Abstract
Purpose:
The recent approval of anti-PD-L1 immunotherapy in combination with nab-paclitaxel for metastatic triple-negative breast cancer (TNBC) highlights the need to understand the role of chemotherapy in modulating the tumor-immune microenvironment (TIME).
Experimental Design:
We examined immune-related gene expression patterns before and after neoadjuvant chemotherapy (NAC) in a series of 83 breast tumors, including 44 TNBCs, from patients with residual disease (RD). Changes in gene expression patterns in the TIME were tested for association with recurrence-free (RFS) and overall survival (OS). Additionally, we sought to characterize the systemic effects of NAC through single cell analysis (RNAseq and cytokine secretion) of PD-1HI CD8+ peripheral T cells and examination of a cytolytic gene signature in whole blood.
Results:
In non-TNBC, no change in expression of any single gene was associated with RFS or OS, while in TNBC upregulation of multiple immune-related genes and gene sets were associated with improved long-term outcome. High cytotoxic T cell signatures present in the peripheral blood of patients with breast cancer at surgery were associated with persistent disease and recurrence, suggesting active anti-tumor immunity that may indicate ongoing disease burden.
Conclusions:
We have characterized the effects of NAC on the TIME, finding that TNBC is uniquely sensitive to the immunologic effects of NAC, and local increases in immune genes/sets are associated with improved outcomes. However, expression of cytotoxic genes in the peripheral blood, as opposed to the TIME, may be a minimally invasive biomarker of persistent micrometastatic disease ultimately leading to recurrence.
Keywords: immunogenomics, tumor-infiltrating lymphocytes, neoadjuvant chemotherapy, breast cancer, triple-negative breast cancer, gene expression
Introduction
Combination of conventional chemotherapy with an immunotherapeutic targeting programmed death-ligand 1 (PD-L1), atezolizumab, was recently approved for the treatment of patients with metastatic triple-negative breast cancer (TNBC) based on results from a Phase III clinical trial1. Furthermore, addition of the anti-programmed death-1 (PD-1) monoclonal antibody pembrolizumab to neoadjuvant chemotherapy (NAC) can significantly improve TNBC pathological complete response (pCR) rates2. Thus, existing clinical data indicate that chemotherapy combinations with immunotherapy demonstrate enhanced efficacy compared to chemotherapy alone. However, these results suggest a growing need to better understand how chemotherapy modulates the tumor-immune microenvironment (TIME).
High levels of stromal tumor-infiltrating lymphocytes (sTILs) in the pre-treatment biopsy are predictive of pCR in TNBC patients treated with NAC3. In NAC-treated TNBC patients with residual disease (RD) at surgery or in untreated primary TNBC tumors, higher sTILs in the resected tumor also confer improved prognosis4–7. However, the immunomodulatory effect of chemotherapy on sTILs in patients and how chemotherapy influences the TIME are poorly understood. In a study of patients with non-small cell lung carcinoma, tumors treated with NAC had higher expression of PD-L1 and higher density of CD3+ T cells, suggesting NAC may be immunomodulatory in some tumor types8. Additionally, in a small breast cancer study, post-NAC natural killer cells and IL-6 expression were associated with better response. However, this study was limited by inclusion of only a small number of TNBCs (n=4)9. Intriguingly, a larger study of breast cancer patients (n=60) showed that pre-NAC sTILs and higher pre-NAC expression of cytotoxic T cell markers and cytokines were associated with higher pCR rate. However, this study did not examine long term outcomes in patients with RD, for whom prognosis is worse than those with pCR and only included a limited number of TNBC samples (n=13)10. Furthermore, whether immunomodulatory effects of NAC are loco-regional or systemic (i.e. able to be detected in the peripheral blood) is unknown.
In order to address this gap in knowledge, we examined expression patterns of immune-related genes before and after NAC in a series of 83 breast tumors, including 44 TNBCs, from patients with RD. As patients with pCR generally experience excellent outcomes, we chose to focus on patients with RD, who may benefit from additional therapies. Changes in gene expression patterns in the TIME were tested for association with recurrence-free (RFS) and overall survival (OS). T cell receptor sequencing (TCRseq) was performed on a subset (n=15) of tumors. Additionally, in four patients undergoing NAC, PD-1-high and PD-1-negative CD3+CD8+ peripheral blood mononuclear cells (PBMCs) were profiled using single-cell RNA sequencing (scRNAseq) and multiplexed cytokine secretion assays. Finally, we used a scRNAseq-derived signature of activated cytolytic cells to measure immune activation in the peripheral blood of two cohorts of patients: a Vanderbilt cohort consisting of 34 patients after NAC (collected within 2 weeks prior to surgery) and 24 untreated patients, and a cohort from the Dana Farber Cancer Institute (DFCI) consisting of 30 hormone receptor positive, HER2 negative patients treated with NAC (including bevacizumab) as part of a clinical trial11. We then tested the association of this signature with surgical outcome (pCR or residual disease burden) and post-surgical cancer recurrence.
Intriguingly, higher expression of the gene signature at the time of surgery was associated with higher disease burden (i.e. in those with RD who experienced disease recurrence within three years following surgery or those with the highest residual cancer burden [RCB]). Thus, peripheral cytotoxic activity, guarded by immune checkpoints, may reflect ongoing micrometastatic and primary disease burden, and could be a useful biomarker for disease recurrence and possibly immune checkpoint inhibitor benefit.
Materials and Methods
Patients.
Three cohorts of patients were combined for the tumor profiling study. All included patients received neoadjuvant therapy and had residual disease and matched pre-treatment tissue was required for inclusion. All but 2 (ER+) patients received cytotoxic chemotherapy as part of their regimen. Four patients (ER+) received courses of hormone therapy as part of their neoadjuvant regimen, two of which were in conjunction with cytotoxic chemotherapy. Five patients (HER2+) received HER2-directed therapy as part of their neoadjuvant regimen.
For the ‘Peru’ cohort, clinical characteristics and molecular analysis of the patients (n=48 with matched pre-treatment tissue) were previously described at the Instituto Nacional de Enfermedades Neoplásicas26. Clinical and pathologic data were retrieved from medical records under an institutionally approved protocol (INEN IRB 10-018). For the ‘VICC’ cohort, which included PBMC and whole blood analyses, clinical and pathologic data were retrieved from medical records under an institutionally approved protocol (VICC IRB 030747). For the DARTMOUTH cohort patient samples were collected under a protocol approved by the Dartmouth College Institutional Review Board and the waiver of the subject consent process was IRB-approved. (IRB 28888). Metadata for the primary cohort of patients is presented in Supplementary Table S1. For the peripheral blood study, two cohorts of patients were used (summarized in Table 2). For the Vanderbilt cohort, all blood was collected within 14 days preceding definitive surgery. Metadata Vanderbilt cohort is presented in Supplementary Table S7. For the DFCI cohort11, all blood was collected in the interlude between completion of NAC and definitive surgery.
Table 2:
Demographic information for peripheral blood cohorts
| Blood Cohort n(%) | ||
|---|---|---|
| Vanderbilt | DFCI | |
| Total | 58 | 30 |
| IHC Subtype | ||
| ER+ | 27 (46.55%) | 27 (90%) |
| PR+ | 26 (44.82%) | 27 (90%) |
| HER2+ | 17 (29/31%) | 0 (0%) |
| TNBC | 21 (36.21%) | 0 (0%) |
| Response | ||
| No NAC | 24 (41.38%) | 0 (0%) |
| pCR | 10 (17.24%) | 1 (3.33%) |
| RD | 24 (41.38%) | 29 (96.67%) |
| RCB I not recur | 3 (10%) | |
| RCB I recur | 2 (6.67%) | |
| RCB II not recur | 7 (23.33%) | |
| RCB II recur | 4 (13.33%) | |
| RCB III | 13 (43.33%) | |
| NAC | ||
| No TAXANE | 7 (12.07%) | 0 (0%) |
| TAXANE | 27 (46.55%) | 30 (100%) |
| No NAC | 24 (41.38%) | 0 (0%) |
Tumor-infiltrating lymphocytes quantification.
Stromal tumor-infiltrating lymphocytes were analyzed using full face H&E sections from pre-NAC diagnostic biopsies or post-NAC RD surgical specimens. Samples were scored according to the International TILs Working Group Guidelines27–29. The pre-defined cut point of 30%4 was used for all survival analyses.
NanoString nCounter analysis.
Gene expression and gene set analysis on pre- and post-NAC formalin-fixed tissues were performed using the nanoString Pan-Cancer Immunology panel (770 genes) according to the manufacturers’ standard protocol. Data were normalized according to positive and negative spike-in controls, then endogenous housekeeper controls, and transcript counts were log transformed for downstream analyses. Normalized linear data are presented in Supplementary Table S2. Gene sets were calculated by summing the log2-transformed normalized NanoString counts for all genes contained in a given gene set (Supplementary Table S3). Samples were simultaneously assayed for PAM50 molecular subtyping using a custom-designed 60-gene Elements panel (Supplementary Table S4). Briefly, 10μm sections of diagnostic biopsies or residual tumors were used for RNA preparation (Promega Maxwell 16 RNA FFPE) and 50ng of total RNA >300nt (assayed on a Agilent Tapestation 2200 Bioanalyzer) was used for input into nCounter hybridizations for Pan-Cancer Immunology panels or 500–1000ng RNA for PAM50 analysis. Data were normalized according to positive and negative spike-in controls, then endogenous housekeeper controls, and transcript counts were log transformed for downstream analyses. Subtype prediction was performed in R using the genefu package.
For the 8 gene signature analysis in whole blood, a custom NanoString Elements was constructed to measure the gene expression levels of PDCD1, NKG7, LAG3, GZMH, GZMB, GNLY, FGFBP2, HLA-DRB5, and HLA-G, as well as 3 normalization control genes (PTPRC, RPL13a, TBP). Probe design is shown in Supplementary Table S9. RNA was isolated from whole blood (Promega Maxwell 16 Simply RNA Blood) and 100–200ng was used for input into the nCounter analysis. Data were normalized as above. Linear normalized data are presented in Supplementary Table S8 for the Vanderbilt Cohort.
Isoplexis (single-cell cytokine profiling).
On day 1, cryopreserved PBMCs were thawed and resuspended in complete RPMI media with IL-2 (10 ng/ml) at a density of 1–5 × 106 cells/ml. Cells were recovered at 37°C, 5% CO2, overnight. Plates were prepared by coating with anti-human CD3 (10 μg/ml in PBS, 200–300 μl/well) in a 96-well flat-bottom plate at 4°C, O/N. On day 2, non-adherent cells for each sample were collected and viability was confirmed, with dead cell depletion by Ficoll. For each sample, where sufficient, volume was split in half for each of the following negative isolations: with one half of cells from each sample, CD4 T cells were isolated with CD4+ negative isolation kit following Miltenyi protocol (130-096-533); with the other half of cells from each sample, CD8 T cells were isolated with CD8+ negative isolation kit following Miltenyi protocol (130-096-495). The PD-1+ and PD-1- subsets were from isolated CD4 or CD8 T cells by staining with PE-conjugated anti-PD-1 antibody using the manufacturer’s protocol (Miltenyi, 130-096-164) as follows: 1) stain each subset with 10ul stain :100ul Robosep buffer for every 1×10^7 total cells; 2) incubate at 4°C for 10 mins; 3) rinse cells by adding 1–2 mL of Robosep and C/F at 300xg for 10 mins; 4) aspirate supernatant and resuspend cells pellets in 80ul buffer per 1×107 total cells. PD-1+ cells were then isolated with anti-PE microbeads following the manufacturer’s protocol (Miltenyi, 130-097-054). Cells were resuspended in complete RPMI media at a density of 1 × 106/ml and seeded into wells of the CD3-coated 96-well flat-bottom plate with soluble anti-human CD28 (5 ug/ml). Plates were incubated at 37°C, 5% CO2 for 24 hrs. On day 3, supernatants (100 ul per well) were collected from all wells and stored at −80°C for population assays. T cells were collected and stained with Brilliant Violet cell membrane stain and AlexaFluor-647-conjugated anti-CD8 at RT for 20 min, rinsed with PBS and resuspended in complete RPMI media at a density of 1 × 106/mL
Approximately 30 μl of cell suspension was loaded into the IsoCode Chip and incubated at 37°C, 5% CO2 for additional 16 hours. Protein secretions from ~1000 single cells were captured by the 32-plex antibody barcoded chip and analyzed by fluorescence ELISA-based assay30–35. Polyfunctional T cells that co-secreted 2+ cytokines per cell were evaluated by the IsoSpeak software across the five functional groups: Effector: Granzyme B, TNFα, IFN-γ, MIP1α, Perforin, TNFβ; Stimulatory: GM-CSF, IL-2, IL-5, IL-7, IL-8, IL-9, IL-12, IL-15, IL-21;
Chemoattractive: CCL11, IP-10, MIP-1β, RANTES; Regulatory: IL-4, IL-10, IL-13, IL-22, sCD137, sCD40L, TGFβ1; Inflammatory: IL-6, IL-17A, IL-17F, MCP-1, MCP-4, IL-1β.
TP53 sequencing.
TP53 gene sequencing was performed using either the Foundation Medicine assay as previously reported26 or using the SureMASTR TP53 sequencing assay (Agilent). For the later, purified DNA from FFPE breast tumor sections were amplified and sequenced according to the manufacturer’s standard protocol. Samples were sequenced to a depth of ~10,000 and mutations were called using the SureCall software (Agilent). Mutation allele frequency was set at 5% and only likely functional (early stops, frameshift deletions and known recurrent hotspot single-nucleotide variation mutations) were selected for sample annotation.
T cell receptor sequencing.
TCR sequencing and clonality quantification was assessed in FFPE samples of breast cancer specimens or PBMCs. For FFPE tissue, DNA or RNA was extracted from 10μm sections using the Promega Maxwell 16 FFPE DNA or FFPE RNA kits and the manufacturer’s protocol. For PBMCs, PD-1HI and PD-1NEG CD8+ T cells sorted by fluorescence-activated cell sorting from samples isolated from EDTA collection tubes and processed using a Ficoll gradient. At least 100K cells were collected, centrifuged, and utilized for RNA purification. TCRs were sequenced using survey level immunoSEQ™ (DNA; Adaptive Biotechnologies) and the Immunoverse™ assay (RNA; ArcherDX), as previously described39,40. Sequencing results were evaluated using the immunoSEQ analyzer version 3.0 or Archer Immunoverse analyzer. CDR3 sequences and frequency tables were extracted from the manufacturers’ analysis platforms and imported into R for analysis using the Immunarch package (https://immunarch.com)36 in R. Shannon entropy, a measure of sample diversity, was calculated on the clonal abundance of all productive TCR sequences in the data set. Shannon entropy was normalized by dividing Shannon entropy by the logarithm of the number of unique productive TCR sequences. This normalized entropy value was then inverted (1 − normalized entropy) to produce the ‘clonality’ metric. TCRβ clonotypes and metadata based on the primary cohort (Supplementary Figure 5; Adaptive) are included as Supplementary Dataset 1. TCRβ clonotypes and metadata based on prospectively collected peripheral blood and tumor from Figure 4 (Archer) are included as Supplementary Dataset 2.
Figure 4: Evidence of enhanced T cell functionality in the CD8+ PD-1HIperipheral compartment.

A) Clinical details of 4 patients analyzed prospectively for changes in peripheral blood T cell functionality. NST indicates no special type. B) Polyfunctionality of PD-1HICD4+ and PD-1HICD8+ T cells isolated from PBMCs in 4 patients prior and after NAC (>1000 individual cells/sample/timepoint) was determined by Isoplexis single-cell cytokine profiling. Polyfunctionality is defined as the percentage of cells capable of producing ≥ 2 cytokines following CD3/CD28 stimulation. The percentage of cells in each sample capable of secreting 2, 3, 4, or 5+ cytokines are depicted in stacked bars. Characteristics of each of the 4 patients are shown above the bars. Patients with TNBC (Pt. 1 and Pt. 4) had greater increases in polyfunctionality in the CD8+ compartment with NAC. C) Heatmap representation of log cytokine signal intensity of each cell in each patient sample, pre and post NAC. Each row represents one PD-1HiCD8+ T cell. White indicates no cytokine secreted. D) TCRβ chain repertoire analysis in CD8+ peripheral blood T cells. Upper plots indicate the number of individual T cells sequenced plotted by sample on the left Y axis; number of clonotypes (unique CDR3 amino acid sequences) plotted by sample on the right Y axis. In the lower graph, each sample is divided into the number of clonotypes comprising expanded (hyper-expanded, large, medium, small, and rare) compositions of the detected repertoire (categories divided by orders of magnitude of fraction of total repertoire). E) The fraction of repertoire clonotypes identified in PD-1HI versus PD-1NEG CD8+ T cells (before or after NAC) classified as ‘hyperexpanded’ or ‘large’ (comprising >0.1% of repertoire). P value represents a 2-sample 2-tailed t-test.
Single-cell RNA sequencing.
Peripheral blood mononuclear cells were isolated from EDTA collection tubes, processed using a Ficoll gradient, and cryopreserved in 10% DMSO 90% FBS. Upon thaw, whole live PBMCs were prepared by depleting dead cells using a dead cell removal kit (Miltenyi, cat no: 130-090-101). PD-1HI and PD-1NEG CD8+ T cells were sorted by fluorescence-activated cell sorting from peripheral blood mononuclear cells. Each sample (targeting 5,000 – 10,000 cells/sample) was processed for single cell 5’ RNA sequencing utilizing the 10X Chromium system. Libraries were prepared using P/N 1000006, 1000080, and 1000020 following the manufacturer’s protocol. The libraries were sequenced using the NovaSeq 6000 with 150 bp paired end reads. RTA (version 2.4.11; Illumina) was used for base calling and analysis was completed using 10X Genomics Cell Ranger software v2.1.1. Data were analyzed in R using the filtered h5 gene matrices in the Seurat37,38 package. Briefly, samples were merged, and all cells were scored for mitochondrial gene expression (a marker of dying cells) and cell cycle genes to determine phase. Data were transformed using SCTransform, regressing against mitochondrial gene expression and cell cycle phase. Dimensional reduction was performed using Harmony39. Missing values were imputed using the RunALRA function in the SeuratWrappers package40. For the whole PBMC single cell data, cell types were assigned to individual cells using SingleR41. BlueprintEncodeData was used as a reference42,43.
Statistical analysis.
All statistical analyses were performed in R or Graphpad. For survival curves (RFS and OS), the log-rank statistic was reported (or trend-test for >2 groupings). For gene-level analysis, nominal p-values were calculated using a cox proportional hazards model, and then a adjusted p-value (q-value) was calculated based on the FDR44. All single-cell statistical analyses were calculated in R using the Seurat package37. Shared Nearest Neighbors were calculated using the Harmony reduction, and clusters were identified at a resolution of 0.3, defining 5 total clusters. UMAP was performed for visualization, and missing values were imputed using ALRA40. Visualization and graph generation was performed in R. Some heatmaps were made using the complex heatmap package in R45. P-value cut-offs displayed on plots correspond to “ns” equals p>0.05, * equals 0.01<p≤ 0.05, ** equals 0.001<p≤0.01, *** equals 0.0001<p≤0.001, **** equals p≤0.0001. Code used to generate figures can be accessed at https://github.com/MLAxelrod/Immunologic_changes_with_chemotherapy_inTNBC .
Results
Stromal tumor-infiltrating lymphocytes (sTILs) in residual disease prognosticate improved outcomes in TNBC patients with incomplete response to neoadjuvant chemotherapy (NAC)
We procured matched archived pre-treatment (diagnostic biopsy) and post-treatment (residual disease surgical specimen) tumor specimens from a series of 83 patients, including 44 TNBC patients. Importantly, we refined our study to include only patients who had residual disease at surgery for analysis, thereby excluding patients who achieved pCR. This was a purposeful selection strategy, as patients with pCR usually experience good outcomes, and we instead chose to focus on patients with RD for whom additional risk stratification could identify those who are most likely to benefit from additional therapies. . Metadata for the patients, including treating institution, molecular subtype (PAM50), recurrence-free and overall survival (RFS and OS, respectively), TP53 mutation status, and other molecular and clinical data are summarized in Table 1. Individual patient-level data is available in Supplementary Table S1.
Table 1:
Demographic information for tumor cohort
| DART | EA1311 | PERU | Entire Cohort | |
|---|---|---|---|---|
| n(%) | n(%) | n(%) | n(%) | |
| Total | 24 | 11 | 48 | 83 |
| PAM50 Subtype | ||||
| Basal | 7 (29.17%) | 4 (36.36%) | 31 (64.58%) | 42 (50.60%) |
| Her2 enriched | 2 (8.33%) | 3 (27.27%) | 10 (20.83%) | 15 (18.07%) |
| Luminal A | 6 (25%) | 1 (9.09%) | 1 (2.08%) | 8 (9.64%) |
| Luminal B | 8 (33.33%) | 3 (27.27%) | 1 (2.08%) | 12 (14.46%) |
| Normal | 1 (4.17%) | 0 (0%) | 4 (8.33%) | 5 (6.02%) |
| NA | 0 (0%) | 0 (0%) | 1 (2.08%) | 1 (1.20%) |
| IHC Subtype | ||||
| ER+ | 20 (83.33%) | 5 (45.45%) | 1 (2.08%) | 26 (31.33%) |
| PR+ | 13 (54.17%) | 4 (36.36%) | 1 (2.08%) | 18 (21.69%) |
| HER2+ | 6 (25%) | 2 (18.18%) | 12 (25%) | 20 (24.10%) |
| TNBC | 4 (16.67%) | 5 (45.45%) | 35 (72.92%) | 44 (53.01%) |
| Neoadjuvant Chemotherapy | ||||
| TAXANE | 21 (87.50%) | 7 (63.64%) | 21 (43.75%) | 49 (59.04%) |
| NO TAXANE | 3 (12.5%) | 4 (36.36% | 27 (56.25%) | 34 (40.96%) |
As sTILs have been described and rigorously validated in breast cancer as both a prognostic factor (in surgical specimens for post-surgical outcomes, particularly in TNBC and HER2+ cancers), and a predictive factor (in diagnostic biopsies for benefit from NAC) , we first asked whether these findings were consistent with our study cohort. Using the published cutoff (30%) of sTILs4, we found that higher abundance of sTILs in the post-NAC residual disease in TNBC patients (n=44) was significantly prognostic for both RFS (log-rank p=0.019) and OS (p=0.05; Figure 1A). Representative histology of high (>30%) and low (<30%) sTILs are shown in Supplementary Figure 1. Interestingly, pre-NAC sTILs in the diagnostic biopsy were not prognostic for outcomes in TNBC patients (Supplementary Figure 2A), presumably due to the selection strategy of including only patients who lacked pCR. Consistent with prior literature that the prognostic and predictive effect of sTILs differs by breast cancer subtype12, neither pre-NAC nor post-NAC sTILs were prognostic for OS when considering our entire cohort (n=83). However, post-NAC sTILs were prognostic for RFS (p=0.031) in the whole cohort (Supplementary Figure 2B, 3A). This effect seems primarily driven by TNBC tumors as post-NAC sTILs are not prognostic of either RFS or OS in non-TNBCs (Supplementary Figure 3B). Neither pre-NAC nor post-NAC sTILs were prognostic in either ER+ or HER2+ patients only (Supplementary Figure 4). This may be limited by our sample size as sTILs have been previously shown to be associated with longer RFS in HER2+ cancer patients and shorter overall survival in luminal/HER2- cancer patients12. Stratifying TNBC patients by whether sTILs were qualitatively increased or decreased/equivocal in the surgical resection compared to the diagnostic biopsy did not provide any prognostic capability in this cohort (Supplementary Figure 5A). Interestingly, most patients, regardless of clinical subtype, had a decrease in sTILs over the course of NAC (Supplementary Figure 5B). Importantly, our cohort includes only patients with RD, and there is currently not a validated method for quantifying sTILs in pCR where, by definition, a tumor is no longer present. Thus, in our cohort, abundance of sTILs has the strongest prognostic effect for the post-NAC surgical resection specimen in TNBC tumors with an incomplete response to NAC. These findings, consistent with both retrospective studies and analyses from randomized controlled trials, prompted us to perform more detailed molecular studies aimed at understanding how NAC influences the TIME.
Figure 1: Immunologic changes in breast tumors after neoadjuvant chemotherapy.

A) High levels of sTILs are associated with RFS (left; n=41) and OS (right; n=42) after surgery in TNBC. Patients are stratified based on post-NAC sTILs ≤ 30% or > 30%, scored as recommended by the International TILs Working Group27,28, according to the predefined cut point4. B) Heatmap demonstrating gene expression patterns for 770 immune-related genes (NanoString Pan-Cancer Immune Panel) across all patients (TNBC and non-TNBC; n=83 total patients, 166 samples). C) Heatmap of gene expression patterns as detailed in panel B, instead depicting the change in expression of each gene in matched paired (pre- and post-NAC; n=83) samples. Red data points represent an upregulation, while blue data points represent a downregulation in the post-NAC residual disease compared to the pre-treatment diagnostic biopsy.
Suppression of immunologic gene expression with NAC in TNBC
To measure transcriptional changes occurring in the tumor-immune microenvironment (TIME) induced by NAC, we performed gene expression profiling for a series of 770 immune-related genes using nanoString (Pan-Cancer Immune Panel), before and after NAC in the entire cohort (n=83). Transcriptional patterns and hierarchical clustering for all data primarily segregated tumors based on receptor status (ER/PR/HER2) and/or molecular subtype, with most luminal/hormone receptor-positive tumors appearing in the first cluster, most HER2-positive tumors in the second cluster, and most basal-like/TNBC tumors in the third cluster (Figure 1B). Examining the data as the change in gene expression for each gene after NAC in a patient-matched fashion (Δ expression; post-NAC minus pre-NAC) yielded similar patterns, with a trend of most TNBC patients having generalized decreased immune gene expression patterns after NAC (Figure 1C). In order to test for effects driven by differences in sampling (i.e. pre-NAC samples are biopsies whereas post-NAC samples are surgical resection samples) or treating institution, we performed principal component analyses. We did not detect significant clustering by time of sampling or cohort (Supplementary Figure 6).
NAC-induced immunologic gene expression is a positive predictor of outcome in TNBC
While the TIME change in sTIL abundance did not prognosticate outcome in TNBC patients, we asked if changes in individual immune-related genes are associated with outcome. We performed iterative Cox proportional hazards models, using the delta (Δ) of each gene (post-NAC minus pre-NAC) in an independent univariate analysis, for both RFS and OS. All analyses are reported using a nominal p-value as well as a false discovery rate (FDR; Benjamini-Hochberg method) q-value for association with RFS or OS. After correction for FDR (q <0.10), upregulation of 11 genes were associated with improved RFS, while upregulation of only one gene was significantly associated with worse RFS (CDH1, which encodes e-cadherin) in our TNBC cohort. Interestingly, e-cadherin is known to interact with killer cell lectin-like receptor G1 (KLRG1), an inhibitory receptor expressed by memory T cells and NK cells13. In contrast, upregulation of a larger number of genes was associated with improved OS (n=189) or reduced OS (n=15) at FDR q<0.10 (Figure 2A). These genes are listed in Supplementary Table 5. Kaplan-Meier visualization examples of strongly prognostic genes (negative prognostic: CDH1; positive prognostic: CD70) reinforced the prominent association of TNBC disease outcomes with changes in immune gene expression during NAC (Figure 2B). Conversely, no changes in immune-related gene expression were significantly associated with RFS or OS in non-TNBC patients at q<0.10 (Supplementary Figure 7A).
Figure 2: Identification of immune-associated genes associated with RFS and OS in TNBC after chemotherapy.

A) Individual genes (changes pre- to post-NAC) were tested iteratively in a univariate cox-proportional hazards model for their association with RFS (left) or OS (right) after chemotherapy and surgery. Individual genes are colored for their statistical significance (red: nominal p-value <0.05; green: q-value (FDR) <0.10; black: not significant). Selected top genes are labeled but are limited in number for clarity. Genes with negative coefficients (left of the center line) are associated with better outcome, while genes with positive coefficients (right of the center line) are associated with worse outcome. B) Representative Kaplan-Meier plots for selected detrimental (CDH1; e-cadherin) and beneficial (CD70) genes are shown. Strata are defined by tertiles, and generally represent upregulation during NAC (blue), no change/equivocal (green), and downregulation (red). P-values represent the log-rank test for trend.
Dimensional reduction through collapsing individual genes into pathways or defined functions can improve interpretation of high-dimensional data. Thus, we collapsed the gene expression data into bioinformatically-categorized immune signatures (sum-scores, defined as the summation of the log2 expression values for all genes in a category). Organization of the data in this manner and testing the signatures (n=70) for association with RFS and OS yielded a surprising finding - all significant (q<0.10) gene sets (n=40 for RFS and n= 60 for OS; listed in Supplementary Table 6) identified in this analysis were associated with good outcome (Figure 3A). Many of the top-scoring gene sets were associated with T cells, including “T cell polarization”, “T cell immunity”, “T cell activation”, among others. Although manual inspection revealed some overlap in these gene sets, they were largely composed of signature-exclusive genes (Supplementary Table 3). Kaplan-Meier visualization examples of strongly prognostic gene sets (“NK cell functions” and “T cell activation”) reinforced the considerable association of changes in immune gene sets during NAC with outcomes (Figure 3B). Interestingly, no gene or gene set was significantly associated with RFS or OS in non-TNBC patients at q<0.10 (Supplementary Figure 7A-B). When the non-TNBC group is separated into ER+ and HER2+ groups, no gene or gene set was significantly associated with outcome (Supplementary Figure 8). Thus, these data suggest that NAC, exclusively in TNBC, could promote immunologic activity leading to improved outcomes in a subset of patients. However, these effects may be related to factors beyond TNBC biology, as hormone-receptor-positive and HER-2 positive patients receive additional endocrine or HER-2 directed therapy in the adjuvant setting, complicating associations with RFS and possibly OS. Nonetheless, immune-related signatures, particularly those derived from T cells, appeared to be strongly associated with improved outcomes in TNBC.
Figure 3: Upregulation of immune-associated gene sets after chemotherapy are associated with improved RFS and OS in TNBC.
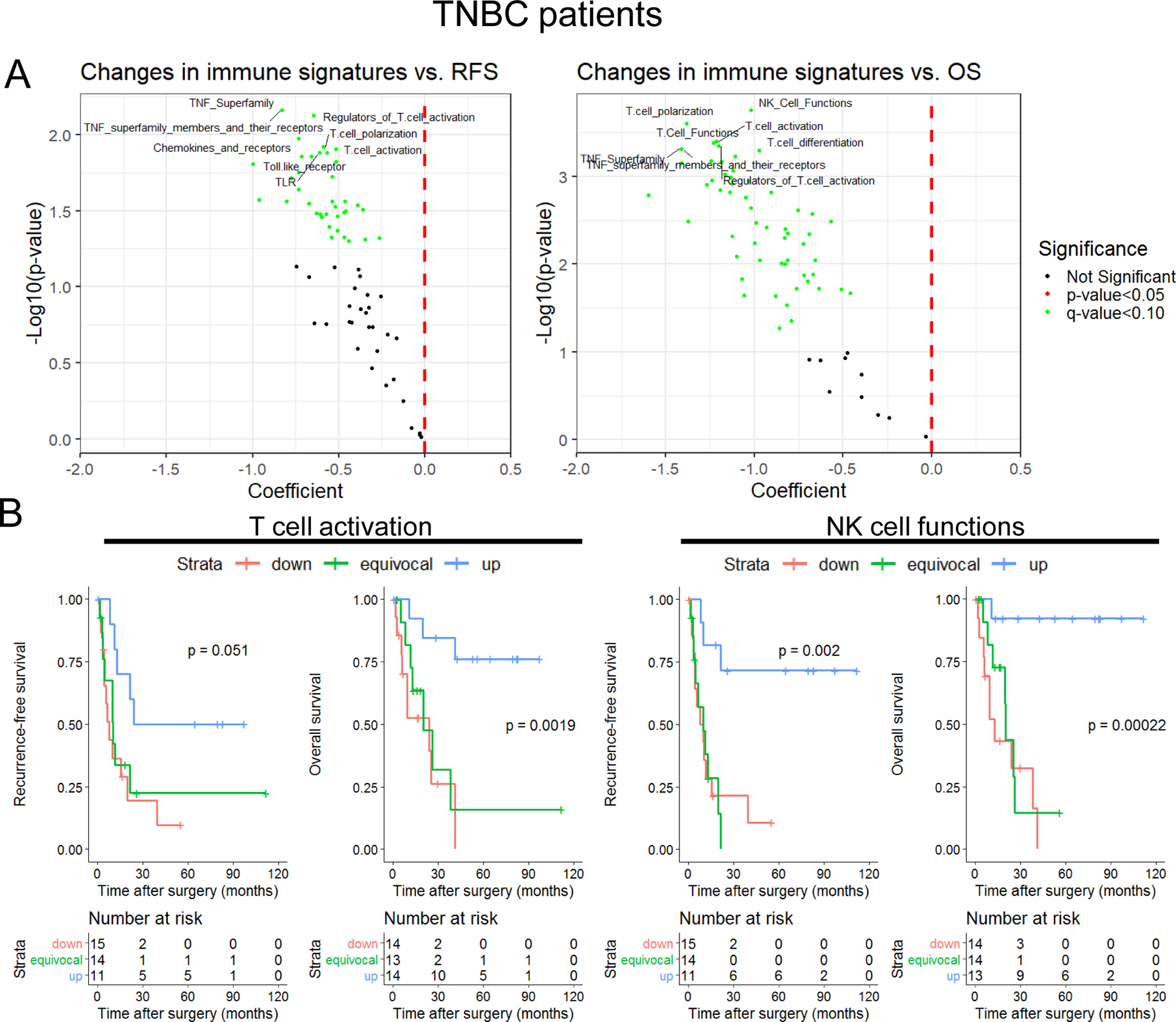
A) Gene set scores were calculated by summing expression levels of all gene set member genes across each candidate gene set (n=70). Changes pre- to post-NAC was then calculated for each TNBC patient (n=44) and each gene set score was tested iteratively in a univariate cox-proportional hazards model for association with RFS (left) or OS (right) after chemotherapy and surgery. Individual gene sets are colored for their statistical significance (red: nominal p-value <0.05; green: q-value (FDR) <0.10; black: not significant). Selected top gene sets are labeled but are limited for clarity. Gene sets with negative coefficients are associated with better outcome, while gene sets with positive coefficients are associated with worse outcome. B) Representative Kaplan-Meier plots for selected gene set changes with beneficial associations are shown (left: T cell activation; right: NK cell functions). Strata are defined by tertiles, and generally represent upregulation during NAC (blue), no change/equivocal (green), and downregulated (red). P-values represent the log-rank test for trend.
Changes in T cell clonality and function in tumors and peripheral blood induced by NAC
A robust T cell response is characterized by oligoclonal expansion of antigen-specific T cells. Therefore, we next asked whether clonality of T cells in the TIME was altered during NAC. In a subset of samples (n=15; 8 TNBC, 7 non-TNBC), we performed T cell receptor (TCR) β chain sequencing using the ImmunoSeq assay to estimate the number of unique T cell clones (diversity), and the presence of expanded T cell clones in the TIME before and after NAC. Given the breadth of sTILs fractions observed among breast tumors as well as caveats associated with comparison of samples derived from diagnostic core needle biopsies vs. surgical resections, we first verified that the number of productive T cells was associated with estimation of sTILs determined on adjacent sections. We detected a strong association between these parameters (R2=0.6; p<0.0001; Supplementary Figure 9A), raising confidence in the assay results. In this sample set, NAC did not universally alter productive clonality (Supplementary Figure 9B), a measurement of the number of times the same (productive) TCRβ sequence is represented in the sample, which is a descriptor of T cell clonal expansion. When stratified by breast cancer subtype, there was no significant change in productive clonality with NAC (one-sample t-test). However, TNBC tumors demonstrated a qualitative trend toward decreased clonality after NAC, while non-TNBC tumors trended toward increased clonality after NAC. The difference between these two subgroups approached significance (p=0.054; two-sample t-test; Supplementary Figure 9C). There was no association of change in clonality with change in sTILs, suggesting that changes in sTIL abundance after NAC are not necessarily due to expansion of existing clones (Supplementary Figure 9D).
To further explore changes in T cell clonality and function in response to chemotherapy, we prospectively collected PBMCs from four breast cancer patients (including two TNBCs) before and after NAC (Figure 4A). In addition, the post-NAC residual disease (or in one case, pCR residual scar) was analyzed in tandem. Based on previous findings demonstrating that tumor-reactive T cells are enriched in the CD8+ PD-1HI population of peripheral T cells14, we purified CD4+ and CD8+ cells from each sample by fluorescence-activated cell sorting (FACS), further stratifying by PD-1-negative (PD-1NEG) and PD-1HI (top 20% expressers of CD8+ or CD4+ cells) status (gating scheme shown in Supplementary Figure 10). Using a functional fluorescence ELISA-based assay of cytokine (32-plex antibody barcoded chip, Supplementary Figure 11A) secretion following CD3/CD28 stimulation, we determined that PD-1HI peripheral T cells had functional capacity, secreting multiple cytokines following activation, and these effects were particularly pronounced in CD8+ T cells (Supplementary Figure 11B). In 2/2 TNBC patients, the percentage of ‘polyfunctional’ PD-1HI CD8+ T cells – those capable of expressing multiple cytokines after TCR stimulation – were increased following NAC (Figure 4B). In contrast, 2/2 ER+ breast cancer patients experienced a drop or stasis in the functionality of the PD-1HI CD8+ population of cells following NAC (Figure 4B). Of note, the patient with ER+HER2+ disease has a near complete loss of T cell functionality after NAC. Cytokines produced by individual PD-1HI CD8+ cells in TNBC patients were primarily effector (e.g., Granzyme B, IFN-γ, MIP-1α, TNF-α, and TNF-β) and chemo-attractive (MIP-1β) cytokines (Figure 4C). PD-1HI CD4+ T cells also produced primarily effector cytokines including IFN-γ and TNF-α (Supplementary Figure 11C).
PD-1HI CD8+ and PD-1NEG CD8+ T cells from pre-NAC and post-NAC blood (except patient 4, for whom a sufficient pre-NAC sample was not available) were also analyzed by TCR sequencing. While the number of detected T cells was consistent among all samples, the clonotypes detected (unique TCRs) were considerably lower in PD-1HI CD8+ T cells (Figure 4D). This suggests that there are more repetitive sequences detected in the PD-1HI population, indicating clonal expansion. Consistent with this observation, the proportion of the overall TCR repertoire occupied by expanded clonotypes (large or hyperexpanded clonotypes consisting of greater than 0.1% or 1% of the total repertoire, respectively) was substantially higher in the PD-1HI than in PD-1NEG CD8+ T cell fractions (Figure 4E). We additionally sequenced the TCR repertoire in the post-NAC residual disease, although the number of T cells sequenced in these samples were limited due to fixation of tissue and small T cell abundance as a function of total RNA in the bulk samples, and thus should be interpreted with caution. Nonetheless, we found that the similarity (Jaccard index, normalized to size of repertoire detected) of tumor-infiltrating TCRs in the post-NAC sample was universally more similar to the PD-1HI CD8+ peripheral TCR repertoires, compared to the PD-1NEG CD8+ repertories (Supplementary Figure 12). This suggests that the PD-1HI peripheral compartment is enriched for similarity to TILs relative to the PD-1NEG peripheral compartment.
Single-Cell RNAseq of peripheral PD-1HI CD8+ T cells identifies a unique population of cytolytic effector cells
Next, we utilized scRNAseq to describe the post-NAC peripheral PD-1HI CD8+ T cell populations at the time of surgery in the blood of two TNBC patients: one with residual disease (Pt. 1) and one with matrix-producing metaplastic TNBC who experienced pCR (Pt. 4). Uniform Manifold Approximation and Projection (UMAP) analysis was performed on Harmony-normalized samples to adjust for inter-sample technical variation, and we stratified cells based on 5 clusters identified through the Louvain algorithm (Figure 5A–B). A heatmap of the top 10 most differentially expressed genes by cluster is presented in Figure 5C. Although the composition of the cells was largely similar, we identified one cluster (‘cluster 0’) which was enriched in Pt. 4. Examination of genes differentially expressed in this cluster of cells suggested a cellular identity concordant with that of highly cytotoxic memory (TBX21-expressing) T cells, which had an abundance of MHC-I (HLA-A/B/C) and MHC-II (e.g., HLA-DRA, HLA-DRB5) family member expression as well as expression of cytolytic and immune checkpoint genes (e.g., LAG3, FCRL615, and higher transcriptional expression of PDCD1; Figure 5C–D). Verification of the pattern of expression of key cytolytic and killer-identity genes [GNLY (granulysin), GZMB (granzyme B), and FGFBP2 (killer-secreted protein 37)] showed that these genes were almost exclusively expressed in cluster 0 (Supplementary Figure 13A). This analysis also demonstrated purity-of-sort in that all clusters expressed CD8A and PDCD1, but not CD4 (Supplementary Figure 13B).
Figure 5: Single-cell RNA sequencing of CD8+ PD-1HI peripheral T cells from 2 patients with TNBC after NAC demonstrate high expression of cytolytic markers.
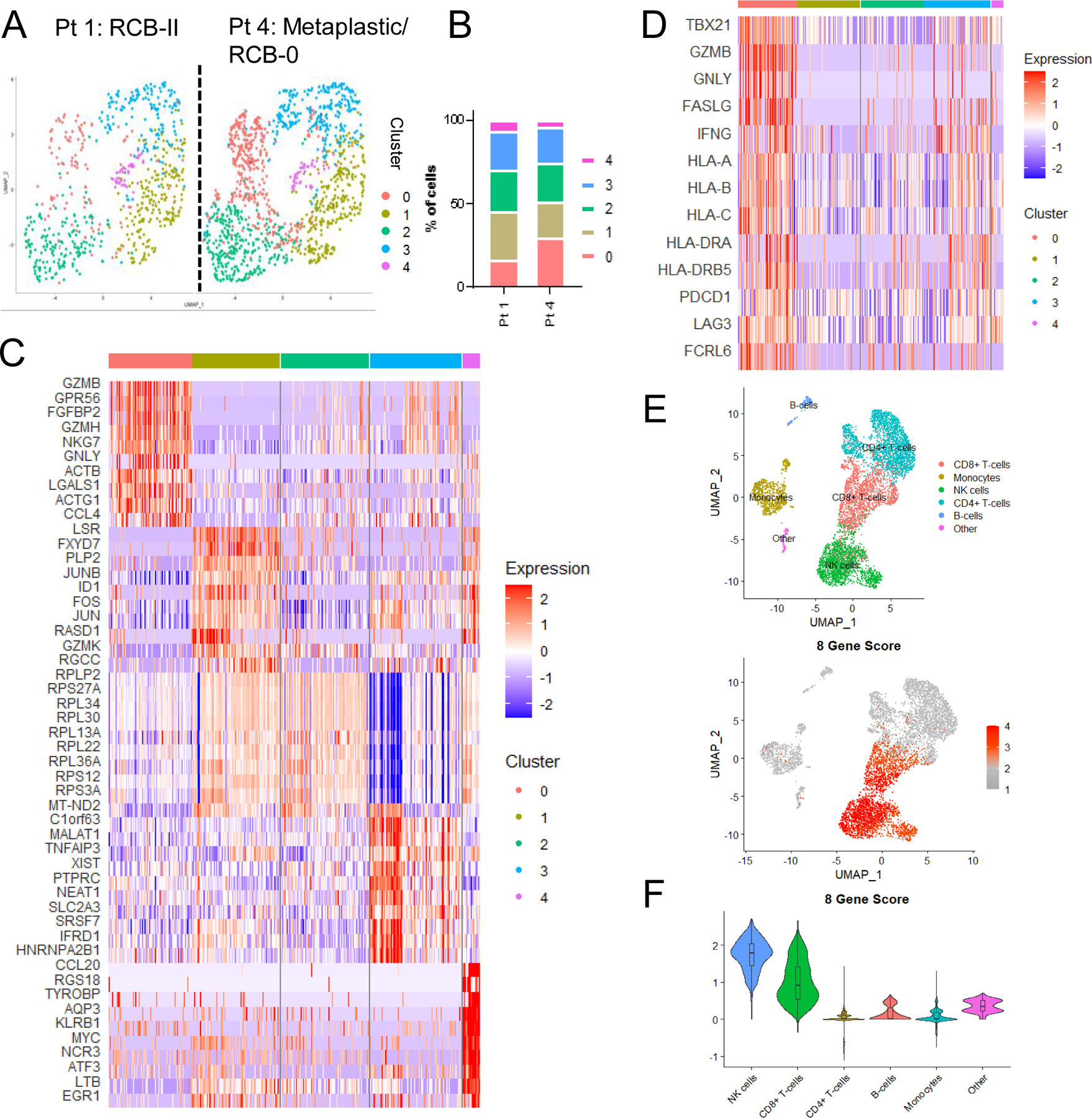
A) UMAP plots of 1,964 PD-1HICD8+ peripheral T cells across 2 patients (672 and 1,292 respectively) are shown. Five (5) clusters (0–4) were defined. B) Percent of cells sequenced comprising each cluster are plotted. C) Heatmap identifying top 10 most differentially expressed transcripts across clusters. D) A selection of genes defining cluster 0 are highlighted. Data depicted include combined cells from both Pt.1 and Pt.4. E) UMAP plots of 7,062 PBMCs from 2 TNBC patients (3,525 cells from patient 4 and 3,537 cells from patient 5). Cell type annotations are defined by SingleR. 8 gene score is defined by expression of FGFBP2 + GNLY + GZMB + GZMH + NKG7 + LAG3 + PDCD1 – HLA-G. F) Violin plots showing expression of the 8 gene score by cell type. Overlaid box plots show mean and interquartile range for each cell type.
These data led us to propose two competing hypotheses: 1) Cluster 0 genes, reflective of cytolytic CD8+ T cells, are a positive prognostic factor reflective of robust anti-tumor immunity as evidenced by their enrichment in the metaplastic TNBC patient with pCR; or 2) Cluster 0 genes are reflective of ongoing disease including the micrometastatic component that cannot be sampled from the primary tumor. The second hypothesis is supported by the observations that pCR is less prognostic of RFS and OS in metaplastic disease16,17 and that rates of recurrence following chemotherapy are higher for metaplastic disease than non-metaplastic TNBC16,18,19. Interestingly, matrix-producing metaplastic breast cancer (Pt. 4) has been shown to be associated with pCR to NAC, but often can still recur despite pCR16,20. Follow-up for this individual patient was immature at the time of reporting, and thus recurrence, and therefore presence of micrometastatic disease at the time of sampling, cannot be ruled out.
To gain a deeper understanding of cluster 0 genes, we performed additional single cell RNA sequencing on post-NAC whole PBMCs from two TNBCs patients with pCR (patient 4, described above, and patient 5, not used in any prior analysis). UMAP was used for dimensionality reduction on Harmony-normalized samples. SingleR was used to computationally assign cell type annotations (Figure 5E). Analysis of the top 5 differentially expressed transcripts by cluster (Supplementary Figure 14A) and expression of key cell type identity markers (Supplementary Figure 14B) validated annotations by SingleR. Low confidence cell type annotations were collapsed into the category “other” and make up a minor fraction of cells (Supplementary Figure 15A). Seven genes enriched in cluster 0 (PDCD1, NKG7, LAG3, GZMH, GZMB, GNLY, FGFBP2) and one gene strongly de-enriched in patient 4 (HLA-G) were chosen as an 8 gene signature for downstream applications, including validation in a larger cohort of patients (Supplementary Figure 15B–C). Expression of this 8 gene score (PDCD1 + NKG7 + LAG3+ GZMH + GZMB + GNLY + FGFBP2 − HLA-G) was the highest in CD8+ T cells (as expected, given the derivation from PD-1HIC8+T cells) and natural killer cells (Figure 5F). This finding was similar in both patients (Supplementary Figure 16A–B). Strong expression of this signature in post-NAC whole PBMCs led us to test whether this signature was predictive of response in archived whole blood samples from breast cancer patients.
Cytolytic gene expression signatures are present in blood and associated with increased likelihood of recurrence
To determine whether cytolytic signatures representative of cluster 0 genes were associated with disease outcome, we evaluated archived whole blood from a series of 58 breast cancer patients. All samples were collected within 14 days preceding surgical resection for primary breast cancer, with 34 samples having received NAC, in addition to 24 samples from untreated patients. As described above, a series of eight genes enriched in cluster 0 (PDCD1, NKG7, LAG3, GZMH, GZMB, GNLY, FGFBP2, HLA-DRB5), one gene enriched in Pt 1 (RCB-II) over Pt 4 (pCR; HLA-G) (Supplementary Figure 15B-C), and three normalization control genes (PTPRC, RPL13a, TBP)21 were selected for a 12-gene custom NanoString gene expression analysis. HLA-G has been described as an immune checkpoint which can dampen anti-tumor immune responses22–24, and thus we expected HLA-G expression to be inversely correlated with the other selected genes, as is the case in our scRNAseq dataset. One of these genes performed poorly (HLA-DRB5), likely due to frequent polymorphisms in the gene leading to highly variable probe binding and was therefore omitted from further analysis. Information on the presence of pCR/RD at surgery, ER/PR/HER2 status, and clinical follow-up (recurrence at 1000 days after surgery for RD patients) was collected (Table 2 and Supplementary Table S7).
Nearly all tested genes demonstrated a pattern supporting the hypothesis that gene expression in whole blood is associated with ongoing disease, being highest (or lowest in the case of HLA-G) in untreated patients (who have ongoing tumor burden by virtue of not having received therapy prior to surgery) and those with RD compared to those with pCR. However, it is important to note that there is a high degree of heterogeneity in the untreated patients, possibly reflecting heterogeneity in tumor size, strength of anti-tumor immune response at baseline, or anti-tumor response following systemic therapy. Furthermore, among patients with RD, higher expression (or lower in the case of HLA-G) tended to be observed in patients who had early recurrences in the first 3 years following surgery (and thus may have had micrometastatic disease at the time of surgery). Several of these genes (FGFBP2, GNLY, PDCD1, LAG3, and NKG7) were also significantly differentially expressed or approached statistical significance across the outcome groups (Kruskal-Wallis test). Comparisons were particularly striking between the group of patients with RD who experienced early disease recurrence and the group with pCR following NAC (post-hoc Dunn test; Figure 6A). A composite score of PDCD1 + NKG7 + LAG3+ GZMH + GZMB + GNLY + FGFBP2 − HLA-G also demonstrated statistically significant associations with presence of ongoing disease (Figure 6B). Interestingly, expression levels of these genes did not always correlate with one another, indicating heterogeneity in their expression patterns and some degree of independence (Figure 6C). Trends in gene expression were similar for TNBC and non-TNBC patients, but in-depth subgroup analyses were limited by sample size. Thus, peripheral anti-tumor immunity in blood may be a useful measure of persistent residual primary or micrometastatic disease and could identify patients likely to benefit from additional therapy.
Figure 6: An 8-gene activated T cell signature derived from whole blood at surgery is associated with pCR and prognosticates recurrence in RD patients.
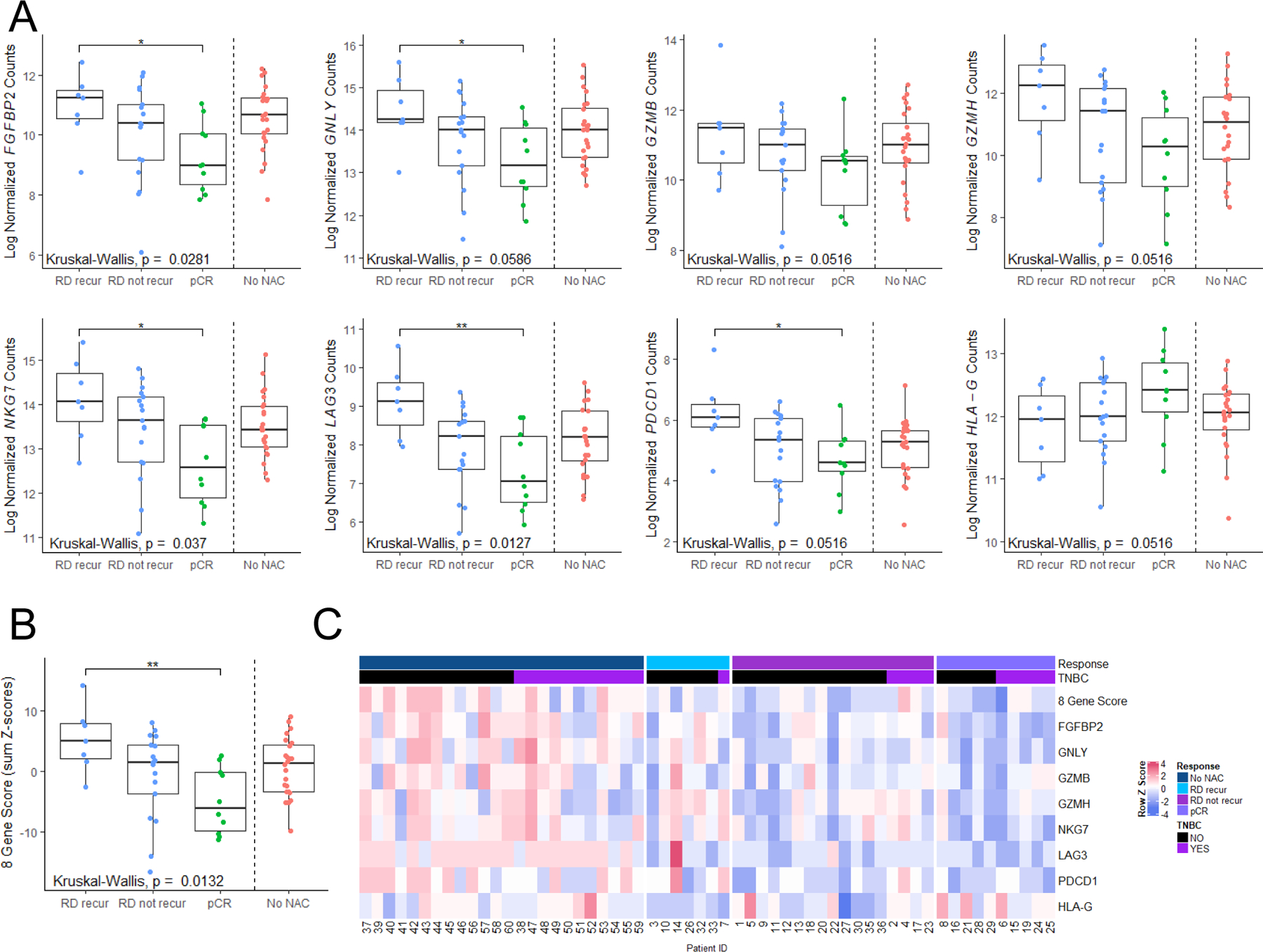
A) Individual gene plots of 8 analyzed genes by nanoString from RNA derived from whole blood sampled within 14 days leading up to definitive surgery. Datapoints are stratified by untreated patients (No NAC), those experiencing pCR (pCR), those with RD that did not recur (RD not recur) and those with RD that recurred (RD recur) within 3 years after surgery. Box plots represent the interquartile range. P values represent Kruskal-Wallis tests. * indicates p<0.05 by post-hoc Dunn test. B) A composite gene signature derived as PDCD1 + NKG7 + LAG3+ GZMH + GZMB + GNLY + FGFBP2 – HLA-G (sum of Z-scores), stratified by outcome, as in (A). C) Heatmap showing row-standardized (Z-score) gene expression for genes assayed across all patients.
To extend these findings, we evaluated gene expression in a second cohort of archival blood from breast cancer patients treated at DFCI (n=30), with blood collected in the interlude between completing NAC and definitive surgery. Notably, this cohort had differing baseline characteristics (summarized in Table 2). All of the patients were hormone receptor positive (a subtype known to have a lower pCR rate than TNBC patients), HER2 negative, and all were treated with bevacizumab. Intriguingly, expression of the 8 gene composite score was also correlated with enhanced disease burden in this cohort, being highest in patients with RCB III (regardless of recurrence) and lowest in patients with RCB 0/I/II who did not have a breast cancer recurrence within 3 years (Supplementary Figure 17). These findings suggest that peripheral blood gene expression may be predictive of response in diverse groups of breast cancer patients.
Discussion
With the recent approval of immunotherapy in combination with chemotherapy in mTNBC, and promising early results in the neoadjuvant setting, an improved understanding of how chemotherapy re-shapes anti-tumor immunity, both in the tumor and in the peripheral compartment, is needed. Perhaps the most widely studied marker to approximate anti-tumor immunity is sTILs, which are predictive of improved NAC response when measured in the primary untreated tumor and are prognostic of good outcomes in the residual disease of patients lacking pCR. Moreover, sTILs have primarily been a useful biomarker in TNBC as opposed to hormone receptor-positive cancers, but this inference is complicated by the routine use of endocrine-targeted agents in the adjuvant setting, that can impact post-surgical outcomes. However, this likely confounder leaves space for a biological contribution, as numerous differences between molecular and clinical subtypes exist. Even when considering only TNBC, quantification of sTILs is an imperfect biomarker, which does not precisely inform on the immuno-biology of the tumor.
In this study, we present a molecular analysis of the TIME in response to NAC in 83 breast cancer patients, specifically focusing on patients lacking a pCR, as these patients have worse outcomes. We found that changes in tumor immunity seem to be most prevalent in TNBC, often resulting in decreases in expression of immune-related genes. However, an upregulation of immune-related gene expression in tumors following NAC was associated with a strikingly improved outcome after surgery, specifically in TNBC. Of these genes, those involved in cytotoxic effector cells were among the most robustly associated with outcome. Furthermore, we found that cytokines expressed by PD-1HI CD8+ T cells in the peripheral blood were increased dramatically in TNBC patients following NAC.
Analysis of TCR clonotypes infiltrating into tumors suggested that chemotherapy may preferentially increase the recruitment of new T cell clones into the tumor, rather than expanding the T cells already present. This effect was consistent with that observed in the peripheral blood, where PD-1HI CD8+ T cells, while highly clonal compared to PD-1NEG cells, did not substantially change in clonality during NAC; these observations reflect a lack of clonal expansion in response to NAC, as we found in the TIME.
Assessment of peripheral blood represents a unique opportunity to monitor anti-tumor immunity through minimally invasive means. Using scRNAseq, we identified a population of cytolytic effector T cells in blood that expressed elevated levels of exhaustion/checkpoint genes. A gene expression signature derived from this population was used to test the hypothesis that these highly cytolytic, but potentially exhausted cells may be reflective of an ongoing disease process, and therefore a peripheral approximation of disease burden. This hypothesis was confirmed in two validation sets totaling 88 breast cancer patients and serves as a proof-of-principle for the use of this signature as a possible biomarker of outcome.
There are several limitations of our study. We intentionally chose to focus on patients lacking a pCR for the tumor study, as these patients have the worst outcomes after surgery, and because of the caveats and difficulty in defining or assessing anti-tumor immunity after chemotherapy in surgical specimens that lack tumor tissue. Future studies will need to be done to determine whether alterations in immune-related genes are seen over the course of NAC in patients experiencing pCR. Furthermore, our sample size was limited in our exploration of peripheral blood PD-1Hi CD8+ T cells by single cell RNA sequencing to two patients. Using this method, we identified a unique cluster of cytotoxic cells and derived a gene signature. We used further single cell RNA sequencing to identify that this gene signature is expressed predominately by circulating CD8+ T cells and NK cells. Therefore, expression of this gene signature in whole blood is not limited to PD-1Hi CD8+ T cells. We tested expression of this gene signature in whole blood of two different cohorts of breast cancer patients and found that expression was highest in those with residual disease who recurred or had the largest residual cancer burden. This provides proof-of-concept that peripheral blood gene signatures may predict response, but optimization of signature genes is likely needed to improve prediction.
Importantly, there has been a paucity of studies looking at the effect of chemotherapy on peripheral blood25, with little data on disease outcomes. This study provides a novel assessment and framework for an improved understanding of how chemotherapy alters anti-tumor immunity both in the TIME and the peripheral compartment. These data represent a unique opportunity to better understand patient populations most likely to benefit from the addition of immunotherapy to chemotherapy, particularly in the neoadjuvant setting. Furthermore, our findings demonstrating the association of expression of key cytolytic and immune-activation genes in the peripheral blood with presence of residual disease and recurrence represent a possible biomarker platform. Peripheral gene expression signatures may identify high-risk populations with potentially exhausted T cells and either primary or micrometastatic disease who may benefit from additional immunotherapeutic strategies.
Supplementary Material
Translational Relevance:
Indications for immunotherapy, alone or in combination, are expanding, including in breast cancer. However, the immunologic landscape of breast cancer and how chemotherapy, the current standard of care for triple negative breast cancer (TNBC), influences local and systemic immune responses are incompletely characterized. Herein, we show that increases in expression of immune-related genes and gene sets in the tumor over the course of chemotherapy are associated with improved prognosis in TNBC, but not other subtypes of breast cancer. Conversely, a gene expression signature of immune activation and cytotoxicity in the peripheral blood was associated with persistent disease following chemotherapy and disease recurrence following surgery. Examining immune-related signatures locally and systemically may serve as biomarkers of patients likely to benefit from additional immunotherapeutic approaches.
Acknowledgements:
Funding for this work was provided by Susan G. Komen Career Catalyst Grant CCR14299052 (JMB), NIH/NCI R00CA181491 (JMB), NIH/NCI SPORE 2P50CA098131-17 (JMB), Department of Defense Era of Hope Award BC170037 (JMB), and the Vanderbilt-Ingram Cancer Center Support Grant P30 CA68485. Additional funding was provided by NIH T32GM007347 (MLA) and F30CA236157 (MLA); F30CA216966 (KS); R01CA200994 (TWM) and R01CA211869 (TWM) and Dartmouth College Norris Cotton Cancer Center Support Grant P30CA023108 (TWM).
Footnotes
Conflict of Interest Statement: Justin Balko receives research support from Genentech/Roche, Bristol Myers Squibb, and Incyte Corporation, has received consulting/expert witness fees from Novartis, and is an inventor on provisional patents regarding immunotherapy targets and biomarkers in cancer. Roberto Salgado receives research support from Roche, Puma and Merck, has received travel funding from Roche Merck and Astra Zeneca, and is on the advisory board of BMS and Merck. Todd W Miller has received research support from Takeda Pharmaceuticals. Ingrid Mayer has received research funding from Pfizer and Genentech and is on the advisory board for Genentech, Novartis, Lilly, GSK, Immunomedics, Macrogenics, Seattle Genetics, and Astra-Zeneca. Vandana Abramson has received research support from Genentech and Pfizer and consulting fees from Daiichi Sankyo, AbbVie, and Esai. Jing Zhou is employed by and has equity ownership in IsoPlexis; Sean Mackay is cofounder of, has equity ownership in and holds patents with IsoPlexis. Sara Tolaney receives institutional research funding from Novartis, Genentech, Eli Lilly, Pfizer, Merck, Exelixis, Eisai, Bristol Meyers Squibb, AstraZeneca, Cyclacel, Immunomedics, Odonate, Sanofi, and Nektar. Sara Tolaney has served as an advisor/consultant to Novartis, Eli Lilly, Pfizer, Merck, AstraZeneca, Eisai, Puma, Genentech, Immunomedics, Nektar, Paxman, Athenex, Oncopep, Daiichi-Sankyo, G1 Therapeutics, Silverback Therapeutics, Kyowa Kirin Pharmaceuticals, AbbVie, Sanofi, Seattle Genetics, Celldex, Bristol-Myers Squibb and Nanostring. Ian Krop has received research support (paid to his institution) from Genentech/Roche and Pfizer, has received fees from Novartis and Merck for Data Monitoring Board participation, and has received consulting fees from Bristol Meyers Squibb, Daiichi/Sankyo, Macrogenics, Context Therapeutics, Taiho Oncology, Genentech/Roche , Seattle Genetics, Celltrion, and AstraZeneca. All other authors declare no potential conflicts of interest.
References:
- 1.Schmid P, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. The New England journal of medicine 379, 2108–2121 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Schmid P, et al. LBA8_PRKEYNOTE-522: Phase III study of pembrolizumab (pembro) + chemotherapy (chemo) vs placebo (pbo) + chemo as neoadjuvant treatment, followed by pembro vs pbo as adjuvant treatment for early triple-negative breast cancer (TNBC). Annals of Oncology 30(2019). [Google Scholar]
- 3.Denkert C, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 33, 983–991 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Loi S, et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 37, 559–569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luen SJ, et al. Prognostic implications of residual disease tumor-infiltrating lymphocytes and residual cancer burden in triple-negative breast cancer patients after neoadjuvant chemotherapy. Annals of oncology : official journal of the European Society for Medical Oncology 30, 236–242 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Loi S, et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin Cancer Res 22, 1499–1509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieci MV, et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: a retrospective multicenter study. Annals of oncology : official journal of the European Society for Medical Oncology 25, 611–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parra ER, et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in non-small cell lung carcinomas as determined by multiplex immunofluorescence and image analysis approaches. Journal for immunotherapy of cancer 6, 48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim R, et al. Immune correlates of the differing pathological and therapeutic effects of neoadjuvant chemotherapy in breast cancer. Eur J Surg Oncol 46, 77–84 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Li X, et al. Immune profiling of pre- and post-treatment breast cancer tissues from the SWOG S0800 neoadjuvant trial. Journal for immunotherapy of cancer 7, 88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tolaney SM, et al. Role of vascular density and normalization in response to neoadjuvant bevacizumab and chemotherapy in breast cancer patients. Proceedings of the National Academy of Sciences of the United States of America 112, 14325–14330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denkert C, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. The Lancet. Oncology 19, 40–50 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Rosshart S, et al. Interaction of KLRG1 with E-cadherin: new functional and structural insights. European journal of immunology 38, 3354–3364 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Gros A, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nature medicine 22, 433–438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DB, et al. Tumor-specific MHC-II expression drives a unique pattern of resistance to immunotherapy via LAG-3/FCRL6 engagement. JCI Insight 3(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han M, et al. Metaplastic breast carcinoma: a clinical-pathologic study of 97 cases with subset analysis of response to neoadjuvant chemotherapy. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 32, 807–816 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kaminsky A, Bhargava R, McGuire KP & Puhalla S Single institution experience with neoadjuvant chemotherapy for metaplastic breast cancer (MBC). Journal of Clinical Oncology 30, 1038–1038 (2012). [Google Scholar]
- 18.Bae SY, et al. The prognoses of metaplastic breast cancer patients compared to those of triple-negative breast cancer patients. Breast cancer research and treatment 126, 471–478 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Aydiner A, et al. Metaplastic Breast Carcinoma Versus Triple-Negative Breast Cancer: Survival and Response to Treatment. Medicine (Baltimore) 94, e2341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Hilli Z, et al. Metaplastic breast cancer has a poor response to neoadjuvant systemic therapy. Breast cancer research and treatment 176, 709–716 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledderose C, Heyn J, Limbeck E & Kreth S Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res Notes 4, 427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumont C, et al. CD8(+)PD-1(−)ILT2(+) T Cells Are an Intratumoral Cytotoxic Population Selectively Inhibited by the Immune-Checkpoint HLA-G. Cancer immunology research 7, 1619–1632 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P & LeMaoult J HLA-G: An Immune Checkpoint Molecule. Adv Immunol 127, 33–144 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Lin A & Yan WH Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol 9, 2164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foulds GA, et al. Immune-Phenotyping and Transcriptomic Profiling of Peripheral Blood Mononuclear Cells From Patients With Breast Cancer: Identification of a 3 Gene Signature Which Predicts Relapse of Triple Negative Breast Cancer. Front Immunol 9, 2028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balko JM, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 4, 232–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendry S, et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendry S, et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv Anat Pathol 24, 235–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salgado R, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 26, 259–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma C, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nature medicine 17, 738–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proceedings of the National Academy of Sciences of the United States of America 112, E607–615 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma C, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer discovery 3, 418–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi J, et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 132, 804–814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Q, et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. Journal for immunotherapy of cancer 5, 85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisi G, et al. Persistence of adoptively transferred T cells with a kinetically engineered IL-2 receptor agonist. Nature communications 11, 660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ImmunoMind. ImmunoMind Team. immunarch: An R Package for Painless Analysis of Large-Scale Immune Repertoire Data. . (ImmunoMind Team, 2019). [Google Scholar]
- 37.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nature biotechnology 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korsunsky I, et al. Fast, sensitive, and accurate integration of single cell data with Harmony. bioRxiv, 461954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linderman GC, Zhao J & Kluger Y Zero-preserving imputation of scRNA-seq data using low-rank approximation. bioRxiv, 397588 (2018). [Google Scholar]
- 41.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature immunology 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martens JH & Stunnenberg HG BLUEPRINT: mapping human blood cell epigenomes. Haematologica 98, 1487–1489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 45.Gu Z, Eils R & Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


