Abstract
Cytolytic T cells (CTLs) play a pivotal role in surveillance against tumors. Induction of CTL responses by vaccination may be challenging, as it requires direct transduction of target cells or special adjuvants to promote cross-presentation. Here, we observed induction of robust CTL responses through electroporation (EP)-facilitated, DNA-launched nanoparticle vaccination (DLnano-vaccines). EP was observed to mediate transient tissue apoptosis and macrophage infiltration, which were deemed essential to the induction of CTLs by DLnano-vaccines through a systemic macrophage depletion study. Bolus delivery of protein nano-vaccines followed by EP, however, failed to induce CTLs, suggesting direct in vivo production of nano-vaccines may be required. Following these observations, new DLnano-vaccines scaffolding immunodominant melanoma Gp100 and Trp2 epitopes were designed and shown to induce more potent and consistent epitope-specific CTL responses than the corresponding DNA monomeric vaccines or CpG-adjuvanted peptide vaccines. DNA, but not recombinant protein, nano-vaccinations induced CTL responses to these epitopes and suppressed melanoma tumor growth in mouse models in a CD8+ T cell–dependent fashion. Further studies to explore the use of DLnano-vaccines against other cancer targets and the biology with which they induce CTLs is important.
Keywords: DNA vaccines, in vivo assembly, nanoparticles, cytotoxic T cells, melanoma
Introduction
Cytolytic T cells (CTLs) are an extremely important component of the adaptive immune system, which can selectively kill target cells through release of cytokines, granzyme and perforin, as well as mediating target cell apoptosis through Fas and Fas-Ligand interactions (1). As such, induction of broad endogenous antigen-specific CTLs through vaccination may be an important approach to treat cancer (2). However, induction of CTLs through vaccination can be challenging, as it requires antigen-presenting cells (APCs) to present HLA-restricted epitopes to MHC I and provide costimulatory signals (3). Vaccinating with viral vectors encoding target antigens is an approach to drive CTL responses, potentially through direct transduction of target cells (4), and special adjuvants can be used to facilitate cross-presentation of target antigens, which to varying degrees promote phago-lysosomal escape and retro-translocation of target antigens into the cytosol (5). Alternatively, dendritic cells (DCs) can be loaded ex vivo with saturating concentrations of peptides and adoptively transferred back into the host to prime CD8+ T cells (6). However, each approach has certain drawbacks: (i) relatively few adjuvants have been demonstrated to help prime CD8+ T-cell responses in clinic (7), (ii) viral-vectored vaccines can be limited by pre-existing immunity against the viral vector (8), and (iii) there are manufacturing issues with large-scale production of DC-based vaccines.
DNA vaccination has also been observed to elicit CD8+ T-cell responses both in preclinical animal models and in clinical trials (9), plausibly through a combination of direct transfection of target cells (10) and cross-presentation of DNA-encoded antigens (11). We previously reported a new strategy to use DNA to launch in vivo-produced nanoparticle vaccines (DLnano-vaccines), which result in significant improvements in the rate of seroconversion, magnitude of humoral response, dose-sparing, and protection from viral challenge (12). We have also observed that DLnano-vaccines induce significantly improved CD8+ T-cell responses compared to DNA-encoding monomeric antigens (12). In this work, we compared induction of CTL responses by DLnano-vaccines and the corresponding protein nano-vaccines, DNA-encoded monomeric vaccines, and CpG-adjuvanted peptide vaccines in mice. Mechanistic studies were pursued to determine the role of electroporation, transient tissue apoptosis, and APC infiltration in the priming of CTL responses by DLnano-vaccines. Finally, DLnano-vaccines capable of eliciting epitope-specific CTL responses were designed and evaluated in immunogenicity studies and a prophylactic B16-F10 melanoma challenge model. This work provides a demonstration of the capability of DLnano-vaccines to drive CTLs against desired antigens and should be explored against additional cancer targets.
Materials and Methods
Design of DNA-launched nanoparticles
DNnano_LS_GT8 and DLnano_LS_HA(NC99) were developed in prior work (12). The self-assembling nanoparticles scaffolding Trp2 and Gp100 peptides were engineered using structure-guided design. In our preliminary experiments, Lumazine Synthase on its own does not express well in mammalian cell lines, and we hypothesized that it could not be used on its own to scaffold anti-tumor peptide antigens. We engineered a new version of DLnano_LS_GT8 capable of displaying peptide antigens. In this construction, the heavily glycosylated GT8 domain facilitates solubilization and secretion of designed nanoparticles and could potentially be replaced by other heavily glycosylated domains. To ensure epitope accessibility and homogenous nanoparticle assembly, N-linked glycans in proximity to the C-terminus of GT8 were removed by mutations in the PNGS sequence (N112D, N165D and N170D), and a 9–amino acid linker (GGSGSGGGS)was also incorporated to the C-terminus of GT8 upstream of the scaffolded antitumor peptides (Gp10025:: EGPRNQDWL and Trp2188: SVYDFFVWL).
DNA design and plasmid synthesis
Protein sequences for IgE Leader Sequence and eOD-GT8-60mer were as previously reported (Supplementary Table S1)(13,14). Protein sequence for HA1_NC99 was obtained from GenBank (accession number AY289929.1)(Supplementary Table S1). DNA-encoding protein sequences were codon- and RNA-optimized as previously described (13). The optimized transgenes were synthesized de novo (GenScript) and cloned into a modified pVAX-1 backbone (Invitrogen, Catalog: V26020) under the control of the human CMV promoter and bovine growth hormone poly-adenylation signal.
Production of recombinant protein eOD-GT8-60mer, HA(NC99)-60mer, LS_Trp2188-60mer, and LS _Gp10025-60mer
2 × 109 Expi293F cells (Invitrogen) in 1L Expi Expression medium (Invitrogen) were transfected with 300 μg pVAX-1 plasmid vector encoding the eOD-GT8-60mer, HA(NC99)-60mer, LS_Trp2188-60mer or LS_Gp10025-60mer with PEI (Sigma Aldrich)/OPTI-MEM (Invitrogen) and harvested 6 days post-transfection. Transfection supernatant was first purified with affinity chromatography using the AKTA pure 25 system and gravity flow columns filled with GNL Lectin beads (GE Healthcare). The eluate fractions from the affinity purification were pooled, concentrated with Amicon Ultra-15 Centrifugal Filter Unit with 30kDa cut-off (Milipore), and dialyzed into 1X PBS buffer before being loaded onto the Superose 6 Increase 10/300 GL size-exclusion chromatography (SEC) column (GE healthcare) for purification. Identified eluate fractions were then collected and concentrated to 1 mg/mL in PBS as previously described (12).
Study approval and animal use
The study does not involve any human subjects requiring Institutional Review Board approval. All animal experiments were carried out in accordance with animal protocols 201214, 201115, and 201221 approved by the Wistar Institute Institutional Animal Care and Use Committee (IACUC).
Immunizations:
For DNA-based immunization, 6-to-8-week-old female C57BL/6, BALB/c, or B6.129S(c)-Batf3tm1Kmm/J (BATF3-KO) mice (Jackson Laboratory) were immunized with DNA vaccines via intramuscular injections into the tibialis anterior muscles, coupled with or without intramuscular EP with the CELLECTRA 3P device (Inovio Pharmaceuticals), as previously described (15). For DNA immunizations using DLnano_LS_GT8 and DLnano_LS_HA(NC99) as antigens (except in dose comparison study, Supplementary Figure S1E-H), 25 μg of plasmid DNA was used, a standard DNA dose that has been utilized in prior studies (12,16). For DNA immunizations involving DLnano_LS_Trp2188, DLnano_LS_Gp10025, DLmono_Trp2188, and DLmono_Gp10025, 10 μg of individual plasmid DNA was used either alone or in combination. For vaccinations involving recombinant protein, a high protein dose was used in this study compared to prior studies (17). 6 to 8-week-old female BALB/c were immunized subcutaneously with 10 μg of recombinant eOD-GT8-60mer protein co-formulated with either 50 μL Sigma Adjuvant System (RIBI) (SigmaAldrich), 50 μg poly (I:C) (InvivoGen), or 20 μg CpG ODN 1826 VacciGrade (InvivoGen), with or without EP. Alternatively, BALB/c or C57BL/6 mice were immunized with 10 μg protein HA(NC99)-60mer and 4 μg of LS_Trp2188-60mer and LS_Gp10025-60mer in 50 μL PBS co-formulated with 50 μL Sigma Adjuvant System (SigmaAldrich). For the high dose protein versus DNA comparison study reported in Supplementary Figure S1E-H, 50 μg DLnano_LS_GT8 was administered with EP, or 50 μg protein eOD-GT8-60mer co-formulated with RIBI was administered without EP. For peptide vaccinations, C57BL/6 mice received intramuscular vaccinations of 10 μg Trp2188 or 10 μg Gp10025 peptides (GenScript) co-formulated with 20 μg CpG ODN 1826 VacciGrade in sterile saline. For all immunizations reported in Figure 1 or Supplementary Figure S1, the mice were immunized twice, three weeks apart and euthanized two weeks post the second vaccination for spleen collection. The sera were collected on days 0, 7, 14, 21, and 35 days post the first vaccination by bleeding through the submandibular vein. For immunizations reported in Figure 2H-I, or Supplementary Figure S2K, the mice were immunized once and euthanized two weeks post the vaccination for spleen collection. For all immunogenicity studies in Figure 3C-D and Supplementary Figure S3D, S3F-I, the mice were immunized twice two weeks apart and euthanized two weeks post the second vaccination for spleen collection.
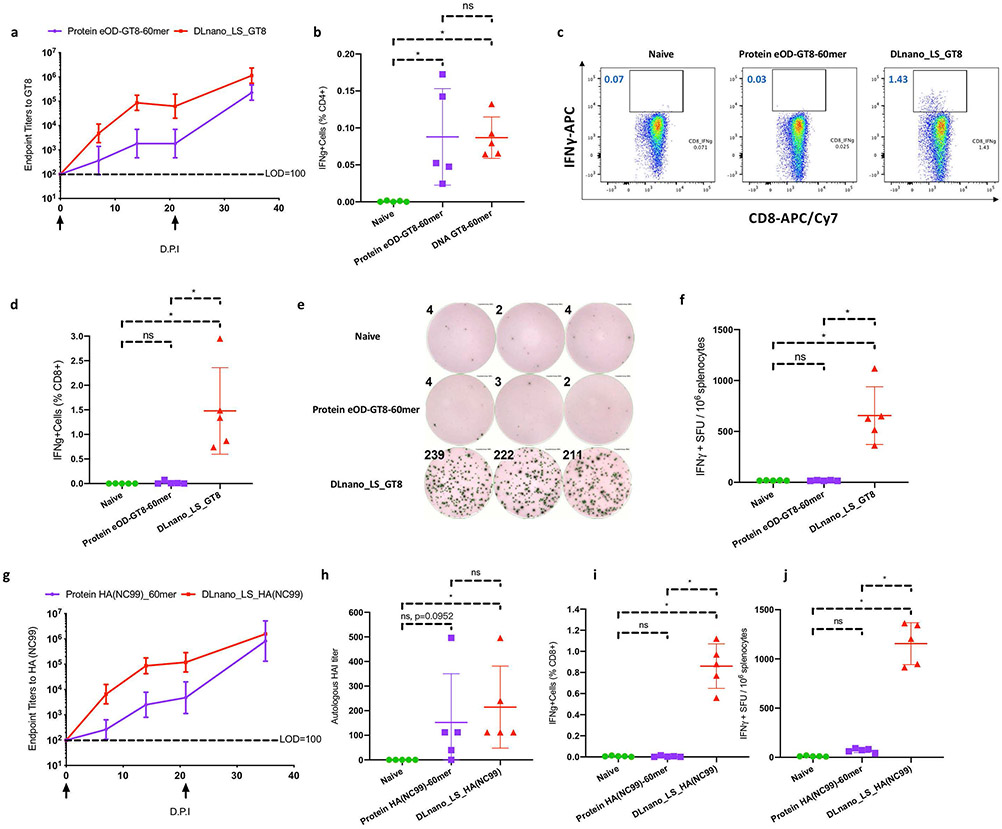
Figure 1. Comparison of immune responses induced by DLnano_LS_GT8 and protein eOD-GT8-60mer, as well as DLnano_LS_HA(NC99) versus HA(NC99)-60mer in BALB/c mice.
Mice received either 25 μg DNA vaccination with EP or 10 μg RIBI-adjuvanted protein vaccination without EP twice, three weeks apart, and were euthanized two weeks post second vaccination for cellular analysis. (A) Endpoint titers to GT8-monomer induced by DLnano_LS_GT8 compared to RIBI-adjuvanted protein eOD-GT8-60mer at 0, 7, 14, 21, and 35 d.p.i. LOD: Limit of Detection. (B) CD4+ IFNγ responses to the LS domain induced by DLnano_LS_GT8, protein eOD-GT8-60mer, or in naïve mice 35 d.p.i. (C) Flow cytometry plots and (D) combined statistics for CD8+ IFNγ responses to the GT8 domain induced by DLnano_LS_GT8, protein eOD-GT8-60mer, or in naïve mice at 35 d.p.i. (E) IFNγ ELISpot images with numbers indicating spot counts and (F) combined statistics for overall splenic T-cell responses to the GT8 domain induced by DLnano_LS_GT8, protein eOD-GT8-60mer or in naïve mice at 35 d.p.i. SFU: spot-forming units. (G) Endpoint titers to NC99 hemagglutinin induced by DLnano_LS_HA(NC99) compared to RIBI-adjuvanted protein HA(NC99)-60mer at 0, 7, 14, 21, and 35 d.p.i. (H) HAI titers against autologous H1 A/NewCaledonia/20/1999 induced by DLnano_LS_HA(NC99), protein HA(NC99)-60mer, or in naïve mice at 35 d.p.i. (I) ICS determination of CD8+ T-cell IFNγ responses to the HA domain induced by DLnano_LS_HA(NC99), protein HA(NC99)-60mer, or in naïve mice at 35 d.p.i. (J) IFNγ ELISpot assays for overall splenic T-cell responses to the HA domain induced by DLnano_LS_HA(NC99), protein HA(NC99)-60mer or in naïve mice at 35 d.p.i. Two independent experiments performed for each panel with similar observations made; N=5 mice/group; each dot represents an animal. Error bar represents standard deviation. Arrow below the plot (A,G) represents an immunization. Two-tailed Mann-Whitney Rank test used to compare groups; p-values were adjusted for multiple comparison for (B), (D), (F), (H), (I), and (J); *p-value<0.05; ns: not significant.
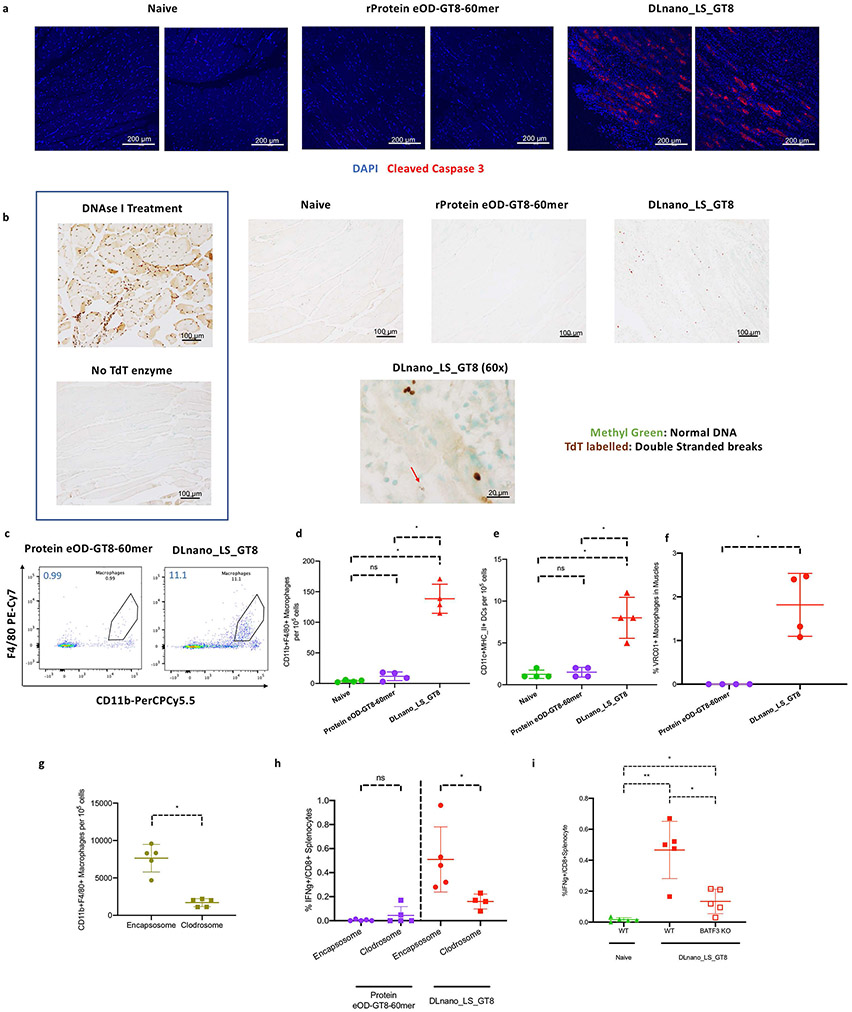
Figure 2. Determination of the role of tissue apoptosis and APC infiltration upon DNA vaccination in C57BL/6 mice.
(A) Immunofluorescence staining for cleaved caspase-3 (red) or nuclei (blue) in muscle sections from naïve mice or those treated with protein eOD_GT8-60mer without EP or DLnano_LS_GT8 with EP harvested 4 d.p.i. (B) TUNEL assay to determine presence of double-stranded DNA breaks (brown) or intact DNA (green) in muscle sections from naïve mice or those treated with protein eOD_GT8-60mer without EP or DLnano_LS_GT8 with EP harvested 4 d.p.i. Red arrow in the 60x image points to an apoptotic neucleus. (C) Flow cytometry plots and (D) combined statistics for the frequency of muscle-infiltrating CD11b+F4/80+ macrophages in naïve mice or those treated with protein eOD-GT8-60mer or DLnano_LS_GT8 7 d.p.i. (E) Flow determination of muscle-infiltrating CD11c+MHCII+ DCs in naïve mice or those treated with protein eOD-GT8-60mer or DLnano_LS_GT8 7 d.p.i. (F) Flow determination for GT8-uptake by VRC01-FITC staining for muscle macrophages in naïve mice or mice treated with protein eOD-GT8-60mer or DLnano_LS_GT8 7 d.p.i. (G) Comparison of changes in frequencies of muscle-infiltrating macrophages 4 d.p.i in mice treated with DLnano_LS_GT8. Mice also received three doses of IV clodrosome or control encapsosome on Days −3, 0 and 3 relative to DNA vaccination. (H) CD8+ T-cell responses induced by DLnano_LS_GT8 or protein eOD-GT8-60mer in mice that did or did not receive systematic macrophage depletion with clodrosome. Mice received 25 μg DNA vaccination with EP or 10 μg RIBI-adjuvanted protein vaccination without EP, and were euthanized two weeks post-vaccination for splenic IFNγ+CD8+ T cells. (I) CD8+ T-cell responses in naïve mice or in C57BL/6 or BATF3KO mice vaccinated with DLnano_LS_GT8. Mice were vaccinated and euthanized with the same dose and schedule as described in (H). One independent experiment was performed for each panel in the figure. N=4 mice/group (C-F) or N=5 mice/group (G-I); each dot represents an animal. Error bar represents standard deviation. Two-tailed Mann-Whitney Rank test used to compare groups; p-values were adjusted for multiple comparison for panels (D), (E), and (I); *p-value<0.05; **p-value<0.005; ns: not significant.
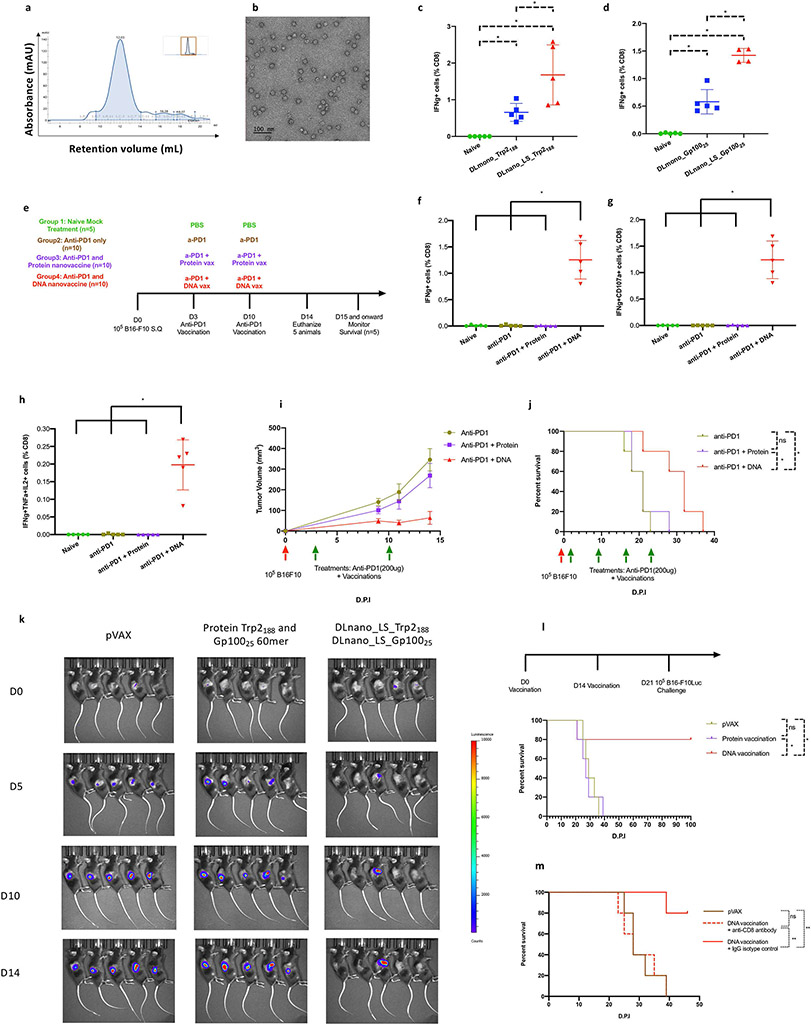
Figure 3. Characterization of the functional importance of CD8+ T-cell priming by DNA-launched versus protein nanoparticle vaccination in a melanoma model in C57BL/6 mice.
(A) SEC trace for the designed LS_Trp2188-60mer. (B) nsEM image of SEC purified LS_Trp2188-60mer nanoparticles. (C-D) Comparison of immunogenicity of DLnano-vaccines versus monomeric DNA vaccines. Mice were vaccinated twice with 10 μg of each DNA vaccine, two weeks apart, and euthanized two weeks post second vaccination for cellular analysis. (C) Comparison of CD8+ IFNγ T-cell responses to Trp2188 peptide in the spleens of naïve or immunized mice. (D) Comparison of CD8+ IFNγ T-cell responses to Gp10025 peptide in the spleens of naïve or immunized mice. (E) Treatment and vaccination schemes used to study CD8+ T-cell responses to both Trp2188 and Gp10025 peptides in naïve mice and B16F10 tumor-bearing mice that received anti–PD-1 treatment alone or in combination with protein (4 μg) or DNA vaccination (10 μg) of LS-GT8–scaffolded 60mer nanoparticles presenting Trp2188 and Gp10025 epitopes. (F-H) Induced IFNγ+, IFNγ+CD107a+, or IFNγ+TNFα+IL2+ CD8+ T-cell responses to Trp2188 in the spleens of naïve tumor-free mice or B16-F10–bearing mice that received treatments as described in (E). (I) Tumor growth and (J) overall survival in mice challenged with 105 B16-F10 cells and then received treatments as described in (E). (K) IVIS imaging to determine in vivo tumor growth in the prophylactic tumor model where mice first received two vaccinations of pVAX vector, combination of protein Trp2188 and Gp10025-60mer, or combination of DLnano_LS_Trp2188 and DLnano_LS_Gp10025 and were then challenged with 105 B16-F10-Luc cells seven days post second immunization. (L) Survival curves for mice shown in (K). (M) Survival curves for mice that first received two vaccinations, two weeks apart, of pVAX vector or combination of DLnano_LS_Trp2188 and DLnano_LS_Gp10025. The mice were then given either anti- CD8 or rat IgG2b isotype control and one day later challenged with 105 B16-F10-Luc cells at seven days post the second immunization. One independent experiment was performed for each panel. N=5 mice/group; each dot represents a mouse. Green arrows below the plot (I,J) represents treatments. Error bar represents standard deviation. Two-tailed Mann-Whitney Rank test used to compare groups for (C), (D), (F), (G), and (H); log-rank tests were used to compare survivals between two groups for (J), (L) and (M); p-values were adjusted for multiple comparison for (C), (D), (F), (G), (H), (J), (L), and (M); *p-value<0.05; **p-value<0.005; ns: not significant.
Mechanistic studies (Figure 2 and Supplementary Figure S2):
C57BL/6 mice received intramuscular injections of 80 μg of DLnano_LS_GT8 co-formulated with 12 U hyaluronidase (SigmaAldrich) with or without intramuscular EP or 20 μg of protein eOD-GT8-60mer co-formulated 1:1 with Sigma Adjuvant System with or without intramuscular EP. For Supplementary Figure S2c-S2d, mice sera were collected by cheek bleeds 0, 1, 4, and 10 d.p.i for measurments of muscle enzymes in the sera. For in vivo macrophage depletion, 6-to-8-week-old female C57BL/6 mice received one or three intravenous injections (on days −3, 0, 3 with respect to DNA vaccination) of 750 μg clodronate disodium formulated in clodrosome (150 μL injection, Encapsula Nanoscience) via retro-orbital injections; control mice each received 150 μL of encapsosome (Encapsula Nanoscience) IV at the same timepoints. For the one injection study (Supplementary Figure S2h-S2j), spleens were collected one day post clodrosome/encapsosome treatments; for the three injection studies, muscles were collected 4 days post DNA vaccination for Figure 2g, and spleens were collected from a different set of mice 14 days post DNA vaccination for Figure 2h. For Figure 2i and Supplementary Figure S2k, the BATF3KO-mice or the control WT C57BL/6 mice received single 25 μg DLnano_LS_GT8 vaccination and were euthanized for sera and spleen collection 14 days post DNA vaccination.
Tumor challenge:
B16-F10 (ATCC, Catalog: CRL-6475; authenticated by ATCC) were maintained in 10% FBS (Lampire)/DMEM (Corning) under low passage (less than 10) and B16-F10-Luc2 cells (ATCC, Catalog: CRL-6475-LUC2; authenticated by ATCC) were maintained in 10% FBS/DMEM enriched with 10 μg/mL blasticidin (Gibco) with routine testing for mycoplasma contaminations and other mouse pathogens. Cells were trypsinized and strained through 70-μm strainer to generate single-cell suspensions. They were then administered subcutaneously to mice (105 cells to each mouse in 100 μL PBS). The tumor size was measured every two days with a digital caliper, and tumor volume was determined with the formula V=0.5W2L (V=tumor volume, W= tumor width, L=tumor length). Mice with tumor volumes greater than 2000 mm3 or with any dimension exceeding 2 cm were euthanized for humane purposes or until 80 days post tumor inoculation (experimental endpoint). For recombinant anti–PD-1 (RMP1-14, Bio X Cell, Catalog: BE0146) administration in the melanoma therapeutic treatment model, 200 μg of antibody was injected intraperitoneally in 100 μL PBS to each mouse weekly starting three days post tumor inoculation. For mechanistic study to determine the role of CD8+ T cells in mediating in vivo control of tumor growth (Figure 3M and Supplementary Figure S3I), prophylactically vaccinated mice received 200 μg anti-mouse CD8α (2.43, BioXcell, Catalog: BE0061) or 200 μg Rat IgG2b isotype control (LTF-2, BioXcell, Catalog: BE0090) in 100 μL sterile PBS one day prior to tumor inoculation.
ELISAs
GT8-binding ELISA:
96-well half area plates (Corning) were coated at room temperature for 8 hours with 1 μg/mL MonoRab anti-His antibody (GenScript, Catalog: A01857), followed by overnight blocking with blocking buffer containing 1x PBS, 5% skim milk (Sigma), 10% goat serum (Milipore), 1% BSA (Sigma), 1% FBS (Lampire), and 0.2% Tween-20 (Sigma). The plates were then incubated with 2 μg/mL of his-tagged GT8-monomer at room temperature for 2 hours, followed by addition of mice sera serially diluted with PBS with 1% FBS and 0.1% Tween and incubation at 37°C for 2 hours (diluted 100 to 7,812,500-fold). BALB/c mice sera from 0, 7, 14, 21, and 35 d.p.i were analyzed in Figure 1 and BATF3-KO mice sera from 0, and 14 d.p.i were analyzed in Supplementary Figure S2k. The plates were incubated at room temperature for 1 hour with anti-mouse IgG H+L HRP (Bethyl) at 1:20,000 dilution, followed by addition of TMB substrates (ThermoFisher) and then quenched with 1M H2SO4. Absorbance at 450 nm and 570 nm were recorded with Synergy 2 plate reader (BioTEK). Endpoint titer was defined as the highest dilution at which the OD of the post-immune sera exceeds the cut-off (mean OD of naïve animals plus standard deviations of the OD in the naïve sera multiplied with standard deviation multiplier f at the 99% confidence level).
HA-binding ELISA:
96-well half area plates were coated at 4°C overnight with 2 μg/mL of recombinant HA(ΔTM)(H1N1/A/New Caledonia/20/1999; Immune Technology), and blocked at room temperature for 2 hour with the buffer as described above. The plates were subsequently incubated with serially diluted mouse sera at 37°C for 2 hours (diluted 100 to 7,812,500-fold). BALB/c mice sera from 0, 7, 14, 21, and 35 d.p.i were analyzed in Figure 1. This was followed by 1-hour incubation with anti-mouse IgG H+L HRP (Bethyl, Catalog: A90-116P) at 1:20,000 dilution at room temperature and developed with TMB substrate. Absorbance at 450 nm and 570 nm were recorded with BioTEK plate reader and endpoint titers calculated as above.
Antigenic profile characterization of designed GT8, Tpr2, and Gp100-nano-vaccines
Corning half-area 96-well plates were coated with 2 μg/mL of GT8-monomer, eOD-GT8-60mer, Trp2-60mer, and Gp100-60mer at 4°C overnight. The plates were then blocked with the buffer as described in the “GT8-binding ELISA” section for 2 hours at room temperature, followed by incubation with serially diluted broadly neutralizing HIV antibody, VRC01 (NIH AIDS reagent), from 1000 ng/mL to 0.5 ng/mL at room temperature for 2 hours. The plates were then incubated with anti-human Fc (cross-adsorbed against rabbits and mice; Jackson Immunoresearch) at 1:10,000 dilution for 1 hour, followed by addition of TMB substrate for detection. Absorbance at 450 nm and 570 nm were recorded with BioTEK plate reader.
HAI assay
BALB/c mice sera at 35 d.p.i were treated with receptor-destroying enzyme (RDE, 1:3 ratio; SEIKEN) at 37°C overnight for 18–20 hours, followed by complement and enzyme inactivation at 56°C for 45 minutes. RDE-treated sera were subsequently cross-adsorbed with 10% rooster red blood cells (Lampire Biologicals) in PBS at 4°C for 1 hour. The cross-adsorbed sera were then serially diluted (16 to 2048-fold) with PBS in a 96-well V-bottom microtiter plates (Corning). Four hemagglutinating doses (HAD) of A/New Caledonia/20/99 (BEI) were added to each well, and the serum–virus mixture was incubated at room temperature for 1 hour. The mixture was then incubated with 50 μL 0.5% v/v rooster red blood cells (Lampire) in 0.9% saline for 30 minutes at room temperature. The HAI antibody titer was scored with the dot method, and the reciprocal of the highest dilution that did not cause agglutination of the rooster red blood cells was recorded.
ELISpot Assay
Spleens were collected from immunized BALB/c mice 35 d.p.i or C57BL/6 mice 28 d.p.i and homogenized into single-cell suspensions with a tissue stomacher in 10% FBS/1% penicillin-streptomycin (Sigma) in RPMI 1640. Red blood cells were subsequently lysed with ACK lysing buffer (ThermoFisher), and the percentage of viable cells were determined with Trypan Blue exclusion using Vi-CELL XR (Beckman Coulter). 200,000 cells were then plated in each well of mouse IFNγ ELISpot plates (MabTech), followed by addition of peptide pools that span both the lumazine synthase, GT8 or HA domains, or individual Trp2 (SVYDFFVWL) and Gp10025 peptide (EGPRNQDWL) at 5 μg/mL of final concentration for each peptide (GenScript). The cells were then stimulated at 37°C for 16-18 hours, followed by development according to the manufacturer’s instructions. Spots for each well were then imaged and counted with ImmunoSpot Macro Analyzer.
Intracellular cytokine staining
Single-cell suspension from spleens of immunized animals (BALB/c mice 35 d.p.i or C57BL/6 mice 28 d.p.i) were prepared as described before and stimulated with 5 μg/mL of peptide pools that span both the lumazine synthase, GT8 or HA domains, or individual Trp2 (SVYDFFVWL) and Gp10025 peptide (EGPRNQDWL) (GenScript) for 5 hours at 37°C in the presence of 1:500 protein transport inhibitor (ThermoFisher) and anti-mouse CD107a-FITC (ThermoFisher). The cells were then incubated with live/dead Fixable Violet Dead Cell Stain Kit (for 405 nm excitation) for 10 minutes at room temperature, and surface stained (anti-mouse CD4-BV510, Biolegend, Catalog: 100559; anti-mouse CD8–APC-Cy7, Biolegend, Catalog: 100714) at room temperature for 30 minutes. The cells were then fixed and permeabilized according to manufacturer’s instructions for BD Cytoperm Cytofix kit and stained with anti-mouse IL2–PE-Cy7 (BioLegend, Catalog: 503832), anti-mouse IFNγ-APC (BioLegend, Catalog: 505810), anti-mouse CD3e–PE-Cy5 (BioLegend, Catalog: 100310), and anti-mouse TNFα-BV605 (BioLegend, Catalog: 506329) at 4°C for 1 hour. The cells were subsequently analyzed with LSR II 18-color flow cytometer. The data was analyzed with FlowJo V10.6.1. ICS gate for CD8+IFNγ+ population is set as in Figure 1C and Supplementary Figure S4.
Immunofluorescence
To detect presence of cleaved caspase-3 using immunofluorescence, four days after C57BL/6 mice were intramuscularly immunized with DLnano_LS_GT8 or protein eOD-GT8-60mer, tibialis anterior muscles of the mice were harvested and preserved in 4% paraformaldehyde (PFA)/PBS for 4 hours at room temperature and then stored overnight in 70% EtOH/H2O at 4°C. The tissues were then serially dehydrated for paraffin embedding, and blocked in 3% BSA/PBS for 1 hour at room temperature, permeabilized with 0.5% Triton X-100 (Fisher)/PBS, followed by overnight staining with rabbit anti-cleaved caspase 3 (1:200, Cell Signaling, Catalog: 9664S). The sections were then washed and stained with anti-Rabbit Alexa Fluor 594 antibody (1:200, Invitrogen, Catalog: A11037) for 1 hour at room temperature, then counterstained with 0.5 μg/mL DAPI and imaged with Leica SP5 confocal microscopes (with the following settings: 3% laser power for 405nm laser, and 1.25% laser power for 488nm laser; line accumulation: 3; frame average: 3; instrument gain: 100%).
Serum CK and LDH assay
Mouse serum creatine kinase and lactose dehydrogenase was diluted 10-fold in PBS and the enzyme activities were measured 0, 1, 4, 7, and 10 days following intramuscular administrations of 25 μg DLnano_LS_GT8 or 10 μg RIBI-adjuvanted protein eOD-GT8-60mer, or from untreated control mice, using colorimetric assays according to manufacturer’s protocol (Abcam Ab 155901 and Ab 65393). Changes in absorbance at 450 nm was measured at 37°C under kinetics mode over time using a BioTEK plate reader.
TUNEL assay
A TUNEL assay was performed on fixed muscle specimen according to manufacturer’s protocol (Abcam, Kit Catalog: ab206386). Briefly, four days post treatment with DNA or protein vaccine, tibialis anterior muscles were harvested and fixed in 4% PFA/PBS for 3 hours at 4°C. The tissues were then dehydrated and paraffin-embedded, and the sample sections were then rehydrated by serial transfer of the tissues from 100% ethanol to 70% ethanol/water. The specimens were then permeabilized by treatment with 50 μL Proteinase K at 20 μg/mL (Abcam) at room temperature for 20 minutes. Endogenous peroxidase was then quenched by treatment of tissue sections with 3% H2O2 in methanol. The sections were then incubated with 40uL TdT enzyme (diluted 1:40 from the TdT enzyme stock, Abcam) in equilibration buffer at room temperature for 1.5 hours in a humidified chamber, and the reactions were stopped with the stop solution provided in the kit. The sections were then blocked with the provided blocking buffer at room temperature for 10 minutes and labelled with provided detection conjugate diluted 1:25 in blocking buffer at room temperature for 30 minutes. The sections were then developed with DAB substrate at room temperature for 15 minutes and counterstained with methyl green before they were imaged with a Nikon Eclipse 80i microscope under 20x and 60x magnification (with autoexposure settings).
Flow analyses of muscle tissues post DNA or protein vaccination
Following intramuscular protein or DNA vaccinations in C57BL/6 mice 4 or 7 d.p.i (as described above), transfected muscles were harvested. Cells were extracted from muscles using a 30-minute digestion in Hanks’ Balanced Salt Solution [Gibco] supplemented with 0.5% Type IV collagenase (Life Technologies), 0.2% BSA, and 0.025% trypsin (Corning) at 37°C. The cells were then stained with Ultra-Violet Reactive (Live/Dead dye, 1:333 in PBS), and then with a 1:200 dilution of anti-CD11b–PerCPCy5.5 (Biolegend, Catalog: 101228), anti-CD11c–PE (Biolegend, Catalog: 301606), anti-MHC Class II–APC-Cy7 (Biolegend, Catalog: 107628), anti-F4/80–PE-Cy7 (Biolegend, Catalog: 123114), and anti-CD206–APC (Biolegend, Catalog: 141708) in 2% FBS/PBS at room temperature for 1 hour before they were permeabilized with BD CytoPerm/CytoFix buffer for 20 minutes at 4°C and subsequently labelled with FITC-lightning link labelled VRC01 (Expedon) at 4°C for 1 hour. The cells were then resuspended in 1x BD Permeabilization Wash buffer for analyses on LSR II 18-color flow cytometer. The data is analyzed with FlowJo V10.6.1. ICS gate for macrophages is set as in Figure 2C and Supplementary Figure S2E.
Negative-stain electron microscopy (EM) of purified nanoparticles
The nanoparticles produced in Expi293 cells were purified using agarose-bound lectin beads (Agarose Galanthus Nivalis Lectin, Vector Laboratories) followed by size-exclusion chromatography (GE Healthcare) using the Superose 6 Increase 10/300 GL Column. The proteins were further dialyzed into Tris-buffered saline (TBS). A total of 3 μL of purified proteins was adsorbed onto glow-discharged carbon-coated Cu400 EM grids (Electron Microscopy Sciences). The grids were then stained with 3 μL of 2% uranyl acetate, blotted, and stained again with 3 μL of the stain followed by a final blot. Image collection and data processing was performed on a FEI Tecnai T12 microscope equipped with a Oneview Gatan camera at 90,450X magnification at the camera and a pixel size of 1.66 Å.
In vivo imaging
For B16-F10-Luc2 challenge study, tumor-challenged mice received 150 mg/kg administration of VivoGlo™ Luciferin (Promega) at each timepoint (0, 5, 10, and 14 days post tumor inoculation) formulated in sterile PBS, and then imaged with an IVIS Spectrum CT for Bioluminescence with the autoexposure settings (or for 60 seconds, whichever was shorter) 10 minutes post injection.
Statistics
Mice in the experiments were randomly assorted into cages by Animal Facility staff and not further randomized. Data acquisition and analysis were not blinded. Power analysis was performed with R based on our preliminary data to determine the smallest sample size that would allow us to achieve a power of 0.9 with a pre-set α-value of 0.05. All statistical analyses were performed with PRISM V8.2.1 and R V3.5.1. Each individual data point was sampled independently. Two-tailed Mann Whitney Rank tests were used to compare differences between groups. Log Rank test was used to compare survival between two groups in challenge survival studies. Bonferroni corrections were used to adjust for multiple comparisons.
Results
DNA-launched nanoparticle vaccines induce CD8+ T-cell responses to HIV and influenza antigens
First, we compared adaptive immune responses induced by DLnano-vaccines and protein nano-vaccines. We utilized two model antigens: a priming HIV antigen eOD-GT8-60mer and an influenza hemagglutinin (H1 A/NewCaledonia/20/1999)60mer antigen scaffolded by lumazine synthase (18,19). DNA-Launched nanoparticle Lumazine Synthase decorated with an anti–HIV-1 immunogen eOD-GT8 (DLnano_LS_GT8, Supplementary Figure S1A) and RIBI-adjuvanted protein eOD-GT8-60mer induced similar antibody titers in BALB/c mice after two immunizations (Figure 1A). Epitope mapping conducted in prior studies identified that CD4+ T-cell responses elicited by LS-scaffolded DLnano-vaccines were predominantly directed to the LS domain (20). In this study, CD4+ T-cell responses to the LS domain after two vaccinations, measured by intracellular cytokine staining (ICS), were similar between DLnano_LS_GT8 and protein eOD-GT8-60mer (Figure 1B). We also observed robust induction of CD8+ T-cell responses to the GT8 domain in mice immunized with DLnano_LS_GT8 but not in those vaccinated with protein eOD-GT8-60mer (Figure 1C-F).
We further determined if this observation was limited by our choice of RIBI as the protein adjuvant and performed a head-to-head comparison of 25 μg DLnano_LS_GT8 versus 10 μg protein eOD-GT8-60mer adjuvanted with either RIBI, 50 μg Poly(I:C), or 20 μg CpG ODN (21,22). We observed only DLnano_LS_GT8, but not protein eOD-GT8-60mer administered with any form of adjuvant, was capable of inducing CD8+ T-cell responses (Supplementary Figure S1B-C), even though both DLnano_LS_GT8 and protein eOD-GT8-60mer induced CD4+ T-cell responses to varying degrees (Supplementary Figure S1D). We next determined if this observation might be dose-limited and compared DLnano_LS_GT8 and RIBI-adjuvanted protein eOD-GT8-60mer vaccinations both at 50 μg doses. Only DLnano_LS_GT8, but not protein eOD-GT8-60mer, was observed to induce CTL responses (Supplementary Figure S1E-F), despite induction of CD4+ T-cell and humoral responses by both routes of vaccination (Supplementary Figure S1G-H).
We also determined the role of EP in mediating CD8+ T-cell responses by vaccinating mice with DLnano_LS_GT8 or RIBI-adjuvanted protein eOD-GT8–60mer with or without EP. Although EP increased humoral responses for DLnano_LS_GT8 and protein eOD-GT8-60mer (Supplementary Figure S1I-J), EP could only adjuvant CD8+ T-cell responses in the DLnano_LS_GT8 groups, and no responses were observed in the protein eOD-GT8-60mer groups with or without EP (Supplementary Figure S1K). Antigens supplied by protein versus DNA-launched eOD-GT8-60mer existed in different forms. Protein was given exogenously as a depot and existed as soluble factors (23). DNA-launched nano-vaccines, however, were expressed by with hosts’ own myocytes and were cell-associated (12). This finding suggested that induction of CD8+ T-cell immunity, in this context, required cell-associated antigens expressed by host cells from DNA-cassettes.
We determined if this observation may apply to an alternative nanoparticle-scaffolded influenza antigen. Indeed, two vaccinations of DLnano_LS_HA(NC99) and RIBI-adjuvanted protein HA(NC99) _60mer induced similar binding antibody titers to recombinant NC99 hemagglutinin (Figure 1G), as well as hemagglutination inhibition titers against the autologous A/NewCaledonia/20/1999 virus (Figure 1H). CD4+ T-cell responses were similarly induced by DLnano_LS_HA(NC99) and protein HA(NC99)_60mer (Supplementary Figure S1L). However, only DLnano_LS_HA(NC99), but not protein HA(NC99)_60mer, induced CD8+ T-cell responses to HA (Figure 1I, Supplementary Figure S1M, Figure 1J).
Induction of tissue apoptosis and APC infiltration are important for priming CTLs by DLnano-vaccines
We hypothesized that the distinct modality of antigen uptake and presentation for DLnano-vaccines compared to protein nano-vaccines may attribute to unique induction of CTL responses by DLnano-vaccines. Previous studies observe that co-administration of DNA-vaccines with DNA-cassettes encoding pro-apoptotic genes lead to significantly improved CTL responses (24). We, therefore, decided to examine tissue apoptosis and APC infiltration following vaccination. We first assessed the extent of muscle tissue damage upon vaccination using immunofluorescent staining against cleaved caspase-3 (25). In C57BL/6 mice, four days post injection (d.p.i), hypercellularity and expression of cleaved caspase-3 were only observed in the muscles of mice immunized with DLnano_LS_GT8 combined with EP, but not in those vaccinated with protein eOD-GT8-60mer co-formulated in RIBI without EP or in naïve mice (Figure 2A). Alternatively, using a TUNEL assay (26) four d.p.i., brown nuclei, suggestive of double-stranded DNA breaks and cellular apoptosis, were only observed in muscle tissues treated with DLnano_LS_GT8 and electroporation but not in muscle sections from naïve mice or those treated with RIBI-formulated protein eOD-GT8-60mer without EP (Figure 2B). EP was determined to be instrumental to the induction of myocyte apoptosis, as cleaved caspase-3 expression was observed four d.p.i when either protein eOD-GT8-60mer or DLnano_LS_GT8 was delivered with EP (Supplementary Figure S2A). Without EP, low cleaved caspase-3 expression was observed only for DLnano_LS_GT8 but not for protein eOD-GT8-60mer.
We performed a time course experiment and observed that DLnano_LS_GT8–induced tissue apoptosis was transient, peaking at 6 d.p.i and was fully resolved by 14 d.p.i (Supplementary Figure S2B). We also observed that tissue apoptosis was limited locally to the injection sites, as indicators of tissue damage such as serum lactose dehydrogenase (LDH) and creatine kinase (CK) were not elevated in mice immunized with DLnano_LS_GT8 or protein eOD-GT8-60mer relative to untreated control mice (Supplementary Figure S2C-D).
We next examined the consequences of induced tissue apoptosis on tissue APC infiltration. Seven d.p.i, we observed increased influx of CD11b+F4/80+ macrophages into muscles of mice treated with DLnano_LS_GT8 and EP compared to protein eOD-GT8-60mer formulated in RIBI without EP (Figure 2C-D). Infiltrating macrophages were observed to be predominantly pro-inflammatory CD11c+CD206− M1 macrophages rather than anti-inflammatory CD11c−CD206+ M2 macrophages (Supplementary Figure S2E-F). Increased influx of CD11c+MHCII+ DCs was also observed for mice treated with DLnano_LS_GT8 compared to those treated with protein eOD-GT8-60mer. However, infiltrating DCs were significantly less abundant than macrophages (Figure 2E). Staining of APCs with GT8-specific VRC01 demonstrated that infiltrating macrophages induced by DLnano_LS_GT8, but not those induced by protein eOD-GT8-60mer, had taken up the GT8 antigen (Figure 2F, Supplementary Figure S2G).
To determine the functional roles played by macrophages in priming of CD8+ T-cell responses by DLnano-vaccines, we systemically depleted the macrophage population in C57BL/6 mice prior to DNA vaccination with intravenous (IV) injection of clodrosome (27). A single IV infusion of clodrosome specifically depleted CD11b+F4/80+ macrophages but not CD11c+MHCII+ DCs in the spleens of animals one d.p.i (Supplementary Figure S2H-J). Depletion of infiltrating macrophages in the muscles post vaccination required increased doses of clodrosome on −3, 0, and 3 d.p.i (with respect to DNA vaccination), which significantly reduced macrophage infiltration into the muscles by approximately four-fold (Figure 2G). This depletion scheme significantly attenuated induced CD8+ T-cell responses following one vaccination of DLnano_LS_GT8 14 d.p.i in C57BL/6 mice by approximately three-fold (Figure 2H).
It has previously been reported that monocytic populations can transfer sequestered peptides through gap junctions to conventional (c)DCs, which can more efficiently prime CD8+ T cells (28). We investigated the role of CD11c+CD8α+ cDCs in cross-presentation of DLnano-immunogens using BATF3KO transgenic mice, which lack splenic development of CD8α+ cDCs (29). Single immunization of DLnano_LS_GT8 induced similar humoral responses to GT8 in BATF3KO and wild-type C57BL/6 mice (Supplementary Figure S2K), even though induced CD8+ T-cell responses were significantly attenuated in BATF3KO mice (Figure 2I). Taken together, the data highlight the importance of both phagocytic macrophages and CD8α+ cDCs in priming CD8+ T-cells.
DNA-launched Trp2188 and Gp10025 nano-vaccines control melanoma growth mice
To demonstrate the functional relevance of CTL priming by DLnano-vaccines in the treatment of cancer, we designed nano-vaccines displaying immunodominant CD8+ T-cell epitopes. The self-assembling nanoparticles were engineered using a structure-guided process as described in the materials and methods. We designed nanoparticles presenting 60 copies of Trp2188 peptide (DLnano_LS_Trp2188) and Gp10025 peptide (DLnano_LS_Gp10025). Homogenous in vitro assemblies of DLnano_LS_Trp2188 and DLnano_LS _Gp10025 were observed by SEC (Figure 3A, Supplementary Figure S3A) and negative-stain electron microscopy, nsEM (Figure 3B, Supplementary Figure S3B), respectively. We also demonstrated the multivalent nature of the designed DLnano_LS_Trp2188 and DLnano_LS _Gp10025 nanoparticles, which bound to VRC01 with affinity similar to that of eOD-GT8-60mer, and higher than that of GT8-monomer (Supplementary Figure S3C).
Groups of C57BL/6 mice were immunized with designed DNA-launched nanoparticle Trp2188 or Gp10025 vaccines individually and were observed to induce significantly improved CD8+ T-cell responses to the Trp2188 and Gp10025 peptides compared to DNA vaccines encoding monomeric GT8-scaffolded Trp2188 and Gp10025, respectively (DLmono_Trp2188 and DLmono_Gp10025, Figure 3C-D). When DLnano_LS_Trp2188 and DLnano_LS_Gp10025 were administered in separate sites in a single animal, the co-administration resulted in improved elicitation of CD8+ T-cell responses to both Trp2 and Gp100 peptides compared to DLmono_Trp2188 and DLmono_Gp10025 (Supplementary Figure S3D). We evaluated the potency of the designed DLmono_Trp2188, DLmono_Gp10025, DLnano_LS_Trp2188 and DLnano_LS_Gp10025 in a therapeutic B16-F10 melanoma model (Supplementary Figure S3E). We observed DLnano_LS_Trp2188 and DLnano_LS_Gp10025 treatments, in the absence of anti–PD-1, significantly extended median survival compared to pVAX treatment. Potency of DLnano_LS_Trp2188 and DLnano_LS_Gp10025 treatments could be further enhanced when anti–PD-1 was co-administered. DLnano_LS_Trp2188 and DLnano_LS_Gp10025 combined with anti–PD-1 significantly improved survival compared to DLmono_Trp2188 and DLmono_Gp10025 alone, extending the median survival by 9 days.
We next compared CTL responses induced by DLnano-vaccines to CpG-adjuvanted peptide vaccines. We observed that both DLnano_LS_Trp2188 and CpG-adjuvanted Trp2188 peptide vaccination induced peptide-specific CD8+ T-cell responses, although responses induced by DLnano_LS_Trp2188 vaccination were significantly higher (Supplementary Figure S3F). For the Gp10025 peptide, prior studies suggest that Gp10025 peptide vaccination induces weaker and less consistent CD8+ T-cell responses, unless combined with lymph node targeting agents or TLR7 agonists (30,31). Our study indicated that vaccination with only DLnano_LS_Gp10025, but not CpG adjuvanted Gp10025 peptide, induced robust peptide-specific CD8+ T-cell responses (Supplementary Figure S3G-I).
We next compared induction of CD8+ T-cell responses to Trp2188 and Gp10025 epitopes in B16-F10 melanoma–bearing mice, which received treatments as shown in the scheme (Figure 3E)(32). We observed CD8+ T-cell responses to Trp2 only in mice treated with DNA vaccinations but not those receiving anti–PD-1 treatment alone or anti–PD-1 plus protein vaccination (Figure 3F). Induced Trp2-specific CD8+ T-cells exhibited effector phenotypes (IFNγ+CD107a+)(Figure 3G) and were also polyfunctional (IFNγ+TNFα+IL2+)(Figure 3H). Similar observations were also made for Gp100-specific CD8+ T-cells (Supplementary Figure S3J-L). In this therapeutic model, DNA, but not protein, vaccination of Trp2188 and Gp10025-60mer was observed to suppress B16-F10 tumor growth, and significantly prolonged median survival of mice by 11 days (Figure 3I-J). The effect was even more pronounced when mice were vaccinated prior to tumor inoculation. Two vaccinations of DLnano_LS_Trp2188 and DLnano_LS_Gp10025 prior to B16-F10-Luc challenge completely prevented tumor growth in 80% (4/5) mice, whereas tumor growth was observed in all pVAX control mice or mice treated with protein Trp2188 and Gp10025-60mer (Figure 3K). Although all mice in the pVAX and protein groups died, 80% of mice in the DNA group had tumor-free survival (Figure 3L). Finally, protection by DLnano-vaccination was observed to be CTL-mediated, as indicated by CD8+ T-cell depletion (Supplementary Figure S3M, Figure 3M). Taken together, the data suggest DLnano-vaccination can elicit CD8+ T-cell responses to confer protection against melanoma both in prophylactic and therapeutic settings.
Discussion
CD8+ T cells can play an extremely important role in surveillance against intracellular pathogens and tumors. In cancer, presence of tumor-infiltrating CD8+ T cells correlates with improved prognosis in patients (33,34). Such observations have led to the use of T cell–based therapies, such as DC vaccines or in vitro expansion and adoptive transfer of tumor-infiltrating lymphocytes (TILs), in cancer patients, achieving varying degrees of success (35,36). Alternatively, several reports also describe the antitumor activity of antigen-specific CD4+ T-cells. TH1 cells, for instance, are responsible for immune responses against tumors by either enhancing CD8+ T-cell response or activating macrophages to phagocytose cancerous cells. CD4+ CTLs are another subset of CD4+ T cells that have acquired cytolytic activity and demonstrate clear antitumor activity (37,38). Our work here mainly focuses on designing DNA-launched nanoparticle vaccines scaffolding CD8+ T-cell epitopes, but nano-vaccines scaffolding CD4+ T-cell epitopes should also be closely examined in future work.
Induction of CTL responses by vaccination can be challenging and not readily achieved by vaccination with protein or inactivated virus (39). DNA vaccines have been shown to elicit CD8+ T-cell responses both in preclinical animal models and in clinical trials (9,16). Here, we observed that EP resulted in transient tissue apoptosis and macrophage infiltration, and significantly enhanced CTL responses by DLnano-vaccines. Bolus protein nanoparticle vaccinations and EP, however, did not result in CTL induction, possibly suggesting that direct in vivo production of nano-vaccines in host’s cells would be required. The work also demonstrated the importance of both EP and macrophages in CTL induction by DLnano-vaccines. However, the study did not directly demonstrate that electroporation-induced muscle-infiltrating macrophages was critical because the clodrosome treatments used here systemically depleted macrophages, including, but not limited to, the EP-induced muscle-infiltrating macrophages. Future study to characterize the biology of these muscle-infiltrating macrophagesis important. We also demonstrated the importance of CD8+ T-cell priming by DNA-launched nanoparticle vaccines in a melanoma model. We designed a generalizable nanoparticle platform for displaying CD8+ T-cell epitopes to various tumor antigens. As a proof-of-principle, we engineered two nanoparticle vaccines scaffolding 60 copies of CD8+ epitopes from Trp2 and Gp100. DNA-cassettes encoding anti-melanoma LS-nanoparticles scaffolding Trp2 and Gp100 epitopes could elicit significantly improved CTL responses to both targets compared to conventional monomeric DNA vaccines, CpG-adjuvanted peptide vaccines, and protein nano-vaccines, and conferred 80% protection when the vaccine was prophylactically administered before tumor challenge. Finally, DLnano-vaccines could induce CTL responses to whole antigens, such as GT8, influenza hemagglutinin, or to selected peptides. Whole antigen or neoantigen peptide-oriented DLnano-vaccines may be designed for various cancer targets and presented as a viable and attractive strategy in the exciting era of cancer immunotherapy.
Supplementary Material
Acknowledgements
We would like to thank Animal Facility staff at Wistar Institute for providing care to the animals. We thank Imaging Facility Core at Wistar Institute for assistance with in vivo imaging experiment. We thank Histotechnology Core at Wistar Institute for assistance with sectioning/ preparation of sample specimens. We thank the Flow Core at the Wistar Institute for assistance on the flow experiments. The following reagent was obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: VRC01 antibody from Xueling Wu, Zhi-Yong Yang, Yuxing Li, Gary Nabel and John Mascola.
Funding sources
This research is supported by NIH IPCAVD Grant U19 Al109646-04, NIH/NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051, Inovio Pharmaceuticals Virus Grant 5181101374 awarded to D.B.W, W. W. Smith Charitable Trust 68112-01-383 awarded to D.W.K, and by Wistar Monica H.M. Shander Memorial Fellowship awarded to Z.X.
Footnotes
Conflicts of interests
Z.X., D.B.W and D.W.K. have a pending patent US.62784318. M.C.W, K.E.B, and L.M.H are employees of Inovio Pharmaceuticals and as such receive salary and benefits including ownership of stock and stock options from the company. D.B.W. has received grant funding, participates in industry collaborations, has received speaking honoraria, and has received fees for consulting, including serving on scientific review committees and board series. Remuneration received by D.B.W. includes direct payments, stock or stock options, and in the interest of disclosure he notes potential conflicts associated with his work with Inovio and possible others.
References
- 1.Halle S, Halle O, Forster R. Mechanisms and Dynamics of T Cell-Mediated Cytotoxicity In Vivo. Trends Immunol 2017;38:432–43 [DOI] [PubMed] [Google Scholar]
- 2.Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol 2019;234:8509–21 [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 2013;13:227–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan WG, Jin HT, West EE, Penaloza-MacMaster P, Wieland A, Zilliox MJ, et al. Comparative analysis of simian immunodeficiency virus gag-specific effector and memory CD8+ T cells induced by different adenovirus vectors. J Virol 2013;87:1359–7223175355 [Google Scholar]
- 5.Gros M, Amigorena S. Regulation of Antigen Export to the Cytosol During Cross-Presentation. Front Immunol 2019;10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena M, Bhardwaj N. Re-Emergence of Dendritic Cell Vaccines for Cancer Treatment. Trends Cancer 2018;4:119–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng K, El-Boubbou K, Landry CC. Binding of HIV-1 gp120 glycoprotein to silica nanoparticles modified with CD4 glycoprotein and CD4 peptide fragments. ACS Appl Mater Interfaces 2012;4:235–43 [DOI] [PubMed] [Google Scholar]
- 8.Fausther-Bovendo H, Kobinger GP. Pre-existing immunity against Ad vectors: humoral, cellular, and innate response, what's important? Hum Vaccin Immunother 2014;10:2875–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trimble CL, Morrow MP, Kraynyak KA, Shen X, Dallas M, Yan J, et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet 2015;386:2078–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akbari O, Panjwani N, Garcia S, Tascon R, Lowrie D, Stockinger B. DNA vaccination: transfection and activation of dendritic cells as key events for immunity. J Exp Med 1999;189:169–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu TM, Ulmer JB, Caulfield MJ, Deck RR, Friedman A, Wang S, et al. Priming of cytotoxic T lymphocytes by DNA vaccines: requirement for professional antigen presenting cells and evidence for antigen transfer from myocytes. Mol Med 1997;3:362–71 [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Wise MC, Chokkalingam N, Walker S, Tello-Ruiz E, Elliott STC, et al. In Vivo Assembly of Nanoparticles Achieved through Synergy of Structure-Based Protein Engineering and Synthetic DNA Generates Enhanced Adaptive Immunity. Adv Sci (Weinh) 2020;7:1902802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Wise MC, Choi H, Perales-Puchalt A, Patel A, Tello-Ruiz E, et al. Synthetic DNA delivery by electroporation promotes robust in vivo sulfation of broadly neutralizing anti-HIV immunoadhesin eCD4-Ig. EBioMedicine 2018;35:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, et al. Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies. Cell 2016;166:1459–70 e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amante DH, Smith TR, Mendoza JM, Schultheis K, McCoy JR, Khan AS, et al. Skin Transfection Patterns and Expression Kinetics of Electroporation-Enhanced Plasmid Delivery Using the CELLECTRA-3P, a Portable Next-Generation Dermal Electroporation Device. Hum Gene Ther Methods 2015;26:134–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duperret EK, Wise MC, Trautz A, Villarreal DO, Ferraro B, Walters J, et al. Synergy of Immune Checkpoint Blockade with a Novel Synthetic Consensus DNA Vaccine Targeting TERT. Mol Ther 2018;26:435–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokatlian T, Read BJ, Jones CA, Kulp DW, Menis S, Chang JYH, et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 2019;363:649–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016;351:1458–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Kulp DW. Protein engineering and particulate display of B-cell epitopes to facilitate development of novel vaccines. Curr Opin Immunol 2019;59:49–56 [DOI] [PubMed] [Google Scholar]
- 20.Xu Z, Chokkalingam N, Tello-Ruiz E, Walker S, Kulp DW, Weiner DB. Incorporation of a Novel CD4+ Helper Epitope Identified from Aquifex aeolicus Enhances Humoral Responses Induced by DNA and Protein Vaccinations. iScience 2020:101399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salem ML, Diaz-Montero CM, El-Naggar SA, Chen Y, Moussa O, Cole DJ. The TLR3 agonist poly(I:C) targets CD8+ T cells and augments their antigen-specific responses upon their adoptive transfer into naive recipient mice. Vaccine 2009;27:549–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speiser DE, Lienard D, Rufer N, Rubio-Godoy V, Rimoldi D, Lejeune F, et al. Rapid and strong human CD8+ T cell responses to vaccination with peptide, IFA, and CpG oligodeoxynucleotide 7909. J Clin Invest 2005;115:739–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan T, Verma P, Rao DN. Novel adjuvants & delivery vehicles for vaccines development: a road ahead. Indian J Med Res 2013;138:779–95 [PMC free article] [PubMed] [Google Scholar]
- 24.Chattergoon MA, Kim JJ, Yang JS, Robinson TM, Lee DJ, Dentchev T, et al. Targeted antigen delivery to antigen-presenting cells including dendritic cells by engineered Fas-mediated apoptosis. Nat Biotechnol 2000;18:974–9 [DOI] [PubMed] [Google Scholar]
- 25.Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ 1999;6:99–104 [DOI] [PubMed] [Google Scholar]
- 26.Kyrylkova K, Kyryachenko S, Leid M, Kioussi C. Detection of apoptosis by TUNEL assay. Methods Mol Biol 2012;887:41–7 [DOI] [PubMed] [Google Scholar]
- 27.Weisser SB, van Rooijen N, Sly LM. Depletion and reconstitution of macrophages in mice. J Vis Exp 2012:4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang M-N, Nicholson LT, Batich KA, Swartz AM, Kopin D, Wellford S, et al. Antigen-loaded monocyte administration induces potent therapeutic anti-tumor T cell responses. The Journal of Clinical Investigation 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008;322:1097–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moynihan KD, Holden RL, Mehta NK, Wang C, Karver MR, Dinter J, et al. Enhancement of Peptide Vaccine Immunogenicity by Increasing Lymphatic Drainage and Boosting Serum Stability. Cancer Immunol Res 2018;6:1025–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Stipdonk MJ, Badia-Martinez D, Sluijter M, Offringa R, van Hall T, Achour A. Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100. Cancer Res 2009;69:7784–92 [DOI] [PubMed] [Google Scholar]
- 32.Moynihan KD, Opel CF, Szeto GL, Tzeng A, Zhu EF, Engreitz JM, et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat Med 2016;22:1402–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu S, Hiratsuka H, Koike K, Tsuchihashi K, Sonoda T, Ogi K, et al. Tumor-infiltrating CD8(+) T-cell density is an independent prognostic marker for oral squamous cell carcinoma. Cancer Med 2019;8:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer 2011;105:93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohaan MW, van den Berg JH, Kvistborg P, Haanen J. Adoptive transfer of tumor-infiltrating lymphocytes in melanoma: a viable treatment option. J Immunother Cancer 2018;6:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu R, Forget MA, Chacon J, Bernatchez C, Haymaker C, Chen JQ, et al. Adoptive T-cell therapy using autologous tumor-infiltrating lymphocytes for metastatic melanoma: current status and future outlook. Cancer J 2012;18:160–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res 2014;2:91–8 [DOI] [PubMed] [Google Scholar]
- 38.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 2010;207:637–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moorthy VS, Ballou WR. Immunological mechanisms underlying protection mediated by RTS,S: a review of the available data. Malar J 2009;8:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.