Abstract
Purpose
Nowadays Computer-Aided Diagnosis (CAD) models, particularly those based on deep learning, have been widely used to analyze histopathological images in breast cancer diagnosis. However, due to the limited availability of such images, it is always tedious to train deep learning models that require a huge amount of training data. In this paper, we propose a new deep learning-based CAD framework that can work with less amount of training data.
Methods
We use pre-trained models to extract image features that can then be used with any classifier. Our proposed features are extracted by the fusion of two different types of features (foreground and background) at two levels (whole-level and part-level). Foreground and background feature to capture information about different structures and their layout in microscopic images of breast tissues. Similarly, part-level and whole-level features capture are useful in detecting interesting regions scattered in high-resolution histopathological images at local and whole image levels. At each level, we use VGG16 models pre-trained on ImageNet and Places datasets to extract foreground and background features, respectively. All features are extracted from mid-level pooling layers of such models.
Results
We show that proposed fused features with a Support Vector Classifier (SVM) produce better classification accuracy than recent methods on BACH dataset and our approach is orders of magnitude faster than the best performing recent method (EMS-Net).
Conclusion
We believe that our method would be another alternative in the diagnosis of breast cancer because of performance and prediction time.
Keywords: Histopathological images, Breast cancer, Histology, Image classification, Deep learning, Computer-aided diagnosis
Introduction
Breast Cancer (BC) is responsible for a high mortality rate of cancer in women worldwide. According to Cancer in Australia 2018 report,1 it is predicted that the breast cancer is the most commonly diagnosed in Australia, followed by prostate cancer. In breast cancer treatment, early diagnosis is imperative to prevent further complications. Various diagnostic tests, such as physical examination, mammography, magnetic resonance image (MRI), and ultrasound, have been used for years. However, Histopathological image analysis of tissue from needle biopsy is considered as the most effective test [38]. In this test, the tissue is stained with hematoxylin-eosin (H&E) to make the structures of interest (nucleus and cytoplasm of cells) visible under the microscope and pathologist analyze the microscopic image for any abnormalities related to cancer.
Because the analysis of large and complex histopathological images by experts is time-consuming and prone to human bias [38], computer-aided diagnosis (CAD) tools based on machine learning algorithms such as classification are used for efficiency with promising accuracy [3, 17, 25, 36]. Recently, deep learning-based methods [5, 18, 21, 23, 28] and deep ensemble learning methods [7, 24, 26, 38] have been extensively used in histopathological images classification because of their superior performance over traditional methods. These methods exploit the concept of transfer learning and use fine-tuned pre-trained deep learning models to classify images or just to extract their features to use in other machine learning models. Fine-tuning pre-trained models to classify histopathological images are prone to overfitting due to the limited availability of such datasets. This could lead to counterproductive results if we do not perform proper data augmentation and hyperparameters tuning in the training step. This is because deep learning needs big data to achieve the appropriate patterns of the image.
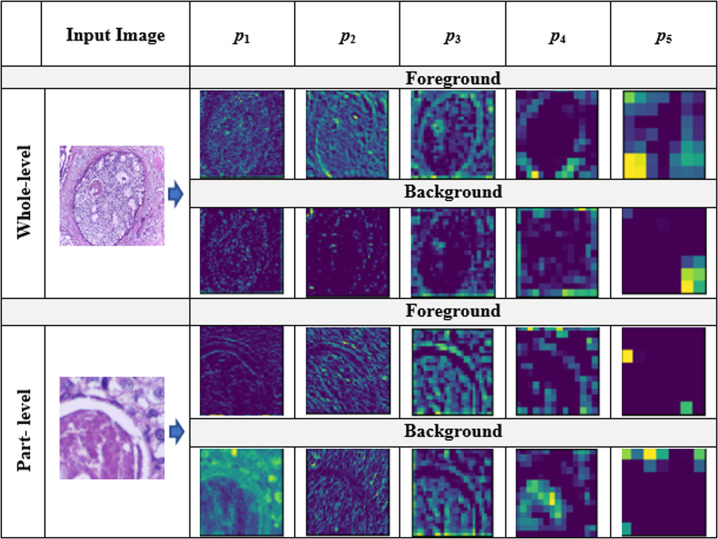
In the meantime, feature extraction should also be performed carefully depending on the nature of images because all images may not be discriminated well by the same types of deep features extracted from pre-trained deep learning models. Specifically, we need to choose an appropriate layer of a pre-trained deep learning model for feature extraction depending on the nature of images of the study. Meanwhile, identifying areas of interest (cellular regions) in high-resolution histopathological images is crucial as they offer important clues for better classification. However, it is very challenging to capture all those clues with one type of feature. Most existing methods extract features either from certain segments of an image (part-level) [37, 38] or the entire image (whole-level) [1, 2] only. In the meantime, they mostly focus on structures of interest (foreground) in an image ignoring the layout details (background). Meanwhile, they do not consider more than one type of feature. To this end, we believe that information from all these four components (foreground features at whole-level, foreground features at part-level, background features at whole-level, and background features at part-level) can be useful in making predict decisions. This could be effective because we can analyze the same image using four different techniques. We can visualize four different kinds of visual features for each pooling layer ( to ) of VGG16 in Fig. 1. Note that and impart more discriminating information than other remaining layers.
Fig. 1.
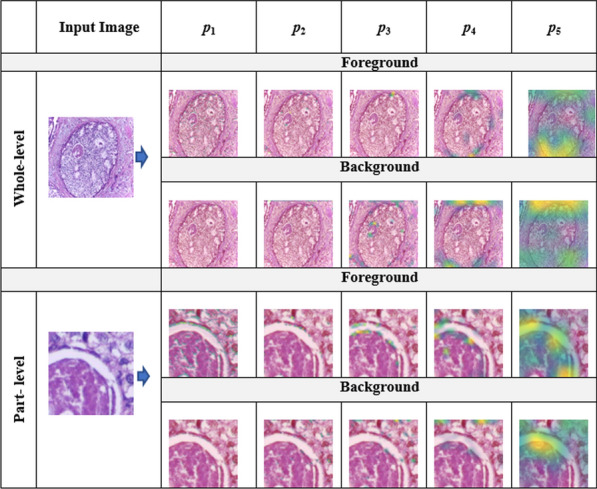
Grad-CAM visualization of five pooling layers of VGG16 for the input image at whole-level and part-level using both foreground and background features, where to represent the five pooling layers of VGG16 models
In this work, we propose to combine the four types of features—foreground and background features extracted at both part-level and whole-levels. To capture scattered areas of interest in high-resolution microscopic images, a cropping strategy is used to divide the image into multiple sub-images. We also consider the entire image just in case if some interesting regions are masked by cropping. Similarly, for both parts and whole images, we extract (i) foreground features—details of structures in the images, and (ii) background features—information about the layouts of structures in images. Specifically, foreground features capture details of structures in images such as shape, size, etc., whereas background features capture the peripheral information such as layout details present in the image. Similarly, to achieve the representative features of all sub-images at part-level, we aggregate part-level features of all sub-images using the average pooling technique, where we perform the element-wise average operation of the feature vectors of patches.
The main contributions of this paper are as follows:
Propose a new feature extraction technique to represent histopathological images by the fusion of four types of features - foreground and background features at image parts and the whole image levels. We use a simple and popular deep learning model called VGG16 [29] to extract features. VGG16 has been shown to be effective to extract features for the classification of various types of images including scene images [30], histopathological images [1, 18], etc. Also, it has only five pooling layers that makes us easier to analyse compared to other deep learning models such as GoogleNet [32], ResNet-50 [13], Inception-V3 [34], etc. We use the VGG16 model pre-trained with the ImageNet [10] dataset to extract foreground features and VGG16 model pre-trained with the Places [39] dataset to extract background features.
To the best of our knowledge, we are the first to identify the importance of background features, which are achieved from Places pre-trained VGG16 model, for histopathological image representation. We also believe that foreground and background features are complementary to each other for the better representation of histopathological images.
Evaluate the effectiveness of the fusion of the four types of features in the breast cancer histopathological image classification with the Support Vector Machine (SVM) using the BreAst Cancer Histopathology (BACH) dataset [2]. We empirically analyze the classification performance of the proposed fused features extracted from different pooling layers of the VGG16 models as they produce different feature maps of the histopathological images. We observed that the combination of features from pooling layers 3 and 4 produce the best classification result. Our CAD approach based on the proposed fusion of different types of features in the Support Vector Classifier (SVM) [9] results in better classification performance than existing CAD models for breast cancer histopathological image classification. In terms of run time, our approach is several orders of magnitude faster than the best performing state-of-the-art method called EMS-Net [38].
The rest of the paper is organized as follows. “Related works” section reviews related works of histopathological image representation and classification. “Materials and methods” section explains the materials and method for representation and classification. “Result and discussion” section explains results and discussion about our proposed method. Finally, “Conclusion” section concludes the paper with potential future directions.
Related works
In this section, we review related works of histopathological image representation and classification from two different perspectives, namely: traditional computer vision-based methods (“Traditional computer vision-based methods” section) and deep learning-based methods (“Deep learning-based methods” section).
Traditional computer vision-based methods
Inspired by the use of traditional low-level computer vision-based features in various image processing tasks, several researchers [3, 17, 25, 31, 36] have used them in histopathological image analysis. These features are extracted from the fundamental information of images such as intensity, colors, orientations, etc. Firstly, Qi et al. [25] presented a novel technique to segment the overlapping cells using traditional methods such as object localization, mean shift clustering, and contour extraction. Similarly, Vink et al. [36] presented a modified AdaBoost algorithm to make two detectors that grab different characteristics of nuclei in the nucleus detection purpose. Furthermore, Khan et al. [17] established the geodesic geometric mean of regional co-variance descriptors to represent the histopathological microscopy images. Likewise, Barker et al. [3] utilized shape, color, and texture information of local tiles of histopathological images for classification purposes. Similarly, considering the importance of multi-instance learning in this domain, Sudharshan et al. [31] proposed a multiple instance learning (MIL) framework based on traditional descriptors. Although these traditional computer vision algorithms are easy to implement, they have a limited classification performance, especially for complex images with inter-class similarity and intra-class variations such as histopathological images of breast tissues. Such methods are more suitable for specific types of images such as texture images, which are the spatial arrangement of colors or intensities.
Deep learning-based methods
Deep learning [20], a machine learning model based on large neural networks, has been revolutionized image processing tasks including classification. Many intermediate layers in a deep learning model capture different hierarchical features for images that could provide useful information to discriminate images belonging to different classes. Broadly Deep Learning Models (DLM) can be categorized generally into two types: (1) user-defined models and (2) pre-trained models. If we design our deep learning model ourselves by adding several stacked layers and their associated activations with backpropagation, it is called user-defined deep learning models. These types of modes are appropriate if we have enough data to train them, otherwise, they suffer from the overfitting problem because deep learning models learn patterns using a large number of data. Similarly, there are several deep learning models already designed by previous researchers which are pre-trained with large image collections such as ImageNet [10], Places [39], etc. The main advantage of these pre-trained models is transfer learning. Such models can be used as base models to transfer knowledge from the original domain (e.g., ImageNet) to new target domains (e.g., histopathological images) either by fine-tuning them for image classification in the target domain directly or use them to extract features to represent target domain images. There are several state-of-the-art pre-trained deep learning models available.
Because of the superior performance of deep learning models in computer vision and image processing, researchers also started using them in breast cancer diagnosis [1, 5, 7, 12, 12, 15, 18, 19, 21, 23, 24, 26, 28, 37, 38]. Initially, to leverage the benefit of deep learning with machine learning, Araujo et al. [1] combined DLM and SVM to classify breast cancer histology images. In their model, DLM was used as a feature extractor model, whereas SVM was used as a classifier. Mehta et al. [21] designed Y-Net by adding a parallel branch to U-Net [11] for the simultaneous segmentation and classification. Similarly, to take the advantage of multi-instance learning with deep learning, Campanella et al. [5] presented a concept of deep multiple instances learning (MIL) for the classification and localization of histopathological images. Histopathological images are bigger and thus, patch-level information can provide discriminating clues for the classification. To capture the patch-level information to some extent, Nazeri et al. [23] established a two-stage DLM for the patch-wise network and image-wise network to classify breast cancer-related images into four classes. Furthermore, Kwok et al. [19] classified the histopathological images using the pre-trained Incep.-Resnet-V2 [33] model, where the patches of such images were extracted for the training and classification. Roy et al. [28] proposed a patch-based convolutional neural network for the classification of histopathological images. Gandomkar et al. [12] proposed a model, called MuDeRN, based on the ImageNet pre-trained ResNet model [13] to classify histopathological images. Jiang et al. [15] designed a novel convolutional neural network to classify the breast cancer histopathological images. Yan et al. [37] utilized fine-tuned Inception-V3 [34] to extract the features from the intermediate layers and fused them using long short term memory (LSTM) for the classification of breast cancer histopathological images. To utilize the efficacy of pre-trained models without fine-tuning in achieving the prominent features for the image representation tasks, Abhinav et al. [18] utilized VGG-16 model to extract the features of the histopathological images. They extracted features from five pooling layers after performing global average pooling (GAP) operation on each of them. And then, performed classification using Random Forest [4] and SVM [35].
Furthermore, the combination of different deep learning models (DLMs) helps to leverage different types of information in images. Some of the recent works have used such ensemble learning. Firstly, Makarchuk et al. [24] extracted features of the microscopy patches using ResNet34 [13] and DenseNet [14] and trained using XGBoost classifier [6] for the classification purposes. This helps to learn different discriminating information from these two deep learning models. Similarly, Rakhlin et al. [26] also extracted features using several DLMs and classified using gradient boosted trees algorithm [16]. Similarly, Chennamsetty et al. [7] also designed an ensemble of three DLMs, each of which was trained on different configurations that won the BACH challenge [2]. Recently, Yang et al. [38] presented an ensemble model, called EMS-Net, of DLMs using multi-scale features and fine-tuning of several pre-trained models. Their model requires labor-intensive fine-tuning tasks on various pre-trained models to obtain the optimal set of pre-trained models.
In summary, the existing deep learning methods used in the classification of breast cancer histopathological images have two limitations. First, existing methods used as feature extractors extract features based on either whole-level or part-level only, which have a limited classification ability. This is because such methods are not able to capture interesting semantic regions over high-resolution images. Second, no matter whether the model is standalone or ensemble, it suffers from overfitting and underfitting because of limited data. Moreover, the fine-tuning tasks are time-consuming on such deep learning models. In breast cancer diagnosis, there is a necessity to work on a limited amount of data without compromising its classification accuracy.
Materials and methods
In this section, we explain the materials to be used in our method. Also, we present the step-wise procedure of our method.
Dataset
The ICIAR (International Conference on Image Analysis and Recognition) 2018 BreAst Cancer Histopathology (BACH) dataset [2] is used in our experiment. This dataset contains 400 labeled H&E stained histopathological images of breast tissues with a size of pixels. As we are using a patch of size and stride in patch extraction (“Patch extraction” section) as done by Yang et al. [38], each image results in 54 sub-images (i.e., ) for part-level features extraction. The dataset contains four classes as normal, benign, in situ, and invasive, where each category contains 100 images. We prepared 5 sets of random train/test split in the ratio of 4:1 per class and report the average accuracy. This provides a total of 320 images as training and a total of 80 images as testing. We use the same split ratio as used by Yang et al. [38] recently. Example images from each of the four classes are shown in Fig. 2.
Fig. 2.
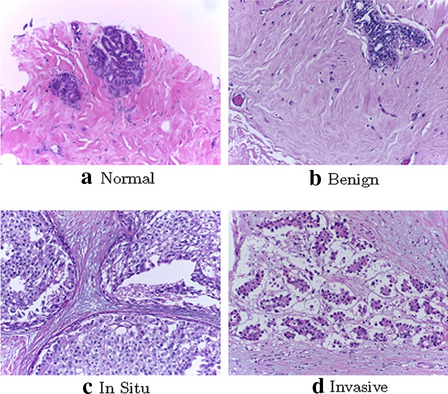
Sampled example H&E images from four classes of the BACH dataset: a normal, b benign, c in situ, and d invasive
Implementation
We use the Keras [8] implementation in Python [27] of the pre-trained VGG16 models. Similarly, we implement all our feature extraction and classification logic in Python. We used the Support Vector Machine (SVM) implemented in Sklearn2 with radial basis function (RBF) kernel. We tune the cost parameter C in the range and fix gamma parameter to 1e−04. Additionally, we evaluated our proposed features using Multilayer Perceptron Algorithm (MLP) [22], Random Forest (RF) [4], and XGBoost [6]. The detailed optimal parameter settings of these three algorithms are mentioned in Table 1. All the experiments are conducted on a computer with NVIDIA GeForce GTX 1050 GPU with 4GB GDDR5 VRAM.
Table 1.
Detailed optimal parameters of three algorithms
| Algorithm | Description |
|---|---|
| MLP | Network: (728,256,4), optimizer: Adam, Epoch: 50, and batch size: 8 |
| RF | Number of estimators=100, bootstrap=True, max_features=’sqrt’ |
| XGBoost | Objective=“reg:linear”, random state=42 |
VGG-Net
VGG-Net is one of the simple, yet powerful pre-trained deep learning model proposed by Simonyan et al. [29]. It has five variants starting from VGG-A to VGG-E. All variants consist of three types of layers (convolution layers, Pooling layers, and Fully connected layers). Among such five variants, one of the models, called VGG-D (VGG16), has been mostly used in the image analysis. VGG16 model has 16 weight layers, among which five layers are Pooling layers. Pooling layers reduce the features by using either max pooling or avg pooling operation on the Convolution layers. Meanwhile, Convolution layers extract the features by using different Kernel size coupled with appropriate activation functions such as Sigmoid, Tanh, Relu, etc.
Because the VGG16 model has only five Pooling layers that provide meaningful semantic information, it is very easy and convenient to utilize them quickly for image analysis. Furthermore, it has shown that such models provide prominent performance for various image analysis ranging from scene image [30] to recent medical image analysis tasks [1, 18]. Note that deep learning models such as VGG16 impart visual information according to the data they are trained with. For example, VGG16 models pre-trained with ImageNet [10] and Places [39] provide the foreground and background information, respectively. These two pieces of information represent two different kinds of information related to the input image, which would play an important role in its better representation.
Proposed method
We believe that more information about interesting regions and their orientation high-resolution histopathological images can be captured by combining different types of features (see detail in Figs. 1, 3). Hence, we propose to extract hybrid features by combining four types of features—foreground (structure interesting regions such as shape, size, etc.) and background (layout of interesting regions) feature at both part-level and whole-level. Using VGG16 models that have been pre-trained with ImageNet and Places, we extract both features (foreground and background) at both levels. Also, we believe that mid-level features (those from intermediate layers) of VGG16 models are more appropriate for histopathological images. Low-level features are more generic, which may not be suitable in this case because histopathological images are not like texture images requiring low-level information. Similarly, the high-level features are more specific to objects or scenes in the real-world which are not applicable in the case of histopathological images. Empirically, we find that the combination of features from the third and fourth pooling layers produce better classification accuracy (analysis of features from different pooling layers is in “Analysis of pooling layers” section). So, we combine foreground and background features from the third and fourth pooling layers at whole and part levels. The overall pipeline of the proposed method is shown in Fig. 4.
Fig. 3.
Diagram showing the features map of the corresponding input image at whole-level and part-level using both foreground and background features, where to represent the five pooling layers of VGG16 models
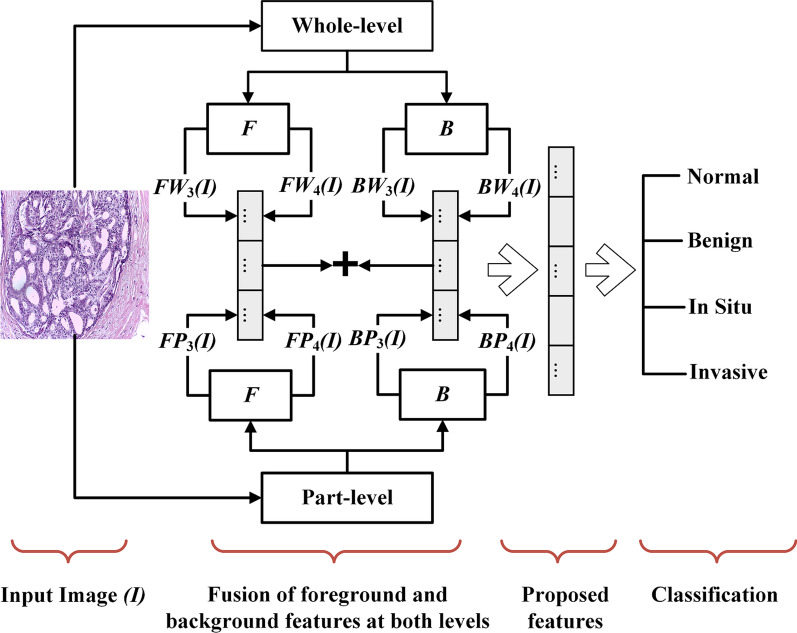
Fig. 4.
Block diagram of the proposed method, where and represents the foreground features extracted from jth pooling layers () at whole-level and part-level, respectively. Similarly, and represents the background features extracted from the jth pooling layer at whole-level and part-level, respectively
We discuss the step-wise procedure to achieve our proposed hybrid features in the next four subsections: patch extraction (“Patch extraction” section), part-level feature extraction (“Part-level feature extraction” section), whole-level feature extraction (““Whole-level feature extraction” section” section), features aggregation to achieve the final features (“Features aggregation” section).
Patch extraction
To capture part-level features of an image, we extract non-overlapping patches of the image using similar approach as employed by Yang et al. [38]. Given an input image of size (), a patch of size is moved with a stride of s on the input image resulting in N patch images.
| 1 |
In this paper, we used the patch size of and stride size of because our pre-trained model VGG16 has been trained on images of that size. Equation 1 yields N patches per image, each with pixels. This helps to extract features related to local level interesting regions. The block diagram to extract patches and corresponding features in the part-level is shown in Fig. 5. We represent N non-overlapping uniform patches of an image I as .
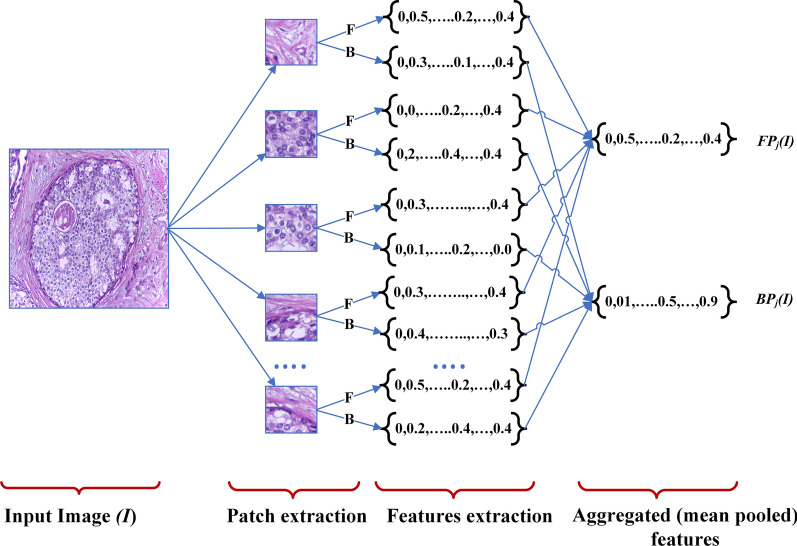
Fig. 5.
Patch-level features extraction for the proposed method. Note that the diagram utlizes the jth pooling layer to extract the aggregated foreground () and background features () using both foreground (F) and background (B) information
Part-level feature extraction
After the extraction of uniform non-overlapping local patches from each image, we extract corresponding deep features of each patch and aggregate them to achieve the combined patch-level features representing an image as in Fig. 5. We extract the patch-level features based on foreground and background information from VGG16 pre-trained on ImageNet (VGG16I) and Places (VGG16P), respectively. To achieve the aggregated foreground features and background features of all N patches of an image I from the jth pooling layer, we perform mean pooling to leverage both higher and lower activation values, as seen in Eqs. (2)–(3). and represent the Foreground and Background features of image I at Part-level from the jth pooling layer, respectively. and represent deep features of patch image extracted from the jth pooling layers of VGG16 models pre-trained on the ImageNet and Places datasets, respectively. Note that the order of patches does not affect the end result (aggregated features) because we apply average pooling in our method. Thus, the patch extraction and its arrangement of features can be done in any order for the aggregation purpose.
| 2 |
where .
| 3 |
where .
Whole-level feature extraction
To extract deep features from whole images, we first resize images to pixels because pre-trained VGG16 models were trained on images. Then, we feed such resized images into VGG16 models and extract both foreground and background features accordingly. We use whole-level images to capture information about interesting regions that are not achieved or missed by part-level techniques. Equations (4)–(5).
| 4 |
| 5 |
where and represent the Foreground and Background features of image I at Whole-level from jth pooling layer, respectively.
Features aggregation
To achieve the final hybrid features of image I (H(I)), we aggregate all part-level and whole-level features (Eqs. (2)–(5)) from the 3rd and 4th pooling layers using simple and easy technique, Concat pooling (Eq. 6), as suggested by Sitaula et al. [30]. Concat pooling is the best to perform aggregation of different types of features because it preserves multiple information uniformly than other methods such as min, max, mean, etc.
| 6 |
This aggregation results in the fusion of eight types of features, both foreground and background features at parts and whole levels for the features extracted from the 3rd and 4th pooling layers of VGG16 models. Because the size of features from the 3rd layer of VGG16 model is 256 and that from the 4th pooling layers is 512, the size of the final hybrid features H is 3072.
Result and discussion
In this section, we present the results of experiments conducted to evaluate the performance of our proposed hybrid deep features in the classification of breast cancer histopathological images using the Support Vector Machine (SVM) classifier [9].
Comparison with state-of-the-art methods
We compared the performance of our method with four recent state-of-the-art methods for the classification of breast cancer histopathological images based on the classification accuracy and computation time. Additionally, we listed the accuracy of our method using four machine learning algorithm, including SVM algorithm. The classification accuracy and computation time results are provided in Tables 2 and 3, respectively. The listed results are the average over five train-test sets. To minimize the bias, we compared our results with the published results of the state-of-the-art methods on the BACH dataset.
Table 2.
Classification accuracy (%) of our method and recent existing state-of-the-art methods
| Methods | Accuracy (%) |
|---|---|
| DCNN+SVM, 2017 [1] | 77.8 |
| Pre-trained VGG-16, 2018 [2] | 83.0 |
| Ensemble of three DCNNs, 2018 [2] | 87.0 |
| Kwok et al., 2018 [19] | 87.0 |
| Makarchuk et al., 2018 [24] | 90.0 |
| EMS-Net, 2019 [38] | 91.7 |
| Our hybrid features + SVM | 92.2 |
| Our hybrid features + MLP | 85.2 |
| Our hybrid features + RF | 80.2 |
| Our hybrid features + XGBoost | 82.7 |
Bold value indicates the best accuracy
Table 3.
Computation time (seconds) for features extraction, training, and testing
| Method | Feat. extraction | Train. | Test. |
|---|---|---|---|
| EMS-Net, 2019 [38] | – | 608,400 | 400 |
| Ours | 1483.84 | 0.35 | 0.06 |
Bold values indicate the best computation time
In Table 2, we can see that our method outperformed all the previous methods. EMS-Net [38] had competitive accuracy to our method (worse by 0.50%), whereas other contenders were significantly worse with differences between 5% to 15%. With our proposed features, we observed that SVM surpasses the three other classifiers (MLP, RF, and XGBoost) with at least 7% higher accuracy.
Furthermore, while comparing our method with the closest contender very recent state-of-the-art method of EMS-Net [38] in terms of computation time (Table 3), our method was several orders of magnitude faster. Their run time results for both training and testing are not comparable. Our method was able to achieve better classification accuracy than EMS-Net in significantly lower run time. It is interesting to note that our GPU configuration (NVIDIA GeForce GTX 1050 GPU with 4GB GDDR5 VRAM) is not as good as the one reported in the EMS-Net paper (NVIDIA GeForce GTX 1080Ti GPU with 32GB memory). Despite working in such a low power machine, we can achieve orders of magnitude faster run time than EMS-Net.
In summary, our proposed feature extraction method provides an effective representation of breast tissue histopathology images for cancer diagnosis in terms of accuracy and computation time. Because EMS-Net used an ensemble of fine-tuned pre-trained models for image classification directly, it requires heavy data augmentation to avoid overfitting as the size of training images is quite small in this case. However, we used pre-trained deep learning models for feature extraction and it does not require data augmentation. While being very simple and fast, our approach is very effective in achieving state-of-the-art classification accuracy. Furthermore, our features can be used with any other classifiers. Thus, we believe that it can be very useful to health practitioners in breast cancer diagnosis.
Analysis of pooling layers
We analyzed the effectiveness of features extracted from each of the five pooling layers of VGG16 models in the classification accuracy. The detail results are presented in Table 4. The results show that all types of features (FP, FW, BP, BW, ) extracted from the 4th pooling layer produced better classification accuracy than the other remaining layers. This proves our hypothesis that mid-level features are more appropriate to represent histopathological images. Among different types of features extracted from the 4th pooling layer, the combined features () produced better results than any one type of feature. This shows the benefit of combining different types of features. Other interesting observations in Table 4 are: (i) foreground features produced better results than corresponding background features indicating that foreground information provides more class discrimination power, and (ii) part-level features produced better results than corresponding whole-level features indicating that part-level (local) information provide more class discrimination power.
Table 4.
Classification accuracies (%) of features extracted from the five pooling layers
| Features type | Pooling layer | ||||
|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | |
| FP | 61.0 | 68.5 | 81.2 | 88.0 | 86.5 |
| FW | 57.2 | 66.2 | 79.0 | 83.0 | 78.0 |
| BP | 61.5 | 66.2 | 78.5 | 85.0 | 80.5 |
| BW | 57.7 | 64.7 | 76.7 | 78.5 | 68.5 |
| Combined | 63.2 | 83.2 | 85.7 | 90.7 | 85.2 |
Bold values indicate the best accuracy
FP and FW represent the foreground features using corresponding pooling layer at part-level and whole-level, respectively. Similarly, BP and BW represent background features using the corresponding layer at part-level and whole-level, respectively. Combined features in the last row are the combination of all four types of features ()
We also analysed if adding combined () features from other pooling layers (1st,2nd,3rd or 5th) with those extracted from the 4th pooling layer improves the classification accuracy. The results are presented in Table 5. It is interesting to see that the concatenation of features from layers {1st, 4th}, {2nd, 4th} and {3rd, 4th} produced better accuracy than just using features from the 4th layer, whereas {5th, 4th} produced the worse result. The concatenation of combined features from the 3rd and 4th layers produced the largest improvement of 1.5% over the accuracy produced by features from the 4th layer only.
Table 5.
Classification accuracy (%) of concatenation of combined () features achieved from the 4th pooling layer and each of the other four pooling layers ({jth, 4th} where )
| {1st, 4th} | {2nd, 4th} | {3rd, 4th} | {5th, 4th} |
|---|---|---|---|
| 91.0 | 91.5 | 92.2 | 87.5 |
Bold value indicates the best accuracy
Class-wise analysis of our method
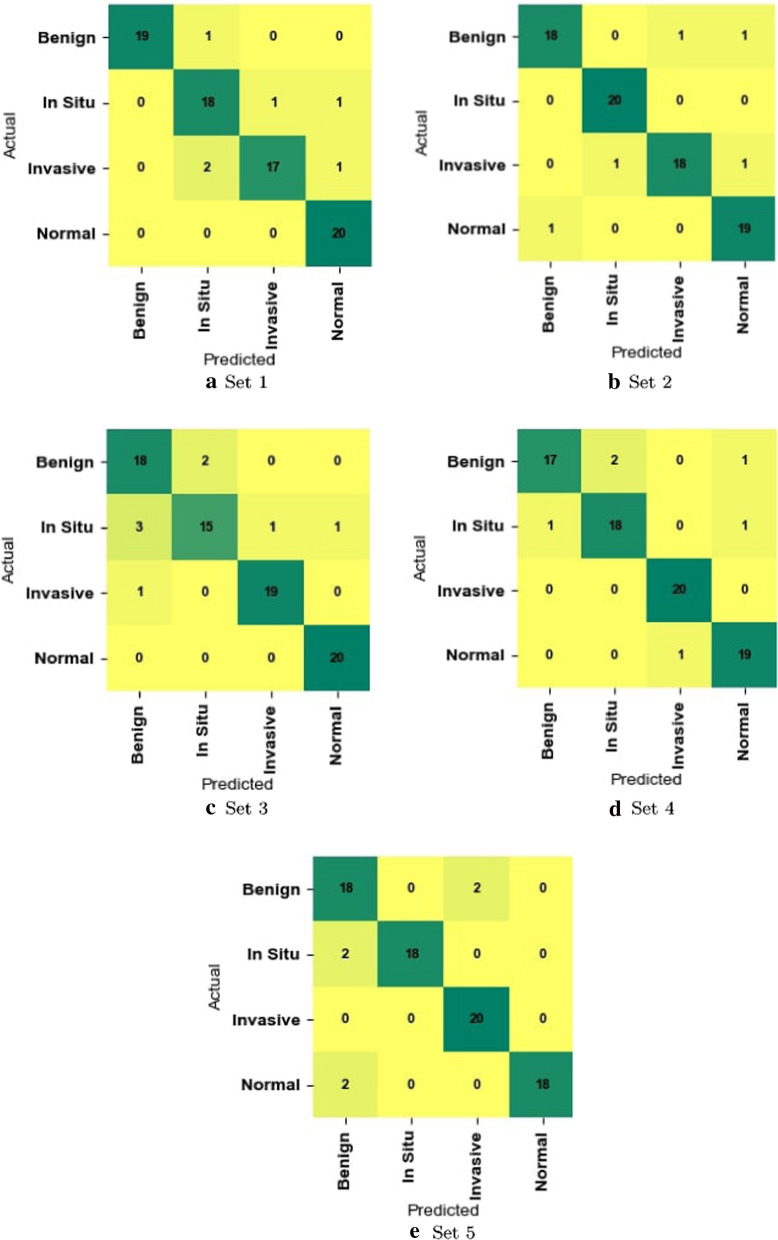
To analyze the effectiveness of our method in differentiating each of the four classes in the BACH dataset, we calculated class-wise Precision, Recall, and F1-score which are reported in Table 6. We also used the confusion matrix to see the distribution of images into different classes or categories (Fig. 6). These results show that our proposed method is consistently good in discriminating all four classes.
Table 6.
Average performance (%) using per class Precision, Recall, and F1-score on the testing set of BACH dataset
| Class | Precision | Recall | F1-score |
|---|---|---|---|
| Normal | 90.0 | 90.0 | 90.0 |
| Benign | 91.0 | 89.0 | 90.0 |
| In situ | 94.0 | 94.0 | 93.0 |
| Invasive | 93.0 | 96.0 | 94.0 |
Bold values indicate the best performance
Fig. 6.
Confusion matrix of our method on the testing split of a Set 1, b Set 2, c Set 3, d Set 4, and e Set 5
Conclusion
In this paper, we proposed a combined model for feature extraction to represent histopathological images capturing information in the images from different perspectives. We combine foreground and background features at image parts and whole levels extracted from two mid-level (third and fourth) pooling layers of pre-trained VGG16 models. Foreground and background feature to capture information about areas of interest and their layout in microscopic histopathological images of breast tissues. Part-level and whole-level features are useful to detect interesting regions scattered in high-resolution histopathological images at local and whole image levels. We demonstrate the effectiveness of our proposed features in the classification of H&E stained histopathological images of breast tissues using the Support Vector Machine (SVM) classifier. Our experimental results in the BACH breast cancer dataset show that our method produces better classification accuracy compared to four existing state-of-the-art classification models proposed for breast cancer histopathological image classification. Its performance is fairly consistent in differentiating all classes (i.e., detecting different types of cancer). Our proposed method is orders of magnitude faster than the best performing state-of-the-art method of EMS-Net. Also, we are planning to design a new patch-based deep learning model for the classification of histopathological images. Note that augmentation in patch level could provide significant information than whole level because whole image level augmentation mostly shifts the ROIs. Additionally, we will focus on the domain-specific augmentation techniques to increase the datasets for training our model.
Because of its effectiveness and efficiency, we believe that our proposal will be useful to support pathologists and clinicians in fast and early detection of breast cancer. The main implication of our study is that histopathological images need to be analyzed from different perspectives as interesting clues important for diagnosis might be anywhere in the complex high-resolution microscopic images.
However, the technique we proposed in this paper may not be suitable for other types of medical images such as x-ray scans, CT scans, Colonoscopy scans owing to their differing layout structures and visual contents. So, in the future, we would like to extend our idea to other types of medical images.
Acknowledgements
We would like to thank ICIAR2018 grand challenge for providing a histopathological image dataset to use in our research. Dr Sunil Aryal is supported by the Air Force Office of Scientific Research grant under Award Number FA2386-20-1-4005.
Funding
There are no financial supports to complete this work.
Data availability
The datasets are publicly available.
Code availability
Source codes will be made publicly available upon acceptance of the paper.
Compliance with ethical standards
Conflict of interest
We would like to confirm that there are no known conflict of interests exist.
Footnotes
https://www.aihw.gov.au/reports/cancer/cancer-in-Australia-2019 (Accessed date: 15/02/2020).
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chiranjibi Sitaula, Email: csitaul@deakin.edu.au.
Sunil Aryal, Email: sunil.aryal@deakin.edu.au.
References
- 1.Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, Polónia A, Campilho A. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE. 2017;12(6):e0177544. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aresta G, Araújo T, Kwok S, Chennamsetty SS, Safwan M, Alex V, Marami B, Prastawa M, Chan M, Donovan M, et al. Bach: Grand challenge on breast cancer histology images. Med Image Anal. 2019;56:122–139. doi: 10.1016/j.media.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Barker J, Hoogi A, Depeursinge A, Rubin DL. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med Image Anal. 2016;30:60–71. doi: 10.1016/j.media.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 5.Campanella G, Silva VWK, Fuchs TJ. Terabyte-scale deep multiple instance learning for classification and localization in pathology. arXiv preprint. arXiv:180506983 (2018).
- 6.Chen T, Guestrin C. Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp 785–794 (2016).
- 7.Chennamsetty SS, Safwan M, Alex V. Classification of breast cancer histology image using ensemble of pre-trained neural networks. In: Proceedings of the international conference on image analysis and recognition (ICIAR), pp 804–811 (2018).
- 8.Chollet F, et al. Keras. https://github.com/fchollet/keras (2015).
- 9.Cristianini N, Shawe-Taylor J. An introduction to support vector machines and other kernel-based learning methods. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- 10.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In: Proceedings of the IEEE conference on computer vision pattern and recognition (CVPR) (2009).
- 11.Falk T, Mai D, Bensch R, Çiçek O, Abdulkadir A, Marrakchi Y, Böhm A, Deubner J, Jäckel Z, Seiwald K, et al. U-net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16(1):67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 12.Gandomkar Z, Brennan PC, Mello-Thoms C. Mudern: multi-category classification of breast histopathological image using deep residual networks. Artif Intell Med. 2018;88:14–24. doi: 10.1016/j.artmed.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 13.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of IEEE conference on computer vision and pattern recognition (CVPR), pp 770–778 (2016).
- 14.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of IEEE conference on computer vision and pattern recognition, pp 4700–4708 (2017).
- 15.Jiang Y, Chen L, Zhang H, Xiao X. Breast cancer histopathological image classification using convolutional neural networks with small se-resnet module. PLoS ONE. 2019;14(3):e0214587. doi: 10.1371/journal.pone.0214587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q, Liu TY. Lightgbm: a highly efficient gradient boosting decision tree. In: Proceedings of the advances in neural information processing systems, pp 3146–3154 (2017).
- 17.Khan AM, Sirinukunwattana K, Rajpoot N. Geodesic geometric mean of regional covariance descriptors as an image-level descriptor for nuclear atypia grading in breast histology images. In: Proceedings of the international workshop on machine learning in medical imaging (MLMI), pp 101–108 (2014).
- 18.Kumar A, Singh SK, Saxena S, Lakshmanan K, Sangaiah AK, Chauhan H, Shrivastava S, Singh RK. Deep feature learning for histopathological image classification of canine mammary tumors and human breast cancer. Inf Sci. 2020;508:405–421. doi: 10.1016/j.ins.2019.08.072. [DOI] [Google Scholar]
- 19.Kwok S. Multiclass classification of breast cancer in whole-slide images. In: Proceedings of the international conference on image analysis and recognition (ICIAR), pp 931–940 (2018).
- 20.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 21.Mehta S, Mercan E, Bartlett J, Weaver D, Elmore JG, Shapiro L. Y-net: joint segmentation and classification for diagnosis of breast biopsy images. In: Proceedings of the international conference on medical image computing and computer-assisted intervention (MICCAI), pp 893–901 (2018).
- 22.Murtagh F. Multilayer perceptrons for classification and regression. Neurocomputing. 1991;2(5–6):183–197. doi: 10.1016/0925-2312(91)90023-5. [DOI] [Google Scholar]
- 23.Nazeri K, Aminpour A, Ebrahimi M. Two-stage convolutional neural network for breast cancer histology image classification. In: Proceedings of the international conference image analysis and recognition (ICIAR). Springer, pp 717–726 (2018).
- 24.Pimkin A, Makarchuk G, Kondratenko V, Pisov M, Krivov E, Belyaev M. Ensembling neural networks for digital pathology images classification and segmentation. In: Proceedings of the international conference on image analysis recognition (ICIAR), pp 877–886 (2018).
- 25.Qi X, Xing F, Foran DJ, Yang L. Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans Biomed Eng. 2011;59(3):754–765. doi: 10.1109/TBME.2011.2179298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakhlin A, Shvets A, Iglovikov V, Kalinin AA. Deep convolutional neural networks for breast cancer histology image analysis. In: Proceedings of the international conference on image analysis and recognition (ICIAR), pp 737–744 (2018).
- 27.Rossum G. Python reference manual. Technical report. Amsterdam: Stichting Mathematisch Centrum; 1995. [Google Scholar]
- 28.Roy K, Banik D, Bhattacharjee D, Nasipuri M. Patch-based system for classification of breast histology images using deep learning. Comput Med Imaging Graph. 2019;71:90–103. doi: 10.1016/j.compmedimag.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint. arXiv:14091556 (2014).
- 30.Sitaula C, Xiang Y, Basnet A, Aryal S, Lu X. Hdf: Hybrid deep features for scene image representation. In: Proceedings of the international joint conference on neural networks (IJCNN), pp 1–8 (2020).
- 31.Sudharshan P, Petitjean C, Spanhol F, Oliveira LE, Heutte L, Honeine P. Multiple instance learning for histopathological breast cancer image classification. Expert Syst Appl. 2019;117:103–111. doi: 10.1016/j.eswa.2018.09.049. [DOI] [Google Scholar]
- 32.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR), pp 1–9 (2015).
- 33.Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv preprint. arXiv:160207261 (2016a).
- 34.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826 (2016b).
- 35.Vapnik V. The nature of statistical learning theory. Berlin: Springer; 2013. [Google Scholar]
- 36.Vink JP, Van Leeuwen M, Van Deurzen C, de Haan G. Efficient nucleus detector in histopathology images. J Microsc. 2013;249(2):124–135. doi: 10.1111/jmi.12001. [DOI] [PubMed] [Google Scholar]
- 37.Yan R, Ren F, Wang Z, Wang L, Zhang T, Liu Y, Rao X, Zheng C, Zhang F. Breast cancer histopathological image classification using a hybrid deep neural network. Methods. 2020;173:52–60. doi: 10.1016/j.ymeth.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Ran L, Zhang S, Xia Y, Zhang Y. Ems-net: ensemble of multiscale convolutional neural networks for classification of breast cancer histology images. Neurocomputing. 2019;366:46–53. doi: 10.1016/j.neucom.2019.07.080. [DOI] [Google Scholar]
- 39.Zhou B, Khosla A, Lapedriza A, Torralba A, Oliva A. Places: an image database for deep scene understanding. arXiv preprint. arXiv:161002055 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets are publicly available.
Code availability
Source codes will be made publicly available upon acceptance of the paper.