Abstract
Circular RNAs (circRNAs) are a diverse class of RNAs with varying sizes, cellular abundance, and biological functions. Investigations from the past decade have revealed that circRNAs are ubiquitously found in eukaryotes and have defined the different biological roles of circRNAs to illuminate this previously unrecognized class of molecules. In the context of the immune system, immune responses and immune-related diseases alter circRNA expression. More recently, several oncogenic double-stranded DNA viruses have been found to encode circRNAs. In this review, we summarize the current understanding of circRNAs and their emerging functions in immune regulation and autoimmune disorders, and discuss the identification and potential roles of viral circRNAs during infections. Finally, we present promising areas for future investigations in the nascent field of circRNAs.
Keywords: Virus-Encoded CircRNA, Self/Non-Self, N6-Methyladenosine (m6A), DNA Virus, Autoimmune Disease
Overview of CircRNAs
CircRNAs are covalently closed single-stranded RNAs that lack ends. Originally thought of as mere byproducts of aberrant splicing [1], circRNAs are now appreciated to have important biological roles [2]. CircRNAs have been found to regulate gene expression, inhibit protein activity, and encode protein, among other functions [2]. Next-generation sequencing coupled with advanced computational algorithms have revealed that many circRNAs are abundant and evolutionarily conserved across species [3]. In recent years, significant progress to advance our understanding of the biogenesis and identification of circRNAs has been made, but many questions about their purpose remain. In this review, we provide an overview of the current understanding of circRNAs, with a particular focus on their functions in immune cells and immune response. We introduce recent groundbreaking investigations that reveal new roles for eukaryotic circRNA in innate immunity regulation, and we discuss the discovery and potential function of circRNAs that are encoded by DNA viruses.
CircRNA Biogenesis is Directed by Intronic Complementary Sequences or RNA-Binding Proteins
The spliceosome (see Glossary) complex forms circRNAs from precursor messenger RNAs (pre-mRNAs) by “back-splicing” a splice donor to a splice acceptor that is 5’ to the donor site instead of “forward-splicing” to produce linear RNA [4]. While some circRNAs are a mix of exons and introns [5] and a few have only introns [6], the vast majority of mature circRNAs are composed exclusively of exons [3]. Computational and experimental studies have found that circRNA formation depends on cis-regulatory elements and trans-acting factors that bring the downstream splice donor into close proximity to the upstream splice acceptor, to facilitate back- instead of forward-splicing. Flanking introns that base pair to each other increase the efficiency of back-splicing of the intervening exon(s) to generate circRNA (Box 1) [7–9]. A parallel approach uses RNA-binding proteins, such as Quaking (QKI) and muscleblind (MBL), which form homodimers to guide the splice donor and acceptor sites together to facilitate preferential circRNA formation [10, 11]. Together, intronic complementary sequences and dimeric proteins enable the generation of cellular circRNAs that span a wide range of sizes, from a single exon to more than 10 exons [12].
Box 1.
Experimental evidence that intronic complementary sequences promote circularization of the intervening sequence comes from studies on the enzyme adenosine deaminase acting on RNA (ADAR). ADAR recognizes RNA double-stranded secondary structures and targets adenines for conversion into inosine in those regions. While adenine typically pairs with uracil, the deamination reaction changes the Watson-Crick-Franklin base pairing between intron complementary sequences to cause the RNA secondary structure to become single-stranded. The lack of base pairing increases the proximity of the splice donor and acceptor, which would favor forward- over back-splicing. When ADAR enzyme levels increase, circRNA formation decreases [9]. These findings demonstrate that the process of generating circRNAs depends on the proximity of the splice donor and acceptor for back-splicing.
CircRNA Identification Requires Specific Experimental Approaches and Bioinformatics Algorithms
CircRNAs are expressed ubiquitously across the kingdoms of life that splice RNA, including eukaryotes and archaea [2]. The first circRNAs identified in nature were the genomes of viroids (plant pathogens) [13] and the hepatitis delta virus [14]. Following these initial discoveries, only a handful of endogenous circRNAs were found over the next few decades. CircSry and circMbl were among the first ones detected, due to their high cellular abundance in mouse testes and Drosophila heads, respectively [15, 16]. Even the prevalence of next-generation sequencing in the 21st century did not immediately reveal the presence of circRNAs. While sequencing approaches identified many new RNA species, circRNAs remained the dark matter of the transcriptome. The experimental approaches and bioinformatics algorithms employed were based on assumptions about RNA biology, which underlie the four main reasons why circRNAs were not readily detected until relatively recently. First, many RNA isolation protocols for next-generation sequencing begin with poly(A) enrichment. This step captures mRNAs and long non-coding RNAs (lncRNAs), but omits RNAs that lack poly(A), including circRNAs. Second, most circRNAs are expressed at low levels, estimated to be at 5–10% of their corresponding linear RNA products [3, 8, 12, 17, 18]. Therefore, researchers either need to increase the sequencing depth and/or enrich for circRNAs to improve their detection. Total RNA depleted of rRNA and/or treated with exonuclease RNase R is often used for sequencing experiments to identify circRNAs. Third, circRNAs span a wide range of sizes, so selection that specifically enriches for small (i.e. miRNAs) or long RNAs (i.e. lncRNAs), will not capture the full diversity of circRNAs. Fourth, even in the absence of poly(A) enrichment, sequencing results that revealed a back-splice junction were often discarded. Scientists presumed that a rearrangement of exon sequences stemmed from a sequencing artifact or an error in the bioinformatics processing. Therefore, genome-wide identifications of circRNAs from sequencing data require specialized experimental approaches and bioinformatics algorithms [19].
CircRNAs Have Diverse Biological Roles
Most circRNAs may be a result of dysregulated splicing and are transcriptional noise. However, several observations suggest that cells actively control the formation of circRNAs and at least some circRNAs have physiological functions. CircRNAs have cell-specific expression, where circRNAs are enriched in the central nervous system but only slightly present in liver and muscle tissues [20]. Furthermore, many circRNAs are evolutionarily conserved throughout eukaryotes, while some are specific to metazoans and humans [3]. Studies have also shown that circRNA expression is differentially regulated compared to their linear RNA counterparts [18, 21], which suggest that circRNAs have dynamic regulation and function independent of their linear RNA counterparts.
One of the first demonstrated roles of circRNAs was as a microRNA (miRNA) sponge [18, 21]. The ability of circRNAs to base pair with other nucleic acids enables circRNAs to bind and sequester miRNAs from activity. The circRNA CDR1as, also known as ciR-7, has more than 70 binding sites antisense to miR-7 and one binding site antisense to miR-671 [18, 21]. CDR1as interacts with the miRNA seed sites to prevent the miRNA from acting on its targets but does not undergo cleavage by protein Argonaute 2 (AGO2). In vivo knockout experiments demonstrate that CDR1as loss leads to downregulation of miR-7 in neuronal tissues, which suggests that CDR1as is important for maintaining miR-7 expression [22] On the other hand, miR-7 is also involved in regulating CDR1as levels in neurons. Studies have found that lncRNA Cyrano can repress miR-7 via target-directed miRNA degradation. Unchecked miR-7 levels resulting from lncRNA Cyrano loss-of-function prevents the accumulation of CDR1as by enhancing the miR-671–directed slicing of CDR1as [23, 24]. These findings demonstrate a delicate regulatory network between circRNAs, lncRNAs, and miRNAs to control the proper function of these noncoding RNAs. Another abundant circRNA, circSry, has the capability to sponge sixteen copies of miR-138 at a time to regulate the translation of the corresponding mRNAs [21]. However, these highly-expressed circRNAs may be exceptions with regard to the number of miRNA binding sites that they encode. The majority of circRNAs do not have an increased number of miRNA binding sites compared to their linear RNA counterparts, suggesting that most circRNAs may not function as miRNA sponges [25].
CircRNAs can also regulate the gene expression of linear RNAs. MBL induces the circularization of exon 2 to form circMbl from its corresponding mRNA, which competes with canonical pre-mRNA splicing [11]. Therefore, the levels of circMbl and mature mRNA are negatively correlated. When there is excess protein, MBL binds to the flanking introns to promote circMbl biosynthesis. As the protein levels decrease, MBL is unavailable for binding, which enables the mature mRNA to be canonically spliced and translated. The process by which MBL induces circMbl formation may be a unique situation in how circRNAs regulate linear RNA levels post-transcriptionally.
Early investigations into whether circRNAs are translated have concluded that circRNAs do not associate with polysomes, which suggests that circRNAs are non-coding RNAs [3]. However, more recent studies have revealed that some circRNAs interact with translating ribosomes [26–31]. Since circRNAs are generally expressed in a cell type- and tissue-specific manner [3, 17, 20], the translation of circRNAs could also exhibit similar patterns. The initial analysis of ribosome profiling data and non-poly(A)-selected RNA-seq data for U2OS (human bone osteosarcoma epithelial) cells did not reveal ribosome protected fragment reads that correspond to a known circRNA [12]. However, ribosome footprinting from Drosophila heads where circRNAs are abundantly expressed showed that circRNAs are indeed associated with translating ribosomes [27]. Moreover, a recent investigation into the human translatome has demonstrated that circRNAs encode proteins smaller than 100 amino acids (microproteins) in the human heart [32]. The microprotein primary sequences correspond to translation across the back-splice junction of circRNAs. The functions of these circRNA-encoded microproteins still require elucidation.
The mechanisms driving how circRNAs are translated also invite additional investigation. The internal ribosome entry site (IRES), originally discovered in picornavirus mRNAs, is a well-characterized mechanism to initiate cap-independent translation in certain cellular mRNAs [33–35]. More recently, the RNA modification N6-methyladenosine (m6A) was also shown to recruit ribosomes and enable cap-independent translation of mRNAs [36, 37]. Since circRNAs lack a 5’ end, they rely on cap-independent translation initiation. Studies have shown that both m6A [28, 38] and IRES [39, 40] recruit ribosomes to circRNA templates for translation. While cellular factors, including eukaryotic initiation factor 3 (eIF3) and ATP-binding cassette subfamily F member 1 (ABCF1), have been shown to mediate cap-independent translation of mRNA [36, 41], circRNA translation require other proteins. The m6A-driven translation of circRNAs needs the initiation factor eukaryotic translation initiation factor 4 gamma 2 (eIF4G2) and m6A reader YTH domain-containing family protein 3 (YTHDF3) [28, 38]. These findings establish that circRNAs containing m6A or IRES may be translated, but whether the main function of cellular circRNAs is to encode for proteins and the roles of the resulting proteins remain unknown.
CircRNAs in Immune Response and Immune Cells
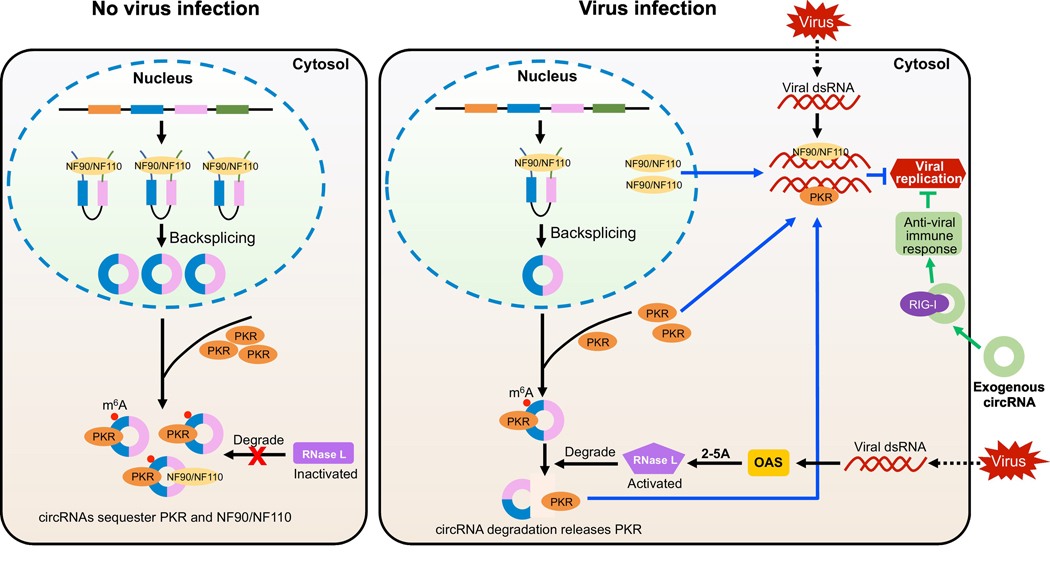
Although rapid progress has been made toward understanding the functions of circRNAs in cancers [42], neurological diseases [43], and cardiovascular disorders [44], few studies have reported on the roles of circRNAs in the regulation of the immune response. A greater understanding of circRNAs can reveal fundamental principles in immunology and RNA biology as well as offer new targets for treating diseases. Of the studies conducted, a recent investigation showed that cellular circRNAs can inhibit protein kinase R (PKR), a key enzyme in antiviral signaling [45] (Figure 1). PKR recognizes double-stranded RNA (dsRNA) longer than 30 base pairs (bp) and initiates an immune response [46, 47]. Short dsRNAs less than 30 bp in length can bind the PKR monomer but prevent its activation [48, 49]. Most cellular circRNAs have one or more intra-molecular imperfect RNA duplexes that range from 16 to 26 bp, which enable them to interact with PKR to suppress its activity. Upon viral infection or poly(I:C) treatment, endonuclease RNase L degrades cellular circRNAs to free PKR for innate immune response [45]. These results reveal that cellular circRNAs, as a class, function to inhibit aberrant PKR activity in the absence of viral infection.
Figure 1 |. CircRNAs in Immune Responses.
Under normal conditions, the immune factors NF90/NF110 promote the formation of circRNAs by stabilizing the intronic complementary sequences. After back-splicing, endogenous circRNAs are marked by the m6A modification and exported to the cytoplasm where they can be recognized as self to bind immune regulators NF90/NF110 and PKR. Upon viral infection, nuclear NF90/NF110 translocate to the cytoplasm, which decreases circRNA production. Thus, fewer cytoplasmic circRNAs are available to interact with NF90/NF110 and PKR. Meanwhile, double-stranded viral RNAs activate 2’,5’-oligoadenylate synthetase (OAS) to produce 2’−5’-oligoadenylates (2–5A) and stimulate endonuclease RNase L to degrade endogenous circRNAs. Following the release of PKR, the receptor then binds to viral RNA and inhibits virus replication. Exogenous circRNAs without m6A modification are recognized by immune receptor RIG-I to trigger an immune response to suppress viral infection.
Immune Responses to Endogenous and Exogenous CircRNA
The immune factors NF90/NF110 promote circRNA biogenesis by stabilizing the intronic RNA pairs in the nucleus. Upon viral infection, NF90/NF110 translocate to the cytoplasm, which suppress the production of nascent circRNAs [50]. Cytoplasmic NF90/NF110 bind to viral mRNAs in antiviral defense to inhibit viral replication (Figure 1), connecting circRNA production with the activation of the innate immune response. In contrast to endogenous circRNAs functioning as suppressors of immune stimulation, exogenous circRNAs elicit potent immune signaling [51]. Nucleic acid sensor retinoic acid-inducible gene I (RIG-I) sense in vitro synthesized circRNA to trigger a strong antiviral immune response (Figure 1). The activation is independent of the 5’ triphosphate, double-stranded RNA structure, or the sequence identity of the exogenous circRNA. The immune response stimulated by delivery of exogenous circRNA to HeLa cells protects the cells from subsequent viral infection [51]. In vivo vaccination of C57BL/6 mice with circRNA and ovalbumin (OVA) promotes OVA-specific CD8+ T cell and antibody responses [52]. To probe the duration of immune protection following vaccination, B16-melanoma cells that express OVA were injected in circRNA-vaccinated mice. Mice with prior exposure to exogenous circRNA had decreased tumor growth and increased survival. These studies demonstrate that exogenous circRNAs can be used as vaccine adjuvants to boost the host’s innate and adaptive immune responses, and thus provide maximum protection against tumors or pathogens [52].
Other investigations have proposed that the immune response generated from the in vitro transcribed circRNAs is due to 5’-triphosphate linear RNA impurities in the delivered sample [53]. The 5’-triphosphate is an established RIG-I ligand [54] and multiple groups have shown that immune activation by 5’-triphosphate linear RNA is more potent than that induced by exogenous circRNA [51, 53]. In the case with the resulting immune response from circRNAs, distinctions between the studies in the isolation of exogenous circRNA by treatment with exonuclease RNase R and alkaline phosphatase may explain the contrasting findings. Gel extraction and purification by high-performance liquid chromatography are complementary approaches to RNase R treatment for isolation of exogenous circRNA produced by in vitro transcription. While one study showed that additional purification steps decreased the immune response from exogenous circRNA transfection [53], another group revealed that there was no change in RIG-I activation resulting from further circRNAs purification [52]. The varying experimental approaches, assays, and analyses (summarized in Table 1) may have resulted in the differing conclusions. Overall, these studies demonstrate that circRNA immunogenicity is variable and can be actively modulated, inviting further research to more systematically and comprehensively classify the properties and situations that determine the immunogenicity of individual circRNAs.
Table 1.
Comparison of Two Investigations into the Immunogenicity of Exogenous CircRNAs
| Experiment | “N6-Methyladenosine Modification Controls Circular RNA Immunity” [52] | “RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo” [53] |
|---|---|---|
| Type of linear RNA compared to circRNA | 5’ hydroxyl linear RNA | 5’ triphosphate linear RNA |
| RNase R treatment of in vitro transcribed circRNA | 2–3 hours | 15 min |
| Phosphatase treatment | 2 hours | 15 min |
| Experiment readout | Measure immune gene expression | Measure cytokine and chemokine in culture medium |
| RNA transfection | Forward, used Lipofectamine 2000 | Reverse, used Lipofectamine MessengerMax |
| RNA transfection efficiency | Measured levels of RNA delivered for normalization | Did not measure levels of RNA delivered |
Additional investigations have revealed that cells distinguish between endogenous and exogenous circRNAs based on the m6A modification status of these circRNAs [52]. The m6A modification is introduced to RNA co-transcriptionally under the guide of gene-body H3K36me3 histone modification [55]. Most endogenous circRNAs contain constitutive m6A modifications to avoid eliciting an immune response, but the methylation patterns are distinct from their cognate linear RNAs [52, 56]. Moreover, while m6A-modified linear RNAs do not trigger RIG-I-mediated innate immune signaling [57], the presence of m6A modification on circRNAs is insufficient to fully mask their immunogenicity [52]. The m6A reader protein YTH domain-containing family protein 2 (YTHDF2) is also required to completely suppress the innate immune response from circRNAs. YTHDF2 is known to recruit m6A-modified mRNAs to processing bodies for degradation and mediate rapid degradation of YTHDF2-bound circRNAs by the RNase P/MRP complex [56, 58, 59]. The mechanism of how YTHDF2 binding masks endogenous circRNA immunogenicity to evade RIG-I sensing requires further research.
CircRNAs in Immune Cells
In addition to playing a role in regulating antiviral response in non-immune cells, circRNAs may also control the differentiation and function of immune cells. Thousands of circRNAs were identified in primary human leukocytes: naïve B cells, hematopoietic stem cells (HSCs), and neutrophils [60]. The comprehensive analysis of human hematopoietic progenitors and differentiated lymphoid and myeloid cells showed that circRNA expression is widespread and produced in a manner specific to the cell-type and developmental stage [61]. Moreover, the global expression of circRNAs increases upon cell maturation. Enucleated blood cells, such as platelets and erythrocytes, tend to accumulate more circRNAs [61]. However, the functional potential of circRNAs in these immune cells and precursor stem cells remains elusive. Intriguingly, a recent study reported that a highly-expressed constitutive circRNA—circular RNA antagonist for cGAS (cia-cGAS)—located in the nucleus of long-term HSCs (LT-HSCs), could bind nuclear cGAS to prevent its sensing of self DNA and thus protect dormant LT-HSCs from DNA sensor cGAS-mediated exhaustion [62]. This demonstrates that an individual circRNA may function to maintain immune precursor cells homeostasis by regulating the balance between self-renewal and differentiation of HSCs.
In addition to having roles in HSCs, circRNAs may also contribute to the activation and function of macrophages. Macrophages are important cells of the immune system that engulf and digest pathogens, foreign materials, and dead cells. This process plays an essential role in tissue homeostasis and inflammation [63]. A study identified alterations in the circRNAs expression profile during macrophage polarization by isolating bone marrow-derived macrophages from BALB/c mice, stimulating them with lipopolysaccharide (LPS)/interferon-gamma (IFN-γ) or interleukin-4 (IL-4), and analyzing the circRNA expression by microarrays. The authors reported that hundreds of circRNAs are differentially expressed by macrophages under two different polarization conditions, suggesting that circRNAs may play important roles in regulating or maintaining macrophage polarization [64]. Moreover, during LPS stimulation to activate macrophages, circRasGEF1B controls the expression of intercellular adhesion molecule 1 (ICAM-1) to positively regulate macrophage activation [65]. ICAM-1 recruits leukocytes to inflamed sites [66] and promotes cell-to-cell interactions during antigen presentation [67]. Additionally, circZC3H4 regulates macrophage activation in response to silicon dioxide (SiO2) exposure [68]. These investigations underscore that circRNAs play a critical role in fine-tuning the activation and function of macrophages. In addition to macrophages, circRNAs have been examined in many other immunocytes. For example, hundreds of circRNAs have been identified as being differentially expressed in neutrophils between healthy subjects and patients with asymptomatic Moyamoya disease [69]. Comprehensive circRNA profiling has revealed that circRNA100783 is involved in chronic CD28-associated CD8+T cell aging [70]. However, many of the roles of circRNAs in immunocytes are still not known due to the lack of detailed functional studies in these cell types.
Functions and Applications of CircRNAs in Immune-Related Diseases
Several circRNAs have been associated with immune-related diseases, such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE). RA is a chronic and systemic autoimmune disease with unknown etiology [71]. By analyzing the expression of circRNAs in peripheral blood mononuclear cells (PBMCs) through high-throughput circRNA microarray or RNA-seq, multiple groups independently have identified several differentially expressed circRNAs between RA patients and healthy controls [72–75]. Several circRNAs have been further validated and proposed as potential biomarkers for RA diagnosis, although the mechanisms of these differentially expressed circRNAs require further investigation. The detailed study of circRNAs in RA may lead to a better understanding of RA pathogenesis and the development of therapeutic targets.
SLE is another chronic autoimmune disease characterized by immune complex deposits, multiple autoantibody production, and widespread organ damage driven by local inflammation [76]. Similar to the investigations in RA, numerous studies have measured the circRNA expression profiles in PBMCs or T cells from patients with SLE and healthy controls by circRNA microarray or RNA-seq [77–80]. Several circRNAs, including hsa_circ_0044235, circPTPN22, and hsa_circRNA_407176, have been found to be significantly decreased in SLE patients and proposed as potential negative biomarkers for SLE diagnosis. Given the challenges with accurate diagnosis of SLE in a timely manner, the discovery of these indicators is useful for early disease treatment. Moreover, a study has found that decreasing hsa_circ_0045272 expression in Jurkat cell lines significantly promotes early apoptosis in vitro, providing a biological pathway for their observation that hsa_circ_0045272 expression is depleted in SLE T cells [77]. Of note, the discovery that many cellular circRNAs form short intra-molecular duplex structures to inhibit PKR (Figure 1) demonstrated the mechanisms linking aberrant circRNA levels to the pathogenesis of SLE [45]. Previous findings have established that the overexpression of PKR in activated T cells is associated with SLE by showing that the number and expression level of circRNAs is globally reduced in different cell types from SLE patients [81]. Lower levels of circRNAs in SLE patients enable PKR to be activated instead of interacting with cellular circRNA [45]. This study identifies circRNAs as a protein sponge for PKR and uncovers an unexpected pathophysiological connection between circRNA function and SLE.
Human Oncogenic DNA Viruses Encode CircRNAs
Infection with oncogenic DNA viruses Epstein-Barr virus (EBV), Kaposi’s sarcoma virus (KSHV), and human papillomavirus (HPV) has been associated with the development of a variety of human malignancies and accounts for approximately 13% of human cancers [82, 83]. These DNA viruses have overlapping open reading frames with few introns, allowing the virus to maximize its repertoire of encoded proteins from a compact genome [84]. During infection, DNA viruses deliver their genomes into the host’s nucleus for transcription and splicing of viral genes using cellular machinery [85]. Since the host splicing complex processes bicistronic and polycistronic RNAs, the spliceosome has the potential to back-splice viral RNAs to generate circRNAs. To find viral and host circRNAs in infected samples, several groups applied RNase R treatment and next-generation sequencing. They identified multiple circRNAs from HPV, as well as herpesviruses, EBV, and KSHV (summarized in Figure 2 and Table 2) [30, 86–89]. In addition to the double-stranded DNA viruses, single-stranded RNA viruses and retro-transcribing viruses likely also encode circRNAs. A systematic survey of RNA sequencing data from the infection of 23 viral species was conducted, which detected numerous virally-encoded circRNAs [90]. The identification of viral circRNAs has expanded the transcriptome repertoires of these viruses in both lytic and latent stages. Researchers have probed the cellular localization, viral life cycle stage, and functions of the abundant viral circRNAs, but many questions regarding their roles in viral biology, the associated malignancies, and the molecular signals that promote their formation remain to be answered.
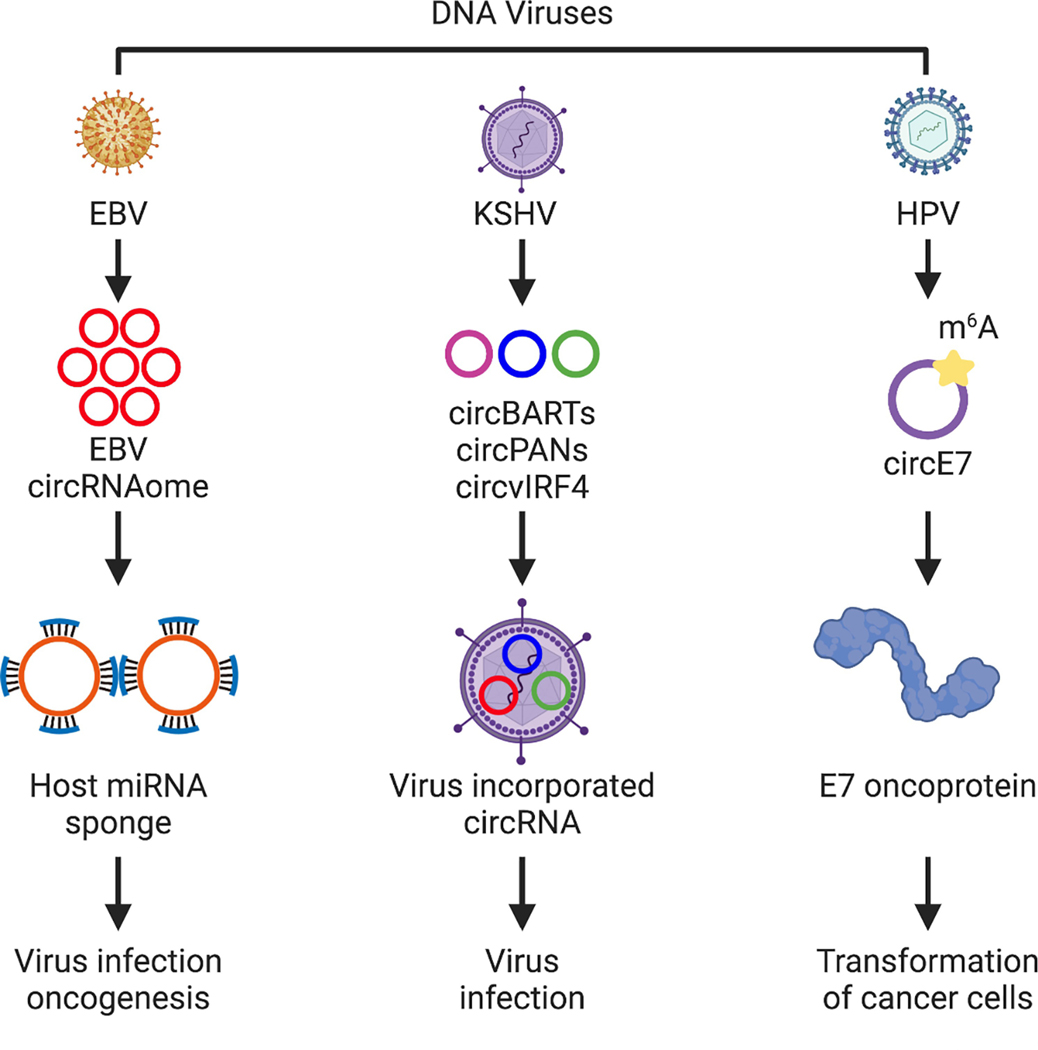
Figure 2 |. Potential Functions of CircRNAs Encoded by DNA Viruses.
DNA viruses, EBV, KSHV, and HPV, encode circRNAs. EBV contains multiple circRNAs from several gene loci of the EBV genome. The EBV circRNAome may function as sponges for host miRNAs to regulate EBV infection and oncogenesis. KSHV generates several circRNAs that can be incorporated into virus particles, which may play a role in virus infection. CircE7 derived from HPV can be translated in a cap-independent manner to produce an oncoprotein that plays an important role in the transformed growth of cancer cells.
Table 2.
CircRNAs encoded by DNA Viruses and their Functions.
| Virus | CircRNA | Sequencing method | Function | Ref |
|---|---|---|---|---|
| EBV | circRPMS1 | rRNA depletion RNA | miRNA sponge for human and | [86] |
| sequencing | EBV-encoded miRNAs | |||
| circBART_1 | RNase R-resistant | Unknown | [88] | |
| circBART_2 | RNA-seq | |||
| circBHLF1 | ||||
| circLMP2 | ||||
| circEBNA_W1_C1 | RNase R-resistant | Function as host miRNA sponge | [89, 92] | |
| circEBNA_W2_C2 | RNA-seq | regulating virus infection, cell cycle, | ||
| circEBNA_U | and oncogenesis | |||
| circRPMS1 multiple variants | ||||
| circLMP2 multiple variants | ||||
| circBHLF1 | ||||
| circBGLF1_BHLF1 | ||||
| circBDLF1_BHLF1 | ||||
| circBXLF1_BHLF1 | ||||
| circBILF2_BHLF1 | ||||
| circBHRF1 | ||||
| KSHV | circvIRF4 | RNase R-resistant | Incorporation into viral particles for | [88, 94] |
| circPAN/K7.3 multiple | RNA-seq | preformed delivery, suggesting a | ||
| variants | potential function in early infection | |||
| Kcirc3 (ORF4, ORF6) | RNase R-resistant | Located within ORFs of viral lytic | [87] | |
| Kcirc29 (K7, PAN) | RNA-seq | genes, upregulated upon induction | ||
| Kcirc38 (ORF21, ORF22) | of the lytic cycle, and alter cell | |||
| Kcirc54 (ORF34) | growth | |||
| Kcirc55 (ORF34, 35, 36) | ||||
| Kcirc57 (ORF34, 35, 36, 37) | ||||
| Kcirc97 (ORF60, 61, 62) | ||||
| HPV | circE7 | Circular RNA | circE7 can be translated through | [30] |
| detection from NCBI | cap-independent mechanisms, | |||
| and TCGA RNA-Seq | which is essential for the | |||
| data | transformed growth of CaSki | |||
| cervical carcinoma cells | ||||
EBV-Encoded CircRNAs
EBV is one of the nine types of herpesvirus known to infect humans and is causally associated with B cell lymphoma, gastric cancer, and nasopharyngeal carcinoma [91]. Analysis of RNase R-treated viral samples has identified several viral circRNAs derived from different loci of EBV genome, including circBART [88] and circRPMS1 [89]. CircBART has two different isoforms: exonic or exon-intron, both of which were detected at relatively high levels in EBV-positive posttransplant lymphoproliferative diseases and in the latent B cell lymphoma line BC1 [88]. Cellular fractionation assays have shown that the exon-only circBART localizes to the cytoplasm, whereas the intron-retaining circBARTs remain in the nucleus [88], which are consistent with the characteristics of host circRNAs [3, 5, 60]. Another EBV-encoded circRNA, circRPMS1, which is back-spliced from the latency long non-coding RPMS1 locus, also has several alternative back-splicing isoforms with different expression levels. Certain circRPMS1 isoforms, such as circRPMS1_E4_E3a, were detected in both EBV latency and reactivation conditions [89]. The computational pipelines RegRNA2.0 and RNAhybrid have predicted that circRPMS1 is a target of eight viral and 47 human miRNAs, suggesting that circRPMS1 can serve as a miRNA sponge to regulate the expression of other genes [86]. In addition, EBV infection in the B cell line LCLd3 decreases the expression of several host miRNAs that have the potential to interact with EBV circRNAs [92]. The targets of these miRNAs that are potentially inhibited by EBV circRNAs are mainly involved in cell cycle and virus infection regulation, which suggest that EBV-encoded circRNAs may regulate EBV infection and oncogenesis through sponging host miRNAs [92].
EBV has several latency phases, which are defined by the viral proteins and RNAs produced during those periods. Many studies have demonstrated that the EBV latency macromolecules contribute to cancer development [91]. Common circRNAs have been found across EBV latency types, which suggests that these viral circRNAs may play fundamental roles during latent infection and contribute to virus-induced oncogenesis [89]. While circRNAs from the EBNA latency locus likely do not sponge miRNAs since they do not have viral or human miRNA seed sites, some of the circRNAs expressed from this locus may be translated. For example, circEBNA_U contains a single six-amino-acid open reading frame (ORF). The EBNA U exon has a previously-identified IRES that overlays this short ORF, so circularization may regulate translation of the downstream spliced EBNA reading frames. In addition, two EBV-encoded circRNAs have been identified in EBV-positive stomach cancer biopsies, which provide evidence for circRNA expression in vivo and present the potential for their use in cancer diagnostics [89]. While multiple EBV-encoded circRNAs have been identified from virally-infected cells and tissues, the exact pathophysiological functions of these circRNAs remain elusive. Future studies are warranted to elucidate the roles of viral circRNAs in EBV biology and associated diseases.
KSHV-Encoded CircRNAs
KSHV, also known as human herpesvirus 8, can cause multiple disorders, including Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease [93]. The KSHV genome expresses mRNAs and noncoding RNAs during both latent and lytic stages. Recently, KSHV has been shown to encode and express multiple circRNAs [82, 87]. Most of the circRNAs are spliced from an abundant transcript in an early lytic phase, the polyadenylated nuclear (PAN) RNA [82]. The abundance of circPANs is comparable to their counterpart linear RNA levels, suggesting that these circRNAs may play important roles for the viral life cycle. Another KSHV-encoded circRNA, circvIRF4, is more abundant than its corresponding linear transcript in the primary effusion lymphoma lines BCP-1 and BBG1. In other B cell lines, the expression of circvIRF4 is minimally induced and less abundant than the parental linear vIRF4 mRNA [94]. These results are consistent with previous findings that the majority of circRNAs in eukaryotes are less abundant than their corresponding linear transcripts [95]. KSHV circPANs can be detected in both the nucleus and cytosol; while the fully processed form of circvIRF4 is exported to the cytoplasm, the intron-retaining form is retained in nucleus. Even though the exonic circIRF4 and circPANs localize to the cytoplasm, none of them have been found in polysome fractions, which suggests that they are unlikely to be translated [94]. Moreover, all KSHV circRNAs are incorporated into KSHV virions [94], but whether virion-packaged KSHV circRNAs have a functional role in facilitating KSHV infection or modulating innate immune response during KSHV infection requires additional investigation.
HPV-Encoded CircRNAs
Over a hundred different subtypes of HPV have been discovered; fourteen of them are classified as “high-risk” types. Using ten HPV subtypes as reference genomes for the analysis of RNA-seq datasets from HPV-infected tissues, a study identified transcripts from HPV16 and HPV35 strains containing back-splice junctions [30]. The circRNAs encoded by HPV16 have similar expression levels to the spliced linear mRNAs and the most abundant circRNA encodes the E7 open reading frame. The 472-nucleotide circE7 predominantly resides in the cytoplasm and is translated to produce the E7 oncoprotein. Since the RNA modification m6A has been implicated in promoting cap-independent translation of circRNAs [28, 38], the authors explored the possibility that m6A enables the translation of circE7. Indeed, the studies have found that circE7 contains m6A modifications, which are essential for E7 protein expression. The protein produced by circE7 translation controls transformation in CaSki cervical carcinoma cells and in tumor xenografts [30]. This research suggests that circRNAs derived from HPV are functional and may contribute to HPV oncogenesis.
Viral Infection Changes the Host-Encoded CircRNAs Transcriptome
The expression landscape of circRNAs in host cells changes upon viral infection. Host circRNAs induced by foreign attack may have antiviral or immune functions to regulate viral pathogenesis [86–88]. Following KSHV infection, hundreds of differentially expressed human circRNAs have been identified in both primary human umbilical vein endothelial cells (HUVECs) and the infected B cell line MC116 [87]. However, only 2% of the upregulated human circRNAs overlap in both HUVECs and MC116 cells, which suggests that expression of host circRNAs in response to KSHV infection displays a cell-type specific pattern. Due to their low cellular abundance, the functional potential of individual circRNAs remains to be seen. High expression of circRNAs is likely necessary for its functionality. The human circRNA hsa_circ_0001400 is the most abundant circRNA induced following viral infection. The increase in hsa_circ_0001400 significantly down-regulates levels of the viral genes LANA and RTA, whereas the entry of viral genome into cells remains unaffected. hsa_circ_0001400 also increases the expression of TNF-α coding gene upon infection, but other interferon-stimulated genes and pro-apoptotic genes remain unchanged [87]. This human circRNA may stimulate host defense pathways to enhance the antiviral response. Human circRNAs may also regulate viral transcription by altering chromatin structure, which is one putative mechanism for how hsa_circ_0001400 inhibits expression of both KSHV genes: RTA and LANA. Due to their ability to form base-pairs with other nucleic acids, host encoded circRNAs can target viral factors in a sequence-specific manner to produce a more directed immune response compared with pattern recognition receptors. Some endogenous circRNAs can also convey antiviral information to surrounding cells by extracellular vesicle-mediated export [87].
In addition to providing antiviral functions, host-derived circRNAs may also have roles as dependency factors [96]. Infection by hepatitis C virus, a single-stranded RNA virus in the Flaviviridae family, changes the expression of several circRNAs in liver cells. Intriguingly, the increase in circPSD3 expression upon hepatitis C virus infection has a pronounced effect on viral RNA abundances in both hepatitis C virus- and Dengue virus-infected cells. CircPSD3 inhibits the cellular nonsense-mediated decay pathway, which is a powerful antiviral response in virus-infected cells [96]. Previous studies have also documented that the immune factors NF90/NF110 are involved in regulating host circRNAs expression in response to RNA virus infection [50]. Additional investigation is needed to gain a comprehensive understanding of how viral infection alters host circRNAs expression.
Concluding Remarks
RNAs such as mRNAs, rRNAs, and transfer RNAs are grouped by their functions, but circRNAs are classified by their topology—the lack of 5’ and 3’ ends. CircRNAs are a diverse class of RNAs with varying expression levels, cellular localization, and biological functions. While the identities and structures of circRNAs are increasingly revealed, their mechanisms, regulation, and cellular roles are still relatively unknown and subjects of ongoing investigation. Whether the main function of circRNAs is to increase transcriptome diversity or to actively interact with cellular components remains to be seen.
Future research should include deeper investigations to reveal the mechanisms of circRNA function and applications of circRNAs to improve health (see Outstanding Questions). Studies should also determine more comprehensively the RNA modifications in addition to m6A that are found on circRNAs, and probe how the post-transcriptional modifications control the functions and properties of circRNAs. Investigations to reveal whether other viruses in addition to EBV, KSHV, and HPV encode circRNAs should be conducted to better understand the viral life cycle and host immune response. These are challenging tasks given the low expression levels of most circRNAs and the genetic tools currently available to solely perturb the circRNA under investigation without disrupting its counterpart linear RNA. Thus, technology development to better enrich, annotate, and study circRNAs present in a given sample will be greatly useful. Finally, the ability to use circRNAs as biomarkers or novel therapeutic targets is highly attractive given their unique characteristics: high stability, protein encoding, and ability to induce immune responses. We anticipate that in the coming years, many studies will be focused on these areas.
Outstanding Questions:
What biological roles do circRNAs have in the development, progression, and treatment of autoimmune diseases, such as rheumatoid arthritis and systemic lupus erythematosus?
What RNA modifications, in addition to m6A, are found on circRNAs? How do the post-transcriptional modifications control the functions and properties of circRNAs?
What other viruses encode circRNAs? What are the functions of the virally-encoded circRNAs?
Can circRNAs be used as biomarkers for disease diagnosis or progression?
How can circRNAs be engineered into novel therapies?
What new technologies and analyses can identify circRNAs in a comprehensive manner? What tools enable the investigation of circRNAs?
Highlights:
Circular RNAs (circRNAs) are a recently identified class of ubiquitously expressed RNAs found in eukaryotes, archaea, and viruses.
DNA viruses Kaposi’s sarcoma-associated herpesvirus, Epstein–Barr virus, and human papillomavirus encode circRNAs. Some circRNAs possess pro-viral functions as miRNA sponges and templates for translation.
CircRNAs are differentially expressed in immune cells and in autoimmune diseases, including rheumatoid arthritis and systemic lupus erythematosus.
Emerging data suggest that circRNAs have biological roles in the immune system as transcriptional regulators, protein sponges, and templates for translation.
Acknowledgements
We thank our lab for helpful discussion and Dr. Yongsheng Shi, Dr. Jie Jane Chen, and Dr. Nancy Ruddle for critical reading of the manuscript. This work is supported by the National Institute of Health (5K12CA215110) and the American Cancer Society (IRG17–172-57) to Y.G.C, and the Trudeau Fellowship to L.Y.
Glossary
- Back-splicing
A spliceosome-mediated non-canonical process that joins a downstream splice donor to an upstream splice acceptor to produce a covalently closed circular product: circRNA
- Internal ribosome entry site (IRES)
An RNA element that recruits the ribosome to initiate translation of the RNA in a cap-independent manner
- MicroRNA (miRNA) sponge
A type of RNA transcript containing multiple complementary binding sites to a miRNA that prevents the miRNA from interacting with their target mRNAs
- N6-methyladenosine (m6A)
An RNA modification that is methylated on the adenine base at the nitrogen-6 position; the most prevalent internal modification on eukaryotic RNA that is involved in many important biological processes
- RNase R
A 3’ to 5’ exoribonuclease originally identified in E. coli that efficiently degrades nearly all linear RNAs but not circRNAs. RNase R is typically used to confirm the circularity of RNA and to enrich circRNAs for investigation
- Spliceosome
A multi-subunit ribonucleoprotein complex that catalyzes the removal of introns from transcribed pre-mRNAs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cocquerelle C et al. (1993) Mis-splicing yields circular RNA molecules. FASEB J 7 (1), 155–60. [DOI] [PubMed] [Google Scholar]
- 2.Li X et al. (2018) The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 71 (3), 428–442. [DOI] [PubMed] [Google Scholar]
- 3.Jeck WR et al. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 (2), 141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X et al. (2019) A unified mechanism for intron and exon definition and back-splicing. Nature 573 (7774), 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z et al. (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22 (3), 256–64. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y et al. (2013) Circular intronic long noncoding RNAs. Mol Cell 51 (6), 792–806. [DOI] [PubMed] [Google Scholar]
- 7.Liang D and Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28 (20), 2233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XO et al. (2014) Complementary sequence-mediated exon circularization. Cell 159 (1), 134–147. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov A et al. (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10 (2), 170–7. [DOI] [PubMed] [Google Scholar]
- 10.Conn SJ et al. (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160 (6), 1125–34. [DOI] [PubMed] [Google Scholar]
- 11.Ashwal-Fluss R et al. (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56 (1), 55–66. [DOI] [PubMed] [Google Scholar]
- 12.Guo JU et al. (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15 (7), 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger HL et al. (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 73 (11), 3852–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kos A et al. (1986) The hepatitis delta (delta) virus possesses a circular RNA. Nature 323 (6088), 558–60. [DOI] [PubMed] [Google Scholar]
- 15.Capel B et al. (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73 (5), 1019–30. [DOI] [PubMed] [Google Scholar]
- 16.Houseley JM et al. (2006) Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered 97 (3), 253–60. [DOI] [PubMed] [Google Scholar]
- 17.Salzman J et al. (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9 (9), e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 (7441), 333–8. [DOI] [PubMed] [Google Scholar]
- 19.Chen L et al. (2020) The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybak-Wolf A et al. (2015) Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 58 (5), 870–85. [DOI] [PubMed] [Google Scholar]
- 21.Hansen TB et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495 (7441), 384–8. [DOI] [PubMed] [Google Scholar]
- 22.Piwecka M et al. (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357 (6357). [DOI] [PubMed] [Google Scholar]
- 23.Kleaveland B et al. (2018) A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 174 (2), 350–362 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen TB et al. (2011) miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30 (21), 4414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You X et al. (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18 (4), 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legnini I et al. (2017) Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 66 (1), 22–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pamudurti NR et al. (2017) Translation of CircRNAs. Mol Cell 66 (1), 9–21 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y et al. (2017) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27 (5), 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M et al. (2018) A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9 (1), 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J et al. (2019) Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun 10 (1), 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M et al. (2018) A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37 (13), 1805–1814. [DOI] [PubMed] [Google Scholar]
- 32.van Heesch S et al. (2019) The Translational Landscape of the Human Heart. Cell 178 (1), 242–260 e29. [DOI] [PubMed] [Google Scholar]
- 33.Hellen CU and Sarnow P (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15 (13), 1593–612. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert WV et al. (2007) Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317 (5842), 1224–7. [DOI] [PubMed] [Google Scholar]
- 35.Weingarten-Gabbay S et al. (2016) Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351 (6270). [DOI] [PubMed] [Google Scholar]
- 36.Meyer KD et al. (2015) 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163 (4), 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J et al. (2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526 (7574), 591–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Timoteo G et al. (2020) Modulation of circRNA Metabolism by m(6)A Modification. Cell Rep 31 (6), 107641. [DOI] [PubMed] [Google Scholar]
- 39.Fan X et al. (2019) Pervasive translation of circular RNAs driven by short IRES-like elements. 473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CY and Sarnow P (1995) Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268 (5209), 415–7. [DOI] [PubMed] [Google Scholar]
- 41.Coots RA et al. (2017) m(6)A Facilitates eIF4F-Independent mRNA Translation. Mol Cell 68 (3), 504–514 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu S et al. (2018) The emerging functions and roles of circular RNAs in cancer. Cancer Lett 414, 301–309. [DOI] [PubMed] [Google Scholar]
- 43.Lu S et al. (2019) Current status and potential role of circular RNAs in neurological disorders. J Neurochem 150 (3), 237–248. [DOI] [PubMed] [Google Scholar]
- 44.Altesha MA et al. (2019) Circular RNA in cardiovascular disease. J Cell Physiol 234 (5), 5588–5600. [DOI] [PubMed] [Google Scholar]
- 45.Liu CX et al. (2019) Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 177 (4), 865–880 e21. [DOI] [PubMed] [Google Scholar]
- 46.Schlee M and Hartmann G (2016) Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol 16 (9), 566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia MA et al. (2007) The dsRNA protein kinase PKR: virus and cell control. Biochimie 89 (6–7), 799–811. [DOI] [PubMed] [Google Scholar]
- 48.Nallagatla SR et al. (2011) Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol 21 (1), 119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng X and Bevilacqua PC (2004) Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10 (12), 1934–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X et al. (2017) Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell 67 (2), 214–227 e7. [DOI] [PubMed] [Google Scholar]
- 51.Chen YG et al. (2017) Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell 67 (2), 228–238 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen YG et al. (2019) N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell 76 (1), 96–109 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wesselhoeft RA et al. (2019) RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Mol Cell 74 (3), 508–520 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hornung V et al. (2006) 5’-Triphosphate RNA is the ligand for RIG-I. Science 314 (5801), 994–7. [DOI] [PubMed] [Google Scholar]
- 55.Huang H et al. (2019) Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567 (7748), 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou C et al. (2017) Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep 20 (9), 2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durbin AF et al. (2016) RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 7 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park OH et al. (2019) Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell 74 (3), 494–507 e8. [DOI] [PubMed] [Google Scholar]
- 59.Wang X et al. (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salzman J et al. (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7 (2), e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicolet BP et al. (2018) Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res 46 (16), 8168–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia P et al. (2018) A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 48 (4), 688–701 e7. [DOI] [PubMed] [Google Scholar]
- 63.Ginhoux F and Jung S (2014) Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 14 (6), 392–404. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y et al. (2017) Microarray analysis of circular RNA expression patterns in polarized macrophages. Int J Mol Med 39 (2), 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng WL et al. (2016) Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol 13 (9), 861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Long EO (2011) ICAM-1: getting a grip on leukocyte adhesion. J Immunol 186 (9), 5021–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lebedeva T et al. (2005) ICAM-1 co-stimulates target cells to facilitate antigen presentation. Curr Opin Immunol 17 (3), 251–8. [DOI] [PubMed] [Google Scholar]
- 68.Yang X et al. (2018) Silica-induced initiation of circular ZC3H4 RNA/ZC3H4 pathway promotes the pulmonary macrophage activation. FASEB J 32 (6), 3264–3277. [DOI] [PubMed] [Google Scholar]
- 69.Ma Q et al. (2019) Circular RNA profiling of neutrophil transcriptome provides insights into asymptomatic Moyamoya disease. Brain Res 1719, 104–112. [DOI] [PubMed] [Google Scholar]
- 70.Wang YH et al. (2015) Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing 12, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coutant F and Miossec P (2020) Evolving concepts of the pathogenesis of rheumatoid arthritis with focus on the early and late stages. Curr Opin Rheumatol 32 (1), 57–63. [DOI] [PubMed] [Google Scholar]
- 72.Ouyang Q et al. (2017) Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Rheumatoid Arthritis Patients. Cell Physiol Biochem 42 (2), 651–659. [DOI] [PubMed] [Google Scholar]
- 73.Zheng F et al. (2017) Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep 16 (6), 8029–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang X et al. (2019) Aberrant dysregulated circular RNAs in the peripheral blood mononuclear cells of patients with rheumatoid arthritis revealed by RNA sequencing: novel diagnostic markers for RA. Scand J Clin Lab Invest 79 (8), 551–559. [DOI] [PubMed] [Google Scholar]
- 75.Wen J et al. (2020) RNA-seq reveals the circular RNA and miRNA expression profile of peripheral blood mononuclear cells in patients with rheumatoid arthritis. Biosci Rep 40 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D’Cruz DP et al. (2007) Systemic lupus erythematosus. Lancet 369 (9561), 587–96. [DOI] [PubMed] [Google Scholar]
- 77.Li LJ et al. (2018) Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology 155 (1), 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo G et al. (2019) Hsa_circ_0000479 as a Novel Diagnostic Biomarker of Systemic Lupus Erythematosus. Front Immunol 10, 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo Q et al. (2019) Identification of circular RNAs hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus. Int J Mol Med 44 (4), 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miao Q et al. (2019) RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus 28 (4), 520–528. [DOI] [PubMed] [Google Scholar]
- 81.Grolleau A et al. (2000) Impaired translational response and increased protein kinase PKR expression in T cells from lupus patients. J Clin Invest 106 (12), 1561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Damania B (2007) DNA tumor viruses and human cancer. Trends Microbiol 15 (1), 38–44. [DOI] [PubMed] [Google Scholar]
- 83.de Martel C et al. (2020) Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 8 (2), e180–e190. [DOI] [PubMed] [Google Scholar]
- 84.Schlub TE and Holmes EC (2020) Properties and abundance of overlapping genes in viruses. Virus Evol 6 (1), veaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ajiro M and Zheng ZM (2014) Oncogenes and RNA splicing of human tumor viruses. Emerg Microbes Infect 3 (9), e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang JT et al. (2019) Identification of virus-encoded circular RNA. Virology 529, 144–151. [DOI] [PubMed] [Google Scholar]
- 87.Tagawa T et al. (2018) Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci U S A 115 (50), 12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toptan T et al. (2018) Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A 115 (37), E8737–E8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ungerleider N et al. (2018) The Epstein Barr virus circRNAome. PLoS Pathog 14 (8), e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai Z et al. (2020) VirusCircBase: a database of virus circular RNAs. Brief Bioinform. [DOI] [PubMed] [Google Scholar]
- 91.Shannon-Lowe C and Rickinson A (2019) The Global Landscape of EBV-Associated Tumors. Front Oncol 9, 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qiao Y et al. (2019) Epstein-Barr virus circRNAome as host miRNA sponge regulates virus infection, cell cycle, and oncogenesis. Bioengineered 10 (1), 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sunil M et al. (2010) Update on HHV-8-Associated Malignancies. Curr Infect Dis Rep 12 (2), 147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abere B et al. (2020) Kaposi’s Sarcoma-Associated Herpesvirus-Encoded circRNAs Are Expressed in Infected Tumor Tissues and Are Incorporated into Virions. mBio 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y et al. (2016) The Biogenesis of Nascent Circular RNAs. Cell Rep 15 (3), 611–624. [DOI] [PubMed] [Google Scholar]
- 96.Chen T-C et al. (2020) Host-derived Circular RNAs Display Proviral Activities in Hepatitis C Virus - Infected Cells. 2020.01.24.917971. [DOI] [PMC free article] [PubMed] [Google Scholar]