Abstract
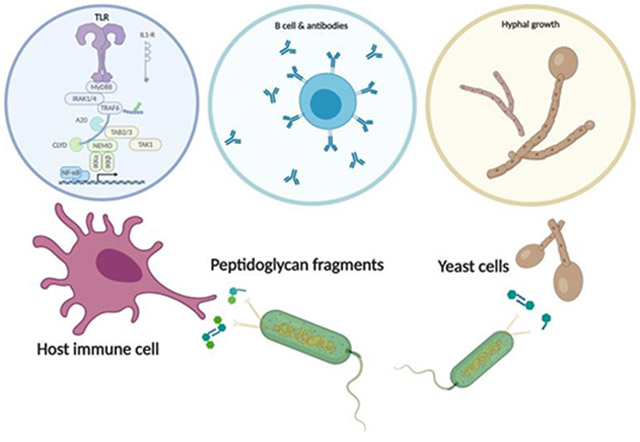
The interaction between host immunity and bacterial cells plays a pivotal role in a variety of human diseases. The bacterial cell wall component peptidoglycan (PG) is known to stimulate an immune response, which makes PG a distinctive recognition element for unveiling these complicated molecular interactions. Pattern recognition receptor (PRR) proteins are among the critical components of this system that initially recognize molecular patterns associated with microorganisms such as bacteria and fungi. These molecular patterns are mostly embedded in the bacterial or fungal cell wall structure and can be released and presented to the immune system in various situations. Nonetheless, detailed knowledge of this recognition is limited due to the diversity among the PG polymer and its fragments; the subsequent responses by multiple hosts add more complexity. Here, we discuss how our understanding of the role and molecular mechanisms of the well-studied PRR, the NOD-like receptors (NLRs), in the human immune system has evolved in recent years. We highlight the instances of other classes of proteins with similar behavior in the recognition of PG that have been identified in other microorganisms such as yeasts. These proteins are particularly interesting because a network of cellular interactions exists between human host cells, bacteria and yeast as a part of the normal human flora. To support our understanding of these interactions, we provide insight into the chemist’s toolbox of peptidoglycan probes that aid in the investigations of the behaviors of these proteins and other biological contexts relevant to the sensing and recognition of peptidoglycan. The importance of these interactions in human health for the development of biomarkers and biotherapy is highlighted.
Graphical Abstract
Introduction
The initial recognition of a bacterial pathogen and the human cell is something like a handshake. The human cell is “feeling” for things that are different from self. These first interactions are governed by the innate immune system.1 The intricate system uses a series of receptors to do this handshake and there are a multitude of signals that the hand could grab. Bacterial peptidoglycan is one of those signals that the immune system is searching to find in this complex landscape. This bacterial polymer proves to be an excellent recognition element for the innate immune system, as bacterial cells wrap their cells in it and human cells do not. However, there is complexity to this system, as not all bacterial peptidoglycans are exactly the same. The diversity among peptidoglycan means that there must also exist a multitude of ways in which the polymer and its fragments are detected and then subsequently responded to by various hosts. A fundamental understanding of recognition and activation of peptidoglycan by the host unleashes a framework for unveiling the molecular mechanisms of a plethora of pathological diseases, and physiological indications that remain underexplored. Characterizing the molecular recognition of peptidoglycan (PG) by the human host will uncover not only immunological parameters but also physiological and metabolic parameters that are at the forefront of human health and disease.
In this perspective, we will provide a brief review of the current findings of PG recognition and its implications for human health, as we introduce a relatively new player in the recognition and sensing of PG: Candida albicans (C. albicans). We will compile existing knowledge of peptidoglycan chemistry and make recommendations based on our own contributions to the field, its implications and applications in innate immunity while introducing its future in adaptive immunity. We also offer an introduction to the chemical functionalization of peptidoglycan fragments and their applications for peptidoglycan recognition and sensing.
Structure and Diversity of Peptidoglycan
Peptidoglycan (PG) is the primary structural component of the bacterial cell wall.2 It is the exterior surface of Gram-positive (Gram (+)) bacterium and the surface layer just below the outer membrane in Gram-negative (Gram (−)) bacterium. It is a heteropolymer of linear glycan strands crosslinked by short pentapeptide bridges.2,3 The glycan strands are formed by repeating units of the disaccharide β-1,4-linked N-acetylglucosamine and N-acetylmuramic acid (GlcNAc-MurNAc), and the pentapeptide bridges are attached to the lactyl moiety of MurNAc via an amide linkage.4 The typical peptide structure is: 1) l-alanine, 2) d-glutamate, 3) an amine functionalized amino acid, 4) d-alanine and 5) d-alanine.5 Importantly, the second and third amino acid are linked via the carboxylic acid in glutamate’s side chain, not the α-carboxylic acid (Figure 1). Two of the most prominent examples are the following: in most Gram (−) bacteria, the peptide moiety at the third position is m-DAP (meso-diaminopimelic acid), and connects the glycan strands through direct m-DAP-d-Ala cross linkage, whereas in Gram (+) bacteria the third amino acid is lysine and is cross-linked to other glycan strands via glycine bridges.6
Figure 1:
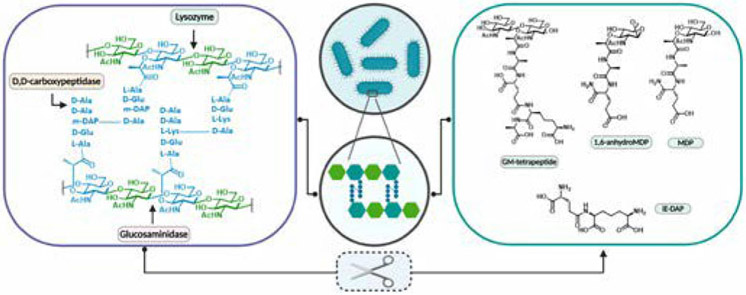
Schematic representation of peptidoglycan and corresponding fragments upon enzymatic digestion. Left: Cross-linked peptidoglycan. Depicted in green is N-acetyl glucosamine and in blue N-acetyl muramic acid with the corresponding representative peptide linkage. Right: representative set of possible PG fragments upon digestion with enzymes such as Lysozyme and Carboxypeptidases. Note that MDP is a synthetic fragment and has not been shown to be produced in a biological context. The remaining fragments have been observed upon peptidoglycan enzymatic digestion with various enzymes, for example muramidase, lysozyme, and lytic transglycosylases.12,13
Though the glycan backbone is generally conserved in bacteria, the peptide moiety exhibits considerable diversity among Gram (−) and Gram (+) bacteria (Figure 1).7 Substitution of m-DAP by other amino acids such as lanthionine and its stereochemical variant ll-DAP has been reported for Gram (−) bacteria Fusobacterium nucleatum8 and Porphyromonas gingivalis9 respectively. Similarly, in the Gram (+) bacteria Herpetosiphen aurantiacus10 and Ornithinmicrobium humiphilum11 m-DAP is completely replaced by l-ornithine.
The rigid structure of PG lends to its diverse functions that play critical roles for the protection and integrity of the bacterial cell.14 Premature or unwanted degradation of PG results in bacterial cell lysis.14 The presence of PG is multifaceted, as it serves to preserve and define cell integrity, as well as scaffold cell components such as lipopolysaccharides.15 The role of PG is very broad and has scientific relevance from antibacterial development16, the human microbiome17, to human innate immune signaling.1
PG has long been the target of antibiotics, since its integrity determines the survival of the bacterial cell.18 The well-known class of antibiotics, β-lactams, confers its antibiotic mechanism by inhibiting the synthesis of PG by binding directly to biosynthetic enzymes. This ultimately blocks the formation of the peptide crosslinks (Figure 1), thereby disrupting the formation of PG and subsequently resulting in osmotic lysis of the bacterial cell.19 As a survival mechanism, bacteria have adapted a plethora of ways to modify their PG backbones.
Such modifications are in the form of chemical variations of the glycan strands, for example, O-acetylation, N-glycolylation and de-N-acetylation of GlcNAc, MurNAc or both20 (Figure 2). O-acetylation has been found in pathogenic bacterial species Staphylococcus aureus and Neisseria gonorrhoeae16, and is responsible for lysozyme resistance among these species. The N-glycolyl modification has been shown in Mycobacterium tuberculosis and is added via a monooxygenase during biosynthesis; this addition of oxygen to the side chain increases the immunostimulatory properties of the PG.21,22 Another major modification, de-N-acetylation, is a prominent survival strategy of the human pathogen Listeria monocytogenes.23 De-N-acetylation of Listeria monocytogenes PG enhances the pathogen’s ability to survive the destruction of bacteriolytic activity of lysozyme.23
Figure 2:
Pathogen Recognition Receptor interplay in bacterial cell wall recognition. Homologous PG receptors exist for mammals, plants, and insects. A common domain found in these receptors is the evolutionary and structurally conserved Leucine Rich Repeat protein domain, which is not only responsible for the sensing and detection of PG fragments but is found across innate immune receptors.
In addition to the aforementioned postsynthetic modifications, modifications to the peptidoglycan backbone have also been attributed to flexibility in the assembly pathway. Gram (+) bacteria have evolved resistance to glycopeptide antibiotics such as Vancomycin. These drugs bind the peptidyl D -Ala- D -Ala end of PG precursors located at the cell surface, and also inhibit the transpeptidase activity of Penicillin Binding Protein activity that acts directly on the D -Ala- D -Ala peptide bond of their acyl donor substrate; subsequently inhibiting transglycosylation and transpeptidation. To circumvent this, bacteria have evolved the vanA gene cluster. This gene cluster encodes the biosynthetic enzymes, the VanH dehydrogenase and the VanH ligase24. Together they result in the formation of an ester bond between D -Ala- D -Lac, which subsequently leads to the production of PG ending in a D -Lac residue, instead of the usual D -Ala residue, thus eliminating glycopeptide antibiotic binding.25
The ability to pre and post synthetically modify its PG backbone provides not only a resistance mechanism to bacteria, but also an evolutionary advantage. While it is not certain which phenomena is the superior, it is evident that peptidoglycan modification confers both evolutionary and resistance advantages to bacteria and the two work in tandem in favor of both Gram(−) and (+) bacterium. It remains obvious that these PG diversity elements are critical in understanding bacterial recognition by the immune system.
Peptidoglycan Sensing and Recognition
Peptidoglycan is a dynamic structure that continuously undergoes remodeling during bacterial growth and reproduction, which results in the release of fragments from the bacterial cell wall into the localized environment; a process termed peptidoglycan turnover.26 Bacteria degrade approximately 40 to 50% of their peptidoglycan per generation as part of their normal peptidoglycan remodeling process required for cell wall expansion.27 The heteropolymer has to be at least partially degraded to allow for proper cell division before it is reconstructed to yield mature daughter cells.28,29 During this highly regulated process, small fragments are released into the milieu, and constitute a marker for bacterial presence and activity.30 (Figure 1)
Since peptidoglycan turnover is a highly regulated process, it requires stringent control at the transcriptional level to avoid autolysis and unintentional cell death.26 It is therefore controlled by more than one enzyme. Varying enzymatic control equates to multiple fragments that can be placed in the milieu at any given time, thus the chemical composition of these fragments can vary wildly (Figure 1). For instance, both lysozyme and lytic transglycosylases release disaccharide-peptides, but while the hydrolytic reaction of lysozyme generates a terminal reducing MurNAc, the lytic transglycosylase produces anhydromuropeptides which present a 1,6-anhydro ring at the MurNAc thereby producing an anhydroMurNAc moiety (Figure 1).31 PG turnover/recycling is not limited to Gram (−) or Gram (+) but is indeed a facet of both types of bacteria, further increasing the types of fragments produced.26,32
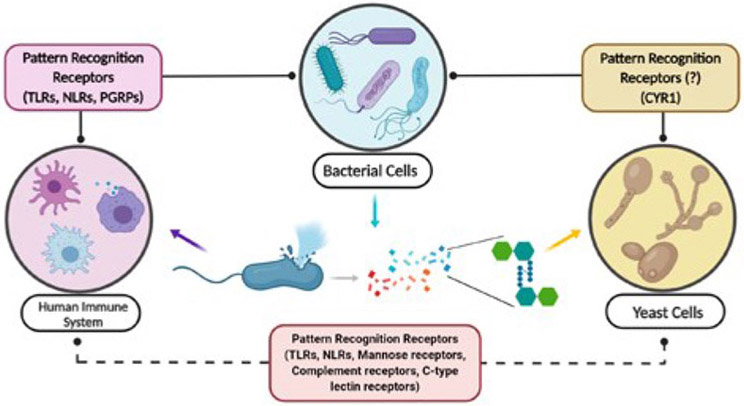
With the array of fragments that can be encountered by the host, one must imagine that nature has evolved inherent sensing mechanisms to generate the appropriate response. Indeed, this is the case as various hosts have evolved peptidoglycan recognition receptors that aid in its detection and subsequent processing. The molecular signatures present in bacteria that are absent in host cells (mammals, plants) are defined as microbe-associated molecular patterns (MAMPs).33 The detection of MAMPs is successful through specific receptors termed Pathogen Recognition Receptors (PRRs).34 Pathogen Recognition receptors are able to bind peptidoglycan and a plethora of other bacterial derived molecules such as lipopolysaccharides (LPS).34 For the purpose of this perspective we will elaborate only on the innate immune receptors and their activation as it pertains to PG alone, specifically MurNAc containing fragments. We note that many excellent reviews have been written on innate immune recognition.1,30,35-37
Plants, insects, and mammals have all evolved a number of germline encoded PRRs and peptidoglycan-recognition receptors.38 Once these receptors are activated, a host response is elicited for the particular class of invading microbe or bacteria. To date, much research has been conducted on Toll-like receptors (TLRs) and nucleotide oligomerization domain (NOD) like receptors (NLRs).39-42 NOD proteins are intracellular and regulatory proteins that respond to a variety of signaling molecules, including PG fragments.42,43 NOD1 and NOD2 are multi-domain proteins consisting of one or two CARD domains respectively, and a centrally located NOD domain followed by a number of C-terminal leucine rich repeats (LRRs).42
Our lab and others have demonstrated that the LRR domain of NOD2 binds to the synthetic bacterial PG fragment MDP.44,45 In addition, other labs have focused on NLRP3 and NOD146-48, which senses GlcNAc49 and m-DAP respectively. In contrast to NLRs, TLRs are integral membrane glycoproteins which are localized to the cell surface (TLR1,2,3) or to intracellular compartments (TLR7,9).41 Similar to Nod Like Receptors, Toll Like Receptors sense pathogens via their LRR domains, with the Pathogen Associated Molecular Patterns binding sites formed by insertions in leucine rich repeat loops. These receptors work in concert to generate the appropriate immune response, with substantial cross-talk between the receptors.50
In addition to triggering immunological responses upon activation, the aforementioned receptors, NLRs and TLRs, all share a common binding domain. Of particular interest in these PG recognition receptors, is the commonality of the LRR domain. Leucine rich repeats are generally 20–29 residues long and contain a conserved 11-residue segment with the consensus sequence LxxLxLxxN/CxL, where x can be any amino acid and leucine residues can also be substituted by valine, isoleucine and phenylalanine.51 Overall, LRRs display a curved shape with parallel β-sheet on the concave side and mostly helical segments on the convex side.52
A less studied LRR that senses bacterial peptidoglycan is the adenylyl cyclase, Cyr1p, from the human commensal C. albicans. Although yeasts, being singled celled organisms, are not considered to have an immune system, Cyr1p functions very much like an innate immune receptor, signaling to the cell that bacterial cell fragments are present and ultimately changing the phenotype of the C. albican’s cell. While much literature can be found on NRLs and TLRs, not much has been presented about the sensing and detection of yeast to bacterial peptidoglycan. Nonetheless, it is highly documented in the medical sector that there is a clear association between C. albicans pathogenesis and bacterial infections.53 C. albicans infections are often isolated with bacterial infections and in fact worsened in the presence of bacteria.54,55 Shing et al. reported that the presence of C. albicans can not only promote Group B Streptococcus urinary tract infections, but can also increase bacterial adherence to bladder epithelium thereby promoting bacterial colonization.56 We now shift our focus to the less characterized “PRR”, Cyr1p, and delve into the ways in which this organism senses and responds to PG.
A Fungal “Pathogen Recognition Receptor”?
C. albicans is a dimorphic fungus that is part of the commensal microbial flora in many healthy individuals.57-59 The microbial flora encompasses the microbiome and mycobiome.60 The microbiome is referred to as the collection of microorganisms that are resident in the human host at any given time.58 It encompasses a diverse ecosystem with an estimated 1,000 bacterial species.58 Juxtaposed to the microbiome is the collection of fungal species resident in the human host.59 This collection of species is referred to as the mycobiome, of which C. albicans is one of the most prominent members.57
As a normal resident of the human body, C. albicans typically coexists with the human host in a symbiotic manner and colonizes several niches in the skin, gastrointestinal and urogenital tracts in almost all healthy individuals.61 It is capable of altering its morphology from that of a budding yeast to filamentous state (hyphae/pseudohyphae) in response to niche disruption via immune incompetence and/or environmental changes.62 This morphological plasticity has severe implications for the symbiotic relationship of C. albicans and the human host. When the host immune defenses are impaired, or when the normal microbial flora is disrupted, the fungus can cause superficial as well as severe systemic infections. This phenomenon is characterized by a morphological transition between growth forms such as budding yeast (commensal state) to filamentous/hyphal form yeast (pathogenic state).62
A key signaling pathway for morphological regulation is the cAMP/protein kinase A (PKA) pathway, which is activated by the adenylyl cyclase Cyr1p.63 In the early 2000s, it was thought that Cyr1p may be involved directly in signal sensing. Klengel et al. reported that the catalytic domain of Cyr1p behaved as a CO2 sensor and mediated CO2 induced filamentous growth, while Hogan et al. reported that a sensory role was suggested for quorum sensing molecules.64,65 While these findings made major headway in the role of Cyr1p and the morphological regulation of C. albicans, the molecular mechanism of signal sensing remained to be elucidated. The ability of Cyr1p to distinguish different stimuli or sense and integrate multiple ones is owed to its several highly conserved domains.63 One such domain, and pertinent to this review, is an evolutionary conserved LRR domain, common in many innate immune receptors as discussed above.1 Elegant work by Wang and coworkers demonstrated that this domain is able to bind PG fragments.66
It was long determined that serum was the most potent activator of the morphological transition from budding yeast to hyphal yeast62, but the active constituent in serum remained unknown. Wang and co-workers discovered that Peptidoglycan fragments, specifically muramyl-dipeptides (MDPs) in serum (Figure 1), was the causative agent of high-inducing hyphal activity.66 Using biotinylated MDP enrichment assays, data was obtained that demonstrated the association between Cyr1p’s LRR and MDP, but direct binding and analytical characterization of this interaction remained unknown. To quantitatively and molecularly characterize this interaction, our lab has successfully expressed and purified an MBP fusion construct of the Cyr1p leucine rich repeat domain and demonstrated via a sensitive, surface plasmon resonance assay that the LRR binds diverse PG fragments with high affinity.67 Briefly, our lab was able to prove that the LRR of Cyr1p binds to muramyl tripeptide (MTP) with a Kd of 176 ± 68nM, and through competition assay demonstrate that binding is strong and specific.67 In order to understand the details of how these data were obtained the next section is dedicated to the development and use of the surface plasmon resonance assay for PG ligands and their various receptors.
Chemical probes and analytical assays to study the interactions of Peptidoglycan and its receptors:
Understanding the peptidoglycan sensing mechanism in detail requires tools to produce and manipulate PG fragments at the molecular level. As mentioned above, the synthetic Nod2 agonist (MDP) has been useful to the immunological community due to its structural simplicity and commercial availability. Recently, chemical glycobiologists have generated a variety of tools to produce expanded peptidoglycan fragments to study the immune system. The native PG fragments were obtained through the collection of high-performance liquid chromatography68,69, or chemical synthesis.
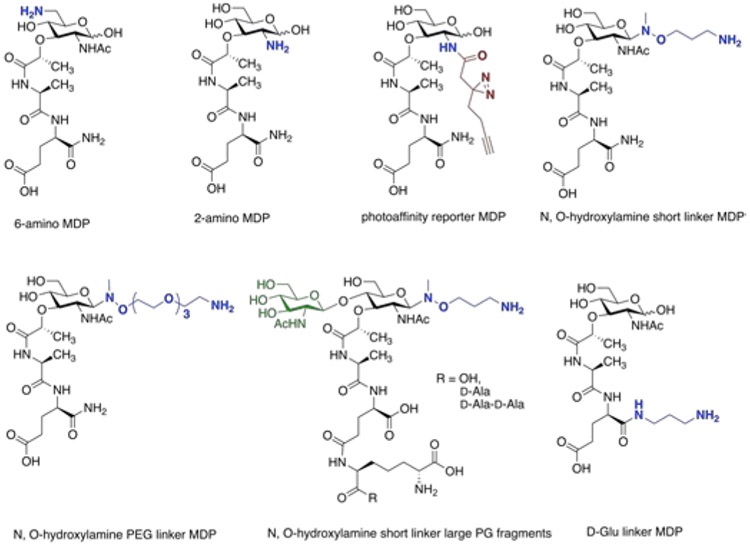
Work by Mobashery and Boons has been critical to advancing the field and inspiring to our lab. 70,72,73 Our laboratory has focused on the production of MDP probes that can be easily modified by the installation of amine groups both on the six and second position of MDP 45,74,75 The amine provides a chemical handle for derivatization, allowing the molecule to be attached to a surface or chemical probes with the exposure of the peptide stem to potential immune receptors (Figure 4). Indeed, the first biochemical evidence for a direct high-affinity interaction between Nod2 and MDP was shown by 6-amino MDP on SPR(as described below).45 To investigate how acetylation/deacetylation of 2-amino MDP modulates molecular recognition, two chemical syntheses have been developed. The first process is a late-stage synthetic approach at the 2-amino position of N-deacetylated MDP, and the second is a form of bioorthogonal modification at carboxylic acid of d-isoGln.76 A number of functional groups were chosen to produce 2-position MDP derivatives to study biological systems. The modification of the 2-position of the carbohydrate was found to be important for stabilizing Nod2 and generating an immune response. A recent study by Howard Hang and coworkers applied these synthetic routes to afford MDP photoaffinity reporters, containing a diazirine for photo-cross-linking in cells and an alkyne tag for bioorthogonal detection of covalently labelled proteins, to evaluate the cross-linking to immune receptors in living cells.77
Figure 4:
MDP chemical probes. The synthetic fragment MDP can be functionalized with various chemical handles allowing for variability in biological assay development. Green =GlcNac. Blue=variable amine linker. Brown=photoactivatable moiety. The disaccharide fragments were generated by lysozyme digestion, reacted with N-hydroxy-ethylamine and detected via mass spectrometric analysis.
Our laboratory has also worked to produce syntheses that would be easily accessible to the non-expert, developing, a one-step amine functionalization that can be introduced at the glycan reducing end of PG fragments with methyl N, O-hydroxylamine linkers (Figure 4). In addition, this modification maintains the original PG fragments’ bioactivity for NF-κB immune response.78 The original chemistry was developed for glycans79 to introduce amine-functionalized linkers on unprotected glycans without opening the sugar ring structure. The significant advantage of this method is the ability to incorporate probes of larger PG fragments beyond MDP, both enzymatically generated or chemically synthesized. It can potentially facilitate the possibility of functionalized PG lysate without further purification.
In addition to the sugar backbone modification on MDP, strategies to functionalize the stem peptide have been explored at the d-Glu group. In Staphylococcus aureus, the modification-amidation of d-Glu on PG does not affect the proinflammatory response.80 Inspired by the available chemical tools for constructing linkers from different angles to MDP, PG derivatives were immobilized on the surface to assess the effects of ligand orientation on the binding affinity of NOD-like receptors (NLRs).44,81 This study reveals the unique recognition mechanism between PG fragments to its innate immune receptors, as described below.
Our lab has been inspired by the recent progressive acceleration in the field of PG synthases, and hypothesized that the enzymes that catalyzed PG biosynthesis in vivo could be used to label bacterial peptidoglycan and synthesize complex PG fragment derivatives in vitro. A library of MurNAc derivative probes containing a bioorthogonal handle was pre-synthesized. After incorporated into PG in living cells, the modified cell wall was labeled with an appropriate fluorophore (Figure 5).82,83 In addition, it enabled the chemoenzymatic synthesis of UDP-MurNAc derivatives,83 which is difficult by tradition chemical methods.70,84 The rapid and scalable access to a variety of functionalized MurNAcs and UDP MurNAcs will open the door to address fundamental questions surrounding PG’s role in immunology and microbiology with other complementary bio-orthogonal labeling strategies. Similarly, a biorthogonal handle was installed on the 6-OH position of MurNAc by taking advantage of PatB, a PG O-acetyltransferase with promiscuous properties; this strategy yielded the ability to selectively label the sixth position of the MurNAc in intact bacterial cells.85 (Figure 5) The tools that our laboratory have synthesized have been useful in revealing biological phenomena and below we describe their specific usage in binding studies.
Figure 5:
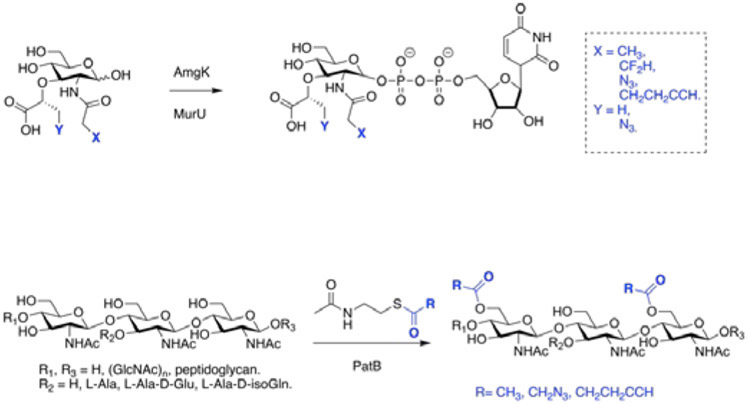
Chemoenzymatic modified PG fragments. Top: Hijacking the biosynthetic pathway of PG, UDP MurNac can be chemoenzymatically modified to contain bioorthogonal probes for biological assays. Modified fragments can be labeled with fluorophores, and varying functionalities. Recycling enzymes Amgk and MurU are native to the PG biosynthetic pathway and synthesize UDP-MurNac using NAM as the building block. Bottom: Peptidoglycan O-acetyltransferase (PatB) catalyzes the O-acetylation in Gram (−) bacteria permitting the installation of biorthogonal handles.
The molecular interactions between the bacterial-derived peptidoglycan fragments and related PRRs have been the focus of several studies in recent years; this includes not only human immune responses but also sensing processes in other microorganisms such as yeasts. Several biosensing methods have been used for this purpose. Surface plasmon resonance (SPR) is among these methods that have been used to analyze the binding process between PG motifs and pattern recognition proteins such as NLRs. The analysis of the kinetic parameters can be achieved by SPR using real time detection of the binding process between two binding partners, with the ligand being immobilized on the surface while the analyte is flowed over the surface.86-92 By formation of self-assembled monolayers (SAMs) on the surface, the attachment of the ligand is usually achieved via different methods including amine, thiol, or aldehyde chemistry, or capturing mechanisms (such as streptavidin-biotin systems).
Using the SPR assay, our lab moved toward studying the binding between the NOD2 LRR domain and its proposed ligand MDP. In this regard, amine functionalized MDP derivatives were chemically synthesized.45 Functionalizing the ligand with an amine group enabled the immobilization of the ligand on the surface of the gold chip, decorated with carboxylic acid containing SAMs, via an amide coupling. NOD2 was then passed over the surface and binding constants were then calculated. Interestingly, the results showed a strong binding affinity, in nanomolar range, for MDP and NOD2.45 We then went on to demonstrate that the LRR alone is sufficient for binding PG fragments using this SPR assay and NOD-LRR constructs.44 When the SPR assay was coupled to mutagenesis experiments, a binding model of the PG-NOD2 interaction was developed. In 2011, Laroui and co-workers showed a direct binding between NOD1 and a DAP derivative using an SPR assay.93 Here, however, the surface of the gold chip was coated with NOD1 and the tri-DAP molecules applied. This study showed a 34.6 μM Kd for binding between DAP and NOD1. It was also shown that truncated NOD1 constructs lacking the LRR domain do not exhibit any significant binding response, suggesting the LRR domain contains the binding site for DAP.
In addition to peptidoglycan binding by LRR protein, our lab has recently demonstrated the preference of LRR proteins NLRP1 and NOD2 for different faces of the PG fragment MDP using the tools described above (Figure 4). Briefly, a repertoire of amine functionalized derivatives with functionality on the C6, C2, C1, and d-isoglutamine positions were tethered to the SPR chip and protein was flowed over the chip at varying concentrations to assess binding (Figure 4). It was determined that NLRP1 can in fact bind both the carbohydrate backbone of MDP as well as the peptide stem individually; albeit with lesser affinity than intact MDP81. More specifically, the SPR data confirmed binding of the LRR with MDP tethered at the C2 position with the highest affinity, implicating that MDP in this orientation provides optimal contact with the LRR for tightest binding.81 LRR binding with the peptide and C6 tethered to the chip displayed similar affinities, indicative that the C2 and C6 positions may be solvent exposed and therefore provides less protein-ligand contact in the binding interface.
This critical and advantageous SPR assay allowed us further insight into the binding preference and pocket of these immune receptors. While the significance of these receptors binding to peptidoglycan fragments are understood, there is little evidence on the binding pocket, or preferences of these receptors. This study is the first to analytically substantiate the binding pocket preference for various faces of the conserved peptidoglycan fragment MDP, found in both Gram (−) and Gram (+) bacteria. In addition, it sets the precedence for further elucidation in the binding pockets of these difficult to purify proteins by providing a glimpse of critical protein-ligand interactions; data that are significantly lacking in the area of immune receptors and bacterial peptidoglycan.
Beyond human immune system proteins, the recognition of PG fragments in yeast cells has also been studied. As mentioned earlier, and by designing an SPR assay for the Cyr1p protein, our lab showed the LRR domain of Cyr1p in C. albicans directly binds to PG fragments such as MDP and MTP with nanomolar affinity.67 This also correlates with the ability of these PG fragments to induce hyphae formation in C. albicans.
In addition to SPR, other biosensing methods such as backscattering interferometry (BSI)94,95, and Biolayer interferometry (BLI)96 are emerging as new tools to study direct molecular interactions involved in recognition of fragments derived from bacterial cell wall structure. BSI is optical method, which also uses the detection of changes in the refractive index; however, in this case, the refractive index is quantified based on the interference fringe pattern formed upon interaction of the back-scattered laser beam in a microfluidic channel. This method has also been used more extensively in recent years to study molecular bindings in a pM-μM range. In our hands, we have used the BSI method as a comparative approach to investigate the binding of PG derivatives in solution and in regards to membrane association of the NOD2 receptor, where we showed that activation of immune responses for a library of PG fragments correlates with their capability of binding to NOD2 receptor in native membrane environment.75 It is noteworthy that BSI requires no tags to be included in either the protein or the ligand. However, BLI and SPR must have either the protein or small molecule attached to a surface.
These tools can be used to study the sensing mechanism for PG fragments in the human immune system, shedding light on potential roles that these immune receptors play in pathology of several human diseases. For instance, our work on studying NOD2 Crohn’s disease (CD) associated variants demonstrated that none of these prominent mutations altered binding to PG. In fact, all major NOD2 CD variants show comparable binding affinity as the WT, suggesting the potential role of these mutations is not necessarily associated with binding to the ligand but the signaling response of NOD2.97 SPR provided critical data to understand the biochemistry behind Crohn’s disease and will be important in studying these types on interactions in the future.
Peptidoglycan beyond the innate immune system
Microbial infections are recognized by the innate immune system both to elicit immediate defense and to generate long-lasting adaptive immunity. The innate immune system is genetically programmed to detect invariant features of invading microbes. Recently, Wolf and her coworkers discovered that hexokinase is an innate immune receptor for the detection of bacterial PG. It is surprising that N-acetylglucosamine, not MDP, is the active component that causes NLRP3 inflammasome activation, released from PG.49
In contrast, the adaptive immune system, composed of T and B lymphocytes, employs antigen receptors that are generated de novo in each organism (Figure 7). This highly specific adaptive immunity is sustained long-term by memory T cells. Gut microbiomes continuously shed PG from their cell walls as they grow and divide (Figure 1), and these off-cast molecules are capable of crossing the gut barrier and entering human blood. Peptidoglycan fragments were first detected in human plasma by the Kodama group.98 Huang et al. developed a monoclonal antibody (2E7) that targets MDP, which was found to be ubiquitously present in the serum of healthy humans, mice and monkeys.99 The presence of various PG fragments has also been identified in the fetal bovine serum that is routinely used in cell culture experiments and serum from healthy mice100, suggesting that a homeostatic function of PG signaling may have been previously underappreciated.
Figure 7: The interaction between bacterial cell wall fragments and the human immune system.
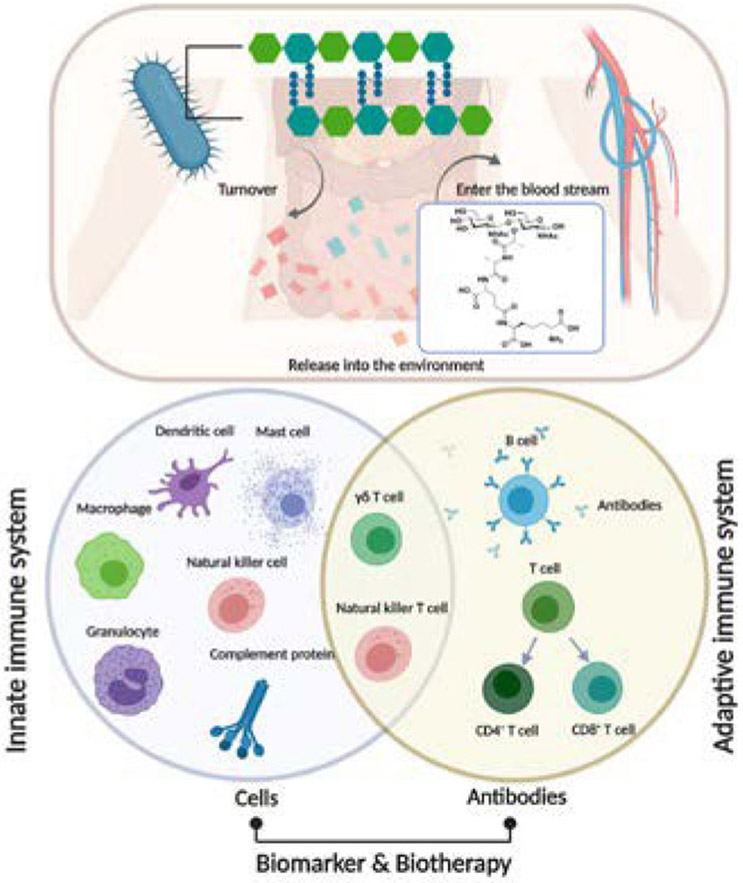
PG turnover and subsequent fragment release to the environment; non-septic PG fragments appear to enter the bloodstream (as shown). The innate immune system response to potential PG antigen is the first line of defense against infection. Many of the cells in the innate immune system (such as dendritic cells, macrophages, mast cells) produce cytokines or interact with other cells directly to activate the adaptive immune system. γδ T cells and Nature Killer cells are lymphocytes without antigen specificity. Therefore, they are considered to be innate cells with some similarities to effector lymphocytes. The adaptive immune system is based on clonal selection of lymphocytes with antigen receptors (B cell receptors and T cell receptors)103 and recent studies suggest that antibodies for specific PG fragment exist. A more detailed understanding of PG related immune response provides new opportunities for improving immunotherapy for autoimmune diseases.
Traditionally, PG sensing has been intensely studied in the context of the PRRs of the innate immune system. In a landmark study, Jutras et al. found patients with Lyme arthritis (LA) develop an adaptive immune response against Borrelia burgdorferi PG (PGBb).27 These authors proved that a specific immunoglobulin G response against PGBb is significantly higher in the synovial fluid samples in LA patients than in the same patient's serum. This study suggests a potential role for specific PG fragments as immunopathogenesis for other Lyme disease manifestations or other autoimmune diseases, such as inflammatory bowel disease (IBD). IBD includes Crohn’s disease and ulcerative colitis, a bacterial related autoimmune disease. The misrecognition of non-self results in mis-activation of the immune system. Its abnormal mucosal immune responses to microflora have been used for IBD biomarker development.101 Anti-Saccharomyces cerevisiae antibodies (ASCA), an antibody with affinity for antigens in the cell wall of the yeast Saccharomyces cerevisiae102, is one of the best-studied serological markers in IBD patients. It is interesting to note that the Saccharomyces cerevisiae cell wall is composed of a layered meshwork of β-glucans, chitin, and mannoproteins other than the peptidoglycan structure. However, as we discussed above the PG fragments can dramatically alter the phenotype of certain yeasts. Thus, an understanding of how the mycobiome and microbiome interact is pivotal in the development of biomarkers for autoimmune diseases.
Conclusions
This mini review focused on a molecular level handshake between the immune system and the bacterial cell wall polymer, peptidoglycan. The molecular interactions centered around PG are multifaceted and involve a plethora of components, signals transducers, molecular switches, etc. In order to properly understand the complex biology and the underlying diseases associated with the immune recognition of this essential bacterial component, the community cannot simply focus on one of these handshakes or interactions. For example, while MDP has been used as the representative of PG, the community needs to broaden their appreciation to the diversity naturally present in the polymer and its relationship to immune recognition. Here we present a challenge to properly characterize all of the handshakes in PG recognition, both innate and adaptive in nature, which will require the skills from immunologists, microbiologists, chemical biologists, organic chemists, biochemists, and geneticists to properly understand this complex signaling network. Information gleaned from these multidisciplinary studies will be invaluable in the development of adjuvants, immunomodulators and anti-inflammatory medications.
Figure 3:
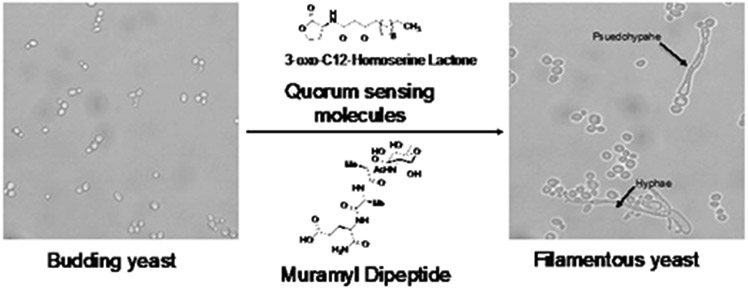
Molecular signals involved in the direct morphological transition from budding yeast to filamentous yeast. Budding yeast is activated by molecules such as 3-oxo-C12-homoserine lactone and Muramyl Dipeptide via the Cyr1p Leucine Rich Repeat domain. Once activated, budding yeast is transitioned into filamentous yeast in the form of hyphae or psuedophyape.100X magnification.
Figure 6:
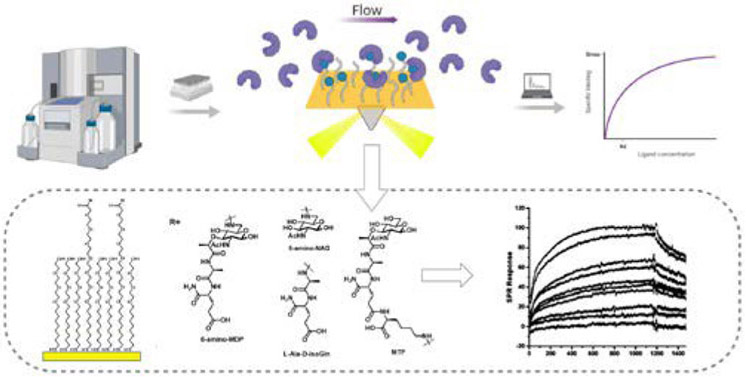
Surface Plasmon Resonance assay. The binding between the immobilized ligand and the analyte is detected as the analyte flow interacts with the ligand. The recorded sensograms can be further processed to obtain the kinetic parameters.
Acknowledgements
This work was supported by National Institute of General Medical Sciences (NIGMS P20GM104316; C.L.G.) and the National Science Foundation (NSF 1554967). GC thanks the National Institute of Health for support (5T32GM008550) through the Chemistry Biology Interface program. C.L.G. is a Camille Henry Dreyfus Teacher-Scholar, Pew Biomedical Scholar and Sloan Fellow and thanks the Dreyfus, Pew and Sloan Foundations.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Janeway CA and Medzhitov R, Annu. Rev. Immunol, 2002, 20, 197–216. [DOI] [PubMed] [Google Scholar]
- 2.Vollmer W, Blanot D and De Pedro MA, FEMS Microbiol. Rev, 2008, 32, 149–167. [DOI] [PubMed] [Google Scholar]
- 3.Rogers HJ, Ann. N. Y. Acad. Sci, 1974, 235, 29–51. [DOI] [PubMed] [Google Scholar]
- 4.Porfírio S, Carlson RW and Azadi P, Trends Microbiol., 2019, 27, 653–654. [DOI] [PubMed] [Google Scholar]
- 5.Vollmer W and Höltje JV, J. Bacteriol, 2004, 186, 5978–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vollmer W and Bertsche U, Biochim. Biophys. Acta - Biomembr, 2008, 1778, 1714–1734. [DOI] [PubMed] [Google Scholar]
- 7.Vollmer W, FEMS Microbiol. Rev, 2008, 32, 287–306. [DOI] [PubMed] [Google Scholar]
- 8.Kato K, Umemoto T, Sagawa H and Kotani S, Curr. Microbiol, 1979, 3, 147–151. [Google Scholar]
- 9.Barnard MR and Holt SC, Can. J. Microbiol, 1985, 31, 154–160. [DOI] [PubMed] [Google Scholar]
- 10.Jürgen UJ, Meissner J, Reichenbach H and Weckesser J, FEMS Microbiol. Lett, 1989, 60, 247–250. [Google Scholar]
- 11.Groth I, Schumann P, Weiss N, Schuetze B, Augsten K and Stackebrandt E, Int. J. Syst. Evol. Microbiol, 2001, 51, 81–87. [DOI] [PubMed] [Google Scholar]
- 12.Girardin SE, Boneca IG, Carneiro LAM, Antignac A, Jéhanno M, Viala J, Tedin K, Taha M-K, Labigne A, Zäthringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ and Philpott DJ, Science (80-. )., 2003, 300, 1584 LP – 1587. [DOI] [PubMed] [Google Scholar]
- 13.V Höltje J and Tuomanen EI, J. Gen. Microbiol, 1991, 137, 441–454. [DOI] [PubMed] [Google Scholar]
- 14.Silhavy TJ, Kahne D and Walker S, Cold Spring Harb. Perspect. Biol, 2010, 2, a000414–a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N and Kahne D, Nat. Rev. Microbiol, 2016, 14, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blundell JK and Perkins HR, J. Bacteriol, 1981, 147, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francino MP, Front. Microbiol, 2016, 6, 1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh C, Nature, 2000, 406, 775–781. [DOI] [PubMed] [Google Scholar]
- 19.Höltje J-V, Microbiol. Mol. Biol. Rev, 1998, 62, 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moynihan PJ, Sychantha D and Clarke AJ, Bioorg. Chem, 2014, 54, 44–50. [DOI] [PubMed] [Google Scholar]
- 21.Mahapatra S, Scherman H, Brennan PJ and Crick DC, J. Bacteriol, 2005, 187, 2341–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen JM, Golchin SA, Veyrier FJ, Domenech P, Boneca IG, Azad AK, Rajaram MVS, Schlesinger LS, Divangahi M, Reed MB and Behr MA, J. Infect. Dis, 2014, 209, 1045–1054. [DOI] [PubMed] [Google Scholar]
- 23.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prévost MC, Balloy V, Chignard M, Philpott DJ, Cossart P and Girardin SE, Proc. Natl. Acad. Sci. U. S. A, 2007, 104, 997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arthur M, Depardieu F, Molinas C, Reynolds P and Courvalin P, Gene, 1995, 154, 87–92. [DOI] [PubMed] [Google Scholar]
- 25.Bugg TDH, Wright GD, Dutka-Malen S, Arthur M, Courvalin P and Walsh CT, Biochemistry, 1991, 30, 10408–10415. [DOI] [PubMed] [Google Scholar]
- 26.Mayer C, Kluj RM, Mühleck M, Walter A, Unsleber S, Hottmann I and Borisova M, Int. J. Med. Microbiol, 2019, 309, 151326. [DOI] [PubMed] [Google Scholar]
- 27.Jutras BL, Lochhead RB, Kloos ZA, Biboy J, Strle K, Booth CJ, Govers SK, Gray J, Schumann P, Vollmer W, Bockenstedt LK, Steere AC and Jacobs-Wagner C, Proc. Natl. Acad. Sci. U. S. A, 2019, 116, 13498–13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boothby D, Daneo-Moore L, Higgins ML, Coyette J and Shockman GD, J. Biol. Chem, 1973, 248, 2161–2169. [PubMed] [Google Scholar]
- 29.Park JT and Uehara T, Microbiol. Mol. Biol. Rev, 2008, 72, 211 LP – 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald C, Inohara N and Nuñez G, J. Biol. Chem, 2005, 280, 20177–20180. [DOI] [PubMed] [Google Scholar]
- 31.Irazoki O, Hernandez SB and Cava F, Front. Microbiol , 2019, 10, 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borisova M, Gaupp R, Duckworth A, Schneider A, Dalügge D, Mühleck M, Deubel D, Unsleber S, Yu W, Muth G, Bischoff M, Götz F and Mayer C, MBio, , DOI: 10.1128/mBio.00923-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackey D and McFall AJ, Mol. Microbiol, 2006, 61, 1365–1371. [DOI] [PubMed] [Google Scholar]
- 34.Wolf AJ and Underhill DM, Nat. Rev. Immunol, 2018, 18, 243. [DOI] [PubMed] [Google Scholar]
- 35.Medzhitov R and Janeway CA, Cell, 1997, 91, 295–298. [DOI] [PubMed] [Google Scholar]
- 36.Mogensen TH, Clin. Microbiol. Rev, 2009, 22, 240–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brubaker SW, Bonham KS, Zanoni I and Kagan JC, Annu. Rev. Immunol, 2015, 33, 257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokes BA, Yadav S, Shokal U, Smith LC and Eleftherianos I, Front. Microbiol, 2015, 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M and Hoffmann JA, Cell, 1996, 86, 973–983. [DOI] [PubMed] [Google Scholar]
- 40.Iwaki D, Mitsuzawa H, Murakami S, Sano H, Konishi M, Akino T and Kuroki Y, J. Biol. Chem, 2002, 277, 24315–24320. [DOI] [PubMed] [Google Scholar]
- 41.Kawasaki T and Kawai T, Front. Immunol , 2014, 5, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franchi L, Warner N, Viani K and Nuñez G, Immunol. Rev, 2009, 227, 106–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyle JP, Parkhouse R and Monie TP, Open Biol., , DOI: 10.1098/rsob.140178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauro ML, D’Ambrosio EA, Bahnson BJ and Grimes CL, ACS Infect. Dis, 2017, 3, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimes CL, Ariyananda LDZ, Melnyk JE and O’Shea EK, J. Am. Chem. Soc, 2012, 134, 13535–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Correa RG, Milutinovic S and Reed JC, Biosci. Rep, 2012, 32, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conforti-Andreoni C, Beretta O, Licandro G, Qian HL, Urbano M, Vitulli F, Ricciardi-Castagnoli P and Mortellaro A, J. Leukoc. Biol, 2010, 88, 1207–1216. [DOI] [PubMed] [Google Scholar]
- 48.V Swanson K, Deng M and Ting JP-Y, Nat. Rev. Immunol, 2019, 19, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M and Underhill DM, Cell, 2016, 166, 624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hajishengallis G and Lambris JD, Nat. Rev. Immunol, 2011, 11, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobe B and Deisenhofer J, Trends Biochem. Sci, 1994, 19, 415–421. [DOI] [PubMed] [Google Scholar]
- 52.Kobe B and V Kajava A, Curr. Opin. Struct. Biol, 2001, 11, 725–732. [DOI] [PubMed] [Google Scholar]
- 53.Malic S, Hill KE, Ralphs JR, Hayes A, Thomas DW, Potts AJ and Williams DW, Oral Microbiol. Immunol, 2007, 22, 188–194. [DOI] [PubMed] [Google Scholar]
- 54.Shirtliff ME, Peters BM and Jabra-Rizk MA, FEMS Microbiol. Lett, 2009, 299, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morales DK and Hogan DA, PLoS Pathog., 2010, 6, e1000886–e1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shing SR, Ramos AR, Patras KA, Riestra AM, McCabe S, Nizet V and Coady A, Front. Cell. Infect. Microbiol, 2020, 9, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seed PC, Cold Spring Harb. Perspect. Med, 2014, 5, a019810–a019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blum HE, Adv. Med. Sci, 2017, 62, 414–420. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, V Lynch S and Knight R, Nat. Med, 2018, 24, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Limon JJ, Skalski JH and Underhill DM, Cell Host Microbe, 2017, 22, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schleifer KH and Kandler O, Bacteriol. Rev, 1972, 36, 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gow NA, Curr. Top. Med. Mycol, 1997, 8, 43–55. [PubMed] [Google Scholar]
- 63.Rocha CRC, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M and Leberer E, Mol. Biol. Cell, 2001, 12, 3631–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hall RA, De Sordi L, MacCallum DM, Topal H, Eaton R, Bloor JW, Robinson GK, Levin LR, Buck J, Wang Y, Gow NAR, Steegborn C and Muhlschlegel FA, PLoS Pathog., 2010, 6, e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davis-Hanna A, Piispanen AE, Stateva LI and Hogan DA, Mol. Microbiol, 2008, 67, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu X-L, Lee RTH, Fang H-M, Wang Y-M, Li R, Zou H, Zhu Y and Wang Y, Cell Host Microbe, 2008, 4, 28–39. [DOI] [PubMed] [Google Scholar]
- 67.Burch JM, Mashayekh S, Wykoff DD and Grimes CL, ACS Infect. Dis, 2018, 4, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Espaillat A, Forsmo O, El Biari K, Bjork R, Lemaitre B, Trygg J, Canada FJ, de Pedro MA and Cava F, J Am Chem Soc, 2016, 138, 9193–9204. [DOI] [PubMed] [Google Scholar]
- 69.Joers A, Vind K, Hernandez SB, Maruste R, Pereira M, Brauer A, Remm M, Cava F and Tenson T, Sci Rep, 2019, 9, 18043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz B, Markwalder JA and Wang Y, J Am Chem Soc, 2001, 123, 11638–11643. [DOI] [PubMed] [Google Scholar]
- 71.Hesek D, Suvorov M, Morio K, Lee M, Brown S, Vakulenko SB and Mobashery S, J Org Chem, 2004, 69, 778–784. [DOI] [PubMed] [Google Scholar]
- 72.Roy Chowdhury A, Siriwardena A and Boons G-J, Tetrahedron Lett., 2002, 43, 7805–7807. [Google Scholar]
- 73.Roy Chowdhury A and Boons G-J, Tetrahedron Lett., 2005, 46, 1675–1678. [Google Scholar]
- 74.Grimes CL, Podolsky DK and O’Shea EK, Bioorg Med Chem Lett, 2010, 20, 6061–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaefer AK, Melnyk JE, Baksh MM, Lazor KM, Finn MG and Grimes CL, ACS Chem. Biol, 2017, 12, 2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Melnyk JE, Mohanan V, Schaefer AK, Hou CW and Grimes CL, J Am Chem Soc, 2015, 137, 6987–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang YC, Westcott NP, Griffin ME and Hang HC, ACS Chem Biol, 2019, 14, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lazor KM, Zhou J, DeMeester KE, D’Ambrosio EA and Grimes CL, Chembiochem, 2019, 20, 1369–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen N and Xie J, Org Biomol Chem, 2016, 14, 11028–11047. [DOI] [PubMed] [Google Scholar]
- 80.Kraus D, Kalbacher H, Buschmann J, Berger-Bachi B, Gotz F and Peschel A, Infect Immun, 2007, 75, 2084–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.D’Ambrosio EA, Bersch KL, Lauro ML and Grimes CL, J Am Chem Soc, 2020, 142, 10926–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liang H, DeMeester KE, Hou C-W, Parent MA, Caplan JL and Grimes CL, Nat. Commun, 2017, 8, 15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeMeester KE, Liang H, Jensen MR, Jones ZS, D’Ambrosio EA, Scinto SL, Zhou J and Grimes CL, J Am Chem Soc, 2018, 140, 9458–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.VanNieuwenhze MS, Mauldin SC, Zia-Ebrahimi M, Winger BE, Hornback WJ, Saha SL, Aikins JA and Blaszczak LC, J Am Chem Soc, 2002, 124, 3656–3660. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Lazor KM, DeMeester KE, Liang H, Heiss TK and Grimes CL, J. Am. Chem. Soc, 2017, 139, 13596–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gedig ET, in Handbook of Surface Plasmon Resonance, Royal Society of Chemistry, 2010, pp. 173–220. [Google Scholar]
- 87.Patching SG, Biochim. Biophys. Acta - Biomembr, 2014, 1838, 43–55. [DOI] [PubMed] [Google Scholar]
- 88.Nguyen HH, Park J, Kang S and Kim M, Sensors (Basel)., 2015, 15, 10481–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pattnaik P, Surface Plasmon Resonance Applications in Understanding Receptor-Ligand Interaction, 2005, vol. 126. [DOI] [PubMed] [Google Scholar]
- 90.Frostell Å, Vinterbäck L and Sjöbom H, Methods Mol. Biol, 2013, 1008, 139–165. [DOI] [PubMed] [Google Scholar]
- 91.Rich RL and Myszka DG, Curr. Opin. Biotechnol, 2000, 11, 54–61. [DOI] [PubMed] [Google Scholar]
- 92.Morton TA and Myszka DG, Methods Enzymol., 1998, 295, 268–282. [DOI] [PubMed] [Google Scholar]
- 93.Laroui H, Yan Y, Narui Y, Ingersoll SA, Ayyadurai S, Charania MA, Zhou F, Wang B, Salaita K, V Sitaraman S and Merlin D, J. Biol. Chem, 2011, 286, 31003–31013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baksh MM, Kussrow AK, Mileni M, Finn MG and Bornhop DJ, Nat. Biotechnol, 2011, 29, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD and Sorensen HS, Science (80-. )., 2007, 317, 1732–1736. [DOI] [PubMed] [Google Scholar]
- 96.Concepcion J, Witte K, Wartchow C, Choo S, Yao D, Persson H, Wei J, Li P, Heidecker B, Ma W, Varma R, Zhao L-S, Perillat D, Carricato G, Recknor M, Du K, Ho H, Ellis T, Gamez J, Howes M, Phi-Wilson J, Lockard S, Zuk R and Tan H, Comb. Chem. High Throughput Screen, 2009, 12, 791–800. [DOI] [PubMed] [Google Scholar]
- 97.Schaefer AK, Wastyk HC, Mohanan V, Hou CW, Lauro ML, Melnyk JE, Burch JM and Grimes CL, Biochemistry, 2017, 56, 4445–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kobayashi T, Tani T, Yokota T and Kodama M, FEMS Immunol. Med. Microbiol, 2000, 28, 49–53. [DOI] [PubMed] [Google Scholar]
- 99.Huang Z, Wang J, Xu X, Wang H, Qiao Y, Chu WC, Xu S, Chai L, Cottier F, Pavelka N, Oosting M, Joosten LAB, Netea M, Ng CYL, Leong KP, Kundu P, Lam KP, Pettersson S and Wang Y, Nat. Microbiol, 2019, 4 766–773. [DOI] [PubMed] [Google Scholar]
- 100.Molinaro R, Flick R, Philpott DJ and Girardin SE, bioRxiv, 2019, 547885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strober W, Fuss I and Mannon P, J. Clin. Invest, 2007, 117, 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peyrin-Biroulet L, Standaert-Vitse A, Branche J and Chamaillard M, Inflamm. Bowel Dis, 2007, 13, 1561–1566. [DOI] [PubMed] [Google Scholar]
- 103.Dranoff G, Nat. Rev. Cancer, 2004, 4, 11–22. [DOI] [PubMed] [Google Scholar]