Abstract
The fungus Candida albicans colonizes the oral mucosal surface of 30–70% of healthy individuals. Due to local or systemic immunosuppression, this commensal fungus is able to proliferate resulting in oral disease, called oropharyngeal candidiasis (OPC). However, in healthy individuals C. albicans causes no harm. Unlike humans mice do not host C. albicans in their mycobiome. Thus, oral fungal challenge generates an acute immune response in a naive host. Therefore, we utilized C. albicans clinical isolates which are able to persist in the oral cavity without causing disease to analyze adaptive responses to oral fungal commensalism. We performed RNA sequencing to determine the transcriptional host response landscape during C. albicans colonization. Pathway analysis revealed an upregulation of adaptive host responses due to C. albicans oral persistence, including the upregulation of the immune network for IgA production. Fungal colonization increased cross-specific IgA levels in the saliva and the tongue, and IgA+ cells migrated to foci of fungal colonization. Binding of IgA prevented fungal epithelial adhesion and invasion resulting in a dampened proinflammatory epithelial response. Besides CD19+ CD138− B cells, plasmablasts, and plasma cells were enriched in the tongue of mice colonized with C. albicans suggesting a potential role of B lymphocytes during oral fungal colonization. B cell deficiency increased the oral fungal load without causing severe OPC. Thus, in the oral cavity B lymphocytes contribute to control commensal C. albicans carriage by secreting IgA at foci of colonization thereby preventing fungal dysbiosis.
Keywords: oropharyngeal candidiasis, commensalism, fungi, host-pathogen interaction, B cell, antifungal immunity
Introduction
The occurrence of fungal infections is rising and a serious threat to the public health (1), yet this problem is relatively underappreciated by the press, the public and funding agencies. In the United States fungal diseases cost more than $7.2 billion annually, including $4.5 billion from >75,000 hospitalizations and $2.6 billion from ~9 million outpatient visits (2). Very few fungal species cause disease in humans. However, some of these opportunistic fungal pathogens are ubiquitous members of the normal human mycobiome. Indeed, the oral cavity hosts various commensal fungal species (3). As part of the human mycobiome the polymorphic fungus Candida albicans colonizes the oral mucosal surface of up to 70% of healthy individuals (4). Due to local or systemic immunosuppression, this fungus is able to proliferate resulting in oral disease, termed oropharyngeal candidiasis (OPC) (5). However, in healthy individuals C. albicans causes no harm. In fact, commensal fungi, such as C. albicans, are required for microbial community structure, metabolic function, and immune priming (6–8). An imbalance of the mycobiome equilibrium, termed fungal dysbiosis, changes the functional composition, structure, and metabolic activities of the host microbial communities (7). Gain of function dysbiosis (9, 10) may lead to mucosal fungal infection such as OPC. The human host evolved finely tuned innate and adaptive immune responses enabling to control fungal commensal organism (5, 11). Unlike humans mice do not host C. albicans in their mycobiome (12). Thus, oral fungal challenge with the commonly used laboratory C. albicans strain SC5314 generates an acute immune response in a naive host (13). Since the adaptive immunity plays a critical role in maintaining immune tolerance toward commensal organisms, such as commensal C. albicans, understanding its relationship with fungi is critical (14). In an adaptive immunity OPC rechallenge model using a derivate of the pathogenic C. albicans strain SC5314, CD4+ Th17 cells protect from mucosal Candida infection but can be compensated by other IL-17-producing cells in CD4-deficient hosts (15, 16). However, as a commensal, C. albicans is in constant interaction with the host epithelium (17). Therefore, lack of incessant fungal exposure limited our advances in understanding antifungal adaptive immune responses at mucosal surfaces in this rechallenge model. Recently, Schonherr et al. showed that prolonged oral C. albicans colonization depends on the fungal isolate and can be accomplished without immunosuppression of the host thus mimicking the scenario in humans (18). Strikingly, fungal persistence in the oral cavity is independent of a suppressed antifungal immunity since regulatory T cells depletion or deletion of the immune regulatory cytokine IL-10 did not alter the protective type 17 immunity (19). However, tissue-resident memory (TRM) Th17 cells prevent uncontrolled outgrowth of the commensal fungus (20).
Immunoglobulin A (IgA) is thought to be a bridge between the innate and adaptive immunity. IgA is predominantly induced in response to colonization with commensal organism therefore maintaining homeostasis via immune exclusion (21–23). Among the production of mucosal antibodies, particularly IgA, by tissue-resident B cells is key to controlling the composition of the microbiome (24). IgA is the dominant antibody isotype in the mucosal immune system, which widely exists in the gastrointestinal tract, respiratory tract, vaginal tract, tears, saliva, and colostrum (25). Immune exclusion is the primary mechanism by which secretory low-affinity IgA (sIgA) blocks microorganisms from attaching to mucosal epithelial cells, thereby preventing colonization, damage, and subsequent invasion (26).
In the present study, we utilized C. albicans clinical isolates which are able to persist in the oral cavity without causing disease to analyze adaptive responses to C. albicans colonization. We found that oral fungal colonization upregulates adaptive host responses, including the upregulation of the immune network for IgA production. C. albicans colonization increased the total salivary and tissue IgA levels, thereby preventing adhesion and invasion of the fungus. Furthermore, B cells, plasmablasts, and plasma cells accumulated to foci of fungal colonization at the epithelial surface. Importantly B cell deficiency and antibody-mediated B lymphocyte depletion increased the commensal C. albicans load without causing severe OPC. Thus, in the oral mucosa accumulating B lymphocytes and secreted IgA control commensal C. albicans carriage by preventing fungal outgrowth.
Materials and Methods
Ethics Statement
All animal work was approved by the Institutional Animal Care and Use Committee (IACUC) of the Lundquist Institute at Harbor-UCLA Medical Center.
Organisms and Cell Lines
The C. albicans strains SC5314 (27), 529L (28), and CA101 (18) were used in the experiments and were grown as described previously (29). The Streptococcus oralis strain was purchased from the American Type Culture Collection (ATCC; #35037) and was grown in brain heart infusion broth. The OKF6/TERT-2 oral epithelial cell line (30) was grown as described (31). The OKF6/TERT-2 cells have been authenticated by RNA-Seq (32), and have been tested for mycoplasma contamination. The cell line of murine tongue-derived keratinocytes (TDKs) were kindly provided by S. LeibundGut-Landmann and grown as previously described (33).
Mouse Model of Oropharyngeal Candidiasis
Six week old male C57BL/6J, B6.129S2-Ighmtm1Cgn/J (muMt), and B6.129S7-Rag1tm1Mom/J (Rag1 KO) mice were purchased from Jackson laboratories and housed for 1 week in a pathogen free facility prior infection. The mice were randomly assigned to the infection groups. OPC was induced in mice as described previously (34, 35). Briefly, for inoculation, the animals were sedated, and a swab saturated with 2 × 107 C. albicans cells was placed sublingually for 75 min. For colony-forming unit (CFU) enumeration the tongues were harvested, weighed, homogenized and quantitatively cultured. Wild-type (C57BL/6J) mice were sacrificed after 2, 5, 11, and 20 days of infection. muMT and Rag1 KO mice were sacrificed after 7 days of oral infection with the commensal stains 529L and CA101, respectively. To determine CFUs of commensal outgrowth after systemic immunosuppression wild-type mice were colonized with 529L and CA101. On day 11 and 13 post oral infection mice were given a subcutaneous injection of 25 mg/kg triamcinolone (Kenalog-10, Bristol-Myers Squibb Company). The researchers were not blinded to the experimental groups because the endpoints (oral fungal burden) were an objective measure of disease severity. Saliva was collected at day 11 post infection into chilled tubes after intraperitoneal carbachol injection (100 μl at 10 mg/ml). For antibody depletion, wild-type mice were treated intraperitoneally with 300 μg of anti-mouse B220 (RA3.3A1/6.1, BioXCell) and 300 μg of anti-mouse CD19 (1D3, BioXCell), or isotype controls (2A3, BE0094, BioXCell) on day−1, 4, and 9 relative to infection. For RNA sequencing tongues from 3 Sham-infected and 3 529L colonized mice after 5 and 11 days post infection were processed for analysis.
RNA Sequencing
Total RNA was isolated as described elsewhere (32) and RNA sequencing was performed by Novogene Corporation Inc. (Sacramento, USA). mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. To generate the cDNA library the first cDNA strand was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H−). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Double-stranded cDNA was purified using AMPure XP beads and remaining overhangs of the purified double-stranded cDNA were converted into blunt ends via exonuclease/polymerase. After 3' end adenylation a NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments of 150~200 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA). Finally, PCR amplification was performed and PCR products were purified using AMPure XP beads. The samples were read on an Illumina NovaSeq 6000 with ≥20 million read pair per sample.
Downstream Data Processing
Downstream analysis was performed using a combination of programs including STAR, HTseq, and Cufflink. Alignments were parsed using Tophat and differential expressions were determined through DESeq2. KEGG enrichment was implemented by the ClusterProfiler. Gene fusion and difference of alternative splicing event were detected by Star-fusion and rMATS. The reference genome of Mus musculus (GRCm38/mm10) and gene model annotation files were downloaded from NCBI/UCSC/Ensembl. Indexes of the reference genome was built using STAR and paired-end clean reads were aligned to the reference genome using STAR (v2.5). HTSeq v0.6.1 was used to count the read numbers mapped of each gene. The FPKM of each gene was calculated based on the length of the gene and reads count mapped to it. FPKM, Reads Per Kilobase of exon model per Million mapped reads, considers the effect of sequencing depth and gene length for the reads count at the same time (36). Differential expression analysis was performed using the DESeq2 R package (2_1.6.3). The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the False Discovery Rate (FDR). Genes with an adjusted P-value < 0.05 found by DESeq2 were assigned as differentially expressed (cutoff fold change 1.5, Supplementary Table 1). To allow for log adjustment, genes with 0 FPKM are assigned a value of 0.001. Correlation were determined using the cor.test function in R with options set alternative = “greater” and method = “Spearman.” To identify the correlation between the differences, we clustered different samples using expression level FPKM to see the correlation using hierarchical clustering distance method with the function of heatmap, SOM (Self-organization mapping) and kmeans using silhouette coefficient to adapt the optimal classification with default parameter in R. We used clusterProfiler R package to test the statistical enrichment of differential expression genes in KEGG pathways. The high-throughput sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA) under accession number PRJNA657562.
Adhesion and Invasion Assay
Adhesion of C. albicans and invasion into oral epithelial cells was quantified by a differential fluorescence assay as described previously (31, 37). Briefly, OKF6/TERT-2 cells were grown to confluency on fibronectin-coated circular glass coverslips in 24-well tissue culture plates. C. albicans was incubated with 100 μg/ml sIgA (BioRad) for 30 min. The epithelial cells were infected with 2 × 105 yeast-phase C. albicans SC5314 cells per well (multiplicity of infection; MOI 1) and incubated for 2.5 h, after which they were fixed, stained, and mounted inverted on microscope slides. The coverslips were viewed with an epifluorescence microscope, and the number of endocytosed organisms per high-power field was determined, counting at least 100 organisms per coverslip. Each experiment was performed at least three times in triplicate. To determine the effect of saliva from infected mice on Candida adhesion and invasion saliva from 3 mice was pooled, diluted 1:1 in PBS and incubated with C. albicans before added to 2 × 105 the murine keratinocyte cells for 2.5 h. Fungal adhesion and invasion were determined as described above.
Cytokine and Chemokine Measurements in vitro
Cytokine levels in culture supernatants were determined as previously described (38). Briefly 2 × 105 OKF6/TERT-2 cells in a 24-well plate were infected with C. albicans SC5314 at a MOI of 5. Prior to infection C. albicans was coated with sIgA as described above. After 8 h of infection, the supernatant was collected, clarified by centrifugation and stored in aliquots at −80°C. The concentration of inflammatory cytokines and chemokines in the medium was determined using the Luminex multipex assay (R&D Systems). Each condition was tested in three independent experiments.
IgA ELISA
To determine IgA levels saliva was collected as described above, and diluted 1:10 in PBS. Tongue homogenates were collected as described previously (38) and analyzed for IgA levels using manufactures instructions (IgA Elisa Mouse, Invitrogen).
Immunofluorescence
To determine B220+ and IgA+ cell localization in vivo, 30–50-μm-thick sections of OCT-embedded tongues were fixed with cold acetone. Next, the cryosections were rehydrated in PBS and then blocked using BSA. To detect IgA (FITC, mA-6E1, Invitrogen) or B220 (Alexa 488, RA3-6B2, Biolegend) positive cells sections were incubated with 1:50 diluted antibody overnight. To detect C. albicans, the sections were also stained with an anti-Candida antiserum (Biodesign International) conjugated with Alexa Fluor 568 (Thermo Fisher Scientific) for 1 h. To visualize the nuclei, the cells were stained with DAPI (4′,6-diamidino-2-phenylindole). The sections (z-stack) were imaged by confocal microscopy. To enable comparison of fluorescence intensities among slides, the same image acquisition settings were used for each experiment.
Salivary IgA Binding to S. oralis
108 S. oralis were incubated with 10 μl saliva for 45 min isolated from Sham-infected mice, or mice infected with C. albicans 11 days post infection. Bacteria were washed 3 times before stained with anti-IgA antibody (FITC, mA-6E1). The stained organisms were analyzed on FACSymphony system (BD Biosciences), and the data were analyzed using FACS Diva (BD Biosciences) and FlowJo software (Treestar).
Flow Cytometry of B Lymphocytes
To determine the number of B lymphocytes in the mouse tongues single cell suspension were generated as described previously (39, 40). Briefly, mice were orally infected with C. albicans as described above. After 11 days of infection, the animals were administered a sublethal anesthetic mix intraperitoneally. The thorax was opened, and a part of the rib cage removed to gain access to the heart. The vena cava was transected and the blood was flushed from the vasculature by slowly injecting 10 ml PBS into the right ventricle. The tongue was harvested and cut into small pieces in 100 μl of ice-cold PBS. 1 ml digestion mix (4.8 mg/ml Collagenase IV; Worthington Biochem, and 200 μg/ml DNase I; Roche Diagnostics, in 1x PBS) was added after which the tissue was incubated at 37°C for 45 min. The resulting tissue suspension was then passed through a 100 μm cell strainer. The single-cell suspensions were incubated with rat anti-mouse CD16/32 (2.4G2; BD Biosciences) for 10 min in FACS buffer at 4°C to block Fc receptors. For staining of surface antigens, cells were incubated with fluorochrome-conjugated (FITC, PE, PE-Cy7, allophycocyanin [APC], APC-eFluor 780) antibodies against mouse TER-119 (TER-110, BioLengend), CD326 (G8.8, BioLegend), CD11b (M1/70, BioLegend), Gr-1 (RB6-8C5, BioLegend), CD3 (17A2, BioLegend), CD8a (53-6.7, BioLegend), CD4 (GK1.5, BioLegend), CD45R/B220 (9RA3-6B2, BioLegend), CD19 (6D5, BioLegend), CD138 (281-2, BioLegend). After washing with FACS buffer, the cell suspension was stained with a LIVE/DEAD fluorescent dye (7-AAD; BD Biosciences) for 10 min. For intracellular staining, cell viability was determined using BD Horizon Fixable Viability Stain 780 (BD Biosciences) followed by Cytofix/Cytoperm (BD Biosciences) treatment staining with KI67 antibody (16A8; BioLegend). The stained cells were analyzed on FACSymphony system (BD Biosciences), and the data were analyzed using FACS Diva and FlowJo software. Only single cells were analyzed, and cell numbers were quantified using PE-conjugated fluorescent counting beads (Spherotech).
Results
Commensal C. albicans Isolates Persist in the Oral Cavity and Cause OPC During Systemic Immunosuppression
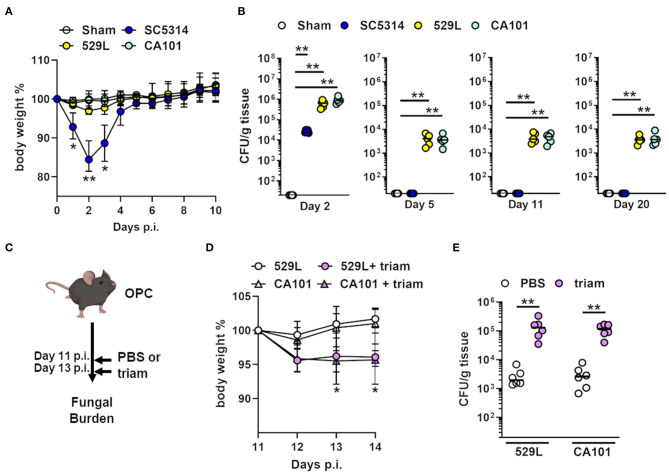
To investigate if oral commensal C. albicans isolates (18, 19) induce adaptive immune responses we infected wild-type mice orally with a pathogenic strain SC5314 and two commensal C. albicans isolates 529L and CA101, respectively. While oral infection with the pathogenic C. albicans strain led to significant body weight loss and rapid clearance from the oral cavity, the commensal strains 529L and CA101 colonized the oral mucosa and persisted for over 20 days without inducing significant weight loss in the host (Figures 1A,B). Although the commensal strains persist in the oral cavity, we noticed a significant decline between day 2 and 5 post infection (Figure 1B) similar what has been reported previously (20). Despite the fact that distinct C. albicans clinical isolates are able to persist in the oral cavity it is unclear if these strains can outgrow and induce severe oral disease. The induction of prolonged OPC in naive mice has been extensively studied using corticosteroids (31, 34, 35, 37, 38). To test the potential of commensal fungal outgrowth during immunosuppression we colonized mice with the C. albicans strains 529L and CA101 for 11 days and induced systemic immunosuppression using triamcinolone (Figure 1C). The administration of triamcinolone resulted in significant body weight loss and >40-fold increase in oral fungal burden (Figures 1D,E). These data indicate that systemic immunosuppression leads to fungal outgrowth of colonizing commensal C. albicans in the oral cavity.
Figure 1.
Commensal C. albicans strains cause OPC during immunosuppression. (A) Body weight (mean± range) of mice orally infected with indicated strains. *P < 0.05, **P < 0.01 (n = 10; Mann–Whitney). (B) Oral fungal burden of wild-type mice infected with indicated strains. Results are median of two independent experiments (n = 5). **P < 0.01 (Kruskal-Wallis). The y-axis is set at the limit of detection (20 CFU/g tissue). (C) Mouse model of immunosuppressed OPC during C. albicans colonization. Triam, triamcinolone. (D) Body weight (mean ± range) of mice starting day 11 during fungal colonization and immunosuppression. Day 11 post infection set as 100%. (E) Oral fungal burden of wild-type mice infected with indicated commensal strains 11 days post oral infection. Results are median of two independent experiments (n = 6). **P < 0.01 (Mann-Whitney). The y-axis is set at the limit of detection (20 CFU/g tissue).
C. albicans Oral Colonization Induces Upregulation of Adaptive Immune Response Signatures
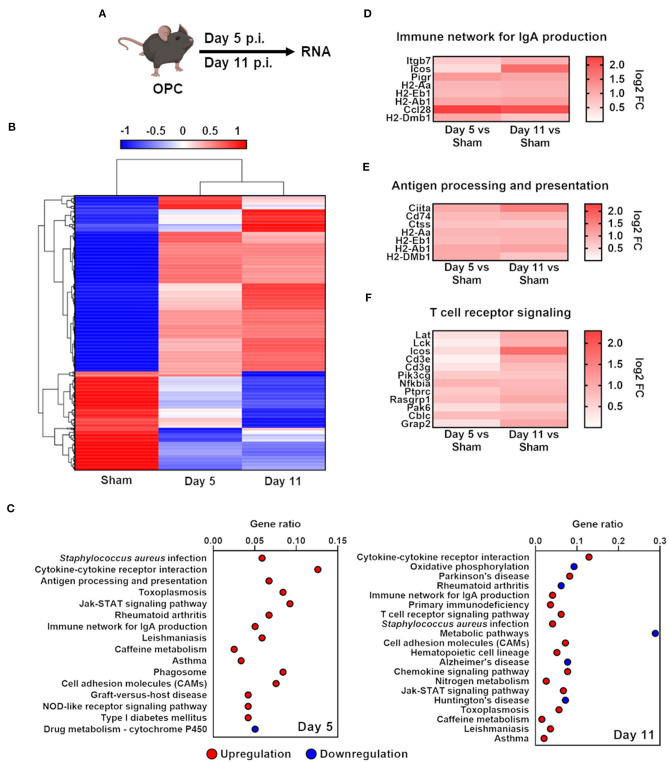
To obtain genome-wide information about the host response to commensal C. albicans colonization in the oral cavity, we performed RNA sequencing. Given the fact that the tested persisting C. albicans strains 529L and CA101 behaved similarly in the mouse model of OPC (Figure 1) we used 529L (41) as a representative commensal strain to assess the transcriptional commensal-specific host response landscape at 5 and 11 days post-infection compared to Sham-infected mice (Figure 2A). Using unsupervised hierarchical clustering, we found oral C. albicans colonization leads to robust changes and dynamic host responses (Figure 2B). C. albicans colonized mice were clustered in one group, with four broad gene clusters. Pathway analysis revealed an upregulation of adaptive host responses due to C. albicans persistence (Figure 2C), including the upregulation of the immune network for IgA production, antigen processing and presentation, and T cell receptor signaling (Figures 2D–F). Thus, oral fungal colonization leads to a robust induction of adaptive immune responses in C. albicans immunological naive mice.
Figure 2.
C. albicans oral colonization induces upregulation of adaptive response pathways in immunocompetent mice. (A) Scheme of infection with 529L or Sham and time points of RNA isolation. (B) Heat map showing hierarchical clustering of C. albicans colonization with 529L in the oral cavity after 5 and 11 days of genes with a fold change of FC >1.5. Red denotes genes with high expression levels, and blue denotes genes with low expression levels. The color ranging from red to blue indicates log10 (FPKM+1) value from highest to lowest. (C) Identified pathways of enriched genes FC > 1.5, adjusted P < 0.05. Shown is the gene ratio (Genes of pathway/all different expressed genes; padj ≤ 0.5). (D–F) Heat map of enriched genes of corresponding pathways.
Oral C. albicans Colonization Upregulates Salivary IgA and Induces Migration of IgA Secreting Cells in the Oral Epithelial Layer
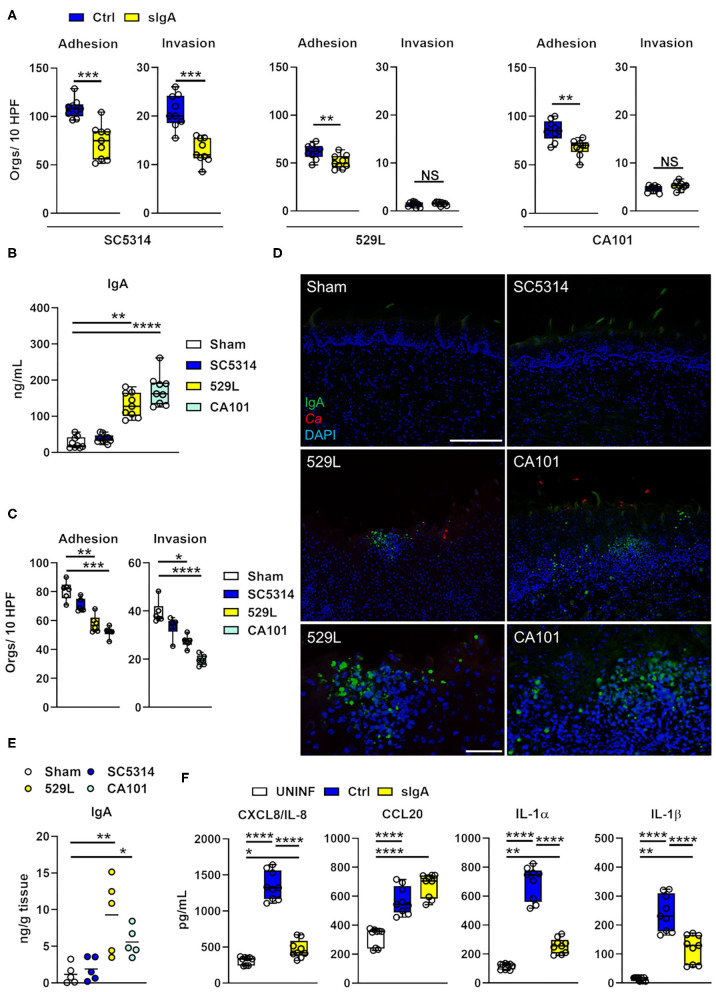
sIgA inhibits the adhesion of C. albicans hyphae to polystyrene (42). Since epithelial adhesion and invasion are required for C. albicans oral infection (4, 43) we determined if the physiological sIgA concentration found in healthy individuals (44) prevents adhesion of C. albicans to and invasion of human oral epithelial cells. Incubation of the pathogenic C. albicans strain SC5314 with sIgA decreased adherence and invasion of OKF6-TERT2 oral epithelial cells (Figure 3A) and adhesion of the commensal strains 529L and CA101, respectively. Of note, the persisting C. albicans strains showed remarkable reduction of epithelial adhesion and invasion compared to the pathogenic strain SC5314 (Figure 3A). High slgA levels are found in various secretory fluids, including saliva (45). Therefore, we measured IgA levels in the saliva of commensal and pathogenic strain infected mice, and found that C. albicans oral commensal colonization with 529L and CA101 increased the abundance of total IgA, while infection with the pathogenic strain SC5314 did not upregulate salivary IgA compared to Sham-infected mice (Figure 3B). Next, we measured if the saliva from Sham- or C. albicans-infected mice is able to prevent fungal adhesion to and invasion of murine tongue-derived keratinocytes (33) using the pathogenic strain SC5314. Saliva from commensal colonized mice was able to prevent epithelial adhesion and invasion compared to saliva from Sham-infected mice or mice infected with the pathogenic strain SC5314 (Figure 3C). The majority of the hosts entire pool of activated B cells is located near the mucosa as well as exocrine glands (46). Thus, we assessed tissue distribution of IgA+ cells in Sham-infected mice, or mice infected either the pathogenic strain SC5314 or the commensal strains 529L and CA101, respectively. IgA+ cells accumulated exclusively in oral epithelial and submucosal layers of mice colonized with the commensal C. albicans strains 529L and CA101 (Figure 3D). Consistent with this observation total IgA levels increased in tissue homogenates of commensal C. albicans colonized mice after 11 days of infection (Figure 3E). By secreting proinflammatory cytokines and chemokines, oral epithelial cells are vital for limiting fungal proliferation during OPC (4, 5). In vivo commensal C. albicans strains fail to induce a strong acute inflammatory response (18). Therefore, we assessed oral epithelial proinflammatory cytokine/chemokine production in the presence and absence of sIgA. We found that binding of sIgA to C. albicans dampend the secretion of the inflammatory mediators CXCL8/IL-8, IL-1α, and IL-1β while CCL20 secretion was unaffected (Figure 3F) suggesting that C. albicans-IgA interactions reduces a subset of the proinflammatory epithelial response. Collectively, our data suggest that fungal colonization upregulates salivary and tissue IgA production by inducing migration of IgA+ cells in close proximity of colonizing fungi and thereby preventing adhesion and invasion of fungi in the oral cavity.
Figure 3.
Oral fungal colonization upregulates mucosal IgA preventing C. albicans epithelial adhesion and invasion. (A) Indicated C. albicans strains were incubated with sIgA prior to infection of the OKF6/TERT-2 oral epithelial cell line. The cells were infected for 2.5 h, after which the number of adhered and invaded organisms was determined using a differential fluorescence assay. Box Whisker plot shows three experiments, each performed in triplicate. Orgs/10 HPF, organisms per 10 high-power fields; Ctrl, control. Statistical significance was determined using Mann-Whitney test (*P < 0.05; **P < 0.01). (B) Total IgA amounts in saliva (diluted 1:10) determined by ELISA. Saliva was collected after 11 days of infection with indicated C. albicans strains. Three independent experiments performed in triplicate. **P < 0.01; ****P < 0.0001 (Kruskal-Wallis). (C) Indicated C. albicans strain was incubated with saliva of infected with prior infection of the murine keratinocyte cell line. The cells were infected for 2.5 h, after which the number of adhered and invaded organisms was determined using a differential fluorescence assay. Box Whisker plot shows three experiments, each performed in dublicate. Orgs/10 HPF, organisms per 10 high-power fields. Statistical significance was determined using Kruskal-Wallis test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (D) Immunofluorescence of IgA in tongues 11 days post oral infection with indicated strains. IgA is shown in green, and C. albicans (Ca) in red. DAPI (blue) visualizes the tissue. Upper, middle panel scale bar 200 μm. Lower panel scale bar 50 μm. (E) Total IgA amounts in tongue homogenates determined by ELISA. Tongues were collected after 11 days of infection with indicated C. albicans strains; N = 5. *P < 0.05; **P < 0.01 (Kruskal-Wallis). (F) Indicated C. albicans strains were incubated with sIgA prior to infection of the OKF6/TERT-2 oral epithelial cell line. The cells were infected for 8 h after which CXCL8/IL-8, CCL20, and IL-1α/β were determined in the supernatant. Box Whisker plot shows three experiments triplicate. Uninf; uninfected; Ctrl, control. Statistical significance was determined using Kruskal-Wallis (*P < 0.05; **P < 0.01; ****P < 0.0001).
C. albicans Oral Colonization Increases Cross-Specific IgA Levels in the Oral Cavity
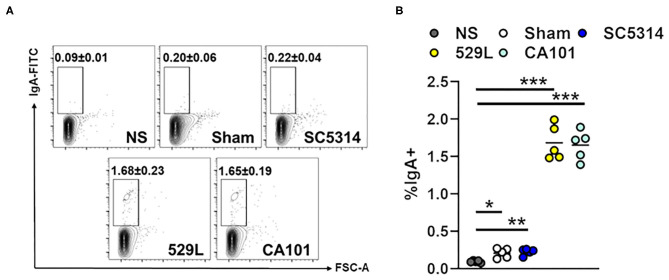
Secretory IgA can have a measurable reactivity to a diverse subset of the microbiota. IgAs interact with commensal organisms by canonical Fab-dependent and non-canonical carbohydrate-dependent binding (47). Therefore, we tested if salivary IgA of infected mice binds to the common oral commensal Streptococcus oralis (48). While IgA from Sham-infected and mice infected with the pathogenic strain SC5314 were able to bind S. oralis (Figures 4A,B) IgA binding to S. oralis increased >7-fold when the bacteria were incubated with saliva from commensal colonized mice. Thus, oral colonization with commensal C. albicans increases total levels of cross-specific IgAs.
Figure 4.
Fungal colonization increases cross-specific IgA levels. (A) Representative flow plots of salivary IgA bound to S. oralis. NS, No Saliva. (B) The percentage of IgA+ S. oralis. Results are median of two independent experiments with 5 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001 (Kruskal-Wallis).
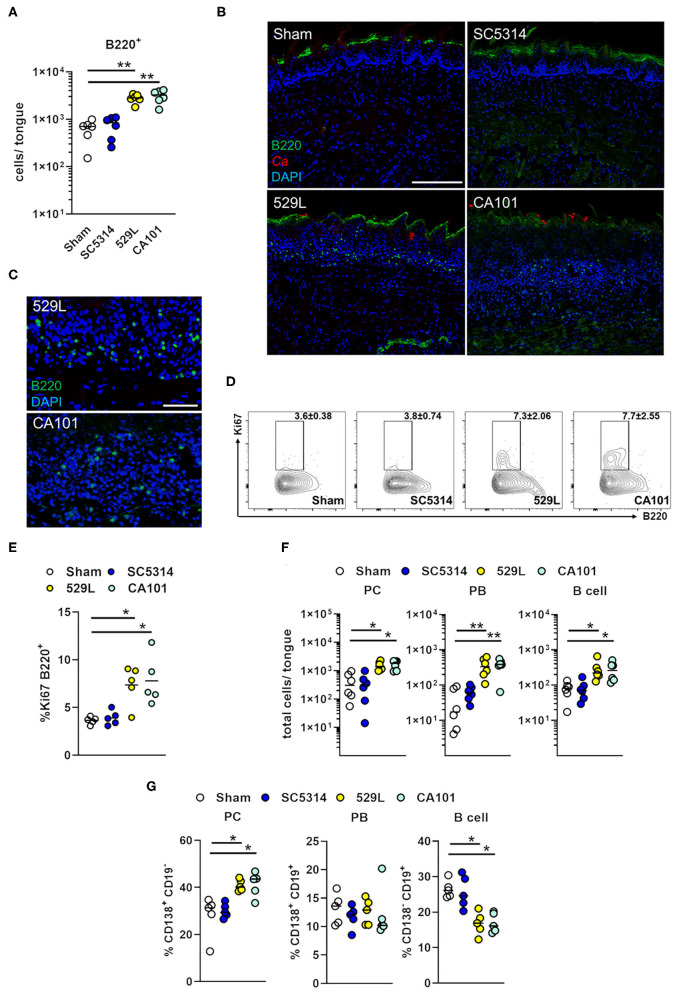
Mucosal B Lymphocytes Expand During Oral Fungal Colonization
Following the initiation of an immune response, B lymphocytes preferentially migrate back to initial sites of antigen encounter (49). We therefore determined B lymphocyte distribution in the oral cavity of infected mice. B lymphocytes (B220+ cells) were enriched during oral commensal colonization with 529L and CA101 in the whole tongue (Figure 5A, Supplementary Figure 1A) localized in the oral epithelial and submucosal layers during fungal colonization (Figures 5B,C). The expansion of B220+ cells could be due to local proliferation (50). Therefore, WT mice were infected orally and intracellular Ki67 was measured by flow cytometry. On day 11, Ki67+B220+ cells were more frequent in the infected oral mucosa of mice infected with the commensal C. albicans strains compared to Sham controls or pathogenic infected mice (Figures 5D,E). Thus, the expansion of B220+ cells during Candida oral colonization can be accounted for by proliferation at the site of fungal persistence. Next we determined B lymphocyte subpopulations including plasma cells (CD19− CD138+), plasmablasts (CD19+ CD138+), and mature B cells (CD19+ CD138−) in Sham-infected mice, mice infected with the pathogenic strain SC5314, or the commensal strains 529L and CA101 (Supplementary Figure 1B). We found that oral colonization with the commensal strains 529L and CA101 increased the tissue distribution of plasma cells, plasmablasts, and mature B cells, while no difference in B lymphocyte populations could be observed when mice were infected with the pathogenic C. albicans strain SC5314 compared to Sham-infected mice (Figure 5F). Although the total B lymphocyte numbers increased during commensal colonization (Figure 5F) the plasma cell population expanded and the CD138− CD19+ B cell frequency decreased in commensal colonized mice compared to Sham-infected mice (Figure 5G). Thus, oral fungal colonization results in expansion of B lymphocytes.
Figure 5.
B lymphocytes accumulate in the oral epithelial and submucosal layers during fungal colonization. (A) B220+ cell infiltration in tongues of immunocompetent wild-type mice after 11 days of infection with indicated C. albicans strains (n = 6). B220+ cells were gated on singlets live CD4− CD8− CD11b− Gr-1− TER-119− EpCam−. Results are median from combined results of two independent experiments. **P < 0.01 (Kruskal-Wallis). (B) Immunofluorescence of B220+ cells in tongues 11 days post oral infection with indicated strains. B220 is shown in green, and C. albicans (Ca) in red. DAPI (blue) visualizes the tissue. Scale bar 200 μm. (C) Scale bar 50 μm. (D) Representative flow plots of Ki67+ B220+ cells in the tongue after 11 days of infection. Cells were gated on singlets live B220+. (E) The percentage of Ki67+ B220+ cells. Results are median of a single experiment with 5 mice per group. *P < 0.05 (Kruskal-Wallis). (F) Total numbers of plasma cells (PC; CD19− CD138+), plasmablasts (PB; CD19+ CD138+), and mature B cells (CD19+ CD138−) were determined after 11 days post infection with indicated strains. Cells were gated on singlets live B220+ CD4− CD8− CD11b− Gr-1− TER-119− EpCam−. Results are median from combined results of two independent experiments (n = 6). *P < 0.5; **P < 0.01 (Kruskal-Wallis). (G) Percentage of plasma cells, plasmablasts, and mature B cells were determined after 11 days post infection with indicated strains. *P < 0.5 (Kruskal-Wallis).
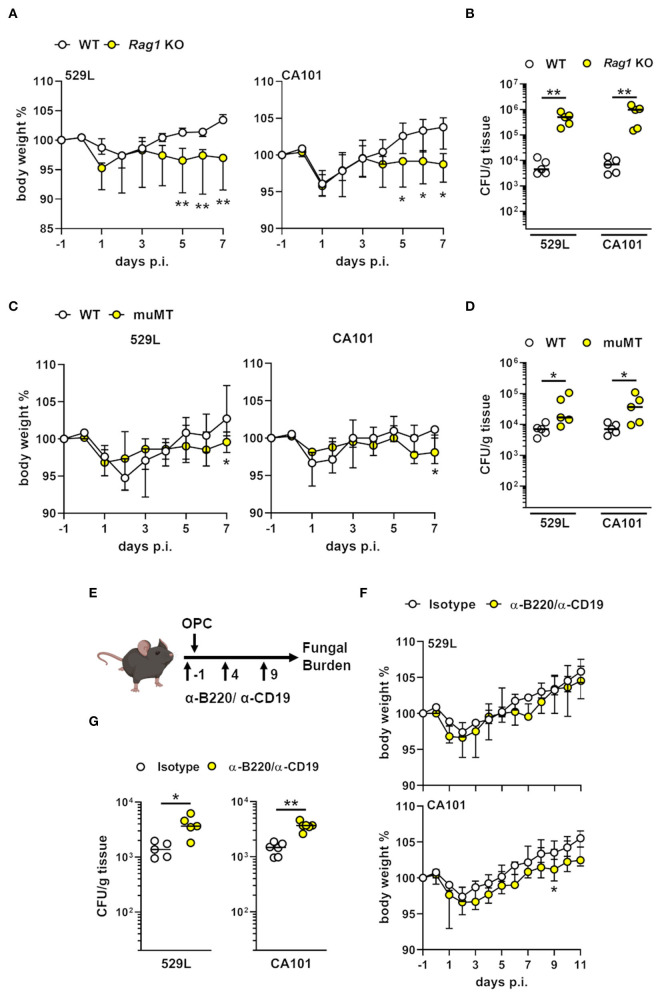
Mucosal B Lymphocytes Control Commensal Fungal Load in the Oral Cavity
Mice with Rag1 deficiency, the absence of endogenous B- and T cells, show increased susceptibility to oral fungal infection by the pathogenic C. albicans strain SC5314 (15). To determine the effect of Rag1 deficiency during commensal colonization we infected WT and Rag1 KO mice with the commensal strains 529L and CA101. After 5 days of infection Rag1 KO mice lost significantly more body weight compared to WT mice (Figure 6A). Next, we determined the oral fungal burden after 7 days of infection. Rag1 KO mice had an increase in fungal burden by >100-fold (Figure 6B) (20). Since Rag1 KO mice lack B- and T cells, we determined the importance of B lymphocytes during oral fungal colonization (51). In a mouse model of fungal asthma, mice lacking the Ig mu-chain (muMT) produce IgG and IgE, but not IgA (52). Therefore, we infected WT and muMT mice with the commensal C. albicans strains 529L and CA101, respectively. muMT lost slightly more body weight after 7 days post infection compared to WT mice (Figure 6C). Since the IgA pathway was already induced after 5 days of commensal colonization (Figure 2C) we determined the oral fungal after 7 days post oral inoculation. muMT mice either colonized with 529L or CA101 had an increased fungal burden by >2 to 5-fold (Figure 6D). In a different approach we treated mice with B220/CD-19 antibodies during commensal oral colonization (Figure 6E) thereby reducing mucosal B220+ cells (Supplementary Figure 2). B220/CD-19 depletion had minimal effect on body weight of colonized mice (Figure 6F), but increased the oral fungal load by >3-fold after 11 days of colonization (Figure 6G). Thus, the absence of B lymphocytes or the lack of IgA results in commensal C. albicans dysbiosis in the oral cavity.
Figure 6.
B lymphocytes contribute to commensal control in the oral cavity. (A) Body weight of Rag1 deficient and wild-type mice during oral commensal C. albicans colonization. *P < 0.05; **P < 0.01 (n = 5; Mann-Whitney). (B) Oral fungal burden of Rag1 KO and WT mice infected with indicated strains. Results are median of a single experiment (n = 5). **P < 0.01 (Mann-Whitney). The y-axis is set at the limit of detection (20 CFU/g tissue). (C) Body weight of muMT and wild-type mice during oral commensal C. albicans colonization. *P < 0.05 (n = 5; Mann-Whitney). (D) Oral fungal burden of muMT and WT mice infected with indicated strains. Results are median of a single experiment (n = 5). *P < 0.05 (Mann-Whitney). The y-axis is set at the limit of detection (20 CFU/g tissue). (E) Scheme B lymphocyte depletion during commensal OPC using anti-B220/CD19 antibodies. (F) Body weight of B lymphocyte depleted and isotype control mice. **P < 0.01 (n = 5–6; Mann-Whitney). (G) Oral fungal burden 11 days post infection of B lymphocyte depleted and isotype control mice infected with commensal C. albicans strains 529L and CA101. Results are median of 5–6 mice per group from two independent experiments. *P < 0.05; **P < 0.01 (Mann-Whitney).
Discussion
Healthy individuals have a protective Candida-specific mucosal immunity which limits fungal proliferation, invasion, and therefore preventing disease (5). Besides the innate immune response, the adaptive immunity to C. albicans is crucial to control mucosal fungal outgrowth (11). T cells are an integral component of the antifungal adaptive immune response and provide direct and indirect means of controlling fungal proliferation. Individuals with mutations in the Th17 pathway suffer from chronic mucocutaneous candidiasis (CMC) (53–55). Similarly to humans, mice exposed to C. albicans generate long-term adaptive Th17 cell responses that confer additional protection from infection (15, 56). In mice oral fungal persistence is independent of a suppressed antifungal immunity but requires tissue-resident memory Th17 cells to maintain stable fungal colonization (19, 20).
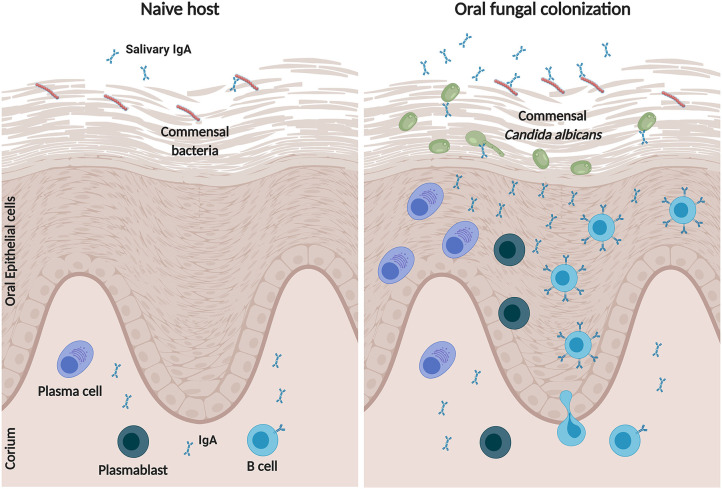
By utilizing persisting commensal C. albicans clinical isolates we show that oral colonization generates mucosal adaptive immune response signatures, including antigen processing and presentation, and T cell receptor signaling. In the oral mucosa monocyte-dependent and tissue-resident dendritic cells (DCs) orchestrate the antigen-specific T cell priming toward pathogenic C. albicans (57). Secretory antibodies of the IgA class released by effector B cells, including plasma cells, form the first line of immune protection against pathogens and antigens at mucosal surfaces linking the innate and adaptive host immunity (22, 58). Furthermore, mucosal IgA governs quantitative and qualitative control of commensal composition (59). Our data indicates that oral persistence of commensal C. albicans stimulates accumulation of B lymphocytes, including plasmablasts and plasma cells, to sites of fungal colonization, where these cells upregulate IgA production (Figure 7). This finding is in agreement with earlier reports showing that that oral mucosal defense against Candida involves innate phagocytes, T and B cell recruitment, as well as local antibody production with a prominent IgA component (60, 61). In a mouse model, oral fungal persistence is associated with a weakened proinflammatory host response compared to the pathogenic C. albicans strain SC5314 thus preventing C. albicans elimination at the onset of colonization (18, 19). While pattern recognition of fungi induces a strong epithelial proinflammatory response (38, 43, 62) the IgA-Candida interaction dampens this robust innate response by inhibiting epithelial adherence and invasion of the fungus. Here we show that immune exclusion is a result of Candida-IgA interactions in vitro. However, the in vivo mechanism remains unclear. A recent study showed that IgA-mediated pathogen cross-linking enchains the organism, thereby preventing separation after division resulting in clumping (63). This enchained growth accelerates pathogen clearance. Therefore, it is possible that IgA not only prevents C. albicans adhesion and epithelial invasion it also traps the fungus resulting in enchained growth and clearance.
Figure 7.
Model of B lymphocyte responses during commensal C. albicans colonization in the oral cavity. In a naive host, salivary and tissue IgA binds to commensal bacteria, which in turn regulates mucosal immunity and microbial compostion. During colonization with commensal C. albicans, IgA+ B cells, in particular mature B cells, plasmablasts and plasma cells, migrate into the oral epithelium and submucosal layers, where they increase the production of polyspecifc-IgAs. The IgA bound to C. albicans will reduce fungal adherence and invasion resulting in a dampened proinflammatory response. Created with BioRender.com.
Reduced salivary flow rate in oral diseased states such as Sjögren's syndrome, or during cytotoxic and radiation therapy increases oral carriage of Candida spp. and is associated with an increase in OPC (64, 65). The saliva, as part of the humoral immune system, contains many molecular factors which restrict microbial growth, including antimicrobial peptides and IgA (66, 67). Plasma cells reside in the salivary glands and produce IgA which is then secreted in the saliva (45). Here we show that oral fungal persistence not only increases the migration of IgA+ B lymphocytes to sites of Candida colonization, fungal persistence also increases IgA amounts in the saliva thus providing a barrier against invading fungi.
Early studies indicated that among patients with CMC over 50% appear to have reduced IgA antibodies (68). The most common humoral immune immunodeficiency is inherited selective IgA deficiency (69). Although selective IgA deficiency is a mild form of immunodeficiency, some patients develop a variety of significant clinical problems, such as CMC (70). Furthermore, Candida infections in individuals with X-linked agammaglobulinemia (XLA), a mutation in the gene encoding for Bruton tyrosine kinase (BTK) which leads to impaired peripheral B cell maturation, have been described (71–73). Lymphoid cancers patients treated with ibrutinib, a BTK inhibitor, develop invasive fungal infections including candidemia (74). In this context, patients targeted by B cell therapy using the anti-CD20 monoclonal antibody rituximab present with candidemia (75). Considering that systemic Candida infections predominantly originate from mucosal barriers (76, 77) B cell responses therefore limit, in part, commensal fungal proliferation and dissemination from mucosal sites. β-glucan, a fungal cell wall component, is able to directly activate B lymphocytes leading to a proinflammatory cytokine response (78), however this mechanism seems dispensable during acute OPC since mice with B cell deficiency are not more susceptibility to pathogenic C. albicans infection (79–81). Because mice lacking B cells exhibited an increase in oral commensal C. albicans carriage without causing severe disease we propose that tissue-resident B lymphocytes, in conjunction with IgA, maintain a stable commensal fungal community in the oral cavity, while T cells in particular Th17 cells prevent commensal breakthrough and severe disease. Thus, mucosal B lymphocytes and their antibody responses monitor fungal exposure in the oral cavity to sustain commensal tolerance and immunity.
The finding that C. albicans oral colonization increases the IgA production leading to increased IgA binding of S. oralis ex vivo suggests that fungal colonization may shape the oral microbial community by inducing B cell expansion and cross-specific antibody secretion. It will be of great interest to analyze the contribution of commensal C. albicans colonization to oral diseases, such as periodontitis or oral lichen planus.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of the Lundquist Institute at Harbor-UCLA Medical Center.
Author Contributions
MS designed the experiments. MS, NM, and NS performed the experiments and analyzed the data. MS wrote the paper. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank S. LeibundGut-Landmann for providing the murine keratinocyte cell line, S.W. French and E. Vitocruz for histopathology assistance, and members of the Division of Infectious Diseases at Harbor-UCLA Medical Center for critical suggestions.
Footnotes
Funding. This work was supported in part by NIH grant R00DE026856 to MS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.555363/full#supplementary-material
Different expressed genes during fungal commensalism. Differential expression analysis was performed using the DESeq2 R package. The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the False Discovery Rate (FDR). Genes with an adjusted P-value < 0.05 found by DESeq2 were assigned as differentially expressed genes that showed ratios log2 ≥± 0.58 were considered to be different regulated.
Gating of infiltrating B lymphocytes. Cells were gated on singlets live B220+ CD4− CD8− CD11b− Gr-1− TER-119− EpCam− and distinguished by CD19 and CD138 expression. Plasma cells (PC; CD19− CD138+), plasmablasts (PB; CD19+ CD138+), and B cells (CD19+ CD138−) were determined.
Tissue B220+ antibody depletion. Flow plot of B220+ cells in the tongue 11 days post infection commensal infection.
References
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. (2012) 4:3004404. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- 2.Benedict K, Jackson BR, Chiller T, Beer KD. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis. (2018) 68:1791–7. 10.1093/cid/ciy776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandara HMHN, Panduwawala CP, Samaranayake LP. Biodiversity of the human oral mycobiome in health and disease. Oral Dis. (2019) 25:363–71. 10.1111/odi.12899 [DOI] [PubMed] [Google Scholar]
- 4.Swidergall M, Filler SG. Oropharyngeal Candidiasis: fungal invasion and epithelial cell responses. PLoS Pathog. (2017) 13:e1006056. 10.1371/journal.ppat.1006056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma A, Gaffen SL, Swidergall M. Innate immunity to mucosal Candida infections. J Fungi. (2017) 3:60. 10.3390/jof3040060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Underhill DM, Pearlman E. Immune interactions with pathogenic and commensal fungi: a two-way street. Immunity. (2015) 43:845–58. 10.1016/j.immuni.2015.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. (2016) 19:865–73. 10.1016/j.chom.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev. Immunol. (2017) 17:635–46. 10.1038/nri.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller AW, Orr T, Dearing D, Monga M. Loss of function dysbiosis associated with antibiotics and high fat, high sugar diet. ISME J. (2019) 13:1379–90. 10.1038/s41396-019-0357-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep. (2019) 9:12918. 10.1038/s41598-019-49452-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson JP, Moyes DL. Adaptive immune responses to Candida albicans infection. Virulence. (2015) 6:327–37. 10.1080/21505594.2015.1004977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jabra-Rizk MA, Kong EF, Tsui C, Nguyen MH, Clancy CJ, Fidel PL, Jr, et al. Candida albicans pathogenesis: fitting within the host-microbe damage response framework. Infect Immun. (2016) 84:2724–39. 10.1128/IAI.00469-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirofski L-A, Casadevall A. Rethinking T cell immunity in oropharyngeal candidiasis. J Exp Med. (2009) 206:269–73. 10.1084/jem.20090093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Zhu L, Qin S. Gut microbiota modulation on intestinal mucosal adaptive immunity. J Immunol Res. (2019) 2019:4735040. 10.1155/2019/4735040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez-Santos N, Huppler AR, Peterson AC, Khader SA, Mckenna KC, Gaffen SL. Th17 cells confer long-term adaptive immunity to oral mucosal Candida albicans infections. Mucosal Immunol. (2013) 6:900–10. 10.1038/mi.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishu S, Hernandez-Santos N, Simpson-Abelson MR, Huppler AR, Conti HR, Ghilardi N, et al. The adaptor CARD9 is required for adaptive but not innate immunity to oral mucosal Candida albicans infections. Infect Immun. (2014) 82:1173–80. 10.1128/IAI.01335-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braunsdorf C, Leibundgut-Landmann S. Modulation of the fungal-host interaction by the intra-species diversity of C. albicans. Pathogens. (2018) 7:11. 10.3390/pathogens7010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schonherr FA, Sparber F, Kirchner FR, Guiducci E, Trautwein-Weidner K, Gladiator A, et al. The intraspecies diversity of C. albicans triggers qualitatively and temporally distinct host responses that determine the balance between commensalism and pathogenicity. Mucosal Immunol. (2017) 8:2. 10.1038/mi.2017.2 [DOI] [PubMed] [Google Scholar]
- 19.Kirchner FR, Littringer K, Altmeier S, Tran VDT, Schönherr F, Lemberg C, et al. Persistence of Candida albicans in the oral mucosa induces a curbed inflammatory host response that is independent of immunosuppression. Front Immunol. (2019) 10:330. 10.3389/fimmu.2019.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchner FR, Leibundgut-Landmann S. Tissue-resident memory Th17 cells maintain stable fungal commensalism in the oral mucosa. Mucosal Immunol. (2020). 10.1038/s41385-020-0327-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macpherson AJ. IgA adaptation to the presence of commensal bacteria in the intestine. Curr Top Microbiol Immunol. (2006) 308:117–36. 10.1007/3-540-30657-9_5 [DOI] [PubMed] [Google Scholar]
- 22.Macpherson AJ, Geuking MB, Mccoy KD. Immunoglobulin A: a bridge between innate and adaptive immunity. Curr Opin Gastroenterol. (2011) 27:529–33. 10.1097/MOG.0b013e32834bb805 [DOI] [PubMed] [Google Scholar]
- 23.Mantis NJ, Rol N, Corthésy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. (2011) 4:603–11. 10.1038/mi.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macpherson AJ, Koller Y, Mccoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. (2015) 36:460–70. 10.1016/j.it.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Jin L, Chen T. The effects of secretory IgA in the mucosal immune system. Biomed Res Int. (2020) 2020:2032057. 10.1155/2020/2032057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corthesy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol. (2013) 4:185. 10.3389/fimmu.2013.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. (1993) 134:717–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman D, Mistry M, Thavaraj S, Challacombe SJ, Naglik JR. Murine model of concurrent oral and vaginal Candida albicans colonization to study epithelial host-pathogen interactions. Microbes Infect. (2007) 9:615–22. 10.1016/j.micinf.2007.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swidergall M, Khalaji M, Solis NV, Moyes DL, Drummond RA, Hube B, et al. Candidalysin is required for neutrophil recruitment and virulence during systemic Candida albicans infection. J Infect Dis. (2019) 220:1477–88. 10.1093/infdis/jiz322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. (2000) 20:1436–47. 10.1128/MCB.20.4.1436-1447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solis NV, Park YN, Swidergall M, Daniels KJ, Filler SG, Soll DR. Candida albicans white-opaque switching influences virulence but not mating during Oropharyngeal Candidiasis. Infect Immun. (2018) 86:e00774-17. 10.1128/IAI.00774-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conti HR, Bruno VM, Childs EE, Daugherty S, Hunter JP, Mengesha BG, et al. IL-17 receptor signaling in oral epithelial cells is critical for protection against Oropharyngeal Candidiasis. Cell Host Microbe. (2016) 20:606–17. 10.1016/j.chom.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altmeier S, Toska A, Sparber F, Teijeira A, Halin C, Leibundgut-Landmann S. IL-1 coordinates the neutrophil response to C. albicans in the oral mucosa. PLoS Pathog. (2016) 12:e1005882. 10.1371/journal.ppat.1005882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. (2012) 7:637–42. 10.1038/nprot.2012.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solis NV, Swidergall M, Bruno VM, Gaffen SL, Filler SG. The aryl hydrocarbon receptor governs epithelial cell invasion during oropharyngeal candidiasis. MBio. (2017) 8:00025–00017. 10.1128/mBio.00025-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. (2008) 5:621–8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 37.Forche A, Solis NV, Swidergall M, Thomas R, Guyer A, Beach A, et al. Selection of Candida albicans trisomy during oropharyngeal infection results in a commensal-like phenotype. PLoS Genet. (2019) 15:e1008137. 10.1371/journal.pgen.1008137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swidergall M, Solis NV, Lionakis MS, Filler SG. EphA2 is an epithelial cell pattern recognition receptor for fungal beta-glucans. Nat Microbiol. (2018) 3:53–61. 10.1038/s41564-017-0059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparber F, Leibundgut-Landmann S. Assessment of immune responses to fungal infections: identification and characterization of immune cells in the infected tissue. Methods Mol Biol. (2017) 1508:167–82. 10.1007/978-1-4939-6515-1_8 [DOI] [PubMed] [Google Scholar]
- 40.Swidergall M, Solis NV, Wang Z, Phan QT, Marshall ME, Lionakis MS, et al. EphA2 is a neutrophil receptor for Candida albicans that stimulates antifungal activity during oropharyngeal infection. Cell Rep. (2019) 28:423–33.e425. 10.1016/j.celrep.2019.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuomo CA, Fanning S, Gujja S, Zeng Q, Naglik JR, Filler SG, et al. Genome sequence for Candida albicans clinical oral isolate 529L. Microbiol Resour Announc. (2019) 8:e00554–e00519. 10.1128/MRA.00554-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.San Millan R, Elguezabal N, Regulez P, Moragues MAD, Quindos G, Ponton J. Effect of salivary secretory IgA on the adhesion of Candida albicans to polystyrene. Microbiology. (2000) 146 (Pt 9):2105–12. 10.1099/00221287-146-9-2105 [DOI] [PubMed] [Google Scholar]
- 43.Swidergall M. Candida albicans at host barrier sites: pattern recognition receptors and beyond. Pathogens. (2019) 8:40. 10.3390/pathogens8010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Aryeh H, Naon H, Szargel R, Horowitz G, Gutman D. The concentration of salivary IgA in whole and parotid saliva and the effect of stimulation. Int J Oral Maxillofac Surg. (1986) 15:81–4. 10.1016/S0300-9785(86)80014-X [DOI] [PubMed] [Google Scholar]
- 45.Brandtzaeg P. Secretory IgA: designed for anti-microbial defense. Front Immunol. (2013) 4:222. 10.3389/fimmu.2013.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mak TW, Saunders ME. 20 - Mucosal and Cutaneous Immunity. In: Mak TW, Saunders ME, editors. The Immune Response. Burlington, VT: Academic Press; (2006). p. 583–609. 10.1016/B978-012088451-3.50022-3 [DOI] [Google Scholar]
- 47.Pabst O, Slack E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. (2020) 13:12–21. 10.1038/s41385-019-0227-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole MF, Bryan S, Evans MK, Pearce CL, Sheridan MJ, Sura PA, et al. Humoral immunity to commensal oral bacteria in human infants: salivary secretory immunoglobulin A antibodies reactive with Streptococcus mitis biovar 1, Streptococcus oralis, Streptococcus mutans, and Enterococcus faecalis during the first two years of life. Infect Immun. (1999) 67:1878–86. 10.1128/.67.4.1878-1886.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egbuniwe IU, Karagiannis SN, Nestle FO, Lacy KE. Revisiting the role of B cells in skin immune surveillance. Trends Immunol. (2015) 36:102–11. 10.1016/j.it.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 50.Kato A, Hulse KE, Tan BK, Schleimer RP. B-lymphocyte lineage cells and the respiratory system. J Allergy Clin Immunol. (2013) 131:933–58. 10.1016/j.jaci.2013.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitamura D, Roes J, Kühn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. (1991) 350:423–6. 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- 52.Ghosh S, Hoselton SA, Schuh JM. μ-chain-deficient mice possess B-1 cells and produce IgG and IgE, but not IgA, following systemic sensitization and inhalational challenge in a fungal asthma model. J Immunol. (2012) 189:1322–9. 10.4049/jimmunol.1200138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puel A, Cypowyj S, Marodi L, Abel L, Picard C, Casanova JL. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol. (2012) 12:616–22. 10.1097/ACI.0b013e328358cc0b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lionakis MS, Iliev ID, Hohl TM. Immunity against fungi. JCI Insight. (2017) 2:93156 10.1172/jci.insight.93156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Casanova JL, Puel A. Mucocutaneous IL-17 immunity in mice and humans: host defense vs. excessive inflammation Mucosal Immunol. (2018) 11:581–9. 10.1038/mi.2017.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bar E, Gladiator A, Bastidas S, Roschitzki B, Acha-Orbea H, Oxenius A, et al. A novel Th cell epitope of Candida albicans mediates protection from fungal infection. J Immunol. (2012) 188:5636–43. 10.4049/jimmunol.1200594 [DOI] [PubMed] [Google Scholar]
- 57.Trautwein-Weidner K, Gladiator A, Kirchner FR, Becattini S, Rülicke T, Sallusto F, et al. Antigen-specific Th17 cells are primed by distinct and complementary dendritic cell subsets in oropharyngeal candidiasis. PLoS Pathog. (2015) 11:e1005164. 10.1371/journal.ppat.1005164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panda S, Ding JL. Natural antibodies bridge innate and adaptive immunity. J Immunol. (2015) 194:13–20. 10.4049/jimmunol.1400844 [DOI] [PubMed] [Google Scholar]
- 59.Mathias A, Pais B, Favre L, Benyacoub J, Corthésy B. Role of secretory IgA in the mucosal sensing of commensal bacteria. Gut Microbes. (2014) 5:688–95. 10.4161/19490976.2014.983763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams DW, Potts AJC, Wilson MJ, Matthews JB, Lewis MAO. Characterisation of the inflammatory cell infiltrate in chronic hyperplastic candidosis of the oral mucosa. J Oral Pathol Med. (1997) 26:83–9. 10.1111/j.1600-0714.1997.tb00026.x [DOI] [PubMed] [Google Scholar]
- 61.Williams A, Williams D, Rogers H, Wei X, Lewis M, Wozniak S, et al. Immunohistochemical expression patterns of inflammatory cells involved in chronic hyperplastic candidosis. Pathogens. (2019) 8:232. 10.3390/pathogens8040232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho J, Yang X, Nikou SA, Kichik N, Donkin A, Ponde NO, et al. Candidalysin activates innate epithelial immune responses via epidermal growth factor receptor. Nat Commun. (2019) 10:2297. 10.1038/s41467-019-09915-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. (2017) 544:498–502. 10.1038/nature22058 [DOI] [PubMed] [Google Scholar]
- 64.Hernandez YL, Daniels TE. Oral candidiasis in Sjogren's syndrome: prevalence, clinical correlations, and treatment. Oral Surg Oral Med Oral Pathol. (1989) 68:324–9. 10.1016/0030-4220(89)90218-1 [DOI] [PubMed] [Google Scholar]
- 65.Nadig SD, Ashwathappa DT, Manjunath M, Krishna S, Annaji AG, Shivaprakash PK. A relationship between salivary flow rates and Candida counts in patients with xerostomia. J Oral Maxillofac Pathol. (2017) 21:316. 10.4103/jomfp.JOMFP_231_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brandtzaeg P. Secretory immunity with special reference to the oral cavity. J Oral Microbiol. (2013) 5. 10.3402/jom.v5i0.20401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swidergall M, Ernst JF. Interplay between Candida albicans and the antimicrobial peptide armory. Eukaryot Cell. (2014) 13:950–7. 10.1128/EC.00093-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehner T, Wilton JM, Ivanyi L. Immunodeficiencies in chronic muco-cutaneous candidosis. Immunology. (1972) 22:775–87. [PMC free article] [PubMed] [Google Scholar]
- 69.Odineal DD, Gershwin ME. The epidemiology and clinical manifestations of autoimmunity in selective IgA deficiency. Clin Rev Allergy Immunol. (2020) 58:107–33. 10.1007/s12016-019-08756-7 [DOI] [PubMed] [Google Scholar]
- 70.Kalfa VC, Roberts RL, Stiehm ER. The syndrome of chronic mucocutaneous candidiasis with selective antibody deficiency. Ann Allergy Asthma Immunol. (2003) 90:259–64. 10.1016/S1081-1206(10)62152-7 [DOI] [PubMed] [Google Scholar]
- 71.Mamishi S, Eghbali AN, Rezaei N, Abolhassani H, Parvaneh N, Aghamohammadi A. A single center 14 years study of infectious complications leading to hospitalization of patients with primary antibody deficiencies. Braz J Infect Dis. (2010) 14:351–5. 10.1016/S1413-8670(10)70074-X [DOI] [PubMed] [Google Scholar]
- 72.Preece K, Lear G. X-linked agammaglobulinemia with normal immunoglobulin and near-normal vaccine seroconversion. Pediatrics. (2015) 136:e1621–4. 10.1542/peds.2014-3907 [DOI] [PubMed] [Google Scholar]
- 73.Xu Y, Qing Q, Liu X, Chen S, Chen Z, Niu X, et al. Bruton's agammaglobulinemia in an adult male due to a novel mutation: a case report. J Thorac Dis. (2016) 8:E1207–12. 10.21037/jtd.2016.10.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varughese T, Taur Y, Cohen N, Palomba ML, Seo SK, Hohl TM, et al. Serious infections in patients receiving ibrutinib for treatment of lymphoid cancer. Clin Infect Dis. (2018) 67:687–92. 10.1093/cid/ciy175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Vollenhoven RF, Emery P, Bingham CO, III, Keystone EC, Fleischmann RM, Furst DE, et al. Long-term safety of rituximab in rheumatoid arthritis: 9.5-year follow-up of the global clinical trial programme with a focus on adverse events of interest in RA patients. Ann Rheum Dis. (2013) 72:1496–502. 10.1136/annrheumdis-2012-201956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nucci M, Anaissie E. Revisiting the source of candidemia: skin or gut? Clin Infect Dis. (2001) 33:1959–67. 10.1086/323759 [DOI] [PubMed] [Google Scholar]
- 77.Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers. (2018) 4:18026 10.1038/nrdp.2018.26 [DOI] [PubMed] [Google Scholar]
- 78.Ali MF, Driscoll CB, Walters PR, Limper AH, Carmona EM. β-glucan-activated human B lymphocytes participate in innate immune responses by releasing proinflammatory cytokines and stimulating neutrophil chemotaxis. J Immunol. (2015) 195:5318–26. 10.4049/jimmunol.1500559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carrow EW, Hector RF, Domer JE. Immunodeficient CBA/N mice respond effectively to Candida albicans. Clin Immunol Immunopathol. (1984) 33:371–80. 10.1016/0090-1229(84)90308-8 [DOI] [PubMed] [Google Scholar]
- 80.Jensen J, Warner T, Balish E. Resistance of SCID mice to Candida albicans administered intravenously or colonizing the gut: role of polymorphonuclear leukocytes and macrophages. J Infect Dis. (1993) 167:912–9. 10.1093/infdis/167.4.912 [DOI] [PubMed] [Google Scholar]
- 81.Jensen J, Warner T, Balish E. The role of phagocytic cells in resistance to disseminated candidiasis in granulocytopenic mice. J Infect Dis. (1994) 170:900–5. 10.1093/infdis/170.4.900 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Different expressed genes during fungal commensalism. Differential expression analysis was performed using the DESeq2 R package. The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the False Discovery Rate (FDR). Genes with an adjusted P-value < 0.05 found by DESeq2 were assigned as differentially expressed genes that showed ratios log2 ≥± 0.58 were considered to be different regulated.
Gating of infiltrating B lymphocytes. Cells were gated on singlets live B220+ CD4− CD8− CD11b− Gr-1− TER-119− EpCam− and distinguished by CD19 and CD138 expression. Plasma cells (PC; CD19− CD138+), plasmablasts (PB; CD19+ CD138+), and B cells (CD19+ CD138−) were determined.
Tissue B220+ antibody depletion. Flow plot of B220+ cells in the tongue 11 days post infection commensal infection.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.