Abstract
Purpose
To utilize a novel mitochondrial function assay with pooled granulosa cells to determine whether mitochondrial function would differ by patient demographics and embryo development.
Methods
This was a prospective pilot study in a hospital-based assisted reproductive program and public university. Mitochondrial metabolic substrate utilization was assessed in pooled granulosa cells from 40 women undergoing in vitro fertilization during 2018 and 2019.
Results
Assessment of mitochondrial substrate metabolism in pooled granulosa cells revealed higher citric acid, L-malic acid, and octanoyl-L-carnitine utilization with higher body mass index (BMI). Utilization of citric acid, cis-aconitic acid, D-alpha-keto-glutaric acid, L-glutamine, and alanine plus glycine was significantly lower as total dosage of FSH administered increased. Utilization of glycogen was significantly higher in patients with a higher percentage of fertilized oocytes. D-alpha-keto-glutaric acid utilization was significantly lower in patients with a higher percentage of good 8-cell embryos. L-glutamine utilization was significantly lower, with a higher percentage of blastocyst formation. Mitochondrial metabolic scores (MMS), which reflect overall mitochondrial activity of the granulosa pool, were significantly higher in patients with higher BMI and with greater numbers of mature oocytes retrieved. MMS in granulosa decreased as total FSH dose administered increased.
Conclusions
Granulosa cell utilization of substrates feeding into the citric acid cycle changed with total FSH dosage and BMI. Fertilization rate, 8-cell embryo quality, and blastocyst formation also associated with different energy substrate usage. Mitochondrial substrate utilization by granulosa cells from individual follicles could be further developed into a useful diagnostic tool.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01946-9) contains supplementary material, which is available to authorized users.
Keywords: Granulosa cells, In vitro fertilization, Mitochondrial function assay, Mitochondrial metabolic score, Mitochondrial substrate utilization, Citric acid cycle
Introduction
Mitochondria are double-membrane organelles, which are believed to have originated from a symbiotic relationship between bacteria and host cells [1]. They are involved in apoptosis, calcium homeostasis, fatty acid oxidation, and metabolism. Mitochondria are the key source of power within eukaryotic cells. They are responsible for generating 88% of a cell’s total energy in the form of adenosine triphosphate (ATP) generated through oxidative phosphorylation via mitochondrial membrane potential [2]. The remaining ATP required by the cell is produced from the processes of glycolysis and the tricarboxylic acid (TCA) cycle through substrate-level phosphorylation. Oxidative phosphorylation is modulated by the cellular demand for ATP and the substrates available for processing within the cytoplasm. Glucose is the main substrate that will produce ATP via the intermediates of glycolysis. Pyruvate and other substrates of the glycolytic process will generate reducing equivalents, nicotinamide adenine dinucleotide hydride (NADH) and flavin adenine dinucleotide hydride (FADH2), which will transfer electrons to the electron transport chain (ETC). The flow of electrons by means of the ETC will provide mitochondrial membrane potential, ultimately producing ATP [3].
Successful embryo development relies upon energy from ATP, supplied mostly by the mitochondria, to drive the cleavage of the fertilized oocyte and blastomeres and other cellular processes during early development. While human oocytes contain the most mitochondria of all cell types within the body, it is problematic to relate mitochondrial DNA (mtDNA) copy number and function to oocyte developmental competence, since the oocytes, and subsequent embryos, are required for clinical use. However, parallels between mtDNA content in human oocytes and granulosa cells (GCs), including both cumulus cells (CCs) and mural granulosa cells (MGCs), have been demonstrated [4, 5]. Additionally, the metabolism of CCs directly affects their enclosed oocytes by providing the proper energy substrates required to complete meiosis and fertilization. Cumulus cells produce ATP and augment oocyte ATP supply during follicular growth [6]. The CCs provide pyruvate, generated from glycolysis, to their enclosed oocytes via gap junctions [7, 8]. Furthermore, mitochondrial function, as measured by ATP production in CCs, relates to proper human oocyte maturation and in vitro fertilization (IVF) outcomes [9–11].
Assessing mitochondrial substrate metabolism in GCs may allow for a non-invasive method for investigating oocyte competence and embryo development. Several studies have evaluated CCs, MGCs, and trophectoderm cells from blastocysts, and it appears there is a high correlation between the mtDNA content found in oocytes and CCs indicating that GC mtDNA might serve as a biomarker for oocyte competence [4, 12–15]. Furthermore, increased GC mtDNA copy numbers are associated with good quality embryos [12, 15]. Therefore, we hypothesized that mitochondrial substrate utilization in pooled GCs, measured using a novel mitochondrial function assay, will show correlations between various patient and oocyte endpoints.
The goal of this study was to evaluate individual metabolic substrates and total substrate utilization by mitochondria based on patient age, serum anti-Mullerian hormone (AMH) level, body mass index (BMI), total FSH dose and the number and percentage of mature oocytes retrieved per patient. We further related individual and total substrate utilization to fertilization rates, good quality 8-cell embryos, and percentage blastocyst formation to determine if one or more substrates could be identified as a potential biomarker for IVF outcome parameters.
To our knowledge, this is the first study to examine a novel mitochondrial function assay to assess the metabolism of a broad range of mitochondrial substrates in GCs. The mitochondrial function assay measures the rate of electron flow through the electron transport chain from 31 metabolic substrates that, when metabolized by cell preparations, produce NADH or FADH2 [16].
Materials and methods
Study population, participants, and GC isolation
This study was a prospective cohort pilot study. In total, 817 pooled CCs and MGCs from 40 patients undergoing infertility treatment at Fertility Center of the Carolinas, Prismahealth - Upstate (Greenville, SC, USA) were analyzed (Fig.1). Of 58 starting patients, those with total viable GC numbers <650,000 per sample were excluded from the study (n = 14) as their viable pooled granulosa cell numbers were insufficient to perform the complete mitochondrial function assay using the MitoPlate S-1 plates (Biolog, Hayward, CA, USA). In addition, patients with sufficient viable GC numbers where the positive assay control did not exhibit color change when compared to the negative control (n = 4) were excluded. Patient demographics and cycle information were collected for each cycle (Table 1 and Supplemental Table 1). Each patient’s ovaries were stimulated using a long luteal protocol or an antagonist protocol. In the antagonist cycles, when one follicle reached 13 or 14 mm or the patient’s estradiol level reached 300 IU/mL, a GnRH antagonist (Cetrotide, EMD-Serono, Rockland, MA, USA or Ganirelix, EMD-Serono, Rockland, MA, USA) was administered. In both protocols, when 3 follicles reached >17 mm by ultrasound, an ovulatory dose of human chorionic gonadotropin (hCG) (Pregnyl, EMD-Serono, Rockland, MA, USA) was delivered. Thirty-six hours later patients underwent oocyte retrieval. Each CC mass was mechanically separated from its oocyte and rinsed in medium (Sperm Wash Medium, Fujifilm-Irvine Scientific, Santa Ana, CA, USA) to remove blood and debris and all CC masses per retrieval were combined in a single tube. The pooled CCs and separately pooled MGCs from each patient were placed on ice prior to processing which occurred within 30 min.
Fig. 1.
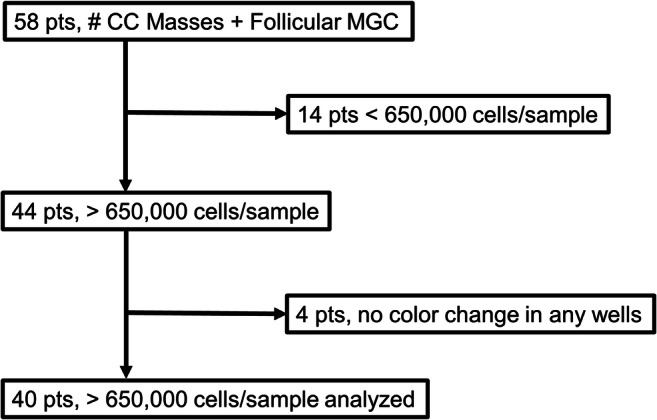
Flow diagram summarizing the study population from oocyte retrieval to the final outcome. Oocytes, cumulus cell (CC) masses, and mural granulosa cells (MGCs) were collected from 58 patients (pts). Individual CC masses from each oocyte collected (n = 817) were combined with the MGCs from each patient to form the granulosa pool. Eighteen patients were excluded from analyses due to insufficient number of cells for assay or no signal in positive control wells of the mitochondrial function assay yielding a final number of patients for analyses n = 40
Table 1.
Patient demographics and cycle information
| n = 40 | Median | Range |
|---|---|---|
| Age | 33 | 22–40 |
| Body mass index | 26 | 17.9–38.3 |
| Anti-Mullerian hormone | 3.41 | 0.55–11.5 |
| Antral follicle count | 20 | 7–30 |
| Human menopausal gonadotropin dose | 675 | 225–1200 |
| Follicle stimulating hormone dose (total) | 1512 | 775–3300 |
| # Oocytes retrieved | 19 | 10–41 |
| # Mature oocytes | 13.5 | 6–26 |
| % Mature oocytes | 73% | 45–100 |
| % Fertilization | 81% | 17–100 |
| % Good 8-cell embryos | 58% | 0–100 |
| % Blastocysts | 55% | 0–100 |
| mtDNA content1 | 0.0009145 | 0.000201–0.006901 |
mtDNA = mitochondrial DNA
1Mitochondrial copy number of total cells used per well
Embryo culture
Mature oocytes were inseminated via intra-cytoplasmic sperm injection. Fertilization was confirmed by the presence of two pronuclei and two polar bodies 16–18 h after insemination. Embryos were group cultured in 50 μL LifeGlobal® global total media drops (Cooper Surgical, Trumbull, CT, USA) under oil (Ovoil, Vitrolife, Englewood, CO, USA) at 37 °C, 6% CO2, 5% O2, 89% N2 in a humidified atmosphere until day 5 or day 6 post retrieval. Embryo morphologies were assessed using the American Society for Reproductive Medicine (ASRM) [17] criteria for day 3 embryos and day 5/6 blastocysts.
Mural granulosa cell preparation and mitochondrial function assay
Follicular fluid containing MGCs from each patient was split between two or more 50 mL tubes and centrifuged for 15 min at 300 xg. A filtered solution of hyaluronidase (50 mg/mL) from bovine testes (Sigma, Raleigh, NC, USA) in media was added to each MGC pellet and the CC tube and incubated at 37 °C for 10 min to dissociate cell clumps. Calcium/magnesium-free phosphate-buffered saline (PBS) (Gibco, Gaithersburg, MD, USA) was added to bring the pellet to approximately 4 mL. MGCs were purified by 35% to 70% discontinuous Percoll gradient in 15 mL tubes and spun in a swinging bucket centrifuge for 30 min at 300 xg. Assay mix (30 μL) containing 2X Biolog Mitochondrial Assay Solution (MAS) (Biolog), 6X Redox Dye (Biolog), 24X saponin (90 μg/mL; Alfa Aesar, Ward Hill, MA, USA) for permeabilization, and sterile water were added to each well of the 96-well MitoPlate S-1 plates (Biolog) and incubated for 1 h per manufacturer’s instructions. MGCs (in the uppermost band in the Percoll gradient) were removed and combined with CCs in 25 mL of calcium/magnesium-free PBS. The mixture was poured through a 70 μm strainer to remove any remaining cell clumps. Pooled GCs were centrifuged for 20 min, and the pellet was resuspended in 1X MAS (Biolog) to a volume of 2 mL. A 0.5 mL aliquot of the GC/MAS mixture was removed, and a 10 μL sample was counted with trypan blue (ThermoFisher Scientific, Carlsbad, CA, USA) using a hemocytometer to determine viable granulosa cells. This remainder of the aliquot was centrifuged for 5 min, and the pellet was placed in 0.5 mL RNAlater solution (Invitrogen-Thermo Scientific, Carlsbad, CA, USA), stored at 4 °C for 24 h, and then snap-frozen and stored in LN2 until transferred to freezer storage at −70 °C until processed. The remaining 1.5 mL GC/MAS mixture was diluted to 3.0 mL with 1X Biolog MAS and a 10 μL sample was counted with trypan blue using a hemocytometer to determine viable granulosa cells. After the 1 h incubation at 37 °C to allow all substrates to dissolve, 30 μL of the viable GC/MAS suspension was added to each well of the MitoPlate S-1 plate. The median number of viable cells used per well and range was 15,350 (7000–53,100). Initial optimization experiments to determine minimal granulosa cell numbers/well and saponin permeabilization conditions needed for the mitochondrial function assay were performed as recommended by the manufacturer. First, saponin concentrations were tested at 30, 60, and 90 μg/mL with 1 million cells (approximately 10,000 cells/well). Cell concentrations per well were graphed to confirm that the difference between the absorbance readings of the negative and positive control wells were within the linear range (Supplemental Fig.1). The manufacturer reports that after passing the detection threshold the assay is linear up to 80,000 cells/well or more with various cell types (Biolog, unpublished data). Samples at approximately 7000 cells/well and greater gave usable readings. The assay plateaus at approximately 5 h after incubation with cells.
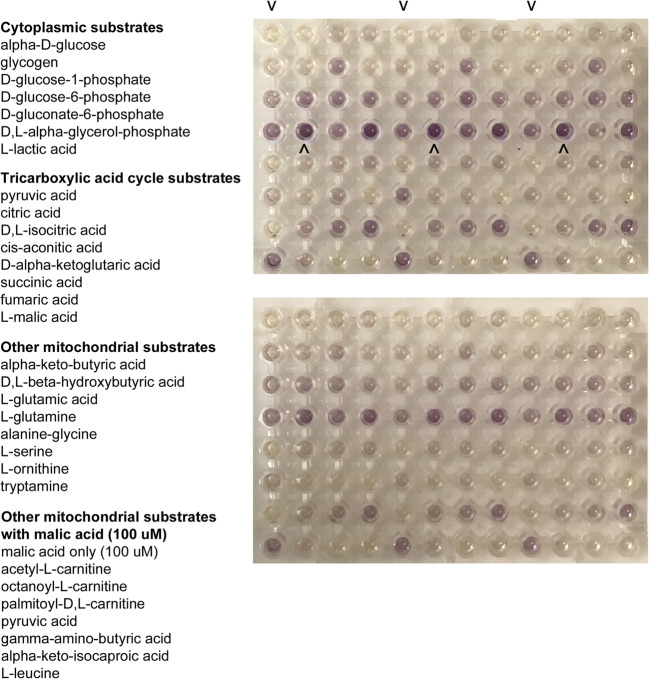
Permeabilized cells were assayed in triplicate for the metabolism of individual substrates using MitoPlate S-1 plates. Metabolism of substrates was assessed by a colorimetric change of a terminal electron acceptor tetrazolium redox dye at an optical density of 590 nm read every 2.5 min for 50 cycles on a kinetic plate reader. Example plates and the substrates are shown in Fig. 2. Increased absorbance readings in the assay reflected greater metabolism/utilization of substrates. The slopes of the optical density absorbance readings were first normalized for the number of viable GCs per well and then to mtDNA copy number per cell (as described below) and the relative absorbance of the negative control wells (no substrate control) containing GC/MAS without any additional substrates. We normalized to both the cell number per well and the mtDNA copy number per cell because the amount of mtDNA per cell may vary, and because we were analyzing mitochondrial substrate utilization instead of cellular substrate utilization. To quantitatively compare the metabolism of each patient, we developed a scoring system based on the aggregate slope ratios of all substrates normalized to the negative control and positive control (succinic acid) wells. The mitochondrial metabolic score was the total of all the slope ratios of each substrate for each individual patient. Slope ratio = [Slope (sample) – Slope (negative control)] / [Slope (positive control) – Slope (negative control)] [18].
Fig. 2.
Two representative mitochondrial functional assays using Mitoplate S-1 and pooled granulosa cells from individual patients. The assay plates contained the individual 31 cytoplasmic and mitochondrial metabolic substrates as the sole energy source in wells, and wells were repeated in triplicate. The substrates are listed to the left. As the cells metabolized the energy source and produced NADH or FADH2, a tetrazolium dye in the medium was proportionately reduced, generating a purple color. Color development was measured by absorbance using a kinetic assay over 2 h. The top two rows indicate no substrate controls (v) and cytoplasmic substrates. Rows 3, 4 contain mitochondrial substrates, including positive control (^) succinic acid. Rows 5, 6 contain other mitochondrial substrates, and 7,8 contain other mitochondrial substrates in the presence of 100 μM malic acid
DNA isolation and quantitative real-time PCR
RNA and DNA were isolated from each pooled CC/MCG sample per patient using the Direct-zol MiniPrep Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. RNA was stored for future studies. DNA isolation omitted the DNAse treatment. Nucleic acid concentration and purity for samples were assessed at the wavelengths of 260 and 280 nm using a biophotometer 6131 spectrophotometer (Eppendorf, Hauppauge, NY, USA). Synthesized primers were cartridge purified (Invitrogen-Thermo Scientific). The nuclear gene for beta-actin (ACTB) was selected as a reference gene to calculate nuclear DNA (nDNA) and the mtDNA gene for mitochondrially encoded ATP synthase 8 (MT-ATP-8) was used to quantify mitochondrial DNA content. ACTB gene primers were forward: 5′- AGCGGGAAATCGTGCGTGAC and reverse 5’-AGGCAGCTCGTAGCTCTTCTC with a 60 °C annealing temperature and 116 bp amplicon. MT-ATP-8 primers were forward 5’-CTAAAATATTAAACACAAACTACCACCTACCTC and reverse 5’-GTTCATTTTGGTTCTCAGGGTTTGTTTAA with annealing at 60 °C and a resulting 92 bp amplicon [13]. Single amplicons of correct size were confirmed initially by agarose gel electrophoresis and single melt curve peaks in each PCR reaction.
Real-time PCR reactions were run with 2 or more wells per sample and the threshold cycle (Ct) values of wells were averaged for each preparation. ACTB was run for 40 cycles and MT-ATP-8 was run for 45 cycles. The mean and standard deviation for ACTB Ct values were 27.8 ± 2.6 and ATP-8 were 25.5 ± 2.4. The reactions included 5 ng starting nucleic acid, 300 nM of each upstream and downstream primer, and 10 μL 2X SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) and sterilized PCR-grade water to a final volume to 20 μL per well. PCR-grade water was substituted for the nucleic acid template as a negative control. PCR amplification was performed using the iCycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories). ACTB was used to quantify nDNA [13]. Target DNA starting quantities were derived from a standard curve made using serial dilutions of its purified amplicon. Relative amounts of mtDNA and nDNA and were determined by qRT-PCR (described below). The ratio of mtDNA:(2x nDNA) was used to determine mtDNA content.
Statistical analysis
The patients’ mitochondrial assay data were analyzed based on age, serum AMH levels, BMI levels, and total FSH dosage. Demographic parameters divided into two groups were compared using paired t-test. In addition, mitochondrial assay data was evaluated by number of mature oocytes, percentage of mature oocytes, fertilization percentage, good 8-cell embryo percentage, and percentage of blastocysts formed. Multiple linear regression models, controlling for the confounding variables, were estimated to predict the normalized average rate of each substrate’s metabolism and mitochondrial metabolic score [19]. Pearson’s correlation analyses were performed to study the relationships between the GC mtDNA copy number, and patient cycle parameters, and embryo cohort development. Box-Cox transformations were used to ensure the normality of the responses in regression models, and the measures used in the correlation analyses. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations were removed to ensure the normality of the response variables. The number of outlying observations removed varied by the response variables. Each substrate was assessed separately, and a mitochondrial metabolic score was created based on the slopes of the normalized absorbance readings of all substrates combined. The mitochondrial metabolic scores were normalized against the mtDNA content in each sample. The individual substrate utilization rates and the mitochondrial metabolic scores were related to patient demographics and normalized to mtDNA content. We defined the statistical significance as P < 0.05, and we considered values 0.05 < P < 0.10 to be trends. Statistical analyses were performed using the statistical software R 3.6.2 (http://www.r.project.org/).
Results
In total, 817 oocytes and associated GCs were harvested from the 40 patients undergoing fertility treatment. The CCs were combined with the MGCs from each patient. Thirty-seven out of 40 patients had at least one fresh or frozen embryo transfer (FET), and currently, 49% of patients who received an ET either have ongoing pregnancies or have delivered a child.
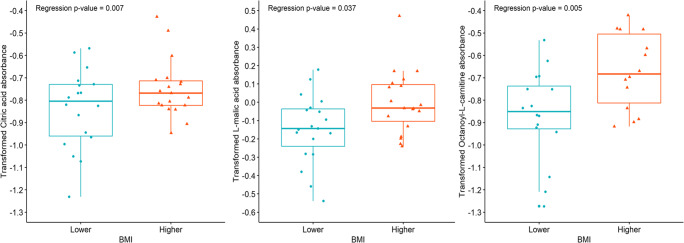
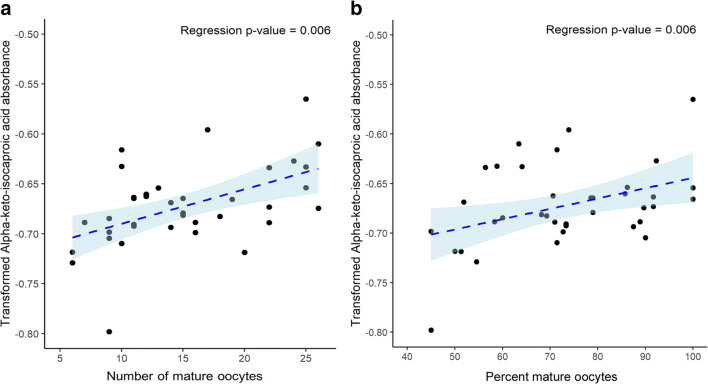
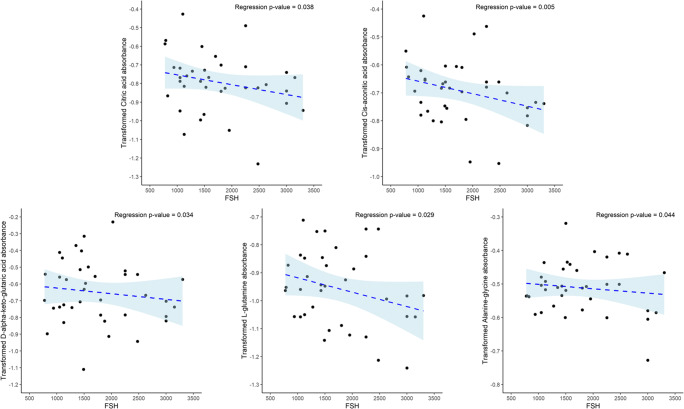
In this pilot study we examined the mitochondrial metabolic profile of granulosa cells of IVF patients using a mitochondrial function assay, which evaluated 31 different substrates. In hypothesis one, we tested whether individual substrate utilization by pooled granulosa mitochondria would differ according to age, serum AMH levels, BMI, total FSH dosage received, and the total number and percentage of mature oocytes retrieved per patient. To test this hypothesis, we analyzed the averaged normalized absorbance readings of each metabolic substrate within the comparison groups using a multiple linear regression model using Box-Cox transformation examining the independent variables of age, BMI, serum AMH, mature oocytes per cohort, antral follicle count, stimulation type, amount of HMG given, and total dose of FSH administered. Age, BMI, and serum AMH were divided into higher and lower groups as follows: Older age ≥ 34 years old and younger age < 34 years old; high BMI ≥ 26 kg/m2 and low BMI < 26 kg/m2; and higher serum AMH ≥ 3.5 ng/mL and lower serum AMH < 3.5 ng/ml. We divided the groups to keep the sample sizes similar and near the standard physiological cutoffs for slightly low for normal and advanced maternal age, slightly high for normal and obese BMI, and low for normal/low AMH versus polycystic ovarian syndrome AMH levels to ensure distinction between the groups. No significant differences in substrate utilization were observed for higher and lower age, higher and lower serum AMH, antral follicle count, stimulation type, amount of HMG given. Based on the estimated multiple linear regression model using observed data, patient mitochondrial utilization of citric acid (P < 0.01), L-malic acid (P < 0.05), and octanoyl-L-carnitine (P < 0.01) were found to be significantly higher in the higher BMI group (Fig. 3). Mitochondrial utilization of cis-aconitic acid (P < 0.10), D,L-beta-hydroxy-butyric acid (P < 0.10), and pyruvic acid with L-malic acid 100 μM (P < 0.10) trended to be higher in patients with higher BMI in pooled GCs, while no other substrates demonstrated utilization differences by BMI (Supplemental Fig. 2). Citric acid and L-malic acid are both components of the TCA cycle and octanoyl-L-carnitine is a medium chain fatty acid that catabolizes to several acetyl-CoA molecules and feeds into the TCA cycle. Individuals with a greater number and percentage of mature oocytes were found to have a significant increase for alpha-keto-isocaproic acid mitochondrial utilization by pooled granulosa (P < 0.01) (Fig. 4). Alpha-keto-isocaproic acid is converted to leucine by transamination reaction and then converted to acetyl-CoA where it enters the TCA cycle. No other substrates demonstrated utilization differences in the model (Supplemental Fig. 3). Patient mitochondrial utilization for citric acid (P < 0.05), cis-aconitic acid (P < 0.01), D-alpha-keto-glutaric acid (P < 0.05), L-glutamine (P < 0.05), and alanine-glycine (P < 0.05) were found to be significantly lower with increasing cumulative dosage of FSH administered (Fig. 5). In addition, mitochondrial utilization of D,L-isocitric acid (P < 0.10), D,L-beta-hydroxy-butyric acid (P < 0.10), L-glutamic acid (P < 0.10), acetyl-L-carnitine (P < 0.10), pyruvic acid with L-malic acid 100 μM (P < 0.10) trended to be lower in pooled GCs of patients with increasing total FSH dosages, while no other substrates demonstrated mitochondrial utilization differences with FSH dose in the model (Supplemental Fig. 4). Cis-aconitic acid and D-alpha-keto-glutaric acid are both components of the TCA cycle. L-glutamine is converted to alpha-ketoglutarate through glutamate dehydrogenase and enters the TCA cycle at that juncture. Alanine and glycine are converted into pyruvic acid and enter the TCA cycle after pyruvic acid is converted to acetyl-CoA.
Fig. 3.
Substrates that demonstrated significant mitochondrial utilization differences based on body mass index (BMI). To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation between BMI groups. BMI was divided into 2 groups with the higher group being patients with a BMI of ≥ to 26 kg/m2 (n = 20) and the lower group being patients with a BMI of <26 kg/m2 (n = 20). Mitochondrial utilization of citric acid (P < 0.01), L-malic acid (P < 0.05), and octanoyl-L-carnitine (P < 0.01) were found to be significantly higher in pooled GCs of patients with higher BMI, after adjusting for the other factors in the model. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers)
Fig. 4.
Substrates that demonstrated significant mitochondrial utilization differences based on A) the number of mature oocytes retrieved and B) the percent mature oocytes retrieved from each patient. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. Individuals with a greater number of mature oocytes and percent mature oocytes retrieved were found to have a significant increase in alpha-keto-isocaproic acid mitochondrial utilization (P < 0.01) in pooled GCs of patients, after adjusting for the other factors in the model. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval
Fig. 5.
Substrates that demonstrated significant mitochondrial utilization differences based on total FSH dose administered. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. Mitochondrial utilization for citric acid (P < 0.05), cis-aconitic acid (P < 0.01), D-alpha-keto-glutaric acid (P < 0.05), L-glutamine (P < 0.05), and alanine-glycine (P < 0.05) in pooled GCs of patients were found to be significantly lower as the total FSH dose administered increased, after adjusting for the other factors in the model. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables.. Data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval
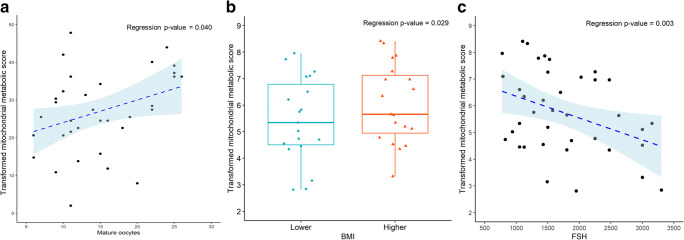
The second hypothesis tested was that a mitochondrial metabolic score generated from the aggregate metabolism of the pooled GCs would differ according to age, serum AMH levels, BMI, total FSH dosage received, and the total number and percentage of mature oocytes retrieved per patient oocyte cohort. This score reflected overall mitochondrial activity. To test this, we analyzed the mitochondrial metabolic scores using a multiple linear regression model using Box-Cox transformation examining the independent variables of age, BMI, serum AMH level, total FSH dosage, number or percentage of mature oocytes, percentage fertilization, percentage good 8-cell embryo development, and percentage blastocyst formation per patient cohort. Based on the estimated multiple linear regression model using observed data, mitochondrial metabolic scores were found to be significantly higher in granulosa of patients with higher numbers of mature oocytes retrieved (P < 0.05; Fig. 6A). Mitochondrial metabolic scores were not significant with higher percentages of mature oocytes retrieved. In a backward selected best fit model using only the BMI and total FSH dosage administered, mitochondrial metabolic scores were found to be significantly higher in pooled GCs of patients with a higher BMI (P < 0.05) (Fig. 6B), and significantly lower as the patient’s FSH dose administered increased (P < 0.01) (Fig. 6C).
Fig. 6.
Mitochondrial metabolic scores, a measure of overall mitochondrial substrate utilization, demonstrated significant differences based on BMI, total FSH dose, and the number of mature oocytes retrieved. Mitochondrial substrate utilization was based on the estimated multiple linear regression model using observed data. A) Mitochondrial metabolic scores were found to be significantly higher in GCs of patients with higher numbers of mature oocytes retrieved (P < 0.05), after adjusting for the other factors in the model. In a backward selected best fit model using only the BMI and total FSH dosage administered, B) mitochondrial metabolic scores were found to be significantly higher in pooled GCs of patients with a higher BMI (P < 0.05). BMI was divided into 2 groups with the higher group being patients with a BMI of ≥ to 26 kg/m2 (n = 20) and the lower group being patients with a BMI of <26 kg/m2 (n = 20). C) Mitochondrial metabolic scores were found to be significantly lower as the patient’s FSH dose administered increased (P < 0.01). Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. For BMI, data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) for mature oocytes retrieved and FSH dose administered, data are presented as a scatter plot of the individual patient values were lambda transformed for normalization showing the trend line and the 95% confidence interval
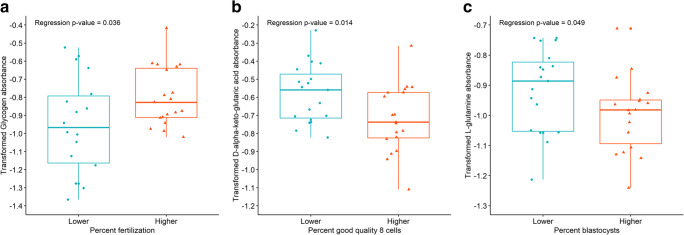
Our third hypothesis proposed that individual substrates metabolized by mitochondria in GCs of patients would differ by overall embryo cohort development. We created a multiple linear regression model using Box-Cox transformation looking at fertilization percentage, percentage good 8-cell embryo development on day 3, and percentage of blastocyst development by day 6. The three groups were divided into higher and lower groups as follows using laboratory key performance indicators: higher fertilization percentage ≥ 80%, and lower fertilization percentage < 80%; higher good 8-cell development >55% and lower good 8-cell development ≤55%; and higher percentage of blastocyst formation >55% and lower percentage of blastocyst formation ≤55%. One patient was excluded from the blastocyst formation data as she had an embryo transfer on day 3 with no additional embryos available for extended culture. Based on the estimated multiple linear regression model using observed data, mitochondrial utilization of glycogen (P < 0.05) was found to be significantly higher in pooled GCs of patients with higher fertilization percentage (Fig. 7A). Glycogen is a cytoplasmic substrate that is converted through several steps to acetyl-CoA where it enters the TCA cycle. No other substrates demonstrated utilization differences for fertilization percentage in the model (Supplemental Fig. 5). In contrast patient mitochondrial utilization of D-alpha-keto-glutaric acid (P < 0.05) was found to be significantly lower (Fig. 7B), and L-serine (P < 0.10) exhibited a higher trend in granulosa of patients with a higher percentage of good quality 8-cell embryos (Supplemental Fig. 6). In addition, granulosa mitochondrial utilization of D-Gluconate-6-phosphate (P < 0.10), and D,L-isocitric acid (P < 0.10) trended to be lower in patients with a higher percentage of blastocyst formation in pooled GCs (Supplemental Fig. 7). In a backward selected best fit model using only the percentage of good 8-cell embryos and percentage of blastocysts, GC mitochondrial utilization of L-glutamine (P < 0.05) was found to be significantly lower in patients with a higher percentage of blastocyst formation (Fig. 7C) and L-glutamine (P < 0.10) trended to be lower in pooled GC from patients with a higher percentage of good quality 8-cell embryos (Supplemental Fig. 6).
Fig. 7.
Substrates that demonstrated significant individual mitochondrial substrate utilization differences based on a patient’s embryo cohort development. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. Cohorts were grouped as follows, higher fertilization percentage ≥ 80% (n = 22), and lower fertilization percentage < 80% (n = 18); higher good 8-cell development >55% (n = 22) and lower good 8-cell development ≤55% (n = 18); and higher percentage of blastocyst formation >55% (n = 17) and lower percentage of blastocyst formation ≤55% (n = 22). One patient was excluded from the blastocyst formation data as she had an embryo transfer on day 3 with no additional embryos available for extended culture. A) Mitochondrial utilization of glycogen (P < 0.05) was found to be significantly higher in GCs of patients with higher fertilization percentage, B) mitochondrial utilization of D-alpha-keto-glutaric acid (P < 0.05) was found to be significantly lower in GCs from patients with a higher percentage of good quality 8-cell embryos, after adjusting for the other factors in the model. C) In a backward selected best fit model using only the percentage of good 8-cell embryos and percentage of blastocysts, mitochondrial utilization of L-glutamine (P < 0.05) was found to be significantly lower in GCs from patients with a higher percentage of blastocyst formation. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers)
A fourth critical hypothesis we wanted to test was that a mitochondrial metabolic score generated from the aggregate metabolism of the pooled GCs would be associated with better fertilization rate and overall embryo development. To test this hypothesis, we analyzed the patient mitochondrial metabolic score using a multiple linear regression model using Box-Cox transformation examining the independent variables of age, BMI, AMH level, total FSH dose, number or mature oocytes per cohort, percentage mature oocytes, percentage fertilization, good 8-cell embryo development, and overall blastocyst formation. Based on the estimated multiple linear regression model using observed data, mitochondrial metabolic scores were not found to be significantly different in any of the study groups.
We also analyzed the DNA content of the granulosa cell pool by demographic endpoints and the groups described above. The only significant difference in mtDNA content was found between groups with lower and higher percentage 8-cell embryos (P < 0.05). There were no differences in granulosa mitochondrial DNA content with age, BMI, serum AMH, FSH dose, or other oocyte endpoints (Supplemental Fig. 8).
Discussion
Our study is the first pilot study to evaluate a wide range of metabolic substrates in granulosa cell mitochondrial function and relate it to patient demographics, fertilization, and embryo development. The key findings of our research are summarized in Fig. 8, with the most predominant differences in substrate utilization occurring with patient BMI and cumulative FSH dose. Being a pilot study with a new assay, the sample size is small and some of the variability may be due to chance owing to the number of substrates examined. A power analysis indicated 40 patients would be sufficient to determine a difference between substrates in the assay for a pilot study. A larger prospective study will need to be conducted to determine the reproducibility of the results. We also acknowledge that our results are based on pooled granulosa cells and analysis of the embryo cohorts rather than individual oocytes/embryos which make some of our discussion speculative in nature.
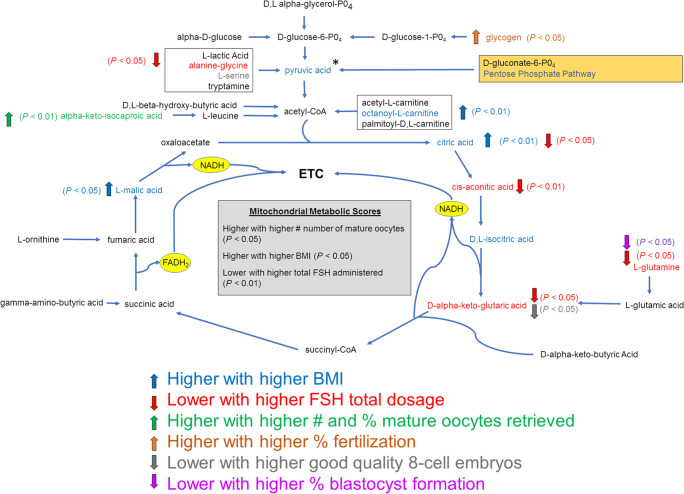
Fig. 8.
Summary diagram showing substrates of the mitochondrial function assay and where they enter the metabolic pathways and the citric acid cycle, and their association with patient and oocyte endpoints. The substrates were metabolized through different biochemical pathways by entering the mitochondria through different transporters and they were then modified by various enzymes and dehydrogenases in granulosa cells to produce NADH or FADH2. BMI = body mass index, ETC = electron transport chain, FADH2 = flavin adenine dinucleotide hydride, FSH = follicle stimulating hormone, NADH = nicotinamide adenine dinucleotide hydride, * indicates substrate in combination with 100 μM L-malic acid
Some studies have shown influences of BMI on IVF outcomes whereas others have not, however a recent metanalysis concluded that high BMI women (≥ 30 kg/m2) compared to normal BMI women (18.5–24.9 kg/m2) exhibited reduced chances of becoming pregnant through in vitro fertilization [20]. Higher BMI ≥ 26 kg/m2 (at or above the median BMI in our study) was associated with higher levels of utilization of medium chain fatty acid octanoyl-L-carnitine and the TCA cycle substrates malic acid and citric acid, as well as a higher overall mitochondrial metabolic score indicating higher overall mitochondrial metabolic activity. There were also upward trends for the usage of substrates that yielded acetyl-CoA and a second early step in the TCA cycle.
The higher substrate utilization and overall mitochondrial activity in the higher BMI group could be interpreted in a few ways. It could indicate the granulosa cells of these patients are more adapted to utilizing the energy from medium chain fatty acids to make acetyl-CoA. It could also mean that the mitochondrial enzymes in the early and late steps of the TCA cycle have higher capacity in the higher BMI group compared to lower BMI group whereas the other TCA cycle reactions are similar. Lipotoxicity caused by excessive lipid accumulation and increased intracellular free fatty acid content leads to mitochondrial dysfunction, which can compromise mitochondrial function through free fatty acid beta-oxidation and oxidative stress [21]. This idea is supported by a study that demonstrated reduced mitochondrial membrane potential in CCs with increasing patient BMI [22]. Furthermore, a recent study showed that fibroblasts from infants born to overweight compared to lean mothers had higher mitochondrial oxygen consumption rates and elevated reactive oxygen species [23]. The modulation of these substrates may indicate that patients with high BMI suffer from compromised mitochondrial function causing mitochondria to work harder in order to produce the same amount of ATP as a person with lower BMI.
Several substrates in the mitochondrial function assay and overall mitochondrial metabolic score were found to be significantly lower in GCs of patients as cumulative administered FSH dosage increased. The initial dose of FSH given was determined in part by the patient’s BMI, antral follicle count, and demographics but the total amount of FSH given was individualized based on follicular development as assessed by ultrasound and circulatory hormone assays, with higher total FSH dose often indicating poor responders [24, 25]. The total FSH dose administered demonstrated only a weak correlation (r = 0.33) with BMI, as there were several patient’s with low BMI, which had larger FSH dosages and vice versa. Our data indicate that a higher total FSH dose required to produce follicle maturation was associated with the suppressed mitochondrial activity. High total FSH dosage in women ≤38 years old has been shown to negatively impact subsequent embryo quality and blastocyst formation rate [26]. In contrast, the FSH dosage had no impact on the mitochondrial expression of a polycistronic RNA transcript in GCs of IVF patients [27].
We did not find any significant differences in mitochondrial metabolism related to age or serum AMH. The reason we may not have seen differences in these groups is that we were unable to interrogate GCs of patients with low oocyte yield, which also normally falls in the low serum AMH and older patient groups. The minimum number of GCs needed to perform the mitochondrial function assay required pooled GCs from at least 10 oocytes. Therefore, the older women and those with lower AMH levels in our study were still within a better prognostic group due to a good number of oocytes being retrieved. If we had been able to use fewer GCs for the plate assay and could have included patients with lower oocyte yield, we may have observed different results.
A secondary goal of this study was to determine if mitochondrial substrates in pooled GCs would relate to a patient’s overall egg cohort development and identify potential mitochondrial substrate biomarkers to possibly predict embryo development in future studies. The premise behind this objective was that one potential explanation for compromised oocytes, and subsequent embryo quality, has been postulated to be deficient in mitochondrial function [28]. Since mtDNA content in the oocytes is associated with mtDNA content in the GCs [4, 12, 15], we hypothesized that the mitochondrial metabolism of the GCs should reflect the mitochondrial metabolism of the oocyte and relate to an oocyte’s overall developmental competence. We performed a correlation analysis between GC mtDNA copy number and patient cycle parameters and embryo cohort development. Embryo viability is related to the amount of energy produced by the mitochondria in the oocyte and surrounding GCs as early embryogenesis demands substantial amounts of energy produced by the mitochondria to drive the cleavage divisions [29–31]. In addition, mitochondria with metabolic dysfunction may impair oocyte quality, overall embryo development, and live birth rates [32].
Mitochondrial utilization of metabolized glycogen was found to be significantly higher in patients with higher fertilization percentage, whereas utilization of individual glucose molecules did not differ by group. A possible explanation for the difference between groups could be differing levels of glycogen phosphorylase (liver type), an enzyme that breaks down glycogen present in granulosa cells. Non-human primate granulosa increase glycogen phosphorylase (liver type) mRNA levels following an ovulatory hCG bolus [33]. Additional studies would be needed to evaluate the activity and amount of this enzyme in human granulosa.
D-alpha-keto-glutaric acid utilization was lower in GCs from patients with a higher percentage of good quality 8-cell embryos compared to those with lower percentage good quality 8-cell embryos. In addition, mitochondrial utilization of L-glutamine was found to be significantly lower in pooled GCs in patients with a higher percentage of blastocyst formation when using a backward selected model looking only at 8-cell embryos and blastocysts. L-glutamine was likely converted to alpha-keto-glutarate and used the same biochemical pathway in the assay. This may indicate that embryos with better developmental potential may display a shift toward lower alpha-ketoglutarate dehydrogenase activity as shown by the decrease in L-glutamine and alpha-keto-glutarate utilization seen in cohorts with better embryo development. Increased mtDNA copy numbers in CCs have been shown to be associated with embryos that developed into high- versus low-quality embryos and embryos more likely to implant than those that did not implant [12, 15, 34]. These differentially utilized substrates may serve as potential mitochondrial biomarkers with further testing in future studies.
Mitochondrial metabolic scores significantly increased as the total number of mature oocytes retrieved increased, but scores were not different with percentage of mature oocytes retrieved. There was a weak correlation between the number of mature oocytes collected and the percent mature oocytes (r = 0.40). This discordance in scores may be due to differences in the percent mature oocytes versus the proportion of GCs from from various size follicles. Mitochondiral function may differ within follicles of various sizes but that was not assessed in this study. It would be expected that cohorts with larger oocyte numbers would have a greater range of follicle diameters corresponding to greater variability in the proportion of mature versus immature GC content per patient. Prior studies have demonstrated higher mean mtDNA levels in fertilized human oocytes compared to both unfertilized oocytes and degenerated oocytes [30, 35, 36]. In our study fertilized oocytes did not exhibit any significant differences. It is possible that a differences were not seen because we were using pooled granulosa cells instead of individual CC masses.
A strength of this study was that we were able to compare multiple substrates at once using the mitochondrial function assay. Another study strength was that we were able to assess mitochondrial function and not just mtDNA content. This study was limited by the minimum number of GCs needed to analyze each substrate in triplicate in the mitochondrial function assay. We found that the threshold concentration of cells needed was approximately 7000 cells per well requiring about 650,000 cells per plate in order to yield absorbance readings that differed from the negative control values. This criterion necessitated the pooling of the MGCs and the CCs to achieve the required number of cells per patient for the assay plate. Since we pooled the GCs this caused us to examine the embryo cohort as a whole, rather than individual embryos, which reduces the clinical utility of using this particular test to assess embryo development. The heterogeneous patient population may have impacted the results in this study as patient diagnosis has been shown to impact granulosa cell profiles in various ways (reviewed in [37]). The impact of the various diagnoses were not assessed in this pilot study due to low sample size, but may be able to be assessed in a larger, future prospective study, which may alter the correlations found here. As this was a pilot study, our goal was to determine if these novel mitochondrial utilization plates would be a viable method to identify and narrow the array of substrates down to a few which could later be optimized using an assay scaled down to analyze individual CC masses. The goal would be to relate those CC results to individual oocytes and resulting embryos in future prospective studies with a larger cohort of patients. For example, specialized assay plates could be made with only the substrates of interest in wells. Other limitations were the use of two stimulation protocols and a heterogeneous patient population. A more ideal study would include patients undergoing a single stimulation protocol and a homogeneous patient population.
The metabolic substrate utilization profile revealed in this study should be unique to granulosa cells and be reproducible with other patients by other clinics. The responsiveness of granulosa cells to gonadotropin hormones, the dynamic nature of their development, and relationship to their oocyte make granulosa different from other somatic cell types. A prior study using a wider metabolic array plate which included many of the substrates in the Mitoplate S-1 assay revealed cancer cells derived from different tissues, brown adipose cells and white adipose cells have distinct profiles unique to the cell type [38]. Furthermore, a recent study by Kuzniewska and colleagues using the same Mitoplate S-1 with murine brain tissue showed no differences between control mice and Fmr1 (fragile X mental retardation syndrome 1) knock-out mice other than succinic acid [39].
Mitochondrial function and its relationship to human reproductive potential is poorly understood, and further research needs to be conducted regarding oocyte developmental competence and embryonic development. Individual follicle CC and possibly MGC mitochondrial activity and specific substrate utilization can provide a non-invasive means of evaluating the energetic potential of its corresponding oocyte. In this study, we found mitochondrial metabolic function was altered with BMI and FSH dosage. We also identified a subset of mitochondrial metabolic substrates that can be explored as novel biomarkers related to embryo development in future studies. We identified substrates of interest related to a smaller cohorts. Octanoyl-L-carnitine, citric acid, and L-malic acid were substrates altered with BMI. Alanine-glycine, citric acid, cis-aconitic acid, D-alpha-keto-glutaric acid, and L-glutamine were substrates altered by total FSH dosage. Alpha-keto-isocaproic acid, glycogen, D-alpha-keto-glutaric acid, and L-glutamine were substrates of interest related to the number and percentage of mature oocytes retrieved, percentage fertilization, good quality 8-cell development and blastocyst formation rate, respectively.
Electronic supplementary material
Supplementary data are available at the Journal of Assisted Reproduction and Genetics online.
Plot of granulosa cell number per well versus the difference in absorbance between the positive and negative controls for the Mitoplate S-1 assays confirm the linear relationship. Data include n = 45 patients including those used in optimization experiments. Patients showing no absorbance difference between the controls at approximately 6500 cells or less were not included. (PDF 230 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on body mass index (BMI). To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation between BMI groups. BMI was divided into 2 groups with the higher group being patients with a BMI of ≥ to 26 kg/m2 (n = 20) and the lower group being patients with a BMI of <26 kg/m2 (n = 20). Mitochondrial utilization of cis-aconitic acid (P < 0.10), D,L-beta-hydroxy-butyric acid (P < 0.10), and pyruvic acid with L-malic acid 100 μM (P < 0.10) trended to be higher in pooled GCs of patients with higher BMI, after adjusting for the other factors in the model. No other substrates showed any mitochondrial utilization differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers). (PDF 8162 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based number (A) and percentage (B) of mature oocytes retrieved. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. No substrates in pooled GCs of patients demonstrated any significant mitochondrial utilization differences after adjusting for the other factors in the model. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval (PDF 8525 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on total FSH dose administered. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. Mitochondrial utilization of D,L-isocitric acid (P < 0.10), D,L-beta-hydroxy-butyric acid (P < 0.10), L-glutamic acid (P < 0.10), acetyl-L-carnitine (P < 0.10), pyruvic acid with L-malic acid 100 μM (P < 0.10) trended to be lower in pooled GCs of patients with higher administered FSH dosages, after adjusting for the other factors in the model. No other substrates showed any mitochondrial utilization differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval (PDF 6449 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based fertilization percentage. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. The two groups were delineated by higher fertilization percentage ≥ 80% (n = 22), and lower fertilization percentage < 80% (n = 18). Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) (PDF 7626 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on good 8-cell embryo development. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. The two groups were delineated by higher good 8-cell development >55% (n = 22) and lower good 8-cell development ≤55% (n = 18). Mitochondrial utilization of L-serine (P < 0.10) trended to be lower in GCs of patients with a higher percentage of good quality 8-cell embryos. In a backward selected best fit model using only the percentage of good 8-cell embryos and percentage of blastocysts, L-glutamine (P < 0.10) trended to be lower in pooled GCs from patients with a higher percentage of good quality 8-cell embryos. No other substrates showed significant differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) (PDF 7199 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on percent blastocyst formation. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. The two groups were delineated by higher percentage of blastocyst formation >55% (n = 17) and lower percentage of blastocyst formation ≤55% (n = 22). Granulosa mitochondrial utilization of D-Gluconate-6-phosphate (P < 0.10) and D,L-isocitric acid (P < 0.10) trended to be lower in patients with a higher percentage of blastocyst formation, after adjusting for the other factors in the model. No other substrates demonstrated significant differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) (PDF 7111 kb)
Pooled granulosa cell mitochondrial DNA (mtDNA) content had no significant association with patient demographics nor oocyte outcomes. Correlation analysis showed mtDNA was similar between all groups. For categorical groups, data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers). For continuous variables, data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval. 8-cells = 8-cell embryos, AFC = antral follicle count, AMH = anti-Mullerian hormone, ANT = antagonist stimulation, BMI = body mass index, FSH = follicle stimulating hormone, HMG = human menopausal gonadotropin, LL = long luteal stimulation, STIM = stimulation type (PDF 3804 kb)
Funding
Supported by an ASPIRE-I grant from the University of South Carolina.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest related to this study.
Ethical approval
“All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and in its later amendments or comparable ethical standards.” This study was approved by the Greenville Health System (IRB number: Pro00072006) and the University of South Carolina Institution Review Board (IRB registration number: 00000240).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci U S A. 1996;93(3):1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutr Metab (Lond). 2010;7:7. doi:10.1186/1743-7075-7-7. [DOI] [PMC free article] [PubMed]
- 3.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137(7):1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucret L, JM CdlB, Moriniere C, Desquiret V, Ferre-L'Hotellier V, Descamps P et al. Relationship between diminished ovarian reserve and mitochondrial biogenesis in cumulus cells. Hum Reprod. 2015;30(7):1653–64. doi:dev114 [pii];10.1093/humrep/dev114. [DOI] [PubMed]
- 5.Pawlak P, Chabowska A, Malyszka N, Lechniak D. Mitochondria and mitochondrial DNA in porcine oocytes and cumulus cells--a search for developmental competence marker. Mitochondrion. 2016;27:48–55. doi: 10.1016/j.mito.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Collado-Fernandez E, Picton HM, Dumollard R. Metabolism throughout follicle and oocyte development in mammals. Int J Dev Biol. 2012;56(10–12):799–808. doi: 10.1387/ijdb.120140ec. [DOI] [PubMed] [Google Scholar]
- 7.Su YQ, Sugiura K, Eppig JJ. Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Semin Reprod Med. 2009;27(1):32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton-McDowall ML, Gilchrist RB, Thompson JG. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction. 2010;139(4):685–695. doi: 10.1530/REP-09-0345. [DOI] [PubMed] [Google Scholar]
- 9.Dalton CM, Szabadkai G, Carroll J. Measurement of ATP in single oocytes: impact of maturation and cumulus cells on levels and consumption. J Cell Physiol. 2014;229(3):353–361. doi: 10.1002/jcp.24457. [DOI] [PubMed] [Google Scholar]
- 10.Huang Z, Wells D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod. 2010;16(10):715–25. doi:gaq031 [pii];10.1093/molehr/gaq031. [DOI] [PubMed]
- 11.Tsai HD, Hsieh YY, Hsieh JN, Chang CC, Yang CY, Yang JG, Cheng WL, Tsai FJ, Liu CS. Mitochondria DNA deletion and copy numbers of cumulus cells associated with in vitro fertilization outcomes. J Reprod Med. 2010;55(11–12):491–497. [PubMed] [Google Scholar]
- 12.Desquiret-Dumas V, Clement A, Seegers V, Boucret L, Ferre-L'Hotellier V, Bouet PE, et al. The mitochondrial DNA content of cumulus granulosa cells is linked to embryo quality. Hum Reprod. 2017;32(3):607–614. doi: 10.1093/humrep/dew341. [DOI] [PubMed] [Google Scholar]
- 13.Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, et al. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril. 2015;104(3):534–541. doi: 10.1016/j.fertnstert.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Fragouli E, Wells D. Mitochondrial DNA assessment to determine oocyte and embryo viability. Semin Reprod Med. 2015;33(6):401–409. doi: 10.1055/s-0035-1567821. [DOI] [PubMed] [Google Scholar]
- 15.Ogino M, Tsubamoto H, Sakata K, Oohama N, Hayakawa H, Kojima T, Shigeta M, Shibahara H. Mitochondrial DNA copy number in cumulus cells is a strong predictor of obtaining good-quality embryos after IVF. J Assist Reprod Genet. 2016;33(3):367–371. doi: 10.1007/s10815-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11(7):1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Racowsky C, Vernon M, Mayer J, Ball GD, Behr B, Pomeroy KO, Wininger D, Gibbons W, Conaghan J, Stern JE. Standardization of grading embryo morphology. J Assist Reprod Genet. 2010;27(8):437–439. doi: 10.1007/s10815-010-9443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderstichel R, Dohoo I, Markham F. Applying a kinetic method to an indirect ELISA measuring Ostertagia ostertagi antibodies in milk. Can J Vet Res. 2015;79(3):180–183. [PMC free article] [PubMed] [Google Scholar]
- 19.Bates D, Machler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. doi: 10.18637/jss.v067/i01.
- 20.Sermondade N, Huberlant S, Bourhis-Lefebvre V, Arbo E, Gallot V, Colombani M, Fréour T. Female obesity is negatively associated with live birth rate following IVF: a systematic review and meta-analysis. Hum Reprod Update. 2019;25(4):439–451. doi: 10.1093/humupd/dmz011. [DOI] [PubMed] [Google Scholar]
- 21.Hauck AK, Bernlohr DA. Oxidative stress and lipotoxicity. J Lipid Res. 2016;57(11):1976–1986. doi: 10.1194/jlr.R066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorshinova VK, Tsvirkun DV, Sukhanova IA, Tarasova NV, Volodina MA, Marey MV, Smolnikova VU, Vysokikh MY, Sukhikh GT. Cumulus cell mitochondrial activity in relation to body mass index in women undergoing assisted reproductive therapy. BBA Clin. 2017;7:141–146. doi: 10.1016/j.bbacli.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abraham M, Collins CA, Flewelling S, Camazine M, Cahill A, Cade WT, Duncan JG. Mitochondrial inefficiency in infants born to overweight African-American mothers. Int J Obes. 2018;42(7):1306–1316. doi: 10.1038/s41366-018-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broekmans FJ. Individualization of FSH doses in assisted reproduction: facts and fiction. Front Endocrinol (Lausanne). 2019;10:181. doi:10.3389/fendo.2019.00181. [DOI] [PMC free article] [PubMed]
- 25.Fedorcsak P, Dale PO, Storeng R, Ertzeid G, Bjercke S, Oldereid N, et al. Impact of overweight and underweight on assisted reproduction treatment. Hum Reprod. 2004;19(11):2523–2528. doi: 10.1093/humrep/deh485. [DOI] [PubMed] [Google Scholar]
- 26.Borges E, Jr, Zanetti BF, Setti AS, Braga DP, Figueira RCS, Iaconelli A., Jr FSH dose to stimulate different patient' ages: when less is more. JBRA Assist Reprod. 2017;21(4):336–342. doi: 10.5935/1518-0557.20170058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito N, Yamashita Y, Ono Y, Higuchi Y, Hayashi A, Yoshida Y, Yamamoto H, Kawabe S, Kamada M, Terai Y, Ohmichi M. Difference in mitochondrial gene expression in granulosa cells between recombinant FSH and hMG cycles under in vitro fertilization and transfer. Reprod Med Biol. 2013;12(3):99–104. doi: 10.1007/s12522-013-0147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentov Y, Yavorska T, Esfandiari N, Jurisicova A, Casper RF. The contribution of mitochondrial function to reproductive aging. J Assist Reprod Genet. 2011;28(9):773–783. doi: 10.1007/s10815-011-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.May-Panloup P, Chretien MF, Jacques C, Vasseur C, Malthiery Y, Reynier P. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod. 2005;20(3):593–597. doi: 10.1093/humrep/deh667. [DOI] [PubMed] [Google Scholar]
- 30.Reynier P, May-Panloup P, Chretien MF, Morgan CJ, Jean M, Savagner F, Barriere P, Malthiery Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod. 2001;7(5):425–429. doi: 10.1093/molehr/7.5.425. [DOI] [PubMed] [Google Scholar]
- 31.Zhao SY, Qiao J, Chen YJ, Liu P, Li J, Yan J. Expression of growth differentiation factor-9 and bone morphogenetic protein-15 in oocytes and cumulus granulosa cells of patients with polycystic ovary syndrome. Fertil Steril. 2010;94(1):261–7. doi:S0015–0282(09)00533–0 [pii];10.1016/j.fertnstert.2009.03.014 . [DOI] [PubMed]
- 32.Dumesic DA, Guedikian AA, Madrigal VK, Phan JD, Hill DL, Alvarez JP, Chazenbalk GD. Cumulus cell mitochondrial resistance to stress in vitro predicts oocyte development during assisted reproduction. J Clin Endocrinol Metab. 2016;101(5):2235–2245. doi: 10.1210/jc.2016-1464. [DOI] [PubMed] [Google Scholar]
- 33.Brogan RS, MacGibeny M, Mix S, Thompson C, Puttabyatappa M, VandeVoort CA, et al. Dynamics of intra-follicular glucose during luteinization of macaque ovarian follicles. Mol Cell Endocrinol. 2011;332(1–2):189–195. doi: 10.1016/j.mce.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taugourdeau A, Desquiret-Dumas V, Hamel JF, Chupin S, Boucret L, Ferre-L'Hotellier V, et al. The mitochondrial DNA content of cumulus cells may help predict embryo implantation. J Assist Reprod Genet. 2019;36(2):223–228. doi: 10.1007/s10815-018-1348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakoshi Y, Sueoka K, Takahashi K, Sato S, Sakurai T, Tajima H, Yoshimura Y. Embryo developmental capability and pregnancy outcome are related to the mitochondrial DNA copy number and ooplasmic volume. J Assist Reprod Genet. 2013;30(10):1367–1375. doi: 10.1007/s10815-013-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2006;85(3):584–591. doi: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Kordus RJ, LaVoie HA. Granulosa cell biomarkers to predict pregnancy in ART: pieces to solve the puzzle. Reproduction. 2017;153(2):R69-R83. doi:REP-16-0500 [pii];10.1530/REP-16-0500. [DOI] [PubMed]
- 38.Bochner BR, Siri M, Huang RH, Noble S, Lei XH, Clemons PA, Wagner BK. Assay of the multiple energy-producing pathways of mammalian cells. PLoS One. 2011;6(3):e18147. doi: 10.1371/journal.pone.0018147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuzniewska B, Cysewski D, Wasilewski M, Sakowska P, Milek J, Kulinski TM et al. Mitochondrial protein biogenesis in the synapse is supported by local translation. EMBO Rep. 2020:e48882. doi:10.15252/embr.201948882. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plot of granulosa cell number per well versus the difference in absorbance between the positive and negative controls for the Mitoplate S-1 assays confirm the linear relationship. Data include n = 45 patients including those used in optimization experiments. Patients showing no absorbance difference between the controls at approximately 6500 cells or less were not included. (PDF 230 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on body mass index (BMI). To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation between BMI groups. BMI was divided into 2 groups with the higher group being patients with a BMI of ≥ to 26 kg/m2 (n = 20) and the lower group being patients with a BMI of <26 kg/m2 (n = 20). Mitochondrial utilization of cis-aconitic acid (P < 0.10), D,L-beta-hydroxy-butyric acid (P < 0.10), and pyruvic acid with L-malic acid 100 μM (P < 0.10) trended to be higher in pooled GCs of patients with higher BMI, after adjusting for the other factors in the model. No other substrates showed any mitochondrial utilization differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers). (PDF 8162 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based number (A) and percentage (B) of mature oocytes retrieved. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. No substrates in pooled GCs of patients demonstrated any significant mitochondrial utilization differences after adjusting for the other factors in the model. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval (PDF 8525 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on total FSH dose administered. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. Mitochondrial utilization of D,L-isocitric acid (P < 0.10), D,L-beta-hydroxy-butyric acid (P < 0.10), L-glutamic acid (P < 0.10), acetyl-L-carnitine (P < 0.10), pyruvic acid with L-malic acid 100 μM (P < 0.10) trended to be lower in pooled GCs of patients with higher administered FSH dosages, after adjusting for the other factors in the model. No other substrates showed any mitochondrial utilization differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval (PDF 6449 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based fertilization percentage. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. The two groups were delineated by higher fertilization percentage ≥ 80% (n = 22), and lower fertilization percentage < 80% (n = 18). Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) (PDF 7626 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on good 8-cell embryo development. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. The two groups were delineated by higher good 8-cell development >55% (n = 22) and lower good 8-cell development ≤55% (n = 18). Mitochondrial utilization of L-serine (P < 0.10) trended to be lower in GCs of patients with a higher percentage of good quality 8-cell embryos. In a backward selected best fit model using only the percentage of good 8-cell embryos and percentage of blastocysts, L-glutamine (P < 0.10) trended to be lower in pooled GCs from patients with a higher percentage of good quality 8-cell embryos. No other substrates showed significant differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) (PDF 7199 kb)
Substrates that demonstrated no significant mitochondrial utilization differences based on percent blastocyst formation. To determine the differences in mitochondrial utilization, substrates were compared using a multiple linear regression model using Box-Cox transformation. The two groups were delineated by higher percentage of blastocyst formation >55% (n = 17) and lower percentage of blastocyst formation ≤55% (n = 22). Granulosa mitochondrial utilization of D-Gluconate-6-phosphate (P < 0.10) and D,L-isocitric acid (P < 0.10) trended to be lower in patients with a higher percentage of blastocyst formation, after adjusting for the other factors in the model. No other substrates demonstrated significant differences. Following the Box-Cox transformation of the data and observing the Q-Q plots, the most outlying observations have been removed to ensure the normality of the response variables. Data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers) (PDF 7111 kb)
Pooled granulosa cell mitochondrial DNA (mtDNA) content had no significant association with patient demographics nor oocyte outcomes. Correlation analysis showed mtDNA was similar between all groups. For categorical groups, data are presented as the median (line inside box), first and third quartile (bottom and top of the box), and highest and lowest data points (top and bottom of whiskers). For continuous variables, data are presented as a scatter plot of the individual patient values lambda transformed for normalization showing the trend line and the 95% confidence interval. 8-cells = 8-cell embryos, AFC = antral follicle count, AMH = anti-Mullerian hormone, ANT = antagonist stimulation, BMI = body mass index, FSH = follicle stimulating hormone, HMG = human menopausal gonadotropin, LL = long luteal stimulation, STIM = stimulation type (PDF 3804 kb)