Abstract
Mutations in the enzyme isocitrate dehydrogenase 1/2 (IDH1/2) are the most common somatic mutations in low-grade glioma (LGG). The Hippo signaling pathway is known to play a key role in organ size control, and its dysregulation is involved in the development of diverse cancers. Large tumor suppressor 1/2 (LATS1/2) are core Hippo pathway components that phosphorylate and inactivate Yes-associated protein (YAP), a transcriptional co-activator that regulates expression of genes involved in tumorigenesis. A recent report from The Cancer Genome Atlas (TCGA) has highlighted a frequent hypermethylation of LATS2 in IDH-mutant LGG. However, it is unclear if LATS2 hypermethylation is associated with YAP activation and prognosis of LGG patients. Here, we performed a network analysis of the status of the Hippo pathway in IDH-mutant LGG samples and determined its association with cancer prognosis. Combining TCGA data with our biochemical assays, we found hypermethylation of LATS2 promoter in IDH-mutant LGG. LATS2 hypermethylation, however, did not translate into YAP activation but highly correlated with IDH mutation. LATS2 hypermethylation may thus serve as an alternative for IDH mutation in diagnosis and a favorable prognostic factor for LGG patients.
Keywords: low-grade glioma, Hippo pathway, Lats2, YAP, IDH1/2, isocitrate dehydrogenase
Introduction
Mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2), mainly Arg132 for IDH1 and Arg140 and Arg172 for IDH2, occur in over 80% of low-grade glioma (LGG) (Parsons et al., 2008; Watanabe et al., 2009; Yang et al., 2012; Cancer Genome Atlas Research Network et al., 2015; Suzuki et al., 2015). While wild type IDH1/2 converts isocitrate to α-ketoglutarate (αKG), gain-of-function mutations of IDH1/2 lead to the production and accumulation of oncometabolite R-2-hydroxyglutarate (R-2HG) (Zhao et al., 2009; Ward et al., 2010). R-2HG drives tumorigenesis by inhibiting αKG-dependent enzymes involved in epigenetic modifications, response to hypoxia, and other biological processes (Zhao et al., 2009; Figueroa et al., 2010; Chowdhury et al., 2011; Xu et al., 2011; Lu et al., 2012; Turcan et al., 2012).
The Hippo pathway consists of a kinase cascade and plays crucial roles in tissue homeostasis and tumorigenesis (Harvey et al., 2013; Yu and Guan, 2013; Moroishi et al., 2015; Yu et al., 2015; Patel et al., 2017; Luo and Yu, 2019). Hippo pathway activation results in the phosphorylation of core Ste20-like kinases MST1 and MST2 (MST1/2), which phosphorylate and activate large tumor suppressor 1/2 (LATS1/2) kinases. LATS1/2, in turn, phosphorylate and inactivate Yes-associated protein (YAP) and WW domain-containing transcription regulator protein 1(WWTR1, also known as TAZ), which function as transcription co-activators and serve as Hippo pathway downstream effectors by regulating expression of genes involved in cell proliferation, death, and differentiation. MST1/2 and LATS1/2 activity is further regulated by diverse regulators and upstream signals. Dysregulation of Hippo pathway has been associated with various cancers (Yimlamai et al., 2015; Yu et al., 2015; Zanconato et al., 2016; Gregorieff and Wrana, 2017; Zhang and Zhou, 2019). LATS2 deficiency, for instance, has been studied in several cancers including glioma (Kawahara et al., 2008; Guo et al., 2017, 2019; Ye et al., 2017; Jin et al., 2018; Pan et al., 2018; Shi et al., 2018, 2019; He et al., 2019; Hsu et al., 2019).
A recent report from The Cancer Genome Atlas (TCGA) Research Network revealed that the promoter of LATS2 is hypermethylated in almost all IDH-mutated LGG clinical samples but not in IDH-wild type samples (Sanchez-Vega et al., 2018). LATS2 promoter hypermethylation in IDH-mutated LGG samples is expected to downregulate LATS2 expression and subsequently activate YAP/TAZ and expression of downstream target genes. However, this hypothesis has not been systematically analyzed and experimentally tested. Here, combining TCGA data with our biochemical assays, we performed a network analysis of the status of the Hippo pathway in IDH-mutant LGG samples and determined its association with cancer prognosis.
Results
Promoter Hypermethylation and Low Expression of LATS2 in IDH-Mutant LGG
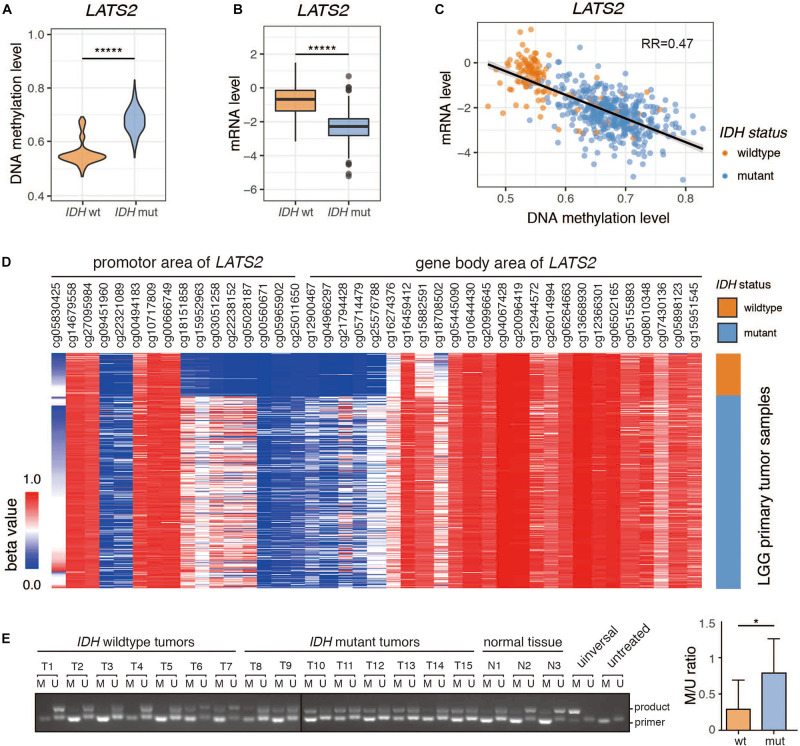
We examined LATS2 methylation level and mRNA expression in LGG dataset from TCGA, and found that LATS2 promoter was hypermethylated and LATS2 mRNA was repressed in IDH-mutant LGG compared to IDH-wild type LGG (Figures 1A,B). Moreover, LATS2 mRNA levels negatively correlated with methylation levels (Figure 1C). The differences in LATS2 gene methylation were mainly located within the promoter region instead of gene body (Figure 1D). We also collected LGG specimens with or without IDH1/2 mutations, and measured LATS2 promoter methylation using a methylation-specific PCR assay (Herman et al., 1996; Oh et al., 2015). Consistent with TCGA data, LATS2 promoter methylation was significantly higher in IDH-mutant LGG (Figure 1E). It is worth noting that while LATS2 promoter hypermethylation had been reported in another cancer with frequent IDH mutations, namely IDH-mutant acute myeloid leukemia (AML), it did not downregulate LATS2 expression as it did in LGG (Supplementary Figure 1A), suggesting a different mechanism or role. Meanwhile, while LATS1 was also hypermethylated, it was not downregulated as LATS2 (Supplementary Figure 2A). Overall, our results indicate that LATS2 is hypermethylated and repressed in IDH-mutant LGG.
FIGURE 1.
Promoter hypermethylation and low expression of LATS2 in IDH-mutant LGG. (A) LATS2 methylation level is increased in IDH-mutant LGG. (B) LATS2 mRNA level is decreased in IDH-mutant LGG. (C) Correlation between LATS2 methylation level and LATS2 mRNA level. RR indicates R squared value of linear regression. (D) Methylation level of different CpG islands in LATS2 promoter and gene body area. (E) Methylation-specific PCR of LGG samples. M: methylation-specific primer; U: unmethylation-specific primers; universal: universally methylated genomic DNA control; untreated: untreated U87 cell genomic DNA; product: ∼130 bp PCR products; primer: primer dimers. Quantitative result on the right. M/U ratio was calculated by comparing the bands from methylation-specific and unmethylation-specific primers of each sample. Mean and standard error were presented (*p < 0.05, *****p < 0.000005, t test).
Hippo Pathway Target Genes Are Not Activated by LATS2 Deficiency in IDH-Mutant LGG
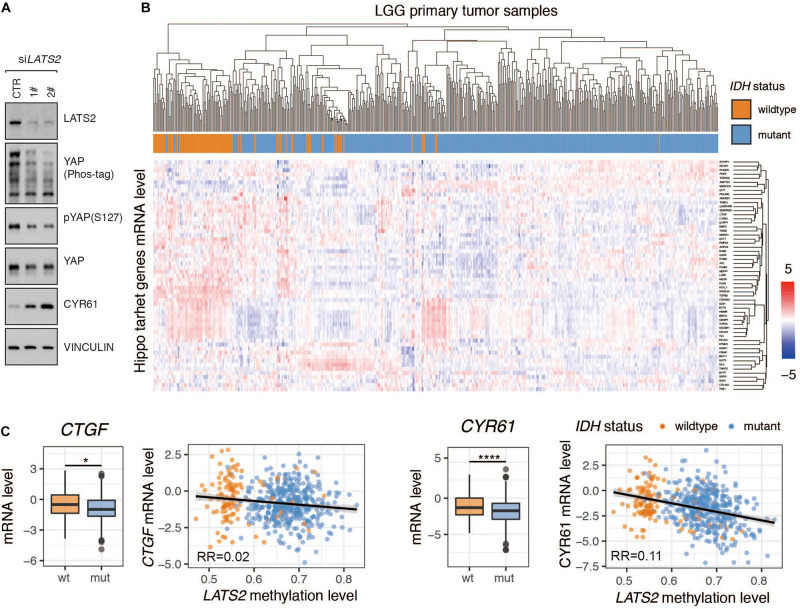
Given that LATS2 is a direct upstream regulator of YAP/TAZ, we examined the effects of LATS2 knockdown on YAP activity. Using two independent siRNAs to target LATS2 in HEK293 cells, we observed that LATS2 knockdown significantly reduced YAP phosphorylation and increased target gene CYR61 expression (Figure 2A). The same result was also observed in glioma cell lines (Guo et al., 2019; Shi et al., 2019). Hence, silencing LATS2 expression in HEK293 cells led to YAP activation.
FIGURE 2.
Hippo pathway target genes are not activated by LATS2 deficiency in IDH-mutant LGG. (A) YAP is activated in HEK293A cells with LATS2 knockdown. (B) cluster analysis of Hippo target gene expression in IDH-wildtype and mutant LGG. The Normalized RESM value was scaled across each gene to yield standard score (Z-score) (C) mRNA levels of Hippo target genes CTGF and CYR61 were decreased in IDH-mutant LGG. Correlation between CTGF/CYR61 mRNA level and LATS2 methylation level is shown. Mean and standard error were presented (*p < 0.05, ****p < 0.00005, t test).
Subsequently, we analyzed if Hippo pathway target genes were activated following LATS2 downregulation in IDH-mutant LGG. Surprisingly, the association between Hippo pathway target gene expression with IDH mutation was weak (Figure 2B). For instance, the mRNA levels of CTGF and CYR61 were reduced in IDH-mutant LGG samples, and correlation analyses showed a nearly negative correlation between Hippo pathway target gene expression and LATS2 methylation (Figure 2C). Hence, it appeared that at least in IDH-mutant LGG, LATS2 downregulation did not translate to YAP activation and YAP-dependent gene expression.
Hippo Pathway Target Genes Are Universally Hypermethylated in IDH-Mutant LGG
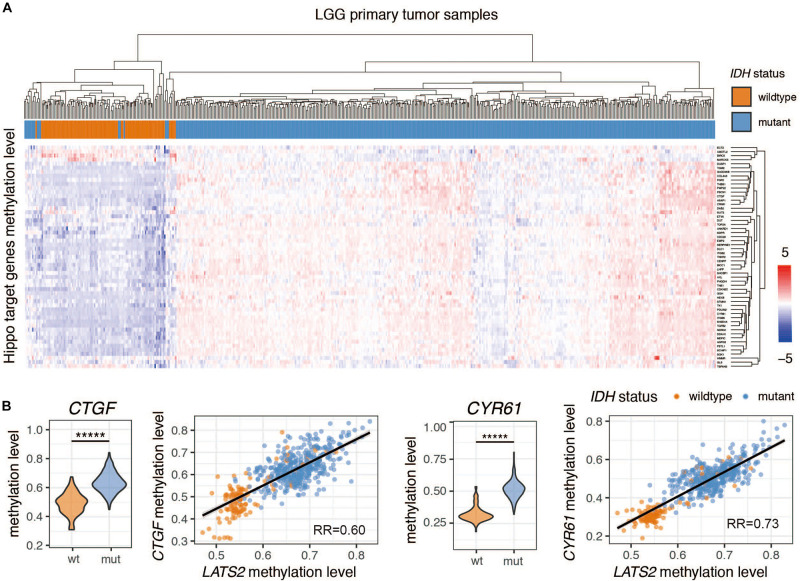
The high methylation levels of CTGF and CYR61 in IDH-mutant LGG suggested that Hippo pathway target genes were also regulated by methylation (Figure 3B). Indeed, a cluster analysis showed that most Hippo pathway target genes were hypermethylated in IDH-mutant LGG samples (Figure 3A). The methylation of these genes was comparable to that of LATS2, as indicated by a tight correlation between methylation levels of LATS2 and those of CTGF or CYR61 (Figure 3B). Thus, the nearly universal hypermethylation of Hippo pathway target genes may explain the ineffectiveness of LATS2 hypermethylation in IDH-mutant LGG to affect YAP activation and target gene expression.
FIGURE 3.
Hippo pathway target genes are universally hypermethylated in IDH-mutant LGG. (A) cluster analysis of Hippo target gene methylation levels in IDH-wildtype and mutant LGG. The Normalized beta value is scaled across each gene to yield standard score (Z-score) (B) Methylation levels of Hippo target genes CTGF and CYR61 are significantly increased in IDH-mutant LGG. Correlation between CTGF/CYR61 methylation level and LATS2 methylation level is shown. Compared by t test (*****p < 0.000005, t test).
Low Expression of YAP/TAZ in LGG
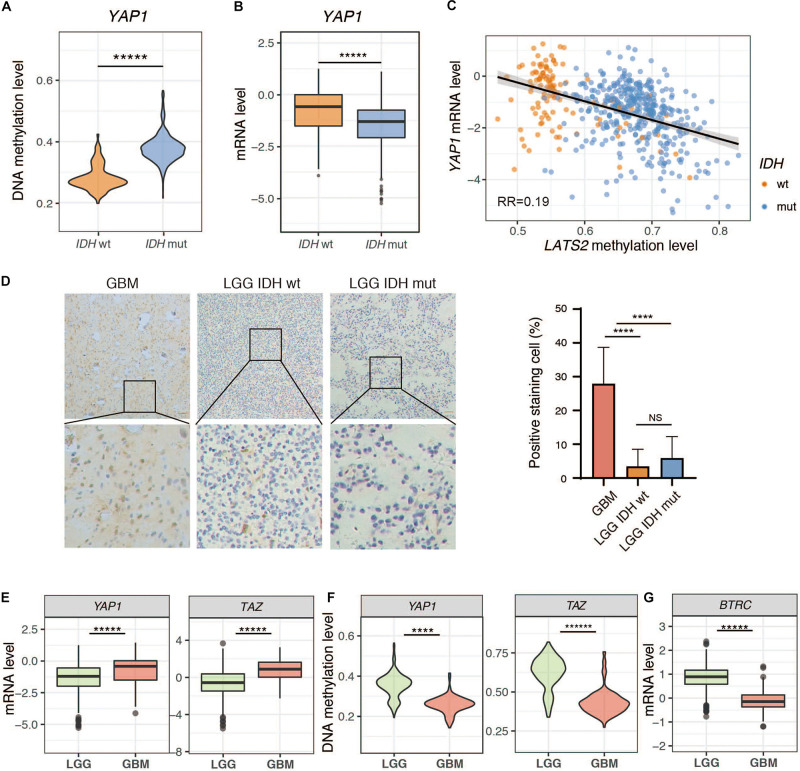
We next analyzed the expression of YAP and TAZ in LGG. Interestingly, both YAP and TAZ were hypermethylated, and were expressed at lower levels in IDH-mutant LGG samples compared to IDH-wild type LGG samples (Figures 4A–C and Supplementary Figure 3). We then assessed YAP expression by immunohistochemistry (IHC) in LGG tumor specimens. Our IHC results indicated, however, that YAP expression was either absent or extremely weak in all LGG samples, regardless of IDH status. In contrast, YAP was highly expressed in glioblastoma (GBM), another common brain tumor (Figure 4D). This could be due to overall higher methylation and lower expression of YAP in LGG compared to GBM (Figures 4E,F), although it could not explain why YAP protein expression showed no significant difference between IDH-wild type and IDH-mutant LGG samples. Hence, it is possible that a posttranslational mechanism may account for low YAP protein levels in LGG. Intriguingly, we found that BTRC, an E3 ligase responsible for YAP degradation (Zhao et al., 2010), was dramatically upregulated in LGG compared to GBM (Figure 4G). On the other hand, several reported deubiquitinases for YAP (Li et al., 2018; Sun et al., 2019; Zhang et al., 2019; Pan et al., 2020; Zhu et al., 2020) were also upregulated (Supplementary Figure 4). Thus, further work is needed to dissect the mechanism(s) for the loss of YAP protein expression in LGG.
FIGURE 4.
Low expression of YAP in LGG. (A) YAP methylation is elevated in IDH-mutant LGG. (B) YAP mRNA level is reduced in IDH-mutant LGG. (C) Correlation between YAP mRNA level and LATS2 methylation level. (D) IHC result of IDH-wildtype or mutant LGG samples and GBM samples. Left: Representative image; Right: Quantitative result. (E) YAP and TAZ mRNA levels in LGG and GBM. (F) YAP and TAZ methylation levels in LGG and GBM. (G) BTRC mRNA level in LGG and GBM. Mean and standard error were presented (****p < 0.00005, *****p < 0.000005, t test).
Dysregulated Expression of Multiple Hippo Pathway Genes in IDH-Mutant LGG
Since LATS2, YAP, and several Hippo pathway target genes were highly methylated in IDH-mutant LGG, we analyzed methylation and gene expression of known Hippo pathway components in LGG. We found that many of them are dysregulated in IDH-mutant LGG (Supplementary Figures 5–8, summarized in Supplementary Figure 9).
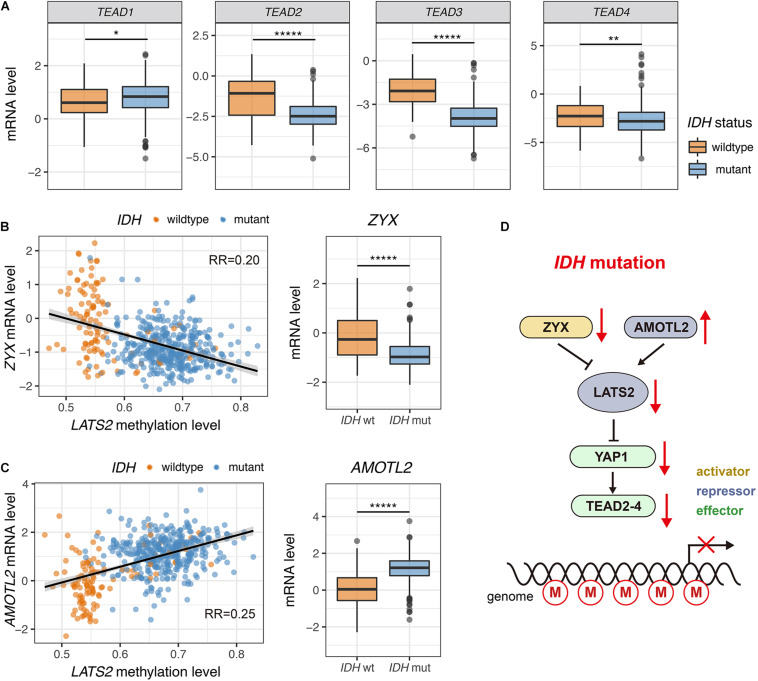
TEA-domain family proteins (TEAD1-4) are the major transcription factors that mediate functions of YAP/TAZ (Ota and Sasaki, 2008; Chen et al., 2010; Lamar et al., 2012; Lin et al., 2017; Holden and Cunningham, 2018). Notably, TEAD2-4 were significantly downregulated in IDH-mutant LGG, while TEAD1 showed a mild upregulation (Figure 5A). Low expression of TEAD genes was correlated with LATS2 hypermethylation (Supplementary Figure 5).
FIGURE 5.
Dysregulated expression of multiple Hippo pathway genes in IDH-mutant LGG. (A) TEAD2-4 are significantly downregulated in IDH-mutant LGG while TEAD1 shows a mild upregulation. (B) Correlation between ZYX mRNA level and LATS2 methylation level (left). ZYX is significantly downregulated in IDH-mutant LGG (right). (C) Correlation between AMOTL2 mRNA level and LATS2 methylation level (left). AMOTL2 is significantly upregulated in IDH-mutant LGG (right). (D) A proposed model of ineffective LATS2 hypermethylation. Mean and standard error were presented (*p < 0.05, ** < 0.005, *****p < 0.000005, t test).
Further, we observed that the expression of known upstream regulators of LATS1/2 were modulated in IDH-mutant LGG samples. For instance, the mRNA levels of Zyxin (ZYX), an inhibitor of LATS2 (Ma et al., 2016), were low in IDH-mutant LGG samples (Figure 5B). On the other hand, the expression of angiomotin like 2 (AMOTL2), an activator of LATS2 (Mana-Capelli and McCollum, 2018), was elevated in IDH-mutant LGG samples (Figure 5C). These changes may also play a role in restricting YAP/TAZ activity in IDH-mutant LGG by inducing activity of residual LATS1/2 (Figure 5D).
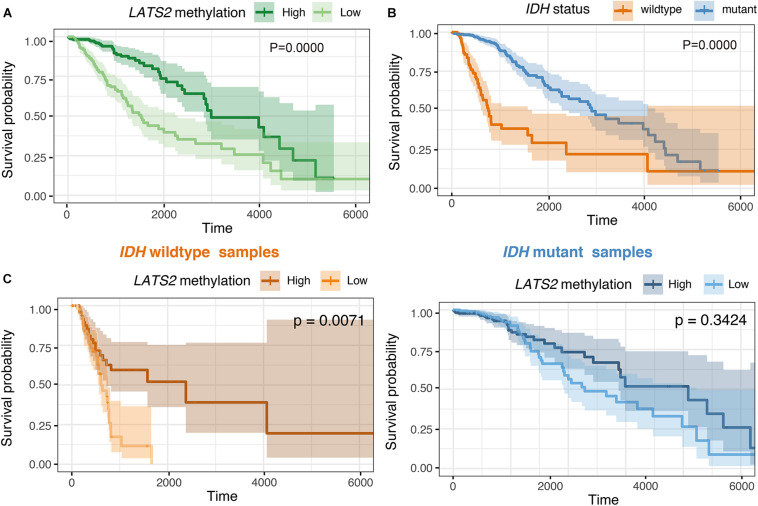
LATS2 Hypermethylation Is a Favorable Prognostic Factor in Overall LGG but Not in IDH-Wild-Type or Mutant Subgroups
Our results thus far indicated that the hypermethylation of LATS2 in IDH-mutant LGG failed to activate YAP/TAZ activity. Next, we interrogated whether hypermethylation of LATS2 could serve as a biomarker with clinical significance. In analyzing survival data of LGG patients, we found that LATS2 hypermethylation is a strong favorable prognostic factor in LGG (Figure 6A). However, when we performed analysis separately in IDH-mutant patients, LATS2 hypermethylation showed no prognostic significance in IDH-mutant subgroups (Figure 6C). In comparing the clinical features between high and low LATS2 methylation groups, we uncovered several characteristics that varied between these two groups including IDH1/2 status (Supplementary Table 2). As IDH mutation was a favorable prognostic factor of LGG (Vuong et al., 2019; Figure 6B), the prognostic significance of LATS2 hypermethylation was likely due to its enrichment in IDH-mutant samples.
FIGURE 6.
LATS2 hypermethylation is a favorable prognostic factor in overall LGG. (A) High LATS2 methylation is a favorable prognostic factor in overall LGG. P value as indicated. (B) IDH mutation is a favorable prognostic factor of overall LGG; (C) High LATS2 methylation level is a favorable prognostic factor in IDH-wildtype LGG but not in IDH-mutant LGG.
To further explore the prognostic value of LATS2 methylation, we did Cox proportional hazard analysis of LATS2 methylation (Table 1). We found LATS2 methylation is a prognostic factor in overall LGG after adjusted for a series of covariates, but lost its prognostic value in either IDH-wildtype or mutant LGG subgroups, indicating its prognostic significance comes from correlation with IDH mutation instead of direct impact on Hippo pathway effectors.
TABLE 1.
Cox proportional hazard model of LAST2 methylation and expression in LGG patients.
| Overall |
IDH wildtype |
IDH mutant |
||||
| Variable | Univariable | Adjusted* | Univariable | Adjusted* | Univariable | Adjusted* |
| HR (95%CI) | HR (95%CI) | HR (95%CI) | HR (95%CI) | HR (95%CI) | HR (95%CI) | |
| LATS2 methylation | 1.64e-05 | 3.63e-04 | 1.32e-05 | 2.72e-02 | 0.37 | 6.69e-03 |
| (1.06e-6, 2.53e-4) | (2.28e-5, 5.78e-3) | (1.09e-8, 1.60e-2) | (2.39e-6, 309.82) | (1.99e-3, 68.60) | (2.64e-5, 1.70) | |
| IDH status wildtype | 4.80 | 3.36 | ||||
| (3.34, 6.90) | (2.18, 5.18) | |||||
*Adjusted for age, gender, neoplasm histologic grade, histological type, and tumor location. HR with p value < 0.05 is marked by bold formatting, CI denotes confidence interval.
LATS2 Hypermethylation Predicts IDH Mutation in LGG
Lastly, we determined whether LATS2 methylation level may work as a biomarker of IDH status. Using CpG island cg03051258 in the promoter area of LATS2 as an example, we applied beta value 0.1 as a threshold to identify LATS2 hyper- and hypo-methylated samples, and IDH-mutant LGG was successfully enriched in the LATS2 hypermethylated group. Using this approach, we could predict IDH mutation in LGG at the sensitivity of 0.95 and specificity of 0.97 (Table 2). Together, these data support that LATS2 hypermethylation is a faithful biomarker of IDH mutations.
TABLE 2.
Methylation level of cg03051258 to predict IDH status.
| cg03051258 | IDH mutant | IDH wildtype | Total |
| beta value > 0.1 | 393 | 3 | 396 |
| beta value < 0.1 | 19 | 88 | 107 |
| Total | 412 | 91 | 503 |
Discussion
The Hippo pathway is known to play critical roles in cancer development, making this signaling network an area of high clinical interest. The TCGA Network project revealed that LATS2 is commonly hypermethylated in IDH-mutant low-grade gliomas, prompting us to explore its role in LGG. Several groups have previously explored a role of LATS2 in gliomas (Guo et al., 2019; Shi et al., 2019). However, a systematic analysis to evaluate its effect on Hippo pathway in IDH-mutant LGG had not been carried out.
Our study found that LATS2 promoter was hypermethylated while LATS2 mRNA was repressed in IDH-mutant LGG samples. Unexpectedly, LATS2 repression failed to activate Hippo pathway target genes, as most of these genes were also hypermethylated in IDH-mutant LGG samples. The universal epigenetic changes caused by IDH1/2 mutation could be a key point to understand this phenomenon. Oncometabolite R-2HG produced by mutated IDH1/2 inhibits the activity of αKG-dependent enzymes including DNA and histone demethylases (Yamane et al., 2006; Chowdhury et al., 2011; Ito et al., 2011; Xu et al., 2011; Turcan et al., 2012; Kohli and Zhang, 2013). In this way, IDH1/2 mutation may cause genome-wide alterations in DNA methylation, including LATS2, Hippo pathway target genes, and additional Hippo pathway component genes.
It is interesting that YAP expression is high in GBM but is extremely low in all LGG regardless of IDH status. This could be a reflection of differentiation status and aggressiveness of tumors, because YAP is frequently activated in less differentiated and malignant cancers (Xu et al., 2009; Fullenkamp et al., 2016; Zanconato et al., 2016). Compared to GBM, LGG is usually well-differentiated and less malignant, and YAP may remain less active in LGG. Moreover, YAP is important in maintaining stemness of progenitor cells (Lian et al., 2010; Beyer et al., 2013; Li et al., 2013). Hence, the difference in YAP activity between GBM and LGG might be inherited from the status of respective cancer progenitor cells.
Along with low YAP expression, additional mechanisms may contribute to the lack of YAP activation in IDH-mutant LGG. For instance, the dysregulated expression of ZYX and AMOTL2 may inhibit LATS1/2 activity, while reduction of TEAD2-4 expression may limit the transcriptional output of YAP.
Although LATS2 hypermethylation was unable to activate YAP in IDH-mutant LGG, it displayed a strong correlation with IDH1/2 mutation and could serve as a favorable prognostic factor for LGG patients. In addition, LATS2 hypermethylation was a faithful biomarker of IDH mutations, and could potentially be used as an alternative for IDH mutation in diagnosis.
In conclusion, our study found LATS2 promoter hypermethylation in IDH-mutant LGG samples which, surprisingly, did not translate into YAP activation, raising the role and involvement, if at all, of the Hippo pathway in the development of LGG. Meanwhile, LATS2 hypermethylation showed a strong correlation with IDH mutation. Hence, LATS2 hypermethylation can serve as an alternative for IDH mutation in diagnosis and a favorable prognostic factor for LGG patients.
Materials and Methods
Data Collection and Processing
TCGA-LGG, TCGA-GBM, TCGA-AML RNA sequence level 3 normalized data, DNA Methylation Level 3 data, clinical data and somatic mutation data were downloaded from GDC Data Portal using package TCGAbiolinks in R (version 3.6.2) environment for further analysis (Colaprico et al., 2016). IDH-mutant samples were composed of samples with IDH1 Arg132 or IDH2 Arg140 and 172 mutations. The level 3 expression data were normalized RMSE value. For each gene, we zero-centered expression data by calculating standard score (Z-score) between each individual sample. Comparison of gene expression between different tumor types were based on pan-cancer normalized expression data from UCSC Xena team (Goldman et al., 2020). Pre-process steps to yield level 3 methylation data (β-value) included background correction, dye-bias normalization. β-values ranged from zero to one, with zero indicating no methylation detected.
Statistical Analysis
The expression and methylation of Hippo-related genes were compared by t test or Mann-Whitney U test. The correlation between expression and methylation status was evaluated by fitting linear models. Survival data were analyzed by Kaplan-Meier analysis and Cox proportional hazard analysis. The Hippo pathway target genes (Supplementary Table 1). were determined according to RNAseq results of our Hippo element knock-out cell lines (data not shown). Hierarchical Clustering of each sample was done by calculating Euclidean distance matrix followed Pearson correlation analysis based on Hippo target features. All the analysis and image drawing (R package ggplot2) was done by R (version 3.6.2).
Patients Samples
This study enrolled patients with GBM (n = 8) and LGG (n = 12, including 5 IDH wildtype and 7 IDH mutant). Glioma frozen tissues and paraffin slides were obtained from Huashan Hospital, Fudan University, Shanghai, China. This study was approved by the Ethics Committee of the Huashan Hospital of Fudan University and informed consents were obtained from all participants. Specimens used for Methylation-specific PCR were taken at the time of surgical resection, snap−frozen in liquid nitrogen and stored at −80°C until use. Formalin-fixed paraffin-embedded (FFPE) tissues were used for immunohistochemical (IHC) staining.
Cell Culture and Gene Knock-Down by siRNA
HEK293A cells were cultured in DMEM (Corning) containing 5% FBS (Gibco) and 50 μg/mL penicillin/streptomycin (P/S). All cells were incubated at 37°C under 5% CO2. siRNAs were purchased from GenePharma and transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s protocol. The following siRNAs were used: siLATS2 1#: UACCAUAAAUACAAUCUUCTT (5′-3′), siLATS2 2#: CCGCAAAGGGTACACTCAATT (5′-3′). HEK293A cells were seeded into 6-well plates and transfected the next day. 60 h later, cells were harvested for immunoblotting.
Immunoblotting
Cells were lysed in 1 × SDS loading buffer containing 50 mM Tris pH 6.8, 2% SDS, 0.025% bromophenol blue, 10% glycerol, and 5% BME. The concentration of total proteins was assayed by BCA method. Protein samples were separated by SDS-polyacrylamide gel electrophoresis (SDS–PAGE) and then transferred onto polyvinylidene fluoride (PVDF) membranes, blocked with 5% non-fat milk in TBST for 1 h at room temperature. The membranes were washed with TBST three times for 5 min and then incubated with primary antibodies (4°C overnight) and HRP-conjugated secondary antibodies. ECL solution and image acquisition equipment (5200S Imager) were from Tanon Science & Technology Co., Ltd. The following primary antibodies were used: anti-LATS2 (CST, 1:1000, 5888S), anti-YAP (CST, 1:1000, 14074S), anti-pYAP (S127) (CST, 1:1000, 4911S), anti-CYR61 (Santa, 1:1000, sc-13011), and anti-vinculin (CST, 1:1000, 13901s).
Immunohistochemistry
Paraffin embedded tissue specimens were sectioned, dewaxed, and rehydrated. Antigen retrieval was performed in 10 mM sodium citrate buffer (pH 6.0) at 95–100°C for 20 min. Endogenous peroxidase activity was blocked by 3% H2O2 for 30 min. Sections were then blocked in 5% BSA for 1 h and incubated with primary antibodies overnight. After extensive washing, the sections were incubated with secondary antibodies at room temperature for 1 h. DAB solution was applied and hematoxylin was used for counterstaining. Anti-YAP (CST, 1:200) was used as a primary antibody. Staining results were visualized with Zeiss Axiocam 208 color. Quantification was conducted to measure the protein expression.
DNA Isolation and Bisulfite Conversion
Genomic DNA of glioma was isolated from frozen LGG tissues using DNA/RNA/protein Extraction Kit (DP423) (Tiangen, Beijing, China) according to the manufacturer’s protocol. EZ DNA Methylation-Startup Kit (Zymo Research) was utilized to perform sodium bisulfite modification of DNA following the manufacturer’s instructions. This converts cytosine residues to uracil in single-stranded DNA while leaving methylated cytosine unchanged.
Methylation-Specific PCR
The methylation status of LATS2 was tested by Methylation-specific PCR utilizing both methylated and unmethylated specific sets of primers: 5′-GTT GGA GTT GTT GTT GGT TTC-3′ (forward) and 5′-CGA ATA TCC CAC TTA AAT CTA CG-3′(reverse) for methylated reaction (PCR products, 131 bp) and 5′-GTT GGA GTT GTT GTT GGT TTT G-3′ (forward) and 5′-AAA TAT CCC ACT TAA ATC TAC ACT-3′ (reverse) for unmethylated reaction (PCR product, 130 bp). PCR amplification was carried out on T-100 Thermal Cycler (Bio-Rad) using Taq DNA polymerase (Vazyme Biotech Co., Ltd., China) in a total volume of 10 μL. 5% DMSO was added to enhance the specificity and yield of PCR reactions. DNA samples were initial denatured at 95°C for 5 min, and was followed by 36 cycles of denaturing at 95°C for 30 s, annealing at 54°C (for methylated reaction) or 59°C (for unmethylated reaction) for 30 s, and extension at 72°C for 45 s. A final extension step at 72°C for 5 min was added for all reactions. Both positive and negative controls were included. Polymerase chain reaction products were subsequently electrophoresed on 2% agarose gels and visualized with image equipment from Tanon Science & Technology Co., Ltd.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://portal.gdc.cancer.gov.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the Huashan Hospital of Fudan University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YG and F-XY designed the study and wrote the manuscript. YG, YuW, YeW, JL, XW, MM, WH, and YL performed experiments and data analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The results here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Funding. This study was supported by grants from the National Key R&D program of China (2018YFA0800304), the National Natural Science Foundation of China (81772965), Science and Technology Commission of Shanghai Municipality (19JC1411100), and Shanghai Municipal Commission of Health and Family Planning (2017BR018) to F-XY.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.586581/full#supplementary-material
References
- Beyer T. A., Weiss A., Khomchuk Y., Huang K., Ogunjimi A. A., Varelas X., et al. (2013). Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5 1611–1624. 10.1016/j.celrep.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Brat D. J., Verhaak R. G., Aldape K. D., Yung W. K., Salama S. R., et al. (2015). Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372 2481–2498. 10.1056/nejmoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Loh P. G., Song H. (2010). Structural and functional insights into the TEAD-YAP complex in the Hippo signaling pathway. Protein Cell 1 1073–1083. 10.1007/s13238-010-0138-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Yeoh K. K., Tian Y. M., Hillringhaus L., Bagg E. A., Rose N. R., et al. (2011). The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12 463–469. 10.1038/embor.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A., Silva T. C., Olsen C., Garofano L., Cava C., Garolini D., et al. (2016). TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44:e71. 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M. E., Abdel-Wahab O., Lu C., Ward P. S., Patel J., Shih A., et al. (2010). Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18 553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullenkamp C. A., Hall S. L., Jaber O. I., Pakalniskis B. L., Savage E. C., Savage J. M., et al. (2016). TAZ and YAP are frequently activated oncoproteins in sarcomas. Oncotarget 7 30094–30108. 10.18632/oncotarget.8979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M. J., Craft B., Hastie M., Repecka K., Mcdade F., Kamath A., et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38 675–678. 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Wrana J. L. (2017). Hippo signalling in intestinal regeneration and cancer. Curr. Opin. Cell Biol. 48 17–25. 10.1016/j.ceb.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Guo C., Liang C., Yang J., Hu H., Fan B., Liu X. (2019). LATS2 inhibits cell proliferation and metastasis through the Hippo signaling pathway in glioma. Oncol. Rep. 41 2753–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cui J., Ji Z., Cheng C., Zhang K., Zhang C., et al. (2017). miR-302/367/LATS2/YAP pathway is essential for prostate tumor-propagating cells and promotes the development of castration resistance. Oncogene 36 6336–6347. 10.1038/onc.2017.240 [DOI] [PubMed] [Google Scholar]
- Harvey K. F., Zhang X., Thomas D. M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13 246–257. [DOI] [PubMed] [Google Scholar]
- He C., Lv X., Huang C., Hua G., Ma B., Chen X., et al. (2019). YAP1-LATS2 feedback loop dictates senescent or malignant cell fate to maintain tissue homeostasis. EMBO Rep. 20:e44948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. G., Graff J. R., Myohanen S., Nelkin B. D., Baylin S. B. (1996). Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U.S.A. 93 9821–9826. 10.1073/pnas.93.18.9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden J. K., Cunningham C. N. (2018). Targeting the hippo pathway and cancer through the TEAD family of transcription factors. Cancers 10:81. 10.3390/cancers10030081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. H., Kuo W. W., Shih H. N., Cheng S. F., Yang C. K., Chen M. C., et al. (2019). FOXC1 regulation of miR-31-5p confers oxaliplatin resistance by targeting LATS2 in colorectal cancer. Cancers 11:1576. 10.3390/cancers11101576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333 1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Cai Q., Wang S., Wang S., Mondal T., Wang J., et al. (2018). Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 9:1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara M., Hori T., Chonabayashi K., Oka T., Sudol M., Uchiyama T. (2008). Kpm/Lats2 is linked to chemosensitivity of leukemic cells through the stabilization of p73. Blood 112 3856–3866. 10.1182/blood-2007-09-111773 [DOI] [PubMed] [Google Scholar]
- Kohli R. M., Zhang Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502 472–479. 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar J. M., Stern P., Liu H., Schindler J. W., Jiang Z. G., Hynes R. O. (2012). The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. U.S.A. 109 E2441–E2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu T., Li Y., Wu C., Luo K., Yin Y., et al. (2018). The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene 37 2422–2431. 10.1038/s41388-018-0134-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Chen Y., Mak K. K., Wong C. K., Wang C. C., Yuan P. (2013). Functional role of Mst1/Mst2 in embryonic stem cell differentiation. PLoS One 8:e79867. 10.1371/journal.pone.0079867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I., Kim J., Okazawa H., Zhao J., Zhao B., Yu J., et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24 1106–1118. 10.1101/gad.1903310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. C., Park H. W., Guan K. L. (2017). Regulation of the Hippo Pathway Transcription Factor TEAD. Trends Biochem. Sci. 42 862–872. 10.1016/j.tibs.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Ward P. S., Kapoor G. S., Rohle D., Turcan S., Abdel-Wahab O., et al. (2012). IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483 474–478. 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Yu F. X. (2019). GPCR-Hippo Signaling in Cancer. Cells 8:426. 10.3390/cells8050426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Cheng H., Gao R., Mu C., Chen L., Wu S., et al. (2016). Zyxin-Siah2-Lats2 axis mediates cooperation between Hippo and TGF-beta signalling pathways. Nat. Commun. 7:11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S., McCollum D. (2018). Angiomotins stimulate LATS kinase autophosphorylation and act as scaffolds that promote Hippo signaling. J. Biol. Chem. 293 18230–18241. 10.1074/jbc.ra118.004187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Hansen C. G., Guan K. L. (2015). The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15 73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. E., Ohta T., Satomi K., Foll M., Durand G., Mckay J., et al. (2015). Alterations in the NF2/LATS1/LATS2/YAP pathway in schwannomas. J. Neuropathol. Exp. Neurol. 74 952–959. 10.1097/nen.0000000000000238 [DOI] [PubMed] [Google Scholar]
- Ota M., Sasaki H. (2008). Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135 4059–4069. 10.1242/dev.027151 [DOI] [PubMed] [Google Scholar]
- Pan B., Yang Y., Li J., Wang Y., Fang C., Yu F. X., et al. (2020). USP47-mediated deubiquitination and stabilization of YAP contributes to the progression of colorectal cancer. Protein Cell 11 138–143. 10.1007/s13238-019-00674-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tong J. H. M., Lung R. W. M., Kang W., Kwan J. S. H., Chak W. P., et al. (2018). RASAL2 promotes tumor progression through LATS2/YAP1 axis of hippo signaling pathway in colorectal cancer. Mol. Cancer 17:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., et al. (2008). An integrated genomic analysis of human glioblastoma multiforme. Science 321 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. H., Camargo F. D., Yimlamai D. (2017). Hippo Signaling in the Liver Regulates Organ Size, Cell Fate, and Carcinogenesis. Gastroenterology 152 533–545. 10.1053/j.gastro.2016.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F., Mina M., Armenia J., Chatila W. K., Luna A., La K. C., et al. (2018). Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Liu Z., Liu Z., Feng X., Hua F., Hu X., et al. (2018). Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine 37 177–187. 10.1016/j.ebiom.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Geng D., Zhang Y., Zhao M., Wang Y., Jiang Y., et al. (2019). LATS2 Inhibits Malignant Behaviors of Glioma Cells via Inactivating YAP. J. Mol. Neurosci. 68 38–48. 10.1007/s12031-019-1262-z [DOI] [PubMed] [Google Scholar]
- Sun X., Ding Y., Zhan M., Li Y., Gao D., Wang G., et al. (2019). Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat. Commun. 10:411. 10.1016/j.devcel.2009.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Aoki K., Chiba K., Sato Y., Shiozawa Y., Shiraishi Y., et al. (2015). Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47 458–468. 10.1038/ng.3273 [DOI] [PubMed] [Google Scholar]
- Turcan S., Rohle D., Goenka A., Walsh L. A., Fang F., Yilmaz E., et al. (2012). IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483 479–483. 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong H. G., Tran T. T. K., Ngo H. T. T., Pham T. Q., Nakazawa T., Fung K. M., et al. (2019). Prognostic significance of genetic biomarkers in isocitrate dehydrogenase-wild-type lower-grade glioma: the need to further stratify this tumor entity - a meta-analysis. Eur. J. Neurol. 26 379–387. 10.1111/ene.13826 [DOI] [PubMed] [Google Scholar]
- Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., et al. (2010). The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17 225–234. 10.1016/j.ccr.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Nobusawa S., Kleihues P., Ohgaki H. (2009). IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 174 1149–1153. 10.2353/ajpath.2009.080958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. Z., Yao T. J., Lee N. P., Ng I. O., Chan Y. T., Zender L., et al. (2009). Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer 115 4576–4585. 10.1002/cncr.24495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S. H., et al. (2011). Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19 17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., et al. (2006). JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125 483–495. 10.1016/j.cell.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Yang H., Ye D., Guan K. L., Xiong Y. (2012). IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin. Cancer Res. 18 5562–5571. 10.1158/1078-0432.ccr-12-1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Li T. S., Xu G., Zhao Y. M., Zhang N. P., Fan J., et al. (2017). JCAD Promotes Progression of Nonalcoholic Steatohepatitis to Liver Cancer by Inhibiting LATS2 Kinase Activity. Cancer Res. 77 5287–5300. 10.1158/0008-5472.can-17-0229 [DOI] [PubMed] [Google Scholar]
- Yimlamai D., Fowl B. H., Camargo F. D. (2015). Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J. Hepatol. 63 1491–1501. 10.1016/j.jhep.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. X., Guan K. L. (2013). The Hippo pathway: regulators and regulations. Genes Dev. 27 355–371. 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. X., Zhao B., Guan K. L. (2015). Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 163 811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M., Piccolo S. (2016). YAP/TAZ at the Roots of Cancer. Cancer Cell 29 783–803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhou D. (2019). Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr. Opin. Cell Biol. 61 64–71. 10.1016/j.ceb.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Du J., Wang S., Shao L., Jin K., Li F., et al. (2019). OTUB2 Promotes Cancer Metastasis via Hippo-Independent Activation of YAP and TAZ. Mol. Cell. 73 7–21.e27. [DOI] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L. (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24 72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., et al. (2009). Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 324 261–265. 10.1126/science.1170944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Yan F., Yuan T., Qian M., Zhou T., Dai X., et al. (2020). USP10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing YAP/TAZ. Cancer Res. 80:0008–5472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://portal.gdc.cancer.gov.