Abstract
The potential of genome editing to improve the agronomic performance of crops is often limited by low plant regeneration efficiencies and few transformable genotypes. Here, we show that expression of a fusion protein combining wheat GROWTH-REGULATING FACTOR 4 (GRF4) and its cofactor GRF-INTERACTING FACTOR 1 (GIF1) substantially increases the efficiency and speed of regeneration in wheat, triticale and rice and increases the number of transformable wheat genotypes. GRF4-GIF1 transgenic plants were fertile and without obvious developmental defects. Moreover, GRF4-GIF1 induces efficient wheat regeneration in the absence of exogenous cytokinins, which facilitates selection of transgenic plants without selectable markers. We also combined GRF4-GIF1 with CRISPR-Cas9 genome editing, generating 30 edited wheat plants with disruptions in the gene Q (AP2L-A5). Finally, we show that a dicot GRF-GIF chimera improves regeneration efficiency in citrus, suggesting that this strategy can be applied to dicot crops.
Editorial summary
A method that increases plant regeneration efficiency extends gene editing to more species and genotypes
Recent studies have reported improvements in the efficiency of regenerating plants from tissue culture by overexpressing plant developmental regulators, including LEAFY COTYLEDON1 1, 2, LEAFY COTYLEDON2 3, WUSCHEL (WUS) 4, and BABY BOOM (BBM) 5. These genes promote the generation of embryo-like structures or somatic embryos, or the regeneration of shoots. For example, overexpression of the maize developmental regulators BBM and WUS2 produced high transformation frequencies from previously non-transformable maize inbred lines and other monocots species 6–8. Another strategy uses different combinations of developmental regulators to induce de novo meristems in dicotyledonous species without tissue culture 9. Still, there remains a need for new methods that provide efficient transformation, increased ease of use, and suitability for a broader range of recalcitrant species and genotypes.
GRF transcription factors are highly conserved in angiosperms, gymnosperms and moss 10. They encode proteins with conserved QLQ and WRC domains that mediate protein-protein and protein-DNA interactions, respectively 11–13. Many angiosperm and gymnosperm GRFs carry a target site for microRNA miR396, which reduces the function of GRFs in mature tissues 14. The GRF proteins form complexes with GIF cofactors that also interact with chromatin remodeling complexes in vivo 15, 16. Multiple levels of regulation control the efficiency of the assembly of functional GRF/GIF complexes in vivo 17. Loss-of-function mutations in GIF genes mimic the reduced organ size observed in GRF loss-of-function mutants or in plants overexpressing miR396 11–13, 18, 19 while overexpression of GIF promotes organ growth and can boost the activity of GRFs 12, 13, 15, 20–22. Furthermore, simultaneous increases in the expression of Arabidopsis GRF3 and GIF1 promotes larger increases of leaf size relative to the individual genes 15. Based on the observation that GRFs and GIFs interact to form a protein complex 15, we decided to evaluate the effect of a GRF-GIF chimeric protein. Here we show that expression of a sequence encoding a chimera of GRF transcription factor and its GIF cofactor substantially increases regeneration efficiency in both monocotyledonous and dicotyledonous species, increases the number of transformable cultivars and results in fertile transgenic plants.
We began by identifying 10 GRFs in the wheat genome (Supplementary Figure 1A) and selected wheat GRF4 based on its homology to OsGRF4, a rice gene that promotes grain and plant growth in rice and wheat 23–27. Among the three wheat GIF cofactors, we selected the closest homologue of Arabidopsis and rice GIF1 (Supplementary Figure 1B), because members of this clade have been shown to control growth in Arabidopsis, rice and maize 12, 13, 21, 22. We then combined GIF1 and GRF4 to generate a GRF4-GIF1 chimera including a short intergenic spacer (Figure 1A) using primers described in Supplementary Table 1 (Online Methods). Transgenic plants overexpressing the GRF4-GIF1 chimera under the maize UBIQUITIN promoter (Ubi::GRF4-GIF1, Supplementary Method 1) were fertile and showed normal phenotypes (Figure 1B). However, they exhibited a 23.9 % reduction in number of grains per spike and 13.7 % increase in grain weight (Supplementary Table 2).
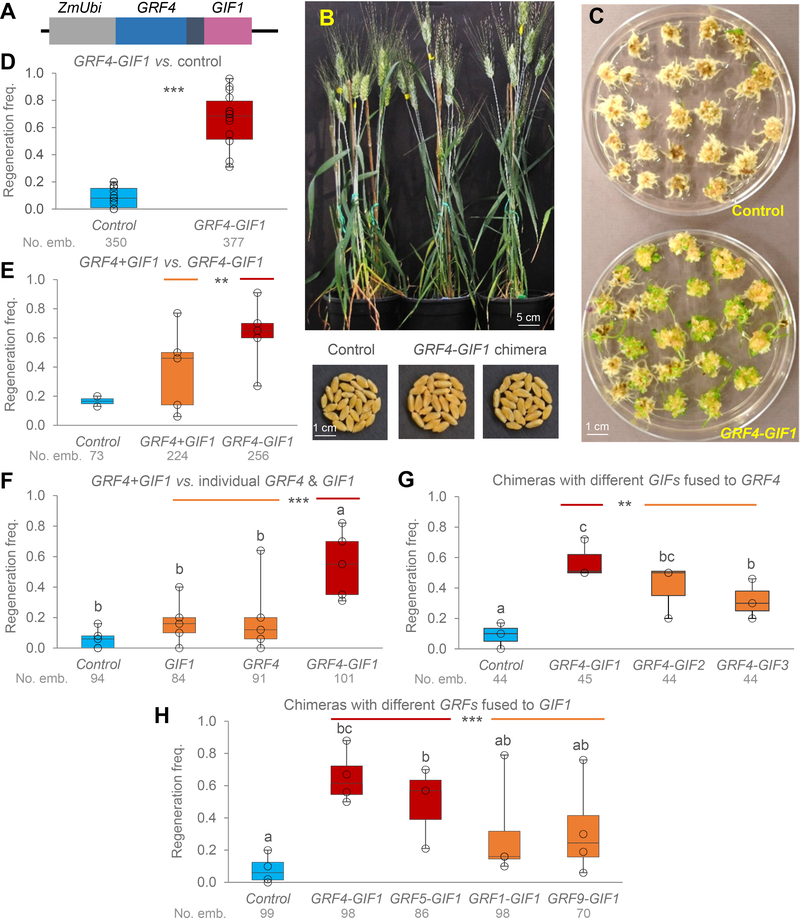
Figure 1. GRF4-GIF1 chimera.
A) Schematic representation of the GRF4 (blue)-GIF1 (pink) chimera. The black region represents a four amino acid spacer. B) The GRF4-GIF1 transgenic wheat plants were normal and fertile. C) Representative transformation showing higher frequency of regenerated shoots during Kronos transformation in the presence of the GRF4-GIF1 chimera than in the control. D-H) Box-plots showing regeneration frequencies of transgenic Kronos plants and their respective controls. The box shows the range from first to third quartiles, and is divided by the median. The whiskers span down to the minimum, and up to the maximum observation. Results from individual experiments are indicated by empty black circles. All experiments include the empty pLC41 vector as control and the wheat GRF4-GIF1 chimera. Numbers below the genotypes are total number of inoculated embryos and different letters above bars indicate significant differences (P < 0.05, Tukey test). D) Control vs. GRF4-GIF1, n= 15 experiments (*** P < 0.001, square root transformation). E) Control, GRF4-GIF1 and vector including GRF4 and GIF1 driven by separate maize UBIQUITIN promoters (GRF4+GIF1), n = 5 experiments (contrast GRF4-GIF1 vs. GRF4+GIF1, ** P = 0.0064, the empty-vector control was included only in two experiments). F) Control, GRF4-GIF1 and vectors including only GIF1 or only GRF4, n = 5 experiments (contrast GRF4-GIF1 vs. combined GRF4 & GIF1 P = 0.0007). G) Control and GRF4 chimeras fused to either GIF1, GIF2 or GIF3, n = 3 experiments (contrast chimeras with GIF1 vs. combined GIF2 and GIF3 ** P = 0.0046). H) Control and chimeras combining different wheat GRF genes fused with GIF1 (n= 4 experiments, except for GRF5 n=3). ** P = 0.0064 in contrast comparing combined GRF4-GIF1 and GRF5-GIF1 chimeras (evolutionary related) with combined GRF1-GIF1 and GRF9-GIF1 chimeras (more distantly related). In all tests, normality of residuals was confirmed by Shapiro-Wilk’s test and homogeneity of variances by Levene’s test (raw-data is available in Supplementary Table 3).
We performed 18 transformation experiments in the tetraploid wheat Kronos (Online Methods) and estimated regeneration frequencies as the number of calli showing at least one regenerating shoot / total number of inoculated embryos (Supplementary Table 3 summarizes regeneration frequencies and number of inoculated embryos). These regeneration efficiencies were used for five different comparisons using experiments as blocks (Figure 1D–H). Across 15 experiments (Supplementary Table 3), the average regeneration efficiency of the GRF4-GIF1 chimera (65.1 ± 5.0 %) was 7.8-fold higher than the empty vector control (8.3 ± 1.9 %, P < 0.001, Figure 1C and D).
We hypothesize that the increased regeneration efficiency of the GRF4-GIF1 chimera is associated with the ability of the GRF-GIF complex to regulate the transition between stem cells to transit-amplifying cells 28 and their capacity to promote cell proliferation in a broad range of organs 19. The wheat GRF4-GIF1 chimera also accelerates the regeneration process, which allowed us to develop a faster wheat transformation protocol that takes 56 d instead of the 91 d required for all the wheat experiments presented in this manuscript (Supplementary Figure 2).
We then compared the effect on regeneration efficiency of having the GRF4 and GIF1 fused in a chimera or expressed separately within the same construct by individual Ubi promoters (not fused) (Supplementary Table 3). In five different experiments, the average regeneration efficiency of the separate GRF4 and GIF1 genes (38.6 ± 12.9 %) was significantly lower (P < 0.0064) than the regeneration efficiency with the GRF4-GIF1 chimera (62.6 ± 10.3 %, Figure 1E). This result demonstrated that the forced proximity of the two proteins in the chimera increased its ability to induce regeneration.
In another five separate transformation experiments (Supplementary Table 3), we observed significantly lower regeneration efficiencies in embryos transformed with the GRF4 gene alone (20.4 ± 11.4 %) or the GIF1 gene alone (17.2 ± 6.6 %) relative to the GRF4-GIF1 chimera (54.6 ± 9.8 %, contrast P = 0.0007, Figure 1F). The regeneration efficiency of the calli transformed with the individual genes was approximately 3-fold higher than the control (6.0 ± 3.0 %) but the differences were not significant in the Tukey test (Figure 1F).
We generated chimeras in which GIF1 was replaced by other GIFs or GRF4 was replaced by other GRFs, and tested their regeneration efficiency in three and four separate experiments, respectively (Supplementary Table 3). The GRF4-GIF1 combination resulted in higher regeneration efficiency than the GRF4-GIF2 and GRF4-GIF3 combination (contrast P = 0.0046), and all three chimeras showed higher regeneration efficiency than the control (Tukey test P < 0.05, Figure 1G). Similarly, the regeneration efficiency induced by chimeras including the closely related GRF4 and GRF5 genes fused with GIF1, was higher than the regeneration observed for chimeras including the more distantly related GRF1 and GRF9 genes fused with GIF1 (contrast P= 0.0064, Figure 1H). Only the chimeras including the GRF4 and GRF5 genes were significantly different from the control (Tukey P < 0.05, Figure 1H).
We then tested the potential of the GRF4-GIF1 chimera to generate transgenic plants from commercial durum, bread wheat and a Triticale line that were recalcitrant to Agrobacterium-mediated or had low regeneration efficiency in previous experiments at the UCD Plant Transformation Facility. With the GRF4-GIF1 chimera we observed high increases in regeneration frequencies in tetraploid wheat Desert King (63.0 ± 17.0 % vs. 2.5 ± 2.5 %, 2 experiments) and hexaploid wheat Fielder (61.8 ± 8.2 % vs. 12.7 ± 10.3 %, three experiments) relative to the control. For the hexaploid wheat varieties Hahn and Cadenza and the Triticale breeding line UC3190, for which we were not able to generate transgenic plants using the Japan Tobacco protocol, we observed regeneration frequencies of 9 to 19 % with the GRF4-GIF1 chimera (versus 0 % with the control, Supplementary Figure 3 and Supplementary Table 4A and B).
High wheat regeneration efficiencies have been reported before using the proprietary Japan Tobacco method in the variety Fielder 29, 30, 31. However, the company warns that these high values require the optimization of multiple factors with narrow optimal windows and that “those values can drop drastically when one of the factors become suboptimal” 29 (Supplementary Table 5). The addition of the GRF4-GIF1 chimera overcame some of the constrains imposed by these narrow optimal windows and allowed us to obtain high transformation efficiencies using a shorter protocol and embryos of a wider range of sizes (1.5 to 3.0 mm) obtained from plants grown in diverse environmental conditions. High regeneration efficiencies were observed even when we used different vectors and genotypes and without embryo excision, a critical step in the Japan Tobacco technology29.
To test the robustness of our method, we transferred our GRF4-GIF1 vector to the John Innes Centre Transformation facility for testing with their recently published wheat transformation method 32. Fielder plants transformed with the GRF4-GIF1 chimera showed a 77.5% regeneration efficiency, compared with 33.3% in the control (Supplementary Table 4A). Taken together, these results indicate that the addition of the GRF4-GIF1 chimera increases the robustness of wheat transformation under different conditions and protocols.
We also tested the wheat GRF4-GIF1 chimera in the rice variety Kitaake (Online Methods). In four independent transformation experiments, we observed a 2.1-fold increase in rice regeneration efficiency (P < 0.00001) in the calli transformed with the wheat GRF4-GIF1 chimera (average 42.8 ± 2.6 %) compared with those transformed with the control vectors (20.3 ± 2.9 %, Supplementary Table 6). These results suggest that the wheat GRF4-GIF1 chimera is effective in enhancing regeneration in another agronomically important monocotyledonous species.
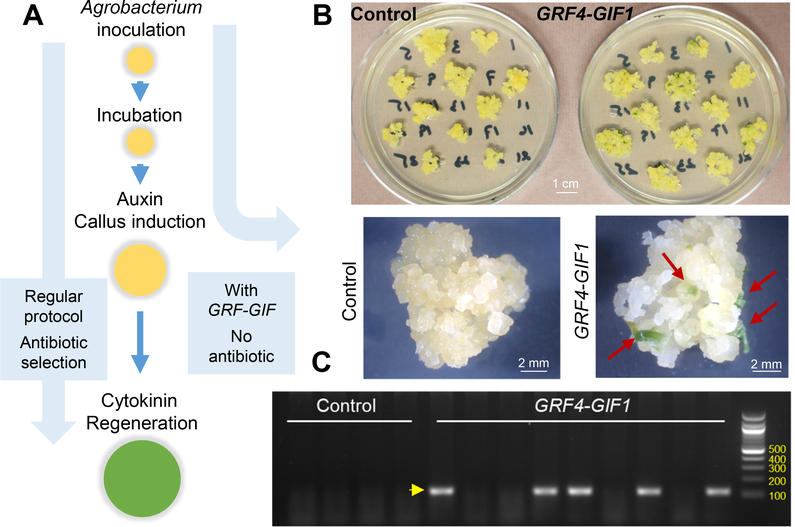
In many plant transformation systems cytokinins are required to regenerate shoots (Figure 2A). Notably, in both laboratories we observed that Kronos and Fielder embryos inoculated with Agrobacterium transformed with the GRF4-GIF1 chimera were able to rapidly regenerate green shoots in auxin media without cytokinin (Figure 2B). We then tested the regeneration efficiency of immature embryos from stable GRF4-GIF1 transgenics (n=27) and non-transgenic (n=26) T1 sister lines in the absence of cytokinin and hygromycin. Under these conditions, the regeneration efficiency of the GRF4-GIF1 transgenic plants (77.8 %) was significantly higher than the non-transgenic sister lines (11.5 %, Supplementary Figure 4). These results indicated that the GRF4-GIF1 chimera can promote either embryogenesis, shoot proliferation, or both, in wheat without the addition of exogenous cytokinin.
Figure 2. The GRF4-GIF1 chimera induces embryogenesis in the absence of cytokinins.
A) Schematic representation of the different steps of wheat transformation. B). Representative calli in auxin media with no hygromycin. Note growing green shoots in callus transformed with the wheat GRF4-GIF1 chimera in the absence of cytokinins (red arrows). Control: pLC41. C) Transgenic specific PCR product (yellow arrow) amplified with primers pLC41–1064 and pLC41–1061 (Supplementary Table S1). In this first experiment (out of three), we identified five transgenic plants among nine regenerated from the GRF4-GIF1 marker-free vector and no transgenic plants among four regenerated from the control.
Based on the previous result, we developed a protocol to select transgenic shoots in auxin media without using antibiotic-based markers. We recovered 40 shoots using a GRF4-GIF1 marker-free vector and 15 for the empty vector across three experiments. Genotyping revealed that 10 out of the 40 (25 %) GRF4-GIF1 shoots were transgenic, while none of the shoots from the control was positive (Figure 2C presents results from the first experiment). These high-regenerating transgenic plants overexpressing the GRF4-GIF1 chimera without selection markers could potentially be used for future transformation experiments to incorporate other genes using selectable markers. This approach could generate separate insertion sites for the GRF4-GIF1 and the second transgene, facilitating the segregation of the GRF4-GIF1 insertion in the next generation.
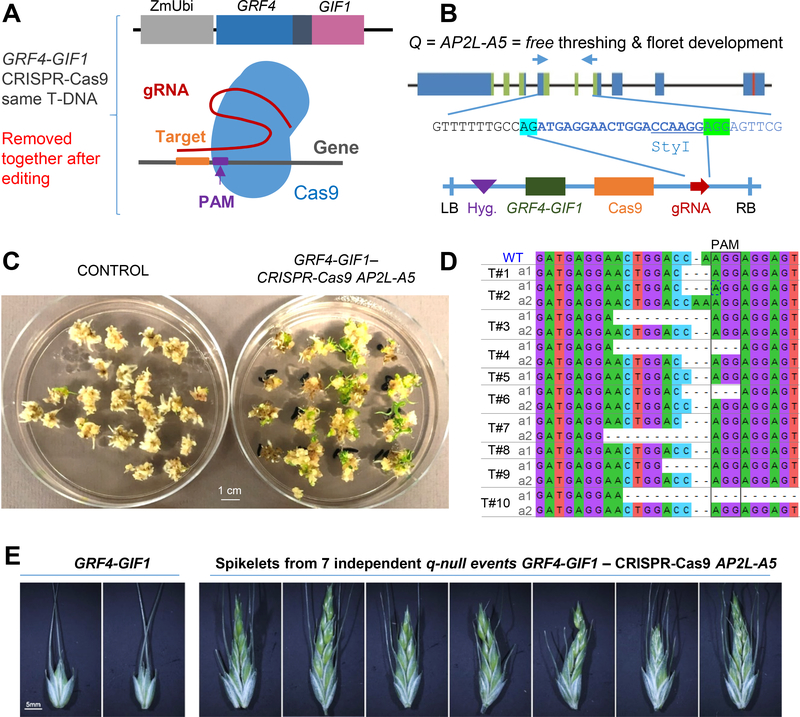
This strategy is not necessary for genome editing, since both the CRISPR-Cas9 and GRF4-GIF1 sequences can be segregated out together after editing the desired region of the genome. Therefore, the GRF-GIF system is ideal to extend genome editing technology to crops with low regeneration efficiencies. As a proof of concept, we generated a binary vector for Agrobacterium transformation that contained a cassette including the GRF4-GIF1 chimera, Cas9 and a gRNA targeting the wheat gene Q (= AP2L-A5) 33 in the same T-DNA (Figure 3A and B). We recovered 30 independent transgenic events out of 32 infected calli (93.7% efficiency, Figure 3C). Disruption of a StyI restriction sites showed Cas9-induced editing in all 30 transgenics (Supplementary Figure 5). We sequenced the PCR products obtained from 10 independent lines and confirmed editing (Figure 3D). Of the ten edited T0 plants transferred to soil, seven showed clear mutant q-null phenotypes (Figure 3E) and the other three died before heading. These T0 transgenic plants were fertile, and the edited Q gene and the CRISPR-Cas9 / GRF4-GIF1 construct are expected to segregate in the T1 progeny, facilitating the selection of edited plants without the transgene.
Figure 3. High frequency of genome edited plants using combined GRF4-GIF1 – CRISPR-Cas9 technology.
A) Technologies combined in a single vector. B) Region of the gene Q (AP2L-A5) targeted with the guide RNA (gRNA) and schematic representation of the vector combining both technologies (LB = left border, Hyg. = hygromycin resistance, RB = right border). C) Kronos shoot regeneration of embryos transformed with an empty vector and with the combined GRF4-GIF1 - CRISPR-Cas9-gRNA-AP2L-A5 construct (93.7 % regeneration efficiency). D) All 10 sequenced transgenic T0 plants showed AP2L-A5 editing. Seven of the 10 plants (T#1 to T#10) carried two different mutations (a1 and a2), documenting high editing efficiency. E) Edited T0 plants showed increased number of florets per spikelet (characteristic of q-null plants).
Lastly, we performed a series of Citrus transformation experiments to test the effect of the GRF-GIF technology in a dicot crop with limited regeneration efficiency and organogenic-based transformation protocols. We generated a citrus and a heterologous grape GRF-GIF chimera using the closest homologs to wheat GRF4 and GIF1 in both species (Supplementary Figure 1A and B). In three independent transformation experiments in the citron rootstock Carrizo (Online Methods), epicotyls were transformed with the citrus and the grape GRF-GIF chimeras. Epicotyls transformed with the citrus GRF-GIF chimera showed a 4.7-fold increase in regeneration frequency relative to those transformed with the empty vector control (Supplementary Figure 6A). The heterologous grape GRF-GIF chimera produced similar increases in citrus regeneration efficiency as the citrus chimera (Supplementary Figure 6B).
We also tested the effect of a miR396-resistant grape GRF-GIF version (henceforth, rGRF-GIF), in which we introduced silent mutations in the GRF binding site for miR396 to avoid cleavage (Supplementary Figure 6B and C). In three independent experiments, we observed that the grape rGRF-GIF chimera produced the highest frequency of transgenic citrus events (7.4-fold increase compared to the control, P < 0.05). A statistical analysis comparing the control versus the three combined GRF-GIF constructs was also significant (P = 0.0136, Supplementary Figure 6D and Supplementary Table 7). In spite of its higher-regeneration frequency, the rGRF-GIF construct would require additional optimization (e.g. an inducible system) because some of the transgenic events produced large calli that were unable to generate shoots (Supplementary Figure 6B).
In summary, expression of a GRF4-GIF1 chimera increased significantly the efficiency and speed of wheat regeneration and the ability to generate large numbers of fertile edited plants, extended the range of transformable genotypes and eliminated the requirement of cytokinin for regeneration, thereby eliminating the need of antibiotic-based selectable markers. The GRF4-GIF1 technology results in fertile and normal transgenic plants without the need of specialized promoters or transgene excision, overcoming some of the limitations of transformation technologies with other morphogenic genes (Supplementary Table 8). Because GRF4-GIF1 likely operates at a later stage of meristem differentiation and stem cell proliferation 28 than Bbm-Wus2 6–8, there is potential to combine both technologies and have synergistic effects in the regeneration efficiency of recalcitrant genotypes. A concurrent and independent work showed that overexpression of Arabidopsis AtGRF5 and AtGRF5 homologs positively enhance regeneration and transformation in monocot and dicot species not tested here 34. We hypothesize that the benefits of the GRF4-GIF1 technology can be rapidly extended to other crops with low regeneration efficiencies by incorporating the GRF4-GIF1 chimera into current protocols. This hypothesis is supported by the high conservation of the GRF and GIF proteins across the plant kingdom and by the higher regeneration frequencies observed for rice and citrus in this study.
Methods
Wheat vectors
We performed all PCRs cloning with Phusion High-Fidelity DNA Polymerase (NEB). We extracted RNA extracted from spikes using the Spectrum Plant Total RNA Kit (Sigma-Aldrich), treated with RQ1 RNase-free DNase (Promega), and then synthesized the cDNA using SuperScript II Reverse Transcriptase (Invitrogen). To clone the coding region of wheat GRF4 and GIF1, we performed PCRs using cDNA generated from Kronos spike. The sequence of the primers specific for GRF4 (Fw-GRF4a/Rev-GRF4a) and GIF1 (Fw-GIF1a/Rev-GIF1a) are indicated in Supplementary Table 1. We first cloned the PCR fragments in pDONR by a B/P gateway reaction and generated the GRF4-GIF1 chimera by overlapping PCR.
In the first step, we amplified the GRF4 and GIF1 coding sequences with primers FW-GRF4a/Rev-GRF4b and Fw-GIF1b/Rev-GIF1b from the pDONR-GRF4 and pDONR-GIF1 clones. The primer Rev-GRF4b generates a 3’ end that overlaps 12 nucleotides with the 5’ end of Fw-GIF1b. Those 12 nucleotides generate a bridge of four alanine amino acids between GRF4 and GIF1. We gel-purified both PCR fragments and used them as template in a second PCR with the primers Fw-GRF4/Rev-GIF1b (Supplementary Table 1). We cloned the resulting product in pDONR. Next, we cloned the GRF4, the GIF1 and the chimera GRF4-GIF1 chimera the binary vector pLC41 by a L/R gateway reaction under the maize UBIQUITIN promoter. We verified the resulting vectors for the individual genes pLC41:GRF4 and pLC41:GIF1, and for the chimera pLC41:GRF4-GIF1 by restriction digestion, and transformed them by electroporation in Agrobacterium strain EHA105 and in a few experiments in strain AGL1 (Supplementary Table 4). Both strains were handled in the same way.
To develop the vector expressing both GRF4 and GRF1 under their own promoters (not fused, Ubi::GRF4-term and Ubi::GIF1-term), we amplified the complete Ubi::GRF4-term cassette by PCR using pLC41:GRF4 as template with primers Fw_HindIII and Rev-HindIII (Supplementary Table 1). We cloned the PCR fragment in pGEMT-easy and then sub-cloned the Ubi::GRF4-term fragment into the HindIII site of pLC41:GIF1.
To generate the different wheat GRF-GIF chimeras, we obtained the coding sequences of GRF1, GRF5, GRF9, GIF2 and GIF3 by gene synthesis. Then, we generated the different chimeras (GRF1-GIF1, GRF5-GIF1, GRF9-GIF1, GRF4-GIF2, and GRF4-GIF3) by overlapping PCR following the same strategy described to generate GRF4-GIF1. All the chimeras were cloned in pLC41 vector by L/R reaction. We verified all the vectors by restriction digestion and transformed by electroporation in Agrobacterium strain EHA105.
To develop the JD635-GRF4-GIF1-Cas9- gRNA-Gene Q vector, we amplified by PCR a cassette including the maize UBIQUITIN promoter, the GRF4-GIF1 chimera and the Nos terminator (primers Fw_ZmUbi-AscI and Rev_NosTerm-AscI). The PCR product was gel-purified and cloned by In-fusion (Takara Bio USA, Inc.) into the AscI site of the pYP25F binary vector, which contains a wheat codon optimized Cas9 (TaCas9) with two nuclear localization signals (NLS), and is a modified version of pDIRECT_25F (https://www.addgene.org/91143/) from Dr. Daniel Voytas group. We validated the vector sequence by Sanger sequencing. Next, we cloned a guide RNA construct targeting the coding region of Gene Q 35 by Golden Gate reaction into two AarI sites of the vector and transformed it into chemical competent E. coli DH5α. We validated the JD635-GRF4-GIF1-Cas9-gRNA-Gene Q vector by Sanger sequencing and transformed by electroporation into Agrobacterium strain EHA105.
Citrus and Vitis vectors
We generated the Citrus and Vitis GRF-GIF chimeras by gene synthesis using the GRF and GIF homologs highlighted in Supplementary Figure 1. We cloned the DNA fragments into pDONR by B/P gateway reaction. We cloned the GRF-GIF chimeras in the binary vector pGWB14 binary vector (L/R gateway reaction under viral 35S promoter) and transformed them by electroporation in Agrobacterium strain EHA105.
We generated a miR396-resistant version of Vitis GRF-GIF (rGRF-GIF) by overlapping PCR. Two PCR reactions were performed with primers Fw-GRF/rGRF-Rev and rGRF-Fw/Rev-GIF (Supplementary Table 1) using pGBW14-vitis GRF-GIF clone as template. The primers rGRF-Fw and rGRF-Rev overlap in 17 nucleotides, and introduce silent mutations in the miR396 target site (Supplementary Figure 6). We gel-purified both PCR fragments and used them as template in a second PCR with the primers Fw-GRF/Rev-GIF (Supplementary Table 1). We cloned the resulting product in pDONR by B/P gateway reaction. Next, we cloned the chimera rGRF-GIF in the binary vector pGWB14 by a L/R gateway reaction under the viral 35S promoter and transformed them by electroporation in Agrobacterium strain EHA105.
Wheat transformation
Wheat transformation followed previously published protocols 29. Briefly, we grew the different wheat and triticale cultivars in a green house or a growth chamber under long-day photoperiod (16 h of 380 μM m−2 s−1 light, 26 °C day and 18 °C night). We harvested immature grains from spikes approximately 2 weeks after anthesis, and surface sterilized for 1 minute in 70 % ethanol followed by 10 minutes in 1.2% (v/v) sodium hypochlorite solution plus 5 μl tween. After surface sterilization, we washed the seeds three times with sterilized water and isolated immature embryos under stereoscopic microscope (embryo sizes 1.5 to 3.0 mm).
We centrifuged the isolated immature embryos in liquid medium and then inoculated with Agrobacterium. We transferred the embryos to co-cultivation medium with the scutellum-side up and incubated at 23 °C in the dark. After 2–3 days, we excised the embryo axis, and transferred them to callus induction medium without selection, where we incubated them at 25 °C in the dark. After 5 days, we transferred the embryos to selection medium with 30 mg/l of hygromycin and incubated them at 25 °C in the dark.
After 3 weeks, we transferred calli to selection medium that contained 100 mg/l of hygromycin. After an additional 3 weeks, we transferred the proliferating tissue to regeneration medium containing 50 mg/l of hygromycin and incubated them at 25 °C under continuous light (30 μM m−2 s−1) for 2 weeks. We transferred the regenerated shoots into rooting medium contained 50 mg/l of hygromycin. Rooted plants were acclimated to soil by transferring them to a 1020 tray containing a 36 sheet inserts filled with Sunshine potting mix and covered with an 11 × 21 × 2 inch clear plastic dome for 10 days under 16 hour of 100 μM light and 26 °C. More recently we developed a shorter transformation protocol to generate GRF4-GIF1 transgenic wheat plants that is summarized in Supplementary Figure 2. Transformation at the John Innes Centre was performed as published before 32.
Rice transformation
Rice transformation followed previously published protocols 36. Briefly, we selected fresh rice seeds, de-husked them and surface sterilized them in a rotating flask containing 20 % (v/v) bleach for 30 min. Then, rinsed the seeds 3 times with sterile water. We placed about 25–50 seeds per plate on callus induction media (MSD, 1x Murashige and Skoog with vitamins medium containing 30 g/l sucrose, 2 mg/l 2,4-dichlorophenoxyacetic acid, 1.2% (w/v) agar, pH 5.6–5.8) without letting the embryo touch the media, wrapped plate with surgical tape end incubated under 16 h light/ 8 h dark at 28 °C. After 10–14 d, we separated the callus from the rest of the germinating seed and transferred to fresh MSD agar plates for another 5 d before co-cultivation.
Agrobacterium culture
We prepared a glycerol freezer stock from a single bacterial colony isolated from a plate. We then inoculated 1 ml LB containing the appropriate antibiotics to maintain the Agrobacterium and the plasmid, and we incubated it overnight at 28 °C at 250 rpm. The following day we added 300 μl of the Agrobacterium culture to 20 ml TY (pH 5.5) containing the appropriate antibiotics and 200 μM acetosyringone. We incubated the culture 28 °C for in a shaking incubator set at 250 rpm until the culture reached an OD600 between 0.1 – 0.2 (approximately 2–4 h).
Transformation and co-cultivation
We placed the calli in Agrobacterium suspension for 30 min, and shook the suspension to ensure uniform access to the calli. After the shaking incubation, we dried the calli on sterile Whatman paper to remove excess bacterial suspension. We transferred the calli onto co-cultivation medium (MSD + S + AS, 1x Murashige and Skoog with vitamins medium containing 30 g/l sucrose, 5% sorbitol, 2mg/l 2,4-dichlorophenoxyacetic acid, 200 μM acetosyringone, 1.6 % (w/v) agar, pH 5.6–5.8) and incubated for 3 d in the dark at 22 °C.
Selection
We transferred the co-cultivated calli to selection media (MSD + CH + PPM, 1x Murashige and Skoog with vitamins medium containing 30 g/l sucrose, 2 mg/l 2,4-dichlorophenoxyacetic acid, 400 mg/L carbenicillin, 200 mg/l timentin, 1ml/l Plant Preservative Mixture, 80 mg/l hygromycin, 1.2% agar, pH 5.6–5.8 ) and incubated the plates under continuous light at 28 °C. We subcultured these calli onto fresh selection media every 8–9 d.
Regeneration and Rooting
After 4–5 weeks on selection media, resistant micro-calli of approximately 2– 5 mm wide started to appear. We picked these off the original callus and transferred them to Petri dishes with regeneration media (BN + S + CH, 1x Murashige and Skoog with vitamins medium containing 30 g/l sucrose, 5 % sorbitol, 3 mg/l BAP, 0.5 mg/l NAA, 400 mg/l carbenicillin, 200 mg/l timentin, 1 ml/l Plant Preservative Mixture, 50 mg/l hygromycin, 1.6 % (w/v) agar, pH 5.6–5.8), and incubated under continuous light at 28 °C. We subculture these calli onto fresh regeneration media every 8–9 days. After 4–5 weeks, the calli that started to turn green, were transferred to regeneration media with reduced hygromycin (BN + S + CH, 1x Murashige and Skoog with vitamins medium containing 30 g/l sucrose, 5 % sorbitol, 3 mg/l BAP, 0.5 mg/l NAA, 400 mg/l carbenicillin, 200 mg/l timentin, 1 ml/l Plant Preservative Mixture, 25 mg/l hygromycin, 1.6 % (w/v) agar, pH 5.6–5.8). When the shoot was properly developed, we transferred the regenerated plants to rooting media (MS + H, 1x Murashige and Skoog with vitamins medium containing 25 mg/l hygromycin, 1.2 % (w/v) agar, pH 5.6–5.) and incubated in 16 h light/ 8 h dark 28 °C. When roots were well developed, we transferred the plants to soil.
Citrus Transformation
We placed seeds of Carrizo citrange rootstock in water to imbibe and then peel off the seed coats making sure not to remove the integument. We surface sterilized seeds in 0.6% (v/v) sodium hypochlorite solution plus 5-μl tween 20 by placing them in a 50 ml centrifuge tube and shaking at 100 rpms for 20 minutes. We rinse the seeds 3x in 150–200 ml of sterile distilled water. We placed seeds on agar solidified 1/2x Murashige and Skoog minimal organics medium (1/2x MSO) containing 15 g/l sucrose, 7 gm TC agar (pH 5.6–5.8), and push seeds slightly into the medium for more uniform germination. Incubate in the dark at 26 °C.
Agrobacterium culture
We prepared a glycerol freezer stock from a single bacterial colony isolated from a plate. We then used 40 μl of the stock to inoculate 20 ml of MGL medium (pH 7.0) containing the appropriate antibiotics to maintain the Agrobacterium and the plasmid, and we incubated overnight at 28 °C at 250 rpm. The following day, we removed 5 ml of the overnight growth and transferred it to 15 ml of TY medium (pH 5.5) containing the appropriate antibiotics and 200 μM acetosyringone. We incubated the culture overnight at 28 °C at 250 rpm and then diluted the overnight culture grown in TY medium to an O.D 600 nm of 0.1 to 0.2.
Co-cultivation
We collected 2–5 week old etiolated epicotyls and place in a petri dish containing 10 ml of the Agrobacterium solution prepared above (0.1–0.2 OD 600). We cut submerged epicotyls into 0.5 cm sections and soak for 10 min. We transferred the epicotyl sections onto co-cultivation medium consisting of Murashige and Skoog minimal organics medium (MSO) modified with 30 g/l sucrose 3.0 mg/l BAP, 0.1 mg/l NAA, and 200-μM acetosyringone pH 5.6–5.8. Incubate at 23 °C in the dark.
Induction
After 2–3 days, we transferred the epicotyl pieces to induction medium consisting of MSO modified with 30 g/l sucrose, 3.0 mg/l BAP, 0.1 mg/l NAA, 400 mg/l carbenicillin, 150 mg/l timentin and 100mg/l kanamycin sulfate, and incubated them in the dark. After 10 days, we subcultured the epicotyl sections to fresh medium of the same formulation and then subcultured them every 21 d. After the second 21-day cycle in the dark, we transferred cultures to light under a 30 μM light and a photoperiod of 16 h light 8 h dark. We continued to transfer every 21 d to fresh medium of the same media until organogenic shoot buds develop at the cut ends.
Elongation
Once shoots began to form, we transferred the developing shoots to elongation medium consisting of MSO modified with 30 g/l sucrose, 0.1mg/l BA, 400 mg/l carbenicillin, 150 mg/l timentin, and 100 mg/l kanamycin sulfate. We incubated as above and subcultured the cultures every 21 d as needed until shoots elongated.
Rooting
Once a shoot reached 2–4 cm in height, we harvested the shoots and transferred them to rooting medium consisting of MSO modified with 30 g/l sucrose, 5 mg/l NAA, 250 mg/l cefotaxime, and 100 mg/l kanamycin. After three to five days, we transferred shoots to MSO modified with 30 g/l sucrose, 0.0 mg/l NAA, 400 mg/l carbenicillin and 100 mg/l kanamycin. Shoots started rooting in 14 days.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability statement
Accession numbers and gene names are available in the phylogenetic tree in Supplementary Figure 1. All wheat gene names are based on genome release RefSeq v1.0. The raw data for the different experiments is available in Supplementary Tables 3–4 and 6–7. The methods for the generation of the different vectors and the transformation protocols are described in Online Methods. The following vectors will be available through Addgene (http://www.addgene.org): JD553 - wheat GRF4-GIF1 in pDONR, JD633 - wheat GRF4-GIF1 in CRISPR vector, JD630 - Vitis GRF4-GIF1 in pDONR, JD638 - Vitis miR396-resistant GRF4-GIF1 in pDONR, JD689 - citrus GRF4-GIF1 in pDONR, JD690 - citrus GRF4-GIF1 in pGWB14, JD631 - Vitis GRF4-GIF1 in pGWB14, and JD639 - Vitis miR396-resistant GRF4-GIF1 in pGWB14.
Supplementary Material
Acknowledgements
This project was supported by the Howard Hughes Medical Institute, NRI Competitive Grant 2017-67007-25939 from the USDA National Institute of Food and Agriculture (NIFA) and the International Wheat Partnership Initiative (IWYP). J.F.P. acknowledges support from the Argentinean Research Council (CONICET) and Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación. P.C.R. was supported by NIH grant #GM122968. S.H. acknowledges support from the Biotechnology and Biological Sciences Research Council Genes in the Environment Institute Strategic Programme BB/P013511/1. J.M.D. was supported by a fellowship (LT000590/2014-L) of the Human Frontier Science Program. M.F.E. is a Latin American Fellow in the Biomedical Sciences, supported by the Pew Charitable Trusts. We thank Y. Wang for the pYP25F binary vector and M. Padilla, R. Rasia, G. Rabasa, B. Van Bockern, and M. Smedley for excellent technical support, and to C. Uauy for coordinating the testing of the GRF4-GIF1 chimera at the John Innes Centre.
Footnotes
Competing interest statement
JFP and JMD are co-inventors in patent US2017/0362601A1 that describes the use of chimeric GRF-GIF proteins with enhanced effects on plant growth (Universidad Nacional de Rosario Consejo Nacional de Investigaciones Científicas y Técnicas). JFP, JD, DMT and JMD are co-inventors in UC Davis provisional patent application 62/873,123 that describes the use of GRF-GIF chimeras to enhance regeneration efficiency in plants. Vectors are freely available for research, but commercial applications may require a paid non-exclusive license. There is a patent application from KWS/BASF (WO 2019 / 134884 A1) for improved plant regeneration using Arabidopsis GRF5 and grass GRF1 homologs. None of the authors of this manuscript is part of the KWS/BASF patent or is related to these companies. The KWS/BASF patent focuses on a different cluster of GRF genes than the one described in our study and does not incorporate the GIF1 cofactor or the generation of GRF-GIF chimeras.
References
- 1.Lotan T et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Lowe K et al. in Plant Biotechnology 2002 and Beyond (Proc. 10th IAPTC&B Congress) (ed. Vasil IK) 283–284 (Orlando, Florida, U.S.A; 2003). [Google Scholar]
- 3.Stone SL et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A 98, 11806–11811 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuo JR, Niu QW, Frugis G & Chua NH The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30, 349–359 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Boutilier K et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon-Kamm B et al. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants-Basel 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe K et al. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 54, 240–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe K et al. Morphogenic regulators Baby boom and Wuschel improve Monocot transformation. Plant Cell 28, 1998–2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher MF et al. Plant gene editing through de novo induction of meristems. Nat Biotechnol 38, 84–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omidbakhshfard MA, Proost S, Fujikura U & Mueller-Roeber B Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Choi DS & Kende H The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Kim JH & Kende H A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 101, 13374–13379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horiguchi G, Kim GT & Tsukaya H The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43, 68–78 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Debernardi JM, Rodriguez RE, Mecchia MA & Palatnik JF Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debernardi JM et al. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J 79, 413–426 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Vercruyssen L et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liebsch D & Palatnik JF MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol 53, 31–42 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Li SC et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J 14, 2134–2146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez RE et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He ZS et al. OsGIF1 positively regulates the sizes of stems, leaves, and grains in rice. Front. Plant. Sci 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimano S et al. Conserved functional control, but distinct regulation, of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-INTERACTING FACTOR 1 orthologs. Development 145 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Zhang D et al. GRF-Interacting Factor1 regulates shoot architecture and meristem determinacy in maize. Plant Cell 30, 360–374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan P et al. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2, 15203 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Hu J et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Che RH et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2, 15195 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Sun PY et al. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integ. Plant Biol 58, 836–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez RE et al. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27, 3354–3366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida Y, Hiei Y & Komari T in Proceedings of the 12th International Wheat Genetics Symposium (eds. Ogihara Y, Takumi S & Handa H) 167–173 (Springer, 2015). [Google Scholar]
- 30.Richardson T, Thistleton J, Higgins TJ, Howitt C & Ayliffe M Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tiss. Organ Cult 119, 647–659 (2014). [Google Scholar]
- 31.Wang K, Liu HY, Du LP & Ye XG Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J 15, 614–623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayta S et al. An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods 15, 121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debernardi JM, Lin H, Chuck G, Faris JD & Dubcovsky J microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144, 1966–1975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong J et al. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant. Sci In press. bioRxiv preprint doi: 10.1101/2020.08.23.263947. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 35.Wang W, Akhunova A, Chao S & Akhunov E Optimizing multiplex CRISPR/Cas9-based genome editing for wheat. bioRxiv doi: 10.1101/051342 (2016). [DOI] [Google Scholar]
- 36.Chern MS et al. Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J 27, 101–113 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers and gene names are available in the phylogenetic tree in Supplementary Figure 1. All wheat gene names are based on genome release RefSeq v1.0. The raw data for the different experiments is available in Supplementary Tables 3–4 and 6–7. The methods for the generation of the different vectors and the transformation protocols are described in Online Methods. The following vectors will be available through Addgene (http://www.addgene.org): JD553 - wheat GRF4-GIF1 in pDONR, JD633 - wheat GRF4-GIF1 in CRISPR vector, JD630 - Vitis GRF4-GIF1 in pDONR, JD638 - Vitis miR396-resistant GRF4-GIF1 in pDONR, JD689 - citrus GRF4-GIF1 in pDONR, JD690 - citrus GRF4-GIF1 in pGWB14, JD631 - Vitis GRF4-GIF1 in pGWB14, and JD639 - Vitis miR396-resistant GRF4-GIF1 in pGWB14.