Counterstain was done with antibody to β-catenin and DAPI. Confocal images were quantitated with image J and Nkx3.1 expression was plotted relative to adjacent β-catenin expression. Data are from a minimum of ten fields. Nkx3.1 expression in Nkx3.1S186A/- mice differed from both Nkx3.1+/+ and Nkx3.1+/− mice by t-test with p<0.01.
Proliferation and TUNEL Staining
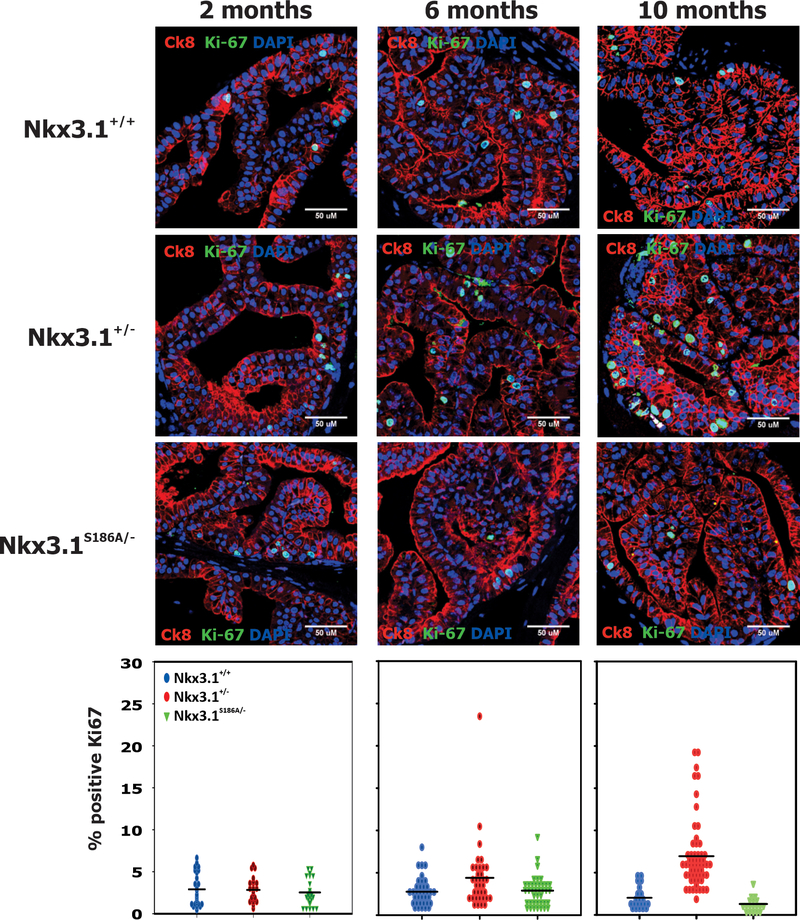
Cell proliferation in the tissue sections was assayed using Ki-67 staining. Nkx3.1+/− mice had increased levels of Ki-67-staining cells compared to either Nkx3.1+/+ or Nkx3.1S186A/- mice. The fraction of Ki-67 cells in the anterior prostate of Nkx3.1+/− mice increased with age (Figure 4). No changes over 10 months were seen in the fraction of Ki-67 staining cells in either Nkx3.1+/+ or Nkx3.1S186A/- mice. In dorsolateral prostate lobes Ki-67 staining in the Nkx3.1+/− mice was greatest at 6 months but declined on all three genotypes by 10 months (Supplementary Figure 6).
Figure 4.
Cell proliferation. Immunostaining for Ki-67 was done with cytokeratin 8 and DAPI as counterstains in sections of anterior prostate lobes. Confocal images were quantitated with image J and the number of Ki-67-positive cells per field as a percentage of all nuclei in the field were recorded. A minimum of ten fields were assayed and all cells were counted. Each point represents the percentage of Ki-67 positive cells in one field. The distribution of Ki-67 positive cells in Nkx3.1+/− mice differed from both Nkx3.1+/+ and Nkx3.1S186A/- mice at both 6 and 10 months by t-test with p<0.01.
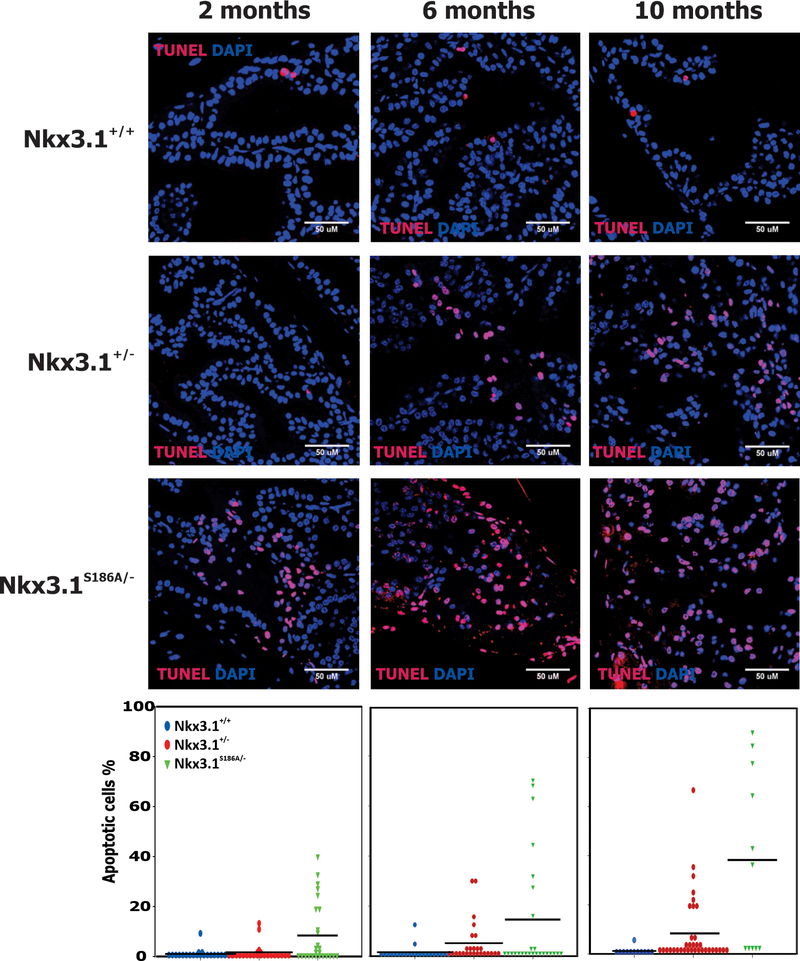
Because NKX3.1 is a mediator of DNA damage response in prostate epithelial cells (11), we utilized TUNEL staining, an assay for DNA ends, as an indicator for DNA damage and repair (22). Representative examples of anterior prostate lobes are shown at 2, 6, and 10 months of age (Figure 5). At every time point a single copy of Nkx3.1(S816A) that showed the same Nkx3.1 staining intensity as two copies of the wild type gene, resulted in a higher level of TUNEL staining. Therefore, it appears that not only protein levels, but Nkx3.1 protein turnover affected the cell’s response to DNA damage. This conclusion was reinforced by TUNEL staining of dorsolateral prostate lobes (Supplementary Figure 7). Despite the finding that Nkx3.1 staining intensity had decreased in the DLP by 10 months, we nevertheless found a level of TUNEL staining as an index of DNA breakage and repair much higher than in either Nkx3.1+/+ or Nkx3.1+/− mice. However, although Nkx3.1(S186A) affected the concentration of DNA ends available for end-labeling, TUNEL staining in these experiments did not assay for either activation of the DNA damage response or apoptosis. We examined prostate sections at all time points for DNA double-strand breakage by staining for γ-histone2AX. We saw no changes in the presence of γ-histone2AX across all genotypes and time points. We also performed staining for cleaved caspase 3 as a direct assay of the apoptotic cascade and found no change at any time point in anterior prostate lobes (Supplementary Figure 8A) with a positive control using exposure to 15Gy γ-irradiation (Supplementary Figure 8B). We also saw no change over time in cleaved caspase 3 staining in dorsolateral prostate lobes (Supplementary Figure 8C).
Figure 5.
TUNEL staining was done on paraffin sections from anterior prostate lobes of Nkx3.1+/+, Nkx3.1+/−, and Nkx3.1S186A/- mice at 2, 6, and 10 months of age. Counterstain was done with DAPI. Confocal images were quantitated with image J and the number of TUNEL-positive cells per field were recorded compared to the total number of nuclei seen in the field. A minimum of ten fields were assayed and each point represents the percentage of TUNEL positive cells in a field. The percentage of TUNEL positive cells in Nkx3.1S186A/- mice differed from both Nkx3.1+/+ and Nkx3.1+/− mice by t-test with p<0.01.
Early Effects of Long Half-Life Nkx3.1(S186A)
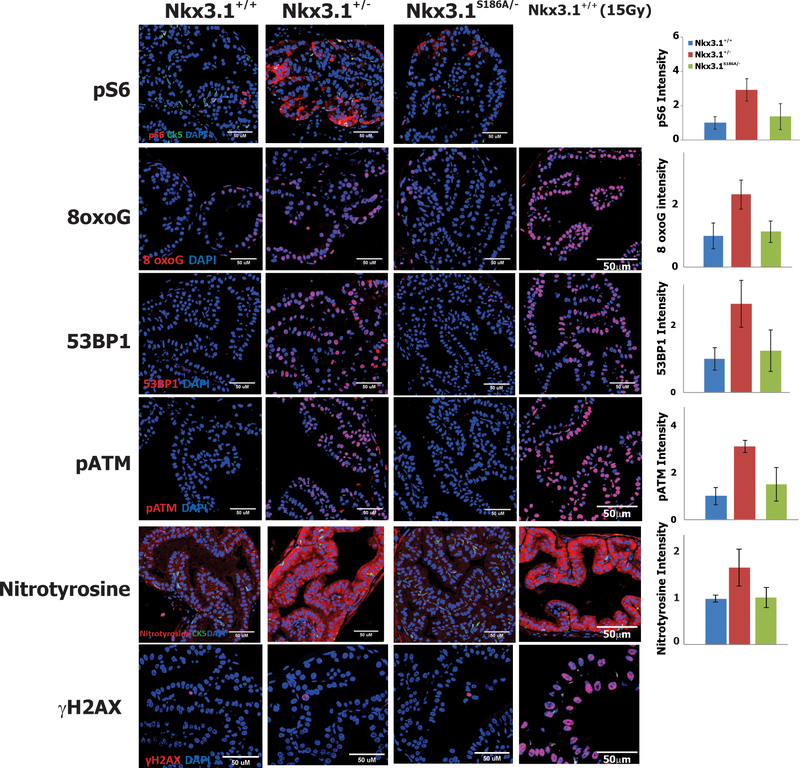
NKX3.1 has been shown to affect numerous cellular processes in both human and murine cells that affect both response to free-radical oxidation and DNA repair (11,12,23–25). To compare the effects of a single copy of Nkx3.1(S186A) with the effects of biallelic, native Nkx3.1 as they both compared to Nkx3.1 heterozygosity prior to the appearance of histologic changes, we examined the expression of a panel of marker proteins by immunohistochemistry in 2-month old mice (Figure 6). As a positive control we included staining of 2-month old wild type murine prostate collected two hours after exposure to 15Gy (far right panels in Figure 6). Phosphorylated ribosomal protein S6, a marker for the activity of cellular metabolism (26), was seen to be more abundant in the Nkx3.1+/− prostate than in either Nkx3.1+/+ or Nkx3.1S186A/−. The prostate in Nkx3.1+/− mice was also more susceptible to oxidation and formation of reactive oxygen species. This was demonstrated by increased levels of 8-oxoguanine adducts (27) and increased levels of nitrotyrosine (28–30). We also found that there was increased presence of 53BP1, a protein expressed in response to DNA damage (31,32), and its downstream mediator of response, phosphorylated ATM. These findings seen in this study only in Nkx3.1+/− mice, all indicate that the mere reduction of Nkx3.1 protein in histologically normal tissue increases cellular stress by increased metabolism and increased oxidative damage that activates cellular defense responses. At two months and, in fact, at no time point, did we observe evidence of double-stranded DNA breaks as indicated by staining for γ-histone2AX as was seen in prostates of irradiated mice at the bottom of Figure 6.
Figure 6.
Metabolic and Stress Response Activation Suppressed by Nkx3.1(S186A). The figure shows confocal immunofluorescent micrographs of anterior prostate lobes from 2-month old mice of the indicated genotype. The micrographs on the far right show the staining of prostates from wild-type mice 2 hours after exposure to 15Gy γ-irradiation. Cytokeratin 5 was employed for pS6 and nitrotyrosine staining to show preservation of histologic architecture. Staining intensities were measured by image J and shown relative to intensity in Nkx3.1+/+ mice in the respective histograms on the far right.
Discussion
NKX3.1 is a critical early cancer suppressor for which mere reduction in protein levels is pathogenic. NKX3.1 levels are under downward pressure from changes in the prostate that occur during aging including inflammatory atrophy (33) and prostatitis (34). Prostatitis is a well-established risk factor for prostate cancer (35–37). Primary prostate cancer samples also exhibit downregulation of NKX3.1 mRNA and NKX3.1 allelic loss (8). The importance of NKX3.1 loss to the initiation of human prostate cancer is underscored by the finding that the single most common genetic lesion in sporadic prostate cancer is loss of NKX3.1 (4). The influence of NKX3.1 in prostate cancer pathogenesis is ubiquitous, even though the gene is rarely affected by the usual lesions of tumor suppressor genes. For example, there has been only one family with heritable risk for prostate cancer that was linked to an NKX3.1 mutation that partially inactivated the protein (38) and only four somatic mutations of NKX3.1 were reported in the initial TCGA cohort of 333 prostate cancer cases (13). Recently a comprehensive analysis of Asian prostate cancer samples showed as well that loss at 8p21.3, the locus of NKX3.1 was consistently found in this cohort as well (39).
NKX3.1 is a multifunctional protein that appears to have been adapted evolutionarily for unique versatility in prostate epithelial cells. The protein by itself is a transcriptional repressor (40) and activates transcription only as a coactivator, such as with serum response factor (41–44). However, through binding of the homeodomain to other proteins NKX3.1 is a cofactor for androgen receptor mediated transcription (45–47). NKX3.1 also is involved in the DNA damage response of prostate cancer cells by virtue of specific damage-induced tyrosine phosphorylation at amino acid 225 that licenses interaction and activation of ATM (12). Moreover, Nkx3.1 affects the expression of numerous proteins that mediate the cellular response to reactive oxygen species (23), cellular differentiation, and DNA repair (48). In the absence of Nkx3.1, although mice are viable and fertile, there are defects in prostatic ductal branching and secretory protein expression (6). Therefore, its loss creates the milieu favoring stochastic generation of DNA damage contributing to prostate carcinogenesis. Of substantial importance to carcinogenesis, is that loss of NKX3.1 expression predisposes to TMPRSS2/ERG gene rearrangement, an early pathognomonic lesion in more than half of all prostate cancers (10,49).
We confirm that loss of a single Nkx3.1 allele causes histologic disruption of the prostate over time. Strikingly, monoallelic expression of Nkx3.1, which produces about 2/3 the normal level of cellular protein (8), alone is sufficient to activate indicators of cellular metabolic activity like pS6, induce evidence of DNA damage such as 8-oxoguanine and nitrotyrosine, and activate cellular stress response as indicated by expression of 53BP1 and phosphorylation of ATM. Thus, the prostate gland is highly sensitive to extant endogenous stressors and requires continuous expression of NKX3.1 and its normal turnover for stability. Prolongation of Nkx3.1 half-life was sufficient to abrogate all these effects in 2-month old mice as well abrogating the later histologic changes resulting from decreased Nkx3.1 expression. However, despite recovery of apparently normal levels of Nkx3.1 transcribed in Nkx3.1S186A/- mice, the nonphysiologic protein turnover induced a robust level of DNA end-labeling suggestive of DNA damage repair regions of epithelial architectural disruption. TUNEL staining is a sensitive assay for free 3’ DNA ends and is known to score physiologic conditions other than apoptosis (22,50). Thus, our mouse model further demonstrates the extraordinary sensitivity of the prostate gland to carefully controlled levels and turnover of this protein and suggests the critical importance of this protein in the pathogenesis of prostate cancer in the aging male.
Lastly, we note that during breeding over 30 months at two institutions, we never identified a single pup with the Nkx3.1S186A/S186A genotype. If this genotype confers embryonic lethality we speculate that this is due to the effects of Nkx3.1 during embryogenesis. Nkx3.1 is expression is widespread in the mouse during embryogenesis and is seen progressively during somite development (51,52). Although this process clearly is able to compensate for complete loss of Nkx3.1, the mutant protein resistant to proteolysis expressed in a homozygous Nkx3.1S186A/S186A genotype seems to disrupt murine development to prevent successful development.
Supplementary Material
Statement of Significance.
Findings show that prolonging the half-life of Nkx3.1 reduces proliferation, enhances DNA end-labeling, and protects from DNA damage, ultimately blocking the proneoplastic effects of Nkx3.1 allelic loss.
Acknowledgements
The authors thank Cory Abate-Shen for comments on the manuscript and for decades of collaboration. Supported by the Falconwood Foundation (E. Gelmann), by NCI grant P01 CA154293 (M.Shen and E. Gelmann), R01CA238005 (M. Shen), K99CA194287 (M.Shen), and by CCSG P30 CA013696-36 via the Microscopy and Animal Shared Resources. This work was also supported by the Tissue Acquisition and Cellular / Molecular Analysis Shared Resource Research of the Arizona Cancer Center supported by the National Cancer Institute of the National Institutes of Health under award number P30 CA02307.
Footnotes
The authors have no competing interests and no conflicts to disclose.
References
- 1.Vocke CD, Pozzatti RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, et al. Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p21–22. Cancer Res 1996;56:2411–6 [PubMed] [Google Scholar]
- 2.Swalwell JI, Vocke CD, Yang Y, Walker JR, Grouse L, Myers SH, et al. Determination of a minimal deletion interval on chromosome band 8p21 in sporadic prostate cancer. Genes ChromosomesCancer 2002;33:201–5 [DOI] [PubMed] [Google Scholar]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, et al. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res 2002;62:2999–3004 [PubMed] [Google Scholar]
- 6.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Norton CR, et al. Roles for Nkx3.1 in prostate development and cancer. Genes and Development 1999;13:966–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009;461:495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asatiani E, Huang WX, Wang A, Rodriguez OE, Cavalli LR, Haddad BR, et al. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res 2005;65:1164–73 [DOI] [PubMed] [Google Scholar]
- 9.Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, et al. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res 2000;60:6111–5 [PubMed] [Google Scholar]
- 10.Bowen C, Zheng T, Gelmann EP. NKX3.1 Suppresses TMPRSS2-ERG Gene Rearrangement and Mediates Repair of Androgen Receptor-Induced DNA Damage. Cancer Res 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowen C, Gelmann EP. NKX3.1 activates cellular response to DNA damage. Cancer Res 2010;70:3089–97 [DOI] [PubMed] [Google Scholar]
- 12.Bowen C, Ju JH, Lee JH, Paull TT, Gelmann EP. Functional activation of ATM by the prostate cancer suppressor NKX3.1. Cell Rep 2013;4:516–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan A, Rao V, De Marzo AM, Bieberich CJ. Regulating NKX3.1 stability and function: Post-translational modifications and structural determinants. Prostate 2016 [DOI] [PubMed] [Google Scholar]
- 15.Markowski MC, Bowen C, Gelmann EP. Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3.1. Cancer Res 2008;68:6896–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song LN, Silva J, Koller A, Rosenthal A, Chen EI, Gelmann EP. The Tumor Suppressor NKX3.1 Is Targeted for Degradation by DYRK1B Kinase. Mol Cancer Res 2015;13:913–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurel B, Ali TZ, Montgomery EA, Begum S, Hicks J, Goggins M, et al. NKX3.1 as a marker of prostatic origin in metastatic tumors. AmJSurgPathol 2010;34:1097–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muhlbradt E, Asatiani E, Ortner E, Wang A, Gelmann EP. NKX3.1 activates expression of insulin-like growth factor binding protein-3 to mediate insulin-like growth factor-I signaling and cell proliferation. Cancer Res 2009;69:2615–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 2014;9:1956–68 [DOI] [PubMed] [Google Scholar]
- 20.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labelling of nuclear DNA fragmentation. JCell Biol 1992;119:493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto N, Akimoto Y, Suzuki T, Kitamura S, Ohta S. Identification of prostatic-secreted proteins in mice by mass spectrometric analysis and evaluation of lobe-specific and androgen-dependent mRNA expression. J Endocrinol 2006;190:793–803 [DOI] [PubMed] [Google Scholar]
- 22.Kanoh M, Takemura G, Misao J, Hayakawa Y, Aoyama T, Nishigaki K, et al. Significance of myocytes with positive DNA in situ nick end-labeling (TUNEL) in hearts with dilated cardiomyopathy: not apoptosis but DNA repair. Circulation 1999;99:2757–64 [DOI] [PubMed] [Google Scholar]
- 23.Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-Function of Nkx3.1 Promotes Increased Oxidative Damage in Prostate Carcinogenesis. Cancer Res 2005;65:6773–9 [DOI] [PubMed] [Google Scholar]
- 24.Erbaykent-Tepedelen B, Karamil S, Gonen-Korkmaz C, Korkmaz KS. DNA damage response (DDR) via NKX3.1 expression in prostate cells. JSteroid BiochemMolBiol 2014;141:26–36 [DOI] [PubMed] [Google Scholar]
- 25.Martinez EE, Anderson PD, Logan M, Abdulkadir SA. Antioxidant treatment promotes prostate epithelial proliferation in Nkx3.1 mutant mice. PLoS One 2012;7:e46792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 2005;19:2199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. MutatRes 2003;531:127–39 [DOI] [PubMed] [Google Scholar]
- 28.Campolo N, Issoglio FM, Estrin DA, Bartesaghi S, Radi R. 3-Nitrotyrosine and related derivatives in proteins: precursors, radical intermediates and impact in function. Essays Biochem 2020;64:111–33 [DOI] [PubMed] [Google Scholar]
- 29.Murata M, Kawanishi S. Oxidative DNA damage induced by nitrotyrosine, a biomarker of inflammation. Biochem Biophys Res Commun 2004;316:123–8 [DOI] [PubMed] [Google Scholar]
- 30.Brennan ML, Wu W, Fu X, Shen Z, Song W, Frost H, et al. A tale of two controversies: defining both the role of peroxidases in nitrotyrosine formation in vivo using eosinophil peroxidase and myeloperoxidase-deficient mice, and the nature of peroxidase-generated reactive nitrogen species. The Journal of biological chemistry 2002;277:17415–27 [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. Embo J 2010;29:574–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward IM, Minn K, van DJ, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. MolCell Biol 2003;23:2556–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, et al. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res 2006;66:10683–90 [DOI] [PubMed] [Google Scholar]
- 34.Khalili M, Mutton LN, Gurel B, Hicks JL, De Marzo AM, Bieberich CJ. Loss of Nkx3.1 expression in bacterial prostatitis: a potential link between inflammation and neoplasia. AmJPathol 2010;176:2259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson WG, De Marzo AM, DeWeese TL, Isaacs WB. The role of inflammation in the pathogenesis of prostate cancer. JUrol 2004;172:S6–11 [DOI] [PubMed] [Google Scholar]
- 36.Roberts RO, Bergstralh EJ, Bass SE, Lieber MM, Jacobsen SJ. Prostatitis as a risk factor for prostate cancer. Epidemiology 2004;15:93–9 [DOI] [PubMed] [Google Scholar]
- 37.De Marzo AM, Meeker AK, Zha S, Luo J, Nakayama M, Platz EA, et al. Human prostate cancer precursors and pathobiology. Urology 2003;62:55–62 [DOI] [PubMed] [Google Scholar]
- 38.Zheng SL, Ju JH, Chang BL, Ortner E, Sun J, Isaacs SD, et al. Germ-Line Mutation of NKX3.1 Cosegregates with Hereditary Prostate Cancer and Alters the Homeodomain Structure and Function. Cancer Res 2006;66:69–77 [DOI] [PubMed] [Google Scholar]
- 39.Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020;580:93–9 [DOI] [PubMed] [Google Scholar]
- 40.Steadman DJ, Giuffrida D, Gelmann EP. DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1. Nucleic Acids Res 2000;28:2389–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju JH, Maeng JS, Lee DY, Piszczek G, Gelmann EP, Gruschus JM. Interactions of the acidic domain and SRF interacting motifs with the NKX3.1 homeodomain. Biochemistry 2009;48:10601–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Fillmore RA, Zimmer WE. Structural and functional analysis of domains mediating interaction between the bagpipe homologue, Nkx3.1 and serum response factor. Exp Biol Med (Maywood) 2008;233:297–309 [DOI] [PubMed] [Google Scholar]
- 43.Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, et al. Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor. JMolBiol 2006;360:989–99 [DOI] [PubMed] [Google Scholar]
- 44.Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The Smooth Muscle gamma -Actin Gene Promoter Is a Molecular Target for the Mouse bagpipe Homologue, mNkx3–1, and Serum Response Factor. JBiolChem 2000;275:39061–72 [DOI] [PubMed] [Google Scholar]
- 45.Song LN, Bowen C, Gelmann EP. Structural and functional interactions of the prostate cancer suppressor protein NKX3.1 with topoisomerase I. The Biochemical journal 2013;453:125–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowen C, Stuart A, Ju JH, Tuan J, Blonder J, Conrads TP, et al. NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res 2007;67:455–64 [DOI] [PubMed] [Google Scholar]
- 47.Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 2015;160:367–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang CC, Chung A, Ku CY, Brill LM, Williams R, Wolf DA. Systems analysis of the prostate tumor suppressor NKX3.1 supports roles in DNA repair and luminal cell differentiation. F1000Res 2014;3:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008;10:177–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charriaut-Marlangue C, Ben-Ari Y. A cautionary note on the use of the TUNEL stain to determine apoptosis. Neuroreport 1995;7:61–4 [PubMed] [Google Scholar]
- 51.Bieberich CJ, Fujita K, He WW, Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. JBiolChem 1996;271:31779–82 [DOI] [PubMed] [Google Scholar]
- 52.West DW, Slattery ML, Robison LM, French TK, Mahoney AW. Adult dietary intake and prostate cancer risk in Utah: a case- control study with special emphasis on aggressive tumors. Cancer CausesControl 1991;2:85–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.